Abstract
RNA binding proteins play essential roles during development and aging, and are also involved in disease pathomechanisms. RNA sequencing and omics analyses have provided a window into systems level alterations in neurological disease, and have identified RNA processing defects among notable disease mechanisms. This review focuses on two seemingly distinct neurological disorders, the RNA binding proteins they are linked to, and their newly discovered functional relationship. When deficient, Fragile X Mental Retardation Protein (FMRP) causes developmental deficits and autistic behaviors while TAR-DNA Binding Protein (TDP-43) dysregulation causes age dependent neuronal degeneration. Recent findings that FMRP and TDP-43 associate in ribonuclear protein particles and share mRNA targets in neurons highlight the critical importance of translation regulation in synaptic plasticity and provide new perspectives on neuronal vulnerability during lifespan.
Keywords: RNA binding proteins, FMRP, TDP-43, mRNA targets, Fragile X syndrome, amyotrophic lateral sclerosis, frontotemporal dementia
1. Introduction
A recent census identified 1,542 RNA binding proteins (RBPs) that can associate with coding, non-coding or micro RNAs in the human genome (Gerstberger et al., 2014b). Of these, about a tenth (~150 RBPs) are listed in the Online Mendelian Inheritance in Man (OMIM) database as linked to neurological and neuromuscular disorders (Gerstberger et al., 2014b). Although the life of an mRNA from transcription to protein synthesis involves multiple processing steps, splicing, transport, localization and translation are among the most affected aspects of RNA metabolism in disease (reviewed in Conlon and Manley, 2017; Gerstberger et al., 2014a).
In recent years, an avalanche of molecular and phenotypic studies showed that FMRP and TDP-43, two RNA binding proteins with seemingly different roles in the nervous system work together closely and regulate common mRNA targets (Coyne et al., 2014; Coyne et al., 2015; Coyne et al., 2017a; Fallini et al., 2012; Majumder et al., 2016; Wang et al., 2008; Yu et al., 2012). Given their involvement in diseases affecting synaptic function during early development (FMRP, Fragile X Mental Retardation Protein) versus aging neurons (TDP-43, TAR-DNA Binding Protein), these intriguing findings provide an opportunity to uncover common and unique aspects of RNA based mechanisms during lifespan. Here we will briefly review the individual roles of FMRP and TDP-43 in neurons, in the context of the clinical phenotypes linked to dysfunction of these RBPs, and highlight their association, overlapping mRNA targets and contribution to different diseases.
2. FMRP and Fragile X syndrome (FXS)
2.1. FXS – Genetic mutations and clinical presentation
Fragile X Syndrome (FXS; OMIM 300624) is a single gene disorder that results from the loss of the fragile X mental retardation gene (FMR1) expression and is characterized by intellectual and other developmental disabilities including cardiac abnormalities, seizures, attention deficit hyperactivity disorder (ADHD), aggression, language, and sleep disturbances (Hagerman et al., 2017; Santoro et al., 2012). Interestingly, approximately 30–54% of FXS patients meet the diagnostic criteria for autism (Clifford et al., 2007; Hall et al., 2008; Kaufmann et al., 2017). FXS is predominantly caused by expansion of CGG repeats located within the 5’UTR. While the average CGG repeat ranges between 22–24 in the general population, expansions >200 repeats cause methylation accompanied by transcriptional silencing of the FMR1 locus and FXS (Nelson et al., 2013). Premutation alleles of FMR1 containing >50 but <200 CGG repeats have also been identified and linked to fragile X premature ovarian insufficiency (FXPOI; OMIM 300624) in females, in their 20’s, and fragile X tremor ataxia syndrome (FXTAS; OMIM 300623) in males, after age 50 (Hagerman and Hagerman, 2004; Hagerman and Hagerman, 2016; Nelson et al., 2013). Being an X-linked disorder, FXS affects 1 in 4,000 males and 1 in 8,000 females; additionally, the genetic anticipation observed in some of affected families can be explained by the dynamics of CGG microsatellite length, and the relationship between repeat number and phenotype (Santoro et al., 2012). These phenotypic relationships have been described in multiple models including fruit flies, zebrafish and mice, further substantiating the complexities of FXS and related disorders, and providing candidate therapeutic leads (Borrie et al., 2017; Davis and Broadie, 2017; Hagerman et al., 2017; Santoro et al., 2012; Tropepe and Sive, 2003; Wu et al., 2017; Zarnescu et al., 2005).
2.2. The FMR1 gene encodes an RNA-binding protein, FMRP, an established translational regulator
FMRP is an RBP with known roles in RNA localization and translation (Brown et al., 1998; Santoro et al., 2012). Structurally, FMRP harbors three types of RNA binding motifs (a Tudor motif, two KH domains and an RGG box) that mediate interactions with mRNA targets (Bardoni et al., 2001; Siomi et al., 1993).
FMRP has been shown to be both a repressor and activator of translation, and it can function as a translation inhibitor, both at the level of initiation (Napoli et al., 2008) and elongation (Darnell et al., 2011). FMRP associates with 4% of brain mRNAs (Sung et al., 2000). Systematic evolution of ligands by exponential enrichment (SELEX) experiments show that FMRP can bind to transcripts directly, via G quadruplex forming sequences or “kissing complex” (Darnell et al., 2001; Darnell et al., 2005). Regulation of translation initiation by FMRP involves interactions with the cap-binding translation factor eIF4E via CYFIP, a 4E-BP (Schenck et al., 2001; Schenck et al., 2003). In addition, FMRP has been shown to bind ribosomes directly, which causes translation inhibition by preventing binding of translation factors (Chen et al., 2014).
