Abstract
For maltose fermentation, budding yeast Saccharomyces cerevisiae operates a mechanism that involves transporters (MALT), maltases (MALS) and regulators (MALR) collectively known as MAL genes. However, functional relevance of MAL genes during sake brewing process remains largely elusive, since sake yeast is cultured under glucose-rich condition achieved by the co-culture partner Aspergillus spp.. Here we isolated an ethyl methane sulfonate (EMS)-mutagenized sake yeast strain exhibiting enhanced maltose fermentation compared to the parental strain. The mutant carried a single nucleotide insertion that leads to the extension of the C-terminal region of a previously uncharacterized MALR gene YPR196W-2, which was renamed as MAL73. Introduction of the mutant allele MAL73L with extended C-terminal region into the parental or other sake yeast strains enhanced the growth rate when fed with maltose as the sole carbon source. In contrast, disruption of endogenous MAL73 in the sake yeasts decreased the maltose fermentation ability of sake yeast, confirming that the original MAL73 functions as a MALR. Importantly, the MAL73L-expressing strain fermented more maltose in practical condition compared to the parental strain during sake brewing process. Our data show that MAL73(L) is a novel MALR gene that regulates maltose fermentation, and has been functionally attenuated in sake yeast by single nucleotide deletion during breeding history. Since the MAL73L-expressing strain showed enhanced ability of maltose fermentation, MAL73L might also be a valuable tool for enhancing maltose fermentation in yeast in general.
Introduction
Generally, yeast alters its gene expression pattern in response to environmental nutritional conditions, notably including sugar and nitrogen sources. Alteration of gene expression in response to environmental sugar availability has been extensively studied using the budding yeast Saccharomyces cerevisiae [1]. Maltose, which is a α-disaccharide comprising two α-glucose monomers, is present at high levels in malt prepared from germinated barley (Hordeum vulgare) seed. Maltose fermentation in S. cerevisiae requires a set of MAL genes (i.e. genes encoding a maltose transporter (MALT), maltase (MALS), and zinc finger-type transcription factors (MALR) that positively regulate MALT and MALS genes [2]. To date, Gibson et al. [3] have reported the sequence of several MALR genes (MAL23, MAL43, and MAL63), and Hu et al. [4] have defined the functional domains of MALR proteins. On the other hand, MAL13 and MAL33 do not seem to be functionally relevant in yeast under laboratory conditions [5, 6]. These MAL genes are crucial for fermentation of maltose, as demonstrated by the observation that yeasts lacking any of these genes fail to grow in medium in which maltose is the sole carbon source [7]. Five structurally similar, but genetically unlinked, MAL loci (MAL1 to MAL4 and MAL6) have been shown to each contain up to three different genes, including: a MALx1 (where x = 1 to 4 or 6) gene encoding a maltose transporter (MALT); a MALx2 gene, encoding a maltase protein (MALS); and a MALx3 gene, a transcriptional regulator (MALR) of other MAL genes [7–11]. These MAL loci are located proximal to the telomeric region of the respective chromosome (MAL1 on chromosome VII, MAL2 on chromosome III, MAL3 on chromosome II, MAL4 on chromosome XI, and MAL6 on chromosome VIII). In medium containing high concentrations of glucose, glucose uptake is promoted via hexose transporters (HXTs) and maltose uptake is simultaneously transcriptionally repressed, a cellular response that is known as a part of the “glucose repression” phenomenon [12]. For example, the maltose transporter-encoding MAL11 gene is repressed in the presence of glucose [13].
In some yeast strains, maltose fermentation ability has been compromised or completely abolished during their evolution or via selection during domestication, as demonstrated by the loss-of-function mutation in, or absence of, one or more MAL loci. The molecular mechanism(s) regulating maltose fermentation have yet to be elucidated in some commercially important yeast strains such as Japanese sake yeast [14]. Among brewing yeasts, beer yeasts show high maltose fermentation ability, including the diversification of various kinds of MAL genes, presumably due to artificial selection under conditions favoring malt-based fermentation. In contrast, Japanese sake yeast strains, which are utilized for fermentation in the glucose-rich environments resulting from the activity of fungal (Aspergillus oryzae) glucoamylases, show only a weak maltose fermentation ability [15]. Although the genome sequence of the Japanese sake yeast K7 strain, which is a major sake yeast, has been reported [16], the basis for the weak maltose fermentation ability of sake yeasts remains elusive. In the present study, we focused on the low maltose fermentation ability of sake yeast, and employed a genetic approach to enhance this property. Here we show that a putative MALR gene YPR196W-2, renamed as MAL73, encodes a novel hypomorphic regulatory protein whose function can be enhanced by a single nucleotide insertion that leads to the expression of MAL73L harboring an extended C-terminal region. In turn, the data also suggest that MAL73L has been functionally attenuated in sake yeast through frameshifts during breeding history. MAL73L expression in the original sake yeast strain resulted in enhanced growth on maltose medium compared to the parental strain. Moreover, MAL73L-expressing strain was capable of fermenting sake slightly faster than a control yeast strain. These results suggest that MAL73L is a genetic tool valuable for improving the maltose fermentation ability of yeast.
Materials and methods
Strains and media
Yeast strains used in this study are X2180-1A (MAT a, ATCC26786) and Kyokai No. 7 (K7) series strains (K7, K9, K1001, K1601, and K1801; the Brewing Society of Japan). Cells were cultured in YPD (2% glucose, 1% yeast extract, and 2% peptone), YPM (2% maltose, 1% yeast extract, 2% peptone), SD medium (2% glucose, 0.67% yeast nitrogen base), SMal medium (2% maltose, 0.67% yeast nitrogen base), SD+Antimycin A (SD medium containing antimycin A; 0.67% yeast nitrogen base (Difco), 2% maltose, 2 mg/L antimycin A), or MA medium (SMal+antimycin A; SMal medium containing antimycin A; 0.67% yeast nitrogen base (Difco), 2% maltose, 2 mg/L antimycin A). Antimycin A is known to inhibit the mitochondrial electron transfer system, thus permitting stringent assessment of maltose fermentation ability [12].
Gene disruption
Primers used in this study are summarized in Table 1. The YPR196W-2Δ mutant was generated by using the CRISPR/Cas9 system [17] as described previously in DiCarlo et al. [18]. Briefly, gRNA (guide RNA) was generated as follows. For the upstream gRNA, the 1st PCR was performed by using the primer pairs gRNA-F + YRP196W-2-N1 and YRP196W-2-N2 + gRNA-R. The two resulting PCR fragments were combined by using the GeneArt Seamless Cloning and Assembly Enzyme Mix (Thermo Fisher Scientific), yielding the desired gRNA. To amplify this gRNA, the 2nd PCR was performed by using the primer pair gRNA-F + gRNA-R with gRNA as a template. The resulting gRNA PCR fragment was introduced into the strain carrying Cas9 plasmid. To generate the downstream gRNA, the 1st PCR was performed by using the primer pairs gRNA-F + YRP196W-2-C1 and YRP196W-2-C2 + gRNA-R. The subsequent steps were as described above for the standard gRNA construction. A PCR fragment containing the hygromycin B resistance-encoding gene (hygB) was amplified by using the primer pair YRP196W-2-hyg-F + YRP196W-2-hyg-R with pYC240 plasmid as the template [19]. These three fragments (upstream gRNA, downstream gRNA, and the hygB fragment) were introduced into cells carrying Cas9 plasmid, and the resulting yeast transformants were selected using 100 μg/ml hygromycin B-containing YPD plates. After verification of the YRP196W-2 gene disruption, transformants were grown in the absence of selection for the Cas9 plasmid, and loss of the plasmid was confirmed by screening.
