Abstract
The activity of RNA is controlled by different types of post-transcriptional modifications, such as the addition of methyl groups and other chemical and structural changes, that have been recently described in human cells by high-throughput sequencing. Herein, we will discuss how the so called epitranscriptome is disrupted in cancer and what the contribution of its writers, readers and erasers to the process of cellular transformation is, particularly focusing in the epigenetic modifications of ncRNAs.
An introduction to RNA modifications
All life forms need mechanisms to regulate RNA levels and activities. An important part of these control belts for RNA occurs at the transcriptional level, but it is becoming apparent that a significant part of RNA homeostasis depends on RNA stability and degradation. This can be particularly true for common human diseases, such as cancer, where an aberrant RNA transcriptome constitutes a hallmark of transformed cells that cannot be fully attributed to alterations in the DNA-associated transcriptional start sites. In this respect, one interesting avenue to be further explored is that molecular lesions in tumors occur not only in the original DNA molecule, but also in RNA itself. More than one-hundred differently modified nucleotides have been described and catalogued in RNA molecules (1). While, some of these have also been described in DNA such as cytosine methylation and hydroxymethylcytosine and recently adenine methylation (2), DNA, however, seems to have a shorter repertoire of modified nucleotides and RNA molecules can show a more flamboyant spectrum that includes, among others, pseudouridine or a hyper-modified 7-deaza-guanosine (queuosine).
From an academic standpoint, the RNA modifications can be divided into two main types: reversible and nonreversible. Among the first, one could count the different types of RNA methylation, such as cytosine and adenosine methylation, whereas in the most permanent RNA changes, one could cite editing and splicing (including the formation of circular RNAs). However, this classic scenario is quickly evolving and previously presumed nonreversible modifications are slowly being discovered as reversible.
Most importantly, the number and types of potential transcripts that are amenable to modification are growing almost exponentially, in part due to the emergence of transcriptomic techniques that use different strategies to provide RNA landscapes for 5-methylcytosine (m5C) (3–5), 5-hydroxymethylcytosine (hm5C) (6), N6-methyladenosine (m6A) (7, 8), N1-methyladenosine (m1A) (9, 10), pseudouridine (ψ) (11–13) or inosine-to-adenine editing (14). Many times, these chemical modifications are dynamic, adapting to the cell environment, however, at the same time, they are locally transmitted during mitosis and meiosis. Thus, in a similar manner as DNA and histone modifications, we can discuss an RNA epigenetic setting or an epitranscriptome. Herein, we will discuss how the ncRNA epitranscriptome is altered in cancer, and the possible functional consequences of these alterations, the existence of defects in the writers (i.e. RNA modifying enzymes), erasers (enzymes that remove these marks and modifications) and readers (proteins that decode these RNA modifications) of the non-coding RNA (ncRNA) epigenetic code, and how this knowledge could be exploited for translational and therapeutic purposes. Figure 1 illustrates the RNA chemical modifications described in human cells; their writers, readers and erasers; and the different effects on the transcript molecule.
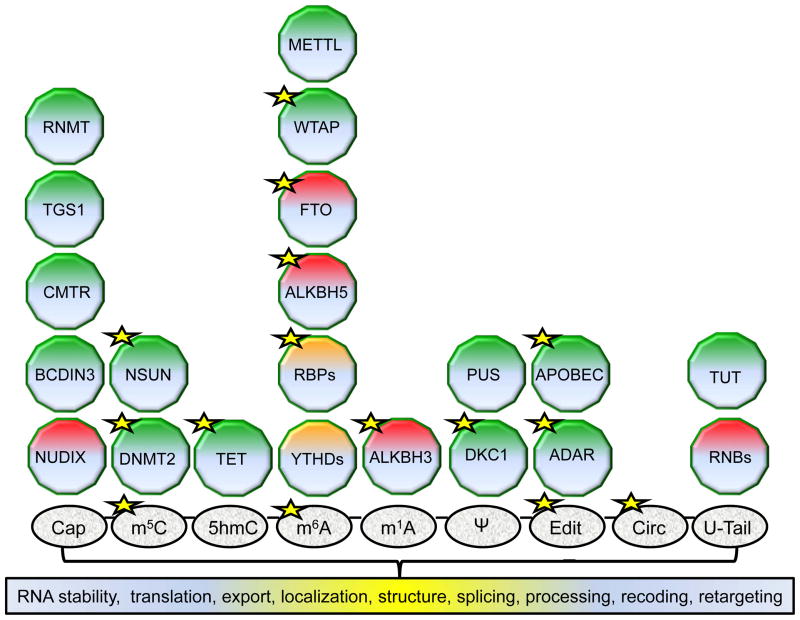
Figure 1. Examples of RNA chemical modifications; their writers, readers and erasers; and different effects on the transcript molecule.
In the middle, a molecule of RNA with different chemical modifications is shown: addition of a 5′-end cap (cap), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), N6-methyladenosine (m6A), N1-methyladenosine (m1A), pseudouridine (ψ), editing (Edit), circularization of RNA (circ) and poly (U) tails (U-Tail). Top, protein(s) associated with each modification: writers (green), readers (orange) and erasers (red), Below, functional consequences of RNA modification. Proteins and RNA chemical marks reported to be involved in tumorigenesis are tagged with a yellow star. ADAR, adenosine deaminase RNA specific represented by ADAR1 and ADAR2; APOBEC, apolipoprotein B mRNA editing enzymes represented by APOBEC3A and APOBEC3B; ALKBH3, alkB homolog 3, alpha-ketoglutarate dependent dioxygenase; ALKBH5, alkB homolog 3, alpha-ketoglutarate dependent dioxygenase; BCDIN3, methylphosphate capping enzyme; CMTR, cap methyltransferases represented by CMTR1 and CMTR2; DKC1, dyskerin pseudouridine synthase 1; DNMT2, DNA methyltransferase-2; FTO, fat mass and obesity associated; methyltransferase-like family represented by METTL3 and METTL14; NUDIX, nudix hydrolases represented by CDP2 and NUDT16; NSUN, NOP2/Sun RNA methyltransferase family; PUS, pseudouridine synthase family; RBPs, RNA Binding Proteins such as HuR and HNRNPA2B1; RNBs, RNase II / RNB family represented by Dis3, Dis3L1 and Dis3L2; RNMT, RNA guanine-7 methyltransferase; TET, tet methylcytosine dioxygenase represented by TET1 and TET2; WTAP, Wilms tumor 1 associated protein; TGS1, trimethylguanosine synthase 1; TUT, terminal uridylyl transferases represented by ZCCHC11 (TUT4), ZCCHC6 (TUT7) and PAPD4 (TUT2); YTHD, YT521-B homology domain-containing proteins composed of the YTHDF family (YTHDF1, YTHDF2, YTHDF3), YTHDC1 and YTHDC2.
