Abstract
Oxaliplatin is a platinum-based chemotherapeutic used for cancer treatment. Its use associates with peripheral neuropathies and chronic gastrointestinal side-effects. Oxaliplatin induces immunogenic cell death by provoking the presentation of damage associated molecular patterns. The damage associated molecular patterns high-mobility group box 1 (HMGB1) protein exerts pro-inflammatory cytokine-like activity and binds to toll-like receptors (namely TLR4). Gastrointestinal microbiota may influence chemotherapeutic efficacy and contribute to local and systemic inflammation. We studied effects of oxaliplatin treatment on 1) TLR4 and high-mobility group box 1 expression within the colon; 2) gastrointestinal microbiota composition; 3) inflammation within the colon; 4) changes in Peyer’s patches and mesenteric lymph nodes immune populations in mice. TLR4+ cells displayed pseudopodia-like extensions characteristic of antigen sampling co-localised with high-mobility group box 1 -overexpressing cells in the colonic lamina propria from oxaliplatin-treated animals. Oxaliplatin treatment caused significant reduction in Parabacteroides and Prevotella1, but increase in Prevotella2 and Odoribacter bacteria at the genus level. Downregulation of pro-inflammatory cytokines and chemokines in colon samples, a reduction in macrophages and dendritic cells in mesenteric lymph nodes were found after oxaliplatin treatment. In conclusion, oxaliplatin treatment caused morphological changes in TLR4+ cells, increase in gram-negative microbiota and enhanced HMGB1 expression associated with immunosuppression in the colon.
Introduction
Platinum-based chemotherapeutic agents are widely used for the treatment of cancer, and oxaliplatin, the third generation drug, is primarily used as the first-line treatment for colorectal malignancies [1,2]. Platinum-based drugs mediate their cytotoxic effects via the formation of nuclear and mitochondrial DNA platinum adducts which ultimately affect cell viability and hinder prospective replication [3–5]. Despite its therapeutic efficacy, the use of oxaliplatin causes unfavourable side-effects which include, but are not limited to, peripheral sensory neuropathy and gastrointestinal dysfunction [2,6–9]. These side-effects are major hurdles for cancer treatment as they result in dose reductions, treatment non-compliance and cessation [7,10,11]. Whilst the peripheral sensory neuropathy caused by oxaliplatin has attracted a large research focus, there are limited studies investigating the effects of this drug on gastrointestinal dysfunction. Only recently, the enteric nervous system (ENS) has gained attention regarding its role in the multifaceted pathophysiology of gastrointestinal dysfunction following chemotherapeutics [8,9,12]. The ENS is an intrinsic and intricate neuronal network embedded throughout the gastrointestinal tract which regulates secretion, absorption, vasomotor tone and motility [13]. The ENS can anatomically be divided into two major plexuses; the submucosal and myenteric. A few studies to date have shown that oxaliplatin induces myenteric neuronal loss, changes in the proportion of neuronal phenotypes, oxidative stress and causes changes in gastrointestinal transit and motility leading to constipation [8,9,14]. However, the mechanisms underlying oxaliplatin-induced changes in the myenteric plexus and cell death remain to be elucidated.
It is well established that anti-cancer agents induce damage to the gastrointestinal mucosa which may cause dysbiosis of commensal microbiota and potentiate inflammation [15–19]. A number of studies have reported microbiota dysbiosis following the anti-cancer chemotherapeutic agents, irinotecan and 5-fluorouracil [20–23]. However, the effects of platinum-based drugs on gastrointestinal microbiota remain largely unexplored. Gastrointestinal inflammation has been associated with persistent alterations in enteric neuron function and neuronal loss [24–26]. The gastrointestinal tract in particular is equipped with lymphoid organs (Peyer’s patches (PPs) and mesenteric lymph nodes (MLNs)) which houses ~70% of the body’s immunocytes, thus, highlighting their important role in discriminating between innocuous and noxious pathogens or danger signals [27]. Further adding to this complexity, oxaliplatin is regarded as a potent inducer of immunogenic cell death. Apoptosis has long been considered to be an immunologically silent or tolerogenic event, however, oxaliplatin treatment has been shown to induce beneficial anti-cancer immune responses through the induction of damage-associated molecular patterns (DAMPs) in colorectal cancer cells [28]. The presentation of DAMPs can prompt the engulfment of dying cells by phagocytes, or apoptotic antigens may be presented to T cells for targeted elimination [28,29]. A classical DAMP is the nuclear-resident non-histone protein high mobility group box 1 (HMGB1) which exerts pro-inflammatory cytokine-like activity once cytoplasmically translocated and/or released into the extracellular environment by damaged cells [28,29]. HMGB1 is a ligand for toll-like receptors and is presented to T cells for priming and activation [29,30]. Both DAMPs and pathogen-associated molecular patterns (PAMPs–microbial endotoxins) can induce an immunological response following anti-cancer chemotherapy [31,32]. It is unknown whether gastrointestinal inflammation may be implicated in enteric neuropathy associated with oxaliplatin treatment, either directly or inadvertently.
Herein, we determined the effects of oxaliplatin treatment on 1) TLR4 and HMGB1 expression within the murine colon; 2) gastrointestinal microbiota composition; 3) inflammation within the colon; and 4) changes in immune populations within the murine PPs and MLNs. It is hypothesized that oxaliplatin treatment will cause gastrointestinal inflammation characterized by an increase in TLR4+ and CD45+ cells within the colon lamina propria and at the level of the myenteric plexus, will cause changes to microbiota composition, and will induce changes in immune populations within the PPs and MLNs which may contribute to inflammatory enteric neuropathy.
Materials and methods
Animals
Male BALB/c mice (n = 40, aged 7–8 weeks, weighing 18-25g) were used in this study. We have previously established an orthotopic mouse model of colorectal cancer in Balb/c mice (35). As cancer and chemotherapy can both differentially affect immune responses, our first aim is to study them exclusive to one another. We have published several papers on the effects of chemotherapy on the enteric nervous system using Balb/c mouse models (8, 9, 14). Our prospective studies are to combine colorectal cancer and chemotherapy. Therefore, Balb/c mouse strain is used in this study. We have based our studies on male mice as fluctuations in hormones during the estrous cycle in female mice may present as confounding factors influencing immune responses.
Mice had access to food and water ad libitum and were kept under a 12 hour light/dark cycle in a well-ventilated room at a temperature of 22°C. Mice acclimatized for up to 1 week prior to the commencement of in vivo intraperitoneal injections. All efforts were made to minimise animal suffering, to reduce the number of animals used, and to utilise alternatives to in vivo techniques, if available. All procedures in this study were approved by the Victoria University Animal Experimentation Ethics Committee (Animal Ethics number 15–011) and performed in accordance with the guidelines of the National Health and Medical Research Council Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
Oxaliplatin treatment
Mice were separated into 2 cohorts (n = 4-10/group): 1) vehicle-treated (sterile water), 2) oxaliplatin-treated (3 mg/kg, Sigma-Aldrich, Australia). All mice received intraperitoneal injections (volume calculated per body weight, maximum of 200 μl/injection) using 26 gauge needles tri-weekly totaling 6 injections. Dosages were calculated per body mass as previously published [33,34]. Mice were culled via cervical dislocation 14 days subsequent to their first intraperitoneal injection, and the colon, PPs, and MLNs were harvested.
Immunohistochemistry
The colon was harvested (n = 4/group), cut along the mesenteric border and pinned to silicone-based petri dishes containing 1x phosphate buffered solution (PBS). Tissues were incubated in Zamboni’s fixative (2% formaldehyde, 0.2% picric acid and 0.1 M sodium phosphate buffer (pH 7.0)) overnight at 4°C. The following day, tissues were washed 3 x 10 minutes in 100% DMSO, followed by 3 x 10 minute washes with 1x PBS. Sections (30μm) were cut and incubated with a mouse blocking reagent (M.O.M. kit, Vector Labs, USA), or 10% normal donkey serum for 1 hour at room temperature, then washed 3 x 10 minutes with PBS and Triton-x100 (PBS-T). Sections were co-labelled with anti-TLR4 (mouse, 1:500, Abcam, USA) and anti-HMGB1 (rabbit, c-terminus, 1:500, Abcam, USA) antibodies or with anti-CD45 (mouse, 1:500, Abcam, USA) antibody alone. Primary antibodies were incubated at room temperature overnight, and were then washed 3 x 10 minutes with PBS-T. The secondary antibody for both TLR4 and CD45 was FITC-conjugated (mouse, 1:200, Abcam, USA) made up in M.O.M. diluents; and the secondary antibody for HMGB1 was AlexaFluor-647-conjugated (rabbit, 1:200, Jackson Immuno-Research, USA). Secondary antibodies were incubated at room temperature for 2 hours and then washed 3 x 10 minutes with PBS-T. Sections were mounted onto glass slides using an anti-fade mounting medium (DAKO, Australia).
Imaging and analysis
Three-dimensional (z-series) images of the colon cross sections were taken using an Eclipse Ti confocal microscope (Nikon, Japan). Excitation wavelengths were set to 473 nm for FITC and 640.4 nm for Alexa Fluor 647. The number of CD45+ cells was counted from 8 images/preparation taken at 20x magnification with a total area of 2 mm2. CD45+ immunoreactivity was measured from 8 images/preparation taken at 20x magnification with a total area of 2 mm2. All images were captured under identical conditions, calibrated to standardise minimum baseline fluorescence, and were converted to binary. Differences in fluorescence from baseline were measured using Image J software (National Institute of Health, USA). All images were coded and analyzed blindly.
Fecal DNA isolation
Fecal pellets were collected from vehicle- and oxaliplatin-treated animals (n = 10/group), and stored at -80°C until time of processing. The PowerFecal DNA Isolation Kit (MO BIO Laboraties Inc, Australia) was used to obtain DNA from the fecal pellets as per manufacturer’s instructions. The isolated fecal DNA was frozen in -80°C until time of high-throughput sequence analysis.
PCR amplification of variable regions 3–4 of the 16S rRNA
PCR amplification of variable regions 3–4 (V3–V4) from the 16S rRNA gene and subsequent Illumina sequencing were performed commercially by the Australian Genome Research Facility (Brisbane, Australia). The 2-stage PCR protocol was followed according to the Illumina 16S guidelines (http://sapac.support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html). Briefly, PCR amplicons were generated using a Forward 341F primer (CCTAYGGGRBGCASCAG) and a Reverse 806 R primer (GGACTACNNGGGTATCTAAT) along with KAPA HiFi HotStart ReadyMix (Roche, Australia). Thermal cycling consisted of 3 min at 95°C, and 25 cycles consisting of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C, followed by 5 min at 72°C. Second stage PCR was completed by using Nextera XT Index Kit (Illumina, Australia). Thermal cycling consisted of 3 min at 95°C, and 8 cycles consisting of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C, followed by 5 min at 72°C. Resulting amplicons were measured by fluorometry, normalised and then pooled with unique indices. This amplicon pool was then run on the Illumina MiSeq platform.
High-throughput sequence analysis of fecal microbiota
Fecal DNA samples underwent high-throughput sequencing on the Illumina MiSeq platform at the Australian Genome Research facility (University of Queensland, Brisbane, Australia). Paired-end reads were assembled by aligning the forward and reverse reads using join_paired_ends.py. Primers were trimmed using Seqtk (version 1.0). Trimmed sequences were processed using Quantitative Insights into Microbial Ecology (QIIME 1.9) 4 USEARCH2,3 (version 8.0.1623) software (Caporaso et al. 2010). Briefly, de-multiplexing and quality filtering were performed using the split_libraries_fastq.py script for each data set. Operational Taxonomic Units (OTUs) were de novo picked at 97% sequence similarity following the usearch pipeline and representative sequences of each cluster were used to assign taxonomy through matching against the Blast 2.2.22 database. Evaluations present at each taxonomic level, including percentage compilations, represent all sequences resolved to their primary identification or their closest relative. Alpha diversity using alpha_diversity.py script was performed for species richness, Good’s coverage, Chao1, Shannon-Wiener’s diversity index and Simpson’s index of diversity. Weighted and unweighted UniFrac distance matrices were obtained through Jack-knifed beta diversities in QIIME and principal coordinate analysis (PCoA) plots were obtained. Sample clustering and statistical analysis were carried out in R environment and SPSS (version 23).
Myeloperoxidase activity
Colon tissues from vehicle- and oxaliplatin-treated animals (n = 3/group) were harvested and homogenized in 4 volumes of MPO assay buffer using a FastPrep24TM5G homogenizer and matrix D lysing tubes (MP Biomedicals, Australia) for 40 seconds. The supernatant was transferred into collecting tubes, and centrifuged at 13,000 x g for 10 minutes at 4°C. Tissue protein levels were quantified using the bicinchoninic acid (BCA) assay (Thermo Scientific, Australia) according to manufacturer’s instructions and absorbance was read at 526 nm using a VarioskanTM Flash Multimode Reader (Thermo Scientific, Australia) using SkanIt software v.2.4.3. The MPO Colorimetric Activity Assay (Sigma-Aldrich, Australia) protocol was followed as per manufacturer’s instructions. Briefly, TNB standards, MPO positive and negative controls and each sample was assayed in triplicate. Samples were normalized to protein content. Absorbance was read at 412 nm using a VarioskanTM Flash Multimode Reader (Thermo Scientific, Australia) using SkanIt software v.2.4.3. All standards and samples were corrected for background absorbance readings. A standard curve plotted and the amount of TNB consumed by the enzyme assay for each sample was determined. MPO activity was calculated as per manufacturer’s instructions: MPO Activity = B Sample Dilution Factor/(Reaction Time) x V. MPO activity is reported as nmol/min/mL = milliunit.
