Abstract
Background
Tissue biopsy-based cancer diagnosis has limitations because of the fact that tumor tissues are in constant evolution and extremely heterogeneous. The current study was aimed to examine whether tumor-educated blood platelets (TEPs) might be a potential all-in-one source for blood-based cancer diagnostics to overcome the limitations of conventional cancer biopsy.
Methods
In the present study, we evaluated the expression pattern of MAGI2 antisense RNA 3 (MAGI2-AS3) and ZNFX1 antisense RNA 1 (ZFAS1) in both plasma and platelets of 101 non-small-cell lung cancer (NSCLC) patients. Receiver operating characteristic (ROC) curve was generated to evaluate their diagnostic potential. In addition, epidermal growth factor receptor (EGFR) mutations were detected in DNA and RNA samples of platelets for companion diagnostics.
Results
Our results showed that the levels of MAGI2-AS3 and ZFAS1 in both plasma and platelets of NSCLC patients were significantly downregulated than those in healthy controls. A positive correlation of long noncoding RNA expression was observed between platelets and plasma (r=0.738 for MAGI2-AS3, r=0.751 for ZFAS1, respectively). By ROC analysis, we found that molecular interrogation of MAGI2-AS3 and ZFAS1 in TEPs and plasma can offer valuable diagnostic performance for NSCLC patients (area under the ROC curve [AUC]MAGI2-AS3= 0.853/0.892, and AUCZFAS1=0.780/0.744 for diagnosing adenocarcinoma and squamous cell carcinoma cases from controls, respectively). Clinicopathologic characteristic analysis further revealed that MAGI2-AS3 level significantly correlated with tumor–node–metastasis (TNM) stage (p=0.001 in TEPs, p=0.003 in plasma), lymph-node metastasis (p=0.016 in TEPs, p=0.023 in plasma), and distant metastasis (p=0.045 in TEPs, p=0.045 in plasma), while ZFAS1 level was only correlated with TNM stage (p=0.005 in TEPs, p=0.044 in plasma). Furthermore, EGFRvIII RNA existed in both TEPs and plasma, but EGFR intracellular mutations cannot be detected in DNA of TEPs isolated from NSCLC.
Conclusion
Our data suggested that TEP is a promising source for NSCLC diagnosis and companion diagnostics.
Keywords: tumor-educated platelets, lncRNA-MAGI2-AS3, lncRNA-ZFAS1, EGFR mutations, NSCLC
Background
Non-small-cell lung cancer (NSCLC), mainly divided into adenocarcinoma (AD) and squamous cell carcinoma (SCC) pathologically,1,2 is the predominant form of lung cancer and accounts for the majority of cancer deaths worldwide.3 Despite much progress being achieved in early detection and treatment, the 5-year survival rate for NSCLC patients is still only 5%–20%.4 Thus, it is an urgent medical need to develop more sensitive diagnostic methods and novel biomarkers, especially for patients with recurrent cancers.5–7
Due to tumor evolution and heterogeneity, traditional tissue biopsy-based cancer diagnostic procedures have limitations in their assessment for cancer diagnosis, genotyping, and prognosis.8 Therefore, clinicians and scientists have tried to use blood-based liquid biopsies as a potential alternative diagnostic method for NSCLC patients.9,10
Liquid biopsy, including tumor-educated blood platelets (TEPs), exosomes, circulating cell-free tumor DNA, and circulating tumor cells, could offer earlier and more convenient diagnosis for patients with NSCLC. Additionally, these techniques are less invasive and may provide a more comprehensive characterization for NSCLC.11-13 The plasma is a pool of cells or cell products (DNA, RNA, and proteins) derived from the tumor or circulating tumor cells and stromal cells of the tumor microenvironment. The analysis of these blood components can, therefore, provide a comprehensive real-time information of the tumor-associated changes in an individual cancer patient.14 However, it has been also reported that tumor-associated biomarkers released into the plasma are susceptible to degradation, while blood platelets are more stable and can offer high sensitivity for the detection of EML4-ALK rearrangements.15 Given the fact that blood platelets, the second most abundant cell type in peripheral blood, can take up tumor-derived microvesicles loaded with tumor-specific cellular compounds (RNA, protein, etc),16 TEPs have been proposed and currently investigated as a potential biomarker source for cancers.17
Long noncoding RNA (lncRNA) is a group of RNA molecules that are longer than 200 nucleotides and do not encode protein,18 and they regulate gene expression through epigenetic regulation, splicing, imprinting, transcriptional regulation, and subcellular transport19 and play important roles in tumorigenesis and tumor progression.20 In recent years, aberrant lncRNA expression has been found to participate in NSCLC development and metastasis. For example, increased HNF1A-AS1 promoted lung AD cell proliferation and metastasis through interacting with DNMT1 and repressing E-cadherin expression.21 LncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a highly conserved nuclear lncRNA, which is upregulated and can serve as a predictive marker for metastasis in lung cancer.22 These data indicated that lncRNAs play important roles in NSCLC pathogenesis, which could provide new insights into the biology of this devastating disease.
The epidermal growth factor receptor (EGFR), a prototype member of the type I receptor tyrosine kinase (TK) family, plays a critical role in NSCLC cell differentiation, proliferation, and treatment response. Genetic alterations of the EGFR, including intracellular mutations such as 19-Del, L858R, T790M, 20-lns, G719X, S768I, and L861Q and extracellular mutation EGFRvIII, predominantly found in ADs,23 are representative biomarkers in determining the appropriate treatment for advanced lung cancer. The identification of EGFR mutations provides guidance for clinical treatment with EGFR TK inhibitors such as gefitinib and erlotinib.24,25
In the present study, we first screened out five lncRNAs with potential diagnostic significance from three gene expression omnibus (GEO) datasets, GSE19188, GSE30219, and GSE27262,26 which included expression information of tumor tissue from patients with AD or SCC. Next, we investigated the expression pattern and diagnostic value of five lncRNAs in platelets and plasma derived from patients with NSCLC to explore whether TEPs could enable NSCLC diagnostic and identify NSCLC types. Furthermore, we evaluated EGFR intracellular mutations in DNA extracted from platelets and plasma of 14 NSCLC patients who have been confirmed with EGFR intracellular mutations in tissue biopsy samples, and we also detected EGFRvIII expression within TEPs RNA for companion diagnostic of NSCLC.
