Graphical Abstract
Distributive conjugal transfer (DCT) in mycobacteria results in transconjugant progeny with mosaic genomes more resembling chromosomes following meiotic recombination. Multiple, donor segments (blue) recombine into the recipient genome (yellow) resulting in both macro- and micro-mosaicism. Transconjugants can become donors if they inherit the mating identity locus. This review describes the unique features of DCT and compares it to other mechanisms of horizontal gene transfer.
Summary
This review discusses a novel form of horizontal gene transfer (HGT) found in mycobacteria called Distributive Conjugal Transfer (DCT). While satisfying the criteria for conjugation, DCT occurs by a mechanism so distinct from oriT-mediated conjugation that it could be considered a fourth category of HGT. DCT involves the transfer of chromosomal DNA between mycobacteria and, most significantly, generates transconjugants with mosaic genomes of the parental strains. Multiple segments of donor chromosomal DNA can be co-transferred regardless of their location or the genetic selection and, as a result, the transconjugant genome contains many donor-derived segments; hence the name DCT. This distinguishing feature of DCT separates it from the other known mechanisms of HGT, which generally result in the introduction of a single, defined segment of DNA into the recipient chromosome (Fig. 1). Moreover, these mosaic progeny are generated from a single conjugal event, which provides enormous capacity for rapid adaptation and evolution, again distinguishing it from the three classical modes of HGT. Unsurprisingly, the unusual mosaic products of DCT are generated by a conjugal mechanism that is also unusual. Here, we will describe the unique features of DCT and contrast those to other mechanisms of HGT, both from a mechanistic and an evolutionary perspective. Our focus will be on transfer of chromosomal DNA, as opposed to plasmid mobilization, because DCT mediates transfer of chromosomal DNA and is a chromosomally encoded process.
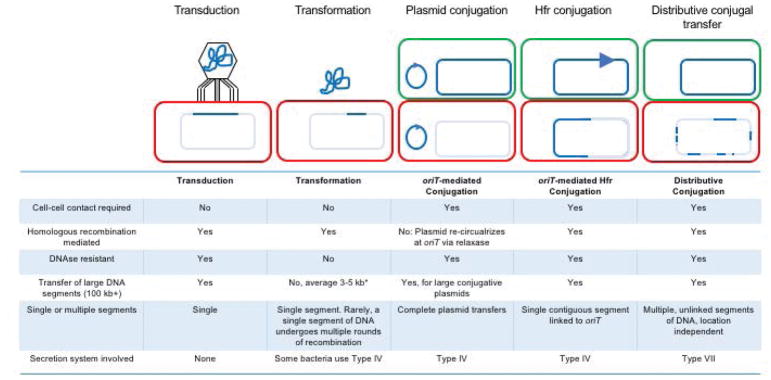
Fig. 1.
Overview of the major mechanisms and products of bacterial HGT. To facilitate comparison between classes, a schematic representation of each is shown above a short list of attributes for that class. Donor cells have a green outline and contain dark blue DNA; recipient cells with a green outline contain pale blue DNA recipient, and varying amounts of transferred donor DNA; the dark blue arrow represents the origin of transfer, oriT, from which transfer is initiated in a 5′ to 3′ direction. *In transformation most acquired DNA segments are small, however, larger fragments (up to 40 kb) can be inherited if their gene content is selected (Blokesch, 2017).
A primer on Mycobacterium species
Mycobacteria are a GC-rich Gram-positive genus belonging to the Actinomycetales. However, in contrast to most Gram-positive bacteria, mycobacteria are surrounded by a lipid-rich envelope that contains an outer (myco)membrane composed of mycolic acid-lipids (Daffe et al., 2014). This mycomembrane endows the mycobacterium with a cell structure more closely resembling that of Gram-negative diderms, and it is thought to constitute an additional barrier against anti-mycobacterial agents (Hoffmann et al., 2008). While most everybody is familiar with the pathogens Mycobacterium tuberculosis and Mycobacterium leprae for the diseases they cause, there are hundreds of mycobacterial species. They are broadly classified by pathogenicity and whether they are fast- or slow-growing.
Many of the slow-growing strains (doubling time around 24hrs) are pathogens, which includes the M. tuberculosis complex (MTBC; M. africanum, M. bovis, M. caprae, M. microti, M. mungi, M. pinnipedii and M. tuberculosis). These very closely related subspecies are greater than 99.9% identical with a phylogenetic structure radiating from a clonal source, with no evidence of recent HGT from outside the MTBC (Boritsch & Brosch, 2016, Brosch et al., 2002, Gagneux, 2018). MTBC subspecies have adapted to the host they infect (sometimes referred to as ecotypes) and can cause TB-like disease; for example, M. bovis infects cattle, and M. caprae infects goats. M. canettii is the outlier of the MTBC for several reasons. M. canettii has a smooth colony morphology, which distinguishes it from the rough colonies of other MTBC members (van Soolingen et al., 1997, Boritsch et al., 2016a). M. canettii is not descended from the clonal founder of the rest of the MTBC, sharing less genomic identity (~97 %) with other members of the MTBC (Boritsch et al., 2014). Most importantly, M. canettii isolates are genetically diverse, not of clonal origin, and their chromosomes contain many remnants of HGT events (Gutierrez et al., 2005, Supply et al., 2013).
Many, but not all, of the fast-growing (2–4 hr doubling time) mycobacterial species are harmless soil saprophytes. The most relevant of these is Mycobacterium smegmatis, which is the model organism used by scientists as a surrogate for its slow-growing pathogenic cousins (Shiloh & DiGiuseppe Champion, 2009). M. smegmatis is the workhorse of mycobacterial genetics and biochemistry because many of its genes are highly conserved in the slow-growing pathogens (2,334 M. tuberculosis genes have orthologs in M. smegmatis with >50% amino acid identity). The strain of M. smegmatis ubiquitously used in research laboratories is a highly transformable derivative called mc2155, with the mc2 designation an homage to the founding institution, Albert Einstein College of Medicine (Snapper et al., 1990, Panas et al., 2014). While this strain serves as the generic M. smegmatis for the research community, sequence comparisons of other M. smegmatis isolates suggests that – as for M. canettii – it is a genetically diverse species that may therefore differ phenotypically. Most relevant for this review is MKD8, an independent isolate of M. smegmatis that we routinely use as a recipient strain in mycobacterial conjugation (Parsons et al., 1998).
DCT generates mosaic genomes
Early studies described the transfer of auxotrophic marker mutations between pairs of independent M. smegmatis isolates (Mizuguchi et al., 1976). Subsequent analyses showed that the recombination process bore the characteristic hallmarks of conjugation (Fig. 2; Derbyshire & Gray, 2014). Transfer required stable, extended (>18 hr), cell-cell contact on solid medium or in a pellicle biofilm but not in planktonic culture (Nguyen et al., 2010, Parsons et al., 1998). DNA transfer was unidirectional, from a donor to a recipient strain and was not observed in same-strain experiments (e.g. differentially marked recipient to recipient transfer). The conjugation classification was further supported by exclusion of other known transfer mechanisms: no phage were detected (excluding transduction); episomal plasmids introduced into the donor strain were not transferred to recombinants (excluding cell fusion); genetic exchange was resistant to DNase treatment and exogenously added DNA was not taken up by the recipient (excluding transformation; Parsons et al., 1998). In addition, naturally occurring plasmids were not detected in the participating strains, and known conjugation genes are absent from their chromosomes. Transfer was of chromosomal DNA, similar to oriT-mediated, Hfr transfer in E. coli. However, genes on the donor chromosome were transferred with equal efficiency regardless of their location (Wang et al., 2005). This is a fundamental difference with Hfr transfer, in which the integrated plasmid oriT leads a procession of genes into the recipient, resulting in a progressive decrease in transfer efficiency with distance from oriT (Wollman et al., 1956)
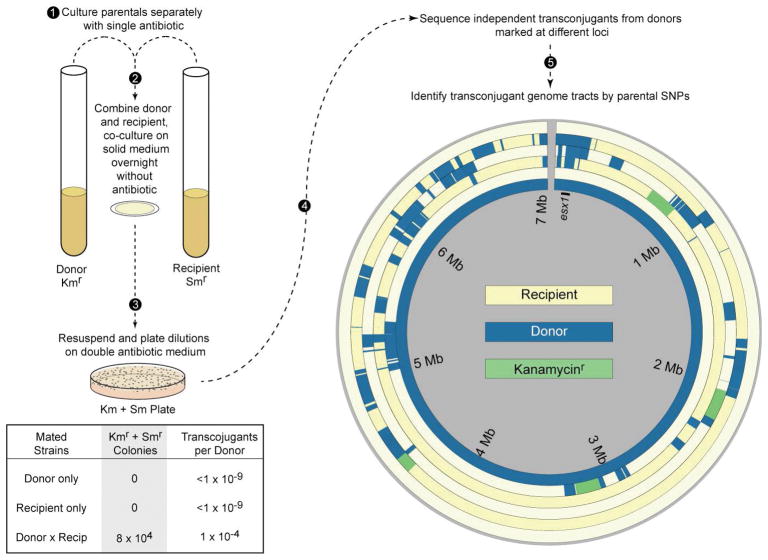
Fig. 2.
