Abstract
Boronic acid liposomes enable triggered content release and cell delivery driven by carbohydrate binding. Dye release assays using hydrophilic and hydrophobic fluorophores validate dose-dependent release upon carbohydrate treatment. Microscopy results indicate dramatic enhancements in cell delivery, showcasing the prospects of boronic acid lipids for drug delivery.
Liposomes have emerged as sophisticated carriers for delivering therapeutic cargo. Indeed, liposome-based drug delivery platforms have been approved by the FDA and many more are in the late-stage of clinical trials.1 To advance liposomal drug delivery, two important areas of research include the enhancement of cell infiltration by liposomes, and the ability to control cargo delivery by triggering the release of contents. For the former, unnatural cationic lipids such as 1,2-dioleoyl-3-trimethylammonium propane (DOTAP) or lipids conjugated to cell penetrating peptides (CPPs), such as octaarginine (R8), are commonly used to infiltrate membranes.2 However, these cationic lipids commonly exhibit significant toxicity.3 Regarding triggered release, approaches exploiting both active and passive stimuli, such as light, redox, pH, enzymes and temperature have been explored, but there are drawbacks in each case.4 Drawbacks of passive release include the minimal variations that are exploited for selective release, such as the slight increase in acidity in cancer cells (pH ~ 6.5-6.9) compared to healthy cells (pH ~ 7.2-7.4).5 Active release protocols are hindered by challenges in delivering external stimuli, such as poor tissue penetration using light-induced release. Herein, we report boronic acid liposomes as a means for enhancing both cell infiltration and content delivery based on carbohydrate binding interactions.
Aberrant glycosylation patterns, both in terms of carbohydrate composition and abundance,6 are linked with diseases such as oncogenic transformation. For example, glycosyltransferase dysregulation leads to increased sialylation of truncated gangliosides and overexpressed complex β-1,6-branched N-linked glycans on human melanoma cells.7 Such cell-type specific complex glycan alternations can provide a handle for selective cell targeting and delivery. The boronic acid molecular recognition unit has been extensively used to bind and separate carbohydrates through reversible formation of boronate esters.8 However, biological application of this sensing group is challenged by the relatively low binding affinity in aqueous media. This can be overcome through multivalent binding interactions, in which avidity effects lead to exponential enhancements in affinity.9 Smith and co-workers have previously shown that boronic acid lipids are effective at driving calcium-dependent liposome fusion10 and at enhancing the binding of cell membranes.11 In this article, we delve into the efficacy of boronic acid liposomes as a means to enhance both cellular infiltration and targeted content release driven by carbohydrate binding.
This project began with the design of boronic acid lipid conjugates to present this recognition group on the surface of resulting liposomes. One such compound is lipid 1, in which the boronic acid is directly attached onto an aminoglycerolipid scaffold. An ortho-(alkylaminomethyl)phenylboronic acid binding unit was chosen since the phenyl group is known to stabilize the boronic acid while the amino moiety enhances carbohydrate binding affinity at physiological conditions.12 The synthesis of 1, shown in Scheme 1, began with racemic 3-aminopropane-1,2-diol (2), the amine of which was first protected as a phthalimide in 3. Next, a Williamson ether synthesis was used to introduce hydrophobic alkyl chains. Ether-linked lipid chains were employed to circumvent potential hydrolysis by lipase enzymes in vivo. The pthalimide was next deprotected to produce 4, which was followed by a reductive amination reaction to produce 1. We additionally designed, synthesized and studied single-chain boronic acid lipid S1 analogous to a fatty acid (Scheme S1). This alternative lipid exhibited similar properties as 1 during release studies, with results reported in the supplementary information.
Scheme 1.
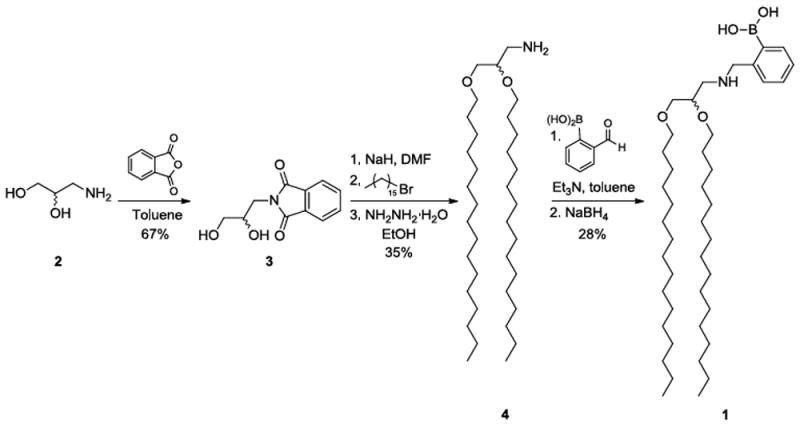
Synthetic route for boronic acid lipid 1.
