Abstract
Alcohol exposure in utero can result in Fetal Alcohol Spectrums Disorders (FASD). Measures of hippocampal neuroplasticity, including long-term potentiation, synaptic and dendritic organization, and adult neurogenesis, are consistently disrupted in rodent models of FASD. The current study investigated whether third trimester-equivalent binge-like alcohol exposure (AE) [postnatal days (PD) 4–9] affects dendritic morphology of immature dentate gyrus granule cells, and brain-derived neurotrophic factor (Bdnf) gene expression and DNA methylation in hippocampal tissue in adult male rats. To understand immediate impact of alcohol, DNA methylation was measured in the PD10 hippocampus. In addition, two behavioral interventions, wheel running (WR) and environmental complexity (EC), were utilized as rehabilitative therapies for alcohol-induced deficits. AE significantly decreased dendritic complexity of the immature neurons, demonstrating the long-lasting impact of neonatal alcohol exposure on dendritic morphology of immature neurons in the hippocampus. Both housing conditions robustly enhanced dendritic complexity in the AE animals. While Bdnf exon I DNA methylation was lower in the AE and sham-intubated animals compared with suckle controls on PD10, alterations to Bdnf DNA methylation and gene expression levels were not present at PD72. In control animals, exercise, but not exercise followed by housing in EC, resulted in higher levels of hippocampal Bdnf gene expression and lower DNA methylation. These studies demonstrate the long-lasting negative impact of developmental alcohol exposure on hippocampal dendritic morphology and support the implementation of exercise and complex environments as therapeutic interventions for individuals with FASD.
Keywords: development, alcohol, plasticity, adult neurogenesis, DNA methylation
INTRODUCTION
Alcohol exposure during the third trimester of pregnancy has a significant impact on the anatomy and function of late-developing brain structures such as the hippocampus. In rodent models of FASD that administer alcohol during the third trimester-equivalent (first two postnatal weeks in rodent models [Dobbing and Sands, 1979]), various measures of hippocampal anatomy and neuroplasticity are impacted including CA1 pyramidal cell number (Bonthius and West, 1990), dendritic morphology (Sakata-Haga et al., 2006), survival of newly generated dentate gyrus (DG) granule cells (Klintsova et al, 2007; Hamilton et al., 2012), induction of CA1 long-term potentiation (LTP) (Puglia and Valenzuela, 2010a,b), and neuronal activity as measured by expression of the protein c-Fos (Murawski et al., 2012). Deficits in structural and functional plasticity likely contribute to observed impairments on various hippocampal-associated behavioral tasks following neonatal alcohol exposure (Thomas et al., 2008; Hamilton et al., 2011; Murawski et al., 2012; Hamilton et al. 2014).
Adult neurogenesis has been consistently shown to be negatively impacted by developmental alcohol exposure (Ieraci and Herrera, 2007; Singh et al., 2009; Gil-Mohapel et al., 2011), with survival, but not generation, of adult-born DG granule cells being significantly affected (Klintsova et al., 2007; Hamilton et al., 2012). These newborn DG neurons undergo “competitive integration and survival” (Kempermann, 2011) during which the maturing cells have a low threshold for excitability (Van Praag et al., 2002; Overstreet-Wadiche and Westbrook, 2006; Kempermann, 2011). Neurons must make a sufficient number of excitatory synaptic connections during this period to successfully integrate and survive. Based on our work demonstrating that cell proliferation and the number of neuronal progenitors is not affected by neonatal alcohol exposure (Hamilton et al., 2012, 2014), while long-term cell survival is compromised, we hypothesized that fewer newly born DG neurons successfully integrate during competitive survival in alcohol-exposed animals, leading to the decrease in adult-born mature neurons observed. One way to determine the health of new neurons is to assess dendritic morphology of immature granule cells: if they exhibit decreased dendritic complexity, the ability to make sufficient number of synaptic connections may be impaired.
Dendritic morphology is just one of the measures of neuroplasticity altered by developmental alcohol exposure, and the identification of factors that contribute to impairments in plasticity is an ongoing challenge for researchers. Brain-derived neurotrophic factor (BDNF), a protein critical for proper cell maturation, dendritic arborization, and LTP, represents a molecular target possibly compromised by developmental alcohol exposure. However, alcohol-induced alterations to BDNF seem to be dependent on alcohol dose, exposure window, brain region, and timing of analysis. Our lab has shown previously that PD4-9 alcohol exposure increased hippocampal BDNF and TrkB protein, Bdnf total gene expression, and generation of Bdnf exon-specific mRNA transcripts in the developing hippocampus (Boschen et al., 2015), which paralleled previous work by the Heaton et al. (2000, 2003). Neonatal alcohol exposure has also been associated with lower levels of BDNF protein and gene expression in the adult rat hippocampus (Fattori et al., 2008; Miki et al., 2008). Based on our reports of compromised hippocampal neuroplasticity in the alcohol-exposed adult rat (Klintsova et al., 2007; Helfer et al., 2009; Hamilton et al., 2012), we predicted reduced basal Bdnf gene expression in the hippocampus.
Epigenetic modifications, which refer to the addition or deletion of chemical groups on chromatin, have recently come to the forefront as a mechanism through which environmental factors could directly impact genes and, in turn, behavior. Developmental alcohol exposure cause a variety of epigenetic modifications depending on the timing of the alcohol administration (Liu et al., 2009; Govorko et al., 2012; Otero et al., 2012; Perkins et al., 2013); however, investigation of gene-specific epigenetic modifications caused by developmental alcohol exposure is still a largely unexplored field (transgenerational alterations to methylation status of the proopiomela-nocortin gene (Pomc) have been reported in a prenatal exposure rat model [Govorko et al., 2012; Bekdash et al., 2013]). DNA methylation can alter downstream gene transcription through recruitment of transcriptional regulator proteins (Moore et al., 2013). Methylation of Bdnf is altered in timing- and exon-specific manner following a variety of early life experiences (reviewed in Blaze and Roth, 2015). Based on previous literature, we predicted that hippocampal DNA methylation would be higher in alcohol-exposed animals than controls.
Two behavioral interventions are beneficial for neuroplasticity and influence BDNF synthesis: aerobic exercise and housing in a complex environment (EC). Voluntary wheel running (WR) and exposure to EC have been investigated as promising therapies to mitigate the structural, functional, and behavioral impairments caused by neonatal alcohol exposure. Both WR and EC have robust benefits on hippocampal adult neurogenesis and dendritic morphology in the hippocampus and prefrontal cortex (Faherty et al., 2005; Kronenberg et al., 2006; Van der Borght et al., 2007; Gelfo et al., 2009; Hamilton et al., 2015). Our lab has demonstrated that 12 days of WR followed by 30 days of EC reverses impairments in new granule cell survival in animals neonatally exposed to alcohol (Helfer et al., 2009; Hamilton et al., 2012). Furthermore, both WR and EC increase BDNF protein and mRNA levels in both humans and rodents and can cause epigenetic modifications to Bdnf (Berchtold et al., 2005; Christie et al., 2005; Griffin et al., 2009; Rasmussen et al., 2009; Kuzumaki et al., 2011), suggesting that enhancements to BDNF could underlie the beneficial effects of WR and EC in neonatally alcohol-exposed animals. We predicted that our behavioral interventions would upregulate Bdnf gene expression with concomitant reductions in DNA methylation levels.
The current study had two primary goals. First, we sought to assess whether PD4-9 alcohol exposure impacts the dendritic morphology of immature neurons in the dorsal DG, and whether exercise alone or exercise followed by housing in a complex environment rescue alcohol-induced deficits in dendritic complexity. Second, we extended our investigation of the impact of PD4-9 alcohol exposure on neonatal BDNF expression (Boschen et al., 2015) into adulthood by assessing Bdnf total and exon-specific gene expression in the hippocampus of adult rats (PD72). Additionally, we examined Bdnf DNA methylation in the adult (PD72) and infant (PD10) hippocampus, as DNA methylation status can be dynamic across the lifespan. Finally, the current study examined whether exposure to either exercise alone (42 days) or exercise (12 days) followed by EC (30 days) alters Bdnf gene expression and DNA methylation.
METHODS
Animals
Briefly, timed pregnant Long-Evans dams were obtained (Harlan Laboratories, Indianapolis, IN). On postnatal day (PD) 3, litters were culled to eight pups each (6 male, 2 female when possible). On PD4, pups were randomly assigned to one of three experimental groups: suckle control (SC), sham-intubated (SI), or alcohol exposed (AE). Following the alcohol exposure procedure, pups were left undisturbed with the dam until PD10 (DNA methylation only) or weaning (PD23). On PD23, rats were placed in standard cages in groups of 3 same-sex animals, counterbalanced for litter and neonatal condition. They remained in these cages until PD30, when each cage was assigned to one of three housing conditions that are described below. Separate cohorts were generated for immunohistochemical and molecular biology assays. In total, 64 male rat pups (11 litters) were generated for immunohistochemistry and 167 male pups (31 litters) were generated for the BDNF assays. The specific sample sizes for each experimental condition used for each measure are listed below.
Alcohol Exposure and BAC Analysis
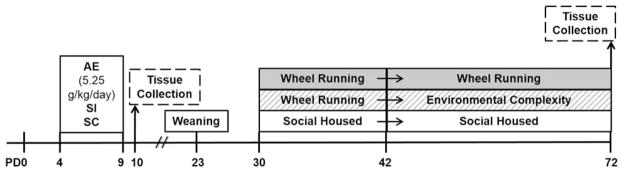
On PD4-9, AE pups were exposed to alcohol in a binge-like manner via intragastric intubation of 11.9% ethanol solution in customized milk formula in a daily dose of 5.25 g/kg/day (Fig. 1). A total of 9 male pups died during or shortly following the intubation procedure. On PD4, blood samples were obtained from AE pups via tail clip for BAC analysis 90 min following the second alcohol exposure. Plasma was analyzed for BAC using an Analox GL5 Alcohol Analyzer (Analox Instruments, Boston, MA). Due to a machine malfunction, plasma from a subset of animals used for immunohistochemistry was not able to be analyzed (8 animals).
Figure 1.
Experimental timeline. PD, postnatal day; AE, alcohol-exposed; SI, sham-intubated; SC, suckle control.