Table 1. Primers used in this study.
| Primer | Sequence |
|---|---|
| Cas9-F | 5’-AATTAACTAAACAGGCCTATGGCTAGTATGCAGAAACTGA-3’ |
| Cas9-R | 5’-TAATTATTACTCAGGCCTTCACTTCTTCTTCTTTGCCTGT-3’ |
| gRNA-F | 5’-CCTTGGAGTAAAAATCAACTATCATC-3’ |
| gRNA-R | 5’-GTATCAGGCTAACGTGCACT-3’ |
| MAL11-RT-F | 5’-TTGGGTCATCATCGATCTGC-3’ |
| MAL11-RT | 5’-TCCGAATGGATCAACCACAG-3’ |
| MAL33-RT-F | 5’-AAGCGGTCCTAACACCATTG-3’ |
| MAL33-RT-R | 5’-ATAGAGCCGCAAGCACTGAT-3’ |
| pYC-Z130-F | 5’-CTCCACCTCAGCCAGAGTTC-3’ |
| pYC-Z130-R | 5’-GACTCCGTAGCCAAAGCATC-3’ |
| TDH3pro-F | 5’-CATTAATGCAGGTTGCGGCCGCACCAGTTCTCACACGGAAC-3’ |
| TDH3pro-R | 5’-AACCAAGGCCTGTTTATGTGTGTTTATTCGAA-3’ |
| TDH3term-F | 5’-TAAACAGGCCTTGGTTGAACACGTTGCC-3’ |
| TDH3term-R | 5’-CGGGCATTTAAATGCGGCCGCCC TCAATCAATGAATCGAAAATGTCA-3’ |
| TDH3-RT-F | 5’-CGGTAACATCATCCCATCC-3’ |
| TDH3-RT-R | 5’-CGGCAGCCTTAACAACCTTC-3’ |
| TDH3-YPR196W-2-F | 5’-ACACACATAAACAGGCCTATGACTTTGGCAAAACAGG-3’ |
| TDH3-YPR196W-2-R | 5’-CGTGTTCAACCAAGGCCTTAAGGAATTATGTCGTCTTCATC-3’ |
| TEF2pro-F | 5’-CATTAATGCAGGTTGCGGCCGCACTTACATATAGTAGATGTCAAGC-3’ |
| TEF2pro-R | 5’-TACTCAGGCCTGTTTAGTTAATTATAGTTCGTTGAC-3’ |
| TEF2term-F | 5’-TAAACAGGCCTGAGTAATAATTATTGCTTCCATATAATATT-3’ |
| TEF2term-R | 5’-CGGGCATTTAAATGCGGCCGCCCTACTGGGTAAATTTTGAAAG-3’ |
| YPR196W-RT-F | 5’-ATGTGATTGCTGTCGCGTTC-3’ |
| YPR196W-RT-R | 5’-GCTTTCTCCGATGGACTTGG-3’ |
| YPR196W-2-N1 | 5’-GTCACACTTTACTCGACGAAGATCATTTATCTTTCACTGC-3’ |
| YPR196W-2-N2 | 5’-TTCGTCGAGTAAAGTGTGACGTTTTAGAGCTAGAAATAGCAA-3’ |
| YPR196W-2-C1 | 5’-TTGAAACATTTTATGATTGCGATCATTTATCTTTCACTGC-3’ |
| YPR196W-2-C2 | 5’-GCAATCATAAAATGTTTCAAGTTTTAGAGCTAGAAATAGCAA-3’ |
| YPR196W-2-hyg-F | 5’-CGTTCCATTGTATTCAATAACTTAAACTAGTGAAGTTCCTATAC-3’ |
| YPR196W-2-hyg-R | 5’-CTCGCCTGAACATAGCTTCTTACACAGCGCTGAAGTTCCTATTC-3’ |
| YPR196W-2-F | 5’-CATTAATGCAGGTTGCGGCCGCATAGAGACACATCTCTTTCCG-3’ |
| YPR196W-2-R | 5’-CGGGCATTTAAATGCGGCCGCCCACTCTATTTATGTTCCGGCAAC-3’ |
| YPR196W-2-RT-F | 5’-CCTTCCCTTCGGTGAACAAC-3’ |
| YPR196W-2-RT-R | 5’-TGGTAGTGGTGGCGCTATTG-3’ |
| YPR196W-2-pro-F | 5’-CGCGTCGACGGTACCGAATTCACTTGATCTGAATATTTTCCTCTCC-3’ |
| YPR196W-2-pro-F | 5’-AACAGCCAAGCTCCGGATCCAGTCATTTGTAAAGTAAAATTCCAATAGA-3’ |
Plasmids
The YCpG-TDH3pt vector was constructed as follows. The TDH3 promoter and terminator were amplified by using primer pairs TDH3-pro-F + TDH3-pro-R and TDH3-term-F + TDH3-term-R, respectively. This construct was designed to introduce a unique StuI site between the promoter and terminator. pJHXSB [20] was digested with AscI, and then these three fragments (THD3 promoter, TDH3 terminator, and linearized pJHXSB fragments) were combined using the GeneArt Seamless Cloning and Assembly Enzyme Mix. The resulting plasmid was digested with AscI, and the fragment containing the TDH3 promoter, terminator and CEN/ARS was ligated with an AscI fragment containing the G418-resistance gene from pYC030 [19] to yield the plasmid YCpG-TDHpt.
To construct the YCp-TEFpro-term vector, the TEF2 promoter and terminator were amplified by using the primer pairs TEF2-pro-F + TEF2-pro-R and TEF2-term-F + TEF2-term-R, respectively. The YCpG-TDH3pt vector was digested with NotI, and then these three fragments were combined by using the GeneArt Seamless Cloning and Assembly Enzyme Mix to generate YCpG-TEF2pt. To construct the TEF2-Cas9 plasmid, Cas9 was amplified by using the primer pair Cas9-F plus Cas9-R with the CAS900A-1 plasmid (System Biosciences) as the template. The YCpG-TEF2pt vector was digested with StuI, and this linearized vector then was combined with the amplified Cas9 gene by using the GeneArt Seamless Cloning and Assembly Enzyme Mix.
To construct YCpG-YRP196W-2, the YRP196W-2 gene was amplified by PCR with the primer pair YRP196W-2-F + YRP196W-2-R using K1801 genomic DNA of as the template. YCpG-TDH3pt was digested with NotI, and then the linearized plasmid and the amplified YRP196W-2 gene were combined by using the GeneArt Seamless Cloning and Assembly Enzyme Mix. To construct YCpG-YRP196W-2L, the YRP196W-2L gene was amplified by PCR with the primer pair YRP196W-2-F plus YRP196W-2-R using K1801M genomic DNA as the template. YCpG-TDH3pt was digested with NotI, and the linearized plasmid and the amplified YRP196W-2L gene were combined using the GeneArt Seamless Cloning and Assembly Enzyme Mix. The identity of the YPR196W-2L clone was confirmed by sequencing. To construct TDH3p-YPR196W-2, the YPR196W-2 ORF was amplified by PCR with the primer pair TDH3-YRP196W-2-F plus TDH3-YRP196W-2-R using K1801 genomic DNA as the template. YCpG-TDH3pt was digested with StuI, and the linearized plasmid and the amplified YRP196W-2 gene were combined using GeneArt Seamless Cloning and Assembly Enzyme Mix, placing the ORF under control of the TDH3 promoter.
To construct MAL11p-lacZ, pYC-Z130 [19] was digested with EcoRI and HindIII, and the MAL11 promoter was amplified by PCR with the primer pair pYC-Z130-F + pYC-Z130-R using X2180-1A genomic DNA as the template. The linearized plasmid and the amplified promoter were combined using the GeneArt Seamless Cloning and Assembly Enzyme Mix.
To construct YPR196W-2p-lacZ, pYC-Z130 was digested with EcoRI and HindIII, and the YPR196W-2 promoter was amplified by PCR with the primer pair YPR196W-2-pro-F + YPR196W-2-pro-R using K1801 genomic DNA as the template. The linearized plasmid and the amplified promoter were combined using the GeneArt Seamless Cloning and Assembly Enzyme Mix. The G418 resistance-encoding marker was then replaced with the hygB selection marker from plasmid pYC240 [19] by cloning via AscI restriction and ligation.
