Abstract
Dimensionality can have a profound impact on stiffness-mediated differentiation of mesenchymal stem cells (MSCs). However, while we have begun to understand cellular response when encapsulated within 3D substrates, the behavior of cells within macro-porous substrates is relatively underexplored. The goal of this study was to determine the influence of macro-porous topographies on stiffness-mediated differentiation of MSCs. We developed macro-porous recombinant elastin-like protein (ELP) substrates that allow independent control of mechanical properties and ligand chemistry. We then used computational modeling to probe the impact of pore topography on the mechanical stimulus that cells are exposed to within these substrates, and finally we investigated stiffness induced biases towards adipogenic and osteogenic differentiation of MSCs within macro-porous substrates. Computational modeling revealed that there is significant heterogeneity in the mechanical stimuli that cells are exposed to within porous substrates and that this heterogeneity is predominantly due to the wide range of possible cellular orientations within the pores. Surprisingly, MSCs grown within 3D porous substrates respond to increasing substrate stiffness by up-regulating both osteogenesis and adipogenesis. These results demonstrate that within porous substrates the behavior of MSCs diverges from previously observed responses to substrate stiffness, emphasizing the importance of topography as a determinant of cellular behavior.
Keywords: Substrate Stiffness, Macro-Porosity, Differentiation, Mesenchymal Stem Cells, Mechanotransduction, Tissue Engineering, Cell-Matrix Interactions
1. Introduction
Mesenchymal stem cells (MSCs) are a heterogeneous population of multipotent cells, isolated from bone marrow, which have the ability to differentiate into bone, cartilage, or fat cells.1–3 These cells have been demonstrated to be an important component of the innate regenerative ability of connective tissues, in addition to playing significant roles in immunomodulation and maintenance of the haematopoietic stem cell niche.2 Due to their differentiation potential and contribution to physiological regeneration, MSCs are frequently utilized in tissue engineering and regenerative medicine. However, for effective clinical translation of MSC-based therapies, it is crucial to achieve robust control of differentiation specificity. Traditionally, biochemical cues have been thought to be the main factor controlling differentiation4. However, over the last decade the importance of mechanical stimulus has attracted much interest, with numerous studies exploring the sensitivity of MSC differentiation to the stiffness of the underlying substrate or matrix.5–10 Accordingly, a comprehensive understanding of the response of MSCs to substrate stiffness is a key aspect to advancing MSC-based tissue engineering therapies.
The mechanosensitivity of MSCs has been long established, and in particular substrate stiffness has been revealed to direct several aspects of MSC behavior, including proliferation, spreading, migration and differentiation.5, 11–13 In the latter case, multiple studies have observed that MSCs will differentiate towards adipogenic lineages on relatively soft substrates, while undergoing osteogenic differentiation on stiffer substrates.6–8 This ability of MSCs to detect and respond to the stiffness of the underlying substrate is thought to be derived from the resistance a cell experiences as it applies tension to the extracellular matrix (ECM) through the actin cytoskeleton.14, 15 Interestingly, modeling of this process has revealed that the mechanical resistance experienced by cells is not solely dependent on the stiffness of the substrate material and that the properties of cell-adhesive ligands presented by the substrate also define the mechanical resistance to cellular tension.16–18 These models have been supported by experimental data revealing that MSC response to substrate stiffness is mediated by ligand identity and density, demonstrating that the interplay between these properties must be considered in any investigation of cell response to substrate stiffness. 18–21
While we have learned much about the roles of substrate stiffness and ligand chemistry in directing MSC fate, it is notable that the majority of these studies have been carried out using two dimensional (2D) substrates. In the few studies that have explored MSC behavior in three dimensional (3D) environments, it has been observed that MSCs encapsulated within hydrogels respond to stiffness in a significantly different fashion to cells upon 2D substrates.10, 22, 23 Most notably, encapsulated cells appear to require the capacity to remodel their surrounding environment; either through cell-mediated degradation or substrate viscoelasticity, in order to generate sufficient tension to allow mechanotransduction.10, 22, 24 Khetan et al. observed that when MSCs were encapsulated within non-degradable hyaluronic acid (HA) hydrogels, only adipogenic differentiation was observed irrespective of stiffness (4.4–91 kPa).22 However, when degradable sequences were introduced to the hydrogels, osteogenic differentiation was observed. Furthermore, Huebsch et al. found that when MSCs were encapsulated within alginate hydrogels with moduli ranging from 1.5–110 kPa they responded similar to 2D studies, in that adipogenic differentiation decreased and osteogenic differentiation increased as the hydrogel stiffness increased.10 However, follow up work by Chaudhuri et al. revealed that the viscoelastic properties of alginate were key to enabling this response.24
Although we have begun to understand some of the differences in cell response to stiffness when encapsulated within homogeneous 3D environments, the behavior of cells within 3D macro-porous scaffolds (i.e. materials containing pore sizes on the order of tens to hundreds of microns) is relatively underexplored. Compared to nano-porous materials, macro-porous scaffolds are often advantageous for clinical translation, as they enable rapid nutrient flux throughout the transplant and promote the migration of both transplanted and endogenous cells.25–27 These substrates have some similarities to 2D substrates, as remodeling of the substrate is not required for cell motility. However, the presentation of matrix stimuli within porous substrates takes on an extra layer of complexity, as the topography of the pores presents a non-uniform mechanical environment while allowing the cells to spread in several planes at once.28–32 Consequently, mechanotransduction in such environments is likely to differ from what has been observed to date.28, 33
As a result of this fundamental gap in knowledge, there is a significant need for the development of novel 3D macro-porous substrates that allow independent control of stiffness and ligand chemistry to enable investigation of MSC response to substrate stiffness in macro-porous environments. Recombinant elastin-like proteins (ELPs) provide an ideal material for carrying out such studies, as their structure can be controlled at the level of the amino acid peptide sequence, allowing control of ligand identity and density without significantly altering the mechanics of the material.34–36 Furthermore, ELPs have been demonstrated to be cyto-compatible, biodegradable, and exhibit less potential for batch-to-batch variation in comparison to harvested, naturally occurring proteins.34, 37 The Young’s moduli of ELP hydrogels has been previously tuned from 0.5 to 50 kPa by altering both the hydrogel weight % and crosslinking density.38 Additionally, the LCST behavior of ELPs significantly streamlines purification in comparison to alternative recombinant protein designs.35 Without the explicit addition of a cell-adhesive ligand within the primary amino acid sequence, ELP hydrogels are relatively resistant to cell adhesion and spreading.35, 39, 40 Here we chose to use ELPs containing either RGD or YIGSR cell-adhesive ligands. RGD is widely used to functionalize hydrogels in studies of mechanotransduction, while previous work has demonstrated that MSCs respond to stiffness differently upon substrates presenting YIGSR instead of RGD.21
With this in mind, the overall goal of this study was to investigate the role of substrate stiffness and ligand chemistry in guiding stem cell fate within 3D macro-porous substrates. The specific objectives were (1) to develop novel, macro-porous ELP substrates that allow independent control of substrate stiffness and ligand chemistry, (2) to use computational models to define the complex mechanical environment within the macro-porous substrates, and (3) to investigate the role of substrate stiffness and ligand identity on osteogenic and adipogenic differentiation of human MSCs within these substrates.
2. Materials and methods
2.1 Design, synthesis and purification of recombinant ELPs
Plasmids encoding for ELPs containing either the cell-adhesive ligand RGD (ELP-RGD) or a scrambled non-adhesive sequence (ELP-RDG) were acquired from stocks constructed previously.35 Recombinant ELP-YIGSR was designed by replacing the cell-adhesive sequence of ELP-RGD with an extended cell-adhesive sequence from the laminin beta one chain (minimal adhesive peptide sequence is YIGSR) and the 5–6 amino acids proceeding and following this sequence (Figure 1A), in order to facilitate conformational flexibility and present a secondary structure that potentially mimics the natural presentation of the adhesive sequence.41 Additionally, serine was substituted for cysteine residues in the flanking sequences to avoid previously reported changes in the lower critical solution temperature (LCST) behavior of YIGSR-containing ELP peptides and allow purification by inverse temperature cycling.42 Plasmids encoding for ELP-YIGSR were purchased (Genscript, NJ, USA) and verified by sequencing (Quintarabio, CA, USA).