Adolescent and Adult Housing Conditions
On PD30, each cage was assigned one of three adolescent/adult housing conditions: standard social housing (SH), continuous access to wheel running for 42 days (WR/WR), or 12 days of wheel running access followed by 30 days of housing in a complex environment (WR/EC) (Fig. 1). Animals were weighed approximately every 8 days and sacrificed on PD72. Standard social housing consisted of three rats housed in opaque cages (17 × 145 × 24 cm). The wheel running condition consisted of the groups of three rats housed in cages with 24 h voluntary access to running wheels attached to standard sized cages (as in previous studies, e.g., Kempermann et al., 1997; Van Praag et al., 1999; Helfer et al., 2009; Kobilo et al., 2011). For SH and WR, animals were housed 3 per cage in the same configuration as PD23-30 housing. Running distance was recorded daily at 9 AM. Rats were housed in this condition either for 42 days (PD30-72) or for 12 days (PD30-42) prior to housing in EC (PD42-72). Each EC cage consisted of a 3-story cage with three ramps, two balconies and a full middle floor. Each cage housed 9–12 male animals (3–4 animals/neonatal condition) and was equipped with a variety of toys, large tubes, and buckets. The toys were changed every second day. Animals were taken directly from their housing condition for sacrifice on PD72.
Immunohistochemistry
On PD72, animals were deeply anesthetized (ketamine/xylazine cocktail), transcardially perfused (0.1M PBS with heparin followed by 4% paraformaldehyde). Brains were sectioned horizontally at 40 μm through the entirety of the hippocampus. Immunohistochemistry to label for the marker doublecortin (DCX) was performed on a pseudo-randomly selected set of section (1/16th) which included the dorsal DG (~−3.1 to −4.3 mm DV Bregma). This study focuses on adult neurogenesis in the dorsal DG due to its more prominent role in spatial navigation and contextual memory compared with the ventral hippocampus, which is more involved with emotional processing due to strong connections with the amygdala (Fanselow and Dong, 2010). Additionally, Ieraci and Herrera (2007) found decreased cell proliferation in dorsal but not ventral DG following a single binge of alcohol on PD7, supporting the differential effect of alcohol along the dorsoventral axis.
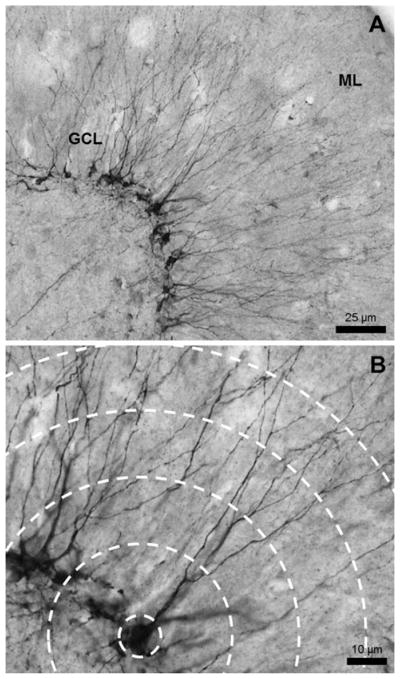
DCX was used to label immature DG granule cells (Fig. 2). The cells labeled were approximately 2–21 days old (Brown et al., 2003), were no longer actively dividing, and had committed to a neuronal fate. The most immature cells were located in the subgranular zone and had no visible dendrites, while older, more mature DCX+ cells had migrated farther into the granule cell layer and had at least one strong dendritic tree. The following protocol was used to immunolabel sections for DCX. Sections were rinsed in 0.1 M Tris-buffered saline (TBS) (Fisher Scientific, Waltham, MA), then treated with 0.6% H2O2 in TBS to exhaust endogenous peroxidase activity for 30 min. Sections were washed again in TBS and incubated in primary antibody made in blocking solution (3% normal donkey serum [Millipore, Billerica, MA], 0.1% Triton-X [Fisher Scientific, Waltham, MA] in TBS) for 24 h at 4°C (1:500, goat anti-DCX C-18, Santa Cruz Labs, Dallas, TX). The next day, tissue was incubated in secondary antibody (1:250; biotinylated anti-goat; Vector Labs, Burlingame, CA) for 1 h at room temperature and an avidin–biotin complex (ABC) diluted in blocking solution (Vector Laboratories, Burlingame, CA) for 1 h. The reaction was visualized using nickel-enhanced diaminobenzidine (DAB). Sections were counterstained with 0.1% Pyronin Y (Electron Microscopy Sciences, Hatfield, PA).
Figure 2.
Representative image of DCX+ immature neurons. (A) The DCX+ cell bodies line the inside of the granule cell layer (GCL) and the dendrites extend outward toward the molecular layer (ML). Image taken with a 20× lens. (B) Dendrites were traced at a high magnification which allows clear visualization of the processes and bifurcations. Image taken with a 40× lens.
Dendritic Structure of Immature Neurons
Immature DG granule cells (DCX+) were traced and Sholl analysis conducted using a computer-based neuron tracing system (NeuroLucida v. 10.52; MBF, Bioscience, Williston, VT). Two sections of dorsal DG were used per animal for the analysis. Fully labeled DCX+ granule cells [Fig. 2(B)] were identified and traced at 400× magnification (40× lens). Cells were included the analysis if they met criteria: (1) fully labeled DCX+ cell in postmitotic stage (presence of a dendrite extending toward the molecular layer [Plümpe et al., 2006]), (2) dendritic tree contained in the analyzed section, and (3) the dendritic tree’s branches were not broken or obscured. These criteria allowed for 6–10 cells to be traced per animal (n =4–5 animals/group from 11 litters). Based on the descriptions put forward by Plümpe et al. (2006), the cells traced in this study were determined to be of the most mature phenotype still labeled with DCX (category F). NeuroLucida software conducted a Sholl analysis which placed concentric spheres extending from the cell soma at 20 μm intervals and analyzed the following measures of complexity: the number of intersections at each radius, total dendritic length per radius, and number of nodes/bifurcations per radius was determined, which allowed for analysis of dendritic complexity. The average length of each cells’ dendritic tree, average number of intersections, length per radius, and number of bifurcations per cell were also analyzed for each animal.
Molecular Biology Assays
On PD10 or PD72, animals were briefly anesthetized with isoflurane and rapidly decapitated with a guillotine. The brain was flash frozen with 2-Methylbutane and sectioned on a brain matrix (1 μm). Sections were stored at −80°C. The hippocampus (both dorsal and ventral) was dissected on dry ice and homogenized. DNA/RNA were extracted using the AllPrep DNA/RNA Mini Kit (Qiagen, Valencia, CA).
For gene expression (PD72 only), reverse transcription was performed to generate cDNA from the RNA (Reverse Transcriptase Kit, Qiagen, Valencia, CA). The cDNA was amplified by real-time PCR (Bio-Rad CFX96) with Taq-man probes (Applied Biosystems) to target Bdnf total mRNA (exon IX) and exon I- and IV-specific transcripts. Tubulin was used as a reference gene. All reactions for each gene target and reference were run in triplicate. Product specificity was verified using gel electrophoresis. The final animal number used for this assay was 7–11/group (from 23 litters).
Methylation status of Bdnf exons I and IV was assessed using methylation specific real-time PCR (MSP) on bisulfite-converted DNA (Qiagen, Valencia, CA) from PD10 or PD72 hippocampal tissue. Primer sets (Sigma-Aldrich; listed below in Table 1; Lubin et al., 2008; Roth et al., 2009, 2015) targeted methylated and unmethylated CG dinucleotides in DNA associated with Bdnf exon I or exon IV. Tubulin was used as a reference gene. All reactions were run in triplicate. Primer specificity was determined by melt curve analysis on all samples and gel electrophoresis on a sub-set of samples. The final sample size for these analyses was 6–10/group (from 23 litters for PD72 and 8 litters for PD10).
Table 1.
Custom Primer Sets for MSP Analysis (Lubin et al., 2008; Roth et al., 2009, 2015)
| Primer | Sequence (5′ to 3′) |
|---|---|
| Bdnf exon I Methylated | Forward: CGGAAAGTATTAGAGGTAGGGTAGC |
| Reverse: TACGAACCCTAAATCTCTAAACGAA | |
| Bdnf exon I Unmethylated | Forward: TGGAAAGTATTAGAGGTAGGGTAGTGA |
| Reverse: TACAAACCCTAAATCTCTAAACAAA | |
| Bdnf exon IV Methylated | Forward: GGTAGAGGAGGTATTATATGATAGTTTACG |
| Reverse: TAAATAAAAAAAACGACAACGCGAA | |
| Bdnf exon IV Unmethylated | Forward: AGGTAGAGGAGGTATTATATGATAGTTTAT |
| Reverse: TAAATAAAAAAAACAACAACACAAA | |
| Tubulin | Forward: GGAGAGTAATATGAATGATTTGGTG |
| Reverse: CATCTCAACTTTCCCTAACCTACTTAA |
Statistical Analyses
Weights for each day of dosing (PD4-9) and housing (PD30, 42, 50, 58, 66, and 72) were averaged across neonatal condition/housing condition for each day. PD4-9 weights were analyzed using a repeated-measures analysis of variance (ANOVA) and PD30-72 weights were analyzed with a two-way (neonatal condition × housing) repeated-measures ANOVA followed by post hoc tests when appropriate. Average PD4 BACs for each animal were calculated based on 2–3 analyses run per animal and the means for the AE group are reported as mg/dL ± standard error of the mean (SEM). Dendritic complexity was analyzed using repeated measures ANOVAs run to analyze changes in morphology between specific animal conditions: (1) AE/SH versus SI/SH versus SC/SH, (2) SC/SH versus SC/WREC versus SC/WRWR, (3) SI/SH versus SI/WREC versus SI/WRWR, and (4) AE/SH versus AE/WREC versus AE/WRWR. For gene expression and MSP data, the comparative Ct method was used to obtain the relative fold change of experimental (AE or SI) versus the average of controls (SC) per plate (Livak and Schmittgen, 2001). A mean value of 1 would indicate no change in transcript level in comparison to the SC group. For PD72, two-way ANOVAs were run on gene expression and MSP data, followed by Tukey’s post hoc tests when appropriate. For PD10 MSP data, one-way ANOVAs were used to test for differences between the neonatal conditions, followed by Tukey’s post hoc test when appropriate. Differences were considered to be statistically significant at p <0.05 and nonsignificant trends at p <0.1 are also reported. For all studies, outliers were identified using Grubb’s test, a commonly used statistical test which removed one outlier from each group (Grubbs, 1969; Hamilton et al., 2012; Diaz et al., 2015; Boschen et al., 2015). Using this test, the following outliers were detected and removed for each measure: Sholl analysis of DCX+ cells—no outliers detected; Bdnf PD72 gene expression—total: 2 outliers (AE/WREC and AE/WRWR), exon I: 3 outliers (SI/SH, SI/WREC, AE/WREC), exon IV: 1 outlier (SI/WRWR); Bdnf PD72 DNA methylation—exon I: 2 outliers (AE/WREC and SC/WREC), exon IV: 1 outlier (AE/WREC); and, Bdnf PD10 methylation: 1 outlier (exon IV, AE animal). Statistics were run using SPSS (v14) or Prism 6 software (GraphPad Inc.).