Phylogenetic analysis
An unrooted tree was constructed using Seaview version 4.4.2 software based on the ClustalW multiple alignment of DNA sequences, using neighbor-joining method with 1000 bootstrap counts [21, 22]. The accession numbers of the genes were as follows: S. cerevisiae MAL23, AF002704.1; S. cerevisiae MAL43, M81157.1; S. cerevisiae MAL63, AF003003.1; S. cerevisiae MAL11, CP020129.1; S. cerevisiae MAL33, CP020124.1; S. cerevisiae YPR196W, CP020138.1; S. carlsbergensis MAL63, M36537.1
Ethyl methanesulfonate (EMS) treatment
Mutagenesis was performed using EMS as described in Methods in Yeast Genetics (Cold Spring Harbor Laboratory Course manual [23]. Briefly, exponentially growing K1801 was collected and then suspended in 5% EMS in Na-potassium buffer (pH 5.5). After 10 min, cells were collected, washed once with sterile water, and plated on an MA plate, which was incubated at 30°C. After 2–3 days, colonies were picked. The survival rate during mutagenesis was in the range of 50–55%.
Reporter assay
Cells carrying a lacZ reporter plasmid were cultured in YPD medium containing 200 μg/mL G418. After the culture achieved exponential growth, cells were harvested and resuspended in YPD or YPM medium containing 200 μg/mL G418 and culturing was continued under the same conditions, with sampling at defined time points. β-galactosidase activity was measured using the yeast β-Galactosidase Assay Kit (Thermo Fisher Scientific).
Test fermentation in SMal medium
Cells were cultured overnight in SD medium, and then cells were pelleted and resuspended in SMal medium at 1 × 107 cells/mL and fermented at 20°C. Sugar content was measured using a refractometer and expressed in Brix unit at the indicated time point.
Real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted as described by Schmitt et al. [24]. cDNA synthesis was performed using Primescript Reverse Transcriptase (Takara), RT-PCR was performed using GoTaq qPCR Master Mix (Promega) on StepOnePlus Real-Time PCR system (Applied Biosystems).The RT-PCR primers used in this study are shown in Table 1.
Small-scale sake test fermentation
Small-scale sake brewing tests were performed with a sake mash consisting of 72.8 g of α-preprocessed rice (Tokushima Seikiku, Tokushima, Japan) derived from a single-step pre-fermentation consisting of 80 g of white rice with a polishing rate at 70% (retaining the inner 70% of the grain), 19.2 g of dried rice koji (a culture of A. oryzae on steamed rice) (Tokushima Seikiku) derived from 20 g of white rice with polishing rate at 70%, 0.1 mL lactic acid (Wako), and 174 mL of water [25]. Each yeast strain was subjected to a two-step pre-cultivation consisting of incubation (at 30°C with rotary shaking for both steps), first in 3 mL of YPD medium for one day and then in 250 mL of YPD medium for a second day. The resulting yeast mycelia were washed twice with sterile water and inoculated into the mash to a density of 1.0 × 107 cells per g of mash; the mixture was further incubated at 15°C for 18 days (from Day 1 to Day 19) without shaking, as described previously [25]. Progression of fermentation was monitored by measuring the culture’s weight at the same time each day; weight loss reflects CO2 emission from mash [26]. For serial sampling of mash at separate time points, multiple mash cultures with the same content were prepared and wholly collected at Day 5, 8, 11, 15, or 19. For sampling at earlier time points, multiple 1/10-scale mash samples were prepared and wholly collected at 0, 12, 24, or 48 h. At the designated time point, each sample was centrifuged at 5000 rpm (4620 g) at 15°C for 15 min. The resulting supernatants were subjected to the analyses described below. For sugar analyses, trichloroacetic acid (TCA) was added to each supernatant, immediately upon decanting, to yield a final concentration of 20% TCA in order to inhibit amylase activity.
Sake metabolic profile
The concentrations of ethanol (and intermediates) during sake fermentation were measured by gas chromatography using a model GC 6890N machine (Agilent Technologies, CA, USA) equipped with a flame ionization detector (FID) and Agilent J&W DB-624 column (45 m × 0.543 mm; film thickness, 3 μm) [25]. Specific gravity (15/4°C), which reflects the sum of the alcohol and extractive content of sake, was measured with a density/specific gravity meter (DA-500; KEM, Kyoto, Japan); readings then were converted into sake meter values using the formula: Sake meter value = 1443/S-1443, where S = Specific gravity (15/4°C). In general, larger values on this scale indicate that fermentation has progressed further.
Acidity was determined by neutralization titration of 10 mL of sake with 0.1 N NaOH. Amino acidity was measured by the formol titration method [27]. The organic acid content was determined by high-performance liquid chromatography (HPLC) using a model LC-10AD (Shimadzu) machine equipped with a conductivity detector (Model CDD-10A; Shimadzu) and a Shim-pack SPR-H column (250 mm × 7.8 mm; Shimadzu). Aliquots (10 μL each) of samples were applied to the column using an automatic sampler SIL-10AD (Shimadzu) and eluted with 4 mM p-toluenesulfonic acid at a flow rate of 0.7 mL/min. The column oven temperature was 40°C. Volatile aromatic compounds were measured by headspace gas chromatography [28] using an Agilent 7694 Headspace Sampler and the GC 6890N (Agilent). Internal standards (consisting of n-amyl alcohol and methyl caprate) were added to aliquots of the samples prior to heating at 50°C for 30 min. Esters and higher alcohols then were separated using a DB-WAX capillary column (0.32 mm i.d. × 30 m, film thickness 0.25 μm; Agilent) after auto-injection of a headspace volume of 1 mL. The following conditions were applied: injection temperature, 200°C; oven temperature, 85°C; detector temperature, 250°C; and carrier gas (He), 2.2 mL/min.
Sugar content in the supernatants was determined by post-column labeling using phenylhydrazine with HPLC [29]. HPLC was performed with a Hitachi HPLC LaChrom Elite sugar analysis system (Hitachi High Technologies, Inc.), equipped with an auto sampler (L-2200), column oven (L-2300), pump (L-2130), FL detector (L-2485) and a column series consisting of a Shodex Asahipak NH2P-50 4E.
Results
The increased maltose fermentation ability of K1801M results from a one-base insertion in YPR196W-2
K7 is a well-known sake yeast, and the K7 genome sequence is available on a public website (http://www.bio.nite.go.jp/dogan/project/view/SC1) [16]. Notably, there are several K7-group strains that share a similar genetic background [30]. Analysis by our laboratory revealed that one of these strains, K1801, exhibits weak but measurable maltose fermentation ability, in contrast to the lack of maltose fermentation detected in other K7-group strains (S1 Fig). K1801 was derived as follows: first, K1601 was obtained from a cross between a K1001 segregant and a K7 segregant. Next, K1801 was obtained from a cross between a K1601 segregant and a K9 segregant (S2 Fig) [30]. To investigate the molecular basis of maltose fermentation by sake yeast in the present study, we chose to further characterize strain K1801, based on the fact that this strain shared the K7 genetic background but exhibited a detectable (albeit weak) background level of maltose fermentation (S1, S2 Figs).
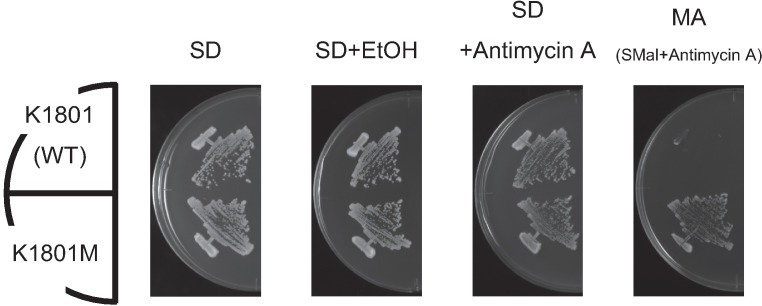
To generate mutants showing high maltose fermentation, K1801 was treated with ethane methane sulfonate (EMS) and spread on MA plates (synthetic maltose medium containing 2 mg/L antimycin A). Antimycin A is known to inhibit the mitochondrial electron transfer system, permitting more precise assessment of the maltose fermentation ability of the resulting isolates [12]. Several mutants showing high maltose fermentation ability were obtained using this screening. One such mutant exhibited significant growth on the MA plate, in contrast to the parental K1801 (Fig 1, S3 Fig). This isolate, which we have designated K1801M, is the subject of further investigation in this study. When growth on SMal solid medium was monitored in parallel to that on MA solid medium (S3 Fig), K1801M exhibited obvious growth on both SMal and MA plates, whereas K1801 exhibited weak growth on the SMal plate and little growth on the MA plate, even at 6 days. Considering that maltose is taken up via a maltose-one-proton symport mechanism [31], and that ATP hydrolysis by the plasma membrane ATPase is required to expel the protons that enter the cell together with maltose [32, 33], the observed growth difference suggested that K1801 can respire maltose weakly, but an extra ATP, derived from the mitochondrial electron transfer system, is required for efficient maltose transport. To exclude the mitochondrial effect in this study, MA plates were used to evaluate maltose fermentation ability in subsequent experiments.