FMRP’s role in mRNA localization and translation is particularly important at post-synaptic sites, as informed by mechanistic studies in animal models as well as dendritic morphology abnormalities described in FXS brains (Nimchinsky et al., 2001). Notable synaptic mRNA targets include mGluR5, GABAB receptors and catalytic subunit of PI3K (reviewed recently in Richter et al., 2015). Recently, work in animal models uncovered a role for FMRP in translation of presynaptic sites and homeostasis (reviewed in Turrigiano, 2008). The challenge remains to understand which mRNA targets, in which cells and at what time during development they are responsible for the observed, pleiotropic FXS phenotypes.
3. TDP-43 in amyotrophic lateral sclerosis (ALS)/fronto-temporal dementia (FTD)
3.1. ALS/ FTD – a spectrum disorder
Frontotemporal dementia (FTD; OMIM: 600274) and amyotrophic lateral sclerosis (ALS; OMIM: 612069) are degenerative syndromes characterized by a certain degree of pathologic and genetic overlap (Hardy and Rogaeva, 2014; Ling et al., 2013). FTD patients are affected by progressive neuronal degeneration and loss in the frontal and temporal cortices, while ALS patients exhibit loss of muscle mass due to the progressive degeneration of the cerebrospinal tracks in both sides of the spinal cord (Hardy and Rogaeva, 2014). In the second half of the 20th century, clinicians started to report cases of ALS patients that present FTD-like symptoms, such as a decline in cognitive function, language impairment and progressive aphasia (Hudson, 1981). Moreover, some FTD patients were found to share ALS phenotypes including a progressive weakness and wasting of muscle tissue. These clinical observations suggest a pathologic overlap between the two neurodegenerative disorders, resulting from the genetic and molecular intersection of common mutation and pathways (Achi and Rudnicki, 2012; Ferrari et al., 2011). Substantiating these observations is the discovery of common mutations in ALS and FTD patients, involved in a wide spectrum of molecular pathways that underlie the pathological phenotypes (Ling et al., 2013; Zago et al., 2011). Notable genes commonly mutated in both ALS and FTD patients include chromosome 9 open reading frame 72 (c9orf72), ubiquilin 2 (UBQLN2), valosin-containing protein (VCP), coiled-coil-helix-coiled-coil-helix domain containing 10 (CHCHD10) and sequestosome 1 (SQSTM1) (Guerreiro et al., 2015). Additionally, TAR-DNA binding protein (TDP-43) and fused in sarcoma (FUS) are ALS and FTD associated RNA/DNA binding proteins with overlapping interactomes that co-localize into cytosolic aggregates and contribute to motor neuron degeneration (Blokhuis et al., 2016; Guerreiro et al., 2015; Nolan et al., 2016). Interestingly, mutations in tardp and fus, two genes encoding RNA binding proteins highlight alterations in RNA metabolism as a key cellular process involved in disease (Ferrari et al., 2011; Ling et al., 2013).
3.2. TDP-43 in ALS/FTD – marker of pathology and mutations
In 2006, TDP-43 has been identified as the major component of the ubiquitin-positive/tau-negative inclusions of frontotemporal lobe and motor neurons (Neumann et al., 2006), and since this discovery, TDP-43 pathology has been identified in 97% of ALS and 45% of FTD cases (Ling et al., 2013). The role of cytoplasmic, insoluble TDP-43 accumulation in disease was demonstrated in vivo in a wide range of models including worms, flies, zebrafish, mice and in vitro in human cell lines (Gendron et al., 2013; Kabashi et al., 2009; Liachko et al., 2010; Wegorzewska et al., 2009; Zhan et al., 2013). With the discovery of causative mutations in its C-terminus domain, and its high association with pathology, TDP-43 has emerged as a common denominator in ALS/FTD, linking TDP-43 to the majority of ALS cases known to date.
In patients, there have been ~ 55 different mutations identified until recently in sporadic (3%) and familial (1.5%) ALS, most of which occur in the C-terminus domain of TDP-43 (Buratti, 2015; Conlon and Manley, 2017; Lattante et al., 2013). Although the mechanism of individual mutations is not known, their presence within the low complexity C terminus domain has been shown to increase the propensity of TDP-43 to aggregate (Johnson et al., 2009). Recently, mutant TDP-43 species derived from ALS tissues have been shown to propagate in a prion-like manner in cultured cells (Nonaka et al., 2013; Smethurst et al., 2016), providing support to the pathological spread theory of disease. Collectively, evidence exists that disease causing mutations alter the cellular localization of TDP-43, and affect protein-protein and protein-RNA interactions in a multitude of ALS/FTD models, under a wide range of experimental conditions (for review see Buratti, 2015).