The experimental outline for DCT and the mosaic genomes it generates. Donor and recipient strains of mycobacteria with different antibiotic markers are expanded under selection to late mid-log densities (1), and equivalent amounts are combined, pelleted, resuspended and spotted on agar for 18–24 hr (2). The coculture polycolony is resuspended and plated on double selection plates to identify transconjugants, and on single selection plates to recover parents for calculating DNA transfer efficiencies (3). Transconjugants arising from independent matings from donors marked at varying chromosomal positions are subjected to whole genome sequencing (4). The many SNPs (>72,000 between the parental reference genomes) facilitate the identification of the parental origin of DNA comprising the transconjugant genomes with great precision. Circos plots of the recipient reference chromosome (outer ring, yellow), four independent transconjugants, and the donor reference chromosome (inner ring, blue). The transconjugants contain both recipient (yellow) and non-selected donor (blue) DNA. Green segments are those containing the selected kanamycin-resistance gene, which were located in different positions in the chromosome of each of the four donor strain derivatives. The three outermost transconjugants were also shown to have acquired the ability to be donors. Consistent with this phenotype, they each contain (blue) donor-derived DNA spanning esx1 and the mid locus (at 0.1 Mb), in contrast to the innermost transconjugant, which still has a recipient phenotype. Transconjugants show the random, complex, mosaicism that can be produced by a single DCT event (5). Modified from (Derbyshire & Gray, 2014)
More recent molecular studies compared the genome sequences of transconjugants with their parental genomes to identify the tracts of DNA transferred in independent events (Gray et al., 2013). These comparative sequence analyses were feasible because the M. smegmatis parental strains used in these experiments differ significantly at the nucleotide level (averaging 1 Single Nucleotide Polymorphism (SNP) per 56 bp). These SNPs allowed definitive determination of the parental origin with nucleotide resolution of transconjugant DNA. Most of the transconjugant genome was of recipient origin and, as expected, contained the donor-derived selectable marker flanked by varying amounts of donor DNA (Fig. 2). Surprisingly, many unlinked – and therefore, unselected – donor DNA segments were co-inherited in the transconjugant genomes. The transconjugant genomes analyzed (n = 22 transconjugants) contained on average 575 kb of donor DNA in 13 segments, ranging in size from 59 bp to 226 kb. The transferred segments were found distributed around the chromosome and, as a result, the chromosome is a mosaic blend of both parental genomes. This defining characteristic led to the descriptive term, Distributive Conjugal Transfer (DCT) and, more importantly for the transconjugant bacterium, it effectively blends the genomes of the parental bacteria.
DCT generates both macro- and micro-mosaicism
From a birds-eye perspective, macro-mosaicism is easy to visualize and can be explained by homologous recombination promoting double cross-over events that replace large segments of the recipient chromosome with transferred DNA (Fig. 3). The transfer of large segments of donor DNA can result in the acquisition of entire genes or operons from the donor (or their loss if they resided in the replaced recipient homologous segment) and, thus, have the potential to dramatically alter the biochemical properties of a cell. However, DCT also concurrently creates microcomplexity; small segments of DNA that contain alternating small (<100 bp) segments of donor and recipient DNA, which are identified by parent-specific SNPs (Fig. 3; Gray et al., 2013). The precise recombination mechanism that creates this microcomplexity is unclear, but the net result is that small numbers of donor SNPs are blended into a defined region of the recipient. If these SNPs map within a gene and introduce amino acid substitutions, then they may result in subtle changes in enzyme activity by emulating spontaneous mutations. These substitutions could, for example, modulate enzyme activity by altering substrate specificity, protein stability or protein conformation, in contrast to the more dramatic qualitative changes caused by macro-mosaicism.
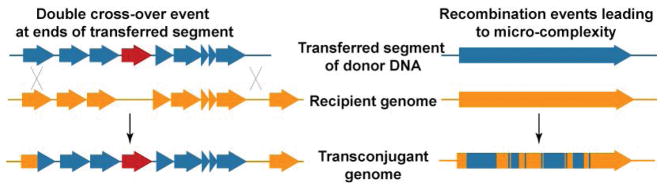
Fig. 3.
Localized recombinant architecture suggests different mechanisms that generate two potential levels of diversity. A single contiguous tract of donor DNA that has recombined into the transconjugant genome exemplifies large chunk exchange, which usually replaces the orthologous tract of recipient DNA and can also result in acquisition of novel genes (red arrow, left panel). Microcomplexity is likely generated by repair mechanisms that alternate between available donor and recipient templates, incorporating SNPs from each in quick succession. Collectively, these disparate recombination architectures can bring in new operons—and the pathways they encode—or make minor adjustments in regulatory elements or individual proteins. Modified from (Derbyshire & Gray, 2014).
Hallmarks of DCT are evident in other mycobacterial species
WGS analyses of other independent isolates of M. smegmatis show that their genomes are also mosaic. The patterns of SNPs present in these genomes indicate they are blends of not only each other, but also segments of DNA of unknown origin (Derbyshire and Gray, unpublished). The mosaicism strongly suggests that DCT is prevalent among environmental M. smegmatis. In contrast to our laboratory DCT matings, where the genome sequences of parents and progeny are all determined, the pedigree of the natural isolates is not known, so the DCT-like mosaicism we observe may arise from other processes. However, we have demonstrated that all of these independent isolates are conjugative, with defined donors and recipients within the group (Parsons et al., 1998). The resulting laboratory-generated transconjugant genomes are mosaic assemblages of their parental strains, confirming that DCT is not an anomaly of our characterized mating pair but is a general property of M. smegmatis (Derbyshire and Gray, unpublished). Since these strains are DCT-proficient, their chromosomal DNA—both core genome variants as well as novel sequences—should be viewed as mobile components of gene flow within the mycobacterial pangenome.
Recent WGS of isolates of M. abscessus (a fast-growing, opportunistic pathogen) and M. canettii revealed that they also have broadly mosaic genomes (Gutierrez et al., 2005, Sapriel et al., 2016, Supply et al., 2013). Again, these observations were facilitated by mapping large numbers of SNPs in the genomes of multiple isolates and comparing blocks of shared SNPs between different isolates. The formation of their mosaic genomes is entirely consistent with the process of DCT. However, without knowledge of the parental genotypes this can only be inferred. This problem plagues analysis of any clinical or environmental isolate where both the origin(s) of the mobile sequences and the elapsed time from the HGT event(s) are unknown. However, two M. canettii isolates have been demonstrated to recombine in experimental conditions identical to those used for DCT in M. smegmatis (Boritsch et al., 2016b). The transconjugants generated have mosaic genomes of the parental pair, confirming that these isolates are proficient in DCT. This has two important implications. One is that DCT has now been experimentally demonstrated in both major clades of mycobacteria: fast- (M. smegmatis) and slow- (M. canettii) growers, suggesting that it is likely an activity found throughout the genus. Secondly, DCT is likely to have played a role in shaping the mosaic genome architectures found in extant mycobacteria and continues to be active in promoting gene flow within the genus. The evidence will be in their genomes and we anticipate many more examples will be identified. We note, for example, that limited sequence analysis of four different isolates of M. avium (a slow-growing opportunistic pathogen) identified a 4 kb region that contained segments of microcomplexity similar to DCT patterns documented in M. smegmatis (Krzywinska et al., 2004).