We first evaluated triggered release from liposomes containing 1 or S1 upon treatment with the polysaccharide heparin as a model carbohydrate. Heparin is an anticoagulant consisting of repeating disaccharide units of sulphated iduronic acid/glucuronic acid and glucosamine residues, and has previously been shown to bind to boronic acids in a multivalent manner.13 Additionally, heparin sulfate proteoglycans have been implicated for driving the cellular entry of cationic liposomes through binding interactions.14 We initially examined the release of hydrophobic contents from liposomes using a Nile red release assay, in which liposomal solubilization of this insoluble dye gives way to fluorescence decreases when the dye is released into aqueous solution and precipitates.15 A cartoon depicting the release of both hydrophobic and hydrophilic contents is shown in Scheme 2.
Scheme 2.
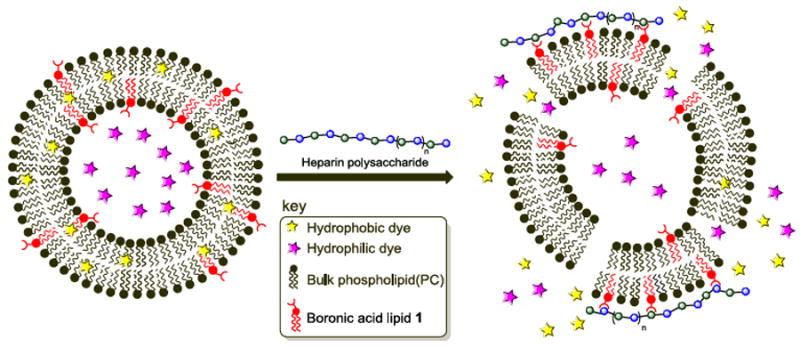
Fluorescence-based dye leakage assays driven by heparin-boronic acid lipid interactions. Hydrophobic (Nile red) or hydrophilic (sulforhodamine B) dyes are separately encapsulated within boronic acid liposome membrane bilayers or aqueous interiors, respectively. Heparin carbohydrate binding is evaluated for the release of contents leading to fluorescence decreases (Nile red) or increases (sulforhodamine B) attributed to membrane distortion upon carbohydrate binding.
Unilamellar liposomes composed of 0% to 25% of boronic acid lipid 1 doped into L-α-phosphatidylcholine (PC, mixed isomers) were first prepared with Nile red included in the formulation. Hydration, freeze-thaw, sonication and extrusion through 200 nm membranes were performed to generate liposomes with uniform size. As seen in Figure 1A, titration of Nile red-loaded liposomes with heparin led to a decrease in fluorescence in a manner that was dependent on the percentage of boronic acid lipid 1 incorporated within the liposomes. Specifically, control liposomes containing 0% of 1 showed minimal background release of Nile red (~5% fluorescence decrease), while this decrease was accentuated with increasing boronic acid lipids (5% 1, ~10% decrease; 10% 1, ~25%; and 20% 1, ~50%). The extent of release appeared to reach a plateau since 20% and 25% of 1 in liposomes yielded similar results. Control Nile red release and dynamic light scattering (DLS) studies in which compound 4 was instead included within liposomes showed minimal change (Figure S1), indicating the necessity of the boronic acid moiety of 1. In addition, titration with horseradish peroxidase (HRP) glycoprotein as an alternative model with different carbohydrate composition17 also yielded greater release using liposomes containing 1 (Figure S2), indicating generality and ruling out simple electrostatic binding. These data provide evidence that the boronic acid units presented on the liposomes bind to carbohydrates, thereby triggering Nile red release. In addition, the threshold-type response we observed suggests the formation of multivalent binding interactions that we expect to play a key role in binding and release.16
Figure 1.