Chemical modifications of ncRNAs and their functions
The role of chemical modifications of RNA has been extensively explored and discussed (1, 15, 16). However, with the exception of some post-transcriptional chemical marks on certain subtypes of ncRNAs, such as transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), that have been known for decades, the focus of research in this area has mainly been on the impact of these modifications on mRNA. By contrast, little attention has been devoted to their effects on other classes of ncRNAs. Part of the explanation for this current lack of knowledge relates, of course, to the most recent timing of ncRNAs discovery and the very recent development of appropriate technologies. Indeed, in recent years, we have witnessed an amazingly exponential growth in knowledge of other types of ncRNAs that exert essential roles in cellular homeostasis and contribute to the natural history of many human diseases, including cancer. These range from small ncRNAs (sncRNAs), such as microRNAs and piRNAs, to long ncRNAs (lncRNA), such as long intergenic ncRNAs (lincRNAs), pseudogene transcripts and circular RNAs (circRNAs). Table 1 shows a summary of the various classes of ncRNAs, describing their characteristics in human cells, and the role of selected RNA chemical modifications according to the transcript. The reader will note that major gaps remain regarding the chemical modifications of several ncRNA subclasses, and very few examples that are reported for those included in Table 1, however we expect that this review will raise the interest of many researchers to act and fill in these unknown gaps. In the following text, we will discuss what is known about these post-transcriptional marks in ncRNAs and how the field could advance.
Table 1.
Epitranscriptomics of ncRNAs.
| A. Chemical modifications in ncRNAs. | ||||||
|---|---|---|---|---|---|---|
| ncRNA type | Size | Location | # humans | Functions | Described chemical modification | Examples of modified ncRNA |
| miRNAs | 19–24 bp | Widespread | >1,500 | Targeting mRNAs, | Editing, m5C, m6A, U-tail, PolyA, 5′-O-m | let-7, miR376a*, miR455-5p, miR381, miR26 |
| piRNAs | 23–31 bp | Clusters | >20,000 | Transposon silencing | 2′-O-m, U-tail | piR30840, piR-L-163 |
| snRNAs | 150 bp | Different loci | Several | RNA splicing | CAP, TMG, 2′-O-m, ψ, U-tail, m5C, m6A | U6 spliceosomal RNA |
| snoRNAs | 60–300 bp | Mostly intronic | >300 | rRNA modifications | CAP, TMG, 2′-O-m, ψ | C/D Box and H/ACA Box snoRNAs |
| tRNAs | 76–90 bp | Widespread | 519 | Aminoacid carriers | m5C, m1A, ψ, Queuosine, mcm5U, editing | AspGUC, GlyGCC, AspGTC, GlyUCC, ValAAC |
| rRNAs | 120–1874 bp | Clusters | 6 | mRNA reading/recoding | m5C, m1A, ψ, editing, 2′-O-m, m1ψ, m7G, m62A, ac4C | 28S RNA, 12S RNA, 5S RNA |
| vault RNAs | 88–98 bp | Chr 5, Chr X | 4 | Vault particle regulation | m5C | vtRNA1.1, vtRNA1.2, and vtRNA1.3 |
| lncRNAs* | >200 bp | Widespread | >30,000 | Many | CAP, m5C, m6A, ψ, splicing, editing, U-tail | HOTAIR, XIST, MALAT1, TERC, RPPH1 |
| Circular RNAs | >200 bp | Mostly intronic | >15,000 | RNA regulation | Splicing, editing | Within UBAC2, SMARCA, SPECC1, HIPK2 |
| B. Role of selected RNA chemical modifications according to the transcript. | ||
|---|---|---|
| Modification | Role in coding RNAs | Role in ncRNAs |
| m5C | Translation efficiency, mRNA structure, genetic recoding | Regulation vault ncRNA processing into small RNAs, tRNA cleavage |
| hm5C | Translation efficiency | Unknown |
| m6A | Translation efficiency, mRNA structure, stability, export and splicing | lncRNA silencing by XIST, t-RNA selection |
| m1A | Translation efficiency | tRNA T-loop, rRNA regulation |
| ψ | Translation efficiency, mRNA structure, genetic recoding | tRNA, rRNA, snoRNA, lncRNA regulation |
lncRNAs include among others lincRNAs, Transcribed-Ultraconserved Regions (T-UCRs), antisense RNAs and pseudogene RNAs.
CAP, 5′-end cap ; m5C, 5-methylcytosine; m6A, N6-methyladenosine; m1A, N1-methyladenosine; ψ, pseudouridine ; 2′-O-m, ribose 2′-O-methylation; 5′-O-m, O-methylation of the 5′ monophosphate; TMG, trimethylguanosine; mcm5U, 5-methoxycarbonylmethyluridine; ψ, 1-methylpseudouridine; m7G, 7-methylguanosine; m62A, N6-dimethyladenosine; ac4C, N4-acetylcytidine
m5C, 5-methylcytosine; hm5C, 5-hydroxymethylcytosine; m6A, N6-methyladenosine; m1A, N1-methyladenosine; ψ, pseudouridine.
