Abstract
Fruit ripening is a complex biological process affecting fruit quality. In tomato the fruit ripening process is delicately regulated by transcription factors (TFs). Among these, the TOMATO AGAMOUS-LIKE 1 (TAGL1) gene plays an important role in both the development and ripening of fruit. In this study, the TAGL1 gene was successfully silenced by virus-induced gene silencing technology (VIGS), and the global gene expression and metabolites profiles of TAGL1-silenced fruits were analyzed by RNA-sequence analysis (RNA-seq) and liquid chromatography–mass spectrometry (LC-MS/MS). The TAGL1-silenced fruits phenotypically displayed an orange pericarp, which was in accordance with the results expected from the down-regulation of genes associated with carotenoid synthesis. Levels of several amino acids and organic acids were lower in the TAGL1-silenced fruits than in the wild-type fruits, whereas, α-tomatine content was greatly increased (more than 10-fold) in the TAGL1-silenced fruits compared to wild-type fruits. The findings of this study showed that TAGL1 not only regulates the ripening of tomato fruits, but also affects the synthesis and levels of nutrients in the fruit.
Introduction
Fruit ripening is a complex developmental process that involves the transformation of the seed-bearing structure of fleshy fruit species into a delicious and nutritive fruit, which appeals to animals and humans, who consume the fruit and act as the dispersers of its seeds [1]. Some general ripening-associated changes are characteristic among different species, including modifications in texture, changes in the sugar content, and alterations in the composition and levels of secondary metabolites such as pigments and flavor [2, 3]. These changes are associated with alterations in multiple biochemical pathways that are regulated by some critical TFs [4, 5].
Tomato (Solanum lycopersicum), one of the world’s most important horticultural crops and an important source of human nutrients, is recognized as an outstanding experimental model to study fleshy fruit development and ripening [3, 4]. The tomato fruit is a typical climacteric fruit with a peak of respiration and ethylene production at the start of ripening [6]. Ethylene is synthesized from S-adenosylmethionine (SAM) by the sequential action of two key ethylene biosynthetic enzymes, namely, 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) and ACC oxidase (ACO) [6,7]. Many transcription factors modulate ethylene synthesis and signal transduction during fruit development and ripening, such as those encoded by the genes RIPENING INHIBITOR (MADS-RIN) [8], COLORLESS NON-RIPENING (CNR) [9], and NON-RIPENING (NOR) [10], and mutations in these genes result in the un-ripened phenotype of the fruit][11]. The RIN protein has been shown to interact with the CArG-box elements in the promoters of genes encoding ACC synthase 2 (ACS2), ACC synthase 4 (ACS4), and cell wall hydrolases such as polygalacturonase (PG), β-galactosidase 4 (TBG4), endo-(1,4)- β- mannanase 4 (MAN4), and α-expansin 1 (EXP1), indicating that it controls ethylene production and fruit softening by directly regulating the transcription of related genes [12,13]. CNR was implicated in the positive regulation of many ripening-related genes, including polygalacturonase gene (PG), pectinesterase gene (PE), xyloglucan endotransglycosylase gene (XET), phytoene Synthase 1 (PSY1), lipoxygenase (LOX), and ACC oxidase 1 (ACO1) [14]. NOR was reported to be involved in the regulation of fruit ripening and quality, possibly by altering the expression of ACO, PSY1, PG2, and others [10]. Other known ripening-associated transcription factors include TOMATO AGAMOUS-LIKE1 (TAGL1) [15, 16, 17, 18], FRUITFULL (FUL1 and FUL2) [19, 20], and APETALA2a (AP2a) [21, 22]. The connections between this highly linked regulatory network and the downstream effectors regulating the color, texture, and flavor of the fruits still remain relatively poorly understood.
TAGL1 belongs to the AGAMOUS clade of the MADS-box genes in tomato. Previous studies have revealed that TAGL1-RNAi plants produced tomato fruits that were unable to ripen normally, had thin pericarp, and displayed a yellowish-orange color and greater firmness, features that are associated with reduced carotenoids and ethylene levels [15, 16]; over-expressing TAGL1 resulted swollen sepals and showed ectopic lycopene production and accumulation of the yellow flavonoid naringenin chalcone [16]. The TAGL1 protein is able to bind to the promoter region of ACS2, directly regulating the activity of ethylene biosynthesis [16]. Moreover, the pericarp cells of the TAGL1-RNAi fruits showed altered cellular and structural properties correlated with the decreased expression of genes regulating both cell division and lignin biosynthesis [23]. However, no study has assessed how TAGL1 transcription regulates fruit nutrition and flavor, which is the sum of the interactions between sugars, acids, and multiple volatile chemicals. Therefore, the present study was designed with the aim of providing an analysis of TAGL1 regulation during fruit ripening using RNA sequencing (RNA-seq) and LC-MS/MS.
Materials and methods
Plant material and growth conditions
Seeds of the tomato cultivar ‘Ailsa Craig’ (AC) were germinated in commercial tomato-cultivation soil. All tomato plants were grown in a greenhouse at 25°C with 75% relative humidity under 16 h light/8 h dark cycles. Flowers were tagged one day post-anthesis (DPA).