3.3. TDP-43 structure and function; RNA targets, RBP partners
TDP-43 protein contains two RNA recognition motifs (RRM1 and 2); RNA binding is required for toxicity (Ihara et al., 2013; Voigt et al., 2010). Although the association between TDP-43 and RNA processing defects is widely accepted, the mechanisms remain poorly understood. Global proteomic analyses highlight the association of TDP-43 with splicing and translation factors (Freibaum et al., 2010; Sephton et al., 2011). Immunoprecipitation experiments followed by deep-sequencing show that pathological accumulation of insoluble TDP-43 results in abnormal sequestration of transcripts, which often contain UG repeats, and are involved in synaptic function, RNA metabolism and neuronal development (Polymenidou et al., 2011; Sephton et al., 2011; Tollervey et al., 2011). Moreover, recent studies found evidence that TDP-43 maintains intron integrity by repressing nonconserved cryptic exons, a process that was found to be impaired in patient tissues (Jeong et al., 2017; Ling et al., 2015; Tan et al., 2016). The RNA binding feature of TDP-43 via RRM1 and RRM2 plays a dual role: it is required for TDP-43 toxicity but it also plays a physiological role in regulating RNA processing. Indeed, under normal conditions, TDP-43 is required for preventing the downregulation of the microtubule stabilizing protein Futsch and maintain synaptic integrity (Godena et al., 2011; Romano et al., 2016). In disease, TDP-43 appears to cause toxicity by sequestering mRNAs and reducing their availability to the translation machinery as shown for futsch and hsc70-4 mRNAs (Coyne et al., 2014; Coyne et al., 2017a). Hsc70-4 is a chaperone required for synaptic vesicle cycling and its downregulation in TDP-43 proteinopathy provides a mechanistic explanation for synaptic dysfunction in ALS (Coyne et al., 2017a). In neuroblastoma cells, TDP-43 interacts directly with ribosomes via RACK1, which impacts translation globally and may have implications in disease (Russo et al., 2017). In summary, evidence is increasingly supporting the notion that TDP-43 proteinopathy results from perturbations in RBP partners interactions (Blokhuis et al., 2016) and RNA processing at multiple steps including RNA splicing, non-coding RNA metabolism, miRNA biogenesis, stress granule formation and translation (recently reviewed in Coyne et al., 2017b). More structural studies are needed to understand how the different domains of TDP-43 may interact with each other and whether the low complexity C terminus domain where the majority of disease associated mutations reside impacts RNA binding and/or interactions with partner proteins.
4. FMRP - TDP-43 form a ribonucleoprotein complex and share mRNA targets
4.1. Protein-protein interactions
Despite being linked to two seemingly distinct neurological disorders, at least in regards to age of onset and clinical presentation, FMRP and TDP-43 have been recently found to physically associate in a complex and share mRNA targets in neurons (Coyne et al., 2015; Majumder et al., 2016). The first evidence that these two RBPs may interact comes from studies of RNA granule transport in cultured mouse hippocampal neurons. Following KCl mediated depolarization, TDP-43 containing RNA granules show increased colocalization with FMRP and Staufen (Wang et al., 2008). These findings support the notion that similar to FMRP (Antar et al., 2004), TDP-43 may adjust its localization and function in response to synaptic activity. Subsequent studies provided further evidence that TDP-43, when overexpressed, colocalizes with FMRP in transport RNA granules in cultured motor neurons derived from mice or flies (Coyne et al., 2015; Fallini et al., 2012).
As suggested by colocalization studies, human FMRP and TDP-43 were found to co-immunoprecipitate from SH-SY5Y or HEK cells (Coyne et al., 2015; Majumder et al., 2016; Yu et al., 2012). In keeping with these observations, overexpressed human TDP-43 was found to assemble in a complex with endogenous FMRP in fly motor neurons (Coyne et al., 2015). Furthermore, full length, purified FMRP and TDP-43 bind in vitro (Coyne et al., 2015). Although additional work is needed to understand the mechanisms and dynamics of binding, co-immunoprecipitation experiments show that the low complexity, Glycine rich, C terminus domain of TDP-43 is required for the interaction with FMRP (Majumder et al., 2016). The C terminus domain of TDP-43 also supports protein interactions with other hnRNPs (Romano et al., 2014) suggesting that it may act as a scaffold for multiple RBP interactions. This in turn provides opportunities for enhancing the repertoire of RNAs that associate with TDP-43 via diverse RBP partners. Other notable TDP-43 partners include PABP and Ataxin 2 that, when knocked-down mitigate TDP-43 proteinopathy (Becker et al., 2017; Elden et al., 2010). These protein-protein interactions, together with findings that FMRP and TDP-43 co-fractionate in polysomes with known translation factors (e.g., CYFIP, Majumder et al., 2016) suggest that just like FMRP, TDP-43 may regulate translation, most likely at the level of initiation.
4.2. Protein-RNA interactions
These reports of physical association suggest the possibility that FMRP and TDP-43 may share mRNA targets. Indeed, systematic evolution of ligands by exponential enrichment (SELEX) identified G-quadruplexes as ligands of both FMRP (Darnell et al., 2001) and TDP-43 (Ishiguro et al., 2016). Furthermore, RNA immunoprecipitations show that just like FMRP, TDP-43 associates with several mRNAs and regulates their localization and translation in neurons. Among these common targets are Rac1 and futsch/MAP1B mRNAs, which regulate spinogenesis and the microtubule cytoskeleton, respectively (Coyne et al., 2014; Godena et al., 2011; Majumder et al., 2012; Majumder et al., 2016; Romano et al., 2016).