Fig 1. K1801M showed improved growth on maltose medium.
Growth of K1801 and K1801M on various maltose-containing media. Each strain was streaked on SD, SD+EtOH (vehicle control), SD+Antimycin A, and MA (SMal+antimycin A) plates. Each plate was incubated at 30°C for 2–3 days.
In the K7 genome sequence database, the genes MAL11, MAL13, MAL31, MAL32, MAL33, YPR196W and YPR196W-2 are annotated as members of the MAL gene family. Comparison of these gene sequences to those of the respective S288C homologs revealed that the K7 MAL33 gene, which encodes a MALR, harbors a single nucleotide deletion (S4 Fig); no such deletion was observed in the paralogous K7 MAL13 and YPR196W genes. Therefore, it is unlikely that MAL13 or YPR196W confer maltose fermentation ability to K1801. In addition, MAL33 gene sequence in K1801 harbors the nucleotide deletion identical to that in K7. These data collectively suggest that MAL33, but not MAL13 nor YPR196W, is functionally associated with the increased maltose fermentation activity in K1801M strain (Fig 1, S3 Fig). In order to compare MALR activity in K1801 with that in K1801M, we introduced into both strains a MAL11p-lacZ reporter plasmid, in which the MAL11 promoter was fused with the lacZ gene. This construct permitted the monitoring of MALR activities, given that expression of MAL11-lacZ depends on trans-activation by MALR proteins [34]. Notably, while MAL11p-lacZ reporter activity was repressed in medium containing glucose, expression of this reporter was induced (in K1801M only) in medium containing maltose (Fig 2). These results indicated that K1801M has a maltose-inducible MALR for MAL11 compared to K1801. However, a candidate MALR, MAL33, in K1801M strain had the same mutation (relative to S288C) as that in the MAL33 gene from K7 and K1801 strains, suggesting that a MALR other than the MAL33 protein mediates the elevated maltose fermentation ability observed in K1801M.
Fig 2. K1801M showed enhanced expression of MAL11p-lacZ reporter gene.
Cells carrying MAL11p-lacZ reporter plasmid were harvested after overnight cultivation in YPD medium, then transferred to YPD or YPM medium, and further cultured at 30°C for 2 h. β-galactosidase activity was measured using yeast β-galactosidase assay kit. X2180-1A, which does not have a functional MALR [5], was used as a negative control.
To clarify the increased trans-activation of MAL11 observed in K1801M, we explored genetic alterations in other MALR-encoding genes in K1801M relative to the K1801 parent. As noted below, we ultimately focused on YPR196W-2. This gene is annotated in the K7 genome database, but not in the Saccharomyces cerevisiae Genome Database (SGD), and was designated YPR196W-2 because the K7 gene shows highest sequence similarity to YPR196W (MALR) among genes in the S288C genome. The Ypr196W-2p sequence was analyzed by BLAST search, and found to be structurally similar to several MALR proteins (including Mal23p, Mal43p, and Mal63p). Gibson et al. [3] reported that MALR proteins like Mal23p, Mal43p, and Mal63p have high trans-activation ability. The MALR genes in the S288C strain and the K7 strain are summarized in Table 2.
Table 2. Chromosomally encoded MALR genes in each yeast.
| S288C strain | MAL13 (Chr7), MAL33 (Chr2), YPR196W (Chr16) |
| K7 strain | MAL13 (Chr7), MAL33 (Chr7), YPR196W (Chr16), YPR196W-2 (Chr7) |
MAL23, MAL43, and MAL63 each encode a polypeptide consisting of 470 amino acids; in contrast, the YPR196W-2 ORF is only 316 amino acids long. YPR196W-2 was the closest to MAL63 (Fig 3A) and multiple alignments of YPR196W-2 and MAL63 revealed that the K7 YPR196W-2 has (compared to MAL63) a single nucleotide deletion at a position corresponding to nucleotide position 917 of the MAL63 ORF (Fig 3B). This deletion results in a frameshift compared to MAL63, such that the YPR196W-2 encodes a MALR protein that is C-terminally truncated relative to Mal63p. The sequence of YPR196W-2 in K1801 was identical to that in the K7 strain. However, some of the YPR196W-2 sequences amplified from K1801M harbored a single nucleotide insertion in the ORF compared to the K1801 sequence. Specifically, for three out of ten clones obtained by PCR from K1801, an adenine insertion was observed at nucleotide 914 of the YPR196W-2 ORF, which therefore encoded a mutant Ypr196W-2p protein 470 amino acids long, a size that is comparable to those of Mal23p, Mal43p, and Mal63p (Fig 3C). The remaining seven clones possessed YPR196W-2 sequences identical to that of K1801. The observed ratio of mutant alleles (3/10) was consistent with the previous proposal that the K1801 strain is a triploid [35]. Indeed, we were able to confirm, using flow cytometry, that both K1801 and K1801M are triploids. These results indicated that a single nucleotide insertion in YPR196W-2 occurred on one of three copies of Chromosome 7 in K1801M. Hereafter, we refer to the mutated allele of YPR196W-2 in K1801M as YPR196W-2L (Long). We also detected other single nucleotide insertions in proximity to ORF nucleotide position 917 in other maltose-assimilative mutants (referred to as K1801M’ and K1801M” in S5 Fig). These results support the notion that extension of the YPR196W-2L ORF by a single nucleotide insertion is associated with enhanced maltose fermentation.
Fig 3. Phylogenetic and sequence analysis of YRP196W-2 in K1801 and K1801M.
a An unrooted phylogenetic tree of selected MALR genes was constructed using the Neighbor-Joining Method (1000 replicates) in the CLUSTALW multiple alignment program [21, 22]. Bar = 0.01 nucleotide substitutions per site. b Sequence comparison of MAL63 [3] and YRP196W-2. c DNA (upper line) and protein (bottom line) sequences of YRP196W-2 and YRP196W-2L. The asterisk indicates the nucleotide insertion detected in K1801M.
Different functionalities of YRP196W-2 and YRP196W-2L
The gene expression of YPR196W-2 and YPR196W-2L was analyzed by real-time polymerase chain reaction (RT-PCR) using total RNA extracted from cells cultured for 6 h in YPD (containing glucose as the sole carbon source) or YPM (containing maltose as the sole carbon source) medium. The level of YPR196W-2 mRNA in K1801 did not differ between YPD and YPM, whereas the level of YPR196W-2L mRNA in K1801M was elevated in YPM compared to that in YPD (Fig 4A). The expression of other MALRs, MAL13, MAL33, YPR196W, in K1801M were same as K1801 (Fig 4A).
Fig 4. Expression of YRP196W-2 /-2L and MAL11 target gene in K1801M.
a Gene expression in K1801 and K1801M. Cells were harvested after overnight cultivation in YPD medium, then transferred to YPD (indicated as Glc) or YPM (indicated as Mal) and cultured at 30°C. After 6 h, total RNA was extracted and RT-PCR was performed using the indicated primers. The mRNA transcript level of each target gene was standardized by normalizing the value to that obtained for the control gene TDH3, and then presented as the value relative to that observed for K1801 grown on YPD. Data are presented as mean and standard deviation from three independent experiments. b Cells harboring a YPR196W-2p-lacZ reporter in a K1801 background were harvested after overnight cultivation in YPD medium, then transferred to YPD or YPM and cultured at 30°C for 6 h. β-galactosidase activity was measured using yeast β-galactosidase assay kit. c Restoration of maltose fermentation ability by YPR196W-2L. Cells carrying the indicated plasmid were streaked on SD, SD+EtOH (vehicle control), SD+Antimycin A, and MA (SMal+antimycin A) plates. Each plate was incubated at 30°C for 2–3 days.