Preparation of vectors
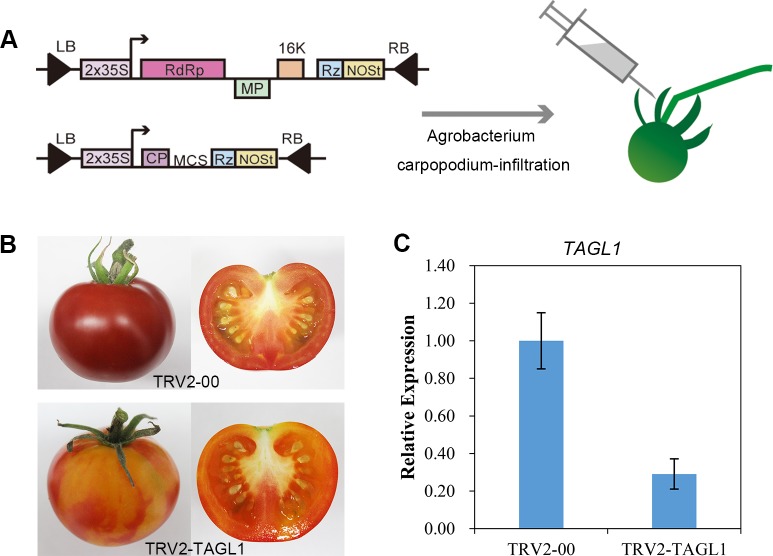
The tobacco rattle virus (TRV)-based vectors pTRV1 and pTRV2 were adopted for virus-induced gene silencing (VIGS). To construct a pTRV2-TAGL1 recombinant, a 420 bp EcoR I/BamH I digested DNA fragment of the TAGL1 gene corresponding to bases 481–900 of the TAGL1 gene sequence (NM_001313930.1) were PCR-amplified from tomato cDNA and inserted into the pTRV2 vector (Fig 1A). The VIGS primers are listed in S1 Table.
Fig 1. VIGS technique applied to the tomato fruits.
(A) Diagram of the VIGS technique used for infecting the tomato fruits. (B) Phenotype of the TAGL1-silenced fruits. Fruit infiltrated with the vector (TRV2-00) was used as control. (C) The silencing efficiency of the TAGL1 gene at the red-ripe stage (RR) using quantitative real time PCR (qRT-PCR).
Virus-induced gene silencing (VIGS) assay
The VIGS assay was carried out as per the previously described protocol of Fu et al [24] with slight modifications. The Agrobacterium GV3101 strain containing pTRV1, pTRV2, or pTRV2-TAGL1 vectors were grown at 28°C in Luria-Bertani medium containing 10 mM MES and 20 mM acetosyringone, with kanamycin, gentamycin, and rifampicin antibiotics. After incubation with shaking for 24 h, the Agrobacterium cells were harvested and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.6, 200 mM acetosyringone) and adjusted to a final OD600 of 6.0. Resuspensions of pTRV1 and pTRV2 or pTRV2-TAGL1 were kept for 3–4 h at 25°C and mixed in a ratio of 1:1 before infiltration. Each mixture of the Agrobacterium strain was injected into the carpopodium of the tomato fruit on about 7–10 DPA with a 1-ml syringe (Fig 1A). Tomato fruits injected with pTRV1 and pTRV2 alone were used as controls. All fruit samples were harvested at approximately 51 DPA when the TAGL1 VIGS fruits were at the red-ripe (RR) stage and produced an obvious visible phenotype. Upon harvesting, the yellow pericarps of TAGL1 silenced fruits were collected, snap-frozen in liquid nitrogen, and stored at -80°C until use.
RNA-seq and data processing
Total RNA was extracted from the TRV2 control red tomato fruit and the orange pericarp section of TAGL1-silenced fruit, (three biological replicates) using an RNeasy MiniKit (Qiagen, GmbH, Germany) at breaker+3 stage. RNA quality was checked on 1% agarose gels. RNA purity and concentration was monitored using a Nano Photometer® spectrophotometer (Implen, CA, USA). Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s instructions, and index codes were added to attribute each sequence to the respective sample. Library quality was assessed on the Agilent Bioanalyzer 2100 system. According to the manufacturer’s instructions, the clustering of the index-coded samples was performed on a cBot Cluster Generation System using Hiseq 4000 PE Cluster Kit (Illumia, San Diego, CA, USA). After cluster generation, the library preparations were sequenced on an Illumina Hiseq 4000 platform and 150 bp paired-end reads were generated.
RNA data processing was carried out as described previously [25]. In short, raw reads were checked for quality and trimmed to remove barcode and adaptor sequences using Cutadapt (https://pypi.python.org/pypi/cutadapt/) and the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/download.html). The quality of the clean reads was checked using the Q < 20 threshold. All clean reads were deposited in the National Center for Biotechnology Information’s (NCBI) Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under the accession number SRP076745. Clean reads from each library were aligned to the tomato reference genome (SGN release version SL2.50, ftp://ftp.sgn.cornell.edu/tomato_genome) using TopHat (http://ccb.jhu.edu/software/tophat/index.shtml). To construct the transcripts, reads with less than two mismatches were used by Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/) with the gene annotation of tomato (ITAG2.5). Genes in TAGL1-RNAi and control lines were considered as differentially expressed genes (DEGs) if the fold-change was ≥ 2 and the Q-value was < 0.05 using DESeq2. The averages of gene expressions in the three replicates were used for DEGs identification. Data shown in S3 Table
Gene Ontology (GO) enrichment analysis and KEGG pathway analysis
WEGO (http://wego.genomics.org.cn/) was used for Gene Ontology (GO) enrichment. The GO enrichment analysis provided all the GO terms that were significantly enriched in the DEGs relative to the genomic background, and the DEGs were filtered according to cellular components, molecular functions, and biological processes. KEGG (Kyoto Encyclopedia of Genes and Genomes; http://www.genome.jp/kegg/) is a main pathway-related database. Based on the comparison of the DEGs to the genomic background, pathway enrichment analysis pinpointed the enriched pathways.