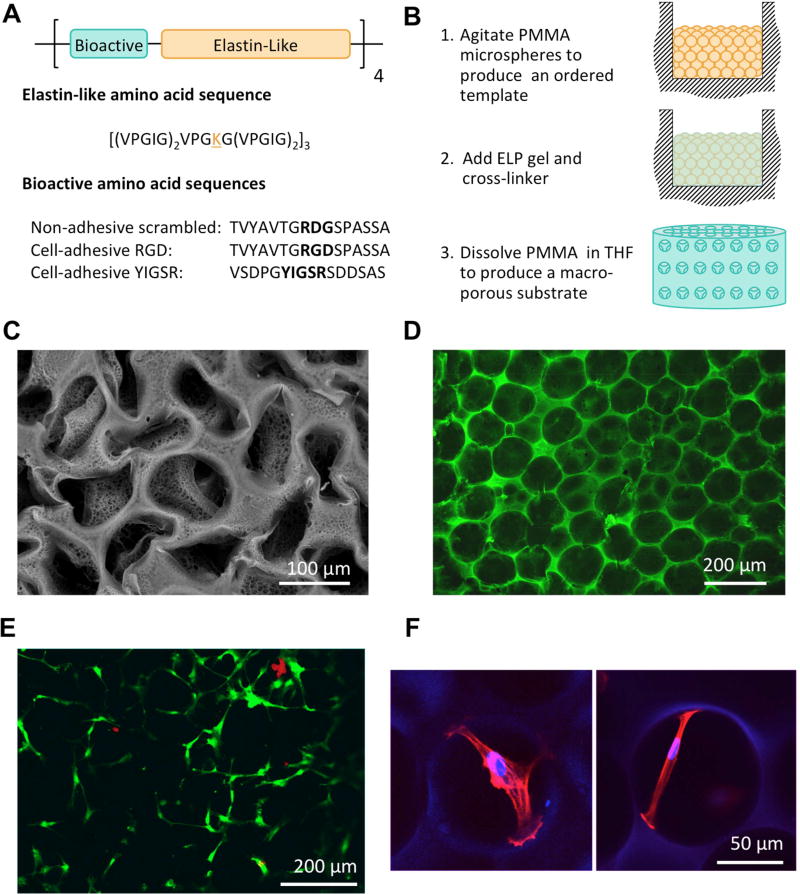
Figure 1.
Porous ELP substrate design, fabrication and biocompatibility. A) ELP peptide sequences were designed with modular repeats of bioactive and elastin-like regions. The bioactive domains used in this study were an extended RGD adhesive sequence from fibronectin, an extended YIGSR adhesive sequence from laminin and a non-adhesive scrambled RDG sequence. B) A schematic of the sacrificial templating technique used to fabricate porous ELP substrates. C) An SEM image of an ELP substrate illustrating the complete removal of PMMA microspheres from the structure. The partial collapse of the structure is due to necessary dehydration of the sample prior to imaging. D) A confocal image of the hydrated pore structure of the ELP substrates. E) A representative image of live (green) and dead (red) hMSCs 24 hours post seeding illustrating that cell viability on ELP substrates is not compromised by the sacrificial templating technique. F) hMSC attachment spans across pores within the porous ELP substrates (actin = red, nucleus/ELP = blue).
All ELPs were recombinantly expressed and purified as previously reported.36, 42 Briefly, a plasmid encoding the specific ELP sequence of interest was transformed into an Escherichia coli host (BL21(DE3)pLysS, ThermoFisher) and expression was induced by activating the T7-lac promoter with isopropyl β-D-1 thiogalactopyranoside (Sigma-Aldrich). After growth, the cells were lysed by alternating freeze/thaw cycles, and the ELP was purified by 3 cycles of centrifugation at alternating temperatures (4 °C and 37 °C), utilizing the LCST behavior of ELPs. The final supernatant was then dialyzed against deionized H2O, lyophilized, and stored at 4 °C. Typical un-optimized protein yields were 83–166 mg/L. The purity of all recombinant proteins was confirmed using protein electrophoresis gels. Additionally, in order to verify the sequence integrity of the purified ELP-YIGSR, amino acid analysis was performed by the Molecular Structure Facility at the University of California, Davis.
2.2 Fabrication of ELP substrates
To produce non-porous hydrogel substrates, ELP solutions were covalently crosslinked using tetrakis (hydroxymethyl) phosphonium chloride (THPC, Sigma-Aldrich).43 Briefly, solutions were prepared containing 62.5 and 125 mg/mL ELP in 1x PBS at 4 °C. These solutions were then mixed with a THPC crosslinking solution to produce hydrogels with a final concentration of 5 wt.% and 10 wt.% ELP. Control of mechanical properties was achieved by varying the molar ratio of the THPC reactive groups to the amine groups within the ELPs (0.5:1 & 1.25:1 Reactive Groups:Amine Groups). Following mixing, 110 µL of these solutions were pipetted into polytetrafluoroethylene (PTFE) molds (well Ø 7 mm) and the crosslinking reaction was allowed to take place for 1 hour at room temperature. The ligand density of the hydrogels was controlled by blending adhesive ELPs (ELP-RGD or ELP-YIGSR) with non-adhesive ELP-RDG to maintain a ligand density of 0.5 mM which is similar to previous studies of MSC response to substrate stiffness.26, 36, 44
In order to produce macro-porous substrates, ELP solutions were gelled around a sacrificial template (Figure 1B) of 100 µm poly(methyl methacrylate) (PMMA) microspheres (Cospheric, CA, USA).30, 32, 45 Briefly, 70 mg of microspheres were loaded into 7 mm diameter wells within a polytetrafluoroethylene mold and agitated to introduce a close packed order. ELP solutions were then prepared as previously described and 46 µL of ELP solution was pipetted on top of each template, and the mold was then centrifuged to perfuse the templates (3 min, 1500 rpm, 4 °C) with the ELP solution. After gelation, the hydrogels were washed in tetrahydrofuran (THF) to dissolve the microspheres (3 changes per day for 3 days). Following the final wash, the hydrogels were rinsed using 70 % ethanol under sterile conditions (x 3), gradually rehydrated in sterile phosphate buffered saline (PBS) and stored at 4 °C until further use.
2.3 Pore structure characterization
Variable-Pressure Scanning Electron Microscopy (VP-SEM) was used to confirm removal of the sacrificial porogens following solvent washing. Prior to imaging, the samples were fixed using 4 % paraformaldehyde (PFA) and washed in deionized H2O. Hydrated samples were then imaged at x 250 magnification using a VP-SEM (Hitachi S-3400N) operated at 15 kV, 50 Pa. The porosity of the substrates was required to build computational models and as an input for the Gibson-Ashby model of the elastic modulus of cellular solids.46 Non-porous and macro-porous substrates were prepared as described previously. The substrate dimensions were measured in a hydrated state, and samples were then flash frozen at −80 °C, lyophilized and weighed. The porosity of the macro-porous substrates was then calculated from the ratio of the porous and non-porous substrate densities. The pore size of the substrates was then measured in order to determine if it was altered by changes in the crosslinking ratio and wt.% ELP. Macro-porous substrates were visualized by staining with a 10 mM solution of NTA-FITC (N925000, Toronto Research Chemicals, ON, Canada) for 1 hour followed by three PBS washes. The substrates were then imaged in a hydrated state using a confocal microscope (Leica SPE). The average pore size (diameter) was quantified from automatically thresholded images using the analyze particles tool in ImageJ (National Institutes of Health, Bethesda, MD, USA) for each image (n > 50 pores/substrate, n=4 substrates/group, pores at image edges were excluded).
2.4 Uniaxial compression testing
Compressive testing was used to determine the effect of wt.% ELP, crosslinking and ligand chemistry on the Young’s modulus of non-porous ELP substrates. Mechanical testing of the samples was carried out using a rheometer (TA Instruments AR-G2, DE, USA) with the ability to perform uniaxial compression tests. Samples were hydrated in PBS for 1 h at 37 °C before testing, and tests were carried out in a bath of PBS on a temperature controlled stage set to 37 °C. Testing was conducted at a strain rate of 10 µm/s. The modulus was defined as the slope of the linear region of the stress-strain curve (n=3).
2.5 Computational modeling of local mechanics
Due to the topographical complexity of porous substrates, experimental quantification of the local stiffness presented by the pore surfaces is problematic (this is expanded upon in section 3.2).32 Consequently, computational models were used to replicate the pore structure of the substrates and determine the non-uniform distribution of local resistance to cellular tension over the surface of a pore.