Since the expression of a major maltose transporter-encoding gene, MAL11, is known to be induced in a MALR-dependent manner [13], we used MAL11 as a reporter gene for MALR activity. Notably, MAL11 expression was significantly higher in K1801M than in K1801 (Figs 2 and 4A). This result indicated that YPR196W-2L induces the expression of MAL11, whose promoter has MAL-activator binding sites [13]. Furthermore, YPR196W-2 itself was also induced by maltose (Fig 4A), a result consistent with the observation that the YPR196W-2 promoter also contains MAL-activator binding sites (S6 Fig; MGCNNNNNNNNNMGS, where M = adenine or cytosine, S = guanine or cytosine, and N = any of the four deoxyribonucleotides [36, 37]). In the YPR196W-2 promoter, two overlapping MAL-activator binding sites were observed at nucleotide positions (relative to the translation start) -178 to -164 and -174 to -160 (S6 Fig, where the two sites are designated MAL BD1 and MAL BD2, respectively). The presence of these MAL-activator binding sites in the YPR196W-2 promoter suggests that YPR196W-2 is itself positively regulated by a MALR. Since the promoter of YPR196W-2/-2L was found to contain putative MAL-binding cis-elements, trans-activation of YPR196W-2/-2L expression by the encoded protein was tested by lacZ reporter assay in K1801. A chimeric reporter gene, in which the lacZ gene was fused with the native promoter of YPR196W-2/-2L (representing 600 bp of sequence extending upstream from the translation start site), was introduced into the yeast along with a plasmid expressing YPR196W-2 or YPR196W-2L under the regulation of the native promoter. In YPD medium, reporter activity in yeast harboring YPR196W-2 or YPR196W-2L was statistically indistinguishable from that obtained in a strain harboring the reporter construct along with empty vector (Fig 4B). Notably, reporter activity was significantly increased in maltose-grown yeast harboring the reporter in combination with YPR196W-2L, but not in maltose-grown yeast harboring the reporter in combination with YPR196W-2.
To exclude the possibility that the maltose fermentation activity of K1801M reflects mutations in genes other than YPR196W-2, YPR196W-2 and YPR196W-2L were separately introduced into the parent K1801 strain, and these transformants were evaluated for growth on MA plates. As expected, the yeast expressing YPR196W-2L showed enhanced maltose fermentation ability, unlike yeast expressing YPR196W-2 (Fig 4C). The improved growth of yeast expressing YPR196W-2L on MA plate mirrored the previous observation that MAL11 was induced to higher levels in K1801M than in K1801 (Fig 2). Together, these results indicated that the basal level of MAL11 expression in K1801 is not sufficient to support growth on the MA plate; in contrast, the mutant YPR196W-2L gene is sufficient for growth on this medium.
These results showed that (1) Ypr196W-2Lp is able to exert autoregulatory trans-activation at the YPR196W-2L promoter, (2) the C-terminal domain of this MALR is required for trans-activation of MAL11, and (3) the gene expression of YPR196W-2L is responsive to maltose.
YPR196W-2 alleles are associated with maltose fermentation ability
As shown in Fig 4C, the introduction of YPR196W-2L was sufficient to confer maltose fermentation ability in the sake yeast K1801. To clarify the contribution to maltose fermentation ability of the endogenous YPR196W-2 alleles, the wild-type genes were deleted in K1801M. As it was technically difficult to selectively disrupt only the genes corresponding to the YPR196W-2 allele (given that YPR196W-2 and YPR196W-2L differ by a single nucleotide polymorphism), these allelic MALR genes were disrupted simultaneously using a CRISPR/Cas9 gene editing system in the triploid K1801M strain. Like K1801, the resulting K1801M ypr196W-2Δ strain (null) showed no maltose fermentation ability on an MA plate (Fig 5A). This result demonstrated that a single nucleotide insertion at YPR196W-2 permits YPR196W-2L to function as a dominant positive regulator for maltose fermentation in K1801M.
Fig 5. The maltose fermentation ability of ypr196W-2Δ sake yeast.
a Growth of K1801 and its derivatives on maltose-containing media. Each strain was streaked on SD, SD+EtOH (vehicle control), SD+Antimycin A, and MA (SMal+antimycin A) plates. Each plate was incubated at 30°C for 2–3 days. b Maltose fermentation test of ypr196W-2Δ strain. Cells were harvested after overnight cultivation in SD medium, then resuspended in SMal medium at 1 × 107 cells/mL and cultured at 20°C. Sugar content in the medium was measured at the indicated times.
Gibson et al. [3] previously demonstrated that a C-terminally truncated Mal63p, 283 amino acids in length, still possessed the ability to induce the maltase-encoding gene, although the level of induction with this shortened protein was lower than that observed with the full length (470-amino acid) Mal63p, indicating that a C-terminally truncated MALR may maintain partial function. Similarly, wild sake yeasts are known to exhibit weak maltose fermentation activity (S1 Fig), suggesting that the C-terminally truncated (316-amino acid) Ypr196W-2p also retains partial function in sake yeast. To test whether the weak maltose fermentation observed in sake yeast depends on the truncated MALR Ypr196W-2p, the sugar consumption rate of yeast cells under maltose fermentation conditions was determined by measuring sugar content using a refractometer. In control experiments, K7 and K1801 strains showed slow maltose consumption (Fig 5B). In contrast, the maltose consumption rate of K1801M was faster than those of the control strains. When YPR196W-2 was disrupted in K1801 or K1801M, the resulting mutants consumed maltose at rates lower than those of the control strains. Similar data were obtained in the K7 genetic background (Fig 5B). These results showed that YPR196W-2 is responsible for the weak maltose fermentation ability of this sake yeast under maltose fermentation conditions. As shown in Fig 5, the K1801M ypr196w-2Δ strain failed to grow on an MA plate, but was viable in maltose-supplemented fermentation conditions, a difference that presumably reflects the decreased availability of the mitochondrial electron transfer system in the presence of antimycin A. This result also is consistent with the case in strain K1801 (Fig 1, S3 Fig). These results indicated that K1801M ypr196w-2Δ still had a weak maltose fermentation ability that is dependent upon the mitochondrial electron transfer system, suggesting that another functional MALR (other than YPR196W-2) that is present in K1801 provides this very weak MALR activity.
To assess the relationship between the levels of the MALR proteins and maltose fermentation, maltose consumption rates were determined in yeasts expressing distinct alleles of this MALR gene under the control of promoters of various strengths. The maltose consumption rate of the cells harboring a construct (TDH3p-YPR196W-2) driving constitutive high-level expression of the truncated protein was lower than that obtained with YPR196W-2L under control of the endogenous YPR196W-2 promoter (YPR196W-2p-YPR196W-2L), but the TDH3p-YPR196W-2 maltose consumption rate was still higher than that obtained with YPR196W-2 under control of the endogenous YPR196W-2 promoter (YPR196W-2p-YPR196W-2) (Fig 6). Taken together, these results indicated the following: (1) exogenous expression of YPR196W-2L is sufficient to confer maltose fermentation ability to K1801 in SMal medium, and (2) the restored C-terminal domain in Ypr196W-2Lp is crucial for full function as a MALR, such that overexpression of the truncated protein is not sufficient to compensate for the attenuated activity of Ypr196W-2p.
Fig 6. Maltose fermentation ability of cells carrying TDH3p-driven YRP196W-2.
Maltose fermentation test of cells carrying TDH3p-YPR196W-2. Cells carrying each plasmid were harvested after overnight cultivation in SD medium, then transferred into SMal medium at 1 × 107 cells/mL and cultured at 20°C. Sugar content in the medium was measured at the indicated times.
K7 sake yeast family members are presumed to share the preponderance of their genetic background with K1801; however, maltose fermentation ability was found to differ among various members of the K7 sake yeast family (S1 Fig). K7-family sake yeast, which share the YPR196W-2 allele shown in Fig 3B, failed to grow on an MA plate, presumably due to the lack of maltose-inducible MALRs. Notably, exogenous introduction of YPR196W-2L into various K7-family strains was sufficient to confer maltose fermentation ability to these sake yeast strains (Fig 7).