While regulating the same mRNA targets seems plausible for two proteins that bind similar structural motifs (e.g., G-quadruplexes) the question remains whether these protein-RNA interactions occur separately or together, and whether this three way association between FMRP, TDP-43 and mRNA bears physiological significance. To address the interdependence between these factors, Majumder et al used a series of elegant knock-down, structure function and in vitro translation studies in cultured cells (Majumder et al., 2016). These experiments showed that the binding of FMRP to Rac1 mRNA was significantly decreased upon TDP-43 depletion, whereas only marginal or no decrease in the binding of TDP-43 was detected upon FMRP depletion in cultured hippocampal neurons. Together with mutagenesis studies followed by RNA immunoprecipitations these results suggest a scenario whereby TDP-43 binds specific UG repeat sequences followed by recruitment of FMRP to the 3’UTR of Rac1 mRNA. Although these findings show that FMRP regulation of Rac1 mRNA translation is dependent on TDP-43, a more robust inhibition is observed when both TDP-43 and FMRP are overexpressed, consistent with a cooperative repression mechanism (Majumder et al., 2016). Pharmacological inhibition studies suggest that the co-repression of Rac1 mRNA by FMRP and TDP-43 occurs at the level of translation initiation but not elongation. Furthermore, FMRP and TDP-43 knock-down experiments in conjunction with immunoprecipitation of CYFIP, a 4E-BP partner of FMRP suggest a model in which, first, TDP-43 binds mRNA via UG sequences, followed by FMRP-CYFIP recruitment and eIF4E binding that collectively inhibits Rac1 mRNA translation at the initiation step (Majumder et al., 2016).
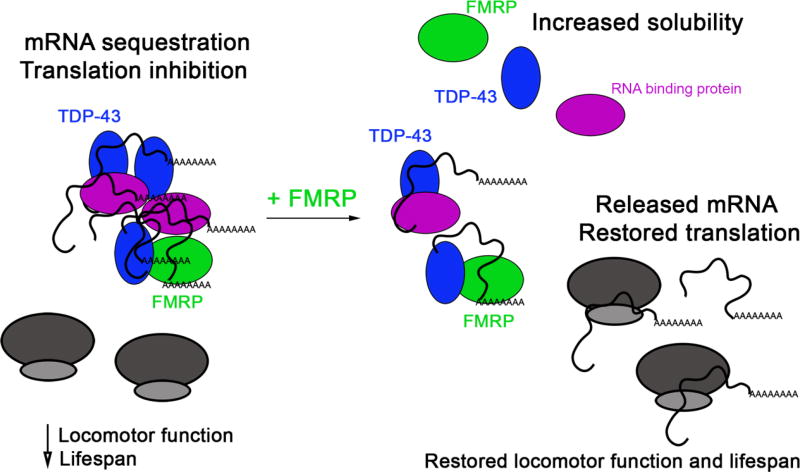
Insights into what these complex protein-RNA interactions may mean for the pathomechanism of disease come from a fly model of ALS based on TDP-43 overexpression (Coyne et al., 2014; Coyne et al., 2015; Estes et al., 2011). RNA immunoprecipitations showed that futsch mRNA, the Drosophila homolog of MAP1B, associates with wild-type and mutant TDP-43 in motor neurons (Coyne et al., 2014). Expression studies and polysome fractionations showed that Futsch protein is upregulated in motor neuron cell bodies as the combined result of decreased futsch mRNA localization at the neuromuscular junction and translation deficits, and this causes a reduction in synaptic microtubule stability. Remarkably, MAP1B, the human homolog of Futsch is also upregulated in motor neuron cell bodies from ALS spinal cords (Coyne et al., 2014). Other studies confirmed the ability of fly and human TDP-43 to downtranslate futsch/MAP1B and further demonstrated the requirement of UG sequences within the 5’UTR of futsch/MAP1B for this regulation (Romano et al., 2016). Since futsch is also a translation target of FMRP, which binds TDP-43, the fly provided an opportunity to study these relationships in vivo. A combination of molecular studies, cellular and polysome fractionations, and genetic interaction experiments suggest a model whereby TDP-43 aggregation and mRNA sequestration in disease are mitigated by FMRP overexpression, via remodeling of RNA granules and release of mRNAs whose translation is restored (Coyne et al., 2015) (see Figure 1). Notably, these molecular changes are accompanied by a rescue of TDP-43 dependent locomotor deficits and an increase in lifespan (Coyne et al., 2015). These findings suggest that remodeling RNA granules, release of sequestered mRNAs and restoring their translation may provide strategies for restoring protein synthesis deficits and mitigating functional phenotypes in ALS/FTD.
Figure 1. FMRP interacts with TDP-43 and remodels TDP-43/mRNA complexes.
Schematic representation the molecular interactions between TAR binding protein (TDP-43; blue) and fragile X mental retardation protein (FMRP; green) with their effects on mRNA sequestration and the resulting phenotypes. TDP43 and FMRP co-localize in cytosolic complexes in normal and pathological conditions. In patients or animal models of amyotrophic lateral sclerosis (ALS) or/and frontotemporal dementia (FTD), cytoplasmic TDP-43 accumulation sequesters mRNAs, which in turn causes decrease of locomotor function and lifespan. Overexpression of FMRP rescues ALS/FTD phenotypes by remodeling protein-RNA complexes (excess FMRP solubilizes TDP-43) and ultimately releasing mRNA from aggregates. Proteostasis can be restored by translation of the released mRNA.