RNA isolation and RT2 profiler PCR arrays
Colons from vehicle (n = 5) or oxaliplatin-treated (n = 4) mice were removed, snap frozen in liquid nitrogen and stored in -80°C until used. Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, California) and further purified using an RNeasy Mini kit (Qiagen, Hilden, Germany), including the on-column DNase digestion step. RNA integrity was determined with an Agilent 2100 Bioanalyzer (Agilent Technologies, USA) using RNA 6000 Nano chips (Agilent Technologies); RNA Integrity Numbers (RIN) of all colon RNA samples were within an appropriate range (vehicle treated: 8.8 ± 0.9, n = 5; oxaliplatin-treated: 9.2 ±0.1, n = 4). Total RNA concentration was determined on a Qubit Fluorometer (Invitrogen) using a Qubit RNA BR Assay. Gene expression was investigated using the pathway specific RT2 Profiler PCR Array ‘Mouse Cancer Inflammation and Immunity Crosstalk’ (Qiagen, Cat. no. PAMM-181Z) according to the manufacturer’s instructions. Arrays were performed using equal quantities of either vehicle or oxaliplatin-treated RNA. Reverse transcription was carried out with the RT2 First Strand Kit (Qiagen) using 0.5μg pooled RNA as template. Equal amounts of cDNA were distributed to each well of the RT2 Profiler Array and real-time PCR was performed in a Biorad CFX96 real-time thermal cycler, using RT2 SYBR Green qPCR Mastermix (Qiagen). PCR cycling comprised an initial denaturation step at 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Melt curve analysis was performed to verify PCR specificity. Arrays were performed in duplicate for each RNA pool. CT values (cycle number at which fluorescence crosses a defined threshold) were obtained using the Bio-rad CFX Manager software, using a constant value across all arrays. The detection limit was set at CT of 35 cycles. CT values were uploaded to and analysed using the web-based Gene Globe Data Analysis Center (Qiagen). Data were normalised to the mean of five reference genes: Glyceraldehyde-3-phosphate dehydrogenase, Gapdh; Beta-2 microglobulin, B2m; Actin-beta, Actb; Glucuronidase-beta, Gusb and Heat shock protein 90 alpha (cytosolic), class B member 1, Hsp90ab1. Fold change was calculated using the ΔΔCT method, as the ratio of normalised gene expression in the oxaliplatin-treated group to normalised gene expression in the vehicle-treated control group.
Flow cytometry
To identify immune cell changes following oxaliplatin treatment the PPs and MLNs were harvested. An n = 5 PPs or MLNs were collected from each animal. PPs and MLNs were placed in 15 mL tubes containing FACS buffer (PBS, 0.1% bovine serum albumin and 0.02% sodium azide) and were kept on ice. Mucosal epithelium lining the PPs and excess adipose tissue attached to MLNs were gently removed using forceps. Manual cell suspensions of the PPs, MLNs and spleen were performed. All samples were then centrifuged at 1500 rpm for 5 minutes at 4°C. The supernatant of each cell suspension was aspirated and the pellet containing the immune cells was then resuspended in 1 mL of FACS buffer and filtered. Manual cell counts were performed, and cells were seeded to 96 U-bottom well plates (BD Biosciences, USA) and centrifuged at 1300 rpm for 3 minutes at 4°C. Subsequent to centrifugation, the 96 U-bottom well plates were then aspirated. A selection of cell surface antibodies was used to identify various immune cell populations (Table 1). Each antibody cocktail was loaded to appropriate wells, and incubated for 20 minutes at 4°C. Subsequent to the incubation period, the cells were washed with FACS buffer and centrifuged at 1300rpm for 3 minutes at 4°C. The plates were aspirated and cells within each well were resuspended in FACS buffer, and then transferred to FACS tubes. BD Biosciences LSR II and FACS CANTO II flow cytometers were used to collect 200,000 cells from each cell suspension. Information was obtained via software FACSDiva (BD Biosciences, USA), and analysis was conducted using FlowJo (Tree Star, USA) or FACSDiva.
Table 1. Antibodies used for FACS experiments.
| Cells | Primary antibody | Conjugate | Host species | Dilution |
|---|---|---|---|---|
| Pan-leukocyte marker | CD45 | PerCP/Cy5.5 | Mouse | 1:400 |
| Pan-T cell marker | CD3 | PerCP/Cy5.5 | Mouse | 1:400 |
| T cell receptor | TCRβ | APC | Rat | 1:250 |
| Granulocytes | GR-1, CD11b | PE-Cy7 | Rat | 1:100 |
| Cytotoxic T cells | CD8 | Pacific Blue | Rat | 1:100 |
| Helper T cells | CD4 | Pacific Orange | Rat | 1:100 |
| B cells | B220 | FITC | Mouse | 1:400 |
| Macrophages | CD11b, Ly6C, Ly6G, CD206, F4/80 | PE | Rat | 1:200 |
| Dendritic cells | CD11c | Pacific Blue | Rat | 1:250 |
| Major histocompatibility complex II | MHC-II | Brilliant Violet 510 | Rat | 1:800 |
| Eosinophils | CD193 | Alexa Fluor 647 | Rat | 1:200 |
| NK cells | CD49b | PE | Rat | 1:100 |
| γδ T cells | γδ-TCR | FITC | Mouse | 1:500 |
| NKT cells | CD1d α-Galactosylceramide tetramer | PE | Rat | 1:500 |
Statistical analysis
For microbiota studies, two-tailed t-tests were used to compare two sets of data, assuming unequal variance. The data generated by mass spectral analyses were normalized with respect to internal standards (RSD = 19.28%), where a magnitude of 1 fold change referred to the concentration of 10 mg/L. Statistical analysis was performed using SIMCA 14 (Umetrics AG, Umeå, Sweden). Statistical analysis for all other experiments included an unpaired t-test with Welch’s correction using GraphPad PrsimTM v6.0 (GraphPad Software, USA). The data were represented as mean ± standard error of the mean (SEM). Statistical significance for all experiments was defined where the P value was less than 0.05.
Results
Animal symptoms
During the course of oxaliplatin treatment, mice failed to gain weight compared to their control counterparts, and display signs of nausea (pica), as well as constipation.
Oxaliplatin treatment causes morphological changes in TLR4+ cells and reduces TLR7, TLR9 and H2-D1 expression in the colon
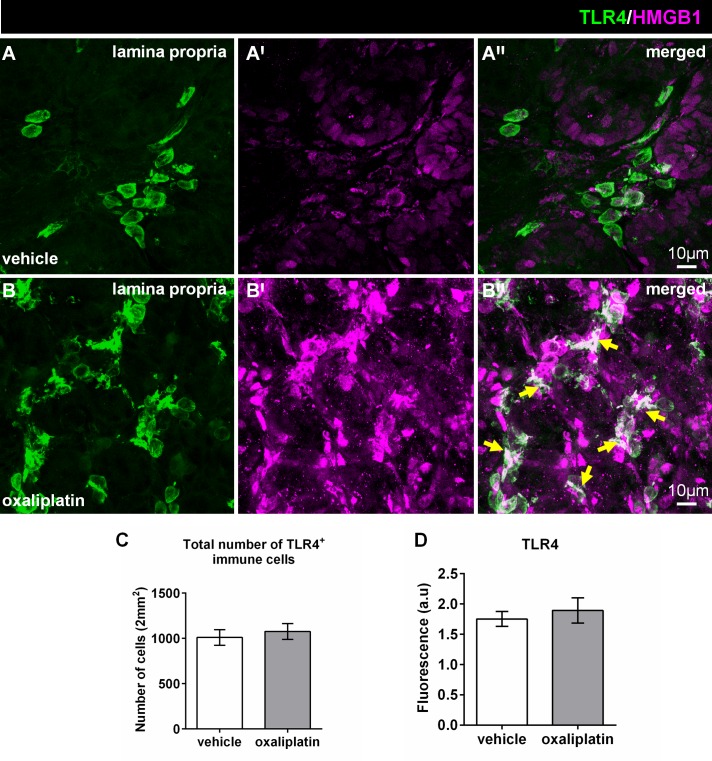
Colon cross-sections from the vehicle and oxaliplatin-treated groups were double-labelled with anti-TLR4 and anti-HMGB1 antibodies to determine any expressional changes. Intense HMGB1 expression within the lamina propria of the colon from the oxaliplatin-treated animals was noted when compared to the vehicle-treated cohort (Fig 1A–1B′′). Furthermore, co-localization of TLR4 and HMGB1 was observed in the lamina propria following oxaliplatin treatment when compared to the vehicle-treated cohort (Fig 1A–1B′′; yellow arrows). There were no differences in the total number of TLR4+ cells between the vehicle-treated (1010 ± 85) and oxaliplatin-treated groups (1077 ± 87), n = 4-5/group (Fig 1C). Furthermore, no changes in the total TLR4+-immunoreactivity amongst vehicle-treated (1.7 ± 0.1) and oxaliplatin-treated cohort (1.8 ± 0.2), n = 4-5/group (Fig 1D) were demonstrated.
Fig 1. Effects of oxaliplatin treatment on HMGB1 expression and co-localisation with TLR4 in the lamina propria of the colon.
Colon cross-sections (30μm thick) from the vehicle and oxaliplatin-treated groups were labelled with anti-TLR4 (green) and anti-HMGB1 (magenta) antibodies (A-B′′). Strong HMGB1 immunoreactivity is observed within the lamina propria (B′) of the colon from the oxaliplatin-treated animals when compared to the vehicle-treated cohort (A′). Greater co-localisation of TLR4 and HMGB1 is evident within the lamina propria (B′′) following oxaliplatin treatment when compared to the vehicle-treated cohort (A′′). The numbers of TLR4+ cells were counted from 8 images per preparation taken at 20x magnification with a total area of 2mm2. No differences in the total number (C), or TLR4+ immunoreactivity (D) was observed following oxaliplatin treatment when compared to the vehicle group. Scale = 10μm; n = 4-5/group.
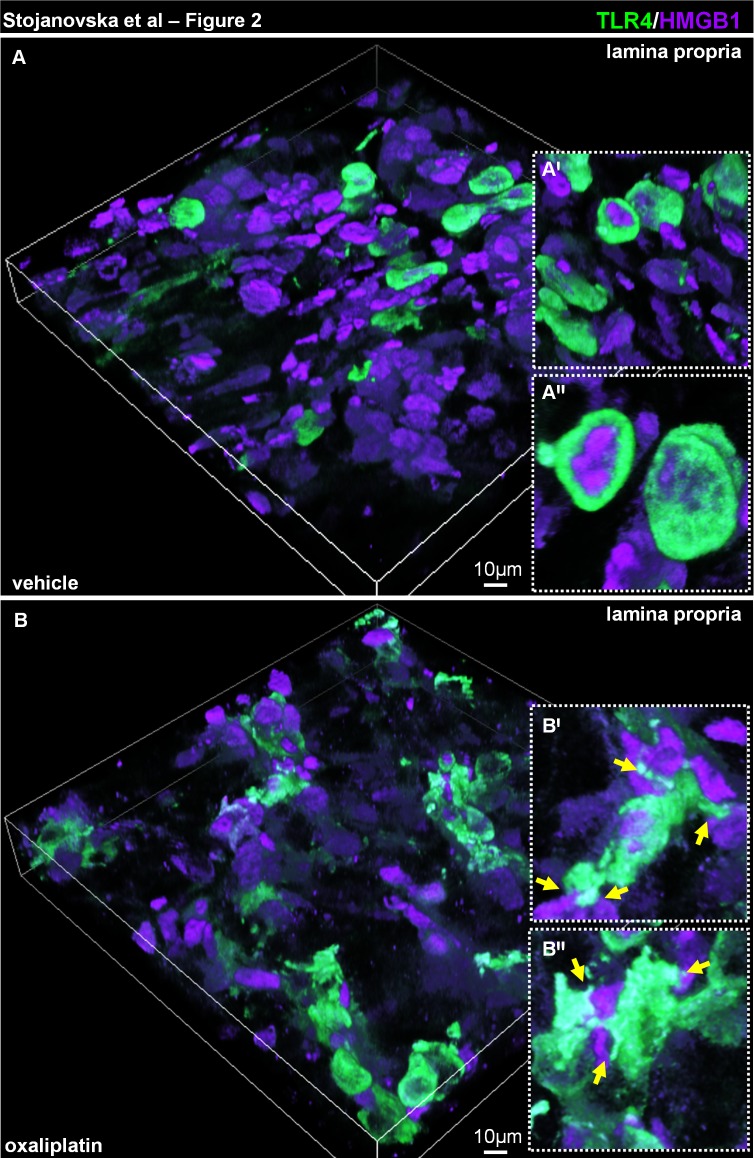
TLR4+ cells within the vehicle-treated cohort display minimal contact/co-localisation with HMGB1 (Fig 2A–2A′′). Whereas, TLR4+ cells co-localising with HMGB1 within the lamina proporia from oxaliplatin-treated group displayed pseudopodia-like extensions characteristic of antigen sampling (Fig 2B–2B′′).
Fig 2. Changes in TLR4+ cell morphology and interaction with HMGB1 in the lamina propria of the colon following oxaliplatin treatment.
Colon cross-sections (30μm thick) from the vehicle (A) and oxaliplatin-treated (B) mice were labelled with anti-TLR4 (green) and anti-HMGB1 (magenta) antibodies and presented as 3D reconstruction of confocal z-series slices. TLR4+ cells within the lamina propria of the colon from the oxaliplatin-treated animals form contacts with HMGB1 through extending processes (B′-B′′, insets with yellow arrows), as opposed to the spherical morphology of TLR4+ cells in the vehicle-treated cohort (A′-A′′, insets), n = 4/group.