Fig 7. Growth in the presence of maltose of K7 family strains carrying YRP196W-2L.
Each strain was streaked on SD, SD+EtOH (vehicle control), SD+Antimycin A, and MA (SMal+antimycin A) plates, and plates were incubated at 30°C for 2–3 days.
Sake fermentation is notable for the incorporation of parallel catalytic pathways, including simultaneous Aspergillus-mediated saccharification and yeast-mediated fermentation. To assess the practical impact on sake fermentation of YPR196W-2L, small scale sake fermentation (total rice amount: 100g) was performed with sake yeast (K1801 strain) transformed with YPR196W-2L or with control empty vector. Based on CO2 emission, yeast expressing YPR196W-2L showed slightly faster fermentation than that harboring control vector (S7 Fig). A large amount of maltose was generated during Days 2–4 of fermentation, presumably due to saccharification of rice starch (Fig 8A lower panel). The amount of maltose in the sake mash was lower in the YPR196W-2L-expressing yeast than in the control yeast, which was consistent with the enhancement of maltose fermentation activity by the YPR196W-2L allele. Moreover, the amount of glucose also was lower with the YPR196W-2L-expressing yeast (compared to the control strain). This result presumably reflects the decreased availability of maltose (for degradation into glucose by A. oryzae) in the sake mash (Fig 8A upper panel). Concomitantly, MAL11 was highly expressed at Day 3 in the YPR196W-2L-expressing strain than in the control yeast (Fig 8B). However, final alcohol production by yeast expressing YPR196W-2L was similar to that of the control (S1 Table). It should be noted that maltose consumption temporally followed glucose consumption, indicating that the glucose repression system was active during sake fermentation. Chemical analysis of organic compounds produced during fermentation showed that there were no remarkable changes in the levels of those compounds when comparing between the strains harboring YPR196W-2L or empty vector (S1 Table). Therefore, sake yeast carrying an ectopic copy of YPR196W-2L is expected to exhibit moderately accelerated fermentation without apparent changes in sake quality. These data showed that YPR196W-2L plays a potential role in maltose fermentation in general.
Fig 8. Cells expressing YPR196W-2L exhibit rapid maltose incorporation compared to control in a small-scale sake test fermentation.
a Sake mash that contained yeast cells with control vector or YPR196W-2L were prepared triplicate as described in the Materials and Methods section. Glucose and maltose content in the test fermentation. Mash was sampled at the indicated time point, and the supernatant was collected by centrifugation and assessed for sugar content. b At Day 3, total RNA was extracted and RT-PCR was performed using the indicated primers. The mRNA transcript level of each target gene was standardized by normalizing the value to that obtained for the control gene TDH3, and then presented as the value relative to that observed for control (empty vector) strain. Data are presented as mean and standard deviation from three independent experiments.
Discussion
In this study, we isolated a mutant sake yeast strain that showed enhanced maltose fermentation ability (Fig 9). A single nucleotide insertion in a MALR gene, YPR196W-2, was responsible for the elevated level of maltose fermentation; introduction of the mutant version of YPR196W-2, designated YPR196W-2L, was sufficient to confer enhanced maltose fermentation to the original sake yeast strain K1801. Conversely, deletion of YPR196W-2 in K1801 lowered the strain’s maltose fermentation ability below that of K1801. YPR196W-2 is structurally similar to MAL63, but (compared to other MALR proteins) lacks a thymine at nucleotide position 917 of the ORF. This single nucleotide deletion presumably occurred in the ancestral strain during differentiation and selection for sake fermentation; the resulting truncated form of Ypr196W-2p is apparently hypomorphic, resulting in decreased maltose fermentation ability. Together with the additional experiments (Figs 4–8), our data demonstrated that YPR196W-2 is the major cause of the attenuated maltose fermentation ability of sake yeast. We propose redesignating YPR196W-2 as MAL73 to indicate its functional association with maltose fermentation.
Fig 9. Summary of this work.
An ancestral brewing yeast is presumed to have harbored a functional Mal63p ortholog. During the evolution or human selection of this ancestral strain, MAL63 experienced the deletion of a residue at nucleotide position 917 of the ORF. The resulting gene, MAL73, encodes a truncated MALR protein with attenuated trans-activation activity. As shown in this study, K1801M, isolated from K1801 on the basis of enhanced maltose fermentation ability, harbors a MAL73 gene in which a single nucleotide was inserted at ORF residue 914, resulting in a frameshifted extension of the ORF. The resulting MAL73L gene encodes a functionally restored MALR with improved trans-activation activity compared to the parental Mal73p.
Brown et al. [38] showed that MAL genes are located in subtelomeric regions and implied that different yeast strains harbor different alleles and copy numbers of the various MAL genes. MALR genes are classified into three clades, including a MAL13-like clade, a YPR196W clade, and a MAL63-like clade. MAL73, the gene characterized in this study, is classified into the MAL63-like clade based on phylogenetic analysis (Fig 3A). Moreover, the Verstrepen group also showed that a difference in DNA-binding domains between Malx3p and Yfl052Wp, notably that at amino acid position 12 (Arg in Malx3p, Cys in Yfl052Wp), was responsible for the distinct DNA-binding specificity of these MALRs [39]. Like Malx3p, Mal73p encodes an Arg residue at position 12, suggesting that MAL73 is evolutionarily derived from a MALx3-type MALR with structural similarity to MAL63, and further indicating that the ancestral sake yeast exhibited a sugar preference for maltose.
Mal73p lacks C-terminal activation domain due to a single nucleotide deletion
The MALR protein has a DNA binding domain at amino acid residues 1–100, a trans-activation domain at residues 60–283, and a maltose responsive domain at the C-terminus [4]. Gibson et al. [3], Hu et al. [4], and Danzi et al. [40] previously reported that mutants that are not inducible by maltose frequently are the result of missense mutations or short deletions within the C-terminal regulatory domain. Moreover, the C-terminal domain is presumed to take part in the formation and/or maintenance of an active state of MALR [41]. The insertion of a single nucleotide insertion in MAL73 resulted in a frameshift that endowed Mal73Lp with an elongated C-terminal domain compared to Mal73p, such that Mal73Lp exhibited enhanced regulatory activity. Our results showed that MAL11 is specifically induced in maltose-grown K1801M, which encodes the elongated Mal73Lp, but not in the maltose-grown parent K1801, which encodes a shorter (relative to Mal63p) protein. This observation is consistent with a previous report that a truncated Mal63p, consisting of 283 amino acids, shows weaker induction of a maltase-encoding gene than the full-length (470-amino acid) Mal63p [3]. Furthermore, MAL73L showed higher self-trans-activation via its own promoter than did MAL73 (Fig 4B). In contrast, overexpression of MAL73 by the TDH3 promoter slightly improved maltose fermentation in K1801 (Fig 6), again demonstrating the functional significance of the C-terminal domain of MALR in trans-activation (Fig 9).
Among the tested sake yeast strains, deletion of MAL73 decreased efficiency of maltose consumption compared to the respective parent (Fig 5B), showing that the weak maltose fermentation ability of sake yeast depends on MAL73. However, these sake yeast strains did not completely lose maltose fermentation ability, retaining a basal level of maltose fermentation higher than that of the laboratory strain X2180-1A. Therefore, we infer that there exist additional (beyond Mal73p) MALRs and/or MALR activation mechanisms in K1801.