Validation of RNA-Seq by qRT-PCR
2 μg total RNA was reverse-transcribed into cDNA with cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) with oligo (dT) primers, and the genomic DNA was removed using TranScript one-step gDNA Removal. Quantitative real-time PCR (qRT-PCR was performed using SYBR Green PCR SuperMix (Trans, China) with a BIO-RAD real-time PCR System CFX96 (Bio-Rad, U.S.A). The reaction condition was set as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. The fluorescence signal was monitored automatically in each cycle. Relative expression levels of specific mRNAs were measured using the 2^(-ΔΔCt) analysis method, and the expression values were normalized using the Actin gene. For each sample, three independent biological replicates were analyzed. Primers used in this validation are listed in the S1 Table.
Metabolite extraction for LC-MS analysis
Metabolite extraction was performed following the method described previously by Chen et al. with some modification [26]. The frozen pericarp samples were ground into powder using a mortar and pestle. 100 mg powder was weighed and suspended in 1.0 mL pure methanol or 75% aqueous methanol for extraction of lipid-soluble metabolites or water-soluble metabolites, respectively, both containing 20 mg/L lidocaine and 20 mg/mL CHAPS. Suspensions were vortexed 1 min for three times for and then stored at 4°C overnight. Following centrifugation at 12000 rpm for 10 min, supernatants containing the lipid-soluble metabolites and water-soluble metabolites were collected and mixed in a ratio of 1:1, and then filtered before LC-MS analysis.
Instrumentation and chromatographic system
A high-performance liquid chromatography (HPLC-20A) unit equipped with a photodiode array detector (Shimadzu, Japan) was used to analyze the metabolites in the tomato extracts. Separation of the metabolites was performed under the following conditions: column, Eclipse XDB-C18 (3.0*50 mm); solvent system water (0.2% formic acid); acetonitrile; gradient program, 95:5V/V at 0 min, 5:95V/V at 12 min, 5:95V/V at 15 min, 95:5 at 15.1 min, 95:5V/V at 22 min; flow rate, 0.2 ml/min; temperature: 45°C; injection volume: 2 μL. Masses of the eluted compounds that ranged from 50 m/z to 1500 m/z were monitored with an Agilent 6460 triple quad LC-MS equipped with an ESI source.
Quantitative detection was performed using UHPLC-ESI-QQQ-MS (Agilent 1290 and 6460 Triple Quadrupole Mass Spectrometry Series, Agilent Corporation, CA, USA). An electrospray ionization (ESI) source working either in positive or negative ion mode was used for all mass spectrometry (MS) analyses, using nitrogen as the drying agent. The MS conditions in the positive mode were as follows: HV voltage 4000 kV; capillary 7 μA, nozzle voltage 500 V, delta EMV 300V, gas flow 5 L/min; gas temperature 400°C; sheath gas flow 11 L/min. Collision energy was optimized based on the standards. Helium was used as the collision gas for collision-induced dissociation (CID). Quantification was done using the multiple reaction monitoring (MRM) mode under unit mass-resolution conditions. The data recorded were processed with MassHunter Software.
Chemicals
MS-grade acetonitrile and methanol were obtained from Sigma-Aldrich (St. Louis, MO, USA). Formic acid (eluent additive used for HPLC-MS analysis) was MS grade (CNW, Germany). The remaining analytical-grade chemicals were obtained from Beijing Chemical Factory (Beijing, China).
Results and discussion
Silencing of TAGL1 gene inhibited fruit ripening
In order to obtain TAGL1-silenced tomato fruit by VIGS, Agrobacterium GV3101 cultures containing either pTRV1: pTRV2-TAGL1 or pTRV1 and pTRV2-00 were injected into the carpopodium of the tomato fruit 7–10 DPA after pollination and fruit phenotypes were observed 20–30 days after injection. When all control fruits injected with pTRV1 and pTRV2-00 had turned red, thirty fruits at the same stage that had been injected with pTRV1 and pTRV2-TAGL1 showed an obviously different phenotype, with orange and red colored regions on the same fruit (Fig 1B). To confirm that the TAGL1 gene was suppressed successfully at the molecular level in TAGL1-silenced fruit, primers specific to the TAGL1 gene outside the region targeted for silencing were designed for real-time PCR. Reverse transcription -PCR showed that the TAGL1 transcripts in the orange sections of the TAGL1-silenced fruits were reduced by more than 71% compared to the red control fruits infiltrated with empty TVR vector (Fig 1C). The result reveals that the down-regulation of TAGL1 causes the unusual ripening phenotype of tomato fruits, which was consistent with the previous finding that TAGL1-RNAi fruits were yellow-orange upon ripening [15, 16]. However, in our experiments, TAGL1-silenced fruits did not show a thin pericarp layer phenotype displayed by RNAi-TAGL1 transgenic fruits [16]. A previous study has reported that the highest expression of the TAGL1 transcripts was observed during flower anthesis and the red ripe stage of tomato fruit [15, 16, 17]. As we injected tomato fruit 7–10 days after pollination, there was no opportunity for TAGL1 expression to be silenced in the tomato flower. We speculate, therefore, that TAGL1 may regulate pericarp development at the stage of ovary development rather than fruit ripening.