One of the most well-known chemical modifications of RNA affects its 5′-end, the so called “5′ cap” (17). Therein, the most characterized cap modification is the addition of an N7-methylguanosine (m7G). Further changes frequently occur and in some small nuclear RNAs (snRNAs) and nucleolar RNAs (snoRNAs), a trimethylguanosine (m2,2,7G or TMG) cap can be added. snRNAs are additionally modified by ribose 2′-O-methylation on the first and second transcribed nucleosides to allow for correct splicing.
5-methylcytosine (m5C) was originally thought to be confined to two particular types of ncRNAs, tRNAs and rRNAs, but the use of bisulfite modification and deep sequencing have identified m5C in many other types of RNA transcripts (3–5). No strict motif for m5C has been described to date (3–5). m5C are common in mRNA 3′ UTRs, whereas Argonaute I–IV binding sites are enriched in the proximity of m5C sites, pointing to a role for m5C in miRNA targeting (3). Modification of lncRNAs by m5C, regulates their processing in smaller regulatory ncRNAs (18), the positioning of 2′-O-methylation of snRNA (19) or the production of tRNA precursors (3–5). The lncRNA XIST is also regulated by m5C where cytosine methylation has been shown to interfere with the binding of the histone modifier PRC2 (20), thus establishing a link between epigenetic modifications in RNAs and histones. Importantly, m5C is not a static mark of RNA. The same enzymes that demethylate DNA are also able to catalyze the oxidative demethylation of m5C to 5-hydroxymethylcytosine (hm5C) in RNA (21). hm5C is preferentially present in polyadenylated RNAs, where it could favor translation in the case of mRNAs (6). It has been reported as an overrepresented motif that tends to be UC-rich and contains UCCUC repeats (6).
RNA can also be modified at adenosines in the form of N6-methyladenosine (m6A) and N1-methyladenosine (m1A). The presence of m6A has also been discovered in DNA (2). m6A is the most abundant internal modification of mRNA (8, 22), but it is also relevant for miRNAs controlling their maturation and expression levels. In this regard, there is an enrichment of the m6A consensus motif at the junction between the hairpin stem and the flanking single stranded RNA regions of pri-miRNAs (23). The depletion of m6A diminished DGCR8 binding to pri-miRNAs, thus preventing the formation of the mature miRNAs as it has been shown in the case of the tumor suppressor miRNA let-7 (24). m6A is also found in lncRNAs, although its functions in this type of transcript is still unclear. In the case of the extremely abundant lncRNA Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) (25), it has been shown that m6A destabilizes the hairpin stem structure (26). m6A is also required for the efficient transcriptional repression mediated by the lncRNA XIST (27). At the global RNA level, transcriptome-wide m6A mapping suggests the motif DRACH (D=A, G or U; H=A, C or U) as a consensus motif for m6A (7,8,28,29). The other methylated adenosine present in RNA, m1A, is typically found in the tRNA T-loop. However, recent data demonstrate that m1A is not restricted to these abundant ncRNAs, but rather, is also observed around the start codon upstream of the first splice site (9) and in the 5′ untranslated regions of mRNAs (10), in sharp contrast with the localization of m6A that is enriched in 3′ UTRs and near stop codons (8). The proposed motifs for m1A also differ greatly from those recognized for m6A (9,10).
Two other RNA nucleotides are also worth mentioning: pseudouridine, the 5′-ribosyluracil isomer of uridine, and queuosine, a rare nucleoside that is structurally similar to guanine. Pseudouridylation is the most abundant modification in ncRNAs (30), found not only in tRNA, rRNA and snoRNAs, but also observed in lncRNAs (11–13), such as the aforementioned XIST and MALAT1. On the other hand, queuosine substitution of guanosine at the wobble position of tRNAs and its presence could alter mRNA decoding (31). Overall, m6A is the most prevalent internal modification found in mRNA (15), and with more than an estimated 20,000 unique sites in lncRNAs (32), whereas m5C is most prevalent in tRNAs and RNAs, with more than 30,000 unique sites in lncRNAs (32). Around 2,000 unique sites have been proposed for pseudouridine in lncRNAs (32), but, for this and for other RNA modifications, emerging epitranscriptomic data and further posterior validation experiments could provide different numbers.
Finally, RNAs can undergo specific modifications at their 3′-ends. Polyadenylation is the obvious modification that ncRNAs undergo to modify their stability, but uridylation can also occur. The best example is the precursor of let-7 (33–35), however there is an increasing amount of data regarding uridylation and adenylation events affecting the activity of miRNAs, as recently reviewed (22). It is probable that the development of new high-throughput technologies will provide an improved landscape of the so called “3′-terminome” of RNA.
Unlike the described RNA modifications, which are reversible and do not interfere with RNA sequence per se, there are more stable changes in RNAs, such as splicing and editing. These are irreversible changes to the RNAs that interfere with the stored information and with their role in cancer, which have been previously reviewed (36,37). A few exciting, recent discoveries in these areas are the cancer-associated alterations of the editing enzymes APOBEC3B (38–40) and ADAR1 (41,42) and the role of circular RNAs in tumorigenesis (43–47), however, they will not be discussed herein.
Chemical modifications of ncRNA in cancer and their associated genes
The above described scenario of balanced ncRNA modifications allow cellular homeostasis to undergo important mayhem in human cancer that we are just now starting to realize. Without having a complete picture of the global epitranscriptome of transformed cells, we have to construct the puzzle from the small pieces that are emerging from each particular RNA modification. At the same time, we can try to go back upstream in the pathway and dissect what went wrong with the genes encoding the proteins responsible for adding, reading or erasing a specific RNA modification.