5. Concluding remarks
In conclusion, similar alterations in RNA processing, specifically at the level of mRNA localization and translation can cause devastating consequences for the nervous system at different times during lifespan (Figure 2). On one hand, FMRP deficiency causes cognitive and behavioral phenotypes in early development while TDP-43 exerts toxicity in middle age or later in life. Yet these two RBPs assemble in the same ribonucleoprotein complex, have the ability to bind each other and even regulate the same mRNA targets, through similar mechanisms, and affect identical cellular processes. How might their effects manifest at strikingly different stages of development? A possible explanation is that FMRP is expressed at higher level than TDP-43 in early development. Although a direct comparison is yet to be done between the two RBPs discussed here, FMRP expression correlates with brain regions affected in disease (Till, 2010; Zangenehpour et al., 2009). At the same time, loss of TDP-43 causes embryonic lethality (Sephton et al., 2010; Wu et al., 2010) therefore expression levels alone cannot provide an explanation for how might TDP-43 dysfunction cause age dependent neurodegeneration. Of note is the fact that although loss of TDP-43 and overexpression models exhibit similar phenotypes (Estes et al., 2011; Feiguin et al., 2009; Kabashi et al., 2009), their mechanisms are likely different (Hazelett et al., 2012). For example, although loss of endogenous TDP-43 and overexpression of human TDP-43 in flies causes comparable phenotypes (Coyne et al., 2015; Estes et al., 2011), FMRP overexpression only mitigates the ALS-like overexpression condition and not the loss of function (Coyne et al., 2015). These findings are consistent with multiple reports that cytoplasmic TDP-43 (whether wild-type, as in sporadic ALS, or disease causing mutant TDP-43) is toxic to motor neurons (Barmada et al., 2010). A plausible explanation is that the accumulation of TDP-43 in the cytoplasm and depletion from its normal location in the nucleus simply takes time. It remains to be determined whether this is caused by age dependent leaky nuclear pores or other cellular stresses, less robust disassembly of cytoplasmic RNA stress granules or accumulation of free radicals. Recently, it has been shown that nuclear pore components associate with RNA stress granules during cellular stress, bolstering the role of cytoplasmic granules in ALS/FTD (Zhang et al., 2018). Thus it would appear that the presence and persistence of TDP-43 containing granules in the cytoplasm provides opportunities for failure of ribostasis through RNA sequestration and translation inhibition, and proteostasis through aggregate formation, both of which correlate with degeneration and death. Although protein aggregates are a hallmark of pathology, their contribution to neuronal death remains controversial (reviewed in Baloh, 2011; Tsao et al., 2012) therefore more work is needed to understand the different types of aggregates at a structural level. Consistent with the presence of “protective” versus “toxic” aggregates, it was recently shown that SOD1 trimers are toxic while fibrils confer protection (Zhu et al., 2018). Additional work on RBP-RNA complexes is needed to better understand lifespan dependent, neuronal and synaptic vulnerability in the nervous system, that ultimately causes different neurological disorders.
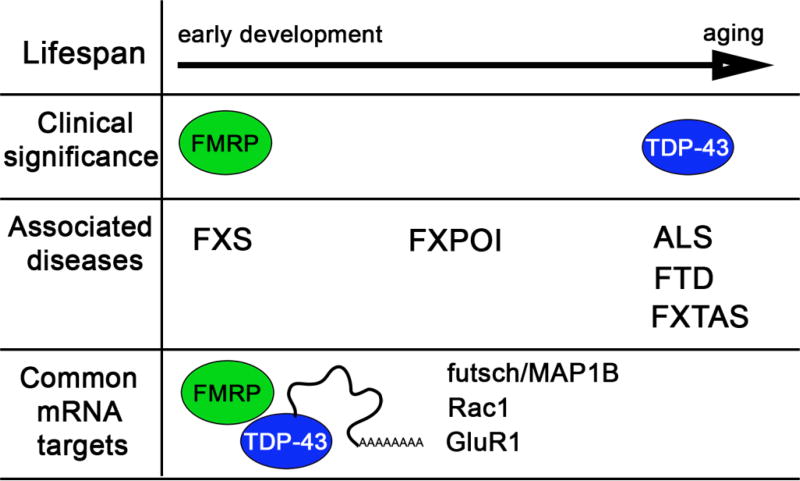
Figure 2. FMRP and TDP-43 cause different clinical syndromes during lifespan.
Diseases associated with FMRP: fragile X syndrome (FXS), Fragile X-associated primary ovarian insufficiency (FXPOI), Fragile X-associated tremor/ataxia syndrome (FXTAS). Diseases associated with TDP-43 proteinopathy: amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Known mRNA targets known to date, as shown.
Highlights.
FMRP and TDP-43 assemble in ribonuclear protein particles
FMRP and TDP-43 share mRNA targets
FMRP and TDP-43 dysfunction cause seemingly different neurological disorders
Acknowledgments
SY is supported by a scholarship from the Beckmann Foundation and DCZ is supported by NIH NS091299 and MDA 418515.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achi EY, Rudnicki SA. ALS and Frontotemporal Dysfunction: A Review. Neurol Res Int. 2012;2012:806306. doi: 10.1155/2012/806306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24:2648–55. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloh RH. TDP-43: the relationship between protein aggregation and neurodegeneration in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. The FEBS journal. 2011;278:3539–49. doi: 10.1111/j.1742-4658.2011.08256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardoni B, Schenck A, Mandel JL. The Fragile X mental retardation protein. Brain Res Bull. 2001;56:375–82. doi: 10.1016/s0361-9230(01)00647-5. [DOI] [PubMed] [Google Scholar]
- Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–49. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, Messing J, Kim HJ, Soriano A, Auburger G, Pulst SM, Taylor JP, Rigo F, Gitler AD. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhuis AM, Koppers M, Groen EJ, van den Heuvel DM, Dini Modigliani S, Anink JJ, Fumoto K, van Diggelen F, Snelting A, Sodaar P, Verheijen BM, Demmers JA, Veldink JH, Aronica E, Bozzoni I, den Hertog J, van den Berg LH, Pasterkamp RJ. Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways. Acta Neuropathol. 2016;132:175–96. doi: 10.1007/s00401-016-1575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrie SC, Brems H, Legius E, Bagni C. Cognitive Dysfunctions in Intellectual Disabilities: The Contributions of the Ras-MAPK and PI3K-AKT-mTOR Pathways. Annu Rev Genomics Hum Genet. 2017;18:115–142. doi: 10.1146/annurev-genom-091416-035332. [DOI] [PubMed] [Google Scholar]
