Abstract
The macrophage is a major phagocytic cell type, and its impaired function is a primary cause of immune paralysis, organ injury, and death in sepsis. An incomplete understanding of the endogenous molecules that regulate macrophage bactericidal activity is a major barrier for developing effective therapies for sepsis. Using an in vitro killing assay, we report here that the endogenous purine ATP augments the killing of sepsis-causing bacteria by macrophages through P2X4 receptors (P2X4Rs). Using newly developed transgenic mice expressing a bioluminescent ATP probe on the cell surface, we found that extracellular ATP levels increase during sepsis, indicating that ATP may contribute to bacterial killing in vivo. Studies with P2X4R-deficient mice subjected to sepsis confirm the role of extracellular ATP acting on P2X4Rs in killing bacteria and protecting against organ injury and death. Results with adoptive transfer of macrophages, myeloid-specific P2X4R-deficient mice, and P2rx4 tdTomato reporter mice indicate that macrophages are essential for the antibacterial, antiinflammatory, and organ protective effects of P2X4Rs in sepsis. Pharmacological targeting of P2X4Rs with the allosteric activator ivermectin protects against bacterial dissemination and mortality in sepsis. We propose that P2X4Rs represent a promising target for drug development to control bacterial growth in sepsis and other infections.
Keywords: Cell Biology
Keywords: Innate immunity
Pharmacological targeting of P2X4Rs with ivermectin protects against bacterial dissemination and mortality in murine sepsis.
Introduction
Sepsis and septic shock are related clinical syndromes, where sepsis is defined as life-threatening organ dysfunction caused by a dysregulated host response to infection, and septic shock is a subset of sepsis, in which the underlying cellular and metabolic abnormalities are profound enough to increase mortality (1). These syndromes are the leading causes of mortality in intensive care units and, together, are the tenth leading cause of death overall in the US (1, 2). It has been estimated that 10% of patients with sepsis and 30% of patients with septic shock die — far more than the number of US deaths from prostate cancer, breast cancer, and AIDS combined (1, 3). Septic patients are generally hospitalized for extended periods, rarely leaving the intensive care unit before 2–3 weeks (4). Accordingly, sepsis represents a major burden to the US health care system, with costs estimated to be approximately $16.7 billion per year (2).
Current concepts suggest that an inability to kill invading bacteria or other pathogens effectively due to dysregulation of immunity is a major cause of multiple organ dysfunction syndrome and death in sepsis (5, 6). The decreased elimination of bacteria and dysregulation of the immune system lead to systemic inflammatory response syndrome, which contributes to the development of organ failure and shock (3, 7–9). Dysregulated host defense and immune response coexisting with increased inflammation also occur in trauma or burn patients that go on to develop the symptoms of sepsis, despite an early lack of detection of infectious agent (10, 11). Current treatment options for sepsis are mainly supportive and include the maintenance of systemic perfusion and administration of antibiotics. Despite these interventions, postmortem studies have revealed that the majority of patients still had infectious foci present (12), thus pointing to a deficit in bacterial clearance.
Macrophages and neutrophils comprise the phagocytic arm of the immune system largely responsible for eradicating bacterial infection. Neutrophil dysfunction and direct neutrophil-mediated organ injury have been proposed to contribute to septic inflammatory organ injury (13). However, impaired cell-autonomous monocyte/macrophage function appears to be primarily responsible for the insufficient antibacterial defenses in the septic host (14–16). In addition, septic monocytes and macrophages are decreased in their ability to respond ex vivo to LPS or other stimuli by producing antibacterial cytokines, such as IL-6 and IL-12 (17, 18).
Intracellular ATP is the universal energy currency of all cells and is critical for all life, from the simplest to the most complex. A plethora of evidence has shown that, in response to stressful situations — such as infection, trauma, hypoxia, cancer, and metabolic stress — ATP is rapidly released into the extracellular space (19–21) where ATP exerts mostly immunostimulatory effect (22). ATP acts by binding to specific cell membrane receptors, which are denoted P2 (23–25). P2 receptors (P2Rs) fall into 2 classes, the ionotropic P2XRs and the metabotropic P2YRs (26). P2XRs are cell membrane cation channels that are gated by extracellular ATP, and the ATP-mediated opening of these channels allows Ca2+ and Na+ influx, as well as K+ efflux. Seven P2XR subtypes have been cloned (P2X1–7), and ATP activates all P2X subtypes. P2X7Rs, which are expressed at high levels on macrophages, are the best-understood host defense/immune-regulating P2XRs (27). P2X7R is the sole receptor mediating the effect of extracellular ATP on NLRP3 inflammasome activation and pyroptosis (24). ATP activation of P2X7Rs also induces the macrophage killing of obligate intracellular bacteria such as Mycobacterium tuberculosis (M. tuberculosis) and Chlamydiae and obligate intracellular protozoans such as Trypanosome gondii and Leishmania by macrophages (28–30).
Although, the role of P2XRs in controlling obligate intracellular bacteria has been studied before, the role of P2XRs in controlling extracellular bacteria that cause sepsis remains unclear. Our results demonstrate that, in contrast to the central role of P2X7Rs in killing obligate intracellular bacteria by macrophages, P2X4Rs kill sepsis-causing bacteria, which are not obligate intracellular pathogens and which include E. coli and Staphylococcus aureus (S. aureus). We conclude that P2X4Rs represent a potentially novel target for drug development to manage extracellular bacteria during sepsis or other infections as Class II antibiotic adjuvants that act on the host (31).
Results
ATP induces bacterial killing by macrophages through P2X4Rs.
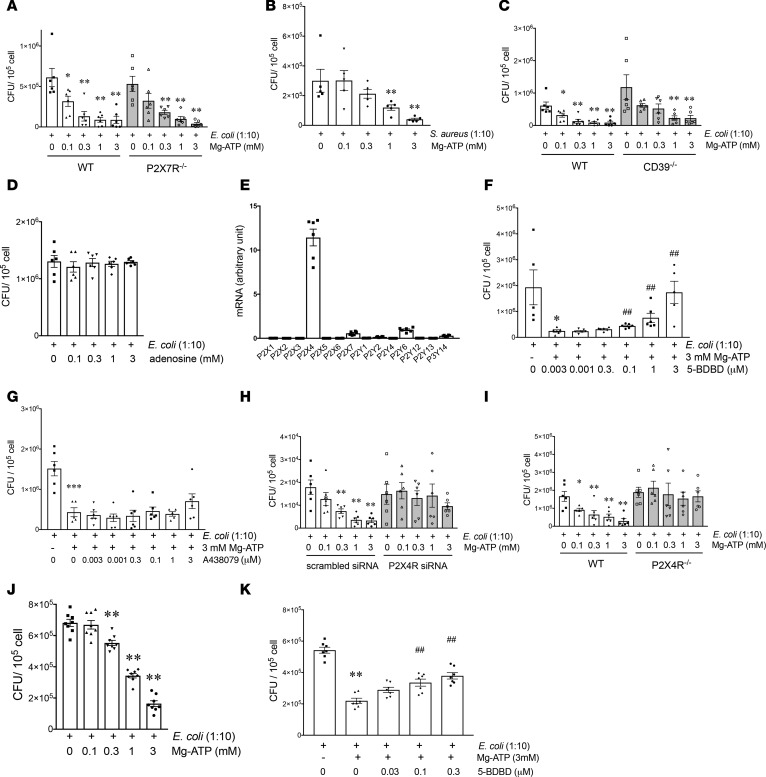
Recently, we showed that exogenous ATP augments macrophage-mediated bacterial killing in murine polymicrobial sepsis (32). To begin to uncover mechanisms, we established an in vitro system using the gentamicin protection assay, as described in Methods. We exposed naive macrophages to E. coli or S. aureus in vitro and then pulsed the macrophages with ATP for 5 minutes after the macrophages had phagocytosed the bacteria. We found that ATP caused a dose-dependent stimulation of both E. coli (Figure 1A) and S. aureus (Figure 1B) killing in WT macrophages when measured 2 hours after the end of the ATP pulse. The ATP-mediated killing of both E. coli and S. aureus was not associated with any macrophage death (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.99431DS1) and ATP also killed bacteria in the absence of gentamicin (Supplemental Figure 1C).
Figure 1. ATP augments bacterial killing in macrophages in a P2X4R-dependent fashion.
(A and B) ATP increases intracellular bacterial killing independently of P2X7Rs. Peritoneal macrophages from WT and P2X7R–/– mice were infected with E. coli (A) or with S. aureus (B) for 90 minutes, which was followed by pulsing the cells with ATP for 5 minutes. Subsequently, after a 2-hour incubation with 400 ng/ml gentamicin, the macrophages were lysed and serial dilutions of their intracellular content were spread onto Luria Bertani (LB) agar plates. *P < 0.05, **P < 0.01 vs. E. coli; n=5–6. (C and D) ATP augments bacterial killing independently of adenosine. Macrophages from WT and CD39–/– mice (a gift from Simon Robson, Beth Israel Deaconess Medical, Harvard University, Cambridge, MA; ref. 85) were infected with E. coli, pulsed with ATP (C) or adenosine (D), and then incubated with gentamicin for 2 hours, which was followed by intracellular CFU counting. *P < 0.05, **P < 0.01 vs. E. coli; n = 4–6. (E) Expression of P2Rs in peritoneal macrophages. Peritoneal macrophages were isolated from WT mice, and RNA was extracted from untreated cells. RNA was transcribed and qPCR was conducted. n = 6. (F–K) P2X4Rs are responsible for the ATP-stimulated increase in bacterial killing in macrophages. Peritoneal macrophages were infected with E. coli for 90 minutes, pretreated with (F) P2X4R antagonist (5-BDBD) or (G) P2X7R antagonist (A438079) for 30 minutes before an ATP pulse for 5 minutes, and killing was determined. As above*P < 0.05, ***P < 0.001 vs. E. coli treatment; ##P < 0.01 vs. ATP/E. coli treatment; n = 5–6. (H) Macrophages were transfected with scrambled siRNA or P2X4 siRNA, and the effect of ATP on bacterial killing was determined. **P < 0.01 vs. E. coli treatment; n=6. (I) Peritoneal macrophages from WT and P2X4R–/– mice were infected with E. coli, pulsed with ATP, and then bacterial killing was determined. **P < 0.01 vs. E. coli treatment; n = 6. *P < 0.05. (J and K) PMA-differentiated human monocytic THP-1 cells were infected with E. coli for 90 minutes and exposed to ATP for 5 minutes, and then killing was determined as described above for peritoneal macrophages (J). In other experiments, THP-1 cells were infected with E. coli and pretreated with 5-BDBD or vehicle for 30 minutes before a 5-minute ATP pulse (K); 2 hours later, intracellular CFUs were determined. **P < 0.01 vs. E. coli treatment; ##P < 0.01 vs. ATP/E. coli treatment. n = 6–7. Data are expressed as mean ± SEM. All results are representatives of 3 experiments. Data obtained by one-way ANOVA followed by Mann Whitney test.