For the processes of RNA capping and decapping modifications, such as m7G or m2,2,7G, and modifiers, such as RNA cap methyltransferases RNTM, TGS1, CMTR1, CMTR2 and BCDIN3 (16) or the decapping enzymes CDP2 and NUDT16 (48) there is a lack of information in the transformed cell setting. More is known about m5C, particularly at the level of the involved RNA methyltransferases. The DNA methyltransferase 2 (DNMT2) turned out to be an RNA methyltransferase (TRDMT1) with activity first described for the methylation of the cytosine located in the position 38 in tRNAAsp (49), a modification that affects its processing into tRNA fragments (tRFs) (50). Interestingly, DNMT2 mutations in cancer cells mostly lead to diminished RNA methyltransferase activity (51). The other type of cytosine-5 RNA methylases is constituted by the NSun family, NSun2 being the most well characterized member (4, 17). Nsun2 methylates cytosines in tRNAs, but also in other ncRNAs including lncRNA and vault associated RNAs (4,17). NSun2 is upregulated in some tumor types by copy-number gains (52) and it is targeted by critical transforming proteins such as C-MYC (53) and Aurora B Kinase (54). However, in other classes of malignancies, such as skin cancer, NSUN2 expression is depleted causing a reduction of protein translation rates and increasing the tumor-initiating population (55). Other proteins in this family include NOP2 (NSun1), NSUN3, NSUN5 and NSUN7. Interestingly, NSUN5 modulates lifespan and stress resistance (56) and NSUN3 deficiency, in addition to m5C reduction, which results in diminished 5-formylcytosine in tRNA (57).
In a manner similar to that of m5C in DNA, Tet methyldioxygenases (TET1, TET2, and TET3) catalyze hydroxylation of m5C to hm5C in RNA (6, 20). Because TET1 mutations and deletions are observed in myeloid leukemias (in association with impaired hm5C in DNA (58)), and because TET1 loss accompanies the presence of aberrant fusion proteins found in hematological malignancies where it can act as a tumor suppressor gene (59), one also wonders if the loss of TET proteins might have a greater impact in RNA demethylation than in DNA demethylation, a possibility that is yet to be explored.
m6A, in addition to the above described presence in ncRNAs with a role in cancer, such as let-7 and MALAT1, has been described to act in cell reprogramming by affecting mRNAs associated with different degrees of pluripotency, where at the same time miRNAs control m6A in these mRNAs by sequence pairing (60). Thus, m6A affects ncRNAs in multiple ways and their related protein machinery could also potentially be involved in cellular transformation. The methylation of m6A is performed by methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14) and the cofactor Wilms’ tumor 1-associating protein (WTAP). For the latter, the connection with cancer is clear, which is a splicing binding partner for the Wilms’ tumor 1 (WT1) protein (61) and regulating cell cycle progression (62). From a functional standpoint, WTAP and METTL3, upon their recruitment mediated by the RBM15 and RBM15B proteins, promote the methylation of XIST and its transcriptional repression activity (27).
While readers of m5C or hm5C in RNA have not yet been identified, these proteins have been characterized for m6A: the YT521-B homology (YTHD) domain-containing proteins that, in humans, is composed of the YTHDF family (YTHDF1, YTHDF2, YTHDF3) (7, 63), YTHDC1 and YTHDC2. The YTHDC1 protein binds m6A residues of Xist and is required for the transcriptional silencing mediated by this ncRNA (27). Interestingly, the Drosophila nuclear protein YT521-B also binds m6A residues in the sex determination factor Sex lethal (Sxl) (64) and contributes to sex determination (64,65). The role of YT521-B in the potentiation of Sxl alternative pre-mRNA splicing (64) has a correspondence in human cells where the YTHDC1 protein has been described as a global regulator of mRNA splicing by its binding to m6A RNA residues (66). Interestingly, even if these proteins exert similar functions among different species, m6A patterns could nevertheless show different patterns, as it has been recently shown in distinct primates (67). These variations could provide another explanation for the phenotypic difference between closely related species. Additional m6A RNA binders are the RNA binding proteins HuR and HNRNPA2B1, which modulate alternative splicing and enhance miRNA production (23, 24).
The m6A modification is not a final fixture of the RNA molecule, but it can be oxidatively reversed by two members of the AlkB family of the Fe(II) and a-ketoglutarate-dependent dioxygenases, FTO (68) and ALKBH5 (69). FTO germline polymorphisms have been associated with melanoma risk (70) and loss-of-function mutations of FTO in a monogenic disorder impair proliferation and promote senescence (71). Conversely, FTO promotes leukemogenesis by reducing m6A levels of mRNA transcripts involved in cell differentiation (72). FTO was also found to be overexpressed in human breast cancer samples. Among the different breast cancer sub-types, the aggressive HER2-positive is the one in which FTO is primarily overexpressed, suggesting a critical role for FTO in carcinogenesis and aggressiveness of breast cancer (73). Interestingly, FTO overexpression triggers an aberrant metabolic state that allows the breast cancer cell line, SUM149, to survive glutamine deprivation stress (74). FTO overexpressed cells are resistant to chemotherapeutic drugs and show a higher ability to metastatic potential. Further, the link to cancer is also found for ALKBH5, where the knockout mice undergo aberrant expression of the p53 functional interaction network (69) and its overexpression stabilizes NANOG levels by decreasing m6A and increasing the number of breast cancer stem cells (75).
For m1A in RNA, neither the corresponding methyltransferases nor the readers for this modification have been characterized, but RNA can be demethylated at this position by other members of the AlkB family of the Fe(II) and a-ketoglutarate-dependent dioxygenases, ALKBH3 (10). It is worth pointing out that ALKBH3 has been recognized as a DNA repair enzyme guarding the genome against alkylation damage (76), but the identified function of ALKBH3 as a RNA demethylase for m1A could open new lines of research in the area of chemotherapy response prediction. In this regard, ALKBH3 repair function is dependent on the DNA helicase ASCC3 (77) and other helicase-binding partners have been associated with chemosensitivity profiles for DNA damaging agents (78), thus a link might exist between the m1A mark, the described helicase activities partner and the observed chemosensitive signatures, which warrants further research.