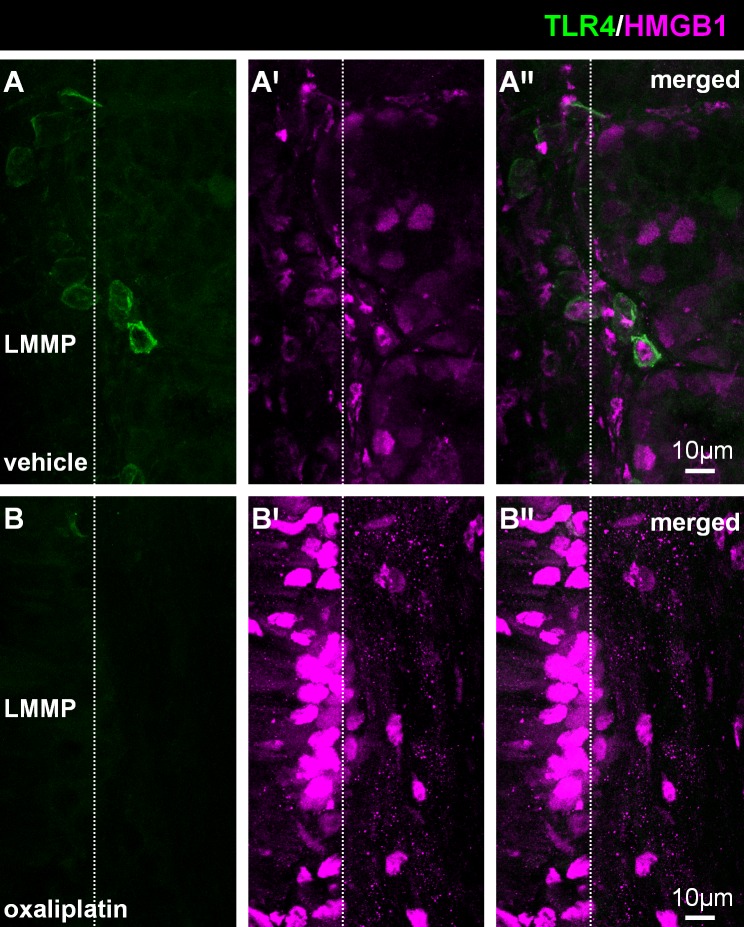
Similar to the lamina propria, there was strong HMGB1 immunoreactivity within the LMMP of the colon from the oxaliplatin-treated group when compared to the vehicle-treated cohort (Fig 3A–3B′′). However, no TLR4+ cells infiltrated the LMMP layer of the colon following either treatment (white arrow; n = 4/group).
Fig 3. Effects of oxaliplatin treatment on HMGB1 expression and co-localisation with TLR4 in the longitudinal muscle-myenteric plexus (LMMP) of colon.
Colon cross-sections (30μm thick) from the vehicle and oxaliplatin-treated groups were labelled with anti-TLR4 (green) and anti-HMGB1 (magenta) antibodies (A-B′′). No TLR4+ cells infiltrated the level of the LMMP in either group (A, B).Strong HMGB1 immunoreactivity is observed within the colon from the oxaliplatin-treated animals (B′) when compared to the vehicle-treated cohort (A′). Scale = 10μm; n = 4/group.
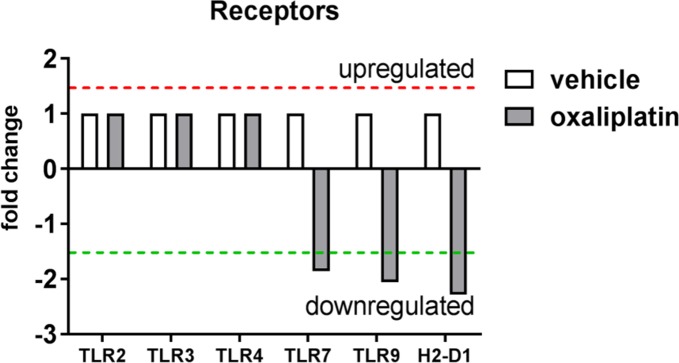
Interestingly, oxaliplatin treatment induced down-regulation of TLR7 mRNA expression (-1.81 fold change) and TLR9 (-2.01 fold change) when compared to the vehicle-treated group (Fig 4). No changes in TLR2, TLR3 or TLR4 expression were noted following oxaliplatin treatment (Fig 4). In addition, oxaliplatin treatment was associated with reduced expression of Histocompatibility 2, D region locus 1 (H2-D1; -2.23 fold change) when compared to the vehicle-treated group, however H2-D1 mRNA expression was low in both groups (CT ≥ 33 cycles).
Fig 4. Effects of oxaliplatin treatment on mRNA expression immune receptors.
To determine whether oxaliplatin treatment induced changes in the expression of receptors within the colon, RT2 Profiler PCR arrays were performed using pooled RNA samples from (vehicle, n = 5; oxaliplatin, n = 4) samples. Oxaliplatin treatment caused a down-regulation of TLR7, TLR9 and H2-D1 mRNA expression when compared to the vehicle-treated cohort. No change in TLR2, TLR3 or TLR4 expression was observed following oxaliplatin treatment.
Oxaliplatin treatment has no effect on richness, diversity and evenness of intestinal microbiota, but causes changes at the genus level
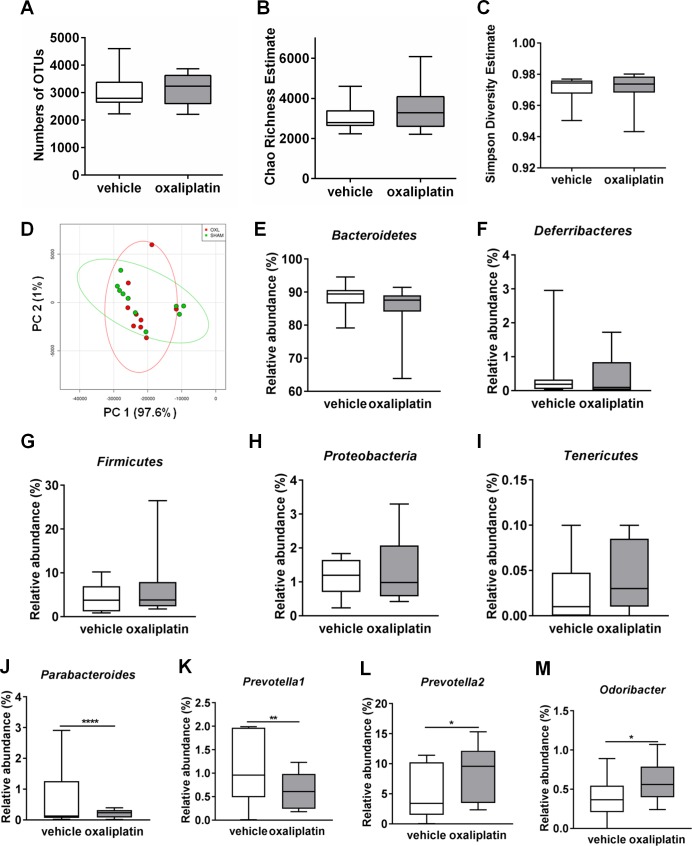
To compare compositional differences in the gut microbiota amongst vehicle-treated and oxaliplatin-treated mice, 16S rRNA sequencing was conducted. Total DNA was isolated from fecal samples (n = 10 mice/group), and PCR amplicons spanning the 16S rRNA V3-V5 hypervariable region were sequenced. Microbiota composition was assessed with regards to operational taxonomic units (OTUs) Chao richness, Shannon diversity index, Simpson’s diversity index, as well as unweighted UniFrac Principal Coordinate Analysis (PCoA).
No significant difference in the numbers of OTUs was observed between the vehicle-treated (3058 ± 233.6) and the oxaliplatin-treated mice (3134 ± 175.6) (Fig 5A). Furthermore, no significant difference in Chao richness was observed amongst the vehicle-treated (3058 ± 233.6) and the oxaliplatin-treated cohort (3562 ± 381.5) (Fig 5B). There were no significant differences in diversity between the vehicle-treated (0.97 ± 0.002) and the oxaliplatin-treated group (0.971 ± 0.003) (Fig 5C). Principle coordinate analysis (PCOA) from oxaliplatin-treated mice microbiome based upon unweighted UniFrac distance did not show significant difference when compared to the vehicle-treated cohort (Fig 5D).
Fig 5. Effects of oxaliplatin treatment on the composition of intestinal microbiota.
Fecal microbiota Chao richness estimate, Simpson diversity estimate and evenness following oxaliplatin treatment were analysed using 16S rRNA sequencing. Oxaliplatin treatment did not cause any significant changes to the number of OTUs (A), Chao richness estimate (B), Simpson diversity estimate (C) or PCoA unweighted UniFrac distance percentages (D). Oxaliplatin treatment did not cause any significant changes to five dominant phyla groups identified in the fecal microbiota: Bacteroidetes (E), Deferribacteres (F), Firmicutes (G), Proteobacteria (H) and Tenericutes (I). Oxaliplatin treatment caused a significant reduction in Parabacteroides (J) and Prevotella1 (K) species, and a significant increase in Prevotella2 (L) and Odoribacter species (M). *P<0.05; **P<0.01; ****P<0.0001; n = 10/group.
In this study, five major phyla groups were identified: Bacteroidetes, Deferribacteres, Firmicutes, Proteobacteria and Tenericutes. Although Bacteroidetes was the most abundant phylum species, no significant differences were observed between the vehicle-treated (88.3 ± 1.2) and oxaliplatin-treated mice (84.9 ± 2.2) (Fig 5E). No significant differences in Deferribacteres phyla were observed amongst the vehicle-treated (0.45 ± 0.25) and the oxaliplatin-treated cohort (0.40 ± 0.16) (Fig 5F) as well as to Firmicutes (1.14 ± 0.15; vehicle treated) compared to (1.35 ± 0.29; oxaliplatin-treated group) (Fig 5G). In addition, there were no significant changes to Proteobacteria abundance following oxaliplatin treatment (1.35 ± 0.29) compared to vehicle-treatment (1.14 ± 0.15) (Fig 5H). Furthermore, no changes in Tenericutes phyla were observed amongst the vehicle-treated (0.03 ± 0.009) and oxaliplatin-treated mice (0.04 ± 0.01) (Fig 5I).
Changes to the composition of the microbiota genera following oxaliplatin treatment was determined using 16S rRNA sequencing. Twelve common species present in both the vehicle-treated and the oxaliplatin-treated cohort were identified. These species included: Bacteroides, Parabacteroides, Prevotella1, Prevotella2, Odoribacter, Mucispirillum, Lactobacillus, Dehalobacterium, Ruminococcus, Sutterella, Bilophila and Desulfovibrio. The particular taxonomy for Prevotella1 and Prevotella2 is yet to be determined. Unknown species ‘unknown’ were also detected in both the vehicle-treated and the oxaliplatin-treated cohorts (Table 2). A significant reduction in Parabacteroides (vehicle-treated: 0.71 ± 0.003; oxaliplatin-treated: 0.21 ± 0.04; P<0.0001) and Prevotella1 species (vehicle-treated: 1.06 ± 0.002; oxaliplatin-treated: 0.64 ± 0.13; P<0.05) was noted in the oxaliplatin-treated group when compared to the vehicle-treated cohort (Fig 5J and 5K, Table 2). Oxaliplatin treatment induced a significant increase in the Prevotella2 species (vehicle-treated: 4.87 ± 0.01; oxaliplatin-treated: 8.58 ± 1.4; P<0.05) and in the Odoribacter species (vehicle-treated: 0.39 ± 0.0007; oxaliplatin-treated: 0.62 ± 0.08; P<0.05) when compared to the vehicle-treated group (Fig 5L and 5M; Table 2). No changes to Bacteroides, Mucispirillum, Lactobacillus, Dehalobacterium, Ruminococcus, Sutterella, Bilophila, Desulfovibrio or ‘unknown’ species were shown.
Table 2. Changes to microbiota at the genus level following oxaliplatin treatment.
| Vehicle | Oxaliplatin | |||||
|---|---|---|---|---|---|---|
| Mean | SEM | N | Mean | SEM | N | |
| Bacteroides | 5.80 | 0.012 | 10 | 5.56 | 1.54 | 10 |
| Parabacteroides | 0.71 | 0.003 | 10 | **** 0.21 | 0.04 | 10 |
| Prevotella1 | 1.06 | 0.002 | 10 | ** 0.64 | 0.13 | 10 |
| Prevotella2 | 4.87 | 0.013 | 10 | * 8.58 | 1.41 | 10 |
| odoribacter | 0.39 | 0.0007 | 10 | * 0.62 | 0.08 | 10 |
| Mucispirillum | 0.45 | 0.002 | 10 | 0.40 | 0.18 | 10 |
| Lactobacillus | 1.01 | 0.002 | 10 | 2.28 | 0.67 | 10 |
| Dehalobacterium | 0.19 | 0.0004 | 10 | 0.17 | 0.04 | 10 |
| Ruminococcus | 0.01 | 0.00005 | 10 | 0.01 | 0.00 | 10 |
| Sutterella | 0.36 | 0.001 | 10 | 0.40 | 0.07 | 10 |
| Bilophila | 0.19 | 0.0006 | 10 | 0.19 | 0.06 | 10 |
| Desulfovibrio | 0.51 | 0.001 | 10 | 0.71 | 0.28 | 10 |
| Unknown | 5.68 | 0.005 | 10 | 6.55 | 0.52 | 10 |
*P<0.05
**P<0.01
****P<0.0001. Mean = % abundance; SEM, standard error of the mean
Lack of immune responses in the colon following oxaliplatin treatment
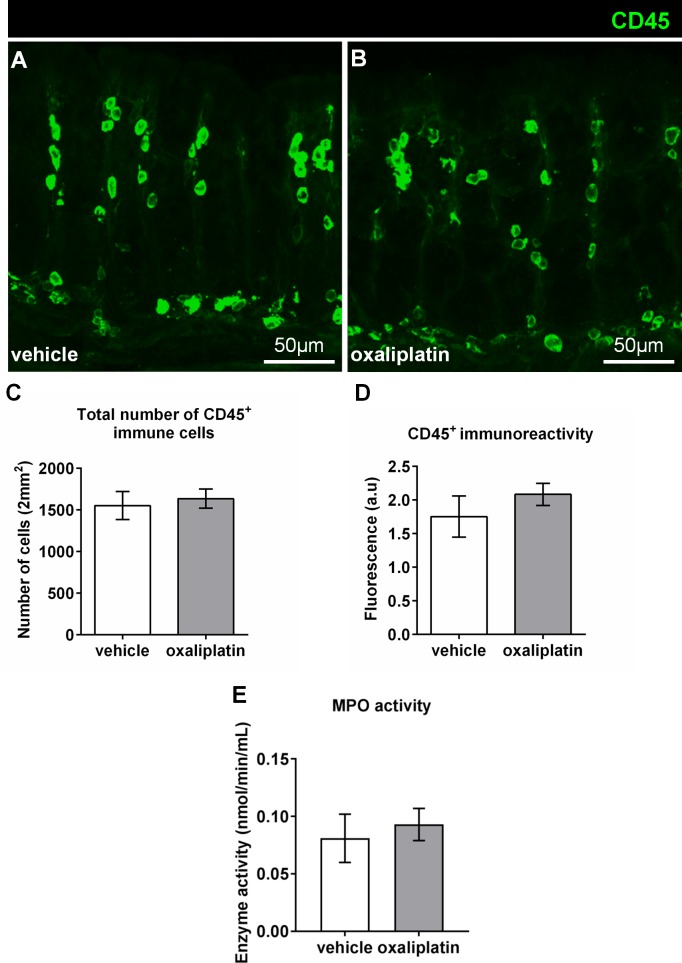
The pan-leukocyte marker anti-CD45 antibody was used to label immunocytes in the colon (Fig 6A and 6B). There were no differences in the total number of CD45+ cells between the vehicle-treated (1553 ± 169) and oxaliplatin-treated groups (1666 ± 143), n = 4/group (Fig 6C). No significant changes in CD45+ immunoreactivity to indicate inflammation in the colon was observed following oxaliplatin treatment (2.08 ± 0.16) when compared to the vehicle-treated cohort (1.75 ± 0.30), n = 4/group (Fig 6D). Furthermore, no CD45+ immune cells were found at the level of the myenteric ganglia. MPO is a peroxidase enzyme with antimicrobial capacity and is typically used as a biomarker for inflammation. No significant difference in MPO activity was shown between the oxaliplatin-treated (0.093 ± 0.01 nmol/min/mL; n = 4) and vehicle-treated groups (0.08 ± 0.02 nmol/min/mL), n = 4/group (Fig 6E).