Can M. tuberculosis mediate DCT like its M. canettii cousin?
M. canettii is a slow-growing pathogen that causes tuberculosis in humans. Notably, while other MTBC members are genetically homogeneous, M. canettii genomes contain large numbers of SNPs (as many as 60,000 SNPs between sequenced isolates) and exhibit mosaicism when compared within the species or with the MTBC (Boritsch et al., 2014, Supply et al., 2013, Blouin et al., 2014, Mortimer & Pepperell, 2014). Based on these analyses, M. canettii is considered an extant member of a genetically diverse progenitor species, M. prototuberculosis, that has undergone extensive HGT, which resulted in the observed genome mosaicism (Gutierrez et al., 2005). M. tuberculosis (and subsequently the MTBC) is hypothesized to have emerged as a particularly successful derivative of M. prototuberculosis, with the subsequent clonal expansion giving rise to the MTBC (Brosch et al., 2002, Gutierrez et al., 2005, Smith et al., 2009, Gagneux, 2018). As extant isolates of M. canettii are capable of DCT, and mosaicism is evident in M. canettii versus M. tuberculosis genomes, it is highly likely that DCT blended the genetic repertoire of M. prototuberculosis strains to generate a more successful human-adapted version that emerged as M. tuberculosis. However, in spite of its possible role in shaping the genome of M. tuberculosis, there is no indication that DCT remains active in this lineage. The low sequence diversity (99.9% identical, with only ~2,000 SNPs between MTBC members) of non-M. canettii MTBC genomes suggests that few, if any, HGT events have occurred in this clade since its clonal beginning. Moreover, the clonal nature of the MTBC would likely preclude active DCT among the species, as all isolates would be of the same mating type (either all donor or recipient), and therefore would not constitute a productive mating pair.
Despite this, there is evidence of recent genetic exchange between M. canettii and M. tuberculosis (Supply et al., 2013). M. tuberculosis and M. canettii genomes are diverse with some isolates containing >60,000 SNPs, when compared with M. tuberculosis. However, sequence comparisons have identified small regions (up to 4 kb) of almost complete sequence identity embedded in a sea of SNPs, which is consistent with HGT having occurred since the two species diverged. While the HGT mechanism underlying these rare events is unknown, given that DCT is active in M. canettii, it must be considered as one possible mechanism, along with transduction by mycobacteriophages. Opportunities for HGT are few because M. tuberculosis resides in the solitary environment of a macrophage and, therefore, it rarely interacts with other mycobacteria, even in co-infections. Thus, in summary, we suggest that DCT was a major player in the initial evolution of M. tuberculosis from its progenitor species, but its continued activity in this species is unlikely.
Genetics points to a significant role for ESX secretion systems in DCT
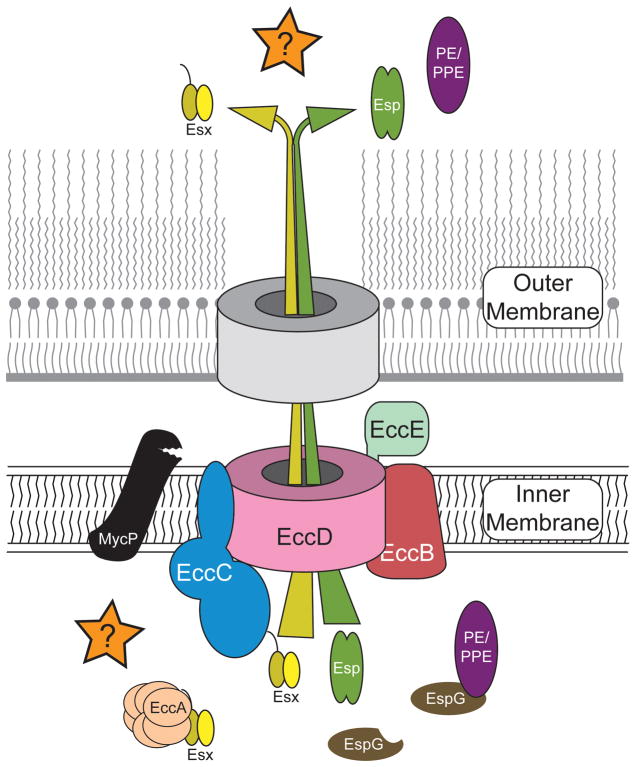
Genetic studies using transposon mutagenesis have identified many genes that affect DCT in both donor and recipient M. smegmatis (Coros et al., 2008, Flint et al., 2004, Nguyen et al., 2010, Nguyen et al., 2009). While these genes have shed little light on the mechanism of transfer per se, they have provided fundamental insights on the modes of DCT regulation and cell-cell sensing. A majority of the genes identified from the transposon screens belong to a class of genes associated with ESX secretion systems (Abdallah et al., 2007, Ates et al., 2016a, Bitter et al., 2009, Groschel et al., 2016). ESX secretion systems (also known as Type VII secretion) are found throughout the Actinobacteria and often occur in multiple copies per genome; M. smegmatis and M. tuberculosis contain three and five esx loci respectively. Each locus contains a similar set of core genes (~10) with non-redundant functions (i.e., the genes from different loci do not cross complement) and all evidence suggests that each system secretes proteins required for distinct cellular functions (Fig. 4). In addition to secreted substrates, the core genes (called ecc for esx conserved component) encode a membrane transporter, chaperones, and ATPases thought to facilitate export through a membrane channel (Figs. 4 and 5). In M. tuberculosis ESX-1, ESX-3 and ESX-5 are required for virulence (Groschel et al., 2016, Tufariello et al., 2016). Only recently have functions been associated with ESX-4: DCT in M. smegmatis and, in Mycobacterium abscessus, survival and pathogenesis in macrophages indicating a broader role for ESX-4 and its effector molecules (Gray et al., 2016, Laencina et al., 2018, and below). No functions have been assigned to ESX-2. The diverse functions of these apparatus are most likely mediated by the unique set of substrates secreted by each system, while the more conserved core proteins assemble to form the secretion machine (Fig. 5).
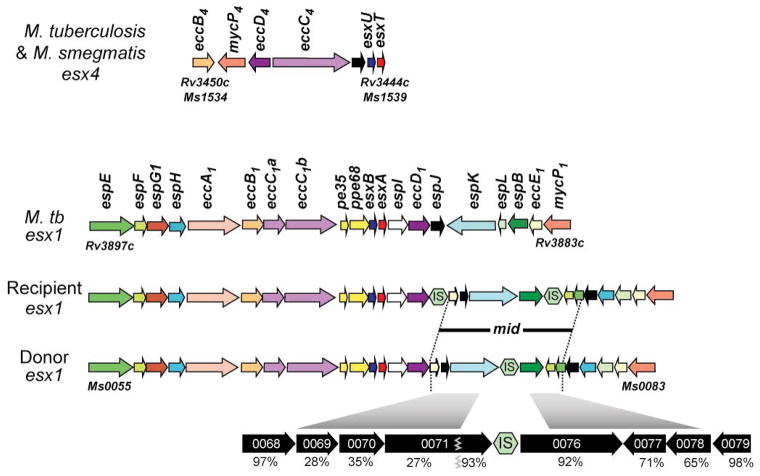
Fig. 4.
esx4 loci share common genes necessary for ESX function. The seven-gene core locus of esx4 is conserved in gene content and order in most mycobacteria, including M. tuberculosis and M. smegmatis donor and recipient strains (top panel). The esx1 locus is more complex and is more heterogeneous in the composition of genes near its 3′ boundary, particularly where the mating identity (mid) genes are located in M. smegmatis (bottom panel). The mid genes are expanded at the bottom to show the level of amino acid identity between donor and recipient strains (IS represents insertion sequence elements). The genes are colour coded to indicate orthologous genes in each ESX system. The M. smegmatis (Ms) and M. tuberculosis (Rv) gene numbers are given to indicate the location of each locus in its respective genome. The gene names follow the classification according to (Bitter et al., 2009). ecc genes are conserved components of all esx loci and are thought to encode core machinery. esp genes are specific to a locus, in this case esx1, and are thought to mediate locus-specific functions. EspB, PE35 and PPE68 are known to be secreted by ESX-1 (and figure 5). EsxB and EsxA form the heterodimer secreted by the ESX-1 system. The equivalent proteins in esx4, EsxU and EsxT, have yet to be shown to be secreted.
