Abstract
Purpose
Locally advanced non‐small cell lung cancer (NSCLC) patients may experience dramatic changes in anatomy during radiotherapy and could benefit from adaptive radiotherapy (ART). Deformable image registration (DIR) is necessary to accurately accumulate dose during plan adaptation, but current algorithms perform poorly in the presence of large geometric changes, namely atelectasis resolution. The goal of this work was to develop a DIR framework, named Consistent Anatomy in Lung Parametric imagE Registration (CALIPER), to handle large geometric changes in the thorax.
Methods
Registrations were performed on pairs of baseline and mid‐treatment CT datasets of NSCLC patients presenting with atelectasis at the start of treatment. Pairs were classified based on atelectasis volume change as either full, partial, or no resolution. The evaluated registration algorithms consisted of several combinations of a hybrid intensity‐ and feature‐based similarity cost function to investigate the ability to simultaneously match healthy lung parenchyma and adjacent atelectasis. These components of the cost function included a mass‐preserving intensity cost in the lung parenchyma, use of filters to enhance vascular structures in the lung parenchyma, manually delineated lung lobes as labels, and several intensity cost functions to model atelectasis change. Registration error was quantified with landmark‐based target registration error and post‐registration alignment of atelectatic lobes.
Results
The registrations using both lobe labels and vasculature enhancement in addition to intensity of the CT images were found to have the highest accuracy. Of these registrations, the mean (SD) of mean landmark error across patients was 2.50 (1.16) mm, 2.80 (0.70) mm, and 2.04 (0.13) mm for no change, partial resolution, and full atelectasis resolution, respectively. The mean (SD) atelectatic lobe Dice similarity coefficient was 0.91 (0.08), 0.90 (0.08), and 0.89 (0.04), respectively, for the same groups. Registration accuracy was comparable to healthy lung registrations of current state‐of‐the‐art algorithms reported in literature.
Conclusions
The CALIPER algorithm developed in this work achieves accurate image registration for challenging cases involving large geometric and topological changes in NSCLC patients, a requirement for enabling ART in this patient group.
Keywords: atelectasis, deformable image registration, deformation, lung, registration
1. Introduction
High precision radiotherapy requires accurate knowledge of the target and surrounding normal tissues’ location and dose. The most commonly seen day‐to‐day change in patient anatomy during locally advanced non‐small cell lung cancer (NSCLC) treatment is tumor regression. Substantial regression of the visible tumor, sometimes greater than 80% of the total volume and ranging from 0.6% to 2.4% shrinkage per day, has been observed in patients treated with radiotherapy just a few weeks after the start of treatment.1, 2, 3, 4 Additional studies have similarly reported average tumor volume reductions of 24.7% halfway through treatment and 44.3% by the end of treatment.5, 6
For centrally located NSCLC tumors, atelectasis is a common co‐pathology which often induces large positional changes.6 As the tumor grows, the central airways can become obstructed, inducing a collapse of the dependent portion of the lung. In CT images, this pathology appears as a consolidation of affected low density lung tissue into a smaller region of frequently uniform, soft tissue intensity. Often, the tumor is located within or adjacent to the collapsed lung at the time of treatment planning and is indistinguishable from the atelectatic lung, making it difficult for the physician to accurately draw the tumor boundaries without additional information.6 If the tumor regresses in response to the radiation, the airways may open again and fully or partially re‐aerate the atelectatic lung. This causes substantial change to the treatment anatomy and possible positional shifts in the tumor.4 Resolution of the non‐tumor pathology may reveal errors in original tumor delineations (eg, over‐ or under‐estimation of tumor extent). Atelectasis appearance/resolution in conjunction with tumor regression is a prime indication for adaptive radiotherapy (ART).5, 7, 8
Image registration can potentially provide the required level of precision to support highly accurate assessment of delivered dose,9 but existing registration algorithms are not designed to deal with the magnitude of day‐to‐day anatomical changes observed in some locally advanced NSCLC patients. Registration in these situations is a challenging and largely unresolved problem due to the presence of tumor regression and atelectasis along with other conditions such as pneumonia and pleural effusion.10 In many cases, it is unclear whether observed shrinkage is of both the tumor and subclinical disease or is only a decrease in image intensity caused by the reduction in tumor density. Therefore, it is important to accurately track adjacent healthy tissue as clinically significant changes occur, rather than just the gross tumor surface, to appropriately accumulate dose throughout treatment.11 When pneumonia or pleural effusion either appears or resolves during the course of treatment, a fundamental assumption of current registration algorithms is violated, namely, that one‐to‐one correspondence exists between the two images being registered such as the planning and weekly images. While atelectasis does not cause new anatomy to appear, the tissue becomes so greatly changed in appearance that the anatomical correspondence can no longer be recognized by common algorithms. These dramatic topological changes of mass and density, up to 24% and 66% of baseline mass and density, respectively, have been shown to induce clinically significant changes in target and OAR doses when the original treatment plan is unaltered.12 Thus, the conditions which contribute to registration failure are the same which necessitate ART.
Under favorable conditions, that is, when no large geometric changes occur and the tumor is located in the lung parenchyma away from the mediastinum, current DIR algorithms used during plan adaptation produce average landmark errors in the range of 1–5 mm in lung tissue.13 When tumor regression, atelectasis resolution, and other non‐tumor pathologies such as pleural effusion and inflammation which commonly accompany the former are present, registration accuracy has not been measured directly, but anecdotally has been found to be “challenging at best”.3 With centrally located tumors, atelectasis resolution, or regression of an invasive tumor which does not displace adjacent normal tissue, feature‐based algorithms that depend on contours tend to fail.14, 15 Algorithms currently used perform poorly since they were not designed to handle the previously discussed cases. This work presents a registration method, named the Consistent Anatomy in Lung Parametric imagE Registration (CALIPER) algorithm, specifically designed to solve the problem of changing tissues, potentially leading to more accurate ART and improved local control and patient survival rates. “Consistent Anatomy” refers to identifying and registering anatomical structures or features that are identifiable in the input images, described in detail in the next few sections. “Parametric” reflects our use of a B‐spline transformation model which greatly reduces the number of parameters needed to describe relatively complex deformations.
2. Methods and materials
2.A. Overview
The goal of this investigation was to develop a multiresolution image registration framework to handle large geometric changes in the thorax. To facilitate the development of the CALIPER algorithm, the importance of different components (eg, atelectasis resolution model, vessel registration, lobe label registration, etc.) on registration accuracy was evaluated in a clinical CT dataset.
2.B. Imaging datasets
Helical CT image sets curated for use in the registrations of this work were gathered from locally advanced NSCLC patients enrolled in various longitudinal CT imaging protocols at Virginia Commonwealth University. Anonymized images of 18 patients were selected from this database using MIM Maestro (MIM Software, Cleveland, OH, USA). Prerequisites for patient selection were multiple CT studies, either breath‐hold or 4DCT, during the course of radiotherapy and the presence of atelectasis. One image pair per patient was selected for registration that included a baseline image obtained around the start of treatment and another acquired mid‐treatment. When multiple mid‐treatment images were available, the image at the time of greatest atelectasis change was selected to present the greatest challenge to the image registration algorithm.