Fig 6. Effects of oxaliplatin treatment on the fluorescence and number of CD45+ immune cells, and MPO activity in the colon.
Colon cross-sections (30μm thick) from the vehicle and oxaliplatin-treated groups were labelled with the pan-leukocyte marker anti-CD45 antibody (green) (A-B: scale bars = 50μm). The numbers of CD45+ cells were counted from 8 images per preparation taken at 20x magnification with a total area of 2mm2. No significant differences in the total number of CD45+ cells (C), or immunoreactivity (D) within the colon is observed between the vehicle-treated and oxaliplatin-treated cohorts, n = 4-5/group. To determine whether oxaliplatin treatment caused inflammation within the colon specific to neutrophils or macrophages a MPO assay was conducted. Oxaliplatin treatment did not induce any significant changes to MPO activity within the colon when compared to the vehicle-treated cohort (E). N = 4/group.
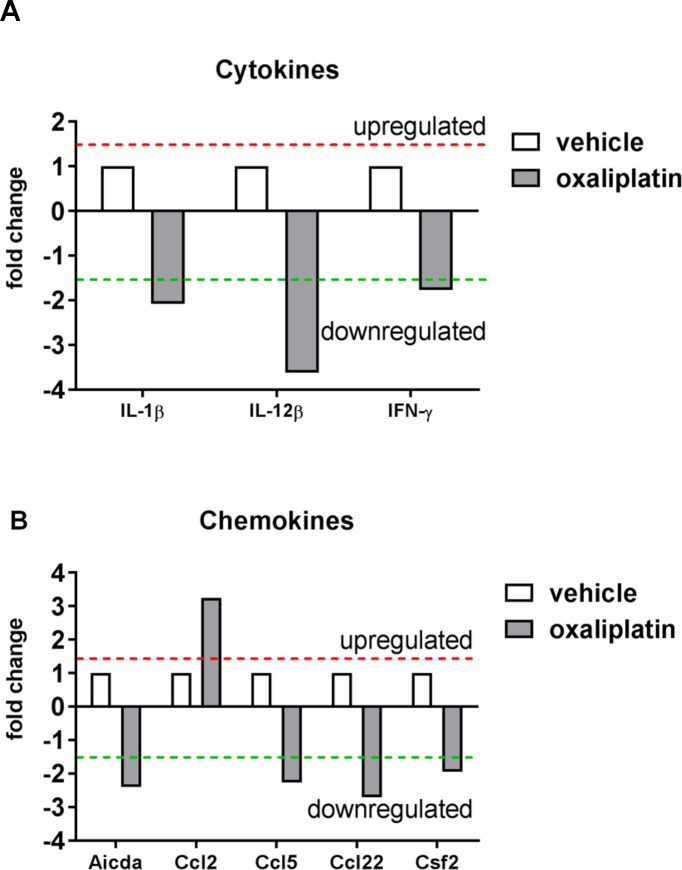
In order to profile changes in gene expression associated with inflammation, RT2 Profiler PCR arrays of colon RNA were performed using pooled RNA samples. Oxaliplatin treatment caused the down-regulation of the cytokines interleukin (IL)-1β (-2.02 fold change) and IL-12β (-3.56 fold change). Reduced interferon gamma mRNA expression (IFN-ᵧ; -1.71 fold change) was also observed following oxaliplatin treatment, however expression was low (CT≥34) in both vehicle and treated groups (Fig 7A). Moreover, oxaliplatin treatment caused higher mRNA expression of the chemokine ligand Ccl-2 (3.25 fold change) and lower expression of Ccl5 (-2.19 fold change) and Ccl22 (-2.63 fold change) (Fig 7B). Lower levels of activation induced cytidine deaminase, Aicda; (-2.32 fold change) and colony stimulating factor 2 (Csf2; -1.87 fold change) were also observed following oxaliplatin treatment, however these genes were also expressed at low levels (CT≥33) in both vehicle and oxaliplatin-treated samples. Overall most genes on the array showed no change in mRNA expression in the oxaliplatin-treated group, when compared to the vehicle-treated group (S1 Table).
Fig 7. Effects of oxaliplatin treatment on cytokine and chemokine mRNA expression.
To determine whether oxaliplatin treatment induced changes in inflammatory mediators within the colon, RT2 Profiler PCR arrays were performed using pooled RNA samples from (vehicle, n = 5; oxaliplatin, n = 4) samples. Oxaliplatin treatment caused the down-regulation of the cytokines IL-1β, IL-12β mRNA expression when compared to the vehicle-treated group (A). Oxaliplatin treatment induced the up-regulation of the chemokine Ccl2, and the down-regulation of Ccl5 and Ccl22 chemokine mRNA expression when compared to the vehicle-treated cohort (B).
Oxaliplatin treatment induces changes in immune cell populations within the MLNs, but not PPs
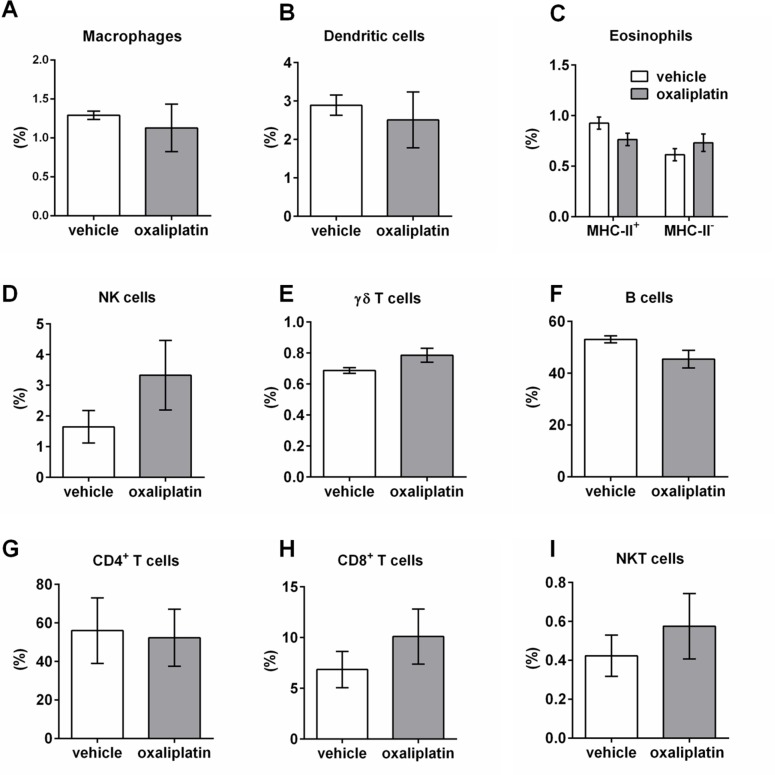
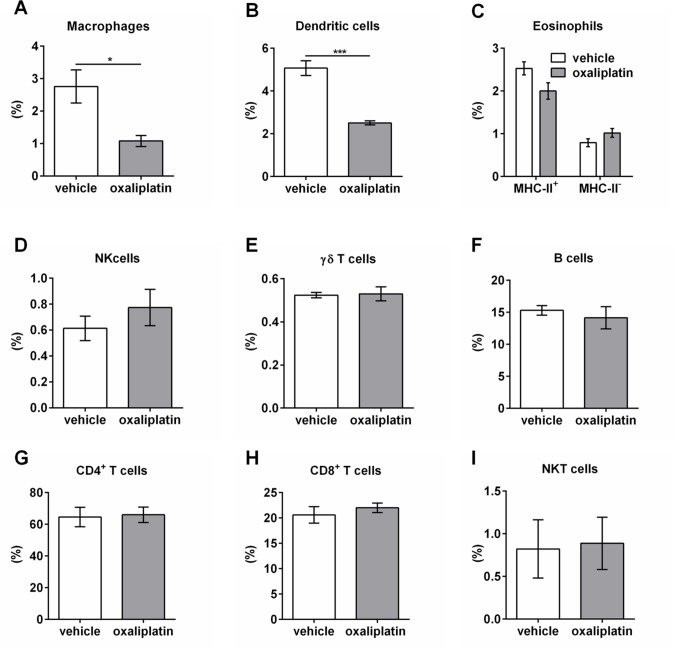
To determine the effects of oxaliplatin treatment on the gastrointestinal immune response, we profiled granulocyte and lymphocyte populations within the PPs and MLNs using FACS. Several immune cell populations were gated by the following: macrophages (F4/80+ MHC-II+); dendritic cells (CD11c+ MHC-II+); eosinophils (CD11B+ MHC-II- Gr-1+ CD193+ and CD11B+ MHC-II+ Gr-1+ CD193+); NK cells (CD49b+ TCR-); γδ T cells (γδ-TCR+ TCRβ-); B cells (CD45+ TCRβ- B220+); CD4+ T cells (CD4+ TCR+); CD8+ T cells (CD8+ TCR+); NKT cells (CD1d α-Galcer tetramer+ TCR. Oxaliplatin treatment did not cause any significant changes to the proportion of immune cells within the PPs when compared to the vehicle-treated cohort (Fig 8A–8I, Table 3; n = 5/group). However, oxaliplatin treatment resulted in a significant reduction in the proportion of macrophages and dendritic cells within the MLNs when compared to vehicle-treated group (Fig 9A and 9B, Table 4). No changes in other immune cell populations were noted (Fig 9C–9I, n = 5/group).
Fig 8. Immune cell populations within the PPs from vehicle and oxaliplatin-treated mice.
A series of gating strategies were used to identify immune cell populations within the PPs: macrophages (F4/80+ MHC-II+); dendritic cells (CD11c+ MHC-II+); eosinophils (CD11B+ MHC-II- Gr-1+ CD193+ and CD11B+ MHC-II+ Gr-1+ CD193+); NK cells (CD49b+ TCR-); γδ T cells (γδ-TCR+ TCRβ-); B cells (CD45+ TCRβ- B220+); CD4+ T cells (CD4+ TCR+); CD8+ T cells (CD8+ TCR+); NKT cells (CD1d α-Galcer tetramer+ TCR+). No significant differences were observed in any immune cell types within PPs (A-I), n = 5/group.
Table 3. Proportions of various immune cell populations within the PPs following vehicle and oxaliplatin treatment.
| Immune cell population | Vehicle | Oxaliplatin |
|---|---|---|
| Macrophages | 1.3 ± 0.1 | 1.1 ± 0.3 |
| Dendritic cells | 2.9 ± 0.3 | 2.5 ± 0.7 |
|
Eosinophils: MHC-II+ MHC-II- |
0.8 ± 0.2 0.6 ± 0.1 |
0.7 ± 0.01 0.7 ± 0.1 |
| NK cells | 1.7 ± 0.5 | 3.3 ± 1.1 |
| γδ T cells | 0.7 ± 0.01 | 0.8 ± 0.04 |
| B cells | 53.1 ± 1.4 |
45.5 ± 3.4 |
| CD4+ T cells | 56.0 ± 17.0 | 52.3 ± 14.8 |
| CD8+ T cells | 6.9 ± 1.8 |
10.1 ± 2.7 |
| NKT cells | 0.4 ± 0.1 |
0.6 ± 0.17 |
Fig 9. Immune cell populations within the MLNs from vehicle and oxaliplatin-treated mice.
A series of gating strategies were used to identify immune cell populations within the MLNs: macrophages (F4/80+ MHC-II+); dendritic cells (CD11c+ MHC-II+); eosinophils (CD11B+ MHC-II- Gr-1+ CD193+ and CD11B+ MHC-II+ Gr-1+ CD193+); NK cells (CD49b+ TCR-); γδ T cells (γδ-TCR+ TCRβ-); B cells (CD45+ TCRβ- B220+); CD4+ T cells (CD4+ TCR+); CD8+ T cells (CD8+ TCR+); NKT cells (CD1d α-Galcer tetramer+ TCR+). Oxaliplatin treatment caused a significant reduction in the proportion of macrophages and dendritic cells (A, B), with no effects on eosinophils, NK cells, γδ T cells, B cells (CD45+ TCRβ- B220+); CD4+ T cells (CD4+ TCR+); CD8+ T cells (CD8+ TCR+); NKT cells (CD1d α-Galcer tetramer+ TCR+) within MLNs (C-I). *P<0.05, n = 5/group.