- Brown V, Small K, Lakkis L, Feng Y, Gunter C, Wilkinson KD, Warren ST. Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J Biol Chem. 1998;273:15521–15527. doi: 10.1074/jbc.273.25.15521. [DOI] [PubMed] [Google Scholar]
- Buratti E. Functional Significance of TDP-43 Mutations in Disease. Adv Genet. 2015;91:1–53. doi: 10.1016/bs.adgen.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile x mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell. 2014;54:407–17. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford S, Dissanayake C, Bui QM, Huggins R, Taylor AK, Loesch DZ. Autism spectrum phenotype in males and females with fragile X full mutation and premutation. J Autism Dev Disord. 2007;37:738–47. doi: 10.1007/s10803-006-0205-z. [DOI] [PubMed] [Google Scholar]
- Conlon EG, Manley JL. RNA-binding proteins in neurodegeneration: mechanisms in aggregate. Genes Dev. 2017;31:1509–1528. doi: 10.1101/gad.304055.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, Siddegowda BB, Estes PS, Johannesmeyer J, Kovalik T, Daniel SG, Pearson A, Bowser R, Zarnescu DC. Futsch/MAP1B mRNA Is a Translational Target of TDP-43 and Is Neuroprotective in a Drosophila Model of Amyotrophic Lateral Sclerosis. J Neurosci. 2014;34:15962–74. doi: 10.1523/JNEUROSCI.2526-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, Yamada SB, Siddegowda BB, Estes PS, Zaepfel BL, Johannesmeyer JS, Lockwood DB, Pham LT, Hart MP, Cassel JA, Freibaum B, Boehringer AV, Taylor JP, Reitz AB, Gitler AD, Zarnescu DC. Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum Mol Genet. 2015;24:6886–98. doi: 10.1093/hmg/ddv389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, Lorenzini I, Chou CC, Torvund M, Rogers RS, Starr A, Zaepfel BL, Levy J, Johannesmeyer J, Schwartz JC, Nishimune H, Zinsmaier K, Rossoll W, Sattler R, Zarnescu DC. Post-transcriptional Inhibition of Hsc70-4/HSPA8 Expression Leads to Synaptic Vesicle Cycling Defects in Multiple Models of ALS. Cell Rep. 2017a;21:110–125. doi: 10.1016/j.celrep.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, Zaepfel BL, Zarnescu DC. Failure to Deliver and Translate-New Insights into RNA Dysregulation in ALS. Front Cell Neurosci. 2017b;11:243. doi: 10.3389/fncel.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–99. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes & development. 2005;19:903–18. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–61. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JK, Broadie K. Multifarious Functions of the Fragile X Mental Retardation Protein. Trends Genet. 2017;33:703–714. doi: 10.1016/j.tig.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rub U, Auburger G, Trojanowski JQ, Lee VM, Van Deerlin VM, Bonini NM, Gitler AD. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–75. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes PS, Boehringer A, Zwick R, Tang JE, Grigsby B, Zarnescu DC. Wild-type and A315T mutant TDP-43 exert differential neurotoxicity in a Drosophila model of ALS. Human molecular genetics. 2011;20:2308–21. doi: 10.1093/hmg/ddr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Bassell GJ, Rossoll W. The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum Mol Genet. 2012;21:3703–18. doi: 10.1093/hmg/dds205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguin F, Godena VK, Romano G, D'Ambrogio A, Klima R, Baralle FE. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–92. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Ferrari R, Kapogiannis D, Huey ED, Momeni P. FTD and ALS: a tale of two diseases. Curr Alzheimer Res. 2011;8:273–94. doi: 10.2174/156720511795563700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. Journal of proteome research. 2010;9:1104–20. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Rademakers R, Petrucelli L. TARDBP mutation analysis in TDP-43 proteinopathies and deciphering the toxicity of mutant TDP-43. J Alzheimers Dis. 2013;33(Suppl 1):S35–45. doi: 10.3233/JAD-2012-129036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Ascano M, Tuschl T. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Adv Exp Med Biol. 2014a;825:1–55. doi: 10.1007/978-1-4939-1221-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014b;15:829–45. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godena VK, Romano G, Romano M, Appocher C, Klima R, Buratti E, Baralle FE, Feiguin F. TDP-43 regulates Drosophila neuromuscular junctions growth by modulating Futsch/MAP1B levels and synaptic microtubules organization. PLoS ONE. 2011;6:e17808. doi: 10.1371/journal.pone.0017808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Bras J, Hardy J. SnapShot: Genetics of ALS and FTD. Cell. 2015;160:798, e1. doi: 10.1016/j.cell.2015.01.052. [DOI] [PubMed] [Google Scholar]