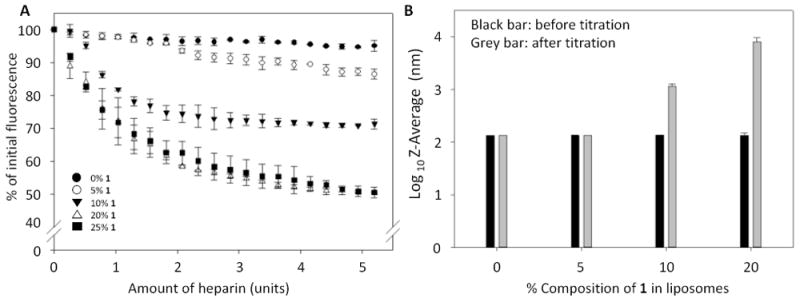
Boronic acid liposome Nile red release and DLS results upon heparin incubation. A. Decreases in Nile red fluorescence attributed to release are dependent upon the percentage of boronic acid lipid 1 in PC liposomes. B. DLS results show that untreated liposomes exhibit the expected sizes, while treatment of liposomes containing higher percentages (10-20%) of 1 with heparin leads to much larger particle sizes attributed to lipid reorganization. Error bars denote the standard errors of at least three replicates.
We anticipated that liposome release would be driven by lipid reorganization upon carbohydrate binding in a manner that perturbs membrane bilayer packing to stimulate content release. To assess this hypothesis, DLS experiments were conducted to probe for changes in the particle sizes of lipid assemblies before and after heparin treatment (Figure 1B). These are reported using an exponential scale. Here, the inclusion of 1 showed no effect on initial liposome size. Upon treatment with heparin, liposomes containing 0 or 5% 1 exhibited minimal changes in size, which matches the slight decrease in Nile red fluorescence observed during release studies. Significant increases in particle sizes were detected following heparin addition to liposomes containing 10-20% of 1. Finally, DLS studies of PC liposomes containing either 0% or 10% of 1 also showed no significant changes over the course of a week at room temperature (data not shown), indicating that these liposomes are robust.
To further explore the issue of changes in assembly, scanning transmission electron microscope (STEM) images were taken before and after heparin treatment. Here, standard liposome images were observed for samples containing 10% of 1 initially, while heparin treatment resulted in much larger and complex membrane assemblies (Figure S3). On the other hand, images of control PC-only liposomes treated with heparin showed liposomes with sizes similar to untreated samples. The DLS and STEM data provide evidence for lipid reorganization driven by heparin binding only when 1 is present, which could be explained by carbohydrate-promoted processes such as fusion, aggregation and/or the formation of different supramolecular lipid assemblies.
We next studied carbohydrate-induced release of the polar fluorescent dye sulforhodamine B.18 In this assay, the dye is encapsulated within the liposome aqueous core at high concentrations such that it is quenched, and size exclusion chromatography (SEC) is used to remove unencapsulated fluorophore. Dye release then leads to an increase in fluorescence intensity in bulk solution (Scheme 2). The extent of dye release in this assay is often variable based on the amount of soluble dye that is entrapped when liposomes form and the variability of liposome concentrations obtained by SEC purification. Thus, we normalized dye release as a percentage of total release caused by final treatment of the liposomes with the detergent Triton X-100. This assay culminated in dose-dependent increases of sulforhodamine B fluorescence based on the percentage of 1 incorporated in the liposomes (Figure 2). These results indicate that boronic acid 1 is also effective for triggered release of polar contents from liposomes, which is more challenging as this requires that polar/charged molecules escape the hydrophobic membrane barrier. Boronic acid lipids 1 and S1 exhibited very similar properties in both the Nile red and sulforhodamine release studies, and thus the results for S1 are shown in the supplementary information (Figures S4 and S5). Similarities in these data suggest that the particular lipid scaffold that anchors the boronic acid in the membrane is not critical for activity. Both lipids do possess ortho-(alkylaminomethyl)phenylboronic acid moieties known to enhance binding affinity at physiological pH.12
Figure 2.