Table 4. Proportions of various immune cell populations within the MLNs following vehicle and oxaliplatin treatment.
| Immune cell population | Vehicle | Oxaliplatin |
|---|---|---|
| Macrophages | 2.8 ± 0.5 | *1.1 ± 0.2 |
| Dendritic cells | 5.1 ± 0.3 | ***2.5 ± 0.1 |
|
Eosinophils: MHC-II+ MHC-II- |
2.5 ± 0.2 0.8 ± 0.1 |
2.0 ± 0.2 1.0 ± 0.1 |
| NK cells | 0.6 ± 0.1 | 0.8 ± 0.1 |
| γδ T cells | 0.5 ± 0.01 | 0.5 ± 0.03 |
| B cells | 15.3 ± 0.8 | 14.2 ± 1.7 |
| CD4+ T cells | 64.6 ± 6.1 | 66.0 ± 4.8 |
| CD8+ T cells | 20.6 ± 1.6 | 22.0 ± 0.9 |
| NKT cells | 0.8 ± 0.3 | 0.9 ± 0.3 |
*P<0.05
***P<0.001.
Discussion
This study is the first to determine the potential for oxaliplatin treatment to induce inflammatory enteric neuropathy within the murine colon. Oxaliplatin is a potent immunogenic cell death inducer, thus, it was hypothesized that mucosal and neuronal damage would be associated with an inflammatory response [12,28,29,35]. However, in this study we have shown that oxaliplatin treatment does not induce inflammation or changes in total TLR4 immunoreactivity, despite noticeable changes in HMGB1 expression which is a TLR4 ligand. It is well established that cytoplasmic and/or released HMGB1 can exert pro-inflammatory cytokine-like activity capable of inducing strong immunological responses [36,37]. In immunogenic cell death HMGB1 is passively released and acts as a potent ‘eat me’ signal, however, oxidative stress and apoptosis can blunt this pro-inflammatory signalling [38–40]. In this study, HMGB1 and TLR4 co-localisation as well as morphological differences in TLR4+ cells within the lamina propria, but not in the LMMP following oxaliplatin treatment were observed. The TLR4+ cells co-localised with HMGB1 show pseudopodia-like morphology which is characteristic of antigen sampling [41]. Under conventional circumstances, antigen presenting cells migrate to their nearest draining lymph nodes (such as PPs and MLNs in this case) upon antigen recognition for the priming and activation of T cells. Despite the HMGB1 and TLR4 interaction, there was no amplification of immune cell populations within the lymphoid organs (PPs and MLNs). Presumably, HMGB1 could still be sampled by antigen presenting cells, but may be regarded as an innocuous/neutral molecule. This is apparent when HMGB1 becomes oxidized [39,42]. Additionally, the pro-inflammatory effects of HMGB1 are also dependent on its redox state [43–46]. We have previously shown that oxaliplatin treatment induces oxidative stress through the upregulation of inducible nitric oxide synthase (iNOS) within the LMMP, as well as an increase in mitochondrial superoxide production and protein nitrosylation in myenteric neurons [9]. Given that oxaliplatin induces an oxidative environment, this particular DAMP may therefore be subjected to oxidisation, and thus, have its inflammatory potential blunted; this needs further investigation. Moreover, we observed the downregulation of TLR7 and TLR9 following oxaliplatin treatment, with no changes to TLR2, TLR3 or TLR4. TLRs are membrane-bound receptors which recognise a myriad of ligands produced by microbiota, as well as DAMPs [47–49]. TLR stimulation by ligands triggers signal transduction pathways and immunological responses [50,51]. TLR2 recognises bacterial lipoproteins and non-enterobacterial LPS [52]. Furthermore, TLR3 recognises double-stranded viral RNA, and TLR4 is stimulated by classical LPS [53,54]. TLR7 recognises single-stranded viral RNA, whereas TLR9 is stimulated by bacterial and viral DNA [51,55,56]. The downregulation of TLR7 and TLR9 observed in this study would impair recognition of certain pathogens and prospective immune responses.
Moreover, the downregulation of H2-D1 was observed following oxaliplatin treatment. The H2-D1 gene is associated with MHC-related molecules and antigen presentation. This downregulation could impact antigen loading and presentation to effector lymphocytes such as CD8+ T cells. Oxaliplatin treatment has been shown to evoke changes in another DAMP, calreticulin, a multifunctional endoplasmic reticulum and nuclear envelope-resident protein [28,57]. A major function for calreticulin is the biogenesis and correct folding and assembly of MHC-related molecules [58,59]. The downregulation of H2-D1 may be associated with changes in calreticulin expression which requires further investigation. The lack of gastrointestinal immune responses following oxaliplatin treatment could be due to defective MHC assembly and antigen presentation.
The mammalian gastrointestinal tract is colonised by diverse microorganisms where a symbiotic relationship between the host and microbiota exists [60]. The composition of microbiota throughout the gastrointestinal tract can vary depending on the location. The colon in particular has the greatest microbial density with an astounding 1x 1012 organisms per gram of feces (dry weight) [61]. In this study, we isolated fecal DNA and identified five major phyla which included: Bacteroidetes, Deferribacteres, Firmicutes, Proteobacteria and Tenericutes. Oxaliplatin did not cause significant changes to the microbiota at the phylum level in terms of OTUs, Chao richness and diversity, however, induced a significant reduction in Parabacteroides and Prevotella1 species, but caused an increase in Prevotella2 and Odoribacter. Parabacteroides, Prevotella and Odoribacter genera stem from the Bacteroides phylum. The Bacteroides phyla and their successive genera stem from the Bacteroidetes family. These bacteria are gram-negative, anaerobic, non-spore-forming rods which are commensal to the gastrointestinal tract, but are known to be opportunistic pathogens in circumstances of intestinal barrier destruction [62]. It is well established that anti-cancer agents cause considerable damage to the gastrointestinal mucosa which can act as a gateway for microbiota-induced inflammation throughout the colon [10,63,64]. Gram-negative bacteria produce endotoxins such as lipopolysaccharides (LPS; bacterial cell wall constituents) which can induce immunological responses and contribute to inflammatory diseases through TLR binding [65–68]. LPS come in ‘classical’ and ‘non-classical’ forms, depending on its structural configuration and degree of acylation [69–71]. Classical LPS are acylated lipid A molecules on short-chain fatty acids which stimulate a number of pro-inflammatory responses through cytokine production (predominantly TNF-α and IL-6) by innate immune cells (neutrophils, monocytes and macrophages) [69,72]. TLR4 is a main receptor for ‘classical’ LPS produced by Escherichia coli, whereas LPS from Bacteroides species are considered ‘non-classical’ and thus, TLR binding may not induce a rapid or strong immunological response [69,73]. Although structurally similar to classical LPS, the non-classical version still contains a lipid A centre, however, with varying degrees of acylation, and is often attached to long-chain fatty acids linked to amino-sugar backbones which is thought to hinder LPS recognition and signalling [65,69]. The inability to produce potent or classical LPS by the Bacteroides family, and the lack of changes to total TLR4 immunoreactivity observed in our present study may explain the absence of inflammation within the colon despite the increased abundance of such species at the genus level following oxaliplatin treatment.
It is known that immune cells within the gastrointestinal mucosa function differently to their counterparts found within the circulation. The mucosal immune system has co-evolved with the gastrointestinal microbiota/antigens and sterile inflammation to downregulate pro-inflammatory responses whilst uncompromising microbicidal or phagocytic activity [74,75]. No inflammation (determined by CD45+ immune cell infiltrate) or differences in total TLR4 immunoreactivity throughout the thickness of the colon was observed following oxaliplatin treatment.
There were no noticeable effects on MPO activity within the colon following oxaliplatin treatment. MPO is a well-established biomarker of inflammation in various conditions such as multiple sclerosis, ischaemic heart disease and acute coronary syndromes, as well as ulcerative colitis [76–80]. MPO is a cytotoxic constituent released by activated myeloid cells such as neutrophils, which also has micobicidal capacity [81–84]. Despite changes in gastrointestinal microbiota at the genus level, as well as increased HMGB1 expression which is known to have pro-inflammatory effects, MPO activity was not altered. Thus, our MPO data provide further evidence that oxaliplatin does not induce gastrointestinal inflammation, which presumably, would not have major effects on the colon myenteric plexus.
Furthermore, we demonstrate that oxaliplatin treatment downregulated gene expression for the pro-inflammatory cytokines IL-1β, IL-12β within the colon. Cytokines can mediate inflammation and trigger extrinsic apoptotic cascades. Although the extrinsic and intrinsic apoptotic pathways are considered to be separate entities, there is some cross over between the two signal transduction pathways [85–87]. Given that pro-inflammatory cytokines are downregulated in the colon following oxaliplatin treatment it is unlikely that the extrinsic apoptotic cascade is implicated in the underlying mechanism of cell death within the myenteric plexus. It has previously been shown that oxaliplatin treatment does not alter IL-1β, IFN-γ expression within rat spinal cord and DRG neurons, nor does it induce immune cell infiltration [88]. However, there are some conflicting studies which have demonstrated a significant increase in pro-inflammatory cytokines IL-1β and TNF-α within the rat spinal cord following oxaliplatin treatment, and a reduction in the anti-inflammatory cytokines IL-4 and IL-10 [89–91]. In the aforementioned studies, acute experiments were conducted on rats which were treated with oxaliplatin through a single 6mg/kg/d, or a 10mg/kg/d for 5 consecutive days. This is in contrast to our current study where we have chronically treated mice tri-weekly for up to 14 days with a 3mg/kg/d. The differences between oxaliplatin dosage, species and experimental time points may contribute to varying results.
Furthermore, we showed that the chemokine Ccl2 was upregulated within the colon following oxaliplatin treatment. Despite the increase in Ccl2 (a monocyte chemoattractant), there were no increases in immune cells in the colon. Ccl2 is a pleiotropic ligand implicated in many pathways. Ccl2 has demonstrated a role in shaping pro-inflammatory/anti-inflammatory macrophage responses as a deficiency leads to skewed pro-inflammatory phenotypes producing high levels of IL-6 and TNF-α [92]. The upregulation of Ccl2 in the colon following oxaliplatin treatment may play a role in dampening pro-inflammatory cytokine production by promoting the polarisation of anti-inflammatory macrophages. Ccl2 is also upregulated in response to gastrointestinal microbiota [93]. Whether the increase in microbiota species observed in this study is implicated in Ccl2 upregulation requires further study. Furthermore, previous research has demonstrated that Ccl2 is upregulated in DRG neurons and microglia following peripheral nerve injury prompting macrophage infiltration and sensory neuropathy [94–96]. Ccl2 can also be upregulated by other cytokines such as s100β [97]. We have previously demonstrated that s100β expression within the myenteric plexus of the ileum is increased following oxaliplatin treatment (Robinson et al., 2016). Whether upregulated s100β can impact Ccl2 levels within the colon requires further investigation. In addition, this study demonstrates that oxaliplatin treatment downregulated Ccl5 and Ccl22. Ccl5 plays a role in lymphocyte trafficking and promoting T cell polarisation towards an IFN-γ-producing Th1 phenotype [98]. Moreover, Ccl22 is implicated in lymphocyte and eosinophil migration [99,100]. The downregulation of Ccl5 and Ccl22 following oxaliplatin treatment may impact lymphocyte and/or eosinophil migration throughout the colon.
Downregulation of Aicda and Csf2 were also observed following oxaliplatin treatment. Aicda regulates B cell proliferation and immunoglobulin class switching [101]. In this study we did not observe any changes to B cell populations within the PPs and MLNs following oxaliplatin treatment. It is unknown whether the downregulation of this gene may affect B cell numbers and function at a later time point. Csf2 is a cytokine involved in macrophage and granulocyte production and maturation [102,103]. Downregulation of Csf2 is consistent with the decrease in macrophages and dendritic cells within the MLNs observed in this study.
We investigated the effects of oxaliplatin treatment on the immunological responses within the PPs and MLNs. PPs contain specialised epithelia known as microfold or M cells which are pivotal induction sites for pathogen or antigen-specific immune responses [104,105]. Dendritic cells within the PPs consistently sample luminal antigens and bacteria, and if loaded with an inflammatory stimulus, prime local T cells to initiate a response [27,106,107]. Intestinal dendritic cells also migrate to T cell areas of MLNs which is thought to play a role in maintaining immunological tolerance [108]. Although there were no demonstrable changes to the proportion of immune populations within the PPs following oxaliplatin treatment, a proportional reduction in both macrophages and dendritic cells within the MLNs was observed. Immunological responses within the lymph nodes would typically occur once antigen presenting cells are antigen-loaded and have migrated to prime T cells to initiate an immune response. However, no demonstrable changes to T cell populations were observed within the PPs or MLNs following oxaliplatin treatment.
Furthermore, oxaliplatin treatment did not induce changes to other immune cell populations investigated in this study which include eosinophils, NK cells, γδ T cells, B cells, CD4+ T cells, CD8+ T cells and NKT cells, all of which have important roles in maintaining gastrointestinal homeostasis as well as initiating and modulating immune responses.
Under normal conditions eosinophils reside within haematopoietic and lymphatic tissues, as well as the gastrointestinal tract mucosa [109,110]. Eosinophils are pro-inflammatory effector cells which release pleiotropic chemokines, cytokines and cytotoxic granules such as eosinophil peroxidase and eosinophil-derived neurotoxin [111,112]. Eosinophils have been implicated in a number of inflammatory conditions in the gastrointestinal tract. These include eosinophilic gastroenteritis, allergic colitis, and inflammatory bowel disease [113–115]. In this study we did not observe changes in the proportion of eosinophils within the PPs and MLNs which is consistent with oxaliplatin-induced immunosuppression observed in the colons from oxaliplatin-treated mice. Thus, eosinophils do not appear to be affected by oxaliplatin treatment, and are unlikely to mediate enteric neuropathy.