RESULTS
Body Weights, BACs, and Running History
Repeated-measures ANOVAs were conducted to compare weights from PD4-9 and PD30, 42, 50, 58, 64, and 72 separately (Table 2). For PD4-9, a Day × Neonatal Condition interaction was found (F(10,900) = 43.289, p <0.0001), as well as main effects of Day (F(5,900) = 6050.731, p <0.0001) and Neonatal Condition (F(2,180) = 21.917, p <0.0001). One-way ANOVAs on PD4 and 9 weights determined that there was no effect of neonatal treatment on PD4 (F <1), however, AE animals weighed less than SI and SC pups on PD9 (main effect of Neonatal Condition: F(2,180) = 27.519, p <0.0001; Tukey’s post hoc: p <0.001 for AE vs. SC and AE vs. SI). On PD9, SI and SC pups did not differ in weight (p =0.982).
Table 2.
Animal Weights for the Neonatal Period (A) and Adulthood (B) Experimental Condition
| A | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Neonatal Condition | PD4 (g ± SD) | PD9 (g ± SD) | BACs (mg/dL ± SEM) | |||
| SC | 10.52 ± 1.18 | 20.16 ± 2.36 | ||||
| SI | 10.71 ± 0.89 | 20.18 ± 1.41 | ||||
| AE | 10.72 ± 0.82 | 17.93 ± 1.71* | 328.2 ± 14.95 | |||
|
| ||||||
| B | ||||||
|
| ||||||
| PD30 (g ± SD) | PD72 (g ± SD) | |||||
|
|
|
|||||
| SH | WREC | WRWR | SH | WREC | WRWR | |
|
| ||||||
| SC | 104.44 ± 6.9 | 104.73 ± 12.1 | 102.94 ± 8.0 | 397.06 ± 27.8 | 393.08 ± 29.9 | 372.38 ± 28.6 |
| SI | 104.22 ± 6.1 | 103.59 ± 6.9 | 103.87 ± 4.2 | 395.39 ± 10.0 | 390.71 ± 23.7 | 381.33 ± 19.1 |
| AE | 101.06 ± 8.8 | 101.81 ± 9.2 | 97.75* ± 5.2 | 389.63 ± 32.5 | 385.47 ± 19.0 | 360.94* ± 22.1 |
AE animals weighed less than SI and SC animals on PD9 and in adulthood (signified by * in the tables; p <0.05). AE, alcohol-exposed; SI, sham-intubated; SC, suckle control; SH, standard social housing; WREC, wheel running followed by environmental complexity; WRWR, wheel running.
For PD30-72 weights, a Day × Neonatal Condition × Housing interaction (F(20,675) = 2.075, p = 0.004), a Day × Housing interaction (F(10,675) = 6.045, p <0.001), Day × Neonatal interaction (F(10,675) = 3.012, p =0.001) were found. In addition, main effects of Day (F(5,675) = 3867.425, p <0.0001), Neonatal Condition (F(2,135) = 4.579, p =0.012), and Housing (F(2,135) = 6.482, p =0.002) were observed. The interaction between Housing × Neonatal Condition was not significant. Post hoc tests (Tukey’s HSD) revealed that AE rats weighed less on average than SI and SC animals (p =0.032 and 0.016, respectively). Long-lasting changes to body weight due to alcohol exposure have never been found previously using our model (Klintsova et al., 2007; Hamilton et al., 2012; Boschen et al., 2014), leading us to conclude that this finding is due to random factors specific to this cohort. In addition, WRWR animals weighed less than rats in the SH group (p =0.001). WREC did not significantly differ from either SH or WRWR animals (p =0.315 and 0.09, respectively). Further analysis revealed that the reduced weight was specific to the AE/WRWR group compared with the SI and SC/WRWR groups (Day × Neonatal interaction: F(10,220) = 3.303, p =0.001 and Neonatal main effect: F(2,44) = 6.674, p =0.003). When AE/SH was compared with both SI and SC/SH animals independently, the weights did not differ (F <1 for Day × Neonatal interaction and Neonatal main effect). The AE/WRWR group was lighter than the control/WRWR groups as early as PD30, prior to WR experience (F(2,44) = 4.589, p =0.015). As the animals are randomly assigned to their adult housing condition, it seems that smaller animals were randomly assigned to the WRWR group on PD30 and this weight difference continued throughout access to WR. No correlation (Pearson’s r2 =0.0003) was found between total Bdnf gene expression and PD72 weight within the WRWR animals, suggesting that weight alone was not driving Bdnf expression.
BACs were analyzed from plasma collected on PD4, 90 min following the second alcohol exposure. The BACs averaged 328.2 mg/dL (± 14.9 SEM). This value is in range of BACs from our previously published work (Hamilton et al., 2012; Boschen et al., 2014, 2015).
Running distance per 24 h was measured per cage (3 animals/cage). For the first 12 days of wheel running access (PD31-42), average running distance was 2.7 miles (± 1.2 SD) per 24 h. For WRWR animals, the average distance ran from PD43-72 was 8.5 miles (± 3.8 SD) per 24 h These running distances are consistent with previously published work from our lab using the WREC intervention (Hamilton et al., 2012; Boschen et al., 2014). The increase in running during the last 30 days in WRWR animals compared with the first 2 days could be due to the longer length/stride of the animals as they aged or increased natural running endurance. It is important to note that individual running distance could not be determined due to there being 3 animals per cage, however, the well-known detrimental impact of social isolation made housing the animals alone during WR unfeasible. Pilot work from our lab has observed no differences in running behavior between the neonatal treatments.
DCX± Dendritic Morphology
Repeated-measures ANOVAs were conducted for each measure of dendritic complexity based on the Sholl analysis results. The three measures determined by the Sholl analysis were: amount of dendritic material (length) per radius, number of intersections per radius, and the number of dendritic bifurcations per radius. For each measure, the repeated-measures ANOVAs compared complexity between the following groups: (1) AE/SH versus SI/SH versus SC/SH, (2) SC/SH versus SC/WREC versus SC/WRWR, (3) SI/SH versus SI/WREC versus SI/WRWR, and (4) AE/SH versus AE/WREC versus AE/WRWR. Representative tracings of SH animals from each neonatal treatment and of AE animals in each housing condition are shown in Figure 3. Statistical output is summarized in Table 3.
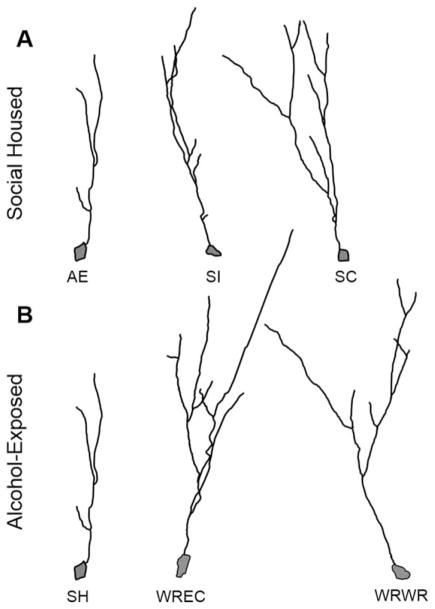
Figure 3.
Representative NeuroLucida tracings of DCX+ immature neurons in the PD72 dorsal dentate gyrus. (A) Dendritic complexity was significantly reduced in AE/SH animals compared with SI/SH and SC/SH rats. (B) In AE animals, WREC and WRWR significantly increased the amount of dendritic material, intersections, and bifurcations. AE, alcohol-exposed; SI, sham-intubated; SC, suckle control; SH, standard social housing; WREC, wheel running followed by environmental complexity; WRWR, wheel running.
Length per Radius
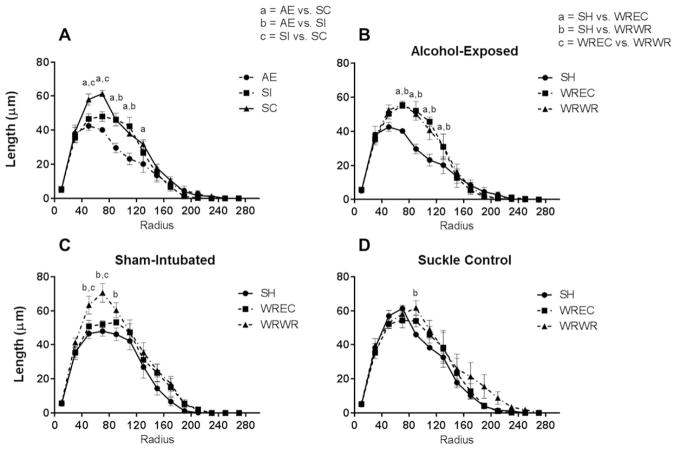
For the comparison of dendritic length between neonatal treatments housed in the SH condition, a radius × neonatal condition interaction was found [F(26,156) = 3.671, p <0.0001; Fig. 4(A)]. Main effects were also found for radius (F(13,156) = 184.8, p <0.0001) and for neonatal condition separately (F(2,12) = 7.851, p =0.0066). Post hoc analysis (Tukey’s multiple comparison test) revealed a significant decrease in total length of dendrites for AE compared with SI for radii 90 and 110 μm from the soma (p <0.0001 for both), significant decreases for AE compared with SC for radii 50, 70, 90, 110, and 130 μm from the soma (p <0.001, 0.0001, 0.0001, 0.001, and 0.01, respectively) and significant decreases in SI compared with SC for radii 50 and 70 μm (p <0.01 for both radii).
Figure 4.