In‐plane image resolution ranged from 0.98 to 1.37 mm with slice thickness of 2–3 mm. Both images for a given pair had the same voxel dimensions except for two patients. Tumor stage ranged from IB to IIIB with 50% of tumors occurring in the lower right lobe of the lung. Mean (SD) tumor volume at the time of baseline imaging was 109.6 (89.2) ml. A total of 56% of patients experienced whole lobe collapse, while the atelectasis for the remaining 44% only affected a portion of the lobe. Atelectasis volume at the time of baseline imaging was 232.42 (181.55) ml. Patients were divided into three groups based on a classification scheme from a previous work.12 Briefly, this scheme was based on the degree of atelectasis volume and/or topology change observed between baseline and mid‐treatment images: full resolution for volume changes > 80% (22% of patients), partial resolution for volume changes between 80% and 20% (50% of patients), and no resolution for changes less than 20% or 15 ml (28% of patients).
The focus of this investigation is deformable image registration of lung parenchyma in the presence of atelectasis resolution. Thus, rigid displacements were removed prior to application of the CALIPER algorithm. Within the MIM software, rigid registration was performed using the standard fusion tool to align the mid‐treatment scan to the baseline scan based on bony anatomy. Since the initial fusion used the entire image volume, additional adjustment of the rigid registration was done using the box‐based alignment tool to focus the fusion on the lungs and mediastinum for improved alignment in the region of interest for this work. The rigidly registered images were exported from MIM using in‐house MATLAB extensions which converted the DICOM images to Meta Image format.
2.C. Lobe labels
Contours for all thoracic structures including individual lobes were delineated by radiation oncologists using the MIM software and were reviewed for consistency between image pairs by a single experienced radiation oncologist who was not involved in the initial contouring. Adjustment of contours was occasionally necessary, particularly for the fissures of the right middle lobe. Contours were saved to binary masks in Meta Image format using in‐house MATLAB extensions for MIM. Once extracted from the database, the binary masks of individual lung lobes were combined to form the lobe labels used for the registrations of this work. The lobe label images consisted of each voxel within a given lung lobe being assigned an integer value. Lobe values were chosen to maximize the intensity difference between adjacent lobes and to provide the largest value to the atelectatic lobe, thus giving atelectasis the greatest influence on the registration.
Since the location of atelectasis varied from patient to patient, lobe label values were assigned on a per‐patient basis. An example of a lobe label pair used for registration is shown in Fig. 1. Left lung lobes were given an intensity of either 3 or 5, while right lobes were assigned values of 1, 3, and 5. A value of 5 was preferentially assigned to lower lobes and 3 to upper lobes due to the increased geometric changes along the diaphragm due to breathing and lung capacity differences between images. In the majority of lobe label images, the highest value of 5 was assigned to the lower lobes of both lungs due to the atelectasis occurring in one of the lower lobes. In cases where the left and right lungs came within a few voxels of each other, usually in the anterior of the chest, reassignment of the contralateral lobe values was necessary to prevent possible misregistration from adjacent lobes having the same value. The manual assignment of lobe values assured the atelectatic lobe of each patient had the greatest influence on the lobe label portion of the registrations.
Figure 1.
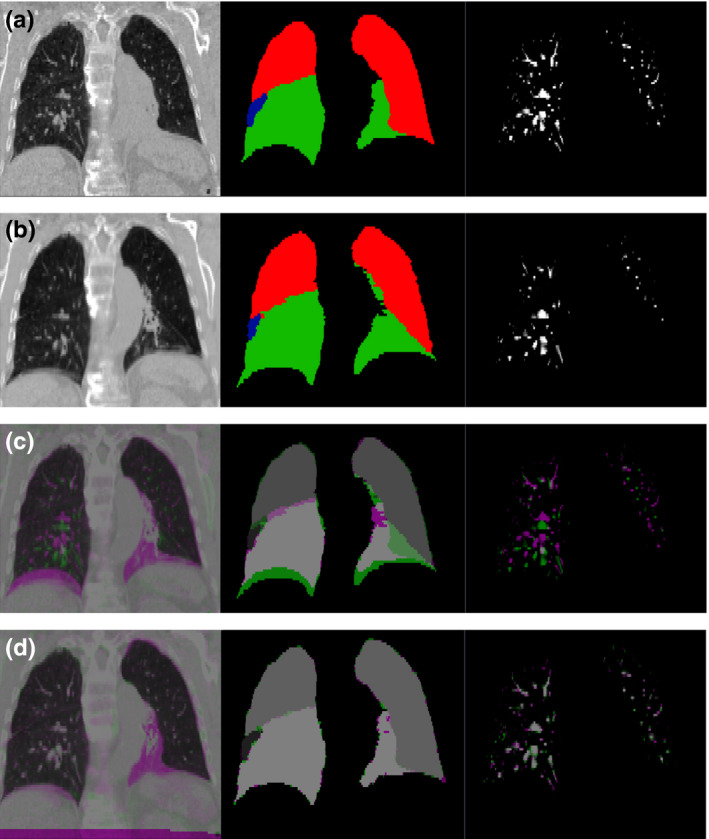
Example of input data and registration results. Input data to the CALIPER algorithm for one subject (Patient 12) is shown for (a) the baseline scan set as the fixed image of the registration and (b) the mid‐treatment scan set as the moving image of the registration. The CT scans are shown in the left column, the lobe label images are shown in the middle column, and the vesselness measure images are shown in the right column. For the lobe label images, green voxels have a value of 5, red voxels have a value of 3, and blue voxels have a value of 1. (c) Complementary color overlays of the fixed (magenta) and moving (green) images are provided to illustrate the degree of initial misalignment due to differences in lung volume and changes in pathology. (d) Finally, the resulting alignment of all three datasets after registration via the CALIPER algorithm is shown. [Color figure can be viewed at wileyonlinelibrary.com]
2.D. Vesselness measure images
Enhancement of lung vasculature has been used to improve the accuracy of lung image registration, particularly in contrast‐poor regions.16, 17 Here, we used a filter approach which enhances tubular structures. The Jerman vesselness measure filter was used to generate the vesselness measure (VM) images for all registration CTs, as it tends to provide more consistent enhancement at bifurcations and across varying vessel diameters.18 The vesselness filter parameters were optimized based on visual inspection of the agreement between vessels in the original image and the resulting VM trees. VM images for all patients were created using the optimal parameters and a mask which only included healthy lobes and excluded atelectasis to assure correspondence between structures of VM image pairs. A resulting VM image is shown in Fig. 1.
2.E. Landmark sets
Using landmarks as a validation measure provides clinically meaningful validation of a registration.19, 20 Therefore, the registrations of this work were assessed primarily by landmark registration error. Physician‐specified landmark correspondences were obtained for all image pairs of the study cohort using the isiMatch software developed by the Imaging Sciences Institute of the University Medical Center Utrecht (Utrecht, Netherlands).21 Landmark sets had a mean (SD) of 169 (31) landmarks per patient. The typical distribution of landmarks throughout the lungs is shown in Fig. 2. The landmark sets were matched by an experienced physician. Due to the large geometric changes of the patient cohort of this study, the automatic matching feature of the isiMatch software was rarely available for use by the observer, so all points were matched manually.