Regarding the other modified nucleotides that are less frequently investigated, pseudouridine formation in RNA involves the pseudouridine synthase (PUS) family, where most of the pseudouridine sites correspond to sequence motifs associated to these enzymes (11,12), and dyskerin (DKC1) (30). DKC1, which modifies rRNA, is mutated in X-linked dyskeratosis congenital, where an increased susceptibility to cancer exists and an impairment in rRNA pseudouridylation is observed before the onset of the disease in hypomorphyc Dkc1 mutant mice (79). A specific defect of the internal ribosome entry site also occurs upon DKC1 loss, affecting the translation of tumor suppressor proteins (80). DKC1 mutations also affect the stability of H/ACA small RNAs and are associated with impaired hematopoietic stem cell differentiation (81). Neither the readers nor erasers of pseudouridine have been described to date.
While still poorly characterized, modified nucleoside and queuosine provide a new perspective of RNA modifications in cancer. Because in this case, although queuosine is incorporated into eukaryotic tRNAs by the tRNA-guanine transglycosylase, only bacteria can synthesize its precursor, queuine, de novo (82). Thus, human cells must rely on nutrition and our gut bacteria to obtain the necessary queuine to substitutes guanosin at the wobble position of tRNAs. Thus, two factors of increasing interest in the cancer arena, diet and microbiome, could be linked by an RNA modification.
Last but not least, alterations in the chemical modification at the 3′-end of ncRNAs and abnormalities in the associated-genes can also contribute to the natural history of cancer. For example, the precursors of the tumor suppressor miRNA let-7 are mono-urydilated by the enzymes ZCCHC11 (TUT4), ZCCHC6 (TUT7) and PAPD4 (TUT2) to facilitate Dicer processing and miRNA maturation (35). Tumoral cells can express the oncogenic protein LIN28 that induces a long U-tail in pre-let-7 oligo-uridylation, which prevents the formation of the mature molecule (33, 35, 83). Interestingly, the poly(U) motifs are enriched close to poly(A) sites, both are regulated by heterogeneous ribonucleoprotein C (HNRNPC) and ELAV-like RBP1 (ELAV1) (84) and alternative polyadenylation signals can clearly affect competing endogenous RNA (ceRNA) networks. The degradation of the poly(U) tail is also tightly regulated by exoribonucleases from the RNase II/RNB family that is represented by the enzymes Dis3, Dis3L1 and Dis3L2 in humans. Therein, the link to cancer can also be invoked because Dis3L2 mutations are associated with the development of Wilms’ tumors (85). Coincidentally or not, the m6A RNA methyltransferase WTAP cooperates with Wilms’ tumor 1 protein to regulate proliferation (61, 62), suggesting that this tumor type (which is also characterized by the aberrant imprinting of ncRNAs) could be particularly associated with a disrupted pattern of RNA chemical modifications. Overall, Table 2 summarizes the most relevant RNA modifiers involved in cancer.
Table 3.
Selected RNA modifiers involved in cancer
| Gene | Activity | Link to cancer (reference) |
|---|---|---|
| DNMT2 | m5C RNA methyltransferase | Diminished activity associated with candidate mutations (51) |
| NSUN2 | m5C RNA methyltransferase | Copy-number gain (52), oncogene targeting (53,54), depletion increase tumor-initiating cells (5) |
| TET1 | DNA and RNA m5C demethylase | Mutations and deletions in leukemia (58), tumor suppressor role (59) |
| WTAP | Cofactor for m6A RNA methyltransferase | Binding partner of the tumor suppressor WT1 (61), regulates cell cycle progression (62) |
| FTO | m6A RNA demethylase | Variants associated with melanoma risk (70), promotes leukemia (72) and breast cancer (73,74) |
| ALKBH5 | m6A RNA demethylase | Knock-out impact the p53 network (69), overexpression increases breast cancer stem cells (75) |
| ALKBH3 | m1A RNA demethylase and DNA repair | Protection against alkylation damage (76) |
| DKC1 | Pseudouridine synthase | Mutated in dyskeratosis congenita (30,79), regulates translation of tumor suppressor proteins (80) |
| Dis3L2 | Exoribonuclease for the poly (U) tail | Mutated in association with the development of Wilms’ tumors (85) |
Perspectives and challenges
The study of the cancer epitranscriptome is in its early stages, even more so if we devote our attention only to the chemical modifications of ncRNAs in transformed cells. However, knowledge in this area has recently made a quantum leap, as we have described above. Not only do we now have more complete deep sequencing coverage of several of the described RNA marks, such as m5C, hm5C, m6A, m1A or pseudouridine, but we have also recognized the involvement in tumors of some of the genes involved in these epitranscriptomic pathways, such as the m6A RNA methyltransferase WTAP, the m6A demethylases FTO and ALKBH5, the m1A demethylase ALKBH3, the pseudouridine forming enzyme DCK1 or the RNA editing enzyme ADAR1. However, the field still needs to carefully address how different the epitranscription profiles are between normal tissues and their paired tumoral samples; how these RNA modifications could shift in carcinogenesis from early benign lesions to metastases and how heterogeneous these marks are within a given tumor. In a few words, future work must address which of these events are critical in tumorigenesis and it must follow the genetic and DNA epigenetic studies that have tackled these key issues of cancer biology in the last twenty-five years.