Fig. 5.
General schematic of an ESX secretion apparatus based on the ESX-5 structure (Ates et al., 2016a, Beckham et al., 2017). The core components (Ecc) and secretion substrates (EsxBA, EspB, PE/PPE) encoded by each paralogous locus are dedicated to that specific apparatus, creating a series of operationally similar, but functionally non-redundant, secretion systems (Bitter et al., 2009, Houben et al., 2014). In ESX-1, EccA is thought to act as a chaperone delivering the secretion substrates to EccC, the membrane-bound ATPase, which then delivers the proteins to the inner membrane channel made by EccD. MycP is a protease, required for processing of some secreted substrates (eg. EspB; Ohol et al., 2010). EspG is a second chaperone dedicated to secretion of PE and PPE proteins (Abdallah et al., 2006, Abdallah et al., 2009). The channel protein and mechanism through which the ESX substrates traverse the outer mycomembrane is unknown. The protein cartoons are not drawn to scale.
Of the three ESX paralogues in M. smegmatis, ESX-1 and ESX-4 are necessary for DCT in the recipient strain; mutations in esx1 or esx4 genes abolished transfer of DNA (Coros et al., 2008, Gray et al., 2016). In the donor strain, the roles of ESX-1 and ESX-4 in DCT differ: esx4 donor mutants transfer at wild-type levels, while esx1 donor mutants are hyper-conjugative. Again, the differential roles in the donor and recipient underscore the functional diversity of ESX secretion systems.
DCT requires a functional ESX-4 apparatus, but only in the recipient
esx4 is the only esx locus found throughout the Actinobacteria and, thus, is considered the progenitor esx locus (Gey Van Pittius et al., 2001, Dumas et al., 2016, Gey van Pittius et al., 2006, Mortimer et al., 2017, Newton-Foot et al., 2016). It is also the smallest esx locus, containing only 7 genes, and missing the core genes, eccA, eccE, and espG, which are found in all other ESX systems and thought to be required for function (Fig. 4). The lack of core genes and an assigned function even led to discussion as to whether esx4 was vestigial, despite its high level of conservation across species (Groschel et al., 2016). However, both transposon and targeted mutagenesis in an M. smegmatis recipient have confirmed that esx4 locus genes are required for DCT (Gray et al., 2016). By contrast, esx4 mutations in the donor strain have no impact on DCT. The donor and recipient loci are almost identical, suggesting that it is the cellular context and not the encoded ESX-4 apparatus that differs between strains.
A second, independent, indication for a role of ESX-4 in DCT was obtained by transcriptional profiling studies (Gray et al., 2016). RNA-seq identified an esx4 mRNA (esxUT) as highly induced under mating conditions (~30-fold), which was confirmed by directed qRT-PCR. Importantly, this esx4 transcript was only induced in the recipient strain and this induction was dependent on coculture; no induction was observed in monocultures of either donor or recipient grown under the same conditions. These rigorous requirements for esx4 expression likely explain why the function of ESX-4 has remained obscure for so long. Coculture did not change the transcript levels of esx1 and esx3 genes in donor or recipient, demonstrating the esx-specificity of contact-dependent esx4 induction. While the functional role of ESX-4 in general, or in DCT specifically, is still not known, what is now clear is that mycobacteria can detect the presence of another mycobacterial strain, and then activate ESX-4 in response to that presence. For the process of DCT, each of these abilities is appropriate to the interaction: donor cell contact is detected by a recipient cell, resulting in induction of ESX-4, which enables the recipient to take up donor DNA during conjugation.
ESX-1 and ESX-4 play important non-redundant roles in DCT
ESX-1 has strain-dependent reciprocal effects on DCT: donor mutants are hyper-conjugative (Flint et al., 2004) but recipient mutants are defective in DCT (Coros et al., 2008). In the recipient, ESX-1 appears to act upstream of ESX-4 by modulating esx4 gene (e.g., esxUT) expression. Remarkably, mutation of ESX-1 affected the level of the esx4 expression response, which was shown to closely correlate with DNA transfer efficiency (Gray et al., 2016). For example, esx1 mutant recipients had an impaired esx4 response to the presence of the donor, and failed to receive DNA during conjugation. Conversely, esx1 mutant donors caused hyper-activation of esxUT in the recipient (~250-fold), providing a molecular basis for the observed hyper-conjugative phenotype: the donor mutation intensifies the recipient response to coculture, thereby increasing DNA transfer efficiency. It is important to note that a mutation in one cell that affects specific gene expression in an adjacent cell is indicative of cell-cell communication. The molecular signal has not yet been identified, but a role for ESX-1 in mediating communication between mycobacteria presents exciting new possibilities in envisioning its function in mycobacterial communities.
ESX-1 and mating identity
Following DCT, a subset of transconjugants are donors (5–10%) indicating they have acquired a donor-conferring locus. This again distinguishes DCT from Hfr transfer in which transconjugants do not become donors. By comparing the genomes of donor converted transconjugants it was possible to map the donor-conferring locus, called mid (mating identity) to esx1 (Gray et al., 2013). This was an unexpected result, as loss-of-function mutations in these esx1 genes from either strain do not switch mating identity. Mating identity, therefore, is an active phenotype determined by products of the mid locus. By repeatedly back crossing donor transconjugants with a wild-type recipient, while continuing to screen for donor function, it was possible to introgress the donor mid locus into the recipient genome; boundaries from independent introgressed lineages delimited mid within esx1 to a six-gene region (Figs. 2 and 4). The functions of the proteins encoded by these genes are unknown, but some are secreted by ESX-1. The mid region is the most divergent esx1 region between donor and recipient strains, suggesting that these secreted proteins identify cells as donor or recipient, or that they are also part of the intercellular communication flow necessary to initiate DCT.
How do ESX-1 and ESX-4 modulate DCT?
There is currently no experimental evidence of protein secretion by ESX-4. EsxU and EsxT are encoded by the bicistronic esxUT transcript within the esx4 locus and form a WXG100 heterodimeric protein pair that is the probable primary ESX-4 secretion substrates. Since esx4 locus genes are not expressed in traditional monoculture conditions, secretion of these (or any esx4-encoded) proteins has not been reported. ESX-1, on the other hand, secretes abundant levels of its WXG100 heterodimer, EsxB/EsxA, in traditional monoculture (Fig. 5), and these are classic diagnostic antigens for M. tuberculosis infection. Beyond the EsxB/A heterodimer, the secretory repertoire of ESX-1 can vary between species. Therefore, while the structural components comprising the ESX-1 apparatus may operate in the same way between species and strains (e.g. M. tuberculosis EsxB and EsxA can be secreted by the M. smegmatis ESX-1 apparatus (Converse & Cox, 2005), we suggest it is the secreted substrates that confer species or strain-specific functions. Consistent with this, ESX-1 may be secreting unique cell-surface mating identifiers and receptor proteins from both the donor and recipient (Fig. 6). In one scenario, a donor-secreted ligand binds to an ESX-1-secreted receptor, and on the recipient surface, this binding initiates a signal cascade that induces esx4, promoting DCT. The absence of a receptor on an esx1-defective donor, reduces competition for the available ligand, leaving more to bind the recipient receptor, increasing recipient esx4 induction, and leading to higher DCT transfer efficiencies. The absence of the recipient receptor in a recipient esx1 mutant prevents ligand binding, esx4 induction and DCT. While the model is speculative, it is clear that the recipient responds to cell-contact with a donor and that a functional ESX-1 apparatus is required in the recipient strain for esx4 expression. This places ESX-1 upstream of ESX-4 in the DCT process and highlights an unprecedented interaction between two ESX apparatus, with each performing non-redundant roles in the same biological pathway.