For further mechanistic studies, we focused on E. coli. Since previous studies have indicated a major role of P2X7Rs in mediating the ATP killing of obligate intracellular bacteria (see above), we tested the role of P2X7Rs in mediating the ATP-induced increase of E. coli killing. Surprisingly, the ATP-stimulated killing of E. coli was independent of P2X7Rs, as ATP increased killing of E. coli both in WT and P2X7R–/– macrophages (Figure 1A).
This observation raised 2 possibilities. The first possibility was that ATP killed E. coli not through a direct effect on P2Rs, but indirectly by being degraded to adenosine, which also has wide-ranging effects on macrophages via adenosine/P1Rs (33, 34). However, the ATP-stimulated bacterial killing was not mediated by the degradation of ATP to adenosine, as ATP also killed E. coli in macrophages lacking CD39 (CD39–/–) (Figure 1C), which initiates ATP degradation to adenosine (22). In addition, adenosine itself failed to increase E. coli killing (Figure 1D). The second possibility was that ATP killed bacteria through another macrophage P2R. To begin to identify this P2R, we first studied which P2Rs are expressed in peritoneal macrophages. Quantitative PCR (qPCR) data showed that peritoneal macrophages expressed several P2Rs, and P2X4Rs were the most abundant (Figure 1E). Thus, we began to study the role of P2X4Rs. Using a pharmacological approach, we observed that a selective P2X4R antagonist (Figure 1F), but not P2X7R antagonist (Figure 1G), prevented the stimulatory effect of ATP on E. coli killing. In addition, shRNA-mediated silencing of P2X4Rs (Supplemental Figure 2) largely reversed the stimulatory effect of ATP on E. coli killing (Figure 1H). Finally, we demonstrated that, in macrophages isolated from P2X4R–/– mice, ATP failed to stimulate E. coli killing, whereas in macrophages from WT littermate controls, ATP caused a dose-dependent increase in killing (Figure 1I). We also confirmed the stimulatory role of P2X4Rs in regulating bacterial killing in human macrophages, as ATP increased killing of E. coli and a P2X4R antagonist inhibited the stimulatory effect of ATP on E. coli killing in PMA-differentiated human monocytic THP-1 cells (Figure 1, J and K). Furthermore, our data show that ATP failed to affect bacterial phagocytosis by macrophages (Supplemental Figure 3A) and had no effect on the migration of macrophages (Supplemental Figure 3B).
In summary, our data indicate that P2X4Rs mediate the stimulatory effect of ATP on E. coli killing in macrophages.
ATP increases bacterial killing independently of increasing phagolysosome fusion.
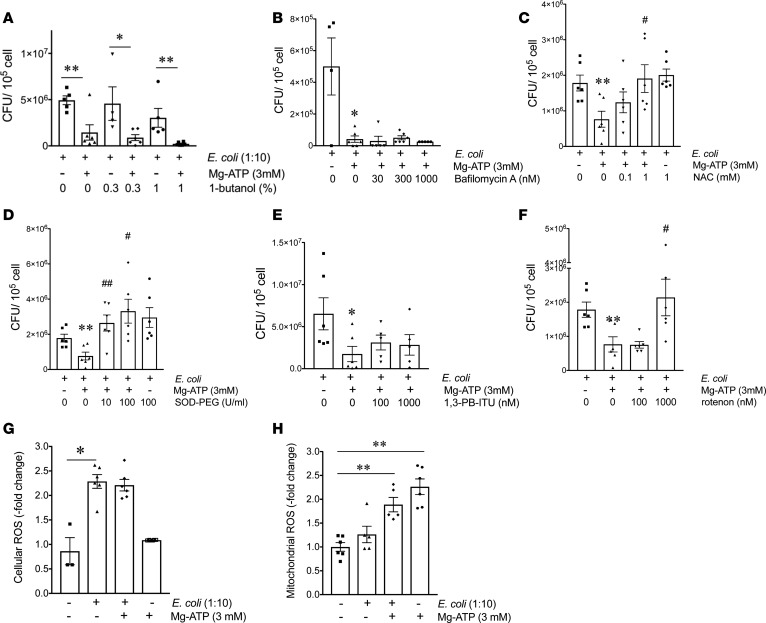
In the next set of experiments, we interrogated the intracellular pathways by which extracellular ATP kills phagocytosed bacteria in macrophages. First, we studied the role of phagolysosome fusion, a major step in the demolition of phagocytosed bacteria (35). ATP promotes phagosome-lysosome fusion by activating phospholipase D (36). Therefore, we tested the role of phospholipase D by using the selective phospholipase D blocker, 1-butanol. Pretreatment with 1-butanol failed to prevent the stimulatory effect of ATP on E. coli killing in macrophages (Figure 2A).
Figure 2. ATP augments bacterial killing by increasing ROS generation.
(A and B) ATP augments bacterial killing in a manner that is independent of phagolysosome fusion. Bacterial killing assay was performed in the presence of (A) 1-butanol or (B) bafilomycin A. Macrophages were infected with E. coli for 90 minutes and then pretreated with 1-butanol or bafilomycin A 30 minutes before ATP treatment for 5 minutes. The cells were then incubated with gentamicin for 2 hours, and intracellular bacterial CFUs were determined at the end of the gentamicin incubation period. *P < 0.05, **P < 0.01 vs. E. coli; n = 4–6. (C–H) ATP increases bacterial killing through enhancing ROS generation. (C–F) Peritoneal macrophages were infected with E. coli for 90 minutes and then treated with (C) N-acetyl-L-cysteine (NAC), (D) superoxide dismutase-polyethylene glycol (SOD-PEG), (E) 1,3-PB-ITU (nitric oxide synthase 2 inhibitor), or (F) rotenone for 30 minutes, followed by a 5-minute ATP pulse. The cells were then incubated with gentamicin for 2 hours, and intracellular CFUs were determined at the end of the incubation period. *P < 0.05, **P < 0.01 vs. E. coli treatment; #P < 0.05, ##P < 0.01 vs. E. coli + ATP treatment; n = 6. (G and H) Measurement of (G) cellular or (H) mitochondrial ROS production. Peritoneal macrophages were incubated with (G) 25 μM 2′,7′-dichlorofluorescin diacetate (DCFDA) (cellular ROS dye) for 45 minutes or with (H) 5 μM MytoSOX dye (mitochondrial ROS dye) for 10 minutes. Thereafter, cells were incubated with E. coli for 90 minutes and then pulsed with ATP for 5 minutes. Following a 1-hour incubation with 400 ng/ml gentamicin, DCFDA- and mytosox-related fluorescence were detected using Victor2 (Perkin Elmer) luminometer. *P < 0.05, **P < 0.01 vs. vehicle treatment; n = 6. Data are expressed as mean ± SEM. All results are representatives of 3 experiments. Data obtained by one-way ANOVA followed by Mann Whitney test.
Phagosome acidification also contributes to bacterial destruction in macrophages, and vacuolar-type H+-ATPase (v-ATPase) is responsible for phagosome acidification (37). To study the role of phagosome acidification, we tested the effect of bafilomycin A, a specific inhibitor of v-ATPase. Bafilomycin A failed to prevent the ATP stimulation of E. coli killing in macrophages (Figure 2B). We conclude that ATP kills bacteria independently of increasing phagolysosome fusion and phagosome/lysosome acidification.
ATP augments bacterial killing by increasing ROS generation.
Toxic ROS and reactive nitrogen species can also eliminate invading extracellular pathogens in the phagolysosome. To study the role of ROS in ATP-mediated bacterial killing, we used the selective oxygen radical scavengers N-acetyl-L-cysteine (NAC) and polyethylene glycol-conjugated superoxide dismutase (SOD-PEG). As Figure 2, C and D, show, both NAC and SOD-PEG prevented the stimulatory effect of ATP on E. coli killing in macrophages. Reactive nitrogen species are produced by NOS2 in macrophages. To study the role of NOS2, we employed the NOS2 inhibitor 1,3-PB-ITU and found that 1,3-PB-ITU failed to prevent the ATP stimulation of bacterial killing (Figure 2E). ROS can originate from several sources, including the mitochondria. We found that mitochondrial ROS may be important in the ATP-mediated killing of bacteria, as (a) ATP increased mitochondrial but not cellular ROS production and (b) rotenone, an inhibitor of mitochondrial ROS production, prevented the ATP augmentation of bacterial killing (Figure 2, F–H). Thus, the ATP stimulation of bacterial killing is mediated through increased ROS generation.
Tissue ATP concentrations are increased in polymicrobial sepsis.
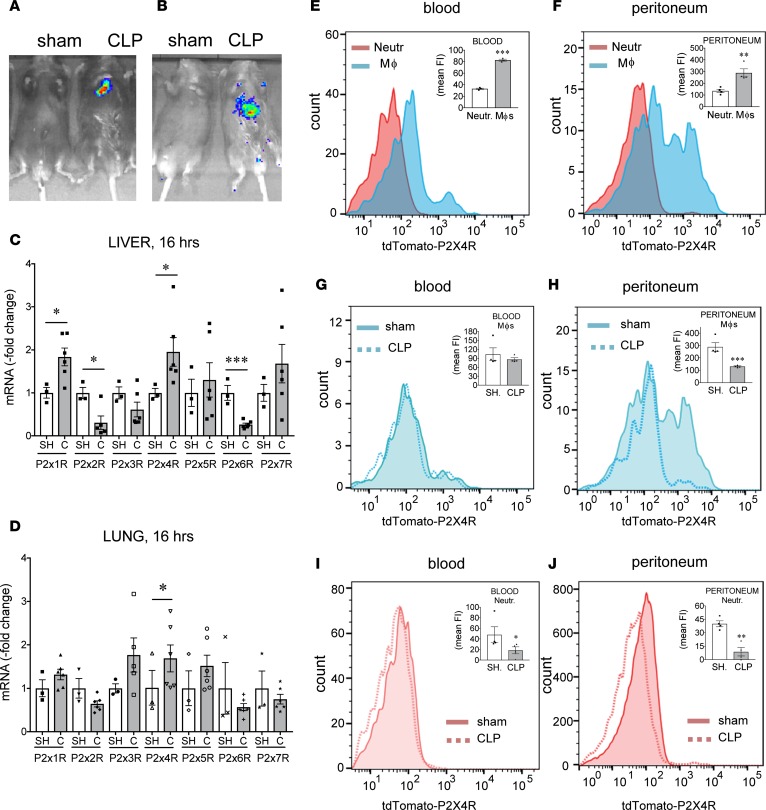
We have previously shown that ATP is released into the extracellular space during sepsis, as we detected increased ATP levels in the plasma of septic mice (32). However, it was unclear what organs or tissues are the major sources of ATP in the body during sepsis. To gain insight into the site of extracellular ATP release during sepsis, we employed potentially novel in vivo imaging technology that takes advantage of cells that express luciferase on their surface. Luciferase emits light in the presence of ATP and its substrate, luciferin, and allows detection of extracellular ATP release when expressed on the outer surface of the cell membrane. In a first approach, transgenic pmeLUC HEK293 cells that express luciferase anchored to the extracellular surface of the cell membrane were injected to mice retro-orbitally, which then allows the reporter cells to seed tissues (38, 39). In a second approach, ATP-reporter transgenic mice (pmeLUC mice) that constitutively and ubiquitously express luciferase on the surface of all cells were used. The mice were subjected to sepsis by cecal ligation and puncture (CLP), a clinically relevant model of polymicrobial sepsis. After CLP, luciferin was injected i.p., and luminescence was detected using an in vivo image monitoring system. Both strategies confirmed that extracellular ATP release occurs during sepsis, which is localized primarily in the upper abdominal area (Figure 3, A and B).