Methods
Platelets and plasma isolation
Whole-blood samples (2 mL) of NSCLC patients and healthy controls were collected from Zhongnan Hospital of Wuhan University under approval of Institutional Review Board, and written informed consent was obtained from all participants. Then, platelets and plasma were isolated from the same sample of whole blood by standard centrifugation within 24 hours to minimize detrimental effects of long-term storage at room temperature and decrease of platelet/plasma RNA quality and quantity. Three hundred microliters of platelets-rich plasma was obtained by centrifugation at room temperature for 20 min at 120 g to remove interference of cells. Then, platelets were isolated from the 300 µL platelet-rich plasma by centrifugation at room temperature for 20 min at 360 g, and resuspended in 300 µL PBS after washing twice. Plasma and platelets were frozen in parallel and stored at −80°C for further use.
GEO lung cancer gene expression data
Three panels of NSCLC in NCBI GEO were used in our lncRNA screening with the accession IDs GSE19188, GSE30219, and GSE27262. Reading and processing of data were done using R package “limm”, genes with adjusted p-value <0.05 and fold change >2 were identified as threshold value to judge differentially expressed lncRNAs, then we proceeded to receiver operating characteristic (ROC) analysis to screen out four lncRNAs that possessed great diagnostic value (we also added an lncRNA that possessed poor diagnostic value).
RNA isolation and cDNA synthesis
Total RNA was isolated from 300 µL platelet suspension and 300 µL plasma using a Liquid Total RNA Isolation Kit (RP4002, BioTeke, Beijing, China) and eluted in 35 µL of prewarmed (65°C) elution according to the manufacturer’s instruction. The concentration of RNA was measured by Nanodrop 2000 spectrophotometer (Thermo Scientific Inc, USA). Then RNA was reverse transcribed to cDNA using PrimeScript™ RT reagent kit with gDNA Eraser (RR047A, Takara, Dalian, China). Reverse transcription conditions were as follows: 42°C for 2 min, and then 37°C for 15 min, 85°C for 5 s. The cDNA was frozen at −20°C for further use.
Quantitative real-time PCR (RT-qPCR) for lncRNAs
The expression of lncRNAs was determined on the Bio-Rad CFX96 (Bio-Rad Laboratories Inc, Hercules, CA, USA) using SYBR-Green I Premix Ex Taq following the manufacturer’s instructions. The GAPDH was used as the endogenous control and was amplified simultaneously with target genes. The synthesized RT-qPCR primers are listed in Table S1. The reactions were performed in a volume of 20 µL (10 µL of SYBR mix, 0.8 µL of 10 µmol sense, 0.8 µL of 10 µmol antisense, 2 µL cDNA and 6.4 µL water). The reactions started at 95°C for 5 min, followed by 42 cycles of 95°C for 30 s, 63.3°C for 30 s, and 72°C for 30 s. All experiments were carried out in duplicate for each data point. The relative gene expression level was calculated using the comparative Ct method formula 2−ΔCt.
DNA extraction and amplification-refractory mutation system (ARMS)-PCR for EGFR intracellular mutations
DNA was isolated from 14 TEPs and plasma samples using QIAamp® Circulating Nucleic Acid kits (QIAGEN, Hilden, Germany) eluted in 50 µL of elution according to the manufacturer’s instruction; EGFR intracellular mutations including 19-Del, L858R, T790M, 20-lns, G719X, S768I, and L861Q were detected using an ADx-ARMS EGFR mutation detection kit (ADx-EG01, AmoyDx, Xiamen, China) by ARMS.
RT-PCR for EGFRvIII
Samples from 110 AD, 95 SCC, and 50 healthy controls were collected and determined EGFR/EGFRvIII existence with RT-PCR in a volume of25 µLsolution (12.5 µL of AmpliTaq Gold® 360 Master mix, 0.5 µL 10 µmol sense, 0.5 µL 10 µmol antisense, 2 µL cDNA, and 9.5 µL water). PCR condition is listed as follows: 95°C 5 min; 95°C 30 s, 55°C 45 s, 72°C 1 min × 42 cycles; 72°C 7 min. The RT-PCR primers are listed in Table S1.
Statistical analysis
Statistical analyses were performed using SPSS version 22.0 (SPSS, Chicago, IL, USA) or Prism6 (GraphPad software, La Jolla, CA, USA). Data were presented as median with interquartile range. The Shapiro–Wilk test was carried out to check the normality of the distribution. The normally distributed numeric variables were evaluated by Student’s t-test, while non-normally distributed variables were analyzed by Mann–Whitney test. Chi-square test was used to analyze the categorical variables. One-way analysis of variance (ANOVA) or nonparametric test was used to validate the different expression levels of lncRNAs among subgroups. Statistical differences were set at *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. p<0.05 was considered statistically significant. To estimate the diagnostic value of the biomarkers, area under the ROC curve analysis was performed.
Results
Identification of potential lncRNAs as biomarkers for NSCLC diagnosis from GEO dataset
To identify relevant datasets, we searched GEO for NSCLC expression profiling studies, and the criteria for choice of datasets are that 1) they were gene profiling studies in patients with NSCLC; 2) they used the same platform; 3) they used NSCLC tissue and normal lung tissue for comparison; and 4) they contained the information of AD or SCC. We finally narrowed down and analyzed three GEO public datasets: GSE19188 containing 45 AD, 27 SCC, and 65 normal lung tissue controls, GSE30219 containing 85 AD, 61 SCC, and 14 normal lung tissue controls, GSE27262 containing 25 AD and 25 normal lung tissue controls. Based on these three GEO datasets, we screened out five lncRNAs including MAGI2-AS3, LOC100507632, FXF1-AS1, LOC100499467, and ZFAS1 listed in Table 1. MAGI2-AS3, LOC100507632, and FOXF1-AS1 were significantly downregulated (p<0.0001 in all three GEO datasets) in NSCLC tissue, while LOC100499467 (p<0.0001 in all three GEO datasets) and ZFAS1 (p<0.05 in all three GEO datasets) were significantly upregulated. In addition, the first four lncRNAs all exhibited great diagnostic value (area under the ROC curve [AUC]>0.900), while the diagnostic value for ZFAS1 was relatively poor.