Global overview of the RNA-seq profile of TAGL1-silenced tomato
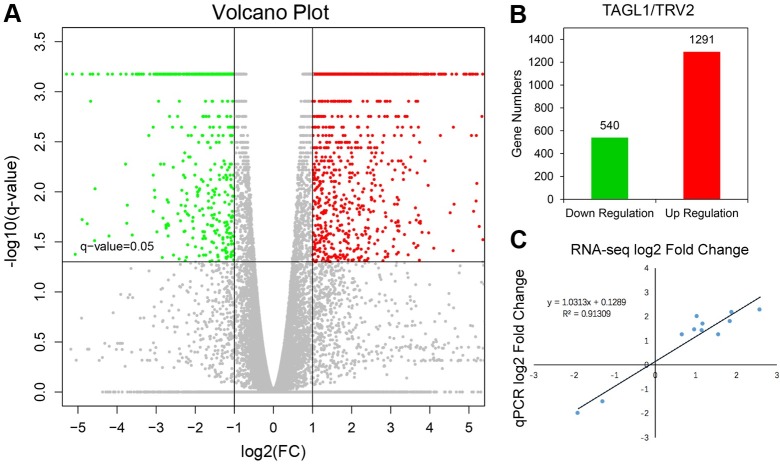
To understand the role of TAGL1 in regulating tomato fruit ripening at the molecular level, TAGL1-silenced (orange section) and control fruit samples(red stage) (TRV2-00) were analyzed via RNA-seq. All clean reads generated in the sequencing experiments were mapped and aligned with the tomato reference genome (ITAG2.5). Within each file, 78.38 ± 1.92% of the reads were found to be uniquely aligned, suggesting that the sequencing results were relatively stable (data not shown). Using the cutoff criteria of expression ratio ≥2.0 and Q-value < 0.05 (BH correction) between TAGL1-silenced and control tissues, 1291 up-regulated and 540 down-regulated genes were identified in TAGL1-silenced samples compared to controls (S3 Table and Fig 2). These results indicate that silencing TAGL1 affects the expression of many genes. To validate RNA-Seq results, 11 ripening or nutrition-associated DEGs were selected and their expression levels were verified by qRT-PCR. The fold changes in the expression patterns of the DEGs in real-time PCR were very similar (r2 = 0.91) (Fig 2, S1 Fig).
Fig 2. Global view of the DEGs in the TAGL1-silenced tomato fruits.
Volcano diagrams of the DEGs. Spots above the threshold line (Q-value = 0.05), indicate that differences are significant. Genes with expressions less than half of that displayed in the control group for Q-value < 0.05 are displayed in the green area, while those with expression at least two-folds greater than that of the control group are displayed in the red area. Genes in the grey area were neither over- nor under-expressed. (B) The number of down/up regulated genes. (C) The expression levels of 11 genes, as determined by RNA-seq and qPCR, are closely correlated. Nine genes were up-regulated, while two genes were down-regulated.
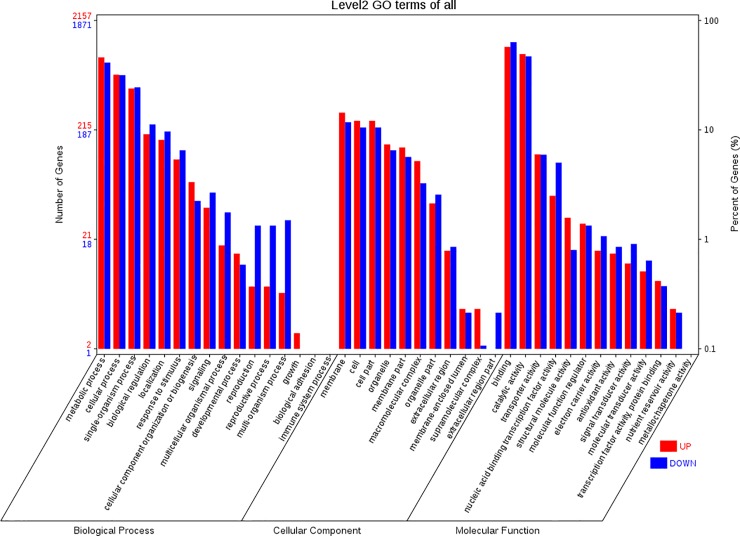
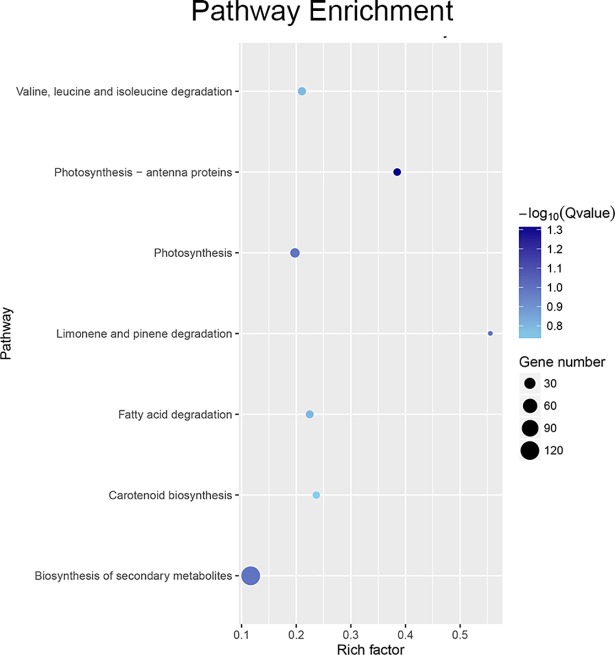
Gene ontology was successfully assigned using WEGO. The SDEGs between the TRV2-TAGL1 and the TRV2-00 were classified into 40 functional groups (Fig 3), of which cellular components accounted for 11 GO terms (the most representative were “membrane” and “cell”), molecular functions accounted for 13 GO terms (the most representative was “binding” and “catalytic”), and biological processes accounted for 16 GO terms (the most representative were “metabolic process”). Results of the KEGG pathway enrichment analysis indicated that TAGL1 plays a role in photosynthesis and biosynthesis of secondary metabolites (Fig 4).