Figure 3. ATP levels and P2X4R expression increase during sepsis.
(A and B) CLP increases ATP levels in the peritoneum. (A) pmeLUC HEK293 cells (1 × 107) were injected retro-orbitally to recipient C57Bl/6J mice before sham operation or CLP. (B) Alternatively, pmeLUC mice were subjected to sham operation or CLP surgery. In both A and B, 3 hours after the CLP, 75 mg/kg luciferin was injected i.p. to animals and whole body luminometry was performed using IVIS 200 preclinical in vivo imaging system. Representative images are shown from sham/CLP-subjected mice from (A) pmeLUC HEK293-injected C57BL/6J mice or (B) pmeLUC transgenic mice. All results are representatives of 3 experiments. (C and D) mRNA expression of P2XRs in the liver and lung of sham/CLP-subjected (SH/C-subjected) C57Bl/6J mice. Mice were subjected to CLP or sham operation and, 16 hours later, liver and lung specimens were harvested. RNA was extracted from the tissues, transcribed, and analyzed by qPCR. *P < 0.05 vs. sham-operated mice; n = 3 (sham) and 6 (clp). ***P < 0.001. (E–J) tdTomato-P2X4R expression on blood and peritoneal macrophages. Peritoneal lavage fluid and blood were harvested from sham- or CLP-operated tdTomato-P2X4R transgenic reporter mice. Recovered cells were stained with anti-CD45, anti-F4/80, and anti-Ly6G antibodies, and tdTomato-P2X4R expression was monitored using flow cytometry. **P < 0.01 and ***P < 0.001 vs. neutrophils; *P < 0.05, **P < 0.01, and ***P < 0.001 vs. sham group. n = 4. Data are expressed as mean ± SEM. All results are representatives of 2 experiments. Data obtained by two-tailed Student’s t test.
P2X4Rs improve survival and decrease bacterial load, the level of inflammatory mediators, and organ injury in CLP-induced sepsis.
To begin to examine the role of P2X4Rs in sepsis, we measured P2X4R mRNA levels in naive animals and mice subjected to CLP. We found that liver and lung levels of P2X4R mRNA increased in response to CLP (Figure 3, C and D). Furthermore, the hepatic mRNA expression of P2X1R increased and P2X2R and P2X6R decreased after CLP (Figure 3C).
Using BAC transgenic mice expressing tdTomato under the control of the P2X4R gene (40) we measured P2X4R expression on monocytes/macrophages and neutrophils, which are the major cell types that ingest and kill bacteria during sepsis. Figure 3, E and F, show that both peritoneal and blood monocytes/macrophages isolated from naive mice display higher P2X4R protein levels when compared with naive peritoneal and blood neutrophils, respectively. P2X4R expression on blood monocytes/macrophages was similar in mice subjected to sepsis and sham operation (Figure 3G). Interestingly, P2X4R expression decreased on peritoneal monocytes/macrophages and on blood and peritoneal neutrophils in septic mice (Figure 3, H–J).
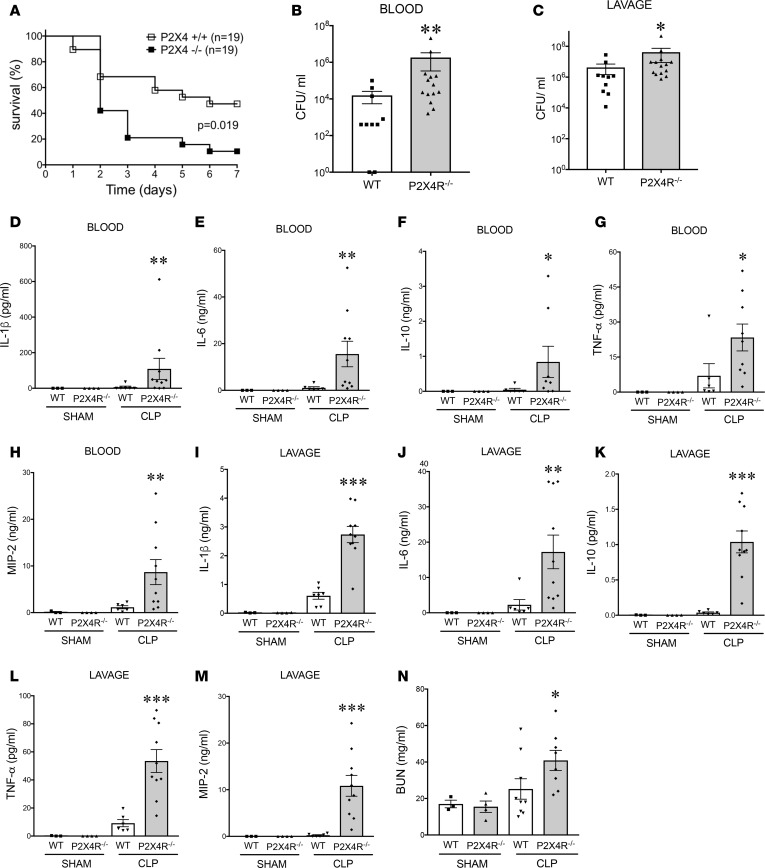
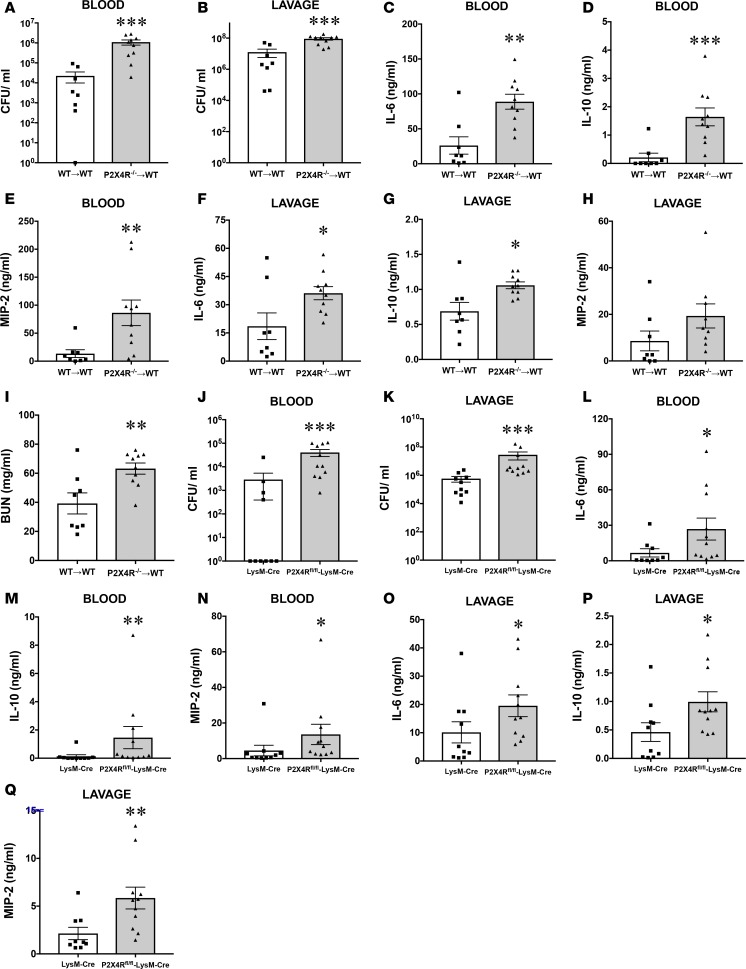
To study the role of P2X4Rs in regulating the host’s response to sepsis, we subjected WT and P2X4R–/– mice to CLP and monitored their survival. We observed that the survival rate of WT mice was higher than that of the P2X4R–/– animals, indicating that P2X4Rs are protective (Figure 4A). We then assessed the host’s immune response at 16 hours after the CLP procedure, at which time bacterial dissemination and inflammation are at their maximum (41). We found that WT mice exhibited decreased bacterial burden in both the blood and peritoneal cavity compared with P2X4R–/– mice (Figure 4, B and C), indicating that P2X4Rs control bacterial burden. We then investigated the inflammatory status of the mice by measuring levels of inflammatory cytokines and chemokines. We found that P2X4R–/– mice had higher levels of inflammatory cytokines and chemokines in blood and peritoneum compared with WT mice (Figure 4, D–M).
Figure 4. P2X4Rs decrease mortality, bacterial load, inflammatory cytokines and chemokines, and organ injury in sepsis.
(A) WT mice have improved survival compared with P2X4R–/– mice. Male WT and P2X4R–/– mice were subjected to CLP, and survival was monitored for 7 days. *P = 0.019 (WT and P2X4R–/–; n = 24 and 25, respectively). (B and C) Bacterial burden was determined by counting the number of CFUs on blood agar plates after serial dilution of blood and peritoneal lavage samples. Blood and lavage fluid were collected at 16 hour after CLP. *P < 0.05; **P < 0.01 vs. WT (WT and P2X4R–/–; n = 10 and 14, respectively, for blood; n = 10 and 14, respectively, for lavage). (D–N) WT and P2X4R–/– mice were subjected to sham or CLP operation and (D and I) IL-1β, (E and J) IL-6, (F and K) IL-10, (G and L) TNF-α, and (H and M) MIP-2 levels were determined with ELISA from blood and peritoneal lavage fluid collected at 16 hours after the operation. (N) Blood urea nitrogen (BUN) was measured in plasma of sham- or CLP-subjected WT and P2X4R–/– mice 16 hours after CLP. *P < 0.05, **P < 0.01, and ***P < 0.001 vs. WT sham- and CLP-operated WT and P2X4R–/–; n = 3, 4 for sham WT and P2X4R–/–; n = 6 and 10 for CLP WT. Data are expressed as mean ± SEM. All results are representatives of 3 experiments. Mortality curves were analyzed using Kaplan-Meier curve and log rank test and in the rest of the experiments one-way ANOVA followed by Mann Whitney test or two-tailed Student’s t test was used
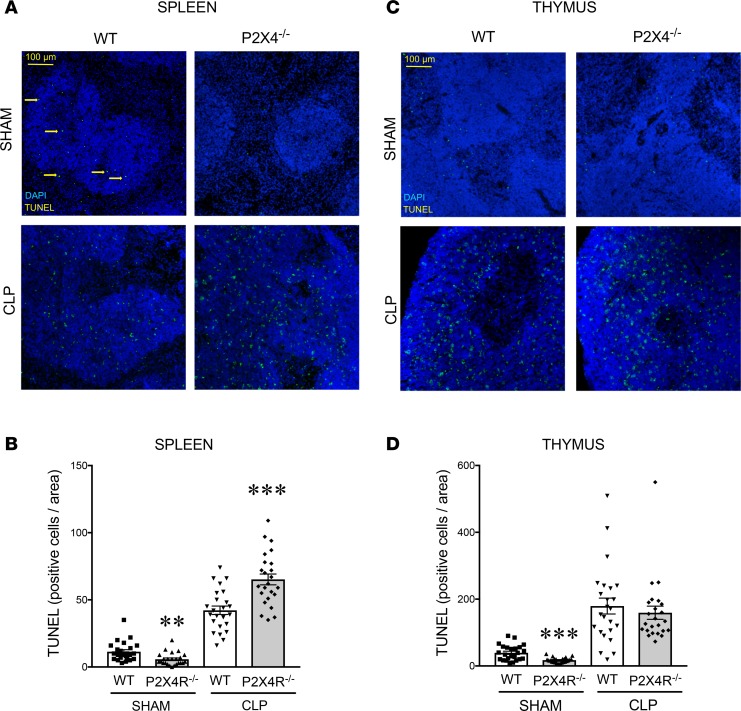
We assessed organ injury to unravel the role of P2X4Rs in regulating host pathophysiology. Septic P2X4R–/– mice exhibited increased kidney injury, as indicated by increases in plasma blood urea nitrogen (BUN) levels (Figure 4N). However, the levels of other organ injury markers, such as aspartate alanine aminotransferase (ALT; liver), aspartate aminotransferase (AST; liver), and creatine kinase (CK; muscle) were comparable between WT and P2X4R–/– mice following CLP (Supplemental Figure 4, A–C). Apoptosis in the septic spleen was higher in P2X4R–/– mice than in their WT littermates, as detected by TUNEL staining (Figure 5, A and B). There was no difference in apoptosis in the thymus between P2X4R–/– and WT mice (Figure 5, C and D). These data indicate that P2X4Rs regulate organ injury and apoptosis in a tissue-specific manner.