Furthermore, no changes in the proportion of NK cells within the PPs and MLNs were observed following oxaliplatin treatment. NK cells are innate lymphocytes present throughout the gastrointestinal tract and associated lymphoid organs [116]. NK cells primarily defend against viral infections, tumors, and microbial species through cell-mediated cytolytic processes involving perforin and granzyme molecules [116–119]. Although oxaliplatin treatment caused mucosal injury, microbial dysbiosis, the proportion of NK cells remained unaffected. Additionally, the presentation of DAMPs following oxaliplatin treatment does not appear the affect the proportion of NK cells within the PPs and MLNs. A study investigating the effects of oxaliplatin treatment on ovarian cancer cells has shown that this platinum-based drug increases NK cell-mediated toxicity [120]. Analysis of NK cells in human peripheral blood of patients receiving low-dose cisplatin and 5-fluorouracil treatment has also shown to prevent NK cell suppression typically observed following colorectal surgery [121]. However, our data suggests that NK cell-mediated cytotoxicity is an unlikely cause for enteric neuropathy following oxaliplatin treatment. No immune cells infiltrated the myenteric plexus (determined by pan-leukocyte CD45+ labelling), and thus, cell-mediated killing of neurons and glia is not apparent.
γδ T cells are intraepithelial lymphocytes which act as immunosurveyers of the gastrointestinal tract [122]. These cells account for upto 50% of intraepithelial lymphocytes within the gastrointestinal tract and play a role in antigen presentation, anti-tumor immunity, microbial defense, neutrophil and macrophage recruitment and cytokine production [123–125]. We did not observe any changes in the proportion of γδ T cells in either the PPs or MLNs. Our data suggests that they are not infiltrating lymphoid organs to present antigens following oxaliplatin therapy, and that they are unlikely to be recruiting myeloid cells such as neutrophils and macrophages given that no observable changes in MPO activity within the colon was found. One study to date has demonstrated that oxaliplatin treatment induces infiltration of IL-17 producing γδ T cells to transplantable tumor sites [126]. It appears that oxaliplatin treatment can sensitise cancer cells to specific γδ T cell-mediated immunity, but this seems unlikely for enteric neurons, given that no infiltrating immune cells were observed.
B cells play an important role in intestinal immunity and mucosal tolerance. They are enriched within gastrointestinal associated lymphoid organs, synthesise IgA which functions to inhibit microbial adherence to mucosal surfaces, and neutralizes toxins, enzymes and antigens [127]. B cells also induce T cell dependent or independent responses. An in vivo study has shown that B cells can impede T cell responses characteristic of immunogenic cell death in mouse tumor model [128]. In our study we did not observe any changes in the proportion of B cells or various T cell subpopulations (CD4+, CD8+, NKT).
As there are a number of CD4+ T cell populations, such as Th1, Th2, Th9, Th17 and Tregs [129], we analyzed them collectively to get an overview of CD4+ T cell proportions following oxaliplatin treatment. CD4+ T cells influence innate and adaptive immune responses through conditioning the milieu with pro-inflammatory or anti-inflammatory cytokines and chemokines [130]. Although NKT cells are a subset of CD4+ T cells they are activated through CD1d-restricted lipid antigens as opposed to classical MHC class I and II molecules [131–133]. The most extensively studied lipid ligand for CD1d is α-galactosylceramide [133]. Both mammalian and bacterial lipids can stimulate NKT cells to stimulating rapid Th1 and Th2 responses. No changes were noted in the proportion of NKT cells within either the PPs or MLNs following oxaliplatin treatment. However, the effects of oxaliplatin treatment on NKT cells in vivo require further studies.
Furthermore, no demonstrable changes in the proportion of CD8+ T cells within the PPs or MLNs were shown following oxaliplatin treatment. CD8+ T cells are activated upon antigen presentation and can exert cytotoxic activity through two major pathways. These include the granule exocytosis or the death receptor (TNF-α and Fas) pathway [134,135]. Previous studies using peripheral blood and colon cancer cell lines have shown marked increases in T cell activation following the presentation of DAMPs induced by oxaliplatin treatment [28,136]. We have previously demonstrated that oxaliplatin treatment evokes the presentation of DAMPs within the myenteric plexus, however, no differences in T cell populations were observed.
Given that the gastrointestinal tract is constantly exposed to a myriad of antigens and pathogens, the local immune system has evolved over time to eradicate noxious stimuli through non-inflammatory host-defense mechanisms [74]. These data suggest that chemotherapy-induced microbiota dysbiosis, mucosal and neuronal damage in this case does not evoke inflammation or immunogenic cell death, thus, ENS damage is most likely mediated through direct drug toxicity.
Conclusion
This is the first study to determine the effects of oxaliplatin treatment on the gastrointestinal microbiota and its effects on immune populations within the PPs and MLNs which mediate local inflammation. Oxaliplatin treatment does not induce severe inflammation throughout the thickness of the colon despite the presentation of DAMPs within in myenteric neurons. These data are suggestive of antigen and microbial-specific tolerance. Thus, it appears that oxaliplatin treatment is not associated with inflammatory enteric neuropathy, and further research is required to determine the mechanism of neuronal damage and death which contributes to gastrointestinal dysfunction.
Supporting information
(DOC)
TLR4+ cells in the colon were images at 100x magnification on a Nikon confocal microscope. We observed consistent differences in the morphology of TLR4+ cells in the oxaliplatin-treated group compared to the vehicle-treated cohort. 8 randomised images (20x magnification) per animal were used to count the number of TLR4+ cells in the colon, as well as immunoreactivity/image. Image J counter plugin was used to mark each cell to ensure they were only counted once, and we measured immunoreactivity/fluorescence by converting the image to 8-bit →binary→measure. We used % area result to determine immunoreactivity per image. The sum of TLR4+ cells and immunoreactivity was averaged between groups.
(XLSX)
All genes included in the array are listed, alongside quality control and normalisation expression. Changes in inflammation-associated gene expression are presented in heatmap and numerical formats. This dataset relates to all PCR data in this study, including Fig 4 (receptors) and Fig 7 (cytokines and chemokines).
(XLSX)
Dataset includes numbers on microbiota phylum, class, order, family, and genus.
(XLS)
Eight randomised images (20x magnification) per animal were used to count the number of CD45+ cells in the colon, as well as immunoreactivity/image. Image J counter plugin was used to mark each cell to ensure they were only counted once, and we measured immunoreactivity/fluorescence by converting the image to 8-bit →binary→measure. We used % area result to determine immunoreactivity per image. MPO activity was measured using the MPO Colorimetric Activity Assay (Sigma-Aldrich, Australia) according to manufacturers instructions.
(XLSX)
This data set contains raw values for all flow cytometry experiments on the various immune cell populations investigated in the PPs and MLNs.
(XLS)
Acknowledgments
This work was supported by Victoria University Research Support grant (KN), the Centre for Chronic Disease (VA), College of Health and Biomedicine (VS), Victoria University.
Data Availability
All data generated during and analysed during the current study are provided as Supporting Information files. All 16S rRNA sequencing data have been deposited to a NCBI-GENEbank public database and can be accessed via SRA RunSelector: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP133585.
Funding Statement
This work was supported by Victoria University Research Support grant (KN), the Centre for Chronic Disease (VA), College of Health and Biomedicine (VS), Victoria University.
References
- 1.Alcindor T, Beauger N (2011) Oxaliplatin: a review in the era of molecularly targeted therapy. Curr Oncol 18: 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McQuade R, Bornstein Joel C, Nurgali Kulmira (2014) Anti-Colorectal Cancer Chemotherapy-Induced Diarrhoea: Current Treatments and Side-Effects. International Journal of Clinical Medicine 5: 393–406. [Google Scholar]
- 3.Canta A, Pozzi E, Carozzi V (2015) Mitochondrial Dysfunction in Chemotherapy-Induced Peripheral Neuropathy (CIPN). Toxics 3: 198 doi: 10.3390/toxics3020198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Podratz JL, Knight AM, Ta LE, Staff NP, Gass JM, et al. (2011) Cisplatin induced Mitochondrial DNA Damage In Dorsal Root Ganglion Neurons. Neurobiol Dis 41: 661–668. doi: 10.1016/j.nbd.2010.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Schumaker LM, Egorin MJ, Zuhowski EG, Guo Z, et al. (2006) Cisplatin preferentially binds mitochondrial DNA and voltage-dependent anion channel protein in the mitochondrial membrane of head and neck squamous cell carcinoma: possible role in apoptosis. Clin Cancer Res 12: 5817–5825. doi: 10.1158/1078-0432.CCR-06-1037 [DOI] [PubMed] [Google Scholar]
- 6.Di Fiore F, Van Cutsem E (2009) Acute and long-term gastrointestinal consequences of chemotherapy. Best Pract Res Clin Gastroenterol 23: 113–124. doi: 10.1016/j.bpg.2008.11.016 [DOI] [PubMed] [Google Scholar]
- 7.Weickhardt A, Wells K, Messersmith W (2011) Oxaliplatin-Induced Neuropathy in Colorectal Cancer. Journal of Oncology 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wafai L, Taher M, Jovanovska V, Bornstein JC, Dass CR, et al. (2013) Effects of oxaliplatin on mouse myenteric neurons and colonic motility. Front Neurosci 7: 30 doi: 10.3389/fnins.2013.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McQuade RM, Carbone SE, Stojanovska V, Rahman A, Gwynne RM, et al. (2016) Role of Oxidative Stress in Oxaliplatin-Induced Enteric Neuropathy and Colonic Dysmotility in Mice. Br J Pharmacol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boussios S, Pentheroudakis G, Katsanos K, Pavlidis N (2012) Systemic treatment-induced gastrointestinal toxicity: incidence, clinical presentation and management. Ann Gastroenterol 25: 106–118. [PMC free article] [PubMed] [Google Scholar]
- 11.Stein A, Voigt W, Jordan K (2010) Chemotherapy-induced diarrhea: pathophysiology, frequency and guideline-based management. Ther Adv Med Oncol 2: 51–63. doi: 10.1177/1758834009355164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stojanovska V, Sakkal S, Nurgali K (2015) Platinum-based chemotherapy: gastrointestinal immunomodulation and enteric nervous system toxicity. Am J Physiol Gastrointest Liver Physiol 308: G223–g232. doi: 10.1152/ajpgi.00212.2014 [DOI] [PubMed] [Google Scholar]
- 13.Furness JB (2012) The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol 9: 286–294. doi: 10.1038/nrgastro.2012.32 [DOI] [PubMed] [Google Scholar]
- 14.Robinson AM, Stojanovska V, Rahman AA, McQuade RM, Senior PV, et al. (2016) Effects of Oxaliplatin Treatment on the Enteric Glial Cells and Neurons in the Mouse Ileum. J Histochem Cytochem 64: 530–545. doi: 10.1369/0022155416656842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zitvogel L, Galluzzi L, Viaud S, Vétizou M, Daillère R, et al. (2015) Cancer and the gut microbiota: An unexpected link. Sci Transl Med 7: 271ps271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poutahidis T, Kleinewietfeld M, Erdman SE (2014) Gut Microbiota and the Paradox of Cancer Immunotherapy. Front Immunol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldszmid RS, Dzutsev A, Viaud S, Zitvogel L, Restifo NP, et al. (2015) Microbiota modulation of myeloid cells in cancer therapy. Cancer Immunol Res 3: 103–109. doi: 10.1158/2326-6066.CIR-14-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Round JL, Mazmanian SK (2009) The gut microbiome shapes intestinal immune responses during health and disease. Nat Rev Immunol 9: 313–323. doi: 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belkaid Y, Hand Timothy W (2014) Role of the Microbiota in Immunity and Inflammation. Cell 157: 121–141. doi: 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stringer AM, Gibson RJ, Logan RM, Bowen JM, Yeoh AS, et al. (2008) Faecal microflora and beta-glucuronidase expression are altered in an irinotecan-induced diarrhea model in rats. Cancer Biol Ther 7: 1919–1925. [DOI] [PubMed] [Google Scholar]
- 21.Lin XB, Dieleman LA, Ketabi A, Bibova I, Sawyer MB, et al. (2012) Irinotecan (CPT-11) Chemotherapy Alters Intestinal Microbiota in Tumour Bearing Rats. PLOS ONE 7: e39764 doi: 10.1371/journal.pone.0039764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stringer AM, Gibson RJ, Logan RM, Bowen JM, Yeoh AS, et al. (2009) Gastrointestinal microflora and mucins may play a critical role in the development of 5-Fluorouracil-induced gastrointestinal mucositis. Exp Biol Med (Maywood) 234: 430–441. [DOI] [PubMed] [Google Scholar]
- 23.Stringer AM, Gibson RJ, Logan RM, Bowen JM, Yeoh AS, et al. (2009) Irinotecan-induced mucositis is associated with changes in intestinal mucins. Cancer Chemother Pharmacol 64: 123–132. doi: 10.1007/s00280-008-0855-y [DOI] [PubMed] [Google Scholar]
- 24.Lomax AE, O'Hara JR, Hyland NP, Mawe GM, Sharkey KA (2007) Persistent alterations to enteric neural signaling in the guinea pig colon following the resolution of colitis. Am J Physiol Gastrointest Liver Physiol 292: G482–491. doi: 10.1152/ajpgi.00355.2006 [DOI] [PubMed] [Google Scholar]