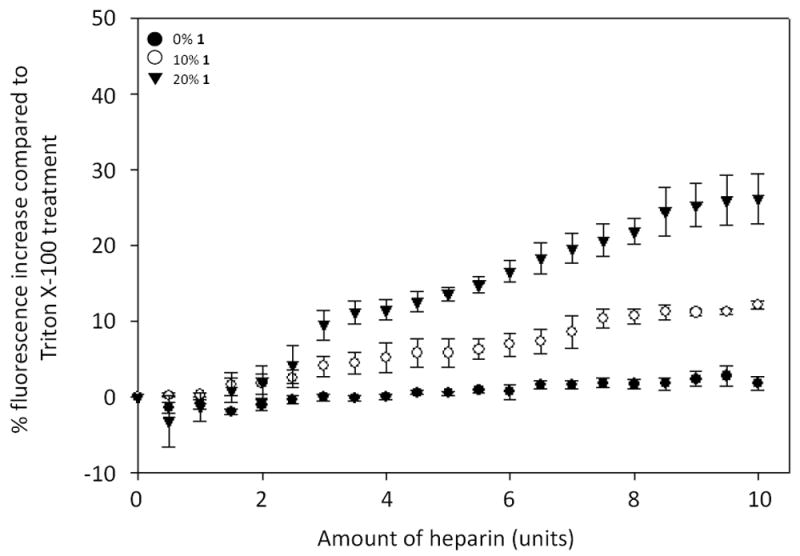
Boronic acid liposome sulforhodamine release results upon heparin titration. Fluorescence increases upon sulforhodamine B release, plotted as a percentage of increases caused by Triton X treatment, are shown to correlate with the percentage of 1 in PC liposomes. Error bars denote the standard errors of at least three replicates.
We next sought to verify that boronic acid liposomes bind to carbohydrates to further justify that release may be caused by molecular recognition. A microplate assay was used to assess the binding of fluorescent liposomes to a commercially available heparin–biotin conjugate immobilized onto streptavidin-coated microplates. We have previously used similar assays to study liposome and lipid binding interactions.19 Liposomes in this study contained either 0% (control) or 20% of 1, 1% rhodamine-labeled phosphatidylethanolamine (Rd-PE) as a fluorescent marker, with the remainder PC. The results (Figure 3) indicate a significant enhancement in surface binding when 1 was present in the liposomes, thereby validating the heparin-binding ability of liposomes containing 1. In an additional control, liposomes containing 1 yielded minimal signal when heparin-biotin was not added (data not shown).
Figure 3.
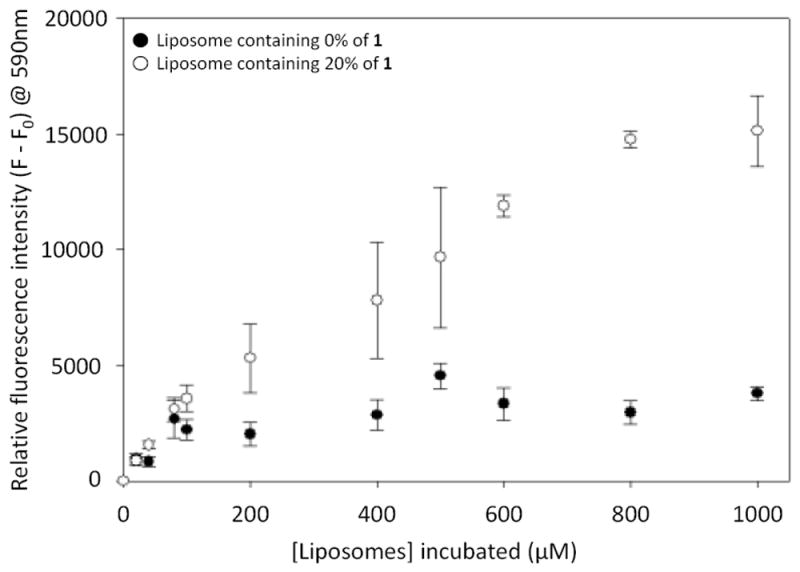
Results from microplate studies indicating the binding of fluorescent liposomes to immobilized heparin–biotin. Error bars indicate standard errors for at least three replicates.