Figure 5. P2X4Rs protect against sepsis-induced apoptosis in the spleen.
Apoptosis in spleen and thymus was examined by fluorescent TUNEL labeling. (A and C) Representative images of TUNEL and DAPI-stained (A) spleen and (C) thymus samples from sham- or CLP-operated WT and P2X4R–/– animals 16 hours after sham/CLP. The slides were scanned with a Zeiss LSM 710 confocal laser-scanning microscope. Arrows indicate apoptotic cells. (B and D) Quantitative analysis of TUNEL-stained slides of (B) spleen and (D) thymus specimens from sham- or CLP-subjected WT and P2X4R–/– mice. Quantification of TUNEL-positive nuclei was done on 8 high-power fields per section using the ImageJ software. **P < 0.01, ***P < 0.001 vs. WT (WT and P2X4R–/–; n = 24 and 24, respectively). Data are expressed as mean ± SEM. Results are representatives of 3 experiments. Data obtained by one-way ANOVA followed by Mann Whitney test was used.
P2X4Rs on macrophages suppress sepsis-induced bacterial dissemination and inflammation.
Macrophages are important in phagocytosing and killing bacteria during sepsis (3). In addition, P2X4Rs mediate the stimulatory effect of ATP on bacterial killing in macrophages in vitro (Figure 1). Thus, we hypothesized that P2X4R signaling on macrophages would be essential for controlling bacterial spread and inflammation in sepsis. To test this hypothesis, we adoptively transferred peritoneal macrophages from donor WT and P2X4R–/– mice to separate groups of recipient WT mice before subjecting the recipient mice to CLP. We observed that P2X4R–/– macrophage–recipient WT mice displayed higher bacterial counts (Figure 6, A and B), with the exception of IL-1β and TNF-α, increased levels of inflammatory cytokines and chemokines (Figure 6, C–H, and Supplemental Figure 5, A–D), and augmented plasma BUN concentrations (Figure 6I), when compared with WT mice receiving WT macrophages. However, ALT, AST, and CK levels were comparable in WT and P2X4R–/– recipient mice (Supplemental Figure 4, D–F). To further assess the cell type–specific role of P2X4Rs, we generated mice in which the P2X4R gene was deleted in myeloid cells (Supplemental Figure 6) (P2X4fl/fl-LysM-Cre mice) by crossing mice expressing the Cre recombinase under the control of the myeloid-specific LysM promoter (LysM-Cre mice) and mice bearing floxed P2X4Rs (P2X4fl/fl). After sepsis induction, we found that P2X4fl/fl-LysM-Cre mice had higher bacterial burden (Figure 6, J and K) and increased levels of cytokines and chemokines (Figure 6, L–Q, and Supplemental Figure 5, E–H) than the control LysM-Cre mice. This finding corroborates the notion that the P2X4R on macrophages suppresses bacterial dissemination and inflammation.
Figure 6. P2X4Rs on macrophages decrease bacterial burden, inflammation, and organ injury during sepsis.
CLP-induced sepsis increased bacterial counts, inflammation, and BUN levels in WT recipients of adoptively transferred P2X4R–/– macrophages (P2X4R–/–→WT) more than in WT recipients of WT macrophages (WT→WT). (A and B) Bacterial burden was determined by counting the number of CFUs on blood agar plates after serial dilution of blood and peritoneal lavage samples. Blood and peritoneal lavage fluid were collected at 16 hours after CLP. (C–H) Cytokine and chemokine levels in WT recipient mice that were adoptively transferred with WT or P2X4R–/– peritoneal macrophages. Cytokines were measured 16 hours after CLP. (I) BUN was measured from plasma of CLP-subjected WT→WT and P2X4R–/–→WT mice 16 hours after CLP. *P < 0.05, **P < 0.01, *** P < 0.001 vs. WT→WT transfer (n = 8–10 recipient mice in each group). (J–Q) Bacterial load and inflammation are higher in myeloid-specific P2X4R–/– mice (P2X4Rfl/fl-LysM-Cre+) when compared with control (LysM-Cre+) mice following sepsis. (J and K) Bacterial burden was determined by counting the number of CFUs on blood agar plates after serial dilution of blood and peritoneal lavage samples. Blood and lavage fluid were collected at 16 hours after CLP. (L–Q) Cytokine and chemokine levels in P2X4Rfl/fl-LysM-Cre+ vs. LysM-Cre+ mice. Cytokines were measured 16 hours after CLP. *P < 0.05, **P < 0.01, and ***P < 0.001 vs. LysM-Cre+ mice (n = 10 mice in each group, except n = 11 in the P2X4Rfl/fl-LysM-Cre+ group of Q). Data are expressed as mean ± SEM. All results are representatives of 2 experiments. Data obtained by two-tailed Student’s t test.
Pharmacological activation of P2X4Rs with ivermectin improves survival and decreases bacterial burden following sepsis.
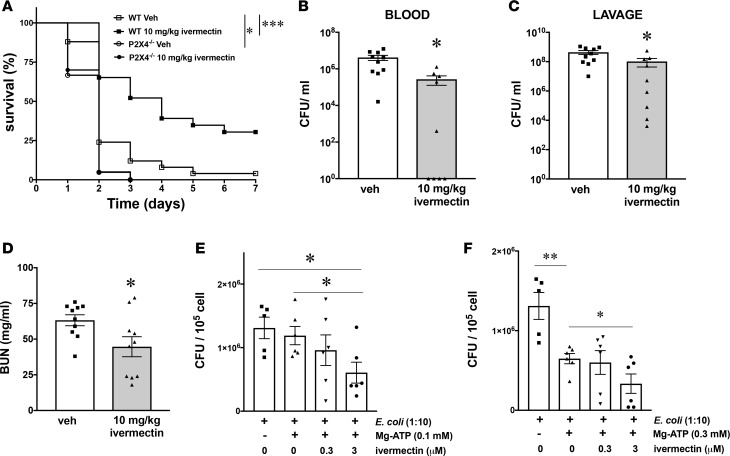
We hypothesized that pharmacological activation of P2X4Rs would have an effect opposite to that of inactivating these receptors genetically, meaning that the pharmacological activation of P2X4Rs would be protective. To examine this hypothesis, we injected mice with ivermectin, an antihelminth drug that is a partial allosteric activator of P2X4Rs (42) 90 minutes after performing CLP. Ivermectin improved survival (Figure 7A), which occurred in a P2X4R-dependent manner, as ivermectin failed to improve survival in P2X4R–/– mice. Ivermectin also decreased bacteria (Figure 7, B and C) and diminished plasma BUN levels (Figure 7D), but plasma levels of ALT, AST, and CK were comparable in ivermectin-treated and vehicle-treated mice (Supplemental Figure 4, G–I). Unexpectedly, ivermectin failed to influence the level of cytokines and chemokines (Supplemental Figure 7, A–J). Furthermore, ivermectin potentiated the ATP stimulation of bacterial killing in macrophages in vitro (Figure 7, E and F). These data confirm the antibacterial and organ-protective effects of P2X4Rs in sepsis.
Figure 7. Ivermectin, an allosteric activator of P2X4Rs improves survival, decreases bacterial burden and organ injury in mice after sepsis, and augments bacterial killing by macrophages.
(A) Ivermectin-treated WT mice showed improved survival compared with vehicle-treated (Veh-treated) WT mice. The survival of ivermectin-treated P2X4R–/– and Veh-treated P2X4R–/– mice was comparable. WT and P2X4R–/– mice were injected with 10 mg/kg ivermectin or its vehicle and subjected to CLP. The survival of mice was monitored for 7 days. *P < 0.05, P2X4R–/– injected with vehicle (n = 21) vs. WT injected with vehicle (n = 25); ***P < 0.001, WT injected with ivermectin (n = 20) vs. WT injected with vehicle (n = 25). (B and C) Bacterial burden was determined by counting the number of CFUs on blood agar plates after serial dilution of blood and peritoneal lavage samples. Blood and lavage fluid were collected at 16 hours after CLP. *P < 0.05 vs. Veh (Veh and 10 mg/kg ivermectin; n = 10 and 9, respectively). (D) BUN was determined from plasma of ivermectin- or Veh-treated mice 16 hours after CLP. *P < 0.05 vs. Veh; n = 10 and 9, respectively. (E and F) Ivermectin increases intracellular killing of E. coli in cultured macrophages. Peritoneal macrophages were infected with E. coli for 90 minutes and then pulsed with ATP for 5 minutes. ATP was then removed, and the macrophages were incubated with ivermectin for 30 minutes. Thereafter, the ivermectin was removed and the cells were incubated in medium containing gentamicin for another 2 hours. The cells were then lysed, and serial dilutions of intracellular content were spread onto LB agar plates. *P < 0.05 (n = 5–6); **P < 0.01 (n = 5–6). Data are expressed as mean ± SEM. All results are representatives of 3 experiments. Mortality curves were analyzed using Kaplan-Meier curve and log rank test and in the rest of the experiments one-way ANOVA followed by Mann Whitney test or two-tailed Student’s t test was used.