Figure 2.
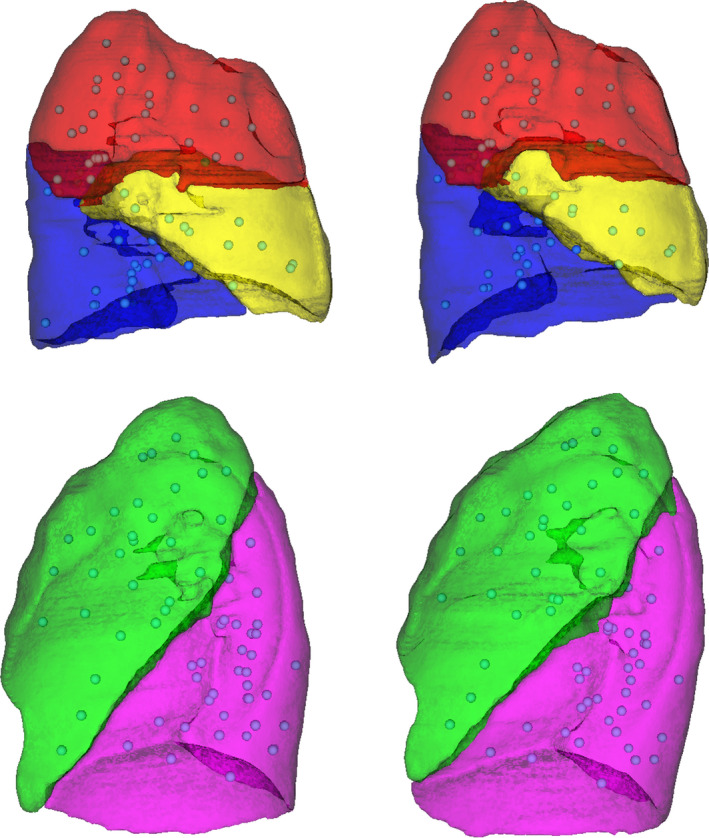
Example of landmark distribution. The distribution of landmarks for a patient with partial lobar atelectasis of the right lower lobe (patient 17) is illustrated. Lateral views of the right lung lobes (top) and left lung lobes (bottom) are shown for the baseline (left) and mid‐treatment (right) images. The landmarks (spheres) were sampled throughout the lung parenchyma of all lobes and were primarily located on vessels. [Color figure can be viewed at wileyonlinelibrary.com]
Accuracy of the landmarks was estimated and improved through an in‐house quality assurance procedure prior to use in quantitating registration accuracy. When points were identified as being incorrectly matched, the observer was asked to review and adjust the specified points. The quality assurance loop was repeated until all point pairs appeared to be correctly matched.
2.F. Image registrations
The registration framework was implemented using the elastix registration software.22 A third‐order B‐splines based transformation model was used with a stochastic gradient descent optimizer and a hybrid cost function utilizing both intensity‐based and feature‐based metrics. A multiresolution registration was implemented which moved from coarse (126 mm) to fine (8 mm) B‐spline grid spacings in five resolutions for added robustness of the optimizer to local minima. The cost function was comprised of a linear combination of up to five components: a healthy lobe registration, atelectatic lobe registration, vesselness measure registration, lobe label registration, and a transformation penalty. Components were turned off/on by removing/including them in the cost function to form the 20 registrations investigated in this study. The ability of several similarity metrics to correctly register atelectasis changes was tested.
The standard sum of squared intensity differences (SSD) and mutual information (MI) metrics, included in the elastix software, were selected due to their wide‐spread use in mono‐ and multimodal image registration, respectively. The sum of squared tissue volume differences (SSTVD) similarity metric,17, 23, 24, 25 which assumes mass, but not density, is preserved by the transformation was recently added to elastix and included in this investigation for its ability to handle intrafractional density changes of lung parenchyma due to respiration. Finally, a hybrid SSD/SSTVD metric referred to as the sum of squared regional volume differences (SSRVD) was developed in‐house which weights the density changes by the average volume change in the region of interest, rather than the voxel‐wise weighting of SSTVD. By weighting intensity in proportion to the average volume change in a region, it was suspected that the metric could better handle atelectasis changes when homogeneous high‐intensity collapse expands to low intensity heterogeneous lung parenchyma.
A total of 20 registrations were performed per patient, each using a different permutation of the cost function, as described in Table 1. Briefly, all registrations used SSTVD within the healthy lung parenchyma (lungs minus atelectatic lobes), as SSTVD yielded the highest registration accuracy of all metrics in computational phantom tests. Registrations were performed with and without the co‐registration of both the corresponding lobe label images and vesselness measure images using a basic SSD similarity metric. Finally, one of several metrics was used to register the atelectatic lobes: SSTVD, SSRVD, SSD, MI, or none. All registrations also used a bending energy penalty on the transformation to reduce the likelihood of non‐physical deformations occurring. The various cost function components were linearly combined to create the full cost function which was minimized using the adaptive stochastic gradient descent optimizer available in elastix.
Table 1.
Registration accuracy for variations of the CALIPER algorithm
| Cost function metrics | Landmark error (mm) | Atelectatic lobe DSC | Failure rate (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Registration | Healthy | Atelectasis | Label | VM | Reg | Mean | P90 | Max | ||
| – | Unregistered | 9.93 | 18.19 | 30.67 | 0.607 | – | ||||
| 1 | SSTVD | – | – | – | BE | 4.31 | 10.63 | 25.54 | 0.648 | 6 |
| 2 | SSTVD | SSTVD | – | – | BE | 4.75 | 11.58 | 24.87 | 0.690 | 22 |
| 3 | SSTVD | SSRVD | – | – | BE | 4.81 | 11.57 | 24.84 | 0.699 | 22 |
| 4 | SSTVD | SSD | – | – | BE | 5.32 | 11.67 | 23.35 | 0.666 | 28 |
| 5 | SSTVD | MI | – | – | BE | 4.56 | 10.98 | 24.82 | 0.708 | 22 |
| 6 | SSTVD | – | SSD | – | BE | 2.78 | 6.03 | 23.16 | 0.891 | 6 |
| 7 | SSTVD | SSTVD | SSD | – | BE | 2.79 | 6.06 | 23.37 | 0.919 | 11 |
| 8 | SSTVD | SSRVD | SSD | – | BE | 2.80 | 6.09 | 23.42 | 0.919 | 11 |
| 9 | SSTVD | SSD | SSD | – | BE | 2.79 | 6.05 | 23.30 | 0.915 | 11 |
| 10 | SSTVD | MI | SSD | – | BE | 2.69 | 5.71 | 23.34 | 0.914 | 11 |
| 11 | SSTVD | – | – | SSD | BE | 4.57 | 11.39 | 27.29 | 0.631 | 11 |
| 12 | SSTVD | SSTVD | – | SSD | BE | 5.01 | 12.90 | 27.46 | 0.674 | 28 |
| 13 | SSTVD | SSRVD | – | SSD | BE | 4.78 | 11.65 | 25.70 | 0.705 | 22 |
| 14 | SSTVD | SSD | – | SSD | BE | 4.92 | 11.64 | 23.60 | 0.665 | 28 |
| 15 | SSTVD | MI | – | SSD | BE | 4.64 | 11.71 | 26.14 | 0.713 | 22 |
| 16 | SSTVD | – | SSD | SSD | BE | 2.58 | 5.35 | 22.35 | 0.877 | 6 |
| 17 | SSTVD | SSTVD | SSD | SSD | BE | 2.60 | 5.46 | 22.99 | 0.911 | 11 |
| 18 | SSTVD | SSRVD | SSD | SSD | BE | 2.61 | 5.52 | 23.01 | 0.912 | 11 |
| 19 | SSTVD | SSD | SSD | SSD | BE | 2.60 | 5.44 | 22.91 | 0.907 | 11 |
| 20 | SSTVD | MI | SSD | SSD | BE | 2.51 | 5.22 | 22.88 | 0.907 | 11 |
Healthy, lobes with healthy parenchyma; atelectasis, lobes containing atelectasis; label, lobe label image; VM, vesselness measure image; Reg, regularization penalty on the transformation; SSTVD, sum of squared tissue volume difference; SSD, sum of squared intensity difference; MI, mutual information; SSRVD, sum of squared regional volume difference; BE, bending energy penalty.