Fig 3. GO analysis classified DEGs in the TAGL1-silenced tomato fruits according to WEGO.
Fig 4. Pathway enrichment analysis of differentially displayed genes from the TAGL1-silenced tomato fruits.
Analysis of constituents in tomato fruit samples
In order to observe the effects of TAGL1 gene silencing on the tomato fruit at the metabolic levels, the presence of 50 chemical compounds were tested and quantified in positive and/or negative modes. The identified compounds were classified into four groups: amino acids, organic acids, phenolics, and solanum alkaloids. By comparing the UPLC retention times and mass spectral data with those of the reference standards, the target peaks were tentatively identified as described below. The metabolites for which commercial standards are unavailable were identified following previously described protocols [26]. Peaks were used to query the MS2 spectral data taken from the literature or to search the databases (MassBank, Moto DB, KOBAS). Best matches were then searched in the Dictionary of Natural Products (DNP) and KEGG for identifying possible structures. More than 20 metabolites were putatively identified.
Silencing of TAGL1 altered fruit metabolism
In order to evaluate the changes in the nutrition and flavor of the TAGL1-silenced fruits, metabolites were analyzed using LC-MS/MS. Fifty metabolites were detected and annotated using standards and/or cross-referencing against libraries, and the production level of 11 metabolites were found to be different between TRV2-TAGL1 (orange section) and TRV2-00 samples (red) (Table 1).
Table 1. Relative quantitation of metabolites in TAGL1-silenced tomato fruits.
| Analytes | Ratio (TAGL1/TRV2) | P-value | Pathway |
|---|---|---|---|
| Aspartic acid1 | 0.467631696 | 0.014933727 | Biosynthesis of amino acids |
| L-Tyrosine1 | 0.202144376 | 0.015775349 | Biosynthesis of amino acids |
| Feruloylputrescine | 0.153963685 | 0.014338758 | Arginine and proline metabolism |
| 5-caffeoylquinic acid | 0.468658398 | 0.001019251 | - |
| α-Tomatine1 | 10.78557867 | 8.33493E-08 | - |
| L-Phenylalanine1 | 0.14565209 | 0.027272425 | Biosynthesis of amino acids |
| L-Valine1 | 0.449745752 | 0.011035025 | Valine, leucine, and isoleucine degradation |
| L-Glutamic acid1 | 0.244228151 | 3.67814E-06 | Biosynthesis of amino acids |
| Isoleucine | 0.26956715 | 0.015007356 | Valine, leucine, and isoleucine degradation |
| L-Leucine1 | 0.203396442 | 0.012507063 | Valine, leucine, and isoleucine degradation |
| C11H23O12P | 0.258873555 | 0.000509657 | - |
| C34H46O14 | 1.5902 | 1.5073E-06 | - |
| 4-Aminobutanoic acid1 | 0.456910298 | 0.001414631 | Arginine and proline metabolism |
1 The analytes were identified by comparison with standards.
Amino acid content of tomato fruits contributes markedly to the taste and nutritional quality. The data showed that the level of seven amino acids dropped in the TAGL1-silenced tomato fruits, with aspartic acid, L-tyrosine, L-glutamic acid, L-phenylalanine, Lvaline, L-leucine1, and isoleucine in the TRV2-TAGL1 fruits reduced to 46.76%, 20.21%, 24.42%, 14.57%, 44.97%, 20.34%, and 26.96% of that in the control fruits, respectively (Table 1). Aspartic acid and glutamic acid are known as amino acids that confer a delicious flavor; phenylalanine and tyrosine are known aromatic amino acids; and leucine, isoleucine, and phenylalanine are essential amino acids for humans. The results indicate that the down-regulation of the TAGL1 gene leads to the reduction in the content amino acid in the tomato fruit.
To understand the mechanism of TAGL1’s regulation of amino acid synthesis, gene expression profiles were analyzed, and 20 DEGs were found to be enriched in the pathway of biosynthesis of the amino acids (Fig 5 and S2 Table). Only three of these genes have been reported before, including gs (encoding glutamine synthetase (GS)) [27], SAM3 (encoding S-adenosylmethionine (SAM) synthase 3) [28], and IPMS2 (encoding 2-isopropylmalate synthase) [29]. GS is a type of chloroplast glutamine synthetase that assimilates ammonia into glutamine, which is a metabolic intermediate in the synthesis of other nitrogen-containing compounds in the plant. In addition, SAM3 catalyzes the formation of S-adenosylmethionine from methionine and could be induced by ozone exposure [30]. IPMS2 is one of a cluster of three genes known to encode an enzyme involved in leucine biosynthesis [31]. Among the other unreported genes, only three were down-regulated, including genes that encode prephenate dehydratase (Solyc11g066890) (probably involved in the first step of the sub-pathway that synthesizes L-phenylalanine from L-arogenate), tyrosine aminotransferase-like protein (Solyc07g053720), and ornithine carbamoyltransferase (Solyc12g089210). While L-glutamic acid was reduced to 24.42% of the control fruits in the TAGL1-silenced fruits, methionine and ornithine were not detected in either the TAGL1-silenced fruits or the control fruits, using LC-MS/MS. Glutamine synthetase catalyzes glutamic acid synthesis from glutamine. From RNA-seq, we found that the expression level of glutamine synthetase gene (Solyc01g080280) was increased about 6-fold in TAGL1-silenced fruit compared with control (S2 Table) which reduced the accumulation of L-Glutamic acid in TAGL1-silenced tomato fruit (Table 1). It is also possible that translational regulation could possibly explain the lack of positive correlation between gene expressions and level of amino acids. Fourteen DEGs were enriched in the pathway of phenylalanine metabolism using KEGG (S2 Table). These results show that TAGL1 positively regulates the biosynthesis of several amino acids through a complex network.