Discussion
Previous studies have highlighted the central role of P2X7Rs in controlling obligate intracellular bacteria (36, 43, 44) in macrophages. Our results here have uncovered a major pathway, the ATP/P2X4R/ROS axis in macrophages, which selectively increases the killing but not phagocytosis or migration of sepsis-causing extracellular bacteria. Although not the subject of the current study, it is also possible that P2X4Rs can also increase, at least under some circumstances, the killing of obligate intracellular bacteria. In fact, Pettengill et al. reported that P2X4Rs augmented the killing of Chlamidia trachomatis (C. trachomatis) in epithelial cells (45). However, based on previous results where P2X7R deficiency almost completely reversed the ATP-stimulated enhancement of M. tuberculosis (36) and C. trachomatis (46) killing in macrophages, a role for P2X4R on macrophages in killing obligate intracellular bacteria may be, at the most, minor. Taken together, the role of P2X4R vs. P2X7R may depend on the type of bacterium and cell type studied. These potentially overlapping functions of P2X4R and P2X7R may be due to the fact that they share the same ancestral gene and were generated by gene duplication (47).
When a macrophage ingests a pathogenic microorganism, the pathogen becomes trapped in the phagosome. The nascent phagosome undergoes a process termed maturation by fusion and fission events with endosomes and lysosomes to generate an acidic and hydrolytic mature phagolysosome (48). Within the phagolysosome, ROS and reactive nitrogen species and enzymes digest and kill the pathogen. Extracellular ATP has been shown to target many of these pathways in the context of infection with obligate intracellular pathogens. For example, ATP — in a P2X7R-dependent manner — increased phagosome-lysosome fusion following M. tuberculosis infection (36) of macrophages. In addition, inhibition of phagolysosome fusion using phospholipase D inhibitors prevented the P2X7R-mediated increased killing of M. tuberculosis (36). However, in our studies, phospholipase D inhibition failed to prevent the ATP-mediated increase in bacterial killing by macrophages, indicating that ATP stimulated bacterial killing independently of effects on phagolysosome fusion.
To destroy invading bacteria, macrophages — as part of the phagocytic/bacterial killing process — produce ROS via phagosomal NADPH oxidase (49). In addition to this, mitochondrial oxidative phosphorylation enzyme complexes also generate ROS to fight infection (50). Although it has been shown that P2X7R activation by ATP augments ROS production after Toxoplasma infection (43), the origin of ROS has not been addressed. Here, we provided evidence that P2X4R activation specifically increases mitochondrial, but not cellular, ROS production, even in the absence of bacterial stimulation. In addition, the mitochondrial function blocker rotenone reversed the ATP stimulation of bacterial killing. These results indicate that mitochondrial ROS mediates, at least in part, the killing effect of ATP. Recently, several intracellular proteins have been identified as important factors in the bacteria-induced mitochondrial ROS generation machinery, including TRAF6, ECSIT (51), Mst1, and 2 kinases (52). Further studies are warranted to decipher the precise mechanisms by which P2X4Rs enhance mitochondrial ROS generation. Although we failed to see increased ROS production at the cellular level using the fluorescent die 2′,7′-dichlorofluorescin diacetate, we cannot exclude a contribution by nonmitochondrial ROS sources, such as the NADPH oxidase to the ATP increase in bacterial killing.
In a previous study, we found increased ATP levels in the plasma of septic mice, indicating increased extracellular ATP release (32). In this study, utilizing both newly created transgenic mice and using injected cellular probes that can report extracellular ATP increases, we confirmed that sepsis induces extracellular release of ATP in tissues. Although the precise organ or tissue releasing ATP is hard to pinpoint due to limitations of the imaging methodology, we found that ATP was primarily released in the upper abdomen. Given the location and size of the signal, it is possible that the liver is the major ATP-releasing organ during sepsis. The most common resident cell types in the liver are hepatocytes and resident macrophages, termed Kupffer cells. However, following tissue injury and inflammation, other inflammatory cells such as neutrophils infiltrate the site of injury, and these cells can also be the source of extracellular ATP. Macrophages release ATP in response to B. anthracis infection in a connexin 43–dependent way (53). Eltzschig et al. (54) showed that neutrophils liberate ATP and that this ATP release is blocked by the inhibition of connexin 43. In another study, N-formyl-met-leu-phe–induced (fMLP-induced) ATP release by neutrophils was pannexin-1 dependent (55). Hepatocytes release ATP in a pannexin-1–dependent manner following saturated free fatty acid–induced hepatic inflammation (56). Cervical epithelial cells release ATP during infection with C. trachomatis (45). In addition to host cells, bacteria can also release ATP, as shown by Wegiel et al. (57). Further experiments will be necessary to uncover the precise mechanisms, and cell and tissue origins of ATP release in sepsis.
Our in vivo sepsis studies utilizing the CLP model (58–60) corroborated the in vitro macrophage data showing that P2X4Rs kill bacteria, as P2X4R–/– mice had an increased bacterial load when compared with WT mice. In addition, P2X4R–/– mice showed decreased survival, indicating that endogenous ATP that is released during sepsis is protective through P2X4Rs. We believe that the in vivo protective effects of P2X4Rs are mediated by signaling through monocyte/macrophage P2X4Rs, as both adoptive transfer of P2X4R–/– macrophages and myeloid-specific P2X4R–/– mice mirrored the deleterious phenotype of P2X4R–/– mice, and P2X4R expression was higher on monocytes/macrophages than neutrophils. Since our results cannot distinguish the role of the particular monocyte (e.g., LyC6+ vs. LyC6–) or macrophage (e.g., adult vs. embryonic) populations, further studies will be necessary to determine the identity of the major P2X4R-expressing protective cell type. It has been shown that P2X4Rs increase the release of inflammatory mediators, such as prostaglandin E2 (61) and brain-derived neurotrophic factor (62), and initiate inflammatory pain (61, 63–65). Although, in this study, we did not investigate the precise cellular and intercellular mechanisms by which P2X4Rs suppress bacterial load and inflammation in detail, mapping out the signaling pathways that mediate the protective effect of P2X4Rs is clearly an important avenue for future studies.
We found that P2X4Rs are downregulated on neutrophils and, to a lesser extent, on macrophages during sepsis. P2X4Rs have been shown to desensitize upon activation; however, this appears to be moderate when compared with other P2XRs (66). At this point, the relevance of this observation is unclear, and further studies will be needed to decipher the role and mechanisms of P2X4R downregulation during sepsis.
Ivermectin was initially introduced commercially as an anthelmintic agent for animal health, and it is effective against a wide range of parasites, including gastro-intestinal roundworms, lungworms, mites, lice, and hornflies (67). Since it was first administrated to cure onchoceriasis in humans in 1988, more than 200 million people use ivermectin annually (67) without major adverse effects. It has been demonstrated that ivermectin is an allosteric activator P2X4R (42, 68, 69). Since selective orthosteric P2X4R agonists are not available, we used the allosteric agonist ivermectin as a pharmacological approach to study the role of P2X4Rs in sepsis. We found that ivermectin increased bacterial killing in macrophages and improved bacterial control and survival of animals following sepsis, which occurred in a P2X4R-dependent manner. Strategies to enhance bacterial killing by macrophages are potentially less harmful than enhancing neutrophil action because macrophages — unlike neutrophils, which are lysed and release their toxic agents during killing — are less likely to cause collateral damage to surrounding cells and tissues. Several agents have been shown to increase killing of extracellular bacteria by macrophages. For example, a screen of microbial natural product extracts identified streptazolin, which stimulated killing of bacteria by broadly enhancing macrophage activation through upregulation of NF-κB (70). Both GM-CSF and IFN-γ enhance macrophage killing of bacteria by generally activating macrophage inflammatory responses (71, 72). The problem with these general, broad approaches is that they increase the production of proinflammatory cytokines, which — if overproduced — are toxic and can cause organ injury and acute septic shock. In contrast, P2X4R activation selectively increases bacterial killing without increasing proinflammatory cytokines, as cytokine and chemokine levels were comparable in ivermectin-treated vs. vehicle-injected animals.
In a previous study, we demonstrated that P2X7Rs can contribute to bacterial killing in sepsis in vivo, which we couldn’t confirm in the current study in vitro (32). Our current results demonstrating in vivo augmentation of bacterial killing by P2X4Rs illustrates, however, that both P2X4Rs and P2X7Rs augment bacterial killing in vivo. There is abundant evidence for physical interactions between P2X4Rs and P2X7Rs in the literature (73, 74). The question of how these receptors interact in sepsis will be the subject of future studies.
In summary, the major innovation of our studies is the discovery of P2X4Rs as potential targets for antibacterial drug discovery. Therefore, we envision using P2X4R agonists or ivermectin as adjuvant therapeutic agents for sepsis, as well as other microbial infections, where macrophages ingest and kill the pathogen. The importance of such a selective, host-targeting strategy is underscored by the decreasing efficacy of direct microbe-targeting molecules, such as antibiotics, due to the emergence of antibiotic-resistant microorganisms.
Methods
Experimental animals, drugs, and cell culture.
The generation of P2X4R–/– mice was described before (75). The generation of P2X4Rfl/fl mice was described before (76). pmeLUC transgenic mice were generated by pronuclear injection of a linearized pmeLUC DNA construct (77) into the pronuclei of B6D2F2 zygotes. Transduced zygotes were then transferred to pseudopregnant foster mothers. P2X7R–/–, P2X4R-tdTomato, LysM-Cre, and C57BL/6J mice were purchased from the Jackson Laboratory and then maintained at the animal facility at Rutgers New Jersey Medical School and Columbia University. Magnesium-ATP (Mg-ATP), ivermectin, rotenone, SOD-PEG, and NAC were from MilliporeSigma; 5-BDBD and A438079 were from Tocris; and bafilomycin A was from Cayman Chemical. PmeLUC HEK293 cells and thioglycollate-elicited peritoneal macrophages from C57BL/6J mice were grown in DMEM supplemented with 10% FBS, 50 U/ml penicillin, 50 μg/ml streptomycin, and 1.5 mg/ml sodium bicarbonate in a humidified atmosphere of 95% air and 5% CO2; all from from Thermo Fisher. Human monocytic THP-1 cells (ATCC) were treated with 80 nM PMA (MilliporeSigma) at 37°C for 16 hours to obtain PMA-differentiated THP-1 macrophages. THP-1 macrophages were cultured in RPMI 1640 medium (Thermo Fisher) supplemented with 10% FBS and 1% penicillin-streptomycin and incubated at 37°C in a humidified 5% CO2 incubator.
Bacterial cultures.
E. coli strain K12 from ATCC was grown in Luria Bertani (LB) medium overnight at 37°C with shaking at 180 rpm. S. aureus Rosenbach strain (ATCC) was grown in tryptic soy broth medium (Calbiochem) overnight at 37°C with shaking at 180 rpm. Cells were centrifuged at 3,000 g for 15 minutes at 4°C and resuspended in PBS (Thermo Fisher) at a density of 6 × 109/ml before treating macrophages.
CLP model of sepsis.
Polymicrobial sepsis was induced by subjecting mice to CLP, as we described before (32, 60, 78). In studies where biochemical, immunological, and bacteriological analyses were performed, the mice were reanesthetized with pentobarbital (50 mg/kg, i.p.) 16 hours after the CLP procedure, and blood, peritoneal lavage fluid, and various organs were harvested. The effect of ivermectin was evaluated using male C57BL/6J mice in a similar fashion to that described for the P2X4R–/– or WT mice. In these experiments, the mice were injected i.p. with 10 mg/kg ivermectin or its vehicle 90 minutes after the CLP operation. Separate sets of WT and P2X4R–/– mice and ivermectin-treated and vehicle-injected mice were used in survival studies.