Registration parameters were manually optimized for each registration separately using an image pair from a subject with full resolution of atelectasis. In particular, the relative weights for each of the cost function components had to be determined. It was anticipated that the optimal parameters for this subject, representing the largest geometric and topological changes present in the study cohort, would translate to accurate registrations for the remaining 17 patients. In addition to landmark‐based registration error, the dice similarity coefficient (DSC) was calculated for the atelectatic lobe since the goal of this work was to obtain high registration accuracy throughout the entire lung volume, not just in the healthy parenchyma. During optimization of the registration parameters, a tradeoff between landmark error and atelectasis DSC was apparent, and the optimal parameters which maximized both accuracy metrics simultaneously were selected. The 20 registrations using optimized parameter sets were performed for all 18 patients.
2.G. Statistical analysis
Statistical analyses were carried out to quantitate how each of CALIPER's cost function components affected registration results. For all tests, the Wilcoxon signed‐rank test was performed due to the small sample sizes involved.
3. Results
3.A. Rigid registration
Rigid registrations based on bony anatomy of the thorax resulted in mean (SD) translations and rotations of 4.10 (2.94) cm and 2.43° (1.27°), respectively, across all 18 image pairs to bring anatomy into starting alignment for the CALIPER algorithm. The large geometric changes present in the patient cohort were evident in the starting displacements prior to deformable registration but after rigid alignment. Across all patients, the mean (SD) of initial landmark errors were 9.93 (5.48) mm, 18.19 (11.73) mm, and 30.67 (18.71) mm for mean, 90th percentile, and maximum landmark errors, respectively. The mean (SD) atelectasis starting DSC was 0.607 (0.233). Over 44% of patients had initial mean landmark displacements greater than 1 cm, and 50% of patients had DSC less than 0.7.
3.B. Deformable registration
Registration results are reported in Table 1. Figure 1 also shows the best registration result, as complementary color overlays of the deformed mid‐treatment images and the baseline counterparts, obtained for patient 12. The equivalent of Fig. 1 for all 18 patients of this study can be found in the Data S1. Figure 3 shows box plots of mean landmark error for each registration, while Fig. 4 shows the same plot for DSC. A clear improvement of both accuracy metrics is noticeable between registrations which used lobe labels (registrations 6–10 and 16–20) from registrations which did not. The registrations using the full cost function of SSTVD for the healthy lobes, SSD for the lobe label and vesselness measure image, and one or none of the atelectatic lobe metrics were found to have the highest accuracy. For these registrations (16 through 20), the mean (SD) of mean landmark error was 2.50 (1.16) mm, 2.80 (0.70) mm, and 2.04 (0.13) mm for no change, partial resolution, and full atelectasis resolution, respectively. The mean (SD) atelectatic lobe DSC score was 0.91 (0.08), 0.90 (0.08), and 0.89 (0.04), respectively. These results demonstrate sub‐slice thickness mean accuracy and excellent alignment of the atelectatic lobe for all degrees of atelectasis resolution. Plausibility of the resulting transformations was assessed by calculating the spatial Jacobian of the deformation vector field. Representative coronal views of the spatial Jacobian map within the lungs and atelectasis for the best registration for each patient are provided in the Data S1.
Figure 3.
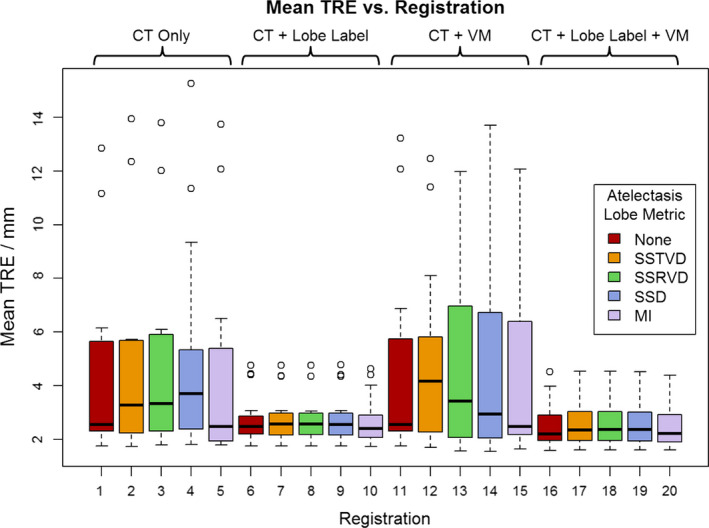
Mean target registration error (TRE) for all registrations averaged across patients. Registrations 1–5 use only the intensity of the CT image while the remaining registrations use some combination of co‐registration: lobe label images, vesselness measure (VM) images, or both. For each combination of input data, one of five similarity metrics was used for registration of the atelectatic lobes. Boxes indicate the interquartile range (IQR) and median, while whiskers note the largest and smallest values within 1.5× IQR. Circles indicate outlier points. TRE values for failed registrations are excluded. [Color figure can be viewed at wileyonlinelibrary.com]
Figure 4.