- 25.Nurgali K, Qu Z, Hunne B, Thacker M, Pontell L, et al. (2011) Morphological and functional changes in guinea-pig neurons projecting to the ileal mucosa at early stages after inflammatory damage. J Physiol 589: 325–339. doi: 10.1113/jphysiol.2010.197707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linden DR, Couvrette JM, Ciolino A, McQuoid C, Blaszyk H, et al. (2005) Indiscriminate loss of myenteric neurones in the TNBS-inflamed guinea-pig distal colon. Neurogastroenterol Motil 17: 751–760. doi: 10.1111/j.1365-2982.2005.00703.x [DOI] [PubMed] [Google Scholar]
- 27.Jung C, Hugot JP, Barreau F (2010) Peyer's Patches: The Immune Sensors of the Intestine. Int J Inflam 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tesniere A, Schlemmer F, Boige V, Kepp O, Martins I, et al. (2010) Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene 29: 482–491. doi: 10.1038/onc.2009.356 [DOI] [PubMed] [Google Scholar]
- 29.Hato SV, Khong A, de Vries IJ, Lesterhuis WJ (2014) Molecular pathways: the immunogenic effects of platinum-based chemotherapeutics. Clin Cancer Res 20: 2831–2837. doi: 10.1158/1078-0432.CCR-13-3141 [DOI] [PubMed] [Google Scholar]
- 30.Klune JR, Dhupar R, Cardinal J, Billiar TR, Tsung A (2008) HMGB1: Endogenous Danger Signaling. Mol Med 14: 476–484. doi: 10.2119/2008-00034.Klune [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, et al. (2013) Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342: 967–970. doi: 10.1126/science.1240527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viaud S, Daillere R, Boneca IG, Lepage P, Langella P, et al. (2015) Gut microbiome and anticancer immune response: really hot Sh*t! Cell Death Differ 22: 199–214. doi: 10.1038/cdd.2014.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renn CL, Carozzi VA, Rhee P, Gallop D, Dorsey SG, et al. (2011) Multimodal assessment of painful peripheral neuropathy induced by chronic oxaliplatin-based chemotherapy in mice. Mol Pain 7: 29 doi: 10.1186/1744-8069-7-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elias D, Matsuhisa T, Sideris L, Liberale G, Drouard-Troalen L, et al. (2004) Heated intra-operative intraperitoneal oxaliplatin plus irinotecan after complete resection of peritoneal carcinomatosis: pharmacokinetics, tissue distribution and tolerance. Ann Oncol 15: 1558–1565. doi: 10.1093/annonc/mdh398 [DOI] [PubMed] [Google Scholar]
- 35.Bezu L, Gomes-de-Silva LC, Dewitte H, Breckpot K, Fucikova J, et al. (2015) Combinatorial strategies for the induction of immunogenic cell death. Front Immunol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinotti S, Patrone M, Ranzato E (2015) Emerging roles for HMGB1 protein in immunity, inflammation, and cancer. Immunotargets Ther 4: 101–109. doi: 10.2147/ITT.S58064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SA, Kwak MS, Kim S, Shin JS (2014) The role of high mobility group box 1 in innate immunity. Yonsei Med J 55: 1165–1176. doi: 10.3349/ymj.2014.55.5.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Y, Tang D, Kang R (2015) Oxidative stress-mediated HMGB1 biology. Front Physiol 6: 93 doi: 10.3389/fphys.2015.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu A, Fang H, Dirsch O, Jin H, Dahmen U (2012) Oxidation of HMGB1 Causes Attenuation of Its Pro-Inflammatory Activity and Occurs during Liver Ischemia and Reperfusion. PLOS ONE 7: e35379 doi: 10.1371/journal.pone.0035379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venereau E, Casalgrandi M, Schiraldi M, Antoine DJ, Cattaneo A, et al. (2012) Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. The Journal of Experimental Medicine 209: 1519 doi: 10.1084/jem.20120189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baranov MV, ter Beest M, Reinieren-Beeren I, Cambi A, Figdor CG, et al. (2014) Podosomes of dendritic cells facilitate antigen sampling. Journal of Cell Science 127: 1052–1064. doi: 10.1242/jcs.141226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang H, Antoine DJ, Andersson U, Tracey KJ (2013) The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J Leukoc Biol 93: 865–873. doi: 10.1189/jlb.1212662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kazama H, Ricci JE, Herndon JM, Hoppe G, Green DR, et al. (2008) Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity 29: 21–32. doi: 10.1016/j.immuni.2008.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magna M, Pisetsky DS (2014) The Role of HMGB1 in the Pathogenesis of Inflammatory and Autoimmune Diseases. Mol Med 20: 138–146. doi: 10.2119/molmed.2013.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ottosson L, Lundbäck P, Hreggvidsdottir H, Yang H, Tracey KJ, et al. (2012) The pro-inflammatory effect of HMGB1, a mediator of inflammation in arthritis, is dependent on the redox status of the protein. Annals of the Rheumatic Diseases 71: A81–A82. [Google Scholar]
- 46.Urbonaviciute V, Meister S, Furnrohr BG, Frey B, Guckel E, et al. (2009) Oxidation of the alarmin high-mobility group box 1 protein (HMGB1) during apoptosis. Autoimmunity 42: 305–307. [DOI] [PubMed] [Google Scholar]
- 47.Janssens S, Beyaert R (2003) Role of Toll-Like Receptors in Pathogen Recognition. Clinical Microbiology Reviews 16: 637–646. doi: 10.1128/CMR.16.4.637-646.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piccinini AM, Midwood KS (2010) DAMPening inflammation by modulating TLR signalling. Mediators Inflamm 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jounai N, Kobiyama K, Takeshita F, Ishii KJ (2012) Recognition of damage-associated molecular patterns related to nucleic acids during inflammation and vaccination. Frontiers in Cellular and Infection Microbiology 2: 168 doi: 10.3389/fcimb.2012.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawasaki T, Kawai T (2014) Toll-Like Receptor Signaling Pathways. Frontiers in Immunology 5: 461 doi: 10.3389/fimmu.2014.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeda K, Akira S (2005) Toll-like receptors in innate immunity. International Immunology 17: 1–14. doi: 10.1093/intimm/dxh186 [DOI] [PubMed] [Google Scholar]
- 52.Werts C, Tapping RI, Mathison JC, Chuang TH, Kravchenko V, et al. (2001) Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat Immunol 2: 346–352. doi: 10.1038/86354 [DOI] [PubMed] [Google Scholar]
- 53.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA (2001) Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413: 732–738. doi: 10.1038/35099560 [DOI] [PubMed] [Google Scholar]
- 54.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, et al. (1999) Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol 162: 3749–3752. [PubMed] [Google Scholar]
- 55.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, et al. (2004) Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A 101: 5598–5603. doi: 10.1073/pnas.0400937101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, et al. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408: 740–745. doi: 10.1038/35047123 [DOI] [PubMed] [Google Scholar]
- 57.Michalak M, Corbett EF, Mesaeli N, Nakamura K, Opas M (1999) Calreticulin: one protein, one gene, many functions. Biochemical Journal 344: 281–292. [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang Y, Dey S, Matsunami H (2014) Calreticulin: Roles in Cell-Surface Protein Expression. Membranes 4: 630–641. doi: 10.3390/membranes4030630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao B, Adhikari R, Howarth M, Nakamura K, Gold MC, et al. (2002) Assembly and antigen-presenting function of MHC class I molecules in cells lacking the ER chaperone calreticulin. Immunity 16: 99–109. [DOI] [PubMed] [Google Scholar]
- 60.Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK (2010) Host–Bacterial Symbiosis in Health and Disease. Adv Immunol 107: 243–274. doi: 10.1016/B978-0-12-381300-8.00008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Hara AM, Shanahan F (2006) The gut flora as a forgotten organ. EMBO Rep 7: 688–693. doi: 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wexler HM (2007) Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 20: 593–621. doi: 10.1128/CMR.00008-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee CS, Ryan EJ, Doherty GA (2014) Gastro-intestinal toxicity of chemotherapeutics in colorectal cancer: The role of inflammation. World J Gastroenterol 20: 3751–3761. doi: 10.3748/wjg.v20.i14.3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Syvak LA, Maidanevych NM, Hubarieva HO, Lial'kin SO, Aleksyk OM, et al. (2012) The toxic effects of chemotherapy on the gastrointestinal tract. Lik Sprava: 25–30. [PubMed] [Google Scholar]
- 65.Molinaro A, Holst O, Di Lorenzo F, Callaghan M, Nurisso A, et al. (2015) Chemistry of lipid A: at the heart of innate immunity. Chemistry 21: 500–519. doi: 10.1002/chem.201403923 [DOI] [PubMed] [Google Scholar]
- 66.Maeshima N, Fernandez RC (2013) Recognition of lipid A variants by the TLR4-MD-2 receptor complex. Front Cell Infect Microbiol 3: 3 doi: 10.3389/fcimb.2013.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Darveau RP (1998) Lipid A diversity and the innate host response to bacterial infection. Curr Opin Microbiol 1: 36–42. [DOI] [PubMed] [Google Scholar]
- 68.Heumann D, Roger T (2002) Initial responses to endotoxins and Gram-negative bacteria. Clin Chim Acta 323: 59–72. [DOI] [PubMed] [Google Scholar]
- 69.Lapaque N, Takeuchi O, Corrales F, Akira S, Moriyon I, et al. (2006) Differential inductions of TNF-alpha and IGTP, IIGP by structurally diverse classic and non-classic lipopolysaccharides. Cell Microbiol 8: 401–413. doi: 10.1111/j.1462-5822.2005.00629.x [DOI] [PubMed] [Google Scholar]
- 70.Ogawa T (1993) Chemical structure of lipid A from porphyromonas (bacteroides) gingivalis lipopolysaccharide. FEBS Letters 332: 197–201. [DOI] [PubMed] [Google Scholar]
- 71.Weintraub A, Zahringer U, Wollenweber HW, Seydel U, Rietschel ET (1989) Structural characterization of the lipid A component of Bacteroides fragilis strain NCTC 9343 lipopolysaccharide. Eur J Biochem 183: 425–431. [DOI] [PubMed] [Google Scholar]
- 72.Hakansson A, Molin G (2011) Gut Microbiota and Inflammation. Nutrients 3: 637–682. doi: 10.3390/nu3060637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alhawi M, Stewart J, Erridge C, Patrick S, Poxton IR (2009) Bacteroides fragilis signals through Toll-like receptor (TLR) 2 and not through TLR4. J Med Microbiol 58: 1015–1022. doi: 10.1099/jmm.0.009936-0 [DOI] [PubMed] [Google Scholar]
- 74.Smith PD, Smythies LE, Shen R, Greenwell-Wild T, Gliozzi M, et al. (2011) Intestinal macrophages and response to microbial encroachment. Mucosal Immunol 4: 31–42. doi: 10.1038/mi.2010.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, et al. (2005) Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest 115: 66–75. doi: 10.1172/JCI19229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loria V, Dato I, Graziani F, Biasucci LM (2008) Myeloperoxidase: A New Biomarker of Inflammation in Ischemic Heart Disease and Acute Coronary Syndromes. Mediators of Inflammation 2008: 135625 doi: 10.1155/2008/135625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olza J, Aguilera CM, Gil-Campos M, Leis R, Bueno G, et al. (2012) Myeloperoxidase Is an Early Biomarker of Inflammation and Cardiovascular Risk in Prepubertal Obese Children. Diabetes Care 35: 2373–2376. doi: 10.2337/dc12-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Masoodi I, Tijjani BM, Wani H, Hassan NS, Khan AB, et al. (2011) Biomarkers in the management of ulcerative colitis: a brief review. Ger Med Sci 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pulli B, Bure L, Wojtkiewicz GR, Iwamoto Y, Ali M, et al. (2015) Multiple Sclerosis: Myeloperoxidase Immunoradiology Improves Detection of Acute and Chronic Disease in Experimental Model. Radiology 275: 480–489. doi: 10.1148/radiol.14141495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forghani R, Wojtkiewicz GR, Zhang Y, Seeburg D, Bautz BRM, et al. (2012) Demyelinating Diseases: Myeloperoxidase as an Imaging Biomarker and Therapeutic Target. Radiology 263: 451–460. doi: 10.1148/radiol.12111593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lau D, Mollnau H, Eiserich JP, Freeman BA, Daiber A, et al. (2005) Myeloperoxidase mediates neutrophil activation by association with CD11b/CD18 integrins. Proceedings of the National Academy of Sciences of the United States of America 102: 431–436. doi: 10.1073/pnas.0405193102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aratani Y, Kura F, Watanabe H, Akagawa H, Takano Y, et al. (2000) Differential Host Susceptibility to Pulmonary Infections with Bacteria and Fungi in Mice Deficient in Myeloperoxidase. The Journal of Infectious Diseases 182: 1276–1279. doi: 10.1086/315843 [DOI] [PubMed] [Google Scholar]