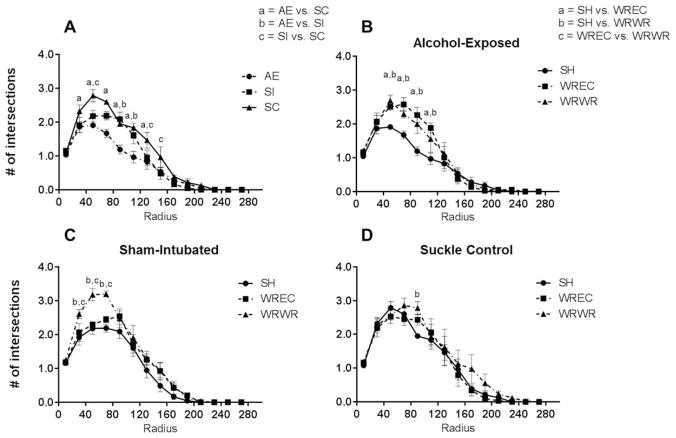
WRWR and WREC rescue alcohol-induced alterations to amount of dendritic material per radius in the PD72 dorsal dentate gyrus. (A) AE significantly decreased amount of dendritic material per radius in immature dentate granule cells. For this graph, significant differences (p <0.05) at each radii are indicated as a =AE versus SC, b =AE versus SI, and c =SI versus SC. (B) In AE animals, WRWR and WREC ameliorated the negative impact of neonatal alcohol exposure on measures of dendritic length. In graphs B–D, significant differences between housing conditions are indicated as a =SH versus WREC, b =SH versus WRWR, and c =WREC versus WRWR. (C and D) In control animals, WRWR increased dendritic material at certain radii. For SC, WRWR increased dendritic material compared with SH only at 90 μm from the soma. Values indicate means ± SEM.
Next, we compared the housing conditions for each neonatal treatment [Fig. 4(B–D)]. For SC animals, a main effect of radius [F(13,140) = 101.7, p <0.0001; Fig. 4(D)] and housing were found (F(2,140) = 6.393, p =0.0022), but the interaction was not significant (F <1). Post hoc analysis (Tukey’s) found a significant increase in dendritic material between SH and WRWR only at radius 90 μm from the soma. For the SI group, a main effect of radius (F(13,140) = 132.5, p <0.0001) and housing condition were observed (F(2,140) = 16.19, p <0.0001), but no significant interaction was found [F(26,140) = 1.328, p =0.1502; Fig. 4(C)]. Post hoc tests revealed a significant increase in length per radius in the WRWR condition compared with both SH and WREC at radii 50 and 70 μm (p <0.05 in all cases) and for WRWR compared with only SH at radius 90 μm from the soma (p <0.01). For AE animals, a significant radius × housing interaction was found [F(26,140) = 2.859, p <0.0001; Fig. 4(B)], as well as main effects for radius (F(13,140) = 122.8, p <0.0001) and housing (F(2,140) = 13.05, p <0.0001). Tukey’s post hoc analysis found significantly increased amounts of dendritic material per radius in WRWR and WREC conditions compared with SH for radii 70, 90, 110, and 130 μm from the soma (p <0.01 at 70 μm, p <0.0001 for 90 um, p <0.001 for 110 μm, and p <0.05 um for 130 μm).
Number of Intersections per Radius
For the comparison of neonatal treatments housed in the SH condition on number of intersections, a radius × neonatal condition interaction was found [F(26,156) = 3.194, p <0.0001; Fig. 5(A)]. Main effects were also found for radius (F(13,156) = 150.4, p <0.0001) and for neonatal condition separately (F(2,12) = 9.42, p =0.0035). Post hoc analysis (Tukey’s) revealed a significant decrease in number of intersections for AE compared with SI for radii 70, 90, and 110 μm from the soma (p <0.05 for all), significant decreases for AE compared with SC for radii 30, 50, 70, 90, 110, and 130 μm from the soma (p <0.05 in all cases) and significant decreases in SI compared with SC for radii 50 and 130 μm (p <0.01 and p <0.05, respectively).
Figure 5.
WRWR and WREC rescue alcohol-induced decreases in intersections per radius in the PD72 dorsal dentate gyrus. (A) AE significantly decreased the number of intersections per radius in immature dentate granule cells (p <0.05). (B) In AE animals, WRWR and WREC mitigated the negative impact of neonatal alcohol exposure on number of intersections. (C and D) In control animals, WRWR increased the number of intersections compared with SH and WREC for SI animals. For SC, WRWR increased intersections compared with SH only at 90 μm from the soma. Values indicate means ± SEM.
We then compared the housing conditions for each neonatal treatment [Fig. 5(B–D)]. For SC animals, a significant main effect of radius (F(13,140) = 75.6, p <0.0001) and of housing were found (F(2,140) = 3.607, p =0.0297), but the interaction was not significant [F <1; Fig. 5(D)]. Post hoc tests found a significant increase in number of intersections per radius in WRWR compared with SH animals at radius 90 μm from the soma (p <0.05). For the SI group, a radius × housing interaction [F(26,140) = 1.706, p =0.0263; Fig. 5(C)], a main effect of radius (F(13,140) = 136.1, p <0.0001), and a main effect of housing condition were observed (F(2,140) = 17.36, p <0.0001). Post hoc tests revealed a significant increase in intersections per radius for WRWR animals compared with both SH and WREC at radii 30, 50, and 70 μm (p <0.05 for 30 μm, p <0.001 for 70 μm, and p <0.01 for 90 μm). For AE animals, a significant radius × housing interaction was found [F(26,140) = 2.488, p =0.0004; Fig. 5(B)], as well as main effects for radius (F(13,140) = 109.2, p <0.0001) and housing (F(2,140) = 13.42, p <0.0001). Post hoc analysis found significantly increased number of intersections per radius in WRWR and WREC conditions compared with SH for radii 50, 70, 90, and 110 μm from the soma (p <0.05 at 50 μm, p <0.01 at 70 μm, p <0.001 for 90 μm, p <0.05 for 110 μm). WREC and WRWR did not differ from one another.
Number of Bifurcations per Radius
We first compared neonatal treatments housed in the SH condition on number of intersections and found a main effect of radius [F(13,156) = 46.46, p <0.0001; Fig. 6(A)] and of neonatal condition (F(2,12) = 9.976, p =0.0028), but no significant interaction was revealed. Post hoc tests (Tukey’s) found a significant decrease in number of bifurcations per radius for AE compared with SI for radii 50 and 70 μm from the soma (p <0.001 and p <0.01, respectively) and significant decreases for AE compared with SC for radii 30 and 50 μm from the soma (p <0.01 and p <0.05, respectively).
Figure 6.
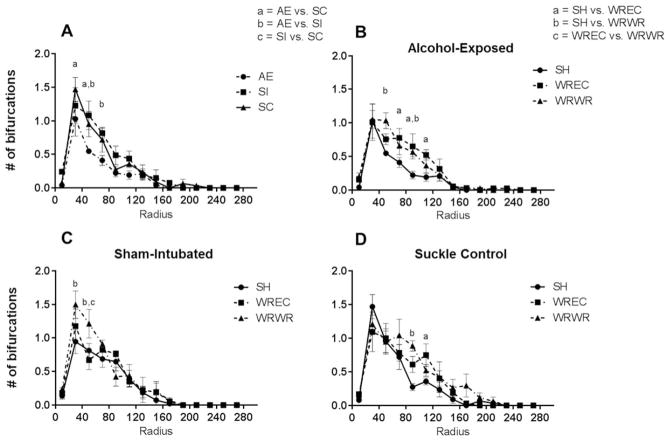
WRWR and WREC rescue alcohol-induced decreases in number of bifurcations per radius in the PD72 dorsal dentate gyrus. (A) AE significantly decreased the number of bifurcations per radius in immature dentate granule cells (p <0.05). (B) In AE animals, WRWR and WREC reversed the negative impact of neonatal alcohol exposure on number of intersections. (C and D) In control animals, WRWR also increased the number of bifurcations, although only at 30 and 50 μm radii in SI and at 90 μm in SC. For SC, WREC increased number of bifurcations at 70 μm as well. Values indicate means ± SEM.
We then compared the housing conditions for each neonatal treatment [Fig. 6(B–D)]. For SC animals, a significant main effect of radius (F(13,130) = 39.25, p <0.0001) and a trending main effect of housing were found (F(2,10) = 3.737, p =0.0614), but the interaction was not significant [F(26,130) = 1.226, p =0.2268; Fig. 6(D)]. Post hoc analysis (Tukey’s) found a significant increase in number of intersections per radius in WRWR compared with SH animals at radius 90 μm from the soma (p <0.001) and in WREC compared with SH at radius 110 μm (p <0.05). For the SI group, a main effect of radius [F(13,140) = 46.33, p <0.0001; Fig. 6(C)] and a trending main effect of housing were observed (F(2,10) = 4.052, p =0.0514). Post hoc tests revealed a significant increase in intersections per radius for WRWR animals compared with both SH and WREC at radius 50 μm (p <0.001 and 0.05, respectively) and an increase in WRWR compared with SH alone at radius 30 μm (p <0.001). For AE animals, no interaction was found, but main effects for radius (F(13,130) = 41.44, p <0.0001) and housing (F(2,130) = 30.88, p <0.0001) were revealed [Fig. 6(B)]. Post hoc tests found significantly increased number of bifurcations per radius in WRWR compared with SH for radii 50 and 90 μm from the soma (p <0.001 at 50 μm and p <0.05 for 90 μm) and increased in WREC compared with SH for radii 70, 90, and 110 μm from the soma (p <0.01, 0.01, and 0.05 respectively). WREC and WRWR did not differ.
Bdnf Gene Expression
Total Bdnf gene expression (by targeting all exon-IX containing transcripts) was measured in the hippocampus on PD72. A main effect of housing was found using a two-way ANOVA (F(2,71) = 24.57, p <0.0001), but no effect of neonatal condition or interaction was revealed. Total Bdnf gene expression was increased in the WRWR condition in all neonatal conditions [p <0.05; Fig. 7(A)]. For all neonatal treatments, gene expression was increased in WRWR compared with both SH and WREC (p <0.05). In all conditions, WREC and SH did not differ from one another. We also examined expression of Bdnf exon I- and IV-specific transcripts. For exon I-specific transcripts, a main effect of housing was found (F(2,73) = 33.08, p <0.0001), but no effect of neonatal condition or interaction was observed. Gene expression was increased in the WRWR condition compared with both SH and WREC in all neonatal conditions [p <0.05, Fig. 7(B)]. WREC and SH did not differ from one another. For exon IV-specific transcripts, again, a main effect of housing was found [F(2,72) = 19.53, p <0.0001; Fig. 7(C)], but no effect of neonatal condition or interaction was found. Gene expression was increased by WRWR compared with both SH and WREC in both SC and AE (p <0.05). For SI, a trend was found that WRWR had higher exon IV gene expression compared with SI was found (p <0.1). In all conditions, WREC and SH did not differ from one another.