2.5.1 Model development
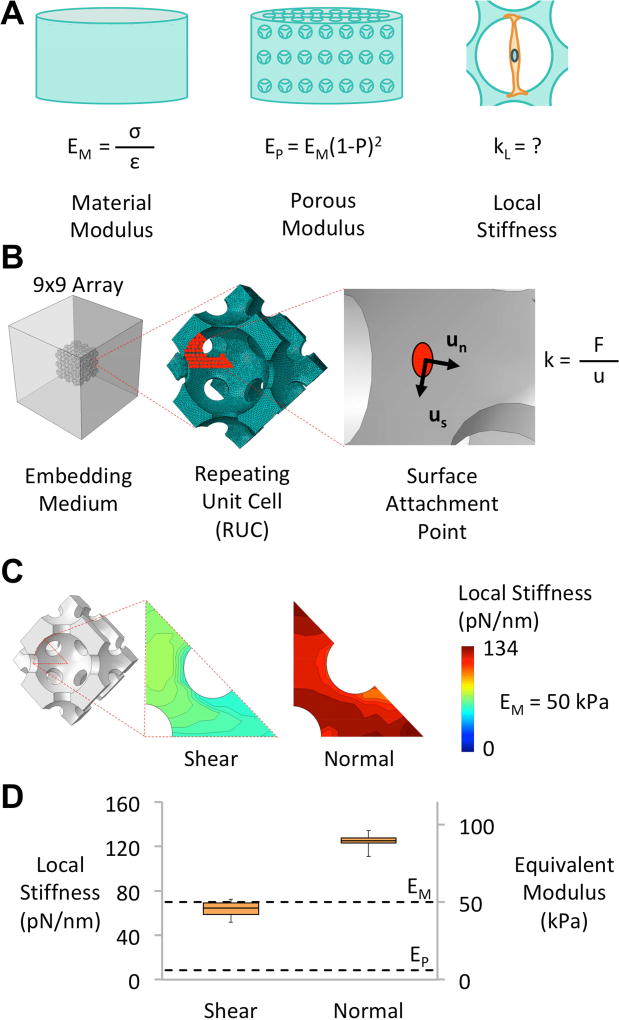
To replicate the ordered structure produced through the porogen-leaching process, this analysis considered an idealized 3D substrate architecture, consisting of repeating units of spherical pores in a face-centered cubic arrangement (Figure 3B). The pore diameter of the substrate was d = 100 µm, while the porosity of the substrate was 60 %. The analysis considered a structure that consisted of 3×3×3 repeating unit-cells, which was embedded in a homogenized medium to avoid boundary effects. As the purpose of this analysis was to characterize the spatial variation of stiffness within the substrate, we considered a number of possible focal adhesion (FA) positions throughout 1/16 of the unit-cell (Figure 3B). Due to symmetry of the unit-cell, these potential locations of FAs are sufficient to describe the spatial stiffness variation throughout the entire substrate. Each substrate model was discretized using approximately 450,000 10-noded tetrahedral elements (C3D10) and solved within the framework of small deformation theory using the ABAQUS finite element code (Simulia, RI, USA). The spatial variation of local stiffness was determined by applying a series of unit micron-displacements to the circular cell-attachment sites, located on the interior surface of each substrate. This represented the local deformation at the cell-substrate interface due to the cellular contractility. Furthermore, in order to replicate the wide ranges of cellular orientations observed in 3D macro-porous substrates, the displacements were applied in both normal and shear orientations with respect to the pore surface.47 Local stiffness (kL, N/m) values were evaluated from the reaction forces generated at control points for each cell attachment site. Further details and validation of this model are provided in supplementary information.
Figure 3.
The computational framework used to determine local stiffness values within porous substrates. A) Material modulus can be determined through uniaxial compression testing, while the modulus of the porous substrate can be derived from the porosity (P) and the material modulus using the Gibson-Ashby cellular solid model.46 However, currently no relationship exists to define the local stiffness at the pore surfaces. B) The model considered attachment points on a 1/16 symmetry of the repeating unit cell. Unit micron displacements in both shear and normal orientations (Un & Us) were then applied to each attachment point and the stiffness (k) was determined based on the reaction force (F) that was generated. C) The spatial variation of shear and normal stiffness across 1/16 of the FCC repeating unit scaffold surface. The stiffness values presented are representative of a substrate with a material modulus of 50 kPa. D) A box plot displaying both the local stiffness and the equivalent modulus presented by a 60 % porous substrate with a material modulus of 50 kPa. The dashed lines signify upper- and lower-bound modulus values, which represent the material and porous modulus of the substrate, respectively.
2.5.2 Equivalent modulus
A further difficulty arises in interpreting the results of the computational models. The local resistance a cell senses when generating tension against a material is a spring constant (kL, N/m) and this is the stimulus captured by our models. However, the majority of experimental work on this topic has reported either the Young’s modulus of the material (EM) or the porous substrate (EP). Therefore, in order to enable direct comparisons between our work and that of others, we calculate an equivalent modulus (EEq), which depicts the local stiffness from our models as the Young’s modulus that an equivalent 2D planar substrate would possess:
| (1) |
where r is the size of a focal adhesion (1 µm).
This is adapted from the work of Ghibaudo et al., who used an equivalent modulus to compare the spring constant stiffness of micro-pillar substrates to the Young’s modulus of planar substrates.48 It is important to note that we are not suggesting that the properties of the material are changing within the porous substrates, we are merely characterizing the mechanical resistance presented to the cells in a manner that allows direct comparison to studies utilizing Young’s modulus as a measure of 2D substrate stiffness.
2.6 hMSC culture and seeding
For LIVE/DEAD experiments, human MSCs (hMSCs) were purchased from Lonza (MD, USA) and expanded as described below. For differentiation experiments, bone marrow aspirates were obtained from the iliac crest of healthy human donors with informed consent and approved by the Clinical Research Ethical Committee at University College Hospital, Galway. MSCs were isolated by direct plating, expanded in culture, and tested for multi-lineage differentiation potential prior to use as described previously.49, 50 All experiments were carried out with four individual MSC isolations. Cells were expanded in standard hMSC growth medium, i.e. low-glucose DMEM (Sigma-Aldrich) supplemented with 10% FBS (Labtech) and 1% penicillin/streptomycin (Sigma-Aldrich) and maintained at 37°C and 5% CO2 in a humidified incubator. Cells were expanded to passage 5 for all experiments. Cells were then detached using 0.25 % trypsin-EDTA acid (Sigma-Aldrich) and suspended at 107 cells/mL. Macro-porous hydrogels were placed into 12-well tissue culture plates and the top surface of each of the substrates was seeded with 10 µL of the cell suspension (1 × 105 cells). The plates were then placed in an incubator for 15 min to allow initial cell attachment. The substrates were then turned over and the opposite surface was seeded with 10 µL of the cell suspension and incubated for a further 15 min. After the second incubation period, 2.5 mL of supplemented medium was added to each well and the plates were returned to the incubator. Both culture flasks and seeded substrates were cultured in a humidified atmosphere at 37 °C and 5 % CO2. Experiments were carried out using the growth medium as described above or mixed induction medium (1:1 adipogenic:osteogenic induction media) which was growth medium supplemented with 250 µM IBMX, 25 µM indomethacin, 5 µg/mL insulin, 25 µg/mL ascorbic acid, 100 nM dexamethasone, and 5 mM β–glycerophosphate. 7, 8, 51
2.7 Cell viability and morphology
A LIVE/DEAD assay kit (Life Technologies) was used to determine the impact of the porogen leaching technique on cell viability. Porous substrates (1.25:1 Reactive Groups:Amine Groups, 10 wt.% ELP, 0.5 mM RGD) were seeded at a density of 6×105 cells/substrate, and the assay was performed at 24 hours post-seeding according to the manufacturer’s instructions. Images were captured using a confocal microscope and analyzed using ImageJ software. Additionally, cellular morphology within the substrates was visualized at this time point. Samples were fixed using 10 % paraformaldehyde and stained with 6-diamidino-2-phenylindole (DAPI, 2 µg/ml, Sigma-Aldrich) to visualize cell nuclei and with rhodamine-conjugated phalloidin (1:200 dilution, Sigma-Aldrich) to visualize F-actin.