- 83.Klebanoff SJ, Kettle AJ, Rosen H, Winterbourn CC, Nauseef WM (2013) Myeloperoxidase: a front-line defender against phagocytosed microorganisms. J Leukoc Biol 93: 185–198. doi: 10.1189/jlb.0712349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Allen RC, Stephens JT (2011) Myeloperoxidase Selectively Binds and Selectively Kills Microbes. Infection and Immunity 79: 474–485. doi: 10.1128/IAI.00910-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elmore S (2007) Apoptosis: a review of programmed cell death. Toxicol Pathol 35: 495–516. doi: 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McIlwain DR, Berger T, Mak TW (2013) Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol 5: a008656 doi: 10.1101/cshperspect.a008656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Parrish AB, Freel CD, Kornbluth S (2013) Cellular Mechanisms Controlling Caspase Activation and Function. Cold Spring Harbor Perspectives in Biology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Makker PGS, Duffy SS, Lees JG, Perera CJ, Tonkin RS, et al. (2017) Characterisation of Immune and Neuroinflammatory Changes Associated with Chemotherapy-Induced Peripheral Neuropathy. PLOS ONE 12: e0170814 doi: 10.1371/journal.pone.0170814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim C, Lee JH, Kim W, Li D, Kim Y, et al. (2016) The Suppressive Effects of Cinnamomi Cortex and Its Phytocompound Coumarin on Oxaliplatin-Induced Neuropathic Cold Allodynia in Rats. Molecules 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jung Y, Lee JH, Kim W, Yoon SH, Kim SK (2017) Anti-allodynic effect of Buja in a rat model of oxaliplatin-induced peripheral neuropathy via spinal astrocytes and pro-inflammatory cytokines suppression. BMC Complement Altern Med 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Janes K, Wahlman C, Little JW, Doyle T, Tosh DK, et al. (2015) Spinal neuroimmune activation is independent of T-cell infiltration and attenuated by A3 adenosine receptor agonists in a model of oxaliplatin-induced peripheral neuropathy. Brain Behav Immun 44: 91–99. doi: 10.1016/j.bbi.2014.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sierra-Filardi E, Nieto C, Domínguez-Soto Á, Barroso R, Sánchez-Mateos P, et al. (2014) CCL2 Shapes Macrophage Polarization by GM-CSF and M-CSF: Identification of CCL2/CCR2-Dependent Gene Expression Profile. The Journal of Immunology 192: 3858 doi: 10.4049/jimmunol.1302821 [DOI] [PubMed] [Google Scholar]
- 93.DePaolo RW, Lathan R, Rollins BJ, Karpus WJ (2005) The Chemokine CCL2 Is Required for Control of Murine Gastric Salmonella enterica Infection. Infection and Immunity 73: 6514–6522. doi: 10.1128/IAI.73.10.6514-6522.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kwon MJ, Shin HY, Cui Y, Kim H, Thi AHL, et al. (2015) CCL2 Mediates Neuron–Macrophage Interactions to Drive Proregenerative Macrophage Activation Following Preconditioning Injury. The Journal of Neuroscience 35: 15934 doi: 10.1523/JNEUROSCI.1924-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Van Steenwinckel J, Reaux-Le Goazigo A, Pommier B, Mauborgne A, Dansereau MA, et al. (2011) CCL2 released from neuronal synaptic vesicles in the spinal cord is a major mediator of local inflammation and pain after peripheral nerve injury. J Neurosci 31: 5865–5875. doi: 10.1523/JNEUROSCI.5986-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gao YJ, Zhang L, Samad OA, Suter MR, Yasuhiko K, et al. (2009) JNK-induced MCP-1 production in spinal cord astrocytes contributes to central sensitization and neuropathic pain. J Neurosci 29: 4096–4108. doi: 10.1523/JNEUROSCI.3623-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang H, Zhang L, Zhang IY, Chen X, Da Fonseca A, et al. (2013) S100B Promotes Glioma Growth through Chemoattraction of Myeloid-Derived Macrophages. Clinical cancer research: an official journal of the American Association for Cancer Research 19: 3764–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Borish LC, Steinke JW (2003) 2. Cytokines and chemokines. Journal of Allergy and Clinical Immunology 111: S460–S475. [DOI] [PubMed] [Google Scholar]
- 99.Pinho V, Oliveira SH, Souza DG, Vasconcelos D, Alessandri AL, et al. (2003) The role of CCL22 (MDC) for the recruitment of eosinophils during allergic pleurisy in mice. Journal of Leukocyte Biology 73: 356–362. [DOI] [PubMed] [Google Scholar]
- 100.Mailloux AW, Young MR (2009) NK-dependent increases in CCL22 secretion selectively recruits regulatory T cells to the tumor microenvironment. J Immunol 182: 2753–2765. doi: 10.4049/jimmunol.0801124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mechtcheriakova D, Svoboda M, Meshcheryakova A, Jensen-Jarolim E (2012) Activation-induced cytidine deaminase (AID) linking immunity, chronic inflammation, and cancer. Cancer immunology, immunotherapy: CII 61: 1591–1598. doi: 10.1007/s00262-012-1255-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Martins A, Han J, Kim SO (2010) The Multifaceted Effects of Granulocyte Colony-Stimulating Factor in Immunomodulation and Potential Roles in Intestinal Immune Homeostasis. IUBMB life 62: 611–617. doi: 10.1002/iub.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hamilton JA (2002) GM-CSF in inflammation and autoimmunity. Trends Immunol 23: 403–408. [DOI] [PubMed] [Google Scholar]
- 104.Owen RL, Pierce NF, Apple RT, Cray WC, Jr. (1986) M cell transport of Vibrio cholerae from the intestinal lumen into Peyer's patches: a mechanism for antigen sampling and for microbial transepithelial migration. J Infect Dis 153: 1108–1118. [DOI] [PubMed] [Google Scholar]
- 105.Siebers A, Finlay BB (1996) M cells and the pathogenesis of mucosal and systemic infections. Trends Microbiol 4: 22–29. [DOI] [PubMed] [Google Scholar]
- 106.Shreedhar VK, Kelsall BL, Neutra MR (2003) Cholera Toxin Induces Migration of Dendritic Cells from the Subepithelial Dome Region to T- and B-Cell Areas of Peyer's Patches. Infect Immun 71: 504–509. doi: 10.1128/IAI.71.1.504-509.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lelouard H, Fallet M, de Bovis B, Meresse S, Gorvel JP (2012) Peyer's patch dendritic cells sample antigens by extending dendrites through M cell-specific transcellular pores. Gastroenterology 142: 592–601.e593. doi: 10.1053/j.gastro.2011.11.039 [DOI] [PubMed] [Google Scholar]
- 108.Huang FP, Platt N, Wykes M, Major JR, Powell TJ, et al. (2000) A discrete subpopulation of dendritic cells transports apoptotic intestinal epithelial cells to T cell areas of mesenteric lymph nodes. J Exp Med 191: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kato M, Kephart GM, Morikawa A, Gleich GJ (2001) Eosinophil infiltration and degranulation in normal human tissues: evidence for eosinophil degranulation in normal gastrointestinal tract. Int Arch Allergy Immunol 125 Suppl 1: 55–58. [DOI] [PubMed] [Google Scholar]
- 110.Straumann A, Simon HU (2004) The physiological and pathophysiological roles of eosinophils in the gastrointestinal tract. Allergy 59: 15–25. [DOI] [PubMed] [Google Scholar]
- 111.Jung Y, Rothenberg ME (2014) Roles and regulation of gastrointestinal eosinophils in immunity and disease. J Immunol 193: 999–1005. doi: 10.4049/jimmunol.1400413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rothenberg ME, Mishra A, Brandt EB, Hogan SP (2001) Gastrointestinal eosinophils. Immunol Rev 179: 139–155. [DOI] [PubMed] [Google Scholar]
- 113.Jawairia M, Shahzad G, Mustacchia P (2012) Eosinophilic Gastrointestinal Diseases: Review and Update. ISRN Gastroenterol 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Torpier G, Colombel JF, Mathieu-Chandelier C, Capron M, Dessaint JP, et al. (1988) Eosinophilic gastroenteritis: ultrastructural evidence for a selective release of eosinophil major basic protein. Clin Exp Immunol 74: 404–408. [PMC free article] [PubMed] [Google Scholar]
- 115.Dvorak AM (1980) Ultrastructural evidence for release of major basic protein-containing crystalline cores of eosinophil granules in vivo: cytotoxic potential in Crohn's disease. J Immunol 125: 460–462. [PubMed] [Google Scholar]
- 116.Ivanova D, Krempels R, Ryfe J, Weitzman K, Stephenson D, et al. (2014) NK Cells in Mucosal Defense against Infection. Biomed Res Int 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hall LJ, Murphy CT, Hurley G, Quinlan A, Shanahan F, et al. (2013) Natural killer cells protect against mucosal and systemic infection with the enteric pathogen Citrobacter rodentium. Infect Immun 81: 460–469. doi: 10.1128/IAI.00953-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zamai L, Ahmad M, Bennett IM, Azzoni L, Alnemri ES, et al. (1998) Natural killer (NK) cell-mediated cytotoxicity: differential use of TRAIL and Fas ligand by immature and mature primary human NK cells. J Exp Med 188: 2375–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Topham NJ, Hewitt EW (2009) Natural killer cell cytotoxicity: how do they pull the trigger? Immunology 128: 7–15. doi: 10.1111/j.1365-2567.2009.03123.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Siew YY, Neo SY, Yew HC, Lim SW, Ng YC, et al. (2015) Oxaliplatin regulates expression of stress ligands in ovarian cancer cells and modulates their susceptibility to natural killer cell-mediated cytotoxicity. Int Immunol 27: 621–632. doi: 10.1093/intimm/dxv041 [DOI] [PubMed] [Google Scholar]
- 121.Ishikawa K, Shimoda K, Shiraishi N, Adachi Y, Kitano S (1998) Low-dose cisplatin-5-fluorouracil prevents postoperative suppression of natural killer cell activity in patients with gastrointestinal cancer. Jpn J Clin Oncol 28: 374–377. [DOI] [PubMed] [Google Scholar]
- 122.Boismenu R (2000) Function of intestinal γδ T cells. Immunologic Research 21: 123–127. doi: 10.1385/IR:21:2-3:123 [DOI] [PubMed] [Google Scholar]
- 123.Sutton CE, Mielke LA, Mills KH (2012) IL-17-producing gammadelta T cells and innate lymphoid cells. Eur J Immunol 42: 2221–2231. doi: 10.1002/eji.201242569 [DOI] [PubMed] [Google Scholar]
- 124.Sheridan BS, Romagnoli PA, Pham QM, Fu HH, Alonzo F, 3rd, et al. (2013) gammadelta T cells exhibit multifunctional and protective memory in intestinal tissues. Immunity 39: 184–195. doi: 10.1016/j.immuni.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Van Acker HH, Anguille S, Van Tendeloo VF, Lion E (2015) Empowering gamma delta T cells with antitumor immunity by dendritic cell-based immunotherapy. Oncoimmunology 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ma Y, Aymeric L, Locher C, Mattarollo SR, Delahaye NF, et al. (2011) Contribution of IL-17-producing gamma delta T cells to the efficacy of anticancer chemotherapy. J Exp Med 208: 491–503. doi: 10.1084/jem.20100269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mantis NJ, Rol N, Corthésy B (2011) Secretory IgA's Complex Roles in Immunity and Mucosal Homeostasis in the Gut. Mucosal Immunol 4: 603–611. doi: 10.1038/mi.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shalapour S, Font-Burgada J, Di Caro G, Zhong Z, Sanchez-Lopez E, et al. (2015) Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature 521: 94–98. doi: 10.1038/nature14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhu J, Yamane H, Paul WE (2010) Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol 28: 445–489. doi: 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Luckheeram RV, Zhou R, Verma AD, Xia B (2012) CD4(+)T cells: differentiation and functions. Clin Dev Immunol 2012: 925135 doi: 10.1155/2012/925135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kitamura H, Iwakabe K, Yahata T, Nishimura S, Ohta A, et al. (1999) The natural killer T (NKT) cell ligand alpha-galactosylceramide demonstrates its immunopotentiating effect by inducing interleukin (IL)-12 production by dendritic cells and IL-12 receptor expression on NKT cells. J Exp Med 189: 1121–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fernandez CS, Cameron G, Godfrey DI, Kent SJ (2012) Ex-vivo alpha-galactosylceramide activation of NKT cells in humans and macaques. J Immunol Methods 382: 150–159. doi: 10.1016/j.jim.2012.05.019 [DOI] [PubMed] [Google Scholar]
- 133.Wu D, Xing G-W, Poles MA, Horowitz A, Kinjo Y, et al. (2005) Bacterial glycolipids and analogs as antigens for CD1d-restricted NKT cells. Proceedings of the National Academy of Sciences of the United States of America 102: 1351–1356. doi: 10.1073/pnas.0408696102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mullbacher A, Lobigs M, Hla RT, Tran T, Stehle T, et al. (2002) Antigen-dependent release of IFN-gamma by cytotoxic T cells up-regulates Fas on target cells and facilitates exocytosis-independent specific target cell lysis. J Immunol 169: 145–150. [DOI] [PubMed] [Google Scholar]
- 135.Andersen MH, Schrama D, thor Straten P, Becker JC Cytotoxic T Cells. Journal of Investigative Dermatology 126: 32–41. doi: 10.1038/sj.jid.5700001 [DOI] [PubMed] [Google Scholar]
- 136.Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, et al. (2012) Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer 12: 860–875. doi: 10.1038/nrc3380 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
TLR4+ cells in the colon were images at 100x magnification on a Nikon confocal microscope. We observed consistent differences in the morphology of TLR4+ cells in the oxaliplatin-treated group compared to the vehicle-treated cohort. 8 randomised images (20x magnification) per animal were used to count the number of TLR4+ cells in the colon, as well as immunoreactivity/image. Image J counter plugin was used to mark each cell to ensure they were only counted once, and we measured immunoreactivity/fluorescence by converting the image to 8-bit →binary→measure. We used % area result to determine immunoreactivity per image. The sum of TLR4+ cells and immunoreactivity was averaged between groups.
(XLSX)
All genes included in the array are listed, alongside quality control and normalisation expression. Changes in inflammation-associated gene expression are presented in heatmap and numerical formats. This dataset relates to all PCR data in this study, including Fig 4 (receptors) and Fig 7 (cytokines and chemokines).
(XLSX)
Dataset includes numbers on microbiota phylum, class, order, family, and genus.
(XLS)
Eight randomised images (20x magnification) per animal were used to count the number of CD45+ cells in the colon, as well as immunoreactivity/image. Image J counter plugin was used to mark each cell to ensure they were only counted once, and we measured immunoreactivity/fluorescence by converting the image to 8-bit →binary→measure. We used % area result to determine immunoreactivity per image. MPO activity was measured using the MPO Colorimetric Activity Assay (Sigma-Aldrich, Australia) according to manufacturers instructions.
(XLSX)
This data set contains raw values for all flow cytometry experiments on the various immune cell populations investigated in the PPs and MLNs.
(XLS)
Data Availability Statement
All data generated during and analysed during the current study are provided as Supporting Information files. All 16S rRNA sequencing data have been deposited to a NCBI-GENEbank public database and can be accessed via SRA RunSelector: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP133585.