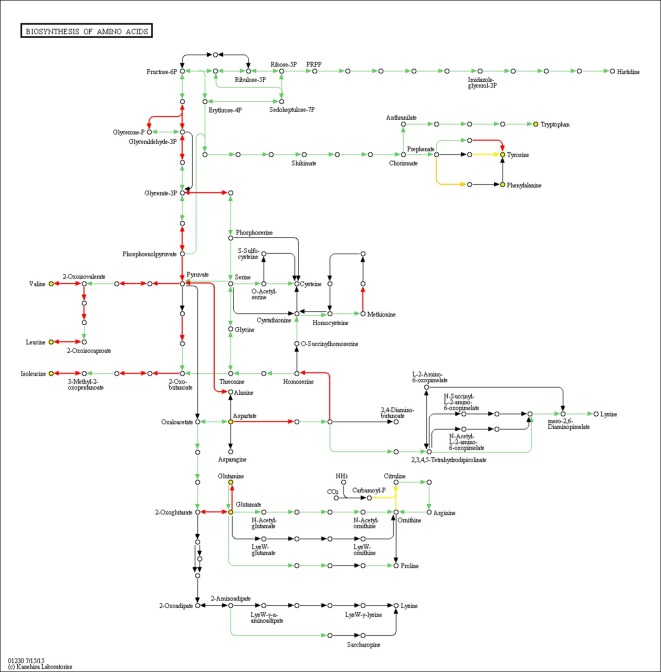
Fig 5. Diagram of amino acid biosynthetic pathways.
The diagram was constructed using KEGG pathway enrichment. Spots represent metabolites and arrows indicate steps. The arrows colored red, yellow, and grey/green indicate up-regulation, down-regulation, and absence of significant differences, respectively.
γ-Aminobutyric acid (GABA), identified as a functional component in reducing blood pressure in the human body, is a four-carbon non-protein amino acid commonly found in bacteria, animals, and plants [32, 33]. GABA is metabolized through a short pathway, called the ‘GABA shunt,’ which is a bypass of the tricarboxylic acid (TCA) cycle, composed of three enzymes, glutamate decarboxylase (GAD), GABA transaminase (GABA-T), and succinic semialdehyde dehydrogenase (SSADH) [34]. The first enzyme, GAD, catalyzes the irreversible decarboxylation of glutamate to produce GABA and CO2; then GABA is reversibly transaminated by the second enzyme, GABA-T, to form succinic semialdehyde (SSA), which is oxidized by the third enzyme, SSADH, to produce succinate [35]. The resulting succinate then flows into the TCA cycle. In tomato, GAD2 and GAD3 catalyze GABA synthesis from glutamate; silencing this gene using transgene technology can effectively reduce GABA levels compared with the control [36]. In our study, GABA was reduced to 46% and L-glutamine was reduced to 24% in the TAGL1-silenced fruits compared to the controls. In addition, the expression level of glutamate decarboxylase (GAD3) (Solyc05g052100) was decreased in the TAGL1-silenced fruits, as evidenced by the RNA-seq data. Although the decreased levels of glutamate contribute to the reduction of GABA, the findings of this study indicate that the expression level of GAD3 is regulated by TAGL1 to control GABA biosynthesis.
5-caffeoylquinic acid (chlorogenic acid) is one of the most abundant and widespread soluble phenolics in vascular plants. Besides indications that it can protect plant cells against oxidative stress, it can also play a role in the resistance to phytopathogens. 5-caffeoylquinic acid biosynthesis in fruits of the family Solanaceae (tomato, tobacco, and potato) was initially thought to occur via transesterification from caffeoyl-CoA and quinic acid by the hydroxycinnamoyl-CoA: quinate hydroxycinnamoyl transferase hydroxycinnamoyl quinic acid [37, 38]. The existence of another route involving direct 3′-hydroxylation of p-coumaryol quinic acid was first suggested by work with carrot cell cultures, and studies of the impact of the level of expression of the hydroxycinnamoyl quinic acid gene in tobacco and tomato plants demonstrated that this route might be predominant in the plants of the Solanaceae family [39].
Analysis of the tomato fruits with VIGS-silenced TAGL1, revealed a significant reduction in 5-caffeoylquinic acid to 47% of the control fruit. Our RNA-seq data showed that the transcript levels of one gene involved in the first proposed pathway for the synthesis of chlorogenic acid in plants, phenylalanine ammonia lyase gene (PAL) (Solyc10g086180), was increased. PAL encodes a protein that is involved in the first step of the sub-pathway that synthesizes trans-cinnamate from L-phenylalanine. We conclude that the decreased level of 5-caffeoylquinic acid content is mainly due to a significant reduction in the amount of phenylalanine, its precursor.
α-tomatine found in tomato is a type of antinutritional factor for humans. However, α-tomatine may also have anticarcinogenic, cardioprotective, and other beneficial effects [39]. There exists a proposed pathway for conversion from cholesterol to α-tomatine: GAME7 (CYP72) hydroxylates the cholesterol at the C22 position, followed by GAME8 (CYP72) hydroxylation at C26 position, and then C22, 26-dihydroxycholesterol is hydroxylated at C16 and oxidized at C22, follow by closure of E-ring by GAME11 (2-oxoglutarate-dependent dioxygenase) and GAME6 (CYP72) to form the furostanol-type aglycone [40]. In our study, the level of α-tomatine was significantly increased in the TAGL1-silenced fruits (10.49 fold). At the same time, the expression level of GAME11 (Solyc02g062490) was increased in the TAGL1-silenced fruit compared to control tomato fruit. It has been reported, using the VIGS technology to silence GAME11, that a putative dioxygenase in the cluster results in a significant reduction in α-tomatine levels and accumulation of several cholestanol-type steroidal saponins in tomato leaves [40], which was consistent with the findings of our study. In summary, TAGL1 VIGS promotes α-tomatine biosynthesis by activating the expression of GAME11.