Table 1.
LncRNAs are screened out from three GEO datasets
| LncRNA | GEO dataset | Adenocarcinoma
|
Squamous cell carcinoma
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | 95% CI | p-value | Se (%) | Sp (%) | AUC | 95% CI | p-value | Se (%) | Sp (%) | ||
| MAGI2-AS3 | GSE19188 | 0.921 | 0.863–0.980 | <0.0001 | 82.2 | 98.5 | 0.985 | 0.967–1.003 | <0.0001 | 100 | 87.7 |
| GSE30219 | 0.945 | 0.895–0.994 | <0.0001 | 88.2 | 92.7 | 0.986 | 0.963–1.008 | <0.0001 | 98.4 | 92.9 | |
| GSE27262 | 0.972 | 0.947–1.012 | <0.0001 | 100 | 92 | − | − | − | − | − | |
| LOC100507632 | GSE19188 | 0.950 | 0.895–1.005 | <0.0001 | 100 | 84.4 | 0.992 | 0.979–1.004 | <0.0001 | 90.8 | 100 |
| GSE30219 | 0.968 | 0.926–1.010 | <0.0001 | 92.9 | 91.8 | 0.993 | 0.980–1.006 | <0.0001 | 100 | 93.4 | |
| GSE27262 | 1.000 | 1.000–1.000 | <0.0001 | 100 | 100 | − | − | − | − | − | |
| FOXF1-AS1 | GSE19188 | 0.928 | 0.864–0.992 | <0.0001 | 93.9 | 91.1 | 0.989 | 0.972–1.005 | <0.0001 | 93.9 | 100 |
| GSE30219 | 0.945 | 0.876–1.015 | <0.0001 | 85.7 | 96.5 | 0.972 | 0.936–1.008 | <0.0001 | 85.7 | 98.4 | |
| GSE27262 | 1.000 | 1.000–1.000 | <0.0001 | 100 | 100 | − | − | − | − | − | |
| LOC100499467 | GSE19188 | 0.887 | 0.809–0.9645 | <0.0001 | 96.9 | 82.2 | 0.995 | 0.987–1.003 | <0.0001 | 96.9 | 100 |
| GSE30219 | 0.857 | 0.740–0.9745 | <0.0001 | 71.4 | 91.7 | 0.968 | 0.927–1.010 | <0.0001 | 85.7 | 96.7 | |
| GSE27262 | 0.992 | 0.977–1.007 | <0.0001 | 96 | 96 | − | |||||
| ZFAS1 | GSE19188 | 0.665 | 0.560–0.771 | 0.0033 | 64.4 | 64.6 | 0.660 | 0.527–0.793 | 0.016 | 63 | 69.2 |
| GSE30219 | 0.668 | 0.540–0.797 | 0.0436 | 61.2 | 78.6 | 0.609 | 0.479–0.739 | 0.0497 | 57.4 | 78.6 | |
| GSE27262 | 0.781 | 0.644–0.9174 | 0.0007 | 92 | 68 | − | − | − | − | − | |
Abbreviations: AUC, area under the ROC curve; GEO, gene expression omnibus; Se, sensitivity; Sp, specificity.
MAGI2-AS3 and ZFAS1 were both downregulated in NSCLC platelets and plasma
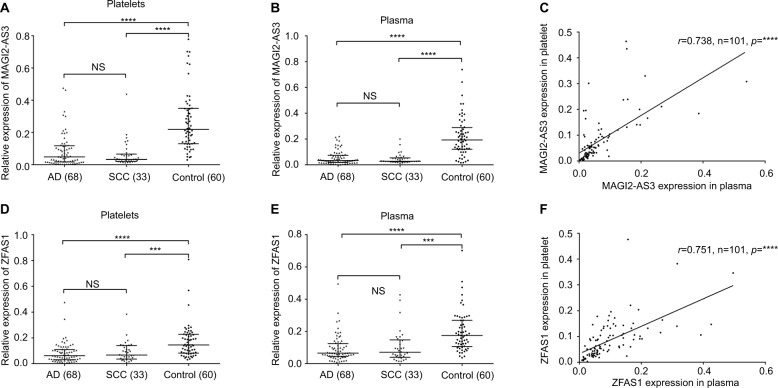
To confirm the finding of bioinformatics analysis, we used 68 ADs and 33 SCCs and 60 healthy controls for detecting these five lncRNAs to test whether TEPs could serve as a liquid biopsy for tumor diagnosis. Our result indicted that we can only detect MAGI2-AS3 and ZFAS1 expression among five lncRNAs from TEP samples. Although LOC100507632, FOXF1-AS1, and LOC100499467 can be detected in A549 NSCLC cell line, we cannot detect their expression because of the low abundance in 300 µL platelet suspension and 300 µL plasma (Figure S1). For MAGI2-AS3, both platelets and plasma were significantly downregulated in line with the result from GEO datasets (Figure 1A, B). The melt curve of MAGI2-AS3 showed that the amplification specificity was good (Figure S2A, B). The result of agarose gel electrophoresis revealed that the amplification product was the target gene MAGI2-AS3 and endogenous gene GAPDH (Figure S2C). In addition, platelets’ MAGI2-AS3 expression was positively correlated with plasma MAGI2-AS3. Spearman’s correlation analysis showed that the correlation coefficient was 0.738 (p<0.0001; Figure 1C). ZFAS1 was significantly downregulated in platelets and plasma samples from NSCLC, which was different from the result from GEO datasets (Figure 1D, E), and spearman correlation analysis showed that the correlation coefficient was 0.751 (p<0.0001; Figure 1F). The melt curve (Figure S2D, E) and result of agarose gel electrophoresis (Figure S2F) both showed that the amplification specificity was good. Based on the results of MAGI2-AS3 and ZFAS1, we proposed that TEPs could also detect tumor biomarkers the same as that using plasma to a certain extent.