Many advances in science depend on the existence of the proper methodology to assess the desired parameters. For ncRNA modifications, the development of the above mentioned epitranscriptomic techniques that mix bisulfite modification-based protocols or immunoprecipitation against the different marks with deep sequencing has been a major achievement. However, some of these approaches do not provide single-nucleotide resolution or they often depend on the batch specificity of the antibodies employed for the analysis. At the same time, most of these methods are unable to discriminate the simultaneous presence of different RNA modifications in the same molecule, thus losing part of the richness and complexity of the epitranscriptome. For example, the same lncRNA can undergo m5C, m6A and pseudouridylation, but do all these modifications take place at the same time? Do they cooperate or antagonize each other? New tools to address these questions are needed. Among these, it is worth highlighting the advances in combination mass spectrometry (86), isoform-characterization sequencing (87) or the use of single-molecule real-time (SMRT) sequencing (88), a third-generation sequencing technique.
Interestingly, part of the information necessary to advance the field may already exist but may be overlooked. Careful data mining of the many RNA-sequencing and fully sequenced genomes and epigenomes publically available can provide further clues about possible aberrant RNA modifications patterns and their possible causes and consequences. For example, a simple review of the data available at The Cancer Genome Atlas (TCGA) has shown underexpression of RNA demethylases and overexpression of mRNA methyltransferase occurs in human cancers, and the use of the cBioPortal suggest the presence of additional mutations in some of these enzymes (89). One list that could grow with the use of other platforms is the COSMIC cancer genome browser, which has been recently updated with new genomic and epigenomic data from 1,001 cell lines (90). Interestingly for some of these cell lines, such as HeLa or MDA-MB-231, epitranscriptomic data are also available and different datasets can be cross-examined. Unfortunately, many of these databases were produced from polyA fractions and, thus, chemical modifications of many ncRNAs cannot be analyzed.
New experimental data, however, will be also fundamental to move forward. Among the more than one-hundred differently modified RNA nucleotides (1), it is not well known how many are present in human cells. For other RNA modifications that have been identified the information is scarce, but exciting. Interesting cases include the O-methylation of the 5′ monophosphate of pre-miRNAs, mediated by BCDIN3D that negatively regulates miRNA maturation (91); the 5-methoxycarbonylmethyluridine (mcm5U) modification of tRNA regulated by the cancer-related enzymes ALKBH8 and hTRM9L (92); the existence of further modifications of an already modified nucleotide, such as the methylation of pseudouridine in rRNA by EMG1 (93); or the presence of intermediate RNA demethylation nucleotides such as formylcytosine, carboxylcytosine, N6-hydroxylmethyladenosine and N6-formyladenosine. It is also relevant to further define the spectrum of modified RNAs: Beyond tRNAs, rRNAs and mRNAs, how many and which miRNAs and lncRNAs are modified? This is a key issue because there is an increasing amount of lncRNAs, such as lincRNAs (94), transcribed-ultra-conserved regions (95, 96) and pseudogenes (97, 98) with key roles in tumorigenesis that could be regulated by RNA modifications, changing their biogenesis, stability, intracellular localization or shifting its interacting RNA targets. Genetic and functional studies, using CRISPR and similar tools, will be also necessary to identify the missing machineries involved in RNA modification, such as the enzymes responsible for circRNA formation, the readers for m5C or the methyltransferases for m1A. In addition, in vivo studies crossing the available genetically modified mice for the writers, readers and erasers of RNA modifications with well-established knock-out and knock-in mice models of recognized tumor suppressor genes and oncogenes will provide another read-out of the contribution of this epigenetic layer to cellular transformation. These experiments are absolutely necessary to distinguish those RNA modifiers that could truly be drivers in carcinogenesis from those events that could be just passengers for tumorigenesis.
Finally, a better definition of the RNA modifications present in human cells, the existence of differences in comparison to cancer tissues, and the characterization of the proteins responsible for the epitranscriptomic patterns, will facilitate the translational use of RNA epigenetics. We can imagine these different scenarios as diagnostic and predictive biomarkers or even as potential targets for innovative therapeutic strategies. In the two first cases, the use of RNA changes in miRNAs and circular RNAs, very stable RNA molecules, for the detection of cancer cells in serum or biological fluids as well as the determination of the epitranscriptome in RNA molecules involved in DNA repair and, thus, in the assessment of chemotherapy sensitivity, are possibilities worth exploring. For treatment, we can start designing synthetic lethal strategies for the recognized defects in the RNA modification machinery, or approaches aimed at inhibiting or activating specific RNA modifying enzymes. Related to this last point, the elucidation of the crystal structures for some of the proteins involved in RNA modifications, such as m6A modifiers and readers (99–101), represents an important step forward. Interestingly, the drug 5-azacytidine, used in leukemia treatments (102), inhibits both DNA and m5C RNA metyltransferases by competing with cytosine, leading to RNA hypomethylation (103,104), thus it is worth exploring how much of the growth-inhibitory effects of DNA demethylating agents relates to a change in m5C RNA levels. On the other hand, we can improve the effects of anticancer RNA medicines by adding appropriate chemical modifications to the RNA drug backbone. Exciting times are coming, stay tuned.
Significance.
Chemical modifications of RNA play a central role in the control of messenger and non-coding RNA activity and, thus, are tightly regulated in cells. In this article, we provide insights about how these marks are altered in cancer cells and how this knowledge can be translated to the clinical setting.
Acknowledgments
GRANT SUPPORT
Research described in the article was funded in part by the National Institutes of Health (NIH) grant R35CA197529-01 and the Ministerio de Economia y Competitividad (MINECO) grant SAF2014-55000-R.