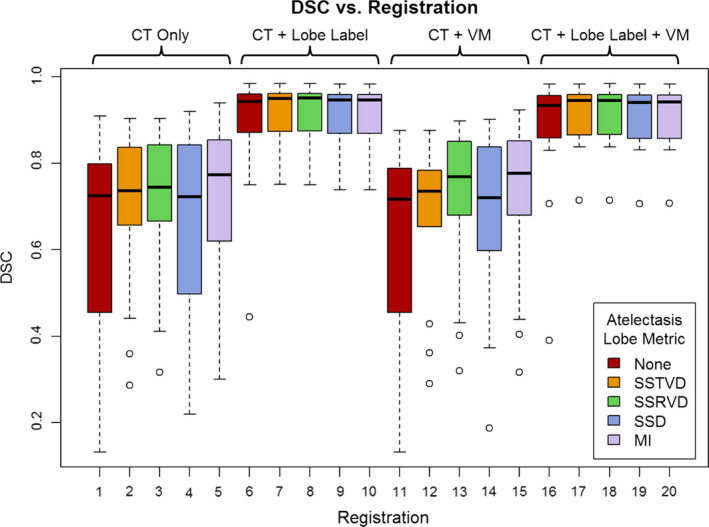
Mean Dice similarity coefficient (DSC) for all registrations averaged across patients. Registrations 1–5 use only the intensity of the CT image as input to the CALIPER algorithm, while the remaining registrations use some combination of co‐registration images: lobe label, vesselness measure (VM), or both. For each combination of input data, one of five similarity metrics was used for registration of the atelectatic lobes. Boxes indicate the interquartile range (IQR) and median, while whiskers note the largest and smallest values within 1.5× IQR. Circles indicate outlier points. DSC values for failed registrations are excluded. [Color figure can be viewed at wileyonlinelibrary.com]
3.C. Distance from pathology investigation
The effect of distance from pathology on registration error was examined. When using the full cost function (registration 16–20), mean and 90th percentile errors were found to have minimal variation with distance from tumor and atelectasis, as shown in Fig. 5. Large variability was present for landmarks within 10 mm of either the tumor or re‐expanding lung, likely due to the small number of landmarks per patient in this region. Mean error was largely unaffected by increasing distance, whereas maximum landmark error increased. Since, in Fig. 5, the landmarks are cumulative with increasing distance from pathology (ie, the ipsilateral region contains all landmarks in the ipsilateral lung, including the landmarks of the 50 mm region, etc.), the increasing max error indicates that the largest errors occurred far from atelectasis.
Figure 5.
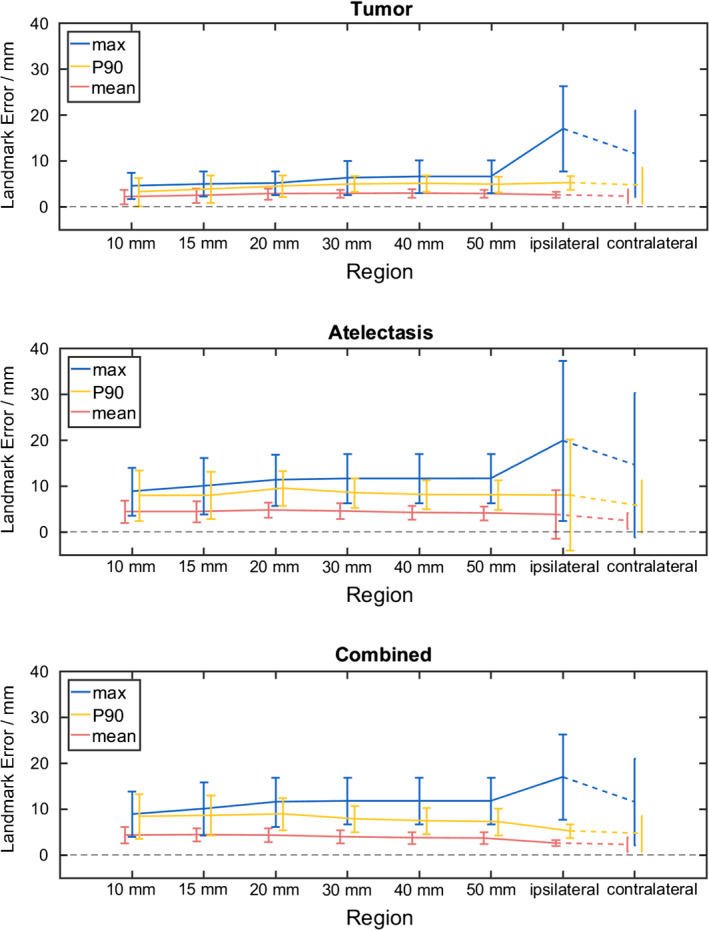
Landmark error as a function of distance from pathology. Landmark error in various regions adjacent to the pathology of interest is shown for the full CALIPER algorithm variants (registrations 16–20) across all patients. Error bars indicate ± one standard deviation. Contralateral lung landmark error is included for comparison only. Investigated pathology regions were tumor (top), atelectasis (middle), and tumor and atelectasis combined (bottom). [Color figure can be viewed at wileyonlinelibrary.com]
3.D. Utility of cost function components
Registrations involving the lobe label SSD component (six through 10 and 16 through 20) were compared against registrations without a lobe label component (one through five and 11 through 15) to determine to impact of adding the co‐registration of label images on accuracy. Mean registration error and DSC score were 4.75 mm and 0.679 without the lobe label and 2.68 mm and 0.907 with the lobe label. Statistically significant improvements in mean landmark error (P = 1.355E‐5), 90th percentile landmark error (P = 2.735E‐6), and atelectatic lobe DSC (P = 0) occurred when the lobe label component was used. The change in maximum error was not significant (P = 0.177). Adding the VM component to the cost functions of both label registrations and non‐label registrations did not result in statistical changes in accuracy metrics.
The inclusion of an intensity‐based similarity metric within the atelectatic lobe was compared to exclusion of the extra cost function component. When no metric was used, the mean landmark registration error was 2.58 mm and the mean atelectatic lobe DSC was 0.877 for the registration including all other components (registration 16). When any of the four candidate metrics were added, landmark error dropped to 2.51 mm and the DSC increased to 0.907. These differences were not significant for mean (P = 0.95), 90th percentile (P = 0.94), and max errors (P = 0.97) or for DSC (P = 0.49). Despite the inconclusive statistical tests, the slight decrease in error and increase in DSC suggested that one of the four atelectatic lobe metrics may be useful, especially since registration speed was not hindered by the additional cost component. The SSRVD metric gave similar results to SSTVD without providing any noticeable advantages; however, there may be special cases where SSRVD could be useful, such as in noisy images (eg, low dose CT) where SSTVD may be sensitive to spurious intensity variation.
The vesselness measure cost function component was not found to have a significant impact on landmark error for mean (P = 0.052), 90th percentile (P = 0.11), and max (P = 0.66) errors or for the atelectatic lobe DSC score (P = 0.14). This could be the result of the SSTVD cost accurately matching vessels within lung parenchyma already, rendering the VM cost ineffective, or gains in accuracy averaging‐out across the patients of this study. However, qualitative improvements were evident for many patients in the study cohort, as illustrated in Fig. 6. This discrepancy may be due to our choice of ground truth. Landmark error may be biased against reflection of VM improvements as landmark placement is primarily made on high‐contrast, easily identifiable vessel bifurcations, which should be more easily registered with SSTVD. The addition of the VM component to SSTVD‐based registration has been shown in other studies to improve registration accuracy particularly near the periphery of the lung, where vessels are small and of low contrast.17 although the addition of the VM component did not noticeably affect the performance of our algorithm when included, we recommend including it for these reasons.
Figure 6.