Figure 1.
MAGI2-AS2 and ZFAS1 levels were downregulated in platelets and plasma samples derived from NSCLC patients.
Notes: (A) MAGI2-AS3 was downregulated in adenocarcinoma and SCC platelets than that in control platelets (p<0.001). (B) MAGI2-AS3 was downregulated in adenocarcinoma and SCC plasma than that in control plasma (p<0.001). (C) Platelets’ MAGI2-AS3 level was positively correlated to the plasma MAGI2-AS3 expression. The correlation coefficient was 0.738 (p<0.0001). (D) ZFAS1 was downregulated in adenocarcinoma and SCC platelets than that in control platelets (p<0.001). (E) ZFAS1 was downregulated in adenocarcinoma and SCC plasma than that in control plasma (p<0.001). (F) Platelets’ ZFAS1 level was positively correlated to the plasma ZFAS1 expression. The correlation coefficient was 0.751 (p<0.0001). The relative expression level was calculated using 2−ΔCT method; error bars represent the median with interquartile range. All data were analyzed using nonparametric test; NS, no significance. ***p<0.001, ****p<0.0001.
Abbreviations: AD, adenocarcinoma; MAGI2-AS3, MAGI2 antisense RNA 3; SCC, squamous cell carcinoma; ZFAS1, ZNFX1 antisense RNA 1.
Correlation between the expression level of MAGI2-AS3 and ZFAS1 and clinical parameters of NSCLC in platelets and plasma
According to the median value of relative lncRNA expression in NSCLC platelets and plasma, 101 NSCLC patients were correspondently classified into two groups; in platelets (plasma), relative high group: expression ratio ≥ median, and relative low group: expression ratio < median. As presented in Table 2, all clinicopathologic data were divided into these two groups based on different grouping criteria and were analyzed by chi-squared test. The clinicopathologic relevance analysis of NSCLC patients demonstrated that the MAGI2-AS3 expression significantly correlated with tumor–node–metastasis (TNM) stage (p=0.001 in platelets, p=0.003 in plasma), lymph-node metastasis (p=0.016 in platelets, p=0.023 in plasma), and distant metastasis (p=0.045 in platelets, p=0.045 in plasma), while ZFAS1 level was only correlated with TNM stage (p=0.005 in platelets, p=0.044 in plasma).
Table 2.
Correlation between MAGI2-AS3 and ZFAS1 level and clinical parameters of NSCLC
| Clinical parameters | Group | n | Platelet
|
MAGI2-AS3
|
p-value | Plasma
|
MAGI2-AS3
|
p-value | Platelet
|
ZFAS1
|
p-value | Plasma
|
ZFAS1
|
p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | Low | High | Low | High | |||||||
| Histology | AD | 68 | 32 | 36 | 0.480 | 33 | 35 | 0.778 | 35 | 33 | 0.394 | 34 | 34 | 0.886 |
| SCC | 33 | 18 | 15 | 17 | 16 | 14 | 19 | 16 | 17 | |||||
| Age | <60 | 48 | 22 | 26 | 0.482 | 21 | 27 | 0.271 | 24 | 24 | 0.776 | 19 | 29 | 0.058 |
| ≥60 | 53 | 28 | 25 | 29 | 24 | 25 | 28 | 31 | 22 | |||||
| Gender | Male | 74 | 37 | 37 | 0.869 | 38 | 36 | 0.463 | 35 | 39 | 0.685 | 37 | 37 | 0.869 |
| Female | 27 | 13 | 14 | 12 | 15 | 14 | 13 | 13 | 14 | |||||
| Smoking | Negative | 40 | 16 | 24 | 0.122 | 18 | 22 | 0.327 | 18 | 22 | 0.567 | 19 | 21 | 0.774 |
| Positive | 61 | 34 | 27 | 32 | 29 | 31 | 30 | 31 | 30 | |||||
| TNM stage | I–II | 25 | 5 | 20 | 0.001a | 6 | 19 | 0.003a | 6 | 19 | 0.005a | 8 | 17 | 0.044a |
| III–IV | 76 | 45 | 31 | 44 | 32 | 43 | 33 | 42 | 34 | |||||
| Invasion | T1–T3 | 73 | 33 | 40 | 0.163 | 36 | 37 | 0.951 | 33 | 40 | 0.283 | 32 | 41 | 0.066 |
| T4 | 28 | 17 | 11 | 14 | 14 | 16 | 12 | 18 | 10 | |||||
| Lymph–node metastasis | NO | 36 | 12 | 24 | 0.016a | 12 | 24 | 0.023a | 17 | 19 | 0.847 | 17 | 19 | 0.733 |
| N1–N3 | 65 | 38 | 27 | 37 | 28 | 32 | 33 | 33 | 32 | |||||
| Distant metastasis | MO | 70 | 30 | 40 | 0.045a | 30 | 40 | 0.045a | 32 | 38 | 0.397 | 32 | 38 | 0.252 |
| MI | 31 | 20 | 11 | 20 | 11 | 17 | 14 | 18 | 13 |
Note:
Overall p<0.05.
Abbreviations: AD, adenocarcinoma; MAGI2-AS3, MAGI2 antisense RNA 3; NSCLC, non-small-cell lung cancer; SCC, squamous cell carcinoma; TNM, tumor–node–metastasis; ZFAS1, ZNFX1 antisense RNA 1.