References
- 1.Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: Form, distribution, and function. Science. 2016;352:1408–12. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heyn H, Esteller M. An Adenine Code for DNA: A Second Life for N6–Methyladenine. Cell. 2015;161:710–13. doi: 10.1016/j.cell.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 3.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, et al. Widespread occurrence of 5–methylcytosine in human coding and non–coding RNA. Nucleic Acids Res. 2012;40:5023–33. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31:458–64. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussain S, Aleksic J, Blanco S, Dietmann S, Frye M. Characterizing 5–methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013;14:215. doi: 10.1186/gb4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, et al. RNA biochemistry. Transcriptome–wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282–85. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- 7.Dominissini D, Moshitch–Moshkovitz S, Schwartz S, Salmon–Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A–seq. Nature. 2012;485:201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 8.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominissini D, Nachtergaele S, Moshitch–Moshkovitz S, Peer E, Kol N, Ben–Haim MS, et al. The dynamic N(1)–methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–6. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, et al. Transcriptome–wide mapping reveals reversible and dynamic N(1)–methyladenosine methylome. Nat Chem Biol. 2016;12:311–6. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León–Ricardo, et al. Transcriptome–wide mapping reveals widespread dynamic–regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–62. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlile TM, Rojas–Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–6. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Zhu P, Ma S, Song J, Bai J, Sun F, et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol. 2015;11:592–7. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 14.Bazak L, Haviv A, Barak M, Jacob–Hirsch J, Deng P, Zhang R, et al. A–to–I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014;24:365–6. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frye M, Blanco S. Post-transcriptional modifications in development and stem cells. Development. 2016;143:3871–81. doi: 10.1242/dev.136556. [DOI] [PubMed] [Google Scholar]
- 16.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramanathan A, Robb GB, Chan SH. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw551. pii:gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, et al. NSun2–mediated cytosine–5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–61. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tycowski KT, Aab A, Steitz JA. Guide RNAs with 5′ caps and novel box C/D snoRNA–like domains for modification of snRNAs in metazoa. Curr Biol. 2004;14:1985–95. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Amort T, Soulière MF, Wille A, Jia XY, Fiegl H, Wörle H, et al. Long non–coding RNAs as targets for cytosine methylation. RNA Biol. 2013;10:1003–8. doi: 10.4161/rna.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, et al. Tet–mediated formation of 5–hydroxymethylcytosine in RNA. J Am Chem Soc. 2014;136:11582–5. doi: 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M, Kim B, Kim VN. Emerging roles of RNA modification: m(6)A and U–tail. Cell. 2014;158:980–7. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6–methyladenosine marks primary microRNAs for processing. Nature. 2015a;519:482–5. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A–Dependent Nuclear RNA Processing Events. Cell. 2015b;162:1299–308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6–methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19:1848–56. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–96. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–72. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge J, Yu YT. RNA pseudouridylation: new insights into an old modification. Trends Biochem Sci. 2013;38:210–8. doi: 10.1016/j.tibs.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaborske JM, DuMont VL, Wallace EW, Pan T, Aquadro CF, Drummond DA. A nutrient–driven tRNA modification alters translational fidelity and genome–wide protein coding across an animal genus. PLoS Biol. 2014;12:e1002015. doi: 10.1371/journal.pbio.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shafik A, Schumann U, Evers M, Sibbritt T, Preiss T. The emerging epitranscriptomics of long noncoding RNAs. Biochim Biophys Acta. 2016;1859:59–70. doi: 10.1016/j.bbagrm.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let–7 precursor MicroRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Newman MA, Mani V, Hammond SM. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA. 2011;17:1795–803. doi: 10.1261/rna.2713611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, et al. Mono–uridylation of pre–microRNA as a key step in the biogenesis of group II let–7 microRNAs. Cell. 2012;151:521–32. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228–37. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avesson L, Barry G. The emerging role of RNA and DNA editing in cancer. Biochim Biophys Acta. 2014;1845:308–16. doi: 10.1016/j.bbcan.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Nik–Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Breast Cancer Working Group of the International Cancer Genome Consortium. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–83. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nik–Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC–dependent mutations in breast cancer. Nat Genet. 2014;46:487–91. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013;19:209–16. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anadón C, Guil S, Simó–Riudalbas L, Moutinho C, Setien F, Martínez–Cardús A, et al. Gene amplification–associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene. 2016;35:4407–13. doi: 10.1038/onc.2015.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu MT, Coca–Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–40. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 44.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon–containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–7. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 47.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic Role of Fusion–circRNAs Derived from Cancer–Associated Chromosomal Translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 48.Grudzien–Nogalska E, Kiledjian M. New insights into decapping enzymes and selective mRNA decay. Wiley Interdiscip. Rev RNA. 2016 doi: 10.1002/wrna.1379.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–8. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 50.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress–induced cleavage. Genes Dev. 2010;24:1590–5. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elhardt W, Shanmugam R, Jurkowski TP, Jeltsch A. Somatic cancer mutations in the DNMT2 tRNA methyltransferase alter its catalytic properties. Biochimie. 2015;112:66–72. doi: 10.1016/j.biochi.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 52.Frye M, Dragoni I, Chin SF, Spiteri I, Kurowski A, Provenzano E, et al. Genomic gain of 5p15 leads to over–expression of Misu (NSUN2) in breast cancer. Cancer Lett. 2010;289:71–80. doi: 10.1016/j.canlet.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc–induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971–81. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 54.Hussain S, Benavente SB, Nascimento E, Dragoni I, Kurowski A, Gillich A, et al. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol. 2009;186:27–40. doi: 10.1083/jcb.200810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–40. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun. 2016;6:6158. doi: 10.1038/ncomms7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039. doi: 10.1038/ncomms12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired hydroxylation of 5–methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–43. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cimmino L, Dawlaty MM, Ndiaye–Lobry D, Yap YS, Bakogianni S, Yu Y, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16:653–62. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 61.Little NA, Hastie ND, Davies RC. Identification of WTAP, a novel Wilms’ tumour 1–associating protein. Hum Mol Genet. 2000;9:2231–9. doi: 10.1093/oxfordjournals.hmg.a018914. [DOI] [PubMed] [Google Scholar]