Conclusion
Gene silencing of TAGL1 in tomatoes using the VIGS technique resulted in a non-ripening phenotype with orange pericarp. The analysis of the metabolites by LC-MS/MS showed the reduction in content of several amino acids and organic acids, as well as the accumulation of α-tomatine in the TAGL1-silenced fruits. The result show that TAGL1 positively regulates the synthesis of amino acids and negatively regulates the synthesis of α-tomatine in tomato fruit. The findings of the present study suggest that TAGL1 controls accumulation of nutritional and flavor components in the tomato fruits by the transcriptional regulation of targeted genes.
Supporting information
(TIF)
(TIF)
(DOCX)
(DOCX)
(CSV)
Acknowledgments
We would like to thank Dr S.P. Dinesh-Kumar (University of California at Davis) for kindly providing pTRV1 and pTRV2 vectors, Donald Grierson (University of Nottingham) and Ayla Norris for revising the manuscript (United States Department of Agriculture, Agricultural Research Service, USA).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China, 31601518 (http://www.nsfc.gov.cn/). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Klee HJ, Giovannoni JJ. Genetics and Control of Tomato Fruit Ripening and Quality Attributes. Annu Rev Genet. 2011; 45(1): 41–59. [DOI] [PubMed] [Google Scholar]
- 2.Seymour GB, Østergaard L, Chapman NH, Knapp S, Martin C. Fruit development and ripening. Annu Rev Plant Biol. 2013; 64: 219–241. doi: 10.1146/annurev-arplant-050312-120057 [DOI] [PubMed] [Google Scholar]
- 3.Pesaresi P, Mizzotti C, Colombo M, Masiero S. Genetic regulation and structural changes during tomato fruit development and ripening. Front Plant Sci. 2014; 5:124 doi: 10.3389/fpls.2014.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlova R, Chapman N, David K, Angenent GC, Seymour GB, de Maagd RA. Transcriptional control of fleshy fruit development and ripening. J Exp Bot. 2014; 65(16):4527–4541. doi: 10.1093/jxb/eru316 [DOI] [PubMed] [Google Scholar]
- 5.Giovannoni J, Nguyen C, Ampofo B, Zhong S, Fei Z. The Epigenome and Transcriptional Dynamics of Fruit Ripening. Annu Rev Plant Biol. 2017; 68:61–84. doi: 10.1146/annurev-arplant-042916-040906 [DOI] [PubMed] [Google Scholar]
- 6.Alexander L, Grierson D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot. 2002; 53(377):2039–2055. [DOI] [PubMed] [Google Scholar]
- 7.Wang KL, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. Plant Cell. 2002; 14 Suppl:S131–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vrebalov J. A MADS-Box gene necessary for fruit ripening at the Tomato Ripening-Inhibitor (Rin) locus. Science. 2002; 296(5566): 343–346. doi: 10.1126/science.1068181 [DOI] [PubMed] [Google Scholar]
- 9.Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King G J, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature Genet. 2006; 38(8): 948–952. doi: 10.1038/ng1841 [DOI] [PubMed] [Google Scholar]
- 10.Yuan XY, Wang RH, Zhao XD, Luo YB, Fu DQ. Role of the tomato Non-Ripening mutation in regulating fruit quality elucidated using iTRAQ protein profile analysis. PLoS ONE. 2016; 11(10): e0164335 doi: 10.1371/journal.pone.0164335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovannoni JJ. Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol. 2007; 10(3):283–289. doi: 10.1016/j.pbi.2007.04.008 [DOI] [PubMed] [Google Scholar]
- 12.Fujisawa M, Nakano T, Ito Y. Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol. 2011; 11: 26 doi: 10.1186/1471-2229-11-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujisawa M, Nakano T, Shima Y, Ito Y. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell. 2013. 25(2):371–386. doi: 10.1105/tpc.112.108118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eriksson E M, Bovy A, Manning K, Harrison L, Andrews J, De Silva J, et al. Effect of the Colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 2004; 136(4): 4184–97. doi: 10.1104/pp.104.045765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vrebalov J1, Pan IL, Arroyo AJ, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J, Irish VF. Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell. 2009; 21(10):3041–62. doi: 10.1105/tpc.109.066936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itkin M1, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 2009; 60(6):1081–95. doi: 10.1111/j.1365-313X.2009.04064.x [DOI] [PubMed] [Google Scholar]
- 17.Pan IL, McQuinn R, Giovannoni JJ, Irish VF. Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J Exp Bot. 2010; 1(6):1795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giménez E, Pineda B, Capel J, Antón MT, Atarés A, Pérez-Martín F, García-Sogo B, Angosto T, Moreno V, Lozano R. Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1gene during reproductive development of tomato. PLoSOne. 2010; 5(12):e14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto Pde B, Angenent GC, de Maagd RA. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell. 2012; 24(11):4437–51. doi: 10.1105/tpc.112.103283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujisawa M, Shima Y, Nakagawa H, Kitagawa M, Kimbara J, Nakano T, Kasumi T, Ito Y. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell. 2014; 26(1):89–101 doi: 10.1105/tpc.113.119453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 2010; 64(6):936–47. doi: 10.1111/j.1365-313X.2010.04384.x [DOI] [PubMed] [Google Scholar]