Diagnostic value of MAGI2-AS3 and ZFAS1 analysis in AD and SCC
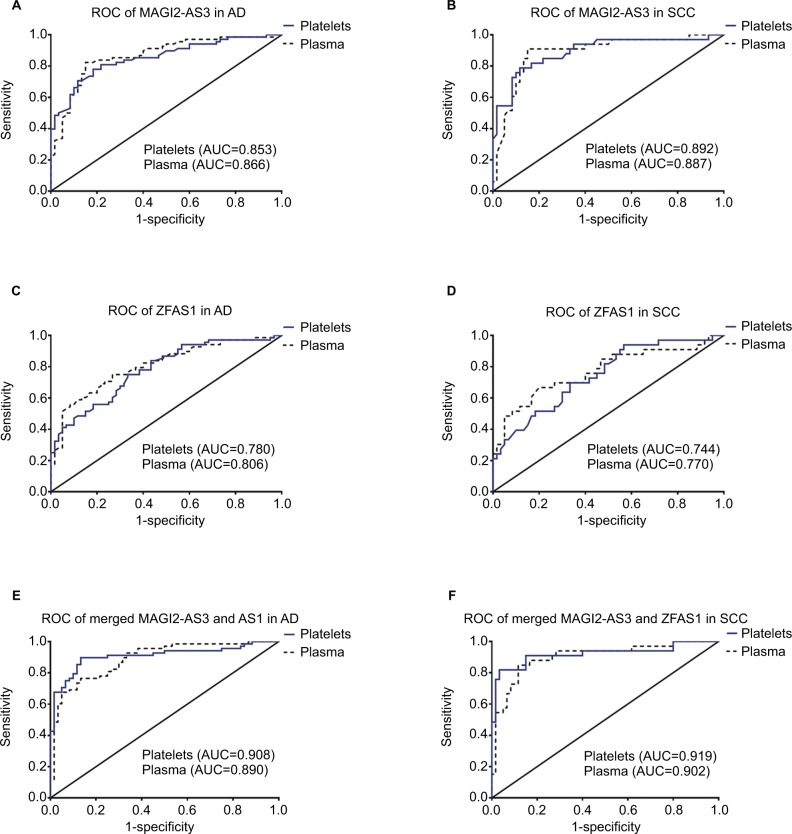
ROC curve analysis was used to evaluate the predicting diagnosis value of lncRNAs for NSCLC. ROC was designed to use two models: AD vs controls, SCC vs controls. AUCs of the platelets and plasma MAGI2-AS3 were 0.853 (95% CI=0.789–0.918, p<0.0001) and 0.866 (95% CI=0.802–0.929, p<0.0001) in AD (Figure 2A), 0.892 (95% CI=0.819–0.965, p<0.0001), and 0.887 (95% CI=0.813–0.961, p<0.0001) in SCC (Figure 2B), respectively. As for ZFAS1, AUCs of platelets and plasma were 0.780 (95% CI=0.701–0.858, p<0.0001) and 0.806 (95% CI=0.731–0.881, p<0.0001) in AD (Figure 2C), and 0.744 (95% CI=0.641–0.848, p<0.0001) and 0.770 (95% CI=0.663–0.878, p<0.0001) in SCC (Figure 2D). Of course, combination of two lncRNAs possessed a better ability for discrimination between NSCLC and controls, both in platelets and plasma, and the AUCs of the combined MAGI2-AS3 and ZFAS1 were 0.908 (95% CI=0.853–0.963, p<0.0001) in platelets and 0.890 (95% CI=0.834–0.946, p<0.0001) in plasma of AD (Figure 2E). The AUCs of the combined MAGI2-AS3 and ZFAS1 were 0.919 (95% CI=0.848–0.990, p<0.0001) in platelets and 0.902 (95% CI=0.833–0.972, p<0.0001) in plasma of SCC (Figure 2F). As for the AUCs of combinations, please see details in Table S2.
Figure 2.
Diagnostic value of MAGI2-AS3 and ZFAS1 in adenocarcinoma and SCC.
Notes: (A) The ROC curve analysis for the diagnostic value of MAGI2-AS3 in platelets from adenocarcinoma (AUC=0.853, 95% CI=0.789–0.918, p<0.0001) and (AUC=0.866, 95% CI=0.802–0.929, p<0.0001) in plasma. (B) The ROC curve analysis for the diagnostic value of MAGI2-AS3 in platelets from SCC (AUC=0.892, 95% CI=0.819–0.965, p<0.0001) and (AUC=0.887, 95% CI=0.813–0.961, p<0.0001) in plasma. (C) The ROC curve analysis for the diagnostic value of ZFAS1 in platelets from adenocarcinoma (AUC=0.780, 95% CI=0.701–0.858, p<0.0001) and (AUC=0.806, 95% CI=0.731–0.881, p<0.0001) in plasma. (D) The ROC curve analysis for the diagnostic value of ZFAS1 in platelets from SCC (AUC=0.744, 95% CI=0.641–0.848, p<0.0001) and (AUC=0.770, 95% CI=0.663–0.878, p<0.0001) in plasma. (E) The ROC curve analysis for the diagnostic value of merged MAGI2-AS3 and ZFAS1 in platelets from adenocarcinoma (AUC=0.908, 95% CI=0.853–0.963, p<0.0001) and (AUC=0.890, 95% CI=0.834–0.946, p<0.0001) in plasma. (F) The ROC curve analysis for the diagnostic value of merged MAGI2-AS3 and ZFAS1 in platelets from SCC (AUC=0.919, 95% CI=0.848–0.990, p<0.0001) and (AUC=0.902, 95% CI=0.833–0.972, p<0.0001) in plasma.
Abbreviations: AD, adenocarcinoma; AUC, area under the ROC curve; MAGI2-AS3, MAGI2 antisense RNA 3; ROC, receiver operating characteristic; SCC, squamous cell carcinoma; ZFAS1, ZNFX1 antisense RNA 1.