Figure 7.
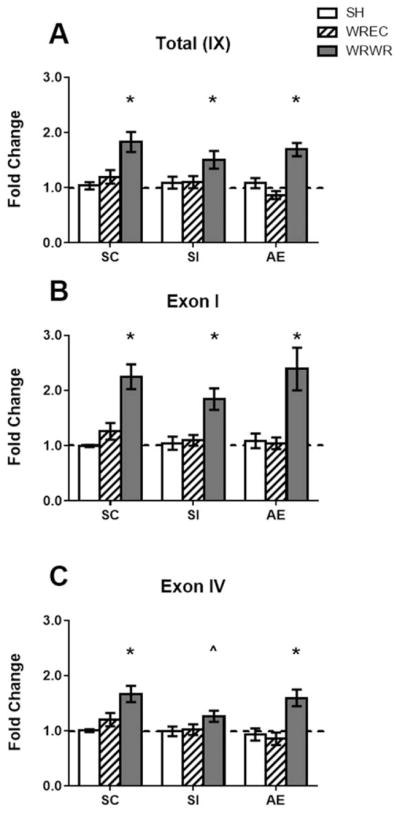
Bdnf gene expression in the PD72 hippocampus. (A) WRWR showed more total Bdnf gene expression compared with SH and WREC animals. (B) Exon I- and (C) exon IV-specific transcripts were significantly elevated in all WRWR animals. A trend toward an increase was found for exon IV-specific gene expression in WRWR pups relative to SH (p <0.1, indicated aŝ on graph). * p <0.05. Data is expressed as a fold change from the SH-suckle control (SC) group (shown as 1 on graphs).
Bdnf Exon I and IV DNA Methylation
DNA methylation status of the Bdnf exon I and IV promoter regions was assessed via MSP in the hippocampus on PD72 and PD10 (Fig. 8). On PD72, a two-way ANOVA found a significant main effect of housing on exon I-associated methylation levels [F(2,55) = 5.564, p =0.0063, Fig. 8(A)]. No main effect of neonatal condition or interaction was found. Tukey’s post hoc analysis revealed that in the SC and SI groups, WRWR was associated with less DNA methylation at exon I (p <0.05) compared with SH. Levels of methylation in the WREC and SH were similar. No significant changes to levels of methylation were found for the AE group. For exon IV methylation, no main effects or interactions were observed [Fig. 8(B)]. Due to our a priori hypothesis that exercise would be associated with lower DNA methylation, we ran a one-way ANOVA on the SC group only for exon IV. A main effect of housing was found (F(2,21) = 4.47, p =0.0241), and post hoc analysis revealed that the WRWR condition was associated with significantly less methylation at exon IV compared with the SC/SH group [Fig. 8(B)].
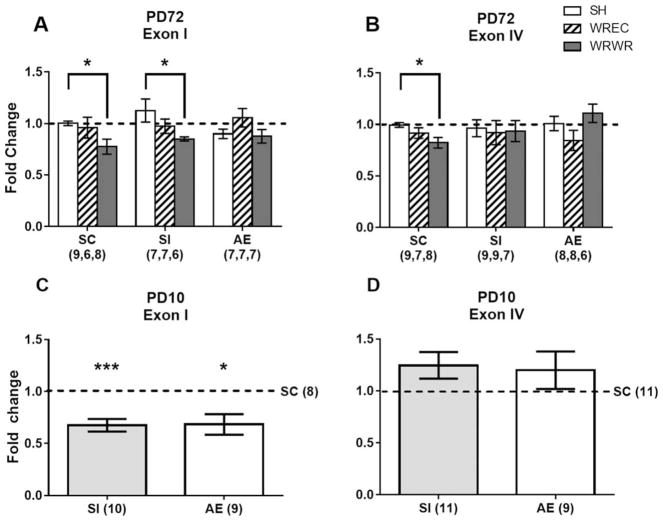
Figure 8.
Bdnf DNA methylation in the PD72 and PD10 hippocampus. (A) On PD72, there was no effect of neonatal condition on methylation status at exon I. WRWR rats showed significantly less Bdnf DNA methylation at exon I compared with SH in the SC and SI groups. (B) There was no effect of neonatal condition on exon IV methylation status. In the SC group, a priori analysis revealed that WRWR was associated with lower DNA methylation compared with the SH condition. (C) On PD10, AE and SI animals showed significantly less Bdnf DNA methylation at exon I, but not exon IV (D), in the hippocampus compared with SC. * p <0.05, *** p <0.001. Data is expressed as a fold change ± SEM from the SC/SH (PD72) or SC (PD10) group (shown as 1 on graphs).
On PD10, analysis of exon I methylation revealed a main effect of neonatal condition [F(2,27) = 6.033, p =0.0068, Fig. 8(C)]. There was less methylation associated with Bdnf exon I DNA methylation in both the AE and SI groups (p <0.05 in both cases) compared with the SC group on PD10. AE and SI groups had similar levels of methylation at exon I (p =n.s.). For exon IV, no significant changes to methylation status due to neonatal condition were observed [F <1; Fig. 8(D)].
DISCUSSION
The current study demonstrated (1) the long-term negative impact of third trimester-equivalent (PD4-9) binge-like alcohol exposure on dendritic complexity of new (DCX+) DG granule cells, (2) the effects of neonatal alcohol exposure on Bdnf gene expression and DNA methylation, and (3) that WRWR and WREC dramatically reversed alcohol-induced deficits in dendritic complexity of immature neurons in AE animals and affected Bdnf expression and DNA methylation in the adult rat hippocampus. Sholl analysis revealed significantly simpler dendritic organization of the immature (DCX+) neurons in the dorsal DG of adult AE animals compared with SI and SC on the measures of total dendritic material, number of intersections, and bifurcations. Additionally, this experiment demonstrated that the influence of our model of FASD on basal Bdnf expression is transient. While this alcohol exposure paradigm increased Bdnf gene expression shortly following the last alcohol administration (PD10, Boschen et al., 2015), the current study showed no effect of neonatal condition on PD72. AE did not alter hippocampal Bdnf total or exon-specific gene expression or DNA methylation status in the social housed (SH) condition. However, Bdnf exon I DNA methylation was decreased on PD10 following our model of FASD, further supporting the transient nature of alcohol’s impact on Bdnf expression. To our knowledge, these results are the first to examine Bdnf mRNA and Bdnf DNA methylation within one model of alcohol exposure.
Another important finding of the current study is that both of the housing interventions implemented within this experiment, wheel running (WR) and environmental complexity (EC), stimulated neuroplasticity in the AE brain. Both WRWR and WREC dramatically reversed alcohol-induced deficits in dendritic complexity of DCX+ immature neurons in AE animals. The amount of dendritic material and the number of intersections and bifurcations per radius were all increased following housing in either intervention. In SC and SI control animals, WRWR increased dendritic complexity of DCX+ neurons, while WREC had little effect on these measures. Interestingly, the current study also found that WRWR and WREC have differential effects on Bdnf expression in the adult rat hippocampus. In all neonatal conditions WRWR, but not WREC, upregulated Bdnf gene expression. Exon I and IV DNA methylation was also decreased by WRWR in the control animals, showing that aerobic exercise can cause epigenetic modifications at these specific Bdnf gene loci. Overall, these findings suggest that exposure to alcohol during development has a transient impact on hippocampal Bdnf expression and that our two behavioral interventions potentially affect neuroplasticity through different pathways.
The effects of alcohol on dendritic complexity are consistent with other literature demonstrating that developmental alcohol exposure models disrupt dendritic organization and complexity. Work from our lab has shown that PD4-9 alcohol exposure causes abnormalities in basilar pyramidal cell dendrites in Layer II/III of the medial prefrontal cortex (mPFC; Hamilton et al., 2010, 2015). Furthermore, prenatal alcohol exposure disrupts dendritic organization in the dorsal hippocampus (Sakata-Haga et al., 2006). The current study analyzed only DCX+ cells; DCX labels a heterogeneous population of neural progenitors and immature neurons, with expression peaking 7–14 days after proliferation (Brown et al., 2003). Previously work from our research group (Hamilton et al., 2011) did not find alcohol-related differences in DCX+ cell number, suggesting that PD4-9 alcohol exposure does not impact the number of immature neurons but does affect dendritic outgrowth. These findings directly relate to the impaired adult-born granule cell survival following neonatal alcohol exposure (Klintsova et al., 2007; Hamilton et al., 2011, 2012, 2014), as an inability to make sufficient number of synaptic connections during competitive survival would impair a cell’s ability to successfully integrate during this process and decrease the number of surviving neurons. The data reported here suggest that the subpopulation of immature neurons about to enter competitive survival and receive glutamatergic input have less complex dendrites, which would impair the ability of these cells to make synapses with the hippocampal circuit. Thus, the dendritic abnormalities in AE animals observed in this study could represent a mechanism through which neonatal alcohol exposure decreases granule cell survival.
Previously published work from our lab demonstrated that our model of FASD increased exon I and IV-driven mRNA transcripts and BDNF protein levels on PD10 (Boschen et al., 2015). The current study expands on these findings by reporting that DNA methylation associated with exon I, but not exon IV, was reduced in the AE group on PD10 [Fig. 8(C,D)]. Our observation of changes in methylation status in AE pups is consistent with the handful of reports that have documented altered DNA methylation in other developmental alcohol models (Liu et al., 2009; Otero et al., 2012; Govorko et al., 2012; Veazey et al., 2015) and activity of DNA methyltransferases (Perkins et al., 2013). Interestingly, while we observed some corresponding methylation and gene expression changes (lower Bdnf exon I methylation and higher exon I gene expression on PD10 [Boschen et al., 2015]; lower Bdnf exon I and IV methylation following WRWR in the SC group [and SI animals for exon I] on PD72 which corresponded with increased exon I and IV-drive gene expression)), that is not always the case (less DNA methylation associated with exon I in the SI group on PD10 which did not correspond with alterations to gene expression [Boschen et al., 2015]; higher expression of Bdnf I and IV-specific mRNA transcripts in both AE and SI animals on PD72 following WRWR without alterations to DNA methylation at these exons [apart from exon I in SI animals]). Similarly, other early life stressors have been shown to impact methylation status of the Bdnf gene without concomitant changes in steady-state mRNA levels (reviewed in Blaze and Roth, 2015). DNA methylation can affect not only baseline gene expression, but also can prime changes to gene transcription in response to environmental stimulation (Baker-Andresen et al., 2013). Thus, alcohol-induced changes to Bdnf methylation status in the early postnatal period could inform both alterations to Bdnf gene expression at this time point, as well as modifications to activity-dependent transcriptional regulation later in life.