We next assessed the ability of lipid 1 to promote liposome cell entry. Boronic acid liposomes were expected to bind to cell surface carbohydrates and enhance proximity to plasma membranes, thereby boosting cell entry pathways such as membrane fusion or pinocytosis (Figure 4A). Confocal fluorescence laser scanning microscopy experiments were used to evaluate the delivery of liposomes containing 0.08% of Rd-PE. Control liposomes with only PC and Rd-PE were compared to study samples doped with 10% of 1. Liposomes were incubated with A375 melanoma cells for one hour at 37°C, washed to remove residual free liposomes, labelled with DAPI, fixed and mounted on glass slides for imaging. Representative images are shown in Figure 4. Cells treated with liposomes containing 10% of 1 yielded a dramatic enhancement in fluorescence (Figure 4C) compared to control liposomes lacking 1 (Figure 4B). These results show that 1 is effective for driving liposomal cell entry. As an initial assessment of toxicity, cells treated with liposomes containing 10% 1 and 0.08% Rd-PE were observed to be healthy after incubation, while liposomes containing 15-20% of 1 with 0.08% Rd-PE led to observable toxicity effects including some cytoplasmic shrinking and nuclear distortion, particularly at longer incubation times, which may result from membrane disruption. While extensive studies are required to assess the clinical viability of this platform, these results showcase boronic acid lipid 1 as a promising agent for cellular delivery.
Figure 4. Cellular delivery studies.
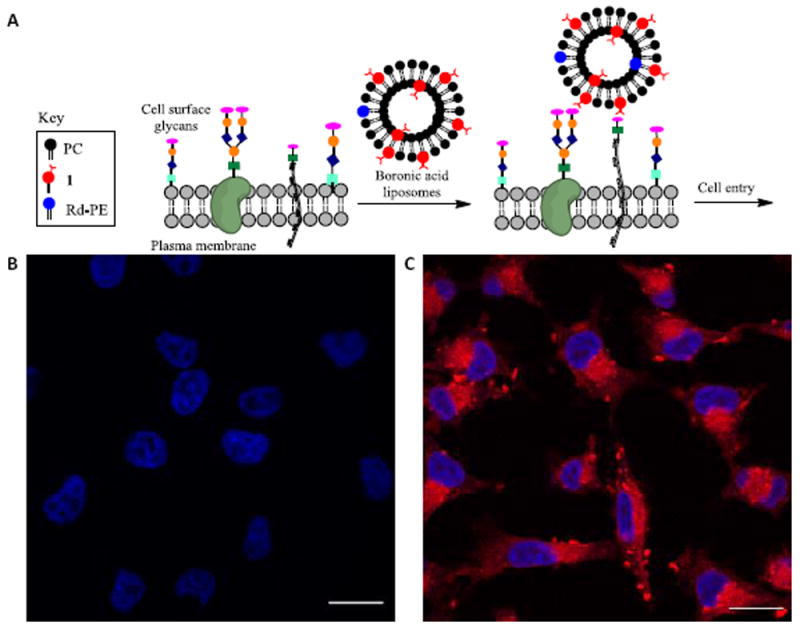
A. Cartoon depicting liposome cell entry driven by binding interactions with cell surface carbohydrates. B. Fluorescence image of cells treated with PC/Rd-PE liposomes. C. Image upon treatment with PC/1(10%)/Rd-PE liposomes. Scale bar depicts 20 μM. DAPI is shown in blue. Rd-PE is shown in red.
In conclusion, we have found that liposomes containing boronic acid lipid 1 are effective for content release and cell entry driven by carbohydrate binding. These liposomes could provide versatility by either delivering therapeutic cargo directly via cell entry or by releasing contents upon in close proximity to cells, both of which are likely to enhance delivery. While this provides a promising means for general cellular delivery applications, it also opens up the possibility of selective delivery to diseased cells based on the specific composition and abundance of cell surface carbohydrates. In particular, boronic acid dimers have been reported that exhibit selective binding to tumor markers such as sialyl Lewis X.20. Future work will be aimed at studying and enhancing cell type-specific cellular delivery using boronic acid moieties similar to these adapted for presentation on liposome surfaces.
Supplementary Material
Acknowledgments
This research was supported by funding from the National Institutes of Health (R15GM120705 and R01GM120642). SDH was supported by a National Science Foundation REU Award (CHE-1560033). The authors also thank Dr. John Dunlap for help with STEM imaging and Dr. Richard Bennett for editing.
Dedicated to the memory of John “Jack” Best
Footnotes
Electronic Supplementary Information (ESI) available: Experimental procedures, spectra for synthetic compounds and supplemental figures.