2.8 Quantitative analysis of hMSC differentiation
hMSC differentiation after 7 days of culture was assessed through quantification of osteogenic (alkaline phosphatase) and adipogenic (triglyceride content) markers normalized to DNA content. Hydrogel samples were washed (x 3) in sterile PBS, transferred to tubes containing 0.5 mL of lysis buffer (10 mM Tris, 1 mM MgCl2 MA, 20 US µM ZnCl2 PT, and 0.02 % Triton X-100 in DI H2O), and put through three freeze/thaw cycles to complete cell lysis. Following this, the double stranded DNA content of the lysate was quantified using a PicoGreen assay kit (Quant-iT PicoGreen dsDNA assay kit, Life Technologies) according to the manufacturer’s instructions. Intracellular alkaline phosphatase (ALP) activity was assayed using 4-MUP reagent (Sigma-Aldrich) with calf intestinal ALP (Sigma-Aldrich) used as a standard.52 Total triglyceride content was assayed using a triglyceride determination kit (Sigma-Aldrich) following the manufacturer’s instructions and using glycerol as a standard.53, 54
2.9 Statistical analysis
Results are expressed as mean and standard deviation. One-way and two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests were used to evaluate the results when appropriate using R (The R Foundation). Statistical significance was indicated by *, ** or *** corresponding to p ≤ 0.05, ≤ 0.01 or ≤ 0.001 respectively. hMSC differentiation data was obtained from 4 independent experiments using different donors, with each individual experiment containing n = 3 independent technical replicates per stiffness and medium type.
3. Results and discussion
3.1. Sacrificial templating produces porous ELP substrates with independently tunable mechanics and ligand chemistry
Both substrate stiffness and ligand chemistry contribute to the mechanical resistance that is sensed by adherent cells.18, 19 Therefore, our first goal was to fabricate porous substrates that allow independent control of these properties. This necessitated the use of a material that could facilitate modulation of mechanical properties over a wide range and incorporation of multiple different ligand chemistries. Furthermore, we required a fabrication technique that resulted in uniform pore architecture, as we aimed to use computational modeling to characterize the effect of the pore architecture on the mechanics sensed by cells. With this in mind, we sought to demonstrate that sacrificial templating could produce cytocompatible ELP substrates with a uniform pore architecture and independently tunable stiffness and ligand chemistry.
Three ELP variants (Figure 1A) were expressed and purified using established protocols, with protein yields similar to previously reported data (83–166 mg/L).35 The novel ELP-YIGSR peptide was successfully produced, and substitution of the cysteine residues resulted in the ability to purify this ELP variant through the inverse temperature cycling process, avoiding the previously reported requirement to use His-tag separation with nickel affinity columns and resulting in higher protein yields.42 Furthermore, the amino acid content of the purified ELP-YIGSR reflected the composition calculated from the plasmid sequence (Table 1). This ligand chemistry was used as an alternative to RGD and allowed investigation of the effect of ligand chemistry on stiffness directed MSC differentiation.
Table 1.
Theoretical and measured amino acid compositions of ELP-YIGSR.
| Residue | % Total Amino Acid
|
|
|---|---|---|
| Theoretical | Measured | |
| METHIONINE* | 0.7 | 0.2 |
| ALANINE | 4.0 | 5.5 |
| SERINE | 6.8 | 5.5 |
| THREONINE | 0.2 | 0.4 |
| GLYCINE | 30.8 | 29.5 |
| HISTIDINE | 1.4 | 1.3 |
| ASPARTIC ACID | 4.7 | 4.8 |
| LYSINE | 3.1 | 4.1 |
| LEUCINE | 1.4 | 1.6 |
| PROLINE | 16.0 | 15.5 |
| TYROSINE | 0.9 | 0.8 |
| ISOLEUCINE | 12.2 | 12.0 |
| ARGININE | 0.9 | 1.1 |
| VALINE | 16.0 | 16.1 |
| GLUTAMINE & GLUTAMIC ACID | 0.9 | 1.5 |
|
| ||
| Total | 100.0 | 100.0 |
Not accurate due to the instability of methionine during acid hydrolysis.
SEM and confocal imaging of the porous substrates revealed that the sacrificial porogen fabrication technique (Figure 1B) was successful in producing a uniform architecture with open, interconnected pores (Figure 1C & D). The substrates had a porosity of 59.8 %, which is within the range of porosities that have been previously reported using similar templating techniques (55–85 %) and as expected is less than the theoretical porosity of perfectly close packed structures (74 %).30, 32, 45, 55 Furthermore, analysis of LIVE/DEAD staining showed no evidence of toxicity after leaching of the microspheres, with 95 % of cells staining positive with calcein-AM (live stain) 24 hours post seeding (Figure 1E). Interestingly, images of cell attachment within the pores show cells taking up morphologies that span across individual pores and not just along individual pore walls (Figure 1F). Similar behavior has been previously observed within freeze-dried collagen scaffolds.47, 56 Providing the cells with an environment which could potentially support this type of spanning cell attachment motivated our initial design choice of 100 µm pores, in addition to the widespread use of this pore size in regenerative medicine.26–28
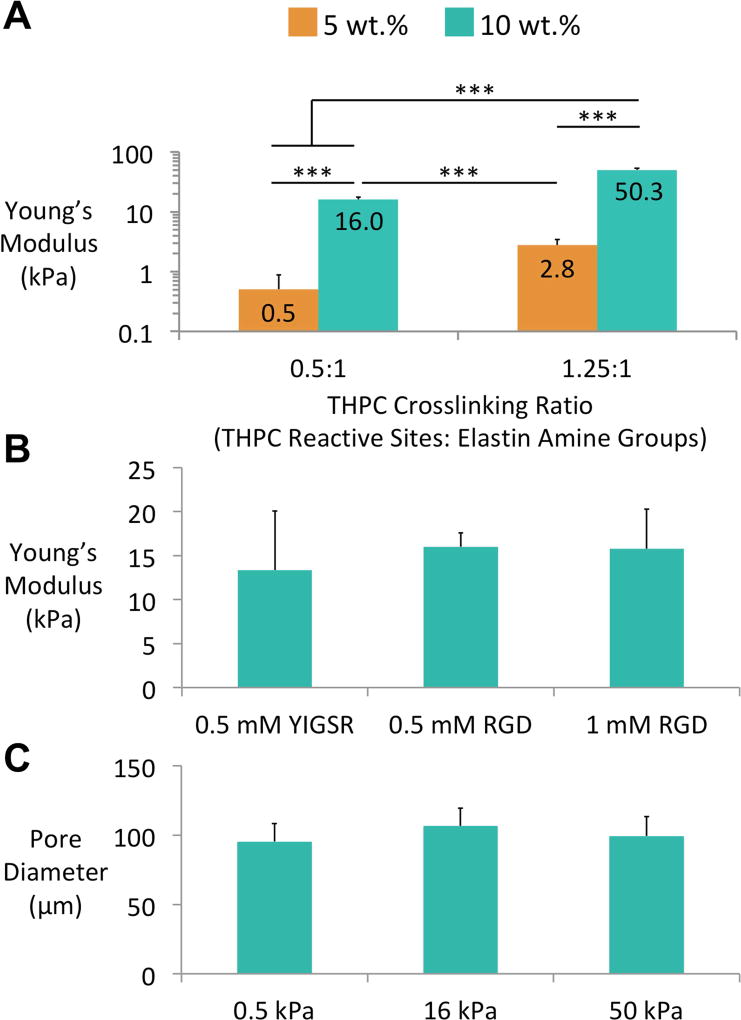
Following the successful fabrication of porous substrates, we assessed the capacity to modulate their mechanical properties over a range suitable for investigation of MSC response to substrate stiffness. Compressive testing of non-porous ELP hydrogels revealed that the Young’s modulus of the ELP hydrogel material could be controlled in a range of 0.5–50.3 kPa by altering both the crosslinking ratio and the wt.% ELP (Figure 2A). This range is similar to those previously used in the literature to investigate MSC response to substrate stiffness.5, 8, 26 Furthermore, modifying the cell-adhesive ligand identity (RGD vs. YIGSR) or ligand density (0.5 vs. 1 mM RGD) of the ELP substrates was found to have no significant effects on the mechanics of the substrates (Figure 2B). This result agrees with previous studies in our laboratory, which have demonstrated that the ligand chemistry and mechanics of our ELP substrates can be independently tuned.35, 36 While the effect of ligand density on cellular behavior was not explored in this study, these data indicate that future studies focusing on this aspect of mechanotransduction can make use of these substrates. Finally, varying the ELP formulation to produce substrates with stiffness ranging from 0.5–50.3 kPa was not found to have a significant effect on the pore diameter of the substrates, which at 95.3–106.6 µm closely matched the diameter of the PMMA microspheres (Figure 2C). Previous work by Marklein et al. has reported an increase in pore size (270–310 µm) as stiffness decreased (7.4-1 kPa), which was thought to be due to swelling of the minimally crosslinked gels.30 This minor discrepancy between our data and that of Marklein et al. is likely due to the differences in the swelling behavior of the peptides and the crosslinking mechanisms used in the two studies. Furthermore, our previous studies of ELP hydrogel swelling have demonstrated no statistically significant differences in swelling behavior across this range of mechanical properties.38
Figure 2.