We did not detect any alterations to the methylation status of DNA associated with exon I or IV in PD72 animals, suggesting that the epigenetic modifications observed on PD10 were short-lived (i.e., transient in nature). Literature showing that developmental alcohol exposure increased DNA methylation and DNA methyltransferase activity in the hippocampus have focused on earlier ages (PD21) (Otero et al., 2012; Perkins et al., 2013), which could influence the directionality of observed changes to methylation status. For example, exposure of infant rats to an aversive caregiving environment alters Bdnf I and IV methylation patterns in an age-dependent manner (Roth et al., 2009; Blaze et al., 2013; Doherty et al., 2016). Importantly, the results of these studies, in combination with the current data, demonstrate that alterations to methylation are highly dependent on the timing of tissue collection, the brain region analyzed, and the target gene in question. Further, the fact that the PD10 SI group showed less methylation at exon I while not also displaying altered gene expression at this exon (Boschen et al., 2015) highlights the complicated interaction between DNA methylation and gene expression. Other epigenetic markers, such as histone modifications, or function of posttranscriptional regulators, such as miRNA or siRNAs, could be differentially affected in AE and SI animals, explaining the disconnect between the methylation and transcription data.
An important finding of the current study is the robust enhancement of immature neuron dendritic complexity in AE animals by WRWR and WREC. Exercise has repeatedly been shown to increase dendritic length and complexity in DG granule cells (Eadie et al., 2005; Redila and Christie, 2006; Stranahan et al., 2007; Wu et al., 2008) and housing in a complex environment also alters dendritic morphology in hippocampus, cortex, and striatum (Faherty et al., 2005; Gelfo et al., 2009). Work from our lab has previously shown that dendritic complexity of mPFC pyramidal cells is enhanced in AE animals by WR on PD42 and WREC on PD72 (Hamilton et al., 2015; Hamilton et al., unpublished data). More complex dendritic trees likely contribute to behavioral and cognitive enhancement observed following exercise and EC (Vaynman et al., 2004; Kohman et al., 2011; Green et al., 2002; Hamilton et al., 2011; Marlatt et al., 2012; Schreiber et al., 2013) and represent an avenue through which WRWR and WREC rescue cell survival and improve behavioral performance in AE rats (Hamilton et al., 2011; Schreiber et al., 2013; Hamilton et al., 2014). Interestingly, while WRWR modestly enhanced dendritic complexity in the SI and SC group, WREC had almost no effect on these measures in control animals (apart from at one radius for number of bifurcations in SC animals). Additionally, the exercise-induced alterations were not as robust in the controls animals as in the AE group, which suggests a ceiling of plasticity for immature neurons in the healthy brain. The lack of effect of WREC on dendritic complexity in the control animals indicates that the AE brain is more susceptible to benefit from this behavioral intervention
The differential effect of WRWR and WREC on Bdnf gene expression in the current study is very interesting, as both of these interventions are beneficial for synaptic plasticity and are reported to upregulate BDNF in the literature (Torasdotter et al., 1996, 1998; Pham et al., 2002; Ickes et al., 2000; Cotman and Berchtold, 2002; Vaynman et al., 2004; Rasmussen et al., 2009; Marlatt et al., 2012). Our research group has demonstrated that WREC rescued survival of adult-born granule cells in the DG, improved contextual fear and trace eyeblink conditioning in AE rats, and increased dendritic complexity in the mPFC (Hamilton et al., 2011, 2012; Schreiber et al., 2013; Hamilton et al., 2014, 2015). Thus, this intervention has a positive impact on neuroplasticity and learning independent of alterations to Bdnf gene expression on PD72. It is possible that BDNF was upregulated by the 12 days of wheel running from PD30-42 and then, following placement of the rats into the EC cage, decreased to baseline while maintaining changes to synaptic plasticity initiated during wheel running. Additional experimental time points (specifically, PD42) would be necessary to empirically test this possibility.
The findings in the SC group on PD72 for exon I were consistent with the expected relationship between DNA methylation and gene expression. SC animals showed less Bdnf exon I methylation following WRWR, which corresponded with increased exon I-driven gene expression in this group. Total Bdnf gene expression was also increased in this condition. Both exercise and EC have been reported as altering Bdnf DNA methylation (Gomez-Pinilla et al., 2011; Kuzumaki et al., 2011). While the relationship between housing condition, DNA methylation, and gene expression was not always clear cut in the SI and AE groups, it is beyond the scope of this study to directly examine the molecular mechanisms contributing to the differences in methylation and gene expression in the neonatal treatments. A variety of factors could influence our findings at each step. Other epigenetic modifications, including DNA hydroxymethylation or histone acetylation, could account for inconsistencies between methylation patterns and gene expression. Furthermore, methylation could be unassociated with changes to baseline gene expression but instead indicate that the transcriptional system is primed to respond to stimulation (Li et al., 2014). DNA methylation could affect gene transcription beyond altering the ability of transcription factors to bind to promoter regions, including control of alternative splicing, insertion or deletion of transposable elements, or shortening the distance between nucleosomes (Baker-Andresen et al., 2013). In addition, methylation of other Bdnf exons could be altered, which is a target for future research. Further work is needed to determine if any of these factors are applicable to the findings reported here.
Interpretation of the current data must also be kept in the context of the tissue and cell-type examined, as gene expression and methylation patterns can vary drastically between neuronal and non-neuronal cells (Iwamoto et al., 2011). Our gene expression and methylation experimentshere examined whole hippocampus, including dorsal and ventral regions and all cell types. Thus, the results might have been different if DG, CA3, and CA1 were analyzed separately (Roth et al., 2014, 2015). In addition, the cells of the hippocampus are heterogeneous and include granule and pyramidal neurons, a variety of inhibitory inter-neurons, and glial cells. Both neurons and astrocytes synthesize BDNF (Miklic et al., 2004); the use of fluorescence-activated cell sorting (FACS) to separate neuronal and non-neuronal cells prior to analysis would give valuable information about BDNF levels in different cell types. Interestingly, the reported decreases in DCX+ dendritic complexity in the AE group occur independently of hippocampus-wide changes to Bdnf gene expression. Additionally, the alterations to dendritic morphology seen in the AE group following WRWR and WREC occur despite no change in hippocampus-wide Bdnf expression in the AE/WREC condition. Finally, WRWR increases Bdnf expression in the SC and SI groups while causing only modest alterations to dendritic complexity. These discrepancies make a direct comparison between the findings for dendritic complexity and Bdnf expression ill-advised. While Bdnf expression was analyzed from whole (dorsal and ventral) hippocampal tissue, dendritic complexity was only analyzed in dorsal DG. Additionally, DCX labels only a subpopulation of immature neurons and dendritic complexity was measured in, essentially, the oldest subpopulation of this subpopulation. Thus, BDNF could still contribute to the results reported here, but a direct relationship cannot be determined from these experiments.
In summary, we found that third trimester-equivalent (PD4-9) binge-like alcohol exposure negatively impacted dendritic complexity of immature neurons in the dorsal DG, but did not alter total or exon-specific gene expression or DNA methylation in the hippocampus on PD72. The results reported here suggest that our model of FASD targets maturing neurons prior to receiving excitatory synaptic input necessary for their long-term survival, possibly disrupting the ability of these neurons to make sufficient number of functional synapses. Impairments to the process of adult neurogenesis likely contribute to hippocampal-associated behavioral deficits observed in animal models of FASD. DNA methylation associated with Bdnf exon I was lower on PD10 in AE and SI animals, suggesting that the effect our FASD model on Bdnf DNA methylation and gene expression (Boschen et al., 2015) is transient. The housing conditions WREC and WRWR robustly enhanced dendritic complexity in the AE brain. WRWR alone increased dendritic complexity in the SC and SI groups. Additionally, WRWR increased Bdnf gene expression and decreased exon I and IV DNA methylation (in control animals). These data give important information about how our behavioral interventions impact synthesis of a critical growth factor involved in a variety of plasticity-related processes. While these experiments are the first to assess multiple levels of Bdnf regulation using the same model of alcohol exposure and a lifespan approach, the results point to many more avenues of research which must be explored. In addition to the future directions discussed above, this study must be replicated in female animals; the current study focused on male offspring alone. Ultimately, these data support continuing to investigate mechanisms underlying the beneficial effects of exercise and EC on alcohol-induced deficits. Importantly, these results also support WREC and WRWR as behavioral therapies in models of FASD as both are beneficial to neuroplasticity in the alcohol-damaged brain.
Table 3.
Summary of Statistical Output for Dendritic Complexity Analyses
| Measure | Planned RM-ANOVA | Interaction | Radius | Neonatal Treatment | Housing Condition | Significant Post Hocs (μm) |
|---|---|---|---|---|---|---|
| Dendritic Material | SH:AE vs. SI vs. SC | *** | **** | ** | N/A | AE vs. SI: 90, 110 |
| AE vs. SC: 50, 70, 90, 110, 130 | ||||||
| SI vs. SC: 50, 70 | ||||||
| SC:SH vs. WREC vs. WRWR | n.s. | **** | N/A | ** | SH vs. WRWR: 90 | |
| SI:SH vs. WREC vs. WRWR | n.s. | **** | N/A | **** | SH vs. WREC: 50, 70 | |
| SH vs. WRWR: 90 | ||||||
| WREC vs. WRWR:50, 70 | ||||||
| AE:SH vs. WREC vs. WRWR | **** | **** | N/A | **** | SH vs. WREC: 70, 90, 110, 130 | |
| SH vs. WRWR: 70, 90, 110, 130 | ||||||
| Intersections per Radius | SH:AE vs. SI vs. SC | **** | **** | ** | N/A | AE vs. SI: 70, 90, 110 |
| AE vs. SC: 30, 50, | ||||||
| 70, 90, 110, 130 | ||||||
| SI vs. SC: 50, 130 | ||||||
| SC:SH vs. WREC vs. WRWR | n.s. | **** | N/A | * | SH vs. WRWR: 90 | |
| SI:SH vs. WREC vs. WRWR | * | **** | N/A | **** | SH vs. WREC: 30, 50, 70 | |
| WREC vs. WRWR: 30, 50, 70 | ||||||
| AE:SH vs. WREC vs. WRWR | *** | **** | N/A | **** | SH vs. WREC: 50, 70, 90, 110 | |
| SH vs. WRWR: 50, 70, 90, 110 | ||||||
| Bifurcations per Radius | SH:AE vs. SI vs. SC | n.s. | **** | ** | N/A | AE vs. SI: 50, 70 |
| AE vs. SC:30, 50 | ||||||
| SC:SH vs. WREC vs. WRWR | n.s. | **** | N/A | n.s.ˆ | SH vs. WREC: 110 | |
| SH vs. WRWR: 90 | ||||||
| SI:SH vs. WREC vs. WRWR | n.s. | **** | N/A | n.s.ˆ | SH vs. WREC: 110 | |
| SH vs. WRWR: 90 | ||||||
| AE:SH vs. WREC vs. WRWR | n.s. | **** | N/A | **** | SH vs. WREC: 70, 90, 110 | |
| SH vs. WRWR: 50 |
Acknowledgments
Contract grant sponsor: National Institutes of Health/NIGMS COBRE: The Delaware Center for Neuroscience Research; contract grant number: 1P20GM103653 – 01A1 to AYK.