Fig. 6.
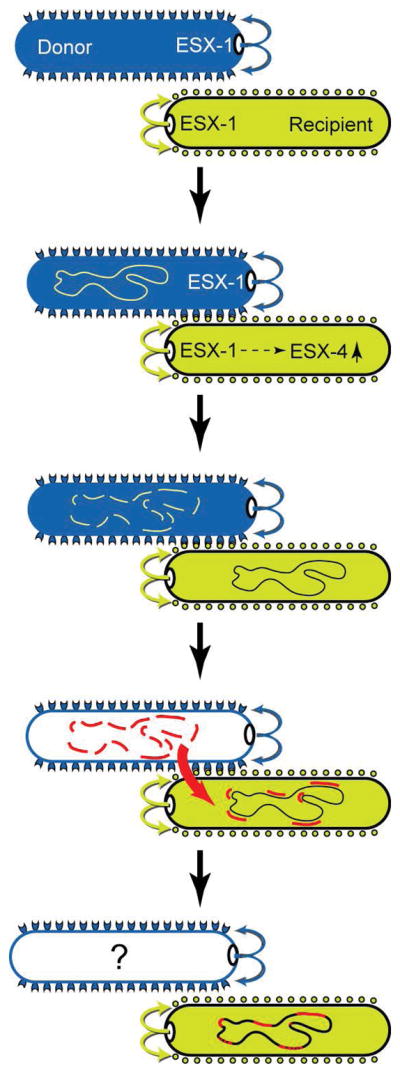
DCT can be conceptualized as a progression of sequential events. DCT begins with contact-dependent signaling between the donor and recipient that includes the polar-localized ESX-1 secretion apparatus from each strain (indicated by curved arrows). We hypothesize that proteins secreted decorate both donor and recipient cell surfaces (cups and circles). Interactions between the secreted proteins (joined cup and circle) trigger donor- and recipient-specific responses. At least one part of the recipient response is to signal induction of esx4, which is required for DCT. The dashed arrow in the recipient reflects the multiple steps between ESX-1 and esx4 induction. Following cell-cell contact, and the expression of genes required to transfer and receive DNA in donor and recipient, respectively, the donor chromosome is fragmented by an unidentified nuclease. (Alternatively, following transfer of the donor chromosome its fragmentation could occur by a nuclease in the recipient). Large, multiple, donor fragments are taken up by the recipient cell, but the DNA import machinery and the fate of the donor are not yet known. The imported donor fragments are integrated into the transconjugant genomes by homologous recombination or template repair mechanisms to create a transconjugant with a mosaic genome. The successful establishment of the transconjugant progeny within a growing population will be determined by the relative fitness conferred by the new combination of genetic variants in that transconjugant relative to the fitness of the parental strains.
What are the consequences of esx4 induction for DCT?
This is a key question to understanding DCT and may help us to understand why the diminutive esx4 locus is well conserved throughout the genus. We suggest several speculative scenarios. One possibility is that ESX-4 secretes the structural or enzymatic components that function as the conduit through which DNA is transported into the recipient. Conceptually, it is likely that a DNA-uptake apparatus in the recipient would be produced only in the presence of a suitable mating partner. This might also explain the minimal complexity of the esx4 locus, in that its activation in response to a specific stimulus suggests that its function is dedicated to that particular stimulus, and therefore has not evolved additional secretory functions. Alternatively, ESX-4 secreted proteins may provide general support for DCT by contributing to cell-cell signaling (ligands or receptors), or remodeling the cell wall to favor DNA transfer. This more general model is attractive as it provides flexibility in explaining the role of ESX-4 in other mycobacteria. For example, in M. abscessus, signaling or remodeling the cell wall would be ways to modify the host response or avoid clearance by host macrophages (Laencina et al., 2018). In further support of a remodeling scenario, mutations in ESX-1 and ESX-5 alter cell wall composition and permeability (Ates et al., 2016b, Garces et al., 2010, Pym et al., 2002). Finally, it is possible that ESX-4 is not a functional secretion apparatus per se, but that it now plays a cytoplasmic role in DCT. While the precedent established by other ESX systems suggests this is unlikely, we cannot rule out the possibility that ESX-4 has non-secretory roles. These hypotheses and the functional dissection of ESX-4 form the focus for future research.
Comparing DCT with other mechanisms of HGT
Atypical conjugation in Streptomyces
Mycobacteria are not the only actinomycete that encodes an atypical conjugation system. Plasmid DNA transfer in Streptomyces occurs by a process more akin to chromosomal segregation that occurs during cell division (Thoma & Muth, 2016). In contrast to oriT-mediated transfer, plasmid DNA is transferred in a double-stranded form (Possoz et al., 2001) and is dependent on the plasmid-encoded TraB protein, which binds a cis-acting locus of transfer, clt (a cluster of 8 bp direct repeats) (Pettis & Cohen, 1994, Reuther et al., 2006). The TraB proteins encoded by different plasmids bind different cognate clt sequences, providing specificity to plasmid mobilization. Plasmid TraB proteins are not relaxases (they bind but do not nick DNA) and, instead, resemble ftsK/SpoIIIE septal-DNA translocator proteins that facilitate chromosomal segregation during cell division or sporulation (Vogelmann et al., 2011). Current models posit that the membrane-associated TraB binds plasmid clt sequences and that its ATPase drives plasmid transfer from the donor into the contacted recipient. Following transfer, the plasmid is spread to other recipient cells within the mycelium by a similar TraB-dependent process (Thoma et al., 2016). While most of the molecular analyses have examined plasmid transfer, older genetic experiments showed that the linear Streptomyces chromosome could also be transferred from donor to recipient. Plasmid encoded TraB is required for this process, which is proposed to bind clt-like sequences on the Streptomyces chromosome (Sepulveda et al., 2011, Vogelmann et al., 2011).
While it is always tempting to find connections, the two actinomycete conjugal systems appear to be as different from each other as they are from E. coli Hfr transfer. Although the Streptomyces and Mycobacterial systems both utilize ftsK/SpoIIIE homologues, the similarities appear to end there. However, the unusual nature of each conjugation system suggests other novel forms of conjugation will be discovered and underscores the need to consider other mechanisms of HGT beyond those discussed in text books.
Bacterial Communication in HGT
DCT is not the first example of cell-cell communication involved in HGT; the most extensively studied system is that involving the conjugative plasmid pCF10 in Enterococcus faecalis (Dunny, 2007, Dunny & Johnson, 2011). In this system, recipient strains secrete small peptides (sex pheromones) that enter the plasmid-carrying donor cell, where the peptides bind and inactivate a repressor protein responsible for preventing expression of the transfer gene operon. Once the recipient becomes a transconjugant, a peptide made from the transferred plasmid is secreted, which inhibits pheromone sensing and thus turns off transfer genes. In E. faecalis, the recipient activates (or communicates with) the donor to trigger transfer, which is the opposite of that observed in M. smegmatis where it appears signals from the donor activate esx4 in the recipient. Bacterial transformation systems also utilize peptides or small molecules to activate competence gene expression (Johnston et al., 2014). However, this self-signaling (quorum sensing) is used to determine the cell density of sister cells, not the presence of the opposite mating type. Regardless of the intercellular communication route, the net goal in HGT processes is presumably to coordinate all the molecular and cellular processes necessary for efficient transfer of DNA only when appropriate contributors are present.