- 62.Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, et al. Identification of Wilms’ tumor 1–associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;88:33292–302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)–methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–99. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–4. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 65.Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540:242–7. doi: 10.1038/nature20568. [DOI] [PubMed] [Google Scholar]
- 66.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–19. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 67.Ma L, Zhao B, Chen K, Thomas A, Tuteja JH, He X, et al. Evolution of transcript modification by N6-methyladenosine in primates. Genome Res. 2017 doi: 10.1101/gr.212563.116.. pii: gr.212563.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6–methyladenosine in nuclear RNA is a major substrate of the obesity–associated FTO. Nat Chem Biol. 2011;7:885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iles MM, Law MH, Stacey SN, Han J, Fang S, Pfeiffer R, et al. GenoMEL Consortium; Q–MEGA and AMFS Investigators. A variant in FTO shows association with melanoma risk not due to BMI. Nat Genet. 2013;45:428–32. doi: 10.1038/ng.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS, et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet. 2009;85:106–11. doi: 10.1016/j.ajhg.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan A, Dang Y, Chen G, Mo Z. Overexpression of the fat mass and obesity associated gene (FTO) in breast cancer and its clinical implications. Int J Clin Exp Pathol. 2015;8:13405–10. [PMC free article] [PubMed] [Google Scholar]
- 74.Singh B, Kinne HE, Milligan RD, Washburn LJ, Olsen M, Lucci A. Important role of FTO in the survival of rare panresistant triple-negative inflammatory breast cancer cells facing a severe metabolic challenge. PLoS One. 2016;11:e0159072. doi: 10.1371/journal.pone.0159072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF–dependent and ALKBH5–mediated m6A–demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047–56. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aas PA, Otterlei M, Falnes PO, Vågbø CB, Skorpen F, Akbar M, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–63. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 77.Dango S, Mosammaparast N, Sowa ME, Xiong LJ, Wu F, Park K, et al. DNA unwinding by ASCC3 helicase is coupled to ALKBH3–dependent DNA alkylation repair and cancer cell proliferation. Mol Cell. 2011;44:373–84. doi: 10.1016/j.molcel.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nogales V, Reinhold WC, Varma S, Martinez–Cardus A, Moutinho C, Moran S, et al. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget. 2016;7:3084–97. doi: 10.18632/oncotarget.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–62. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 80.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, et al. Impaired control of IRES–mediated translation in X–linked dyskeratosis congenita. Science. 2006;312:902–6. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 81.Bellodi C, McMahon M, Contreras A, Juliano D, Kopmar N, Nakamura T, et al. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Rep. 2013;3:1493–502. doi: 10.1016/j.celrep.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Müller M, Hartmann M, Schuster I, Bender S, Thüring KL, Helm M, et al. Dynamic modulation of Dnmt2–dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 2012;43:10952–62. doi: 10.1093/nar/gkv980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let–7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–5. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gruber AJ, Schmidt R, Gruber AR, Martin G, Ghosh S, Belmadani M. A comprehensive analysis of 3′ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res. 2016;26:1145–59. doi: 10.1101/gr.202432.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Astuti D, Morris MR, Cooper WN, Staals RH, Wake NC, Fews GA, et al. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet. 2012;44:277–84. doi: 10.1038/ng.1071. [DOI] [PubMed] [Google Scholar]
- 86.Wetzel C, Limbach PA. Mass spectrometry of modified RNAs: recent developments. Analyst. 2016;141:16–23. doi: 10.1039/c5an01797a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, et al. m(6)A–LAIC–seq reveals the census and complexity of the m(6)A epitranscriptome. Nat Methods. 2016;13:692–8. doi: 10.1038/nmeth.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, et al. Genome–wide mapping of methylated adenine residues in pathogenic Escherichia coli using single–molecule real–time sequencing. Nat Biotechnol. 2012;30:1232–9. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li S, Mason CE. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet. 2014;15:127–50. doi: 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- 90.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–54. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xhemalce B, Robson SC, Kouzarides T. Human RNA methyltransferase BCDIN3D regulates microRNA processing. Cell. 2012;51:278–8. doi: 10.1016/j.cell.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Begley U, Sosa MS, Avivar–Valderas A, Patil A, Endres L, Estrada Y, et al. A human tRNA methyltransferase 9–like protein prevents tumour growth by regulating LIN9 and HIF1–α. EMBO Mol Med. 2013;5:366–83. doi: 10.1002/emmm.201201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wurm JP, Meyer B, Bahr U, Held M, Frolow O, Kötter P, et al. The ribosome assembly factor Nep1 responsible for Bowen–Conradi syndrome is a pseudouridine–N1–specific methyltransferase. Nucleic Acids Res. 2010;38:2387–98. doi: 10.1093/nar/gkp1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann–Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R. CpG island hypermethylation–associated silencing of non–coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liz J, Portela A, Soler M, Gómez A, Ling H, Michlewski G. Regulation of pri–miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell. 2014;55:138–47. doi: 10.1016/j.molcel.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 97.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding–independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Léopold V, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319–32. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feng C, Liu Y, Wang G, Deng Z, Zhang Q, Wu W, Tong Y, Cheng C, Chen Z. Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J Biol Chem. 2014;289:11571–83. doi: 10.1074/jbc.M113.546168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10:927–9. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 101.ŒledŸ P, Jinek M. Structural insights into the molecular mechanism of the m6A writer complex. Elife. 2016:5. doi: 10.7554/eLife.18434.. pii: e18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–9. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 103.Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–32. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- 104.Khoddami V, Cairns BR. Transcriptome-wide target profiling of RNA cytosine methyltransferases using the mechanism-based enrichment procedure Aza-IP. Nat Protoc. 2014;9:337–61. doi: 10.1038/nprot.2014.014. [DOI] [PubMed] [Google Scholar]