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Petros RA, DeSimone JM. Nat Rev Drug Disc. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]; Deshpande PP, Biswas S, Torchilin VP. Nanomedicine. 2013;8:1509–1528. doi: 10.2217/nnm.13.118. [DOI] [PMC free article] [PubMed] [Google Scholar]; Noble GT, Stefanick JF, Ashley JD, Kiziltepe T, Bilgicer B. Trends Biotechnol. 2014;32:32–45. doi: 10.1016/j.tibtech.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Gao H, Zhang Q, Yu Z, He Q. Curr Pharm Biotechnol. 2014;15:210–219. doi: 10.2174/1389201015666140617092552. [DOI] [PubMed] [Google Scholar]; Koren E, Torchilin VP. Trends Mol Med. 2012;18:385–393. doi: 10.1016/j.molmed.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Lv H, Zhang S, Wang B, Cui S, Yan J. J Control Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Alam S, Mattern-Schain SI, Best MD. In: Comprehensive Supramolecular Chemistry. Atwood JL, editor. Vol. 2. Elsevier; 2017. pp. 329–364. [Google Scholar]; Pattni BS, Chupin VV, Torchilin VP. Chem Rev. 2015;115:10938–10966. doi: 10.1021/acs.chemrev.5b00046. [DOI] [PubMed] [Google Scholar]; Zangabad PS, Mirkiani S, Shahsavari S, Masoudi B, Masroor M, Hamed H, Jafari Z, Taghipour YD, Hashemi H, Karimi M, Hamblin MR. Nanotechnol Rev. 2018;7:95–122. doi: 10.1515/ntrev-2017-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillies RJ, Liu Z, Bhujwalla Z. Am J Physiol. 1994;267:C195–C203. doi: 10.1152/ajpcell.1994.267.1.C195. [DOI] [PubMed] [Google Scholar]; van Sluis R, Bhujwalla ZM, Raghunand N, Ballesteros P, Alvarez J, Cerdan S, Galons JP, Gillies RJ. Magn Res Med. 1999;41:743–750. doi: 10.1002/(sici)1522-2594(199904)41:4<743::aid-mrm13>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 6.Kufe DW. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pinho SS, Reis CA. Nat Rev Cancer. 2015;15:540. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 7.Todeschini AR, DosSantos JN, Handa K, Hakomori S-i. J Biol Chem. 2007;282:8123–8133. doi: 10.1074/jbc.M611407200. [DOI] [PubMed] [Google Scholar]
- 8.Springsteen G, Wang BH. Tetrahedron. 2002;58:5291–5300. [Google Scholar]; Jin S, Cheng YF, Reid S, Li MY, Wang BH. Med Res Rev. 2010;30:171–257. doi: 10.1002/med.20155. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhai WL, Sun XL, James TD, Fossey JS. Chem Asian J. 2015;10:1836–1848. doi: 10.1002/asia.201500444. [DOI] [PubMed] [Google Scholar]
- 9.Brooks WLA, Sumerlin BS. Chem Rev. 2016;116:1375–1397. doi: 10.1021/acs.chemrev.5b00300. [DOI] [PubMed] [Google Scholar]
- 10.Zhang ZY, Smith BD. J Am Chem Soc. 1998;120:7141–7142. [Google Scholar]
- 11.Vandenburg YR, Zhang ZY, Fishkind DJ, Smith BD. Chem Commun. 2000:149–150. [Google Scholar]
- 12.Collins BE, Sorey S, Hargrove AE, Shabbir SH, Lynch VM, Anslyn EV. J Org Chem. 2009;74:4055–4060. doi: 10.1021/jo900187a. [DOI] [PubMed] [Google Scholar]; Lauer M, Wulff G. J Chem Soc Perkin Trans. 1987;2:745–749. [Google Scholar]
- 13.Wright AT, Zhong ZL, Anslyn EV. Angew Chem Int Ed. 2005;44:5679–5682. doi: 10.1002/anie.200501437. [DOI] [PubMed] [Google Scholar]; Zhong ZL, Anslyn EV. J Am Chem Soc. 2002;124:9014–9015. doi: 10.1021/ja020505k. [DOI] [PubMed] [Google Scholar]
- 14.Kopatz I, Remy JS, Behr JP. J Gene Med. 2004;6:769–776. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- 15.Liang XL, Yue XL, Dai ZF, Kikuchi J. Chem Commun. 2011;47:4751–4753. doi: 10.1039/c1cc00063b. [DOI] [PubMed] [Google Scholar]; Bayer AM, Alam S, Mattern-Schain SI, Best MD. Chem Eur J. 2014;20:3350–3357. doi: 10.1002/chem.201304094. [DOI] [PubMed] [Google Scholar]
- 16.Liang PH, Wang SK, Wong CH. J Am Chem