Isolation and adoptive transfer of peritoneal macrophages.
Thioglycollate-elicited peritoneal cells (79) from donor WT and P2X4R–/– mice were harvested in PBS. Purified CD11b+ cells were obtained by positive selection using magnetic beads coated with anti-CD11b Ab (Miltenyi Biotech; catalog 130-049-601), according to the manufacturer’s protocol. Purified CD11b+ cells were resuspended in PBS, and 4.5 × 106 cells were injected i.p. to separate groups of recipient WT mice 2 hours before subjecting them to CLP.
Collection of blood, peritoneal lavage fluid, and organs.
Blood samples were obtained aseptically by cardiac puncture through the ribcage using heparinized syringes. The blood samples were placed into heparinized micro centrifuge tubes kept on ice until further processing for bacteriological analysis. The blood samples were centrifuged at 1,000 g for 10 minutes, and then the recovered plasma was stored at –70°C until further use. To collect peritoneal lavage fluid, 2 ml of sterile PBS was injected into the peritoneal cavity via an 18-gauge needle. Then, the peritoneal fluid was recovered through the needle and was placed on ice until processed for bacteriological examination. After making serial dilutions of the peritoneal lavage fluid to determine the number of the CFUs, the peritoneal lavage fluid was centrifuged at 5,000 g for 10 minutes and the supernatant was stored at –70°C until further analysis. Heart, lung, kidney, thymus, liver, and spleen samples were excised and snap frozen using liquid nitrogen or were fixed in formalin for further analysis.
Quantification of bacterial CFUs from blood and peritoneal lavage fluid.
Serial dilutions for bacteriological analysis were made as described previously (41, 58, 60, 78). In brief, blood or peritoneal lavage fluid was diluted serially in sterile PBS. Each dilution (25 μl of each) was aseptically plated and cultured on trypticase blood agar plates (BD Biosciences) at 37°C. After 14–18 hours of incubation, the number of bacterial colonies was counted. The number of cultures is expressed as CFUs per milliliter of blood or peritoneal lavage fluid.
In vivo ATP measurement using pmeLUC HEK293 cells.
pmeLUC HEK293 cells were grown to approximately 75% confluence and were removed from the tissue culture bottle by trypsin. To measure ATP release, 1 × 107 pmeLUC HEK293 cells were injected retro-orbitally before the CLP procedure. This route of injection had been shown to seed all tissues with injected cells (80). Sham operation or CLP was performed on the pmeLUC HEK cell–recipient mice, and 3 hours later, 75 mg/kg luciferin (Promega) was injected i.p. to subject animals to whole body luminometry performed by IVIS 200 preclinical in vivo imaging system.
In vivo ATP measurement using transgenic pmeLUC mice.
Sham operation or CLP was performed on the pmeLUC mice, and 3 hours later, 75 mg/kg luciferin was injected i.p., which was followed by whole body luminometry performed using IVIS 200 preclinical in vivo imaging system.
Determination of BUN, AST, ALT, and CK.
BUN, AST, ALT, and CK were analyzed using a clinical chemistry analyzer system (VetTest8008; IDEXX Laboratories), as we described previously (60).
In vitro bacterial killing assay.
Peritoneal macrophages (1 × 105/well) were seeded on each well of a 96-well plate. Macrophages in the presence of 0.1% FBS were incubated with live E. coli or with live S. aureus (at a macrophage/bacteria ratio of 1:10) for 90 minutes. Nonphagocytosed bacteria were removed by rinsing the cells with 300 μl of PBS 4 times, immediately after which procedure the macrophages were pulsed with various concentrations of ATP for 5 minutes. After 5 minutes, the ATP was removed by washing the cells with 300 μl of PBS 4 times, and then the macrophages were incubated with 400 ng/ml gentamicin (Thermo Fisher)–containing medium for 2 hours to kill the remaining extracellular and cell surface–attached bacteria. Thereafter, gentamicin was removed by washing the cells 3 times with 300 μl of PBS, and the cells were lysed in 100 μl of 100 mM Tris-HCl/1% Triton-X100 (both from Thermo Fisher). Cell lysates were diluted serially in sterile physiological saline. Each dilution (25 μl of each) was aseptically plated and cultured on LB agar plates (BD Biosciences) at 37°C. After 14–18 hours of incubation, the number of bacterial colonies was counted. The number of cultures is expressed as CFUs per 1 × 105 peritoneal cells. In some experiments, the killing assay was performed as described here but in the absence of gentamicin.
Measurement of cell viability using MTT assay and lactate dehydrogenase (LDH) assay.
MTT assay was performed as we described before (81–83). Briefly, peritoneal macrophages were incubated with 0.01% MTT reagent for 30 minutes. Thereafter, supernatants were removed, cells were lysed in DMSO, and OD was measured at 550 nm. LDH measurements were performed using a kit (LDH Activity Assay Kit; MilliporeSigma) and in accordance with the manufacturer’s recommendations (84).
In vitro phagocytosis assay.
Thioglycollate-elicited peritoneal macrophages (1 × 105) obtained from WT and P2X4–/– mice were seeded per well onto 96-well plates and were pulsed with various concentrations of ATP for 5 minutes. After this 5-minute ATP pulse, ATP was removed by washing the cells with 300 μl of PBS 4 times. Thereafter, FITC-labeled E. coli (Molecular Probes) were added to the macrophages at a macrophage/bacteria ratio of 1:10 for 90 minutes. At the end of the incubation period, the medium was removed, and 100 μl of 250 μg/ml trypan blue (Millipore) suspension was added to all wells for 1 minute. The trypan blue solution was then removed by aspiration, and the experimental and control wells were read with a fluorescence plate reader at 480 nm for excitation and 520 nm for emission.
In vitro migration assay.
In vitro migration of thioglycollate-elicited peritoneal macrophages obtained from WT and P2X4–/– mice toward 100 ng/ml MCP-1 (R&D Systems) was measured with QCM Chemotaxis 96-well Cell Migration Assay (MilliporeSigma) using a pore size of 5 μm, according to the manufacturer protocol. In brief, 100 ng/ml MCP-1 in the absence or presence of Mg-ATP (0.1–3 mM) was added in the feeder tray, and 5 × 104 peritoneal macrophages were seeded into the cell migration chamber. Thereafter, the migration chamber was placed into the feeding tray, and then the plates were incubated at 37°C for 4 hours. After this incubation period, transmigrated cells were lysed, and genomic DNA-specific fluorescent dye was added to the lysate. The fluorescence intensity of cell extracts was measured at 480 nm/520 nm.
TUNEL staining of spleen and thymus specimens.
Spleen and thymus samples were fixed in formalin and embedded in paraffin and sections were cut. After the removal of paraffin and rehydration, the sections were treated with proteinase-K (20 μg/ml, Promega) in Tris-EDTA buffer (pH 8.0) for 15 minutes at 37°C, followed by a 10-minute incubation at room temperature. The sections were washed in PBS and incubated for 1 hour with the TUNEL reaction mix (Roche Diagnostics). As a positive control, samples were treated with DNAse (200 U/ml) in 1× DNAse buffer for 10 minutes at room temperature (Thermo Fisher Scientific) before the application of the TUNEL reaction mix. Negative-control sections were prepared by applying the fluorescein-labeled dUTP without the TdT enzyme. After incubation with the TUNEL mix, the sections were washed 3 times in PBS and counterstained with DAPI (1 μg/ml in PBS, Cell Signaling Technologies) for 5 minutes. After washing in PBS, the sections were mounted with ProLong Gold Antifade Reagent (Cell Signaling Technologies). The slides were scanned with a Zeiss LSM 710 confocal laser-scanning microscope. Quantification of TUNEL-positive nuclei was done on 8 high-power fields per section using the ImageJ software.
RNA extraction, cDNA synthesis, and qPCR.
Total RNA was extracted from liver, spleen, kidney, thymus, lung, heart, brain, and peritoneal macrophages using TRI reagent (Molecular Research Center) and reverse transcribed. For detecting P2R mRNA transcripts, a commercially available real-time PCR kit was used (PowerUp SYBR Green Master Mix; Applied Biosystems), and all data were normalized to constitutive ribosomal RNA (18S) values. The Applied Biosystems 7700 sequence detector was used for amplification of target sequences, and quantitation of differences between treatment groups was done using the comparative Ct method.
siRNA-mediated knockdown of P2X4R expression in macrophages.
Peritoneal macrophages were transfected with 300 nM P2X4 siRNA or control siRNA (containing a scrambled DNA sequence) (Santa Cruz Biotechnology Inc.) using electroporation and Amaxa Mouse Macrophage Nucleofactor Kit (Lonza). Eight, 12, and 24 hours after transfection, RNA was isolated and reverse transcribed. P2X4R mRNA levels were measured by qPCR (Supplemental Figure 2) to determine the efficacy of silencing. For in vitro bacterial killing assay, P2X4R siRNA– or scrambled siRNA–transfected peritoneal macrophages were incubated with E. coli 24 hours after transfection.
Measurement of cellular and mitochondrial ROS generation.
Peritoneal macrophages (2 × 104/well were seeded onto black-welled clear-bottomed 96-well plates and were incubated with 25 μM 2′,7′-dichlorofluorescin diacetate (DCFDA; cellular ROS dye; Abcam) for 45 minutes or with 5 μM MytoSOX dye (mitochondrial ROS dye; Invitrogen) for 10 minutes at 37°C in the dark in 1× HBSS. Thereafter, the cells were rinsed with 1× HBSS 3 times and were incubated with E. coli in DMEM for 90 minutes, which was followed by ATP treatment for 5 minutes. Following a 1-hour incubation period with 400 ng/ml gentamicin in DMEM, the cells were washed with HBSS, and DCFDA- and MytoSOX-related fluorescence was detected using Victor 2 (Perkin Elmer) spectrophotometer/luminometer.
FACS analysis of P2X4R expression.
Cells from the peritoneum and RBC-lysed blood of naive tdTomato-P2X4R transgenic mice were recovered, and nonspecific binding to Fc receptors was blocked with LEAF purified CD16/32 antibody (clone 93; catalog 101301; BioLegend), according to the manufacturer’s instructions. Cells were first stained with LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Invitrogen) to identify dead cells, according to the manufacturer’s instructions. Then, the cells were stained with BUV395-conjugated anti-CD45 (clone 30-F11), APC-conjugated anti-F4/80 (clone BM8), and FITC-conjugated anti-Ly6G (clone 1A8). Monocytes/macrophages were defined as live CD45+F4/80+Ly6G– cells, and neutrophils were identified as live CD45+F4/80–Ly6G+ cells (Supplemental Figure S8). tdTomato-P2X4R expression was evaluated in monocytes/macrophages and neutrophils.
Determination of cytokine and chemokine levels.
Concentrations of TNF-α, IL-1β, IL-10, IL-6, and MIP-2 in blood and peritoneal lavage fluid were determined using commercially available ELISA kits (R&D Systems), according to the manufacturer’s instructions.