The authors would like to thank Dr. Jennifer Blaze for her assistance with the gene expression and DNA methylation assays, Shaqran Shareeq and Zubin Hussain for their assistance with spectrophotometry and gel electrophoresis, and all the undergraduate research assistants for help with animal generation and care.
References
- Baker-Andresen D, Ratnu VS, Bredy TW. Dynamic DNA methylation: A prime candidate for genomic metaplasticity and behavioral adaptation. Trends Neurosci. 2013;36:3–13. doi: 10.1016/j.tins.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Bekdash RA, Zhang C, Sarkar DK. Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, DNA methylation, and proopiomelanocortin (POMC) gene expression in β-endorphin-producing POMC neurons of the hypothalamus. Alcohol Clin Exp Res. 2013;37:1133–1142. doi: 10.1111/acer.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Chinn G, Chou M, Kesslak JP, Cotman CW. Exercise primes a molecular memory for brain-derived neurotrophic factor protein induction in the rat hippocampus. Neuroscience. 2005;133:853–861. doi: 10.1016/j.neuroscience.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Blaze J, Roth TL. Exposure to caregiver maltreatment alters expression levels of epigenetic regulators in the medial prefrontal cortex. Int J Dev Neurosci. 2013;31:804–810. doi: 10.1016/j.ijdevneu.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaze J, Roth TL. Evidence from clinical and animal model studies of the long-term and transgenerational impact of stress on DNA methylation. Semin Cell Dev Biol. 2015;43:76–84. doi: 10.1016/j.semcdb.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonthius DJ, West JR. Alcohol-induced neuronal loss in developing rats: Increased brain damage with binge exposure. Alcohol Clin Exp Res. 1990;14:107–118. doi: 10.1111/j.1530-0277.1990.tb00455.x. [DOI] [PubMed] [Google Scholar]
- Boschen KE, Criss KJ, Palamarchouk V, Roth TL, Klintsova AY. Effects of developmental alcohol exposure vs. intubation stress on BDNF and TrkB expression in the hippocampus and frontal cortex of neonatal rats. Int J Dev Neurosci. 2015;43:16–24. doi: 10.1016/j.ijdevneu.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschen KE, Hamilton GF, Delorme JE, Klintsova AY. Activity and social behavior in a complex environment in rats neonatally exposed to alcohol. Alcohol. 2014;48:533–541. doi: 10.1016/j.alcohol.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JP, Couillard-Després S, Cooper-Kuhn CM, Winkler J, Aigner L, Kuhn HG. Transient expression of doublecortin during adult neurogenesis. J Comp Neurol. 2003;467:1–10. doi: 10.1002/cne.10874. [DOI] [PubMed] [Google Scholar]
- Christie BR, Swann SE, Fox CJ, Froc D, Lieblich SE, Redila V, Webber A. Voluntary exercise rescues deficits in spatial memory and long-term potentiation in prenatal ethanol-exposed male rats. Eur J Neurosci. 2005;21:1719–1726. doi: 10.1111/j.1460-9568.2005.04004.x. [DOI] [PubMed] [Google Scholar]
- Diaz MR, Jotty K, Locke JL, Jones SR, Valenzuela CF. Moderate alcohol exposure during the rat equivalent to the third trimester of human pregnancy alters regulation of GABAA receptor-mediated synaptic transmission by dopamine in the basolateral amygdala. Front Pediatr. 2015;2:46. doi: 10.3389/fped.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbing J, Sands J. Comparative aspects of the brain growth spurt. Early Hum Dev. 1979;3:79–83. doi: 10.1016/0378-3782(79)90022-7. [DOI] [PubMed] [Google Scholar]
- Doherty TS, Forster A, Roth TL. Global and gene-specific DNA methylation alterations in the adolescent amygdala and hippocampus in an animal model of care-giver maltreatment. Behav Brain Res. 2016;298:55–61. doi: 10.1016/j.bbr.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eadie BD, Redila VA, Christie BR. Voluntary exercise alters the cytoarchitecture of the adult dentate gyrus by increasing cellular proliferation, dendritic complexity, and spine density. J Comp Neurol. 2005;486:39–47. doi: 10.1002/cne.20493. [DOI] [PubMed] [Google Scholar]
- Faherty CJ, Shepherd KR, Herasimtschuk A, Smeyne RJ. Environmental enrichment in adulthood eliminates neuronal death in experimental Parkinsonism. Mol Brain Res. 2005;134:170–179. doi: 10.1016/j.molbrainres.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Fanselow MS, Dong H-W. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;65:7–19. doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattori V, Abe S, Kobayashi K, Costa LG, Tsuji R. Effects of postnatal ethanol exposure on neurotrophic factors and signal transduction pathways in rat brain. J Appl Toxicol. 2008;28:370–376. doi: 10.1002/jat.1288. [DOI] [PubMed] [Google Scholar]
- Gelfo F, De Bartolo P, Giovine A, Petrosini L, Leggio MG. Layer and regional effects of environmental enrichment on the pyramidal neuron morphology of the rat. Neurobiol Learn Mem. 2009;91:353–365. doi: 10.1016/j.nlm.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Gil-Mohapel J, Boehme F, Patten A, Cox A, Kainer L, Giles E, Brocardo PS, Christie BR. Altered adult hippocampal neuronal maturation in a rat model of fetal alcohol syndrome. Brain Res. 2011;1384:29–41. doi: 10.1016/j.brainres.2011.01.116. [DOI] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Zhuang Y, Feng J, Ying Z, Fan G. Exercise impacts brain-derived neurotrophic factor plasticity by engaging mechanisms of epigenetic regulation. Eur J Neurosci. 2011;33:383–390. doi: 10.1111/j.1460-9568.2010.07508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorko D, Bekdash RA, Zhang C, Sarkar DK. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol Psychiatry. 2012;72:378–388. doi: 10.1016/j.biopsych.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JT, Tran T, Steinmetz JE, Goodlett CR. Neonatal ethanol produces cerebellar deep nuclear cell loss and correlated disruption of eyeblink conditioning in adult rats. Brain Res. 2002;956:302–311. doi: 10.1016/s0006-8993(02)03561-8. [DOI] [PubMed] [Google Scholar]
- Griffin EW, Bechara RG, Birch AM, Kelly AM. Exercise enhances hippocampal-dependent learning in the rat: Evidence for a BDNF-related mechanism. Hippocampus. 2009;19:973–980. doi: 10.1002/hipo.20631. [DOI] [PubMed] [Google Scholar]
- Grubbs FE. Procedures for detecting outlying observations in samples. Technometrics. 1969;11:1–21. [Google Scholar]
- Hamilton GF, Boschen KE, Goodlett CR, Greenough WT, Klintsova AY. Housing in environmental complexity following wheel running augments survival of newly generated hippocampal neurons in a rat model of binge alcohol exposure during the third trimester equivalent. Alcohol Clin Exp Res. 2012;36:1196–1204. doi: 10.1111/j.1530-0277.2011.01726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton GF, Criss KJ, Klintsova AY. Voluntary exercise partially reverses neonatal alcohol-induced deficits in mPFC layer II/III dendritic morphology of male adolescent rats. Synapse. 2015;69:405–415. doi: 10.1002/syn.21827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton GF, Jablonski SA, Schiffino FL, St Cyr SA, Stanton ME, Klintsova AY. Exercise and environment as an intervention for neonatal alcohol effects on hippocampal adult neurogenesis and learning. Neuroscience. 2014;265:274–290. doi: 10.1016/j.neuroscience.2014.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton GF, Murawski NJ, St Cyr SA, Jablonski SA, Schiffino FL, Stanton ME, Klintsova AY. Neonatal alcohol exposure disrupts hippocampal neurogenesis and contextual fear conditioning in adult rats. Brain Res. 2011;1412:88–101. doi: 10.1016/j.brainres.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton GF, Whitcher LT, Klintsova AY. Postnatal binge-like alcohol exposure decreases dendritic complexity while increasing the density of mature spines in mPFC Layer II/III pyramidal neurons. Synapse. 2010;64:127–135. doi: 10.1002/syn.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton MB, Mitchell JJ, Paiva M, Walker DW. Ethanol-induced alterations in the expression of neurotrophic factors in the developing rat central nervous system. Brain Res Dev Brain Res. 2000;121:97–107. doi: 10.1016/s0165-3806(00)00032-8. [DOI] [PubMed] [Google Scholar]
- Heaton MB, Paiva M, Madorsky I, Shaw G. Ethanol effects on neonatal rat cortex: comparative analyses of neurotrophic factors, apoptosis-related proteins, and oxidative processes during vulnerable and resistant periods. Brain Res Dev Brain Res. 2003;145:249–262. doi: 10.1016/j.devbrainres.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Helfer JL, Goodlett CR, Greenough WT, Klintsova AY. The effects of exercise on adolescent hippocampal neurogenesis in a rat model of binge alcohol exposure during the brain growth spurt. Brain Res. 2009;1294:1–11. doi: 10.1016/j.brainres.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ickes BR, Pham TM, Sanders LA, Albeck DS, Mohammed AH, Granholm AC. Long-term environmental enrichment leads to regional increases in neurotrophin levels in rat brain. Exp Neurol. 2000;164:45–52. doi: 10.1006/exnr.2000.7415. [DOI] [PubMed] [Google Scholar]
- Ieraci A, Herrera DG. Single alcohol exposure in early life damages hippocampal stem/progenitor cells and reduces adult neurogenesis. Neurobiol Dis. 2007;26:597–605. doi: 10.1016/j.nbd.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Ueda J, Oldham MC, Ukai W, Hashimoto E, Saito T, Geshwing DH, Kato T. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome research. 2011;21:688–696. doi: 10.1101/gr.112755.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G. Adult Neurogenesis: Stem Cells and Neuronal Development in the Adult Brain. Oxford: Oxford University Press; 2011. [Google Scholar]