- 22.Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell. 2011; 23(3):923–41. doi: 10.1105/tpc.110.081273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pineda B, Capel J, Atare A, Anto T. Functional Analysis of the Arlequin mutant corroborates the essential role of the ARLEQUIN / TAGL1 gene during reproductive development of tomato. PLoS One. 2010; 5(12): e14427 doi: 10.1371/journal.pone.0014427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB. Virus-induced gene silencing in tomato fruit. Plant J:. 2005; 43(2): 299–308. doi: 10.1111/j.1365-313X.2005.02441.x [DOI] [PubMed] [Google Scholar]
- 25.Wang RH, Yuan XY, Meng LH, Zhu BZ, Zhu HL, et al. Transcriptome analysis provides a preliminary regulation route of the ethylene signal transduction component, SlEIN2, during tomato ripening. PLoS One. 2016; 11: e168287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X, et al. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol Plant. 2013; 6(6): 1769–1780. doi: 10.1093/mp/sst080 [DOI] [PubMed] [Google Scholar]
- 27.Scarpeci TE, Marro ML, Bortolotti S, Boggio SB, Valle EM. Plant nutritional status modulates glutamine synthetase levels in ripe tomatoes (Solanum lycopersicum cv. Micro-Tom). J Plant Physiol. 2007; 164(2):137–145. doi: 10.1016/j.jplph.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 28.Dippe M, Brandt W, Rost H, Porzel A, Schmidt J, Wessjohann LA. Rationally engineered variants of S-adenosylmethionine (SAM) synthase: reduced product inhibition and synthesis of artificial cofactor homologues. Chem Commun (Camb). 2015; 51(17):3637–40. [DOI] [PubMed] [Google Scholar]
- 29.de Kraker JW, Luck K, Textor S, Tokuhisa JG, Gershenzon J. Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol. 2007;143(2):970–86. doi: 10.1104/pp.106.085555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hacham Y, Song L, Schuster G, Amir R. Lysine enhances methionine content by modulating the expression of S-adenosylmethioninesynthase. Plant J.2007; 51(5):850–861. doi: 10.1111/j.1365-313X.2007.03184.x [DOI] [PubMed] [Google Scholar]
- 31.Ning J, Moghe GD, Leong B, Kim J, Ofner I, Wang Z, et al. A feedback-insensitive isopropylmalate synthase affects acylsugar composition in cultivated and wild tomato. Plant Physiol. 2015; 169(3):1821–1835. doi: 10.1104/pp.15.00474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akihiro T, Koike S, Tani R, Tominaga T, Watanabe S, Iijima Y., et al. Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant Cell Physiol.2008; 49(9): 1378–1389. doi: 10.1093/pcp/pcn113 [DOI] [PubMed] [Google Scholar]
- 33.Koike S, Matsukura C, Takayama M, Asamizu E, Ezura H. Suppression of gamma-aminobutyric acid (GABA) transaminases induces prominent GABA accumulation, dwarfism and infertility in the tomato (Solanum lycopersicum L.). Plant Cell Physiol 2013; 54: 793–807. doi: 10.1093/pcp/pct035 [DOI] [PubMed] [Google Scholar]
- 34.Seifi HS, Curvers K, Vleesschauwer DDe, Delaere I, Aziz A, Monica H. Concurrent overactivation of the cytosolic glutamine synthetase and the GABA shunt in the ABA-deficient sitiens mutant of tomato leads to resistance against Botrytis cinerea. New Phytol. 2013; 199(2): 490–504. doi: 10.1111/nph.12283 [DOI] [PubMed] [Google Scholar]
- 35.Shelp BJ, Mullen RT, Waller JC. Compartmentation of GABA metabolism raises intriguing questions. Trends Plant Sci. 2012; 17(2): 57–59. doi: 10.1016/j.tplants.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 36.Takayama M, Koike S, Kusano M, Matsukura C, Saito K, et al. Tomato glutamate decarboxylase genes SlGAD2 and SlGAD3 play key roles in regulating gamma-Aminobutyric acid levels in tomato (Solanum lycopersicum). Plant Cell Physiol. 2015; 56: 1533–1545. doi: 10.1093/pcp/pcv075 [DOI] [PubMed] [Google Scholar]
- 37.Negrel J, Javelle F, Morandi D. Detection of a plant enzyme exhibiting chlorogenate-dependant caffeoyltransferase activity in methanolic extracts of arbuscular mycorrhizal tomato roots. Plant Physiol Biochem. 2013; 66: 77–83. doi: 10.1016/j.plaphy.2013.02.009 [DOI] [PubMed] [Google Scholar]
- 38.Mahesh V, Million-Rousseau R, Ullmann P, Chabrillange N, Bustamante J, et al. Functional characterization of two p-coumaroyl ester 3'-hydroxylase genes from coffee tree: evidence of a candidate for chlorogenic acid biosynthesis. Plant Mol Biol. 2007; 64: 145–159 doi: 10.1007/s11103-007-9141-3 [DOI] [PubMed] [Google Scholar]
- 39.Friedman M. Anticarcinogenic, cardioprotective, and other health benefits of tomato compounds lycopene,-tomatine, and tomatidine in pure form and in fresh and processed tomatoes. J Agric Food Chem. 2013; 61(40): 9534–9550. doi: 10.1021/jf402654e [DOI] [PubMed] [Google Scholar]
- 40.Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, et al. Biosynthesis of antinutritional alkaloids in Solanaceous crops is mediated by clustered genes. Science. 2013; 341(6142): 175–179. doi: 10.1126/science.1240230 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(DOCX)
(DOCX)
(CSV)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.