Statistics.
Survival statistics was performed using Kaplan-Meier curve and log rank test. To compare cytokine concentrations, CFU numbers, and all other laboratory parameters, as well as densitometry read outs, 2-tailed Student’s t test or ANOVA followed by Mann Whitney U test was used. Statistical significance was assigned to P values less than 0.05.
Study approval.
All experiments on animals were reviewed and approved by the Rutgers New Jersey Medical School IACUC and by the Columbia University IACUC. All mice were bred and all colonies were maintained in accordance with the recommendations of Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
BC and GH designed the research and analyzed data. ZHN, FDV, DLD, TY, and RM contributed vital reagents. BC, IS, ZVV, JP, SF, ZHN, and PP performed the research. BC and GH wrote the paper. FDV, DLD, TY, RM, and GH reviewed and edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01GM066189 and R01DK113790 (both to GH); NIH grant R01AA022448 (DLD); the Intramural Research Program of the NIH, National Institute on Alcohol Abuse and Alcoholism (to PP); AIRC grant IG 13025 (to FDV); and the 7th Framework Program HEALTH-F2-2007-202231 ATPBone (to FDV).
Version 1. 06/07/2018
Electronic publication
Version 2. 06/12/2018
Added a reference regarding gift of mouse strain
Funding Statement
This work was supported by National Institutes of Health grants R01GM066189 (G.H.) and R01DK113790 (G.H.); the Intramural Research Program of the National Institutes of Health, National Institute on Alcohol Abuse and Alcoholism (P.P.); AIRC grant n. IG 13025, and the 7th Framework Program HEALTH-F2-2007-202231 ‘ATPBone’ (F.D.V.).
Footnotes
Conflict of interest: GH owns stock in Purine Pharmaceuticals, Inc., which has a patent license (62/532,619 and 62/562,770) for the use of P2X4 agonists in infections and sepsis. FDV is a member of the Scientific Advisory Board and receives compensation from Biosceptre Ltd., which is developing anti-P2X7 Abs for therapeutic purposes.
Reference information: JCI Insight. 2018;3(11):e99431. https://doi.org/10.1172/jci.insight.99431.
Contributor Information
Daryl L. Davies, Email: ddavies@usc.edu.
Simonetta Falzoni, Email: simonetta.falzoni@unife.it.
Francesco Di Virgilio, Email: fdv@unife.it.
Rieko Muramatsu, Email: muramatsu@molneu.med.osaka-u.ac.jp.
Toshihide Yamashita, Email: yamashita@molneu.med.osaka-u.ac.jp.
György Haskó, Email: gh2503@cumc.columbia.edu.
References
- 1.Singer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Poll T, Opal SM. Host-pathogen interactions in sepsis. Lancet Infect Dis. 2008;8(1):32–43. doi: 10.1016/S1473-3099(07)70265-7. [DOI] [PubMed] [Google Scholar]
- 3.Stearns-Kurosawa DJ, Osuchowski MF, Valentine C, Kurosawa S, Remick DG. The pathogenesis of sepsis. Annu Rev Pathol. 2011;6:19–48. doi: 10.1146/annurev-pathol-011110-130327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riedemann NC, Guo RF, Ward PA. The enigma of sepsis. J Clin Invest. 2003;112(4):460–467. doi: 10.1172/JCI19523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamim CF, Hogaboam CM, Kunkel SL. The chronic consequences of severe sepsis. J Leukoc Biol. 2004;75(3):408–412. doi: 10.1189/jlb.0503214. [DOI] [PubMed] [Google Scholar]
- 6.Oberholzer A, Oberholzer C, Moldawer LL. Interleukin-10: A complex role in the pathogenesis of sepsis syndromes and its potential as an anti-inflammatory drug. Crit Care Med. 2002;30(1 supp):S58–S63. [PubMed] [Google Scholar]
- 7. Bone RC, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101(6):1644–1655. [DOI] [PubMed] [Google Scholar]
- 8.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348(2):138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 9.Tracey KJ. Understanding immunity requires more than immunology. Nat Immunol. 2010;11(7):561–564. doi: 10.1038/ni0710-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao W, et al. A genomic storm in critically injured humans. J Exp Med. 2011;208(13):2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulloa L, Brunner M, Ramos L, Deitch EA. Scientific and clinical challenges in sepsis. Curr Pharm Des. 2009;15(16):1918–1935. doi: 10.2174/138161209788453248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torgersen C, et al. Macroscopic postmortem findings in 235 surgical intensive care patients with sepsis. Anesth Analg. 2009;108(6):1841–1847. doi: 10.1213/ane.0b013e318195e11d. [DOI] [PubMed] [Google Scholar]
- 13.Kovach MA, Standiford TJ. The function of neutrophils in sepsis. Curr Opin Infect Dis. 2012;25(3):321–327. doi: 10.1097/QCO.0b013e3283528c9b. [DOI] [PubMed] [Google Scholar]
- 14.Galbraith N, Walker S, Carter J, Polk HC. Past, present, and future of augmentation of monocyte function in the surgical patient. Surg Infect (Larchmt) 2016;17(5):563–569. doi: 10.1089/sur.2016.014. [DOI] [PubMed] [Google Scholar]
- 15.Galbraith N, Walker S, Galandiuk S, Gardner S, Polk HC. The significance and challenges of monocyte impairment: for the ill patient and the surgeon. Surg Infect (Larchmt) 2016;17(3):303–312. doi: 10.1089/sur.2015.245. [DOI] [PubMed] [Google Scholar]
- 16.Hotchkiss RS, Monneret G, Payen D. Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect Dis. 2013;13(3):260–268. doi: 10.1016/S1473-3099(13)70001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayala A, Chaudry IH. Immune dysfunction in murine polymicrobial sepsis: mediators, macrophages, lymphocytes and apoptosis. Shock. 1996;6(suppl 1):S27–S38. [PubMed] [Google Scholar]
- 18.Song GY, Chung CS, Jarrar D, Chaudry IH, Ayala A. Evolution of an immune suppressive macrophage phenotype as a product of P38 MAPK activation in polymicrobial sepsis. Shock. 2001;15(1):42–48. doi: 10.1097/00024382-200115010-00007. [DOI] [PubMed] [Google Scholar]
- 19.Junger WG. Immune cell regulation by autocrine purinergic signalling. Nat Rev Immunol. 2011;11(3):201–212. doi: 10.1038/nri2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon JL. Extracellular ATP: effects, sources and fate. Biochem J. 1986;233(2):309–319. doi: 10.1042/bj2330309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.la Sala A, Ferrari D, Di Virgilio F, Idzko M, Norgauer J, Girolomoni G. Alerting and tuning the immune response by extracellular nucleotides. J Leukoc Biol. 2003;73(3):339–343. doi: 10.1189/jlb.0802418. [DOI] [PubMed] [Google Scholar]
- 22.Antonioli L, Pacher P, Vizi ES, Haskó G. CD39 and CD73 in immunity and inflammation. Trends Mol Med. 2013;19(6):355–367. doi: 10.1016/j.molmed.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burnstock G. Pathophysiology and therapeutic potential of purinergic signaling. Pharmacol Rev. 2006;58(1):58–86. doi: 10.1124/pr.58.1.5. [DOI] [PubMed] [Google Scholar]
- 24.Rayah A, Kanellopoulos JM, Di Virgilio F. P2 receptors and immunity. Microbes Infect. 2012;14(14):1254–1262. doi: 10.1016/j.micinf.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Idzko M, Ferrari D, Eltzschig HK. Nucleotide signalling during inflammation. Nature. 2014;509(7500):310–317. doi: 10.1038/nature13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ralevic V, Burnstock G. Receptors for purines and pyrimidines. Pharmacol Rev. 1998;50(3):413–492. [PubMed] [Google Scholar]
- 27.Gu BJ, Zhang WY, Bendall LJ, Chessell IP, Buell GN, Wiley JS. Expression of P2X(7) purinoceptors on human lymphocytes and monocytes: evidence for nonfunctional P2X(7) receptors. Am J Physiol, Cell Physiol. 2000;279(4):C1189–C1197. doi: 10.1152/ajpcell.2000.279.4.C1189. [DOI] [PubMed] [Google Scholar]
- 28.Lammas DA, Stober C, Harvey CJ, Kendrick N, Panchalingam S, Kumararatne DS. ATP-induced killing of mycobacteria by human macrophages is mediated by purinergic P2Z(P2X7) receptors. Immunity. 1997;7(3):433–444. doi: 10.1016/S1074-7613(00)80364-7. [DOI] [PubMed] [Google Scholar]
- 29.Kusner DJ, Adams J. ATP-induced killing of virulent Mycobacterium tuberculosis within human macrophages requires phospholipase D. J Immunol. 2000;164(1):379–388. doi: 10.4049/jimmunol.164.1.379. [DOI] [PubMed] [Google Scholar]
- 30.Coutinho-Silva R, Perfettini JL, Persechini PM, Dautry-Varsat A, Ojcius DM. Modulation of P2Z/P2X(7) receptor activity in macrophages infected with Chlamydia psittaci. Am J Physiol Cell Physiol. 2001;280(1):C81–C89. doi: 10.1152/ajpcell.2001.280.1.C81. [DOI] [PubMed] [Google Scholar]
- 31.Wright GD. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol. 2016;24(11):862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 32.Csóka B, et al. Extracellular ATP protects against sepsis through macrophage P2X7 purinergic receptors by enhancing intracellular bacterial killing. FASEB J. 2015;29(9):3626–3637. doi: 10.1096/fj.15-272450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antonioli L, Blandizzi C, Pacher P, Haskó G. Immunity, inflammation and cancer: a leading role for adenosine. Nat Rev Cancer. 2013;13(12):842–857. doi: 10.1038/nrc3613. [DOI] [PubMed] [Google Scholar]
- 34.Haskó G, Pacher P. Regulation of macrophage function by adenosine. Arterioscler Thromb Vasc Biol. 2012;32(4):865–869. doi: 10.1161/ATVBAHA.111.226852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fairn GD, Grinstein S. How nascent phagosomes mature to become phagolysosomes. Trends Immunol. 2012;33(8):397–405. doi: 10.1016/j.it.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Fairbairn IP, Stober CB, Kumararatne DS, Lammas DA. ATP-mediated killing of intracellular mycobacteria by macrophages is a P2X(7)-dependent process inducing bacterial death by phagosome-lysosome fusion. J Immunol. 2001;167(6):3300–3307. doi: 10.4049/jimmunol.167.6.3300. [DOI] [PubMed] [Google Scholar]
- 37.Ip WK, et al. Phagocytosis and phagosome acidification are required for pathogen processing and MyD88-dependent responses to Staphylococcus aureus. J Immunol. 2010;184(12):7071–7081. doi: 10.4049/jimmunol.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falzoni S, Donvito G, Di Virgilio F. Detecting adenosine triphosphate in the pericellular space. Interface Focus. 2013;3(3):20120101. doi: 10.1098/rsfs.2012.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morciano G, et al. Use of luciferase probes to measure ATP in living cells and animals. Nat Protoc. 2017;12(8):1542–1562. doi: 10.1038/nprot.2017.052. [DOI] [PubMed] [Google Scholar]