EGFR intracellular mutations detection with platelets and plasma DNA
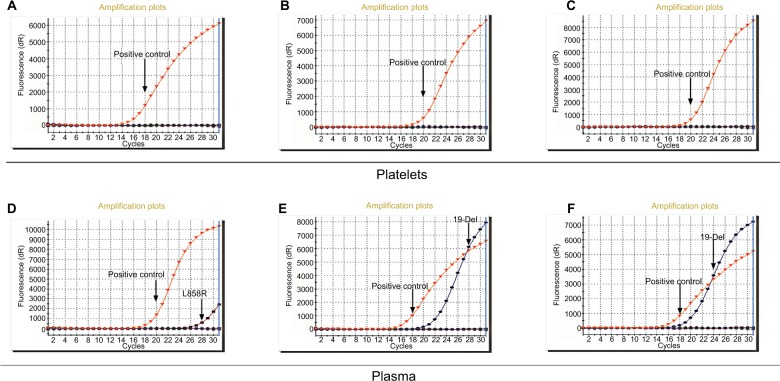
Fourteen NSCLC patients with AD were diagnosed with EGFR intracellular mutations with conventional tissue analysis. We extracted DNA of platelets and corresponding plasma from the 14 patients and detected EGFR intracellular mutations. Three plasma samples (NO. 6, NO. 11, and NO. 12 listed in Table S3) showed positive result in accordance with the result of tissue while there was no positive result in platelets (Figure 3). As for negative result and basic information of 14 patients, please see summary in Table S3.
Figure 3.
ARMS-PCR for EGFR intracellular mutations detection.
Notes: (A), (B), (C) EGFR intracellular mutation cannot be detected in NO.6, NO.11, and NO.12 patient platelets. (D), (E), and (F) EGFR intracellular mutation can be detected in NO.6, NO.11, and NO.12 patient plasma.
Abbreviations: ARMS, amplification-refractory mutation system; EGFR, epidermal growth factor receptor.
Platelets and plasma from NSCLC patients contain mRNA EGFRvIII
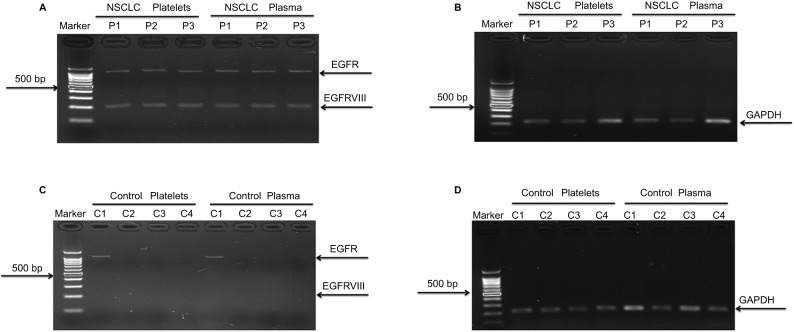
To determine whether circulating blood platelets and plasma isolated from NSCLC patients contain the RNA biomarker EGFRvIII, we compared platelets and plasma from 110 ADs and 95 SCCs to 50 healthy people. Among 205 NSCLC patients, GAPDH was detected in all blood platelets and plasma samples, and EGFRvIII was detected in three (one in ADs and two in SCC) blood platelets and the corresponding plasma samples (Figure 4A, B). In addition, among 50 healthy people, none of the blood platelets and plasma samples detected EGFRvIII expression, but GAPDH was detected in all blood platelets and plasma samples (Figure 4C, D). Our data suggested that EGFRvIII can also be detected in blood platelets and there was no difference with detection in plasma. To verify whether EGFRvIII expression of platelets was contaminated by plasma, we performed the following analysis. First, we mixed the EGFRvIII-positive plasma and EGFRvIII-negative platelets, then separated platelets and detected EGFRvIII expression, and the result showed that there was not EGFRvIII expression in platelets (Figure S3A). Second, we mixed the EGFRvIII-positive platelets and EGFRvIII-negative plasma, then separated plasma and detected EGFRvIII expression, and the result showed that there was not EGFRvIII expression in plasma (Figure S3B). Taken together, these results suggested that the EGFRvIII expression we detected was specific to platelets.
Figure 4.
EGFRvIII expression in platelets and plasma derived from NSCLC patients.
Notes: (A) EGFRvIII expression in 3 NSCLC platelets and plasma. (B) GAPDH expression as positive control in patients’ platelets and plasma, three controls’ results are shown here as an example. (C) No EGFRvIII expression in 50 healthy controls, and four individuals’ results are shown here as an example. (D) GAPDH expression as positive control in healthy individuals’ platelets and plasma. Four individuals’ results are shown here as an example.
Abbreviations: EGFR, epidermal growth factor receptor; NSCLC, non-small-cell lung cancer.
Discussion
For decades, systematic empirical research on NSCLC has identified some prognostic factors and several biomarkers.27 However, the prognosis of NSCLC remains quite poor and the 5-year survival rate for NSCLC patients is low, because most patients are diagnosed at an advanced stage of NSCLC. Recently, the clinical application of liquid biopsy in NSCLC progressively proved a pivotal tool for screening and early detection of cancer,28 including TEPs, exosomes, circulating cell-free tumor DNA, and circulating tumor cells.