DCT generates mosaic progeny in a single event unlike other mechanisms of HGT
The mosaic genomes of transconjugant progeny are startling not only because of the genome-wide blending of parental DNA, but also because the mosaicism results from a single transfer event (Fig. 2). In this regard, transconjugants resemble the progeny of meiotic recombination! Indeed, single sperm sequencing has shown that the number of crossovers per genome in meiosis is about the same as the average number of donor tracts per DCT transconjugant (Wang et al., 2012). The multiple, independent, physical attributes of DNA segments transferred by DCT (Fig. 3) ensures an almost infinite number of possible genotypes for transconjugants of two parental strains. Hence, the range of genetic outcomes from a single DCT event far exceeds that of any other known form of HGT.
Generalized transduction, transformation and classical Hfr-conjugation generally result in the acquisition of a single segment of DNA, which is recombined into the chromosome by homologous recombination (Fig. 1). We note that, in transformation, multiple, usually small, fragments can be co-inherited and often these are clustered in tracts. This is thought to be due to the capture of a single segment of DNA that then undergoes multiple rounds of recombination (Croucher et al., 2012, Golubchik et al., 2012, Mell et al., 2014, Blokesch, 2017, Bubendorfer et al., 2016). Thus, multiple, sequential HGT events need to occur to approximate the genome-wide mosaicism observed in DCT. The rarity of HGT events would likely mean that accruing enough segments to create the mosaic patchwork seen in DCT would require successive HGT events in a lineage over many years. Thus, while most bacteria evolve over extremely long periods by the gradual accrual of spontaneous mutations and serial acquisition of HGT DNA, mycobacteria have the capacity to evolve very quickly by blending their genomes through DCT.
DCT, Transformation and Conjugation
The features of DCT described above indicate that its genetics, mechanism, and products are very distinct from the current text book versions of HGT. Despite having conjugative-like properties (e.g., transfer requires stable contact between donor and recipient cells, DNA transfer is resistant to DNase and only occurs from a donor to a recipient) DCT shares similar characteristics to transformation. DCT and transformation are chromosomally-encoded and primarily mediate the transfer of chromosomal DNA (since the transferred DNAs rely on homologous recombination for chromosomal integration). In each case, the recipient cell encodes the majority of genetic information needed for DNA acquisition, unlike oriT-mediated and Streptomyces conjugation in which the process is donor (plasmid) driven (de la Cruz et al., 2010, Frost et al., 2005). The transfer efficiency of any given gene is the same regardless of its original genomic location for both transformation and for DCT. Finally, while the products of DCT are, in general, quite different from conjugation and transformation there are examples of co-inheritance of unlinked segments of DNA during transformation and regions of microcomplexity (Croucher et al., 2012, Kulick et al., 2008, Mell et al., 2014). In Helicobacter pylori, DNA introduced by transformation contained patches of microcomplexity likely created by mis-match repair of heteroduplexes in the recombined DNA (Kulick et al., 2008, Bubendorfer et al., 2016). Whether a similar mechanism generates the microcomplexity observed in DCT is currently unknown.
Barriers to HGT in mycobacteria
The paucity of mosaicism observed in the sequenced genomes of bacteria suggests that DCT is restricted to mycobacteria. However, as more members of a species are sequenced, more genetic variation will be found, and mosaic patterns of that variation may emerge. After all, with only a single sequenced M. smegmatis genome from strain mc2155 in GenBank, we did not appreciate that it was mosaic until other M. smegmatis strains were sequenced and aligned with it. Moreover, exchange between similar genomes is difficult to detect and a low density of informative SNPs can obscure mosaic patterns. One physical feature of mycobacteria that might require a unique HGT mechanism is their lipid-rich cell envelope (Fig. 5). This highly hydrophobic barrier is likely to prevent uptake of hydrophilic molecules such as DNA by transformation - mycobacteria are not known to be naturally competent. Similarly, the thick envelope may prevent access of conjugative pili to surface receptors necessary for mating-pair formation. Plasmids are rare in mycobacteria compared with other bacteria, and are absent from the MTBC. Only one example of a conjugative mycobacterial plasmid has been described, and the mechanism of transfer of that plasmid is unclear as it encodes both a type VII ESX system and a more traditional type IV conjugation system with a relaxase (Ummels et al., 2014). It is possible that for mycobacteria, DCT fulfills a general need for gene flow in bacterial populations to maintain genomic health and to promote evolution; a need met by traditional HGT mechanisms in other bacterial clades.
Compared with other forms of HGT, the mechanism of DCT is poorly understood. We have learned that ESX secretion systems have central roles and that intercellular communication coordinates participation between donor and recipient. But there is much more to learn. For example, how is the chromosomal DNA fragmented in the donor, is DNA mobility guided by a protein chaperone into the recipient, does the donor cell survive the tryst, and what homologous DNA repair mechanisms replace recipient DNA with the donated DNA in the transconjugant genome? And those are only the mechanistic questions. Can we model the evolutionary impact of DCT on mycobacterial evolution (or reassess the time scales involved), and perhaps allow better interpretation of the gene flow and selective pressures that drive the emergence of new strains? As we learn more about Distributive Conjugal Transfer, we anticipate discovering how DCT has helped shape the Mycobacterium genus and more novel aspects of mycobacterial biology.
Acknowledgments
The authors thank Gunther Muth for insights on Streptomyces conjugation and laboratory members for their many contributions to the research. This work was supported by funding from the NIH R01AI097191 and R21AI07258, and NSF MCB-1614178.
The authors have no conflicts of interest to declare.