- Hagerman PJ, Hagerman RJ. The fragile-X premutation: a maturing perspective. Am J Hum Genet. 2004;74:805–16. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Hagerman P. Fragile X-associated tremor/ataxia syndrome - features, mechanisms and management. Nat Rev Neurol. 2016;12:403–12. doi: 10.1038/nrneurol.2016.82. [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Berry-Kravis E, Hazlett HC, Bailey DB, Jr, Moine H, Kooy RF, Tassone F, Gantois I, Sonenberg N, Mandel JL, Hagerman PJ. Fragile X syndrome. Nat Rev Dis Primers. 2017;3:17065. doi: 10.1038/nrdp.2017.65. [DOI] [PubMed] [Google Scholar]
- Hall SS, Lightbody AA, Reiss AL. Compulsive, self-injurious, and autistic behavior in children and adolescents with fragile X syndrome. Am J Ment Retard. 2008;113:44–53. doi: 10.1352/0895-8017(2008)113[44:CSAABI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Hardy J, Rogaeva E. Motor neuron disease and frontotemporal dementia: sometimes related, sometimes not. Exp Neurol. 2014;262(Pt B):75–83. doi: 10.1016/j.expneurol.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Hazelett DJ, Chang JC, Lakeland DL, Morton DB. Comparison of parallel high-throughput RNA sequencing between knockout of TDP-43 and its overexpression reveals primarily nonreciprocal and nonoverlapping gene expression changes in the central nervous system of Drosophila. G3. 2012;2:789–802. doi: 10.1534/g3.112.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson AJ. Amyotrophic lateral sclerosis and its association with dementia, parkinsonism and other neurological disorders: A review. Brain. 1981;104:217–247. doi: 10.1093/brain/104.2.217. [DOI] [PubMed] [Google Scholar]
- Ihara R, Matsukawa K, Nagata Y, Kunugi H, Tsuji S, Chihara T, Kuranaga E, Miura M, Wakabayashi T, Hashimoto T, Iwatsubo T. RNA binding mediates neurotoxicity in the transgenic Drosophila model of TDP-43 proteinopathy. Human molecular genetics. 2013;22:4474–84. doi: 10.1093/hmg/ddt296. [DOI] [PubMed] [Google Scholar]
- Ishiguro A, Kimura N, Watanabe Y, Watanabe S, Ishihama A. TDP-43 binds and transports G-quadruplex-containing mRNAs into neurites for local translation. Genes Cells. 2016 doi: 10.1111/gtc.12352. [DOI] [PubMed] [Google Scholar]
- Jeong YH, Ling JP, Lin SZ, Donde AN, Braunstein KE, Majounie E, Traynor BJ, LaClair KD, Lloyd TE, Wong PC. Tdp-43 cryptic exons are highly variable between cell types. Mol Neurodegener. 2017;12:13. doi: 10.1186/s13024-016-0144-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–39. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Lin L, Tradewell ML, Dion PA, Bercier V, Bourgouin P, Rochefort D, Bel Hadj S, Durham HD, Velde CV, Rouleau GA, Drapeau P. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Kidd SA, Andrews HF, Budimirovic DB, Esler A, Haas-Givler B, Stackhouse T, Riley C, Peacock G, Sherman SL, Brown WT, Berry-Kravis E. Autism Spectrum Disorder in Fragile X Syndrome: Cooccurring Conditions and Current Treatment. Pediatrics. 2017;139:S194–S206. doi: 10.1542/peds.2016-1159F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattante S, Rouleau GA, Kabashi E. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum Mutat. 2013;34:812–26. doi: 10.1002/humu.22319. [DOI] [PubMed] [Google Scholar]
- Liachko NF, Guthrie CR, Kraemer BC. Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. J Neurosci. 2010;30:16208–19. doi: 10.1523/JNEUROSCI.2911-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349:650–5. doi: 10.1126/science.aab0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–38. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Chen YT, Bose JK, Wu CC, Cheng WC, Cheng SJ, Fang YH, Chen YL, Tsai KJ, Lien CC, Shen CK. TDP-43 regulates the mammalian spinogenesis through translational repression of Rac1. Acta Neuropathol. 2012;124:231–45. doi: 10.1007/s00401-012-1006-4. [DOI] [PubMed] [Google Scholar]
- Majumder P, Chu JF, Chatterjee B, Swamy KB, Shen CJ. Co-regulation of mRNA translation by TDP-43 and Fragile X Syndrome protein FMRP. Acta Neuropathol. 2016;132:721–738. doi: 10.1007/s00401-016-1603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–54. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Nelson DL, Orr HT, Warren ST. The unstable repeats--three evolving faces of neurological disease. Neuron. 2013;77:825–43. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J Neurosci. 2001;21:5139–46. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan M, Talbot K, Ansorge O. Pathogenesis of FUS-associated ALS and FTD: insights from rodent models. Acta Neuropathol Commun. 2016;4:99. doi: 10.1186/s40478-016-0358-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T, Masuda-Suzukake M, Arai T, Hasegawa Y, Akatsu H, Obi T, Yoshida M, Murayama S, Mann DM, Akiyama H, Hasegawa M. Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell reports. 2013;4:124–34. doi: 10.1016/j.celrep.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, Kordasiewicz H, Sedaghat Y, Donohue JP, Shiue L, Bennett CF, Yeo GW, Cleveland DW. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nature neuroscience. 2011;14:459–68. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Bassell GJ, Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci. 2015;16:595–605. doi: 10.1038/nrn4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M, Buratti E, Romano G, Klima R, Del Bel Belluz L, Stuani C, Baralle F, Feiguin F. Evolutionarily conserved heterogeneous nuclear ribonucleoprotein (hnRNP) A/B proteins functionally interact with human and Drosophila TAR DNA-binding protein 43 (TDP-43) J Biol Chem. 2014;289:7121–30. doi: 10.1074/jbc.M114.548859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M, Feiguin F, Buratti E. TBPH/TDP-43 modulates translation of Drosophila futsch mRNA through an UG-rich sequence within its 5'UTR. Brain Res. 2016;1647:50–6. doi: 10.1016/j.brainres.2016.02.022. [DOI] [PubMed] [Google Scholar]