Here, we have examined the use of TEPs and plasma for a noninvasive assessment of lncRNAs and found that TEP is also a promising biosource similar to plasma. We investigated the clinical value of MAGI2-AS3, LOC100507632, FOXF1-AS1, LOC100499467, and ZFAS1. However, we could not detect LOC100507632, FOXF1-AS1, and LOC100499467 which might be due to the low abundance in platelets and plasma, suggesting that not all of the lncRNAs can be detected in platelets. As for MAGI2-AS3, NSCLC patient platelets and plasma were significantly downregulated in line with the results from GEO. For ZFAS1, previous study29 suggested that ZFAS1 is significantly downregulated in human breast cancer, where it serves as a tumor suppressor gene in tumorigenesis and progression, while another study30 showed that ZFAS1 is frequently amplified in hepatocellular carcinoma and functions as an oncogene. Thus, the roles of ZFAS1 in cancers are complex and might play a dual role depending on the tumor. It has previously been demonstrated that other small noncoding RNAs, such as microRNAs, can play a dual role as an oncogene or a tumor suppressor gene according to the cellular context.31 In addition, piRNAs can also play a dual role depending on the tumor. For example, piR-823 has been shown to act as a tumor suppressor in gastric cancer32 and as an oncogene in multiple myeloma.33 In our study, we found that ZFAS1 was significantly downregulated in platelets and plasma, which was in contrast to the high expression in NSCLC tissue samples as demonstrated in previous study,34 and this phenomenon might be explained by the expression of ZFAS1 detected in plasma and platelets which could have come from circulating cells rather than from tumor cells. Meanwhile, we analyzed the diagnostic value of ZFAS1 and MAGI2-AS3 and found that molecular interrogation of platelet lncRNA ZFAS1 and MAGI2-AS3 can offer valuable diagnostics information for NSCLC patients analyzed, of both MAGI2-AS3 (AUC is 0.853 in AD and 0.892 in SCC) and ZFAS1 (AUC is 0.780 in AD and 0.744 in SCC). More importantly, combination of two lncRNAs possessed a better ability for discrimination between NSCLC and controls. We also analyzed the correlation between plasma lncRNA and platelet lncRNA, and we found that there was moderate correlation between both MAGI2-AS3 (r=0.738, p<0.0001) and ZFAS1 (r=0.751, p<0.0001). Furthermore, we found the correlation between the two lncRNAs’ level and the clinicopathologic characteristics of the 101 NSCLC cancer patients and found that the expression level of ZFAS1 is negatively correlated to the TNM stage while MAGI2-AS3 expression significantly correlated with TNM stage, lymph-node metastasis, and distant metastasis. These findings suggested that the use of TEPs as a biosource to diagnose cancer may enable earlier diagnosis of cancer, improve convenience for NSCLC patients, and ultimately serve as supplement for clinical oncologic decision-making.
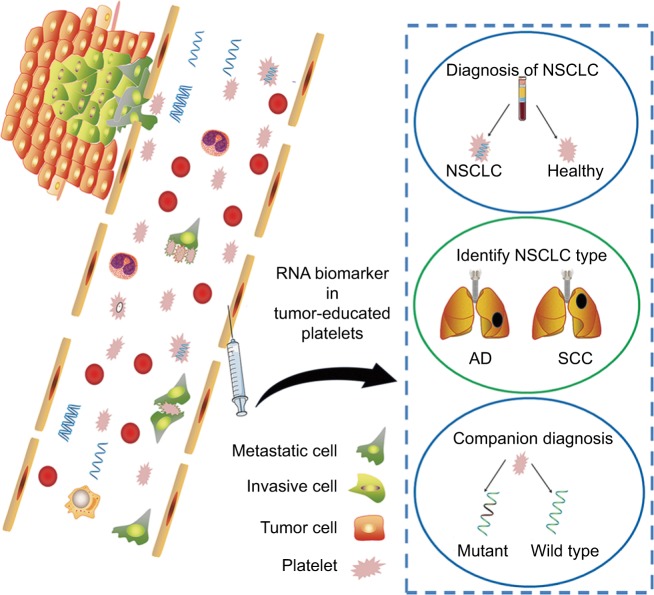
TEP is an emerging source for liquid biopsy, confrontation of platelets with tumor cells through transfer of tumor-associated biomolecules and results in the sequestration of such biomolecules,35,36 as shown in Figure 5. Numerous studies have emphasized the role of platelets in tumor biology, from a diagnostic standpoint of view, and TEPs carry important tumor molecular signature, with the potential to be used as a viable biosource for companion diagnostics and therapy selection.
Figure 5.
A schematic figure summarizing potential role of platelets in NSCLC.
Notes: Platelets communicate with tumor cells through uptaking of tumor-associated biomolecules and thus may serve as a potential diagnostic marker for NSCLC.
Abbreviations: AD, adenocarcinoma; NSCLC, non-small-cell lung cancer; SCC, squamous cell carcinoma.
In the present study, we extracted DNA of 14 platelets and plasma samples to detect EGFR intracellular mutations including 19-Del, L858R, T790M, 20-lns, G719X, S768I, and L861Q by ARMS; our data suggested that only three plasma samples could detect EGFR mutation in accordance with tissue while there was no positive result in platelets, which also indicated there was no contamination of platelet DNA by plasma DNA, and the reasons that we analyzed to explain it are as follows: first, DNA in the blood of cancer patients is derived from cells that disintegrate by necrosis and apoptosis in expanding tumor tissue,37 and this contributed to the results of plasma that were in accordance with the tissue; second, platelets reserve megakaryocyte-derived cytoplasmic pre-mRNA, some of which are spliced into mRNA,38 but most of them are microRNAs accounting for 80%;39 third, platelets can take up tumor-derived membrane vesicles such as exosomes that contain tumor-associated biomarkers;16,35 however, tumor cells might tend to exploit circulating RNA as a means to “communicate” with their regional or distal environment than the DNA derived from necrosis or apoptosis.15,40 Finally, the detection sensitivity of the ADx-ARMS EGFR mutation detection kit we used might be low.
Then, we shifted the focus of research to RNA of platelets. A previous study16 has shown that blood platelets isolated from glioma patients contain the tumor-associated RNA biomarker EGFRvIII, and EGFRvIII has an in-frame deletion of the extracellular domain and is found in numerous types of human tumors. As EGFRvIII has been reported to be tumor specific and has oncogenic potential, it has been shown to be an extremely attractive target for anticancer therapy and is more sensitive to TK inhibition.41 Duan et al42 found EGFR-vIII in eight of 114 (7.0%) in tissues of NSCLC patients. Therefore, we detected the EGFRvIII expression in TEPs and the corresponding plasma isolated from NSCLC; EGFRvIII was detected in three of 205 (1.5%) patients, including 2.1% (2/95) SCC and 0.9% (1/110) AD, and there was no difference with EGFRvIII detection between platelets and plasma. The data suggested that platelets are closely related to NSCLC and platelets contain tumor-derived RNA biomarker, which was consistent with the previous report.16
Conclusion
Our data suggested that TEP is a promising biosource to enable diagnosis of NSCLC, companion diagnostics of NSCLC, and improve convenience for NSCLC patients.