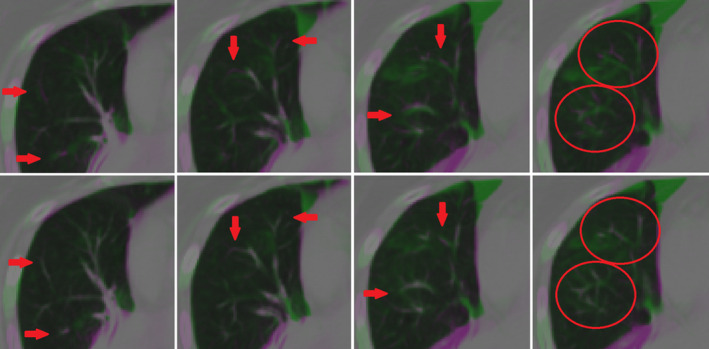
Qualitative improvements in vessel alignment. Vessel alignment appeared to improve from registration with no vesselness measure (VM) cost function component (top) to inclusion of the VM component in the cost function (bottom), despite minimal change to landmark registration error and atelectatic lobe dice similarity coefficient metrics. [Color figure can be viewed at wileyonlinelibrary.com]
4. Discussion
4.A. Comparison to literature
The optimized registrations were performed on all 18 atelectasis resolution subjects. While mean registration error of less than 2 mm was achieved only for a subset of patients, excellent improvement of alignment of the atelectatic lobes resulted from the addition of the lobe label cost function component with DSC > 0.9 in most cases. Considering that the starting DSC was, on average, 0.6, the improvements provided by the CALIPER algorithm of this work are non‐trivial.
Permutations of the CALIPER algorithm of this work were able to achieve mean landmark errors of 2.5–2.6 mm when averaged across all patients. The CALIPER variant with the highest accuracy (2.51 mm mean TRE, 5.22 mm 90th percentile TRE, 0.907 DSC) was achieved using MI similarity metric for the atelectatic lobe in combination with SSTVD for healthy lobes, and SSD for the lobe label and VM images. This degree of accuracy was comparable to registration algorithms recently reported in the literature, summarized in Table 2. In the table, studies are listed in order of increasing relevance to the registrations performed in this work. The first eight studies register inhale to exhale phases of 4DCT scans in which the only changes present are slight density and geometric differences due to respiration. The next two are longitudinal registration studies but with the caveat that high‐resolution images (slice thickness < 1.25 mm) were used for which lower registration accuracy is to be expected compared to standard resolution scans (2–3 mm slice thickness). The final three studies are most comparable to the algorithm developed here, though still none of the reported results in the literature include atelectasis resolution or other substantial large geometric changes. While multiple studies report accuracy on the order of 1 mm, several considerations must be made when comparing to the results reported in this work.
Table 2.
Summary of deformable image registration algorithm accuracy in the lung as reported in peer‐reviewed publications
| Study | Images registered | Pathology | Accuracy metric | Algorithm | Accuracy |
|---|---|---|---|---|---|
| Naini et al.31 | HR 4DCT (inhale to exhale) | Tumor | Tumor centroid | Free‐form | 0.8 mm mean |
| Brock et al.20, a | 4DCT (inhale to exhale) | Tumor | Landmark TRE | Var | 1.0–3.0 mm mean, up to 12 mm max |
| Var B‐splines | 1.6–3.0 mm mean with B‐spline algorithms | ||||
| Kanai et al.19 | 4DCT (inhale to exhale) | Some w/Tumor | Landmark TRE | B‐splines + MI | 1.44 mm mean |
| Kadoya et al.32, a | 4DCT (inhale to exhale) | Some w/Tumor | Landmark TRE | Var (Commercial) | 1.3–6.2 mm mean across all institutions |
| Raystation | 3.3 mm mean | ||||
| MIM | 3.3 mm mean | ||||
| Velocity | 5.0 mm mean | ||||
| Samavati et al.33 | 4DCT (inhale to exhale) | Some w/Tumor | Landmark TRE | Morfeus | 3.0 mm mean |
| Drop | 2.4 mm mean | ||||
| Hybrid | 1.4 mm mean, > 10 mm max | ||||
| Latifi et al.34 | 4DCT (inhale to exhale) | None | Landmark TRE | Optical Flow | 1.6 mm mean |
| Diffeo. Demons | 1.4 mm mean | ||||
| Diffeo. Morphons | 1.4 mm mean | ||||
| Du et al.23 | HR 4DCT (various phases) | Tumor | Landmark TRE | SSTVD B‐splines | 1.5 mm mean, 2.2 mm mean for > 10 mm initial error |
| Yin et al.25 | HR CT (inhale to exhale) | None | Landmark TRE | SSTVD B‐splines |
0.6 mm mean for displacements < 20 mm 1.9 mm mean for displacements > 60 mm |
| SSD B‐splines |
1.8 mm mean for displacements < 20 mm 12.5 mm mean for displacements > 60 mm |
||||
| MI B‐splines |
0.5 mm mean for displacements < 20 mm 8.1 mm mean for displacements > 60 mm |
||||
| Vlachopoulos et al.30 | Warped Longitudinal HR CT | Interstitial Lung Disease | Known DVF | Var | 2.0–2.2 mm mean |
| Nielsen et al.29 | Longitudinal HR CT | Lymphoma Outside Lung | Landmark TRE | Affine | 4.2 mm mean, 13.0 mm max |
| Demons | 1.6 mm mean, 16.0 mm max | ||||
| SSD B‐splines | 1.1 mm mean, 15 mm max | ||||
| Cazoulat et al.16 | Longitudinal 4DCT | Tumor | Landmark TRE | Rigid | 5.8 mm mean overall, 5.8 mm mean near tumor |
| Morfeus | 3.4 mm mean overall, 5.4 mm mean near tumor | ||||
| Morfeus‐VBC | 1.6 mm mean overall, 2.2 mm mean near tumor | ||||
| Cunliffe et al.26 | Longitudinal CT | Fibrosis, Some w/Tumor | Landmark TRE | Plastimatch | 2.2 mm mean overall, 4.6 mm mean near fibrosis |
| MeVis | 1.2 mm mean overall, 2.5 mm mean near fibrosis | ||||
| Stützer et al.28 | Longitudinal CT | Tumor | Landmark TRE | Custom B‐splines | 2.9 mm mean |
Multi‐institutional study; Var, various algorithms tested; TRE, target registration error; HR, high resolution (slice spacing < 1.25 mm); Diffeo., diffeomorphic.
The presence of extensive tissue changes such as atelectasis resolution, pleural effusion, and radiation‐induced damage is known to result in decreased registration accuracy or registration failure for current state‐of‐the‐art algorithms.26, 27, 28 Over 60% of the studies summarized in Table 2 report accuracy of registration between different respiratory phases of a 4DCT study. The DIRlab dataset was commonly used which consists of inhale‐to‐exhale image pairs where the tumor is sometimes not within the lung volume at all and where no atelectasis or other non‐tumor pathology is present.19 For such cases, topology is preserved and large geometric changes are absent; the registration must only account for periodic respiratory motion and slight changes in lung density, as mass is preserved as well.