- Russo A, Scardigli R, La Regina F, Murray ME, Romano N, Dickson DW, Wolozin B, Cattaneo A, Ceci M. Increased cytoplasmic TDP-43 reduces global protein synthesis by interacting with RACK1 on polyribosomes. Hum Mol Genet. 2017;26:1407–1418. doi: 10.1093/hmg/ddx035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol. 2012;7:219–45. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844–9. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Langmann C, Harden N, Mandel JL, Giangrande A. CYFIP/Sra-1 Controls Neuronal Connectivity in Drosophila and Links the Rac1 GTPase Pathway to the Fragile X Protein. Neuron. 2003;38:887–898. doi: 10.1016/s0896-6273(03)00354-4. [DOI] [PubMed] [Google Scholar]
- Sephton CF, Good SK, Atkin S, Dewey CM, Mayer P, 3rd, Herz J, Yu G. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285:6826–34. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, Moore MJ, Yu G. Identification of Neuronal RNA Targets of TDP-43-containing Ribonucleoprotein Complexes. J Biol Chem. 2011;286:1204–15. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–8. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Smethurst P, Newcombe J, Troakes C, Simone R, Chen YR, Patani R, Sidle K. In vitro prion-like behaviour of TDP-43 in ALS. Neurobiol Dis. 2016;96:236–247. doi: 10.1016/j.nbd.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, Conti J, Currie JR, Brown WT, Denman RB. RNAs that interact with the fragile X syndrome RNA binding protein FMRP. Biochem Biophys Res Commun. 2000;275:973–80. doi: 10.1006/bbrc.2000.3405. [DOI] [PubMed] [Google Scholar]
- Tan Q, Yalamanchili HK, Park J, De Maio A, Lu HC, Wan YW, White JJ, Bondar VV, Sayegh LS, Liu X, Gao Y, Sillitoe RV, Orr HT, Liu Z, Zoghbi HY. Extensive cryptic splicing upon loss of RBM17 and TDP43 in neurodegeneration models. Hum Mol Genet. 2016;25:5083–5093. doi: 10.1093/hmg/ddw337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till SM. The developmental roles of FMRP. Biochem Soc Trans. 2010;38:507–10. doi: 10.1042/BST0380507. [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nature neuroscience. 2011;14:452–8. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropepe V, Sive HL. Can zebrafish be used as a model to study the neurodevelopmental causes of autism? Genes Brain Behav. 2003;2:268–81. doi: 10.1034/j.1601-183x.2003.00038.x. [DOI] [PubMed] [Google Scholar]
- Tsao W, Jeong YH, Lin S, Ling J, Price DL, Chiang PM, Wong PC. Rodent models of TDP-43: recent advances. Brain Res. 2012;1462:26–39. doi: 10.1016/j.brainres.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–35. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Herholz D, Fiesel FC, Kaur K, Muller D, Karsten P, Weber SS, Kahle PJ, Marquardt T, Schulz JB. TDP-43-Mediated Neuron Loss In Vivo Requires RNA-Binding Activity. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci U S A. 2009;106:18809–14. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LS, Cheng WC, Hou SC, Yan YT, Jiang ST, Shen CK. TDP-43, a neuropathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- Wu YJ, Hsu MT, Ng MC, Amstislavskaya TG, Tikhonova MA, Yang YL, Lu KT. Fragile X Mental Retardation-1 Knockout Zebrafish Shows Precocious Development in Social Behavior. Zebrafish. 2017;14:438–443. doi: 10.1089/zeb.2017.1446. [DOI] [PubMed] [Google Scholar]
- Yu Z, Fan D, Gui B, Shi L, Xuan C, Shan L, Wang Q, Shang Y, Wang Y. Neurodegeneration-associated TDP-43 Interacts with Fragile X Mental Retardation Protein (FMRP)/Staufen (STAU1) and Regulates SIRT1 Expression in Neuronal Cells. The Journal of biological chemistry. 2012;287:22560–72. doi: 10.1074/jbc.M112.357582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zago S, Poletti B, Morelli C, Doretti A, Silani V. Amyotrophic lateral sclerosis and frontotemporal dementia (ALS-FTD) Arch Ital Biol. 2011;149:39–56. doi: 10.4449/aib.v149i1.1263. [DOI] [PubMed] [Google Scholar]
- Zangenehpour S, Cornish KM, Chaudhuri A. Whole-brain expression analysis of FMRP in adult monkey and its relationship to cognitive deficits in fragile X syndrome. Brain Res. 2009;1264:76–84. doi: 10.1016/j.brainres.2009.01.059. [DOI] [PubMed] [Google Scholar]
- Zarnescu DC, Shan G, Warren ST, Jin P. Come FLY with us: toward understanding fragile X syndrome. Genes Brain Behav. 2005;4:385–92. doi: 10.1111/j.1601-183X.2005.00136.x. [DOI] [PubMed] [Google Scholar]
- Zhan L, Hanson KA, Kim SH, Tare A, Tibbetts RS. Identification of genetic modifiers of TDP-43 neurotoxicity in Drosophila. PLoS One. 2013;8:e57214. doi: 10.1371/journal.pone.0057214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, Bowen KE, Wadhwa H, Yang P, Rigo F, Taylor JP, Gitler AD, Rothstein JD, Lloyd TE. Stress Granule Assembly Disrupts Nucleocytoplasmic Transport. Cell. 2018 doi: 10.1016/j.cell.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Beck MV, Griffith JD, Deshmukh M, Dokholyan NV. Large SOD1 aggregates, unlike trimeric SOD1, do not impact cell viability in a model of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2018 doi: 10.1073/pnas.1800187115. [DOI] [PMC free article] [PubMed] [Google Scholar]