ELP mechanical properties and pore size. A) The Young’s moduli of non-porous ELP substrates can be controlled by altering both the wt.% ELP and the molar ratio of the THPC reactive groups to the lysine within the ELPs. B) The Young’s modulus of non-porous ELP substrates is not affected by alterations in ligand chemistry achieved through mixing the different ELP peptides. C) The average pore diameter of the macro-porous ELP substrates was not affected by changes in the Young’s modulus of the hydrogels. Results are expressed as mean and standard deviation. One-way (B&C) and two-way (A) analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests were used to evaluate the results. ***p < 0.001
Taken together, these results demonstrate that sacrificial templating can be successfully used to produce porous ELP substrates that possess highly uniform architecture and show no evidence of cytotoxicity. Furthermore, the mechanical properties of the substrates are tunable in a range that is relevant to studies of cellular mechanotransduction and unaffected by alterations to the ligand chemistry. The highly customizable nature of these substrates makes them both ideal for investigations of cell-matrix interactions in 3D environments and potentially suitable for a wide range of applications in regenerative medicine.
3.2 Computational modeling reveals significant heterogeneity in the mechanical stimulus presented by porous substrates
Arguably, the main challenge in investigating the response of cells to stiffness in porous substrates is quantification of the mechanical stimulus to which cells are exposed. In the case of 2D substrates, the stiffness that cells experience is largely a function of the material properties alone. However, within porous substrates, cells adhere to and migrate through the internal pore surfaces, which have varying degrees of support (boundary conditions) and thickness.32 It is the local stiffness of these regions that define the mechanical environment that cells sense, which is nonuniform and a function of both the material properties and the pore topography. Furthermore, while cells generate out of plane tensions upon 2D substrates, the distinctive orientations that cells can adopt within porous substrates (Fig. 1F) make it possible for the majority of cellular tension to be applied normal to the substrate surface.57–59 As stiffness is dependent on the angle of the tensile force with respect to the substrate surface, cellular orientation can consequently result in further differences in stiffness sensed between 2D and 3D environments. To date, studies utilizing porous substrates have used either the Young’s modulus of the substrate material or the modulus of the porous substrate as a measure of the stimuli that cells experience (Figure 3A).30, 32, 60 Neither of these proxies capture the non-uniform nature of the local stiffness that cells are exposed to within porous substrates. Unfortunately, there are significant difficulties in making experimental measurements of the local stiffness within porous substrates.32 Measurement of the local stiffness would require probing shear and normal stiffness at multiple points upon the internal surface of intact pores, which is not feasible with standard atomic force microscopy. Furthermore, passive microrheology measurement techniques, which do not require direct physical contact, only quantify the intrinsic moduli of the material rather than the local stiffness.61 With this in mind, we developed computational models to determine the local stiffness within the porous substrates used in this study.
Shown in Figure 3C is the distribution of the stiffness determined for the substrate architecture throughout the 1/16th section of the substrate surface, where the local stiffness ranged from 51.64–72.35 pN/nm in shear and 110.88–134.25 pN/nm in a normal orientation. Figure 3D shows a box plot of the resulting stiffness and the equivalent modulus calculated from these values. As noted in section 2.5.2, the equivalent modulus presents the local stiffness output by our models as the Young’s modulus that an equivalent 2D planar substrate would possess. The upper bound in Figure 3D is calculated based on the material modulus of the substrate (EM = 50 kPa), while the lower bound was determined based on the modulus of the porous substrate (EP = 6.06 kPa). The median equivalent modulus is 46.27 kPa in shear and 89.27 kPa normal to the substrate surface. These data suggest that the local mechanical stimulus presented by our porous substrates is aligned closer to the material-level behavior, rather than the mechanical behavior of the porous structure. However, given that the equivalent moduli range from 0.92–1.8 times the material modulus, the use of material properties as a proxy does not capture the heterogeneity in stimuli resulting from the various cellular orientations that are possible in 3D. It is worth noting that the shear and normal cases that we have modeled serve as minimum and maximum values respectively; cells are exposed to a range of stimuli between these values depending on their orientation. Interestingly, the location of cellular traction within the pore topography had a relatively smaller impact on the distribution of stiffness values sensed by cells. In shear, the quartile values were within 10 % of the median, while the normal stiffness values were within 2 % of the median. This indicates that for the substrates under investigation in this study, cellular orientation rather than topography is the main source of heterogeneity in the mechanical resistance sensed by cells.
The results presented in Figure 3D were then extrapolated upon to build the dataset in Table 2, which reports the mechanics of the particular porous substrates developed in this study and carried forward to our investigation of hMSC response to stiffness. The substrates developed in this study have median equivalent modulus values ranging from 0.46–46.9 kPa in shear and 0.89–89 kPa in a normal orientation, which is suitable for investigation of the relationship between stiffness and hMSC differentiation.5, 7, 8, 26 Similarly, the median local stiffness values also compare well with previous studies suggesting that cells are sensitive to stiffness in a range of approximately 0.1–100 pN/nm.8, 16, 17
Table 2.
The mechanical properties of the porous substrates used in this study. The material modulus is taken from uniaxial compression testing while the porous and local properties are extrapolated from the computational model.
| ELP Formulation | Material Modulus (EM), kPa |
Porous Substrate Modulus (EP), kPa |
Median Local Equivalent Modulus (Eeq), kPa |
Median Local Stiffness (kL), pN/nm |
|||
|---|---|---|---|---|---|---|---|
| wt.% | Crosslinking | Shear | Normal | Shear | Normal | ||
|
| |||||||
| 10 % | 1.25:1 | 50.0 | 6.06 | 46.27 | 89.27 | 64.60 | 124.65 |
| 10 % | 0.5:1 | 16.0 | 2.11 | 14.81 | 28.57 | 20.67 | 39.89 |
| 5 % | 0.5:1 | 0.5 | 0.07 | 0.46 | 0.89 | 0.65 | 1.25 |
In summary, these data represent the first attempt in the literature to explore the relationship between material properties, cellular orientation and local stiffness within porous substrates. Most notably, our models demonstrate that for a substrate with a porosity of 60 % and a pore size of 100 µm, there is significant heterogeneity in the mechanical stimuli that cells are exposed to and that this heterogeneity is predominantly due to the range of cellular orientations that are possible within such pores. It is important to note that the relationships described here are expected to be dependent on both the porosity of the substrates and the specific pore architecture. For example, as the porosity of the substrate increases, it is likely that the local mechanics will move closer to the modulus of the porous substrate and that the range of stiffness values presented within a pore will also increase. Likewise, as the pore size increases, the range of orientations that cells can adopt with in the pores is likely to diminish, and the mechanical stimuli will become more uniform. However, as the porosity of substrates fabricated from microsphere-based templates is relatively fixed due to the physics of packing spheres, the results presented here have applicability to a wide range of studies using similar fabrication techniques. It is envisioned that future work will investigate the relationship between porosity and local mechanics further.
3.3 hMSC differentiation within porous substrates differs significantly from 2D observations
Having characterized the porous ELP substrates and developed a model for relating material properties to local stiffness, we next set out to investigate the interplay between stiffness and ligand chemistry in directing hMSC differentiation within porous substrates. The typical behavior of hMSCs is to transition from favoring adipogenic differentiation to favoring osteogenic differentiation as substrate stiffness increases. 6–8, 19 Additionally, several studies have demonstrated that ligand chemistry can alter cellular perception of substrate stiffness.18–21 However, these trends have predominantly been established using cells grown on top of 2D substrates. In view of the impact of pore structure on local mechanics demonstrated by our models, there is a need to investigate stiffness directed differentiation within porous substrates. Accordingly, hMSCs from 4 donors were seeded upon porous ELP substrates, which were prepared with 3 different material moduli (0.5, 15 and 50 kPa) and 2 different ligand chemistries (0.5 mM RGD or 0.5 mM YIGSR). DNA, ALP, and triglyceride content were then assayed at 14 days post seeding in order to gauge the relative levels of osteogenic and adipogenic differentiation. Additionally, it should be noted that following the conventional experimental design within the field, all experiments were performed in both growth media and mixed induction media conditions (50:50 adipogenic:osteogenic media supplements).7, 8, 10, 51
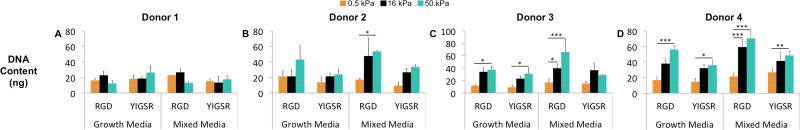
The DNA content within porous substrates is a function of both cellular attachment and proliferation post attachment. Previous studies have observed that increased stiffness can result in an increase in both initial attachment (sometimes termed seeding efficiency) and proliferation within porous substrates.30, 62 In agreement with these observations, we observed significantly higher DNA content when the material modulus increased from 0.5 to 50 kPa (Figure 4C–D, Donor 3&4). However, this result was donor dependent, with half of the donors exhibiting no significant relationship between DNA content and stiffness (Figure 4A–B, Donor 1&2). Additionally, alterations to the ligand chemistry and media type were found to have no effect on this result.