Biological significance of YPR196W-2 allele
The genome of the ancestral yeast strain is inferred to have harbored five paralogous MAL loci. During subsequent descent, those MAL loci presumably evolved in response to changes in natural environments and nutrient supplies, and under human selection for specialized fermentations with various plant materials. Some descendants lost entire loci from their genomes, as was the case of MAL2/4/6 in maltose fermentable yeasts [10, 42]. In other strains, MAL genes were altered in expression or function, as was the case with MAL13 or MAL33 in S288C [5, 6]. In this study, we showed that sake yeast encodes a MALR (Mal73p) that is truncated and exhibits decreased regulatory function (compared to other known MALRs). Presumably, at some point in the past, an ancestral MALR gene suffered a single nucleotide deletion, resulting in a frameshift; this mutation introduced a premature stop codon, yielding a C-terminally truncated MALR protein. Sake fermentation characteristically employs continuous degradation of rice starch into glucose via the action of α-amylase and glucoamylase provided by the fungus A. oryzae. This process occurs in parallel with saccharification, whereby maltose is generated before immediately being further degraded into glucose monomer units (Fig 8A). Together, these processes permit alcoholic fermentation through catalysis by sake yeast. Thus, sake yeast are presumed to have adapted for the specialized situation wherein a large amount of glucose is provided as a carbon source. Liti et al. [43], Schacherer et al. [44] and Peter et al. [45] have described phylogenies of Saccharomyces cerevisiae in which sake yeasts cluster separately as a distinct clade. These results were presumed to reflect, in part, the unique evolution of sake yeast in Japanese archipelago as a consequence of adaptation and selection for sake fermentation. With respect to maltose fermentation, MAL73 gene might be a beneficial trait to retain full ability of maltose fermentation by insertion of a single nucleotide when that ability is required.
Introducing the MAL73L allele into sake yeast facilitated slightly improved maltose fermentation in small sake fermentation as well as in SMal medium. Based on temporal CO2 emission and differential sugar consumption (Fig 8A, S6 Fig), maltose fermentation follows glucose fermentation at Days 2–4 of sake fermentation, suggesting that MAL73L-mediated maltose fermentation initiates earlier than Day 4, presumably due to release from glucose repression. This notion is supported by the observation that MAL73L is inactive in the presence of high glucose concentrations (Fig 4). The slight enhancement of maltose fermentation by MAL73L seems to be practically beneficial for faster sake fermentation; ectopic expression of this allele did not appear to impose obvious metabolic alterations in the characteristic organic compound profile that we evaluated. Our findings indicate that the MAL73L gene may serve as a valuable genetic tool for improvement of maltose fermentation via the insertion of a single nucleotide in a MALR gene.
Notably, all of the sake yeasts surveyed in this work encoded Mal73p as a C-terminally truncated hypomorphic protein (compared to other MALR proteins), suggesting a founder effect derived from an ancestral sake yeast and/or a trace of historical human selection for the hypomorphic allele. Domestication of sake yeasts may have accompanied by genetic traits involving a specific organic compound profile in the fermented products as well as stable fermentation, since such a profile would have significantly altered the flavor and quality of sake during traditional-to-industrialized sake history. An obvious beneficial effect of the hypomorphic allele on sake quality is currently unknown, however future large-scale sake fermentations and subsequent sensory and chemical evaluations would clarify this issue.
Supporting information
Maltose assimilation ability of sake yeast under fermentation conditions. Cells were harvested after overnight cultivation in SD medium, transferred into SMal medium at 1 × 107 cells/mL, and further cultured at 20°C. Sugar content in the medium was measured at the indicated times.
(TIF)
K1801 was bred as follows: first, K1601 was bred by the mating of a K1001 segregant with a K7 segregant. Next, K1801 was bred by the mating of a K1601 segregant and a K9 segregant.
(TIF)
Each strain was streaked on SD, SD+EtOH (vehicle control), SD+Antimycin A, SMal, SMal+EtOH (vehicle control) and MA (SMal+antimycin A) plates. Each plate was incubated at 30°C, and growth was monitored daily over the course of 2–6 days.
(TIF)
The MAL33 (S288C) sequence was downloaded from the Saccharomyces genome database (http://www.yeastgenome.org/), and the MAL33 (K7) sequence was downloaded from the DOGAN–Saccharomyces cerevisiae K7 database (http://www.bio.nite.go.jp/dogan/project/view/SC1)
(TIF)
DNA (upper line) and amino acid (bottom line) sequences of YPR196W-2 homologs from other maltose-assimilative K1801-derived strains (K1801M’ and K1801M”) that were isolated through the mutagenesis screen of K1801 described in the Materials and Methods section. An asterisk is used to indicate the inserted or changed nucleotide in each strain.
(TIF)
MAL-activator binding sites are indicated as MAL BD1 and MAL BD2. The consensus sequence of the MAL-activator binding site is MGCNNNNNNNNNMGS, where M is adenine or cytosine, S is guanine or cytosine, and N is any of the four deoxyribonucleotides.
(TIF)
Sake mash containing yeast cells with control vector or YPR196W-2L plasmid were prepared in triplicate as described in the Materials and Methods section. Weight loss of the mash (reflecting CO2 gas emission) was measured at the same time each day. The upper panel shows total cumulative CO2 gas emission, and the bottom panel shows CO2 gas emission per day. Data are presented as the mean and standard deviation from three independent experiments.
(TIF)
(DOCX)
Acknowledgments
We thank to Dr. Jun Murata (SUNBOR) and Dr. Takashi Matsuyama (TOYOTA Central R&D Labs) for critical reading of the manuscript, and also to Hiromi Toyonaga (SIC), Yuri Yasue (SIC), Miyuki Ogawa (SIC), Yukihiko Narita (SIC), Nozomu Kamada (SIC), Dr. Yukiko Kodama (SIC), Dr. Masaki Okuda (NRIB), and Dr. Nami Goto (NRIB) for technical supports to this work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
TO, FO, HH and EO are employees of Suntory Global Innovation Center Ltd (SIC), a company affiliated to Suntory Holdings Ltd. SIC financially provides salaries of TO, FO, HH and EO and research materials of this study. The specific roles of these authors are articulated in the ‘author contributions’ section. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding received for this study.
References
- 1.Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2014; 38: 254–299. doi: 10.1111/1574-6976.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reece RJ, Ptashne M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science. 1993; 261: 909–911. [DOI] [PubMed] [Google Scholar]
- 3.Gibson AW, Wojciechowicz LA, Danzi SE, Zhang B, Kim JH, Hu Z, et al. Constitutive mutations of the Saccharomyces cerevisiae MAL-activator genes MAL23, MAL43, MAL63, and mal64. Genetics. 1997; 146: 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Z, Gibson AW, Kim JH, Wojciechowicz LA, Zhang B, Michels CA. Functional domain analysis of the Saccharomyces MAL-activator. Curr Genet. 1999; 36: 1–12. [DOI] [PubMed] [Google Scholar]
- 5.Charron MJ, Dubin RA, Michels CA. Structural and functional analysis of the MAL1 locus of Saccharomyces cerevisiae. Mol Cell Biol. 1986; 6: 3891–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow TH, Sollitti P, Marmur J. Structure of the multigene family of MAL loci in Saccharomyces. Mol Gen Genet. 1989; 217; 60–69. [DOI] [PubMed] [Google Scholar]
- 7.Goldenthal MJ, Vanoni M, Buchferer B, Marmur J. Regulation of MAL gene expression in yeast: gene dosage effects. Mol Gen Genet. 1987; 209: 508–517. [DOI] [PubMed] [Google Scholar]
- 8.Barnett JA. The utilization of sugars by yeasts. Adv Carbohydr Chem Biochem. 1976; 32: 125–234. [DOI] [PubMed] [Google Scholar]
- 9.Charron MJ, Michels CA. The naturally occurring alleles of MAL1 in Saccharomyces species evolved by various mutagenic processes including chromosomal rearrangement. Genetics. 1988; 120: 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charron MJ, Read E, Haut SR, Michels CA. Molecular evolution of the telomere-associated MAL loci of Saccharomyces. Genetics. 1989; 122: 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanoni M, Sollitti P, Goldenthal M, Marmur J. Structure and regulation of the multigene family controlling maltose fermentation in budding yeast. Prog Nucleic Acid Res Mol Biol. 1989; 37: 281–322. [DOI] [PubMed] [Google Scholar]