Acknowledgments
This work was supported by Applied Basic Research Program of Science and Technology Bureau Foundation of Wuhan (No. 2016060101010054) and National Natural Science Foundation of China (No. 81672114). This work was also funded by Science and Technology Innovation Fostering Foundation of Zhongnan Hospital of Wuhan University (cxpy20160025) and Wuhan City health and family planning medical talented youth development project.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Pastorino U. Lung cancer screening. Br J Cancer. 2010;102(12):1681–1686. doi: 10.1038/sj.bjc.6605660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seve P, Reiman T, Dumontet C. The role of betaIII tubulin in predicting chemoresistance in non-small cell lung cancer. Lung Cancer. 2010;67(2):136–143. doi: 10.1016/j.lungcan.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Lung Screening Trial Research Team. Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruske-Hohlfeld I. Environmental and occupational risk factors for lung cancer. Methods Mol Biol. 2009;472:3–23. doi: 10.1007/978-1-60327-492-0_1. [DOI] [PubMed] [Google Scholar]
- 7.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol Biomarkers Prev. 2016;25(1):16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 8.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 9.Alix-Panabieres C, Schwarzenbach H, Pantel K. Circulating tumor cells and circulating tumor DNA. Annu Rev Med. 2012;63:199–215. doi: 10.1146/annurev-med-062310-094219. [DOI] [PubMed] [Google Scholar]
- 10.Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4(6):650–661. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32(3–4):623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietras K, Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316(8):1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Ramnath N, Nagrath S. Current status of CTCs as liquid biopsy in lung cancer and future directions. Front Oncol. 2015;5:209. doi: 10.3389/fonc.2015.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joosse SA, Pantel K. Tumor-educated platelets as liquid biopsy in cancer patients. Cancer Cell. 2015;28(5):552–554. doi: 10.1016/j.ccell.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget. 2016;7(1):1066–1075. doi: 10.18632/oncotarget.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nilsson RJ, Balaj L, Hulleman E, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118(13):3680–3683. doi: 10.1182/blood-2011-03-344408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuana Y, Sturk A, Nieuwland R. Extracellular vesicles in physiological and pathological conditions. Blood Rev. 2013;27(1):31–39. doi: 10.1016/j.blre.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enfield KS, Pikor LA, Martinez VD, Lam WL. Mechanistic roles of noncoding RNAs in lung cancer biology and their clinical implications. Genet Res Int. 2012;2012:737416. doi: 10.1155/2012/737416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren S, Wang F, Shen J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49(13):2949–2959. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6(11):9160–9172. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 24.Maemondo M, Inoue A, Kobayashi K, et al. North-East Japan Study Group Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 25.Rosell R, Carcereny E, Gervais R, et al. Spanish Lung Cancer Group in collaboration with Groupe Français de Pneumo-Cancérologie and Associazione Italiana Oncologia Toracica Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Lin J, Liu T, et al. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer. 2014;85(2):110–115. doi: 10.1016/j.lungcan.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Wang LP, Niu H, Xia YF, et al. Prognostic significance of serum sMICA levels in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2015;19(12):2226–2230. [PubMed] [Google Scholar]
- 28.Perez-Callejo D, Romero A, Provencio M, Torrente M. Liquid biopsy based biomarkers in non-small cell lung cancer for diagnosis and treatment monitoring. Transl Lung Cancer Res. 2016;5(5):455–465. doi: 10.21037/tlcr.2016.10.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Askarian-Amiri ME, Crawford J, French JD, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17(5):878–891. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T, Xie J, Shen C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75(15):3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 31.Schotte D, Pieters R, Den Boer ML. MicroRNAs in acute leukemia: from biological players to clinical contributors. Leukemia. 2012;26(1):1–12. doi: 10.1038/leu.2011.151. [DOI] [PubMed] [Google Scholar]
- 32.Cheng J, Deng H, Xiao B, et al. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315(1):12–17. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Yan H, Wu QL, Sun CY, et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29(1):196–206. doi: 10.1038/leu.2014.135. [DOI] [PubMed] [Google Scholar]
- 34.Tian FM, Meng FQ, Wang XB. Overexpression of long-noncoding RNA ZFAS1 decreases survival in human NSCLC patients. Eur Rev Med Pharmacol Sci. 2016;20(24):5126–5131. [PubMed] [Google Scholar]
- 35.Best MG, Sol N, Kooi I, et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015;28(5):666–676. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanikarla-Marie P, Lam M, Menter DG, Kopetz S. Platelets, circulating tumor cells, and the circulome. Cancer Metastasis Rev. 2017;36(2):235–248. doi: 10.1007/s10555-017-9681-1. [DOI] [PubMed] [Google Scholar]
- 37.Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–1665. [PubMed] [Google Scholar]
- 38.Denis MM, Tolley ND, Bunting M, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122(3):379–391. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ple H, Landry P, Benham A, Coarfa C, Gunaratne PH, Provost P. The repertoire and features of human platelet microRNAs. PLoS One. 2012;7(12):e50746. doi: 10.1371/journal.pone.0050746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Azuma M, Danenberg KD, Iqbal S, et al. Epidermal growth factor receptor and epidermal growth factor receptor variant III gene expression in metastatic colorectal cancer. Clin Colorectal Cancer. 2006;6(3):214–218. doi: 10.3816/CCC.2006.n.038. [DOI] [PubMed] [Google Scholar]
- 42.Duan J, Wang Z, Bai H, et al. Epidermal growth factor receptor variant III mutation in Chinese patients with squamous cell cancer of the lung. Thorac Cancer. 2015;6(3):319–326. doi: 10.1111/1759-7714.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]