- 40.Xu J, et al. P2X4 receptor reporter mice: sparse brain expression and feeding-related presynaptic facilitation in the arcuate nucleus. J Neurosci. 2016;36(34):8902–8920. doi: 10.1523/JNEUROSCI.1496-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Németh ZH, et al. Adenosine A2A receptor inactivation increases survival in polymicrobial sepsis. J Immunol. 2006;176(9):5616–5626. doi: 10.4049/jimmunol.176.9.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khakh BS, Proctor WR, Dunwiddie TV, Labarca C, Lester HA. Allosteric control of gating and kinetics at P2X(4) receptor channels. J Neurosci. 1999;19(17):7289–7299. doi: 10.1523/JNEUROSCI.19-17-07289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corrêa G, Marques da Silva C, de Abreu Moreira-Souza AC, Vommaro RC, Coutinho-Silva R. Activation of the P2X(7) receptor triggers the elimination of Toxoplasma gondii tachyzoites from infected macrophages. Microbes Infect. 2010;12(6):497–504. doi: 10.1016/j.micinf.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Biswas D, Qureshi OS, Lee WY, Croudace JE, Mura M, Lammas DA. ATP-induced autophagy is associated with rapid killing of intracellular mycobacteria within human monocytes/macrophages. BMC Immunol. 2008;9:35. doi: 10.1186/1471-2172-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettengill MA, et al. Reversible inhibition of Chlamydia trachomatis infection in epithelial cells due to stimulation of P2X(4) receptors. Infect Immun. 2012;80(12):4232–4238. doi: 10.1128/IAI.00441-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coutinho-Silva R, et al. Inhibition of chlamydial infectious activity due to P2X7R-dependent phospholipase D activation. Immunity. 2003;19(3):403–412. doi: 10.1016/S1074-7613(03)00235-8. [DOI] [PubMed] [Google Scholar]
- 47.Suurväli J, Boudinot P, Kanellopoulos J, Rüütel Boudinot S. P2X4: A fast and sensitive purinergic receptor. Biomed J. 2017;40(5):245–256. doi: 10.1016/j.bj.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pauwels AM, Trost M, Beyaert R, Hoffmann E. Patterns, Receptors, and Signals: Regulation of Phagosome Maturation. Trends Immunol. 2017;38(6):407–422. doi: 10.1016/j.it.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babior BM. NADPH oxidase: an update. Blood. 1999;93(5):1464–1476. [PubMed] [Google Scholar]
- 50.Van den Bossche J, O’Neill LA, Menon D. Macrophage immunometabolism: Where are we (going)? Trends Immunol. 2017;38(6):395–406. doi: 10.1016/j.it.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 51.West AP, et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472(7344):476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geng J, et al. Kinases Mst1 and Mst2 positively regulate phagocytic induction of reactive oxygen species and bactericidal activity. Nat Immunol. 2015;16(11):1142–1152. doi: 10.1038/ni.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ali SR, et al. Anthrax toxin induces macrophage death by p38 MAPK inhibition but leads to inflammasome activation via ATP leakage. Immunity. 2011;35(1):34–44. doi: 10.1016/j.immuni.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eltzschig HK, et al. ATP release from activated neutrophils occurs via connexin 43 and modulates adenosine-dependent endothelial cell function. Circ Res. 2006;99(10):1100–1108. doi: 10.1161/01.RES.0000250174.31269.70. [DOI] [PubMed] [Google Scholar]
- 55.Bao Y, Chen Y, Ledderose C, Li L, Junger WG. Pannexin 1 channels link chemoattractant receptor signaling to local excitation and global inhibition responses at the front and back of polarized neutrophils. J Biol Chem. 2013;288(31):22650–22657. doi: 10.1074/jbc.M113.476283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao F, Waldrop SL, Khimji AK, Kilic G. Pannexin1 contributes to pathophysiological ATP release in lipoapoptosis induced by saturated free fatty acids in liver cells. Am J Physiol Cell Physiol. 2012;303(10):C1034–C1044. doi: 10.1152/ajpcell.00175.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wegiel B, et al. Macrophages sense and kill bacteria through carbon monoxide-dependent inflammasome activation. J Clin Invest. 2014;124(11):4926–4940. doi: 10.1172/JCI72853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Csóka B, et al. A2B adenosine receptors protect against sepsis-induced mortality by dampening excessive inflammation. J Immunol. 2010;185(1):542–550. doi: 10.4049/jimmunol.0901295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Csóka B, et al. CD39 improves survival in microbial sepsis by attenuating systemic inflammation. FASEB J. 2015;29(1):25–36. doi: 10.1096/fj.14-253567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haskó G, et al. Ecto-5′-nucleotidase (CD73) decreases mortality and organ injury in sepsis. J Immunol. 2011;187(8):4256–4267. doi: 10.4049/jimmunol.1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ulmann L, Hirbec H, Rassendren F. P2X4 receptors mediate PGE2 release by tissue-resident macrophages and initiate inflammatory pain. EMBO J. 2010;29(14):2290–2300. doi: 10.1038/emboj.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fujita R, Ma Y, Ueda H. Lysophosphatidic acid-induced membrane ruffling and brain-derived neurotrophic factor gene expression are mediated by ATP release in primary microglia. J Neurochem. 2008;107(1):152–160. doi: 10.1111/j.1471-4159.2008.05599.x. [DOI] [PubMed] [Google Scholar]
- 63.Tsuda M, et al. P2X4 receptors induced in spinal microglia gate tactile allodynia after nerve injury. Nature. 2003;424(6950):778–783. doi: 10.1038/nature01786. [DOI] [PubMed] [Google Scholar]
- 64.Beggs S, Trang T, Salter MW. P2X4R+ microglia drive neuropathic pain. Nat Neurosci. 2012;15(8):1068–1073. doi: 10.1038/nn.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trang T, Beggs S, Wan X, Salter MW. P2X4-receptor-mediated synthesis and release of brain-derived neurotrophic factor in microglia is dependent on calcium and p38-mitogen-activated protein kinase activation. J Neurosci. 2009;29(11):3518–3528. doi: 10.1523/JNEUROSCI.5714-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coddou C, Yan Z, Obsil T, Huidobro-Toro JP, Stojilkovic SS. Activation and regulation of purinergic P2X receptor channels. Pharmacol Rev. 2011;63(3):641–683. doi: 10.1124/pr.110.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crump A, Ōmura S. Ivermectin, ‘wonder drug’ from Japan: the human use perspective. Proc Jpn Acad Ser B Phys Biol Sci. 2011;87(2):13–28. doi: 10.2183/pjab.87.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zemkova H, Tvrdonova V, Bhattacharya A, Jindrichova M. Allosteric modulation of ligand gated ion channels by ivermectin. Physiol Res. 2014;63(suppl 1):S215–S224. doi: 10.33549/physiolres.932711. [DOI] [PubMed] [Google Scholar]
- 69.Jelínkova I, et al. Identification of P2X(4) receptor transmembrane residues contributing to channel gating and interaction with ivermectin. Pflugers Arch. 2008;456(5):939–950. doi: 10.1007/s00424-008-0450-4. [DOI] [PubMed] [Google Scholar]
- 70.Perry JA, Koteva K, Verschoor CP, Wang W, Bowdish DM, Wright GD. A macrophage-stimulating compound from a screen of microbial natural products. J Antibiot. 2015;68(1):40–46. doi: 10.1038/ja.2014.83. [DOI] [PubMed] [Google Scholar]
- 71.Gennari R, Alexander JW, Gianotti L, Eaves-Pyles T, Hartmann S. Granulocyte macrophage colony-stimulating factor improves survival in two models of gut-derived sepsis by improving gut barrier function and modulating bacterial clearance. Ann Surg. 1994;220(1):68–76. doi: 10.1097/00000658-199407000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maródi L, Káposzta R, Nemes E. Survival of group B streptococcus type III in mononuclear phagocytes: differential regulation of bacterial killing in cord macrophages by human recombinant gamma interferon and granulocyte-macrophage colony-stimulating factor. Infect Immun. 2000;68(4):2167–2170. doi: 10.1128/IAI.68.4.2167-2170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schneider M, et al. Interaction of Purinergic P2X4 and P2X7 Receptor Subunits. Front Pharmacol. 2017;8:860. doi: 10.3389/fphar.2017.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hung SC, et al. P2X4 assembles with P2X7 and pannexin-1 in gingival epithelial cells and modulates ATP-induced reactive oxygen species production and inflammasome activation. PLoS One. 2013;8(7):e70210. doi: 10.1371/journal.pone.0070210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sim JA, et al. Altered hippocampal synaptic potentiation in P2X4 knock-out mice. J Neurosci. 2006;26(35):9006–9009. doi: 10.1523/JNEUROSCI.2370-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ozaki T, et al. The P2X4 receptor is required for neuroprotection via ischemic preconditioning. Sci Rep. 2016;6:25893. doi: 10.1038/srep25893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pellegatti P, Falzoni S, Pinton P, Rizzuto R, Di Virgilio F. A novel recombinant plasma membrane-targeted luciferase reveals a new pathway for ATP secretion. Mol Biol Cell. 2005;16(8):3659–3665. doi: 10.1091/mbc.e05-03-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Csóka B, et al. CB2 cannabinoid receptors contribute to bacterial invasion and mortality in polymicrobial sepsis. PLoS One. 2009;4(7):e6409. doi: 10.1371/journal.pone.0006409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Csóka B, et al. A2A adenosine receptors and C/EBPβ are crucially required for IL-10 production by macrophages exposed to Escherichia coli. Blood. 2007;110(7):2685–2695. doi: 10.1182/blood-2007-01-065870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilhelm K, et al. Graft-versus-host disease is enhanced by extracellular ATP activating P2X7R. Nat Med. 2010;16(12):1434–1438. doi: 10.1038/nm.2242. [DOI] [PubMed] [Google Scholar]
- 81.Németh ZH, et al. Adenosine receptor activation ameliorates type 1 diabetes. FASEB J. 2007;21(10):2379–2388. doi: 10.1096/fj.07-8213com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haskó G, Szabó C, Németh ZH, Kvetan V, Pastores SM, Vizi ES. Adenosine receptor agonists differentially regulate IL-10, TNF-α, and nitric oxide production in RAW 264.7 macrophages and in endotoxemic mice. J Immunol. 1996;157(10):4634–4640. [PubMed] [Google Scholar]
- 83.Haskó G, et al. Adenosine inhibits IL-12 and TNF-[alpha] production via adenosine A2a receptor-dependent and independent mechanisms. FASEB J. 2000;14(13):2065–2074. doi: 10.1096/fj.99-0508com. [DOI] [PubMed] [Google Scholar]
- 84.Himer L, et al. Adenosine A2A receptor activation protects CD4+ T lymphocytes against activation-induced cell death. FASEB J. 2010;24(8):2631–2640. doi: 10.1096/fj.10-155192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Enjyoji K, et al. Targeted disruption of cd39/ATP diphosphohydrolase results in disordered hemostasis and thromboregulation. Nat Med. 1999;5(9):1010–1017. doi: 10.1038/12447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.