- 12.Özcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998; 17: 2566–2573. doi: 10.1093/emboj/17.9.2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidgren V, Kankainen M, Londesborough J, Ruohonen L. Identification of regulatory elements in the AGT1 promoter of ale and lager strains of brewer's yeast. Yeast. 2011; 28: 579–594. doi: 10.1002/yea.1888 [DOI] [PubMed] [Google Scholar]
- 14.Baba S, Oguri I, Fukuzawa M, Moriyama K, Iida T, Kobayashi I, et al. Maltose assimilation by Saccharomyces sake. Soc Brew Japan. 1974; 69: 453–455. [Google Scholar]
- 15.Asano T. Studies on Organic Acid Production by Sake Yeast. Seibutsu kogaku. 2007; 85: 63–68. [Google Scholar]
- 16.Akao T, Yashiro I, Hosoyama A, Kitagaki H, Horikawa H, Watanabe D, et al. Whole-genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai no. 7. DNA Res. 2011; 18: 423–434. doi: 10.1093/dnares/dsr029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 201; 337: 816–821. doi: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013; 41: 4336–4343. doi: 10.1093/nar/gkt135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen J, Felding T, Johannesen PF, Piskur J, Christensen CL, Olesen K. Further development of the cassette-based pYC plasmid system by incorporation of the dominant hph, nat and AUR1-C gene markers and the lacZ reporter system. FEMS Yeast Res. 2003; 4: 323–327 [DOI] [PubMed] [Google Scholar]
- 20.Hatanaka H, Omura F, Kodama Y, Ashikari T. Gly-46 and His-50 of yeast maltose transporter Mal21p are essential for its resistance against glucose-induced degradation. J Biol Chem. 2009; 284: 15448–15457. doi: 10.1074/jbc.M808151200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galtier N, Gou M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Compt Appl Biosci. 1996; 12: 543–548. [DOI] [PubMed] [Google Scholar]
- 23.Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics Cold Spring Harbor Laboratory Course manual; 2005 [Google Scholar]
- 24.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990; 18: 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katou T, Kitagaki H, Akao T, Shimoi H. Brewing characteristics of haploid strains isolated from sake yeast Kyokai No. 7. Yeast. 2008; 25: 799–807. doi: 10.1002/yea.1634 [DOI] [PubMed] [Google Scholar]
- 26.Watanabe D, Ota T, Nitta F, Akao T, Shimoi H. Automatic measurement of sake fermentation kinetics using a multi-channel gas monitor system. J Biosci Bioeng. 2011; 112: 54–57. doi: 10.1016/j.jbiosc.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 27.Sørensen SPL, Enzymestudien, Biochem Z. 1907; 7: 45–101. [Google Scholar]
- 28.Yoshizawa K. Head-space gas chromatography for rapid quantification of volatiles in sake. J Brew Soc Japan. 1973; 68: 59–61. [Google Scholar]
- 29.Suzuki H, Kato E, Matsuzaki A, Ishikawa M, Harada Y, Tanikawa K, et al. Analysis of saccharides possessing post-translational protein modifications by phenylhydrazine labeling using high-performance liquid chromatography. Anal Sci. 2009; 25: 1039–1042. [DOI] [PubMed] [Google Scholar]
- 30.Akao T. New Perspectives of Genetic Variability of Sake Yeast Strains in Microevolution and Breeding. Kagaku to Seibutsu. 2014; 52: 223–232. [Google Scholar]
- 31.Serrano R. Energy requirements for maltose transport in yeast. Eur. J. Biochem. 1977; 80:97–102. [DOI] [PubMed] [Google Scholar]
- 32.van Leeuwen CCM, Weusthuis RA, Postma E, van den Broek PCA, van Dijken JP. Maltose/proton co-transport in Saccharomyces cerevisiae. Biochem J. 1992; 284:441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weusthuis RA, Adams H, Scheffers WA, van Dijken JP. Energetics and kinetics of maltose transport in Saccharomyces cerevisiae: a continuous culture study. Appl Environ Microbiol. 1993; 59:3102–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han EK, Cotty F, Sottas C, Jiang H, Michels CA. Characterization of AGT1 encoding a general alpha-glucoside transporter from Saccharomyces. Mol Microbiol. 1995; 17: 1093–1107. [DOI] [PubMed] [Google Scholar]
- 35.Goshima T, Nakamura R, Kume K, Okada H, Ichikawa E, Tamura H, et al. Identification of a mutation causing a defective spindle assembly checkpoint in high ethyl caproate-producing sake yeast strain K1801. Biosci Biotechnol Biochem. 2016; 80: 1657–1662. doi: 10.1080/09168451.2016.1184963 [DOI] [PubMed] [Google Scholar]
- 36.Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, et al. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006; 34: 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sirenko OI, Ni B, Needleman RB. Purification and binding properties of the Mal63p activator of Saccharomyces cerevisiae. Curr Genet. 1995; 27: 509–516. [DOI] [PubMed] [Google Scholar]
- 38.Brown CA, Murray AW, Verstrepen KJ. Rapid Expansion and Functional Divergence of Subtelomeric Gene Families in Yeasts. Current Biol. 2010; 20: 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pougach K, Voet A, Kondrashov FA, Voordeckers K, Christiaens JF, Baying B, Benes V, Sakai R, Aerts J, Zhu B, Dijck PV, Verstrepen KJ. Duplication of a promiscuous transcription factor drives the emergence of a new regulatory network. Nature commun. 2014; 4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danzi SE, Zhang B, Michels CA. Alterations in the Saccharomyces MAL-activator cause constitutivity but can be suppressed by intragenic mutations. Curr Genet. 2000; 38: 233–240. [DOI] [PubMed] [Google Scholar]
- 41.Danzi SE, Bali M, Michels CA. Clustered-charge to alanine scanning mutagenesis of the Mal63 MAL-activator C-terminal regulatory domain. Curr Genet. 2003; 44: 173–183. doi: 10.1007/s00294-003-0429-9 [DOI] [PubMed] [Google Scholar]
- 42.Dubin RA, Charron MJ, Haut SR, Needleman RB, Michels CA. Constitutive expression of the maltose fermentative enzymes in Saccharomyces carlsbergensis is dependent upon the mutational activation of a nonessential homolog of MAL63. Mol Cell Biol. 1988; 8: 1027–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, et al. Population genomics of domestic and wild yeasts. Nature. 2009; 458: 337–341. doi: 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of S. cerevisiae. Nature. 2009; 458: 342–345. doi: 10.1038/nature07670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peter J, De Chiara M, Friedrich A, Yue JX, Pflieger D, Bergström A et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 2018; 556: 339–344. doi: 10.1038/s41586-018-0030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maltose assimilation ability of sake yeast under fermentation conditions. Cells were harvested after overnight cultivation in SD medium, transferred into SMal medium at 1 × 107 cells/mL, and further cultured at 20°C. Sugar content in the medium was measured at the indicated times.
(TIF)
K1801 was bred as follows: first, K1601 was bred by the mating of a K1001 segregant with a K7 segregant. Next, K1801 was bred by the mating of a K1601 segregant and a K9 segregant.
(TIF)
Each strain was streaked on SD, SD+EtOH (vehicle control), SD+Antimycin A, SMal, SMal+EtOH (vehicle control) and MA (SMal+antimycin A) plates. Each plate was incubated at 30°C, and growth was monitored daily over the course of 2–6 days.
(TIF)
The MAL33 (S288C) sequence was downloaded from the Saccharomyces genome database (http://www.yeastgenome.org/), and the MAL33 (K7) sequence was downloaded from the DOGAN–Saccharomyces cerevisiae K7 database (http://www.bio.nite.go.jp/dogan/project/view/SC1)
(TIF)
DNA (upper line) and amino acid (bottom line) sequences of YPR196W-2 homologs from other maltose-assimilative K1801-derived strains (K1801M’ and K1801M”) that were isolated through the mutagenesis screen of K1801 described in the Materials and Methods section. An asterisk is used to indicate the inserted or changed nucleotide in each strain.
(TIF)
MAL-activator binding sites are indicated as MAL BD1 and MAL BD2. The consensus sequence of the MAL-activator binding site is MGCNNNNNNNNNMGS, where M is adenine or cytosine, S is guanine or cytosine, and N is any of the four deoxyribonucleotides.
(TIF)
Sake mash containing yeast cells with control vector or YPR196W-2L plasmid were prepared in triplicate as described in the Materials and Methods section. Weight loss of the mash (reflecting CO2 gas emission) was measured at the same time each day. The upper panel shows total cumulative CO2 gas emission, and the bottom panel shows CO2 gas emission per day. Data are presented as the mean and standard deviation from three independent experiments.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.