Figure 4.
The effect of stiffness and ligand chemistry on substrate DNA content after 7 days of culture. Results are expressed as mean and standard deviation. Two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests were used to evaluate the results. *p < 0.05, **p < 0.01 and ***p < 0.001
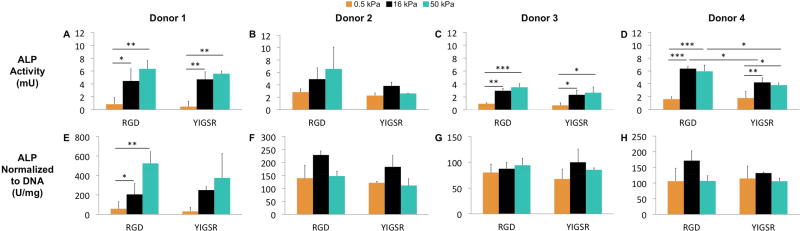
ALP, an enzyme that plays a role in tissue mineralization, was used as an indicator of osteogenic differentiation. Agreeing with previous studies, we found increases in ALP activity as the stiffness increased, with significant increases observed on both the 16 and 50 kPa substrates in comparison to the 0.5 kPa substrates (average increase 4.0-fold and 4.6-fold respectively, Figure 5A–D).8, 26 No significant differences in ALP activity between the 16 and 50 kPa substrates were observed. This response was largely consistent across the four donors, with only one donor showing no response. For the most part, changes to the ligand chemistry had no effect on ALP activity. The notable exception is donor 4, where significantly higher ALP activity was observed upon the RGD substrates in comparison to the YIGSR substrates. Interestingly, normalization of ALP activity to DNA content abolished any relationship between substrate stiffness and ALP activity, with only one donor showing a significant increase in normalized ALP activity with increasing stiffness (Figure 5E–H).
Figure 5.
hMSC expression of ALP after 7 days of culture in growth media with varying substrate stiffness and ligand chemistry. A-D) Total ALP activity. E-H) ALP activity normalized to DNA content. Results are expressed as mean and standard deviation. Two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests were used to evaluate the results. *p < 0.05, **p < 0.01 and ***p < 0.001
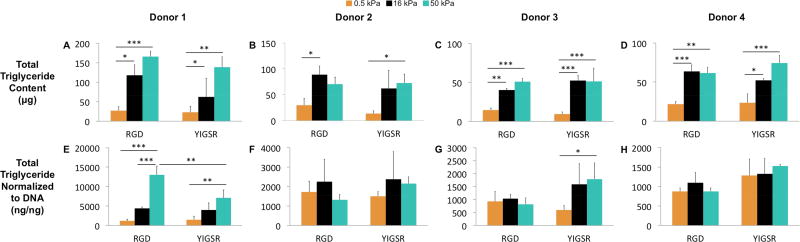
The total triglyceride content within the samples was assayed in order to quantify the increase in lipid content that would be expected with adipogenic differentiation (Figure 6)53, 54 The data in Figure 6 is taken from substrates cultured in mixed induction media, as triglyceride levels were negligible in samples cultured in growth media. Similar to the ALP data, we observed significant increases in triglyceride content on both the 16 and 50 kPa substrates in comparison to the 0.5 kPa substrates (average increase 3.5-fold and 4.4-fold respectively, Figure 6A–D). Again, there were no differences in triglyceride content between the 16 and 50 kPa substrates, and no differences were observed between RGD and YIGSR presenting substrates. Additionally, normalization of triglyceride content to DNA also abrogated any effect of stiffness, with only donor 1 displaying consistent increases in normalized triglyceride content with increasing stiffness (Figure 6E–H).
Figure 6.
hMSC triglyceride content after 7 days of culture in mixed induction media with varying substrate stiffness and ligand chemistry. A–D) Total Triglyceride content. E-H) Total triglyceride normalized to DNA content. Results are expressed as mean and standard deviation. Two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests were used to evaluate the results. *p < 0.05, **p < 0.01 and ***p < 0.001
Taken together, we have observed significant increases in both ALP activity and triglyceride content as the substrate stiffness increases that disappear when the data is normalized to DNA content. Normalization of differentiation markers to DNA with MSCs can be problematic due to the heterogeneous nature of MSC isolations. There are multiple sub-populations of hMSCs within an individual preparation, which have disparate capacities for both proliferation and biases in differentiation towards specific lineages.63–65 It has been noted that the inherent heterogeneity of MSCs may cause bulk observations to mask the behavior of subpopulations.64, 66 Additionally, the non-uniform nature of mechanical stimuli within these macro-porous substrates is likely to further amplify this divergent behavior. As a result of these issues, the lack of significant differences observed in the normalized and aggregate data is not surprising. In the future, the use of new technologies that allow single cell analysis of gene and protein expression may overcome this limitation and provide insight into the behavior of MSCs in macro-porous substrates.66 Nevertheless, it is notable that we have observed statistically significant increases in both osteogenic and adipogenic differentiation markers in conditions where there are no significant increases in DNA content (0.5 kPa vs. 16 kPa, 3 out 4 donors). Furthermore, the lack of additional increases in expression of differentiation markers as the stiffness increases beyond 16 kPa (excluding donor 2) is consistent with previous reports that have observed nonlinear behavior in cellular responses to substrate stiffness.10, 16, 17, 26 It should be noted that previous studies also have observed an eventual decrease in differentiation markers as the stiffness increases further, typically in a range of 90–100 kPa.10, 26 The non-linear nature of cellular response to stiffness may be due to a limit in the cellular capacity to biochemically respond to the mechanical stimulus and/or a limit in the capacity of cells to generate traction against a substrate.16, 17 While differences in the materials and architectures used here make direct comparisons difficult, this nonlinear behavior nevertheless demonstrates that researchers should be cautious in both selecting an appropriate range of stiffness and extrapolating linear behavior from datasets that are limited in the range of stiffness values investigated.
Although we observed a significant correlation between stiffness and expression of differentiation markers, changing the ligand chemistry from RGD to YIGSR had no effect. Previous work has demonstrated that these ligands can have disparate effects on the mechanics of cellular transduction of substrate stiffness.16, 17 However, this effect may to be subtle in comparison to the changes in material properties achieved through altering the crosslinking ratio and peptide wt.%. It has also been suggested that particular ligands can activate signaling pathways that upregulate differentiation towards specific lineages.20, 21 For example, significant increases in osteogenic differentiation markers were observed by Frith et al. for cells on 2D substrates presenting RGD but not upon substrates presenting YIGSR.21 While we did not observe similar results here, it is important to note that previous work has found that cellular response to ligands in 2D is a poor predictor of behavior in 3D.20 Notably, the ligand DGEA only enhanced osteogenic differentiation of MSCs when they were encapsulated within 3D hydrogels presenting the ligand and not when they were seeded upon 2D substrates.20 Similarly, cellular interaction with YIGSR may be enhanced by 3D presentation, potentially explaining why we did not observe similar behavior to Frith et al.