The studies using longitudinal data also had important differences with the current work. Nielsen et al. used lymphoma patients for which large tumors were absent from the lung volume.29 Neither Cazoulat et al. nor Stützer et al. included patients with atelectasis, though the images used by Cazoulat did contain some tumor regression.16, 28 The difference in difficulty from intrafraction to longitudinal registration is illustrated by Stützer's finding of an increase in mean registration error of their algorithm from 1.0 mm using the DIRlab data to 2.9 mm using longitudinal data28. In addition, two of the five studies using longitudinal data, and had high‐resolution CTs available with slice thicknesses less than 1.5 mm. Four patients of this study's cohort had 2 mm slice thickness, while the remaining 14 image pairs had 3 mm slice thickness.29, 30 Finer resolution input data can easily result in lower TRE, seen in the development of the CALIPER algorithm as accuracy decreased when moving from high‐resolution computational phantoms to low‐resolution clinical data.
Large geometric changes, absent from most studies of Table 2, occurred within image pairs of this studies patient cohort. Yin et al. reported an average change in lung volume between images being registered of 3.6% (maximum of 7.2%).25 The longitudinal registration study of Vlachopoulos et al. consisted of patients with similar volume changes of 4.34 ± 2.8% on average.30 The set of six patients used in the study by Cazoulat et al. experienced a mean lung volume change of 8.0% and a mean tumor volume change of 29.5%.16 Across the 18 patients with atelectasis resolution used in the current work, mean (SD) lung volume change was 9.5% (8.8%) for all lobes combined and 10.6% (8.7%) for the healthy lobes. Tumor regression was also substantial, with a mean (SD) change in GTV volume of −39.2% (26.7%). Despite the more challenging registration problem presented by the patient cohort selected for this study, the CALIPER algorithm resulted in accuracies similar to healthy lung registrations of the studies listed in Table 2.
4.B. Limitations
The CALIPER algorithm was successful in the majority, but not for all, of the patient cases. The primary reason for failed registration was a dramatic change in atelectatic lobe shape coupled with poor initial overlap. In such cases, an insufficient number of image samples mapped between the corresponding atelectatic lobes preventing the optimizer from correctly deforming the region corresponding to the atelectasis. Only five patients experienced at least one failed registration. For Patient 5, all registrations which included an atelectatic lobe similarity metric without including the lobe label component failed, amounting to 50% of the registrations. With the exception of cases where atelectatic lobe volume change is accompanied by dramatic shape change, registration failure may be prevented by initial rigid or affine alignment of the atelectatic lobe masks rather than alignment of bony anatomy as was done for the patients of this study.
Patient 4 had all registrations run to completion, but large landmark errors were observed. The ipsilateral lung in this case was too misaligned initially for both the SSTVD metric in the healthy lobes and the intensity metrics for the atelectatic lobe. The lobe label component was not able to improve the results, despite the contralateral lung appearing well‐aligned. As such, Patient 4 was excluded from the results except for the failure rate column of Table 2. Two completed registrations of Patient 1 were also flagged as failures due to their outlying mean landmark errors in excess of 3 cm.
While residual errors for some patients and subsets of landmarks remained large, further investigation was warranted. The landmarks comprising the 10% largest errors for each registration were investigated to determine their location within the lungs. For half of the patients, over 75% of the landmarks comprising the largest errors resided in the ipsilateral lung, as expected. Eight patients experienced between 25% and 50% of the worst errors in the contralateral lung. In one patient, none of the largest errors were in the ipsilateral lung. These results indicated that the pathology was the primary contributor to registration error in only half of the patients. It was clear that registration error near pathology was larger for some patients than others, though not necessarily based on the degree of atelectasis resolution.
The registration algorithm developed in this study was designed for CT‐to‐CT image registration. As adaptive radiotherapy techniques continue to evolve, online plan adaptation based on daily cone beam CT (CBCT) images is becoming more pertinent. Some work has already been done on CBCT‐to‐CT image registration for ART in proton therapy.35 Future work should involve extending the CALIPER algorithm to CBCT image registration.
5. Conclusion
A deformable image registration algorithm was developed to address the outstanding problem of longitudinal registration in the presence of large geometric changes and non‐correspondences of intensity. The algorithm was tested on a set of 18 locally advanced NSCLC patients presenting with atelectasis at the start of treatment and experiencing varying degrees of pathology resolution throughout the course of treatment. The algorithm included a lobe label cost function component for robustness to large geometric changes. A mass‐preserving similarity metric was applied to the healthy parenchyma where tissue correspondences were not in question. Several similarity metrics were applied to the atelectatic lobe to enhance the likelihood of reasonable deformation, with mutual information providing the highest accuracy. Vessel‐enhanced images were co‐registered for fine improvements of the vasculature alignment. Accuracy comparable to registration results reported in the literature was obtained when using all components of the algorithm simultaneously.
While datasets exhibiting atelectasis resolution are often excluded from registration algorithm accuracy assessments, these challenging cases were the focus of this work. Patients experiencing large geometric changes during the course of radiotherapy are prime candidates for adaptive radiotherapy to account for the tissue changes, but accurate deformable image registration is a prerequisite. The registration algorithm described in this work may offer such patients the option of adaptive radiotherapy and enhanced precision of treatment.
Conflicts of interest
Elisabeth Weiss receives research support from Varian Medical Systems and the National Institutes of Health and receives royalties from UpToDate. Gary Christensen maintains research grants from National Institutes of Health and has received a gift from Roger Koch to support research. Geoffrey Hugo receives research support from Philips Healthcare and the National Institutes of Health and has a licensing agreement with Varian Medical Systems.
Supporting information
Data S1. CALIPER registration results per patient.
Acknowledgments
The authors would like to thank Leonid B. Reshko for his contribution with clinical data generation, Kunlin Cao for lending preprocessing code, and Matthew J. Riblett and Yue Pan for their assistance and feedback during the course of this work. This work was supported by a research grant from the National Cancer Institute of the National Institutes of Health under award number R01CA166119.