- Klintsova AY, Helfer JL, Calizo LH, Dong WK, Goodlett CR, Greenough WT. Persistent impairment of hippocampal neurogenesis in young adult rats following early postnatal alcohol exposure. Alcohol Clin Exp Res. 2007;31:2073–2082. doi: 10.1111/j.1530-0277.2007.00528.x. [DOI] [PubMed] [Google Scholar]
- Kobilo T, Liu QR, Gandhi K, Mughal M, Shaham Y, van Praag H. Running is the neurogenic and neurotrophic stimulus in environmental enrichment. Learn Mem. 2011;189:605–609. doi: 10.1101/lm.2283011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohman RA, Rodriguez-Zas SL, Southey BR, Kelley KW, Dantzer R, Rhodes JS. Voluntary wheel running reverses age-induced changes in hippocampal gene expression. PLoS One. 2011;6:e22654. doi: 10.1371/journal.pone.0022654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg G, Bick-Sander A, Bunk E, Wolf C, Ehninger D, Kempermann G. Physical exercise prevents age-related decline in precursor cell activity in the mouse dentate gyrus. Neurobiol Aging. 2006;27:1505–1513. doi: 10.1016/j.neurobiolaging.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Kuzumaki N, Ikegami D, Tamura R, Hareyama N, Imai S, Narita M, Torigoe K, Niikura K, Takeshima H, Ando T, Igarashi K, Kanno J, Ushijima T, Suzuki T, Narita M. Hippocampal epigenetic modification at the brain-derived neurotrophic factor gene induced by an enriched environment. Hippocampus. 2011;21:127–132. doi: 10.1002/hipo.20775. [DOI] [PubMed] [Google Scholar]
- Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harbor perspectives in biology. 2014;6:a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. 2009;4:500–511. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlatt MW, Potter MC, Lucassen PJ, van Praag H. Running throughout middle-age improves memory function, hippocampal neurogenesis, and BDNF levels in female C57BL/6J mice. Dev Neurobiol. 2012;72:943–952. doi: 10.1002/dneu.22009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Kuma H, Yokoyama T, Sumitani K, Matsumoto Y, Kusaka T, Warita K, Wang ZY, Hosomi N, Imagawa T, Bedi SK, Itoh S, Nakamura Y, Takeuchi Y. Early postnatal ethanol exposure induces fluctuation in the expression of BDNF mRNA in the developing rat hippocampus. Acta Neurobiol Exp (Wars) 2008;68:484–493. doi: 10.55782/ane-2008-1714. [DOI] [PubMed] [Google Scholar]
- Miklič Š, Jurič DM, Čaman-Kržan M. Differences in the regulation of BDNF and NGF synthesis in cultured neonatal rat astrocytes. Int J Dev Neurosci. 2004;22:119–130. doi: 10.1016/j.ijdevneu.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawski NJ, Klintsova AY, Stanton ME. Neonatal alcohol exposure and the hippocampus in developing male rats: Effects on behaviorally induced CA1 c-Fos expression, CA1 pyramidal cell number, and contextual fear conditioning. Neuroscience. 2012;206:89–99. doi: 10.1016/j.neuroscience.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero NK, Thomas JD, Saski CA, Xia X, Kelly SJ. Choline supplementation and DNA methylation in the hippocampus and prefrontal cortex of rats exposed to alcohol during development. Alcohol Clin Exp Res. 2012;36:1701–1709. doi: 10.1111/j.1530-0277.2012.01784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet-Wadiche LS, Westbrook GL. Functional maturation of adult-generated granule cells. Hippocampus. 2006;16:208–215. doi: 10.1002/hipo.20152. [DOI] [PubMed] [Google Scholar]
- Perkins A, Lehmann C, Lawrence RC, Kelly SJ. Alcohol exposure during development: Impact on the epigenome. Int J Dev Neurosci. 2013;31:391–397. doi: 10.1016/j.ijdevneu.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TM, Winblad B, Granholm AC, Mohammed AH. Environmental influences on brain neurotrophins in rats. Pharmacology Biochemistry and Behavior. 2002;731:167–175. doi: 10.1016/s0091-3057(02)00783-9. [DOI] [PubMed] [Google Scholar]
- Plümpe T, Ehninger D, Steiner B, Klempin F, Jessberger S, Brandt M, Römer B, Rodriguez GR, Kronenberg G, Kempermann G. Variability of doublecortin-associated dendrite maturation in adult hippocampal neurogenesis is independent of the regulation of precursor cell proliferation. BMC Neurosci. 2006;7:77. doi: 10.1186/1471-2202-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglia MP, Valenzuela CF. Ethanol acutely inhibits ionotropic glutamate receptor-mediated responses and long-term potentiation in the developing CA1 hippocampus. Alcohol Clin Exp Res. 2010a;34:594–606. doi: 10.1111/j.1530-0277.2009.01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglia MP, Valenzuela CF. Repeated third trimester-equivalent ethanol exposure inhibits long-term potentiation in the hippocampal CA1 region of neonatal rats. Alcohol. 2010b;44:283–290. doi: 10.1016/j.alcohol.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen P, Brassard P, Adser H, Pedersen MV, Leick L, Hart E, Secher NH, Pedersen BK, Pilegaard H. Evidence for a release of brain-derived neurotrophic factor from the brain during exercise. Exp Physiol. 2009;94:1062–1069. doi: 10.1113/expphysiol.2009.048512. [DOI] [PubMed] [Google Scholar]
- Redila VA, Christie BR. Exercise-induced changes in dendritic structure and complexity in the adult hippocampal dentate gyrus. Neuroscience. 2006;137:1299–1307. doi: 10.1016/j.neuroscience.2005.10.050. [DOI] [PubMed] [Google Scholar]
- Roth ED, Roth TL, Money KM, SenGupta S, Eason DE, Sweatt JD. DNA methylation regulates neurophysiological spatial representation in memory formation. Neuroepigenetics. 2015;2:1–8. doi: 10.1016/j.nepig.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Matt S, Chen K, Blaze J. Bdnf DNA methylation modifications in the hippocampus and amygdala of male and female rats exposed to different caregiving environments outside the homecage. Dev Psychobiol. 2014;56:1755–1763. doi: 10.1002/dev.21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata-Haga H, Dominguez HD, Sei H, Fukui Y, Riley EP, Thomas JD. Alterations in circadian rhythm phase shifting ability in rats following ethanol exposure during the third trimester brain growth spurt. Alcohol Clin Exp Res. 2006;30:899–907. doi: 10.1111/j.1530-0277.2006.00105.x. [DOI] [PubMed] [Google Scholar]
- Schreiber WB, St Cyr SA, Jablonski SA, Hunt PS, Klintsova AY, Stanton ME. Effects of exercise and environmental complexity on deficits in trace and contextual fear conditioning produced by neonatal alcohol exposure in rats. Dev Psychobiol. 2013;555:483–495. doi: 10.1002/dev.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Gupta S, Jiang Y, Younus M, Ramzan M. In vitro neurogenesis from neural progenitor cells isolated from the hippocampus region of the brain of adult rats exposed to ethanol during early development through their alcohol-drinking mothers. Alcohol. 2009;44:185–198. doi: 10.1093/alcalc/agn109. [DOI] [PubMed] [Google Scholar]
- Stranahan AM, Khalil D, Gould E. Running induces widespread structural alterations in the hippocampus and entorhinal cortex. Hippocampus. 2007;17:1017–1022. doi: 10.1002/hipo.20348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Sather TM, Whinery LA. Voluntary exercise influences behavioral development in rats exposed to alcohol during the neonatal brain growth spurt. Behav Neurosci. 2008;122:1264–1273. doi: 10.1037/a0013271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torasdotter M, Metsis M, Henriksson BG, Winblad B, Mohammed AH. Expression of neurotrophin-3 mRNA in the rat visual cortex and hippocampus is influenced by environmental conditions. Neuroscience Letters. 1996;218:107–110. doi: 10.1016/s0304-3940(96)13127-x. [DOI] [PubMed] [Google Scholar]
- Torasdotter M, Metsis M, Henriksson BG, Winblad B, Mohammed AH. Environmental enrichment results in higher levels of nerve growth factor mRNA in the rat visual cortex and hippocampus. Behav Brain Res. 1998;93:83–90. doi: 10.1016/s0166-4328(97)00142-3. [DOI] [PubMed] [Google Scholar]
- Van der Borght K, Havekes R, Bos T, Eggen BJ, Van der Zee EA. Exercise improves memory acquisition and retrieval in the Y-maze task: Relationship with hippocampal neurogenesis. Behav Neurosci. 2007;121:324. doi: 10.1037/0735-7044.121.2.324. [DOI] [PubMed] [Google Scholar]
- Van Praag H, Kempermann G, Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat Neurosci. 1999;2:266–270. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]
- Van Praag H, Schinder AF, Christie BR, Toni N, Palmer TD, Gage FH. Functional neurogenesis in the adult hippocampus. Nature. 2002;415:1030–1034. doi: 10.1038/4151030a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Gomez-Pinilla F. Hippocampal BDNF mediates the efficacy of exercise on synaptic plasticity and cognition. Eur J Neurosci. 2004;20:2580–2590. doi: 10.1111/j.1460-9568.2004.03720.x. [DOI] [PubMed] [Google Scholar]
- Veazey KJ, Parnell SE, Miranda RC, Golding MC. Dose-dependent alcohol-induced alterations in chromatin structure persist beyond the window of exposure and correlate with fetal alcohol syndrome birth defects. Epigenet Chrom. 2015;8:1. doi: 10.1186/s13072-015-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CW, Chang YT, Yu L, Chen H, Jen CJ, Wu SY, Lo CP, Kuo YM. Exercise enhances the proliferation of neural stem cells and neurite growth and survival of neuronal progenitor cells in dentate gyrus of middle-aged mice. J Appl Physiol. 2008;105:1585–1594. doi: 10.1152/japplphysiol.90775.2008. [DOI] [PubMed] [Google Scholar]