References
- Abdallah AM, Gey van Pittius NC, DiGiuseppe Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CMJE, Appelmelk BJ, Bitter W. Type VII secretion — mycobacteria show the way. Nature Reviews Microbiology. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- Abdallah AM, Verboom T, Hannes F, Safi M, Strong M, Eisenberg D, Musters RJP, Vandenbroucke-Grauls CMJE, Appelmelk BJ, Luirink J, Bitter W. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Molecular Microbiology. 2006;62:667–679. doi: 10.1111/j.1365-2958.2006.05409.x. [DOI] [PubMed] [Google Scholar]
- Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, Jiménez C, Parra M, Cadieux N, Brennan MJ, Appelmelk BJ, Bitter W. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Molecular Microbiology. 2009;73:329–340. doi: 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]
- Ates LS, Houben EN, Bitter W. Type VII Secretion: A Highly Versatile Secretion System. Microbiol Spectr. 2016a:4. doi: 10.1128/microbiolspec.VMBF-0011-2015. [DOI] [PubMed] [Google Scholar]
- Ates LS, van der Woude AD, Bestebroer J, van Stempvoort G, Musters RJP, Garcia-Vallejo JJ, Picavet DI, Weerd Rvd, Maletta M, Kuijl CP, van der Wel NN, Bitter W. The ESX-5 System of Pathogenic Mycobacteria Is Involved In Capsule Integrity and Virulence through Its Substrate PPE10. PLoS Pathog. 2016b;12:e1005696. doi: 10.1371/journal.ppat.1005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham KS, Ciccarelli L, Bunduc CM, Mertens HD, Ummels R, Lugmayr W, Mayr J, Rettel M, Savitski MM, Svergun DI, Bitter W, Wilmanns M, Marlovits TC, Parret AH, Houben EN. Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis. Nat Microbiol. 2017;2:17047. doi: 10.1038/nmicrobiol.2017.47. [DOI] [PubMed] [Google Scholar]
- Bitter W, Houben EN, Bottai D, Brodin P, Brown EJ, Cox J, Derbyshire KM, Fortune SM, Gao LY, Liu J, Gey van Pittius NC, Pym AS, Rubin EJ, Sherman DR, Cole ST, Brosch R. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 2009;5:e1000507. doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch M. In and out-contribution of natural transformation to the shuffling of large genomic regions. Curr Opin Microbiol. 2017;38:22–29. doi: 10.1016/j.mib.2017.04.001. [DOI] [PubMed] [Google Scholar]
- Blouin Y, Cazajous G, Dehan C, Soler C, Vong R, Hassan MO, Hauck Y, Boulais C, Andriamanantena D, Martinaud C, Martin E, Pourcel C, Vergnaud G. Progenitor “Mycobacterium canettii” clone responsible for lymph node tuberculosis epidemic, Djibouti. Emerging infectious diseases. 2014;20:21–28. doi: 10.3201/eid2001.130652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boritsch EC, Brosch R. Evolution of Mycobacterium tuberculosis: New Insights into Pathogenicity and Drug Resistance. Microbiol Spectr. 2016:4. doi: 10.1128/microbiolspec.TBTB2-0020-2016. [DOI] [PubMed] [Google Scholar]
- Boritsch EC, Frigui W, Cascioferro A, Malaga W, Etienne G, Laval F, Pawlik A, Le Chevalier F, Orgeur M, Ma L, Bouchier C, Stinear TP, Supply P, Majlessi L, Daffe M, Guilhot C, Brosch R. pks5-recombination-mediated surface remodelling in Mycobacterium tuberculosis emergence. Nat Microbiol. 2016a;1:15019. doi: 10.1038/nmicrobiol.2015.19. [DOI] [PubMed] [Google Scholar]
- Boritsch EC, Khanna V, Pawlik A, Honoré N, Navas VH, Ma L, Bouchier C, Seemann T, Supply P, Stinear TP, Brosch R. Key experimental evidence of chromosomal DNA transfer among selected tuberculosis-causing mycobacteria. Proceedings of the National Academy of Sciences. 2016b;113:9876–9881. doi: 10.1073/pnas.1604921113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boritsch EC, Supply P, Honore N, Seeman T, Stinear TP, Brosch R. A glimpse into the past and predictions for the future: the molecular evolution of the tuberculosis agent. Mol Microbiol. 2014;93:835–852. doi: 10.1111/mmi.12720. [DOI] [PubMed] [Google Scholar]
- Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van Soolingen D, Cole ST. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubendorfer S, Krebes J, Yang I, Hage E, Schulz TF, Bahlawane C, Didelot X, Suerbaum S. Genome-wide analysis of chromosomal import patterns after natural transformation of Helicobacter pylori. Nat Commun. 2016;7:11995. doi: 10.1038/ncomms11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse SE, Cox JS. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J Bacteriol. 2005;187:1238–1245. doi: 10.1128/JB.187.4.1238-1245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coros A, Callahan B, Battaglioli E, Derbyshire KM. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol Microbiol. 2008;69:794–808. doi: 10.1111/j.1365-2958.2008.06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Harris SR, Barquist L, Parkhill J, Bentley SD. A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog. 2012;8:e1002745. doi: 10.1371/journal.ppat.1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffe M, Crick DC, Jackson M. Genetics of Capsular Polysaccharides and Cell Envelope (Glyco)lipids. Microbiol Spectr. 2014;2:MGM2-0021-2013. doi: 10.1128/microbiolspec.MGM2-0021-2013. [DOI] [PubMed] [Google Scholar]
- de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- Derbyshire KM, Gray TA. Distributive Conjugal Transfer: New Insights into Horizontal Gene Transfer and Genetic Exchange in Mycobacteria. Microbiol Spectr. 2014;2:MGM2-0022-2013. doi: 10.1128/microbiolspec.MGM2-0022-2013. [DOI] [PubMed] [Google Scholar]
- Dumas E, Christina Boritsch E, Vandenbogaert M, Rodríguez de la Vega RC, Thiberge J-M, Caro V, Gaillard J-L, Heym B, Girard-Misguich F, Brosch R, Sapriel G. Mycobacterial Pan-Genome Analysis Suggests Important Role of Plasmids in the Radiation of Type VII Secretion Systems. Genome Biology and Evolution. 2016;8:387–402. doi: 10.1093/gbe/evw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution. Philos Trans R Soc Lond B Biol Sci. 2007;362:1185–1193. doi: 10.1098/rstb.2007.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM, Johnson CM. Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Curr Opin Microbiol. 2011;14:174–180. doi: 10.1016/j.mib.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint JL, Kowalski JC, Karnati PK, Derbyshire KM. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc Natl Acad Sci U S A. 2004;101:12598–12603. doi: 10.1073/pnas.0404892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- Gagneux S. Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol. 2018;16:202–213. doi: 10.1038/nrmicro.2018.8. [DOI] [PubMed] [Google Scholar]
- Garces A, Atmakuri K, Chase MR, Woodworth JS, Krastins B, Rothchild AC, Ramsdell TL, Lopez MF, Behar SM, Sarracino DA, Fortune SM. EspA acts as a critical mediator of ESX1-dependent virulence in Mycobacterium tuberculosis by affecting bacterial cell wall integrity. PLoS Pathog. 2010;6:e1000957. doi: 10.1371/journal.ppat.1000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey Van Pittius NC, Gamieldien J, Hide W, Brown GD, Siezen RJ, Beyers AD. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2001;2:1–18. doi: 10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubchik T, Brueggemann AB, Street T, Gertz RE, Jr, Spencer CC, Ho T, Giannoulatou E, Link-Gelles R, Harding RM, Beall B, Peto TE, Moore MR, Donnelly P, Crook DW, Bowden R. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat Genet. 2012;44:352–355. doi: 10.1038/ng.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Clark RR, Boucher N, Lapierre P, Smith C, Derbyshire KM. Intercellular communication and conjugation are mediated by ESX secretion systems in mycobacteria. Science. 2016;354:347–350. doi: 10.1126/science.aag0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Krywy JA, Harold J, Palumbo MJ, Derbyshire KM. Distributive conjugal transfer in mycobacteria generates progeny with meiotic-like genome-wide mosaicism, allowing mapping of a mating identity locus. PLoS Biol. 2013;11:e1001602. doi: 10.1371/journal.pbio.1001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat Rev Microbiol. 2016;14:677–691. doi: 10.1038/nrmicro.2016.131. [DOI] [PubMed] [Google Scholar]