References
- 1. Siker ML, Tomé WA, Mehta MP. Tumor volume changes on serial imaging with megavoltage CT for non–small‐cell lung cancer during intensity‐modulated radiotherapy: how reliable, consistent, and meaningful is the effect? Int J Radiat Oncol. 2006;66:135–141. [DOI] [PubMed] [Google Scholar]
- 2. Jenkins P, Milliner R, Latimer P. Shrinkage of locally advanced non‐small‐cell lung cancers in response to induction chemotherapy: implications for radiotherapy treatment planning. Int J Radiat Oncol. 2007;69:993–1000. [DOI] [PubMed] [Google Scholar]
- 3. Sonke J‐J, Belderbos J. Adaptive radiotherapy for lung cancer. Semin Radiat Oncol. 2010;20:94–106. [DOI] [PubMed] [Google Scholar]
- 4. Woodford C, Yartsev S, Dar AR, Bauman G, Van Dyk J. Adaptive radiotherapy planning on decreasing gross tumor volumes as seen on megavoltage computed tomography images. Int J Radiat Oncol. 2007;69:1316–1322. [DOI] [PubMed] [Google Scholar]
- 5. Fox J, Ford E, Redmond K, Zhou J, Wong J, Song DY. Quantification of tumor volume changes during radiotherapy for non‐small‐cell lung cancer. Int J Radiat Oncol. 2009;74:341–348. [DOI] [PubMed] [Google Scholar]
- 6. Tennyson N, Weiss E, Sleeman W, Rosu M, Jan N, Hugo GD. Effect of variations in atelectasis on tumor displacement during radiation therapy for locally advanced lung cancer. Adv Radiat Oncol. 2017;2:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho BCJ, Bezjak A, Dawson LA. Image guidance in non‐small cell lung cancer. Semin Radiat Oncol. 2010;20:164–170. [DOI] [PubMed] [Google Scholar]
- 8. Lim G, Bezjak A, Higgins J, et al. Tumor regression and positional changes in non‐small cell lung cancer during radical radiotherapy. J Thorac Oncol. 2011;6:531–536. [DOI] [PubMed] [Google Scholar]
- 9. Jaffray DA, Lindsay PE, Brock KK, Deasy JO, Tomé WA. Accurate accumulation of dose for improved understanding of radiation effects in normal tissue. Int J Radiat Oncol. 2010;76:S135–S139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Møller DS, Khalil AA, Knap MM, Hoffmann L. Adaptive radiotherapy of lung cancer patients with pleural effusion or atelectasis. Radiother Oncol. 2014;110:517–522. [DOI] [PubMed] [Google Scholar]
- 11. Brink C, Bernchou U, Bertelsen A, Hansen O, Schytte T, Bentzen SM. Locoregional control of non‐small cell lung cancer in relation to automated early assessment of tumor regression on cone beam computed tomography. Int J Radiat Oncol. 2014;89:916–923. [DOI] [PubMed] [Google Scholar]
- 12. Guy CL, Weiss E, Jan N, Reshko LB, Christensen GE, Hugo GD. Effect of atelectasis changes on tissue mass and dose during lung radiotherapy: effect of atelectasis changes during radiotherapy. Med Phys. 2016;43:6109–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guckenberger M, Baier K, Richter A, Wilbert J, Flentje M. Evolution of surface‐based deformable image registration for adaptive radiotherapy of non‐small cell lung cancer (NSCLC). Radiat Oncol. 2009;4:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guckenberger M, Richter A, Wilbert J, Flentje M, Partridge M. Adaptive radiotherapy for locally advanced non–small‐cell lung cancer does not underdose the microscopic disease and has the potential to increase tumor control. Int J Radiat Oncol. 2011;81:e275–e282. [DOI] [PubMed] [Google Scholar]
- 15. Guckenberger M, Wilbert J, Richter A, Baier K, Flentje M. Potential of adaptive radiotherapy to escalate the radiation dose in combined radiochemotherapy for locally advanced non‐small cell lung cancer. Int J Radiat Oncol. 2011;79:901–908. [DOI] [PubMed] [Google Scholar]
- 16. Cazoulat G, Owen D, Matuszak MM, Balter JM, Brock KK. Biomechanical deformable image registration of longitudinal lung CT images using vessel information. Phys Med Biol. 2016;61:4826–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cao K, Christensen GE, Ding K, et al. Tracking regional tissue volume and function change in lung using image registration. Int J Biomed Imaging. 2012;2012:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jerman T, Pernuš F, Likar B, Špiclin Ž. Beyond Frangi: an improved multiscale vesselness filter. In edited by S. Ourselin and M.A. Styner (2015), p. 94132A.
- 19. Kanai T, Kadoya N, Ito K, et al. Evaluation of accuracy of B‐spline transformation‐based deformable image registration with different parameter settings for thoracic images. J Radiat Res (Tokyo). 2014;55:1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brock KK. Results of a multi‐institution deformable registration accuracy study (MIDRAS). Int J Radiat Oncol. 2010;76:583–596. [DOI] [PubMed] [Google Scholar]
- 21. Murphy K, van Ginneken B, Klein S, et al. Semi‐automatic construction of reference standards for evaluation of image registration. Med Image Anal. 2011;15:71–84. [DOI] [PubMed] [Google Scholar]
- 22. Klein S, Staring M. elastix the manual, 2015.
- 23. Du K, Bayouth JE, Cao K, Christensen GE, Ding K, Reinhardt JM. Reproducibility of registration‐based measures of lung tissue expansion. Med Phys. 2012;39:1595–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gorbunova V, Sporring J, Lo P, et al. Mass preserving image registration for lung CT. Med Image Anal. 2012;16:786–795. [DOI] [PubMed] [Google Scholar]
- 25. Yin Y, Hoffman EA, Lin CL. Mass preserving nonrigid registration of CT lung images using cubic B‐spline: mass preserving nonrigid registration of CT lung images. Med Phys. 2009;36:4213–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cunliffe AR, Contee C, Armato SG, et al. Effect of deformable registration on the dose calculated in radiation therapy planning CT scans of lung cancer patients a): effect of deformable registration on calculated radiation dose. Med Phys. 2014;42:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Szeto YZ, Witte MG, van Kranen SR, Sonke J‐J, Belderbos J, van Herk M. Effects of anatomical changes on pencil beam scanning proton plans in locally advanced NSCLC patients. Radiother Oncol. 2016;120:286–292. [DOI] [PubMed] [Google Scholar]
- 28. Stützer K, Haase R, Lohaus F, et al. Evaluation of a deformable registration algorithm for subsequent lung computed tomography imaging during radiochemotherapy. Med Phys. 2016;43:5028–5039. [DOI] [PubMed] [Google Scholar]
- 29. Nielsen MS, Østergaard LR, Carl J A new method to validate thoracic CT‐CT deformable image registration using auto‐segmented 3D anatomical landmarks. Acta Oncol. 2015;54:1515–1520. [DOI] [PubMed] [Google Scholar]
- 30. Vlachopoulos G, Korfiatis P, Skiadopoulos S, Kazantzi A. Selecting registration schemes in case of interstitial lung disease follow‐up in CT. Med Phys. 2015;42:4511–4525. [DOI] [PubMed] [Google Scholar]
- 31. Sadeghi Naini A, Pierce G, Lee T‐Y, Patel RV, Samani A. CT image construction of a totally deflated lung using deformable model extrapolation: deflated lung CT image construction using deformable model extrapolation. Med Phys. 2011;38:872–883. [DOI] [PubMed] [Google Scholar]
- 32. Kadoya N, Nakajima Y, Saito M, et al. Multi‐institutional validation study of commercially available deformable image registration software for thoracic images. Int J Radiat Oncol. 2016;96:422–431. [DOI] [PubMed] [Google Scholar]
- 33. Samavati N, Velec M, Brock K. A hybrid biomechanical intensity based deformable image registration of lung 4DCT. Phys Med Biol. 2015;60:3359–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Latifi K, Zhang G, Stawicki M, Elmpt W, Dekker A, Forster K. Validation of three deformable image registration algorithms for the thorax. J Appl Clin Med Phys. 2013;14:19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Veiga C, Janssens G, Teng C‐L, et al. First clinical investigation of cone beam computed tomography and deformable registration for adaptive proton therapy for lung cancer. Int J Radiat Oncol. 2016;95:549–559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. CALIPER registration results per patient.