Collectively, these results indicate surprising differences in the response of hMSCs to substrate stiffness in porous environments relative to 2D planar substrates. While the increase in osteogenic commitment with increasing stiffness agrees with prior observations, we also observed a concurrent increase in adipogenic differentiation with stiffness, conflicting with previous reports in the literature. Though porous substrates expose cells to a wide range of mechanical stimuli for a given material modulus, this does not explain the increased levels of adipogenesis observed in our experiments, as upon the stiffest substrates the lower range of local stiffness is still significantly higher than what has been previously observed to induce adipogenic differentiation. It is worth noting that the mechanical environment is not the only parameter that changes with the move from 2D substrates to 3D porous substrates. It is reasonable to expect that the associated changes in cell shape, cell-cell interactions, and nutrient diffusion may also be affecting the differentiation of hMSCs within these porous substrates.13, 25, 33, 67 Notably, high levels of cell-cell contact promotes adipogenesis and inhibits osteogenesis.13, 51 For example, Cosgrove et al. found that hydrogels that present ligands that mimic cell-cell adhesive motifs alter MSC perception of stiffness by increasing the stiffness threshold at which the YAP/TAZ signaling pathway is activated.13 As YAP/TAZ translocation is thought to be required for stiffness-directed osteogenic differentiation, increasing the threshold at which it is activated suppresses osteogenic differentiation. Macro-porous substrates allow for much greater levels of cell-cell contact in comparison to solid hydrogels, thus offering a possible mechanism behind the increased levels of adipogenesis we have observed. Indeed, a recent study has found that human brown adipose progenitors on relatively stiff (9 kPa) porous substrates show superior adipogenesis in comparison to cells encapsulated within soft (2 kPa) hydrogels.68 Furthermore, there have been previous observations of changes in substrate properties resulting in parallel increases in both adipogenic and osteogenic differentiation. Tong et al. encapsulated hMSCs within hydrogels that allowed crosslinks and ligands to slide along the peptide backbone, facilitating cell-mediated reorganization of the local environment without enzymatic degradation.69 The authors observed increased levels of osteogenic and adipogenic differentiation for cells grown on these sliding hydrogels compared to non-sliding hydrogels at the same level of stiffness. This result indicates that conditions that enhance osteogenic differentiation can also be favorable to adipogenic differentiation. Further research is needed to probe the mechanisms behind the concurrent increases in both adipogenic and osteogenic differentiation markers with increasing stiffness. For example, the use of single-cell assays including quantitative immunostaining of differentiation markers would overcome some of the challenges associated with averaging across a heterogeneous cell population.66, 70 Additionally, the use of advanced microscopy techniques coupled with fluorescence resonance energy transfer (FRET) techniques may allow the relationship between the location of a cell within a pore and the tension it generates to be correlated with differentiation.71, 72
4. Conclusion
Understanding the role of substrate stiffness in guiding differentiation is crucial for the progression of MSC-based regenerative therapies. To that aim, we developed and characterized porous ELP substrates that possess uniform pore architecture, excellent cytocompatibility, and the ability to be fabricated with stiffness values in a range that is suitable for investigating cellular mechanotransduction (0.5–50 kPa). Importantly, the stiffness and ligand chemistry of the substrates were observed to be independently tunable. Furthermore, we have developed a novel framework that takes into account the range of cellular orientations and variable topography within porous substrates to depict the local mechanical stimulus sensed by cells. The results indicate that within such substrates cells are exposed to equivalent moduli ranging from 0.92–1.8 times the material modulus and that the heterogeneous nature of the mechanical stimuli is predominantly due to cellular orientation. Importantly, as the pore architecture utilized in this study is widely employed in regenerative medicine, these results have broad applicability. Lastly, we investigated MSC response to the stiffness of porous substrates and observed a surprising concurrent increase in both adipogenic and osteogenic differentiation markers with increasing stiffness. These results demonstrate that within porous substrates the behavior of MSCs diverges from the typical response to stiffness observed upon 2D substrates, emphasizing the importance of topography as a determinant of cellular behavior.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Lydia-Marie Joubert and the Beckman Cell Sciences Imaging Facility (CSIF) for assistance with and use of the SEM; and Sharon Chen for assistance with macro-porous substrate fabrication. We gratefully acknowledge funding from an Irish Research Council ELEVATE fellowship ELEVATEPD/2013/30 (M.G.H.), the National Institutes of Health U19-AI-116484 (S.C.H.) and the National Science Foundation DMR-1508006 (S.C.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Caplan AI. Mesenchymal stem cells. J. Orthop. Res. 1991;9:641–650. doi: 10.1002/jor.1100090504. [DOI] [PubMed] [Google Scholar]
- 2.Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 3.Friedenstein AJ, Chailakhyan RK, Latsinik NV, Panasyuk AF, Keiliss-Borok IV. Stromal cells responsible for transferring the microenvironment of the hematopoietic tissues: Cloning In Vitro and Retransplantation In Vivo. Transplantation. 1974;17:331–40. doi: 10.1097/00007890-197404000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Murphy WL, McDevitt TC, Engler AJ. Materials as stem cell regulators. Nat. Mater. 2014;13:547–557. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 6.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang C, Tibbitt MW, Basta L, Anseth KS. Mechanical memory and dosing influence stem cell fate. Nat. Mater. 2014;13:645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wen JH, et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trappmann B, et al. Extracellular-matrix tethering regulates stem-cell fate. Nat. Mater. 2012;11:742–742. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 10.Huebsch N, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat. Mater. 2010;9:518–26. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JS, et al. The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-beta. Biomaterials. 2011;32:3921–3930. doi: 10.1016/j.biomaterials.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tse JR, Engler AJ. Stiffness gradients mimicking in vivo tissue variation regulate mesenchymal stem cell fate. PLoS One. 2011;6:e15978. doi: 10.1371/journal.pone.0015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosgrove BD, et al. N-cadherin adhesive interactions modulate matrix mechanosensing and fate commitment of mesenchymal stem cells. Nat. Mater. 2016;15:1297–1306. doi: 10.1038/nmat4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puklin-Faucher E, Sheetz MP. The mechanical integrin cycle. J Cell Sci. 2009;122:179–186. doi: 10.1242/jcs.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 2015;17:955–963. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan CE, Odde DJ. Traction dynamics of filopodia on compliant substrates. Science. 2008;322:1687–91. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- 17.Bangasser BL, Rosenfeld SS, Odde DJ. Determinants of maximal force transmission in a motor-clutch model of cell traction in a compliant microenvironment. Biophys. J. 2013;105:581–592. doi: 10.1016/j.bpj.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elosegui-Artola A, et al. Rigidity sensing and adaptation through regulation of integrin types. Nat. Mater. 2014;13:631–637. doi: 10.1038/nmat3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowlands AS, George PA, Cooper-White JJ. Directing osteogenic and myogenic differentiation of MSCs: interplay of stiffness and adhesive ligand presentation. Am. J. Physiol. Cell Physiol. 2008;295:C1037–44. doi: 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- 20.Mehta M, Madl CM, Lee S, Duda GN, Mooney DJ. The collagen I mimetic peptide DGEA enhances an osteogenic phenotype in mesenchymal stem cells when presented from cell-encapsulating hydrogels. J. Biomed. Mater. Res. Part A. 2015;103:3516–3525. doi: 10.1002/jbm.a.35497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frith JE, Mills RJ, Hudson JE, Cooper-White JJ. Tailored integrin-extracellular matrix interactions to direct human mesenchymal stem cell differentiation. Stem Cells Dev. 2012;21:2442–2456. doi: 10.1089/scd.2011.0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khetan S, et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat. Mater. 2013;12:458–65. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parekh SH, et al. Modulus-driven differentiation of marrow stromal cells in 3D scaffolds that is independent of myosin-based cytoskeletal tension. Biomaterials. 2011;32:2256–64. doi: 10.1016/j.biomaterials.2010.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaudhuri O, et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 2016;15:326–34. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qazi TH, Mooney DJ, Duda GN, Geissler S. Biomaterials that promote cell-cell interactions enhance the paracrine function of MSCs. Biomaterials. 2017;140:103–114. doi: 10.1016/j.biomaterials.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Huebsch N, et al. Matrix elasticity of void-forming hydrogels controls transplanted-stem-cell-mediated bone formation. Nat. Mater. 2015;14:1269–77. doi: 10.1038/nmat4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levingstone TJ, et al. Cell-free multi-layered collagen-based scaffolds demonstrate layer specific regeneration of functional osteochondral tissue in caprine joints. Biomaterials. 2016;87:69–81. doi: 10.1016/j.biomaterials.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Harley BaC, et al. Microarchitecture of three-dimensional scaffolds influences cell migration behavior via junction interactions. Biophys. J. 2008;95:4013–24. [Google Scholar]