- Gutierrez MC, Brisse S, Brosch R, Fabre M, Omais B, Marmiesse M, Supply P, Vincent V. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci U S A. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben EN, Korotkov KV, Bitter W. Take five - Type VII secretion systems of Mycobacteria. Biochim Biophys Acta. 2014;1843:1707–1716. doi: 10.1016/j.bbamcr.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Johnston C, Martin B, Fichant G, Polard P, Claverys JP. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol. 2014;12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- Krzywinska E, Krzywinski J, Schorey JS. Naturally occurring horizontal gene transfer and homologous recombination in Mycobacterium. Microbiology. 2004;150:1707–1712. doi: 10.1099/mic.0.27088-0. [DOI] [PubMed] [Google Scholar]
- Kulick S, Moccia C, Didelot X, Falush D, Kraft C, Suerbaum S. Mosaic DNA imports with interspersions of recipient sequence after natural transformation of Helicobacter pylori. PLoS One. 2008;3:e3797. doi: 10.1371/journal.pone.0003797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laencina L, Dubois V, Le Moigne V, Viljoen A, Majlessi L, Pritchard J, Bernut A, Piel L, Roux AL, Gaillard JL, Lombard B, Loew D, Rubin EJ, Brosch R, Kremer L, Herrmann JL, Girard-Misguich F. Identification of genes required for Mycobacterium abscessus growth in vivo with a prominent role of the ESX-4 locus. Proc Natl Acad Sci U S A. 2018 doi: 10.1073/pnas.1713195115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mell JC, Lee JY, Firme M, Sinha S, Redfield RJ. Extensive cotransformation of natural variation into chromosomes of naturally competent Haemophilus influenzae. G3 (Bethesda) 2014;4:717–731. doi: 10.1534/g3.113.009597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi Y, Suga K, Tokunaga T. Multiple mating types of Mycobacterium smegmatis. Japanese Journal of Microbiology. 1976;20:435–443. doi: 10.1111/j.1348-0421.1976.tb01009.x. [DOI] [PubMed] [Google Scholar]
- Mortimer TD, Pepperell CS. Genomic Signatures of Distributive Conjugal Transfer among Mycobacteria. Genome Biology and Evolution. 2014;6:2489–2500. doi: 10.1093/gbe/evu175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer TD, Weber AM, Pepperell CS. Evolutionary Thrift: Mycobacteria Repurpose Plasmid Diversity during Adaptation of Type VII Secretion Systems. Genome Biol Evol. 2017;9:398–413. doi: 10.1093/gbe/evx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Foot M, Warren RM, Sampson SL, van Helden PD, Gey van Pittius NC. The plasmid-mediated evolution of the mycobacterial ESX (Type VII) secretion systems. BMC Evol Biol. 2016;16:62. doi: 10.1186/s12862-016-0631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K, Piastro K, Gray TA, Derbyshire KM. Mycobacterial biofilms facilitate horizontal DNA transfer between strains of Mycobacterium smegmatis. J Bacteriol. 2010;192:5134–5142. doi: 10.1128/JB.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KT, Piastro K, Derbyshire KM. LpqM, a mycobacterial lipoprotein-metalloproteinase, is required for conjugal DNA transfer in Mycobacterium smegmatis. J Bacteriol. 2009;191:2721–2727. doi: 10.1128/JB.00024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohol YM, Goetz DH, Chan K, Shiloh MU, Craik CS, Cox JS. Mycobacterium tuberculosis MycP1 Protease Plays a Dual Role in Regulation of ESX-1 Secretion and Virulence. Cell Host & Microbe. 2010;7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MW, Jain P, Yang H, Mitra S, Biswas D, Wattam AR, Letvin NL, Jacobs WR., Jr Noncanonical SMC protein in Mycobacterium smegmatis restricts maintenance of Mycobacterium fortuitum plasmids. Proc Natl Acad Sci U S A. 2014;111:13264–13271. doi: 10.1073/pnas.1414207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons LM, Jankowski CS, Derbyshire KM. Conjugal transfer of chromosomal DNA in Mycobacterium smegmatis. Mol Micro. 1998;28:571–582. doi: 10.1046/j.1365-2958.1998.00818.x. [DOI] [PubMed] [Google Scholar]
- Pettis GS, Cohen SN. Transfer of the plJ101 plasmid in Streptomyces lividans requires a cis-acting function dispensable for chromosomal gene transfer. Mol Microbiol. 1994;13:955–964. doi: 10.1111/j.1365-2958.1994.tb00487.x. [DOI] [PubMed] [Google Scholar]
- Possoz C, Ribard C, Gagnat J, Pernodet JL, Guerineau M. The integrative element pSAM2 from Streptomyces: kinetics and mode of conjugal transfer. Mol Microbiol. 2001;42:159–166. doi: 10.1046/j.1365-2958.2001.02618.x. [DOI] [PubMed] [Google Scholar]
- Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol Microbiol. 2002:46. doi: 10.1046/j.1365-2958.2002.03237.x. [DOI] [PubMed] [Google Scholar]
- Reuther J, Gekeler C, Tiffert Y, Wohlleben W, Muth G. Unique conjugation mechanism in mycelial streptomycetes: a DNA-binding ATPase translocates unprocessed plasmid DNA at the hyphal tip. Mol Microbiol. 2006;61:436–446. doi: 10.1111/j.1365-2958.2006.05258.x. [DOI] [PubMed] [Google Scholar]
- Sapriel G, Konjek J, Orgeur M, Bouri L, Frezal L, Roux AL, Dumas E, Brosch R, Bouchier C, Brisse S, Vandenbogaert M, Thiberge JM, Caro V, Ngeow YF, Tan JL, Herrmann JL, Gaillard JL, Heym B, Wirth T. Genome-wide mosaicism within Mycobacterium abscessus: evolutionary and epidemiological implications. BMC Genomics. 2016;17:118. doi: 10.1186/s12864-016-2448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda E, Vogelmann J, Muth G. A septal chromosome segregator protein evolved into a conjugative DNA-translocator protein. Mob Genet Elements. 2011;1:225–229. doi: 10.4161/mge.1.3.18066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh MU, DiGiuseppe Champion PA. To Catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr Opin Microbiol. 2009;13:1–7. doi: 10.1016/j.mib.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NH, Hewinson RG, Kremer K, Brosch R, Gordon SV. Myths and misconceptions: the origin and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol. 2009;7:537–544. doi: 10.1038/nrmicro2165. [DOI] [PubMed] [Google Scholar]
- Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Molecular Microbiology. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, Fiette L, Orgeur M, Fabre M, Parmentier C, Frigui W, Simeone R, Boritsch EC, Debrie AS, Willery E, Walker D, Quail MA, Ma L, Bouchier C, Salvignol G, Sayes F, Cascioferro A, Seemann T, Barbe V, Locht C, Gutierrez MC, Leclerc C, Bentley SD, Stinear TP, Brisse S, Medigue C, Parkhill J, Cruveiller S, Brosch R. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet. 2013;45:172–179. doi: 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma L, Muth G. Conjugative DNA-transfer in Streptomyces, a mycelial organism. Plasmid. 2016;87–88:1–9. doi: 10.1016/j.plasmid.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Thoma L, Vollmer B, Muth G. Fluorescence microscopy of Streptomyces conjugation suggests DNA-transfer at the lateral walls and reveals the spreading of the plasmid in the recipient mycelium. Environmental Microbiology. 2016;18:598–608. doi: 10.1111/1462-2920.13027. [DOI] [PubMed] [Google Scholar]
- Tufariello JM, Chapman JR, Kerantzas CA, Wong K-W, Vilchèze C, Jones CM, Cole LE, Tinaztepe E, Thompson V, Fenyö D, Niederweis M, Ueberheide B, Philips JA, Jacobs WR. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proceedings of the National Academy of Sciences. 2016 doi: 10.1073/pnas.1523321113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ummels R, Abdallah AM, Kuiper V, Aajoud A, Sparrius M, Naeem R, Spaink HP, van Soolingen D, Pain A, Bitter W. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. MBio. 2014;5:e01744–01714. doi: 10.1128/mBio.01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soolingen D, Hoogenboezem T, de Haas PE, Hermans PW, Koedam MA, Teppema KS, Brennan PJ, Besra GS, Portaels F, Top J, Schouls LM, van Embden JD. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int J Syst Bacteriol. 1997;47:1236–1245. doi: 10.1099/00207713-47-4-1236. [DOI] [PubMed] [Google Scholar]
- Vogelmann J, Ammelburg M, Finger C, Guezguez J, Linke D, Flotenmeyer M, Stierhof YD, Wohlleben W, Muth G. Conjugal plasmid transfer in Streptomyces resembles bacterial chromosome segregation by FtsK/SpoIIIE. EMBO J. 2011;30:2246–2254. doi: 10.1038/emboj.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Karnati PK, Takacs CM, Kowalski JC, Derbyshire KM. Chromosomal DNA transfer in Mycobacterium smegmatis is mechanistically different from classical Hfr chromosomal DNA transfer. Mol Microbiol. 2005;58:280–288. doi: 10.1111/j.1365-2958.2005.04824.x. [DOI] [PubMed] [Google Scholar]
- Wollman EL, Jacob F, Hayes W. Conjugation and genetic recombination in Escherichia coli K-12. Cold Spring Harb Symp Quant Biol. 1956;21:141–162. doi: 10.1101/sqb.1956.021.01.012. [DOI] [PubMed] [Google Scholar]