- 29.Corin Ka, Gibson LJ. Cell contraction forces in scaffolds with varying pore size and cell density. Biomaterials. 2010;31:4835–45. doi: 10.1016/j.biomaterials.2010.01.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marklein Ra, Soranno DE, Burdick Ja. Magnitude and presentation of mechanical signals influence adult stem cell behavior in 3-dimensional macroporous hydrogels. Soft Matter. 2012;8:8113. [Google Scholar]
- 31.Haugh MG, Heilshorn SC. Integrating concepts of material mechanics, ligand chemistry, dimensionality and degradation to control differentiation of mesenchymal stem cells. Curr. Opin. Solid State Mater. Sci. 2016;20:171–179. doi: 10.1016/j.cossms.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peyton SR, et al. Marrow-derived stem cell motility in 3D synthetic scaffold is governed by geometry along with adhesivity and stiffness. Biotechnol. Bioeng. 2011;108:1181–93. doi: 10.1002/bit.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker BM, Chen CS. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romano NH, Sengupta D, Chung C, Heilshorn SC. Protein-engineered biomaterials: nanoscale mimics of the extracellular matrix. Biochim. Biophys. Acta. 2011;1810:339–49. doi: 10.1016/j.bbagen.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Straley KS, Heilshorn SC. Independent tuning of multiple biomaterial properties using protein engineering. Soft Matter. 2009;5:114. [Google Scholar]
- 36.Madl CM, Katz LM, Heilshorn SC. Bio-Orthogonally Crosslinked, Engineered Protein Hydrogels with Tunable Mechanics and Biochemistry for Cell Encapsulation. Adv. Funct. Mater. 2016;26:3612–3620. doi: 10.1002/adfm.201505329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maskarinec SA, Tirrell DA. Protein engineering approaches to biomaterials design. Curr. Opin. Biotechnol. 2005;16:422–6. doi: 10.1016/j.copbio.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Madl CM, et al. Maintenance of neural progenitor cell stemness in 3D hydrogels requires matrix remodelling. Nat. Mater. 2017;16:1233–1242. doi: 10.1038/nmat5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu JC, Heilshorn SC, Tirrell DA. Comparative cell response to artificial extracellular matrix proteins containing the RGD and CS5 cell-binding domains. Biomacromolecules. 2004;5:497–504. doi: 10.1021/bm034340z. [DOI] [PubMed] [Google Scholar]
- 40.Liu JC, Tirrell DA. Cell response to RGD density in cross-linked artificial extracellular matrix protein films. Biomacromolecules. 2008;9:2984–2988. doi: 10.1021/bm800469j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heilshorn S. Endothelial cell adhesion to the fibronectin CS5 domain in artificial extracellular matrix proteins. Biomaterials. 2003;24:4245–4252. doi: 10.1016/s0142-9612(03)00294-1. [DOI] [PubMed] [Google Scholar]
- 42.Straley KS, Heilshorn SC. Design and adsorption of modular engineered proteins to prepare customized, neuron-compatible coatings. Front. Neuroeng. 2009;2:9. doi: 10.3389/neuro.16.009.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung C, Lampe KJ, Heilshorn SC. Tetrakis(hydroxymethyl) phosphonium chloride as a covalent cross-linking agent for cell encapsulation within protein-based hydrogels. Biomacromolecules. 2012;13:3912–3916. doi: 10.1021/bm3015279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romano NH, Madl CM, Heilshorn SC. Matrix RGD ligand density and L1CAM-mediated Schwann cell interactions synergistically enhance neurite outgrowth. Acta Biomater. 2015;11:48–57. doi: 10.1016/j.actbio.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stachowiak AN, Bershteyn A, Tzatzalos E, Irvine DJ. Bioactive hydrogels with an ordered cellular structure combine interconnected macroporosity and robust mechanical properties. Adv. Mater. 2005;17:399–403. [Google Scholar]
- 46.Gibson LJ, Ashby MF. Cellular solids: structures & properties. Pergamon Press; 1997. [Google Scholar]
- 47.Freyman TM, Yannas IV, Pek YS, Yokoo R, Gibson LJ. Micromechanics of fibroblast contraction of a collagen-GAG matrix. Exp. Cell Res. 2001;269:140–53. doi: 10.1006/excr.2001.5302. [DOI] [PubMed] [Google Scholar]
- 48.Ghibaudo M, et al. Traction forces and rigidity sensing regulate cell functions. Soft Matter. 2008;4:1836. [Google Scholar]
- 49.Coleman CM, et al. Growth differentiation factor-5 enhances in vitro mesenchymal stromal cell chondrogenesis and hypertrophy. Stem Cells Dev. 2013;22:1968–76. doi: 10.1089/scd.2012.0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raftery RM, et al. Multifunctional biomaterials from the sea: Assessing the effects of chitosan incorporation into collagen scaffolds on mechanical and biological functionality. Acta Biomater. 2016;43:160–169. doi: 10.1016/j.actbio.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 51.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 52.Chaudhuri O, et al. Substrate stress relaxation regulates cell spreading. Nat. Commun. 2015;6:6364. doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li AC, et al. Differential inhibition of macrophage foam-cell formation and atherosclerosis in mice by PPARα, β/δ, and γ. J. Clin. Invest. 2004;114:1564–1576. doi: 10.1172/JCI18730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huh J-E, et al. Arginine enhances osteoblastogenesis and inhibits adipogenesis through the regulation of Wnt and NFATc signaling in human mesenchymal stem cells. Int. J. Mol. Sci. 2014;15:13010–29. doi: 10.3390/ijms150713010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Wang S, Eghtedari M, Motamedi M, Kotov NA. Inverted-Colloidal-Crystal Hydrogel Matrices as Three-Dimensional Cell Scaffolds. Adv. Funct. Mater. 2005;15:725–731. [Google Scholar]
- 56.McCoy RJ, Jungreuthmayer C, O’Brien FJ. Influence of flow rate and scaffold pore size on cell behavior during mechanical stimulation in a flow perfusion bioreactor. Biotechnol. Bioeng. 2012;109:1583–94. doi: 10.1002/bit.24424. [DOI] [PubMed] [Google Scholar]
- 57.Hur SS, Zhao Y, Li YS, Botvinick E, Chien S. Live Cells Exert 3-Dimensional Traction Forces on Their Substrata. Cell. Mol. Bioeng. 2009;2:425–436. doi: 10.1007/s12195-009-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maskarinec SA, Franck C, Tirrell DA, Ravichandran G. Quantifying cellular traction forces in three dimensions. Proc. Natl. Acad. Sci. U. S. A. 2009;106:22108–13. doi: 10.1073/pnas.0904565106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Legant WR, et al. Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proc. Natl. Acad. Sci. U. S. A. 2013;110:881–6. doi: 10.1073/pnas.1207997110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murphy CM, Matsiko A, Haugh MG, Gleeson JP, O’Brien FJ. Mesenchymal stem cell fate is regulated by the composition and mechanical properties of collagen-glycosaminoglycan scaffolds. J. Mech. Behav. Biomed. Mater. 2012;11:53–62. doi: 10.1016/j.jmbbm.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 61.Krajina BA, et al. Dynamic Light Scattering Microrheology Reveals Multiscale Viscoelasticity of Polymer Gels and Precious Biological Materials. ACS Cent. Sci. 2017;3:1294–1303. doi: 10.1021/acscentsci.7b00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haugh MG, Murphy CM, McKiernan RC, Altenbuchner C, O’Brien FJ. Crosslinking and Mechanical Properties Significantly Influence Cell Attachment, Proliferation, and Migration Within Collagen Glycosaminoglycan Scaffolds. Tissue Eng. Part A. 2011;17:1201–1208. doi: 10.1089/ten.TEA.2010.0590. [DOI] [PubMed] [Google Scholar]
- 63.Wu J, Tzanakakis ES. Deconstructing stem cell population heterogeneity: single-cell analysis and modeling approaches. Biotechnol. Adv. 2013;31:1047–62. doi: 10.1016/j.biotechadv.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar RM, et al. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature. 2014;516:56–61. doi: 10.1038/nature13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phinney DG. Functional heterogeneity of mesenchymal stem cells: Implications for cell therapy. Journal of Cellular Biochemistry. 2012;113:2806–2812. doi: 10.1002/jcb.24166. [DOI] [PubMed] [Google Scholar]
- 66.McLeod CM, Mauck RL. On the origin and impact of mesenchymal stem cell heterogeneity: new insights and emerging tools for single cell analysis. Eur. Cell. Mater. 2017;34:217–231. doi: 10.22203/eCM.v034a14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thorpe SD, Nagel T, Carroll SF, Kelly DJ. Modulating Gradients in Regulatory Signals within Mesenchymal Stem Cell Seeded Hydrogels: A Novel Strategy to Engineer Zonal Articular Cartilage. PLoS One. 2013:8. doi: 10.1371/journal.pone.0060764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuss M, et al. Effects of tunable, 3D–bioprinted hydrogels on human brown adipocyte behavior and metabolic function. Acta Biomater. 2018 doi: 10.1016/j.actbio.2018.03.021. doi: https://doi.org/10.1016/j.actbio.2018.03.021. [DOI] [PMC free article] [PubMed]
- 69.Tong X, Yang F. Sliding Hydrogels with Mobile Molecular Ligands and Crosslinks as 3D Stem Cell Niche. Adv. Mater. 2016;28:7257–7263. doi: 10.1002/adma.201601484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spitzer MH, Nolan GP. Mass Cytometry: Single Cells, Many Features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 2013;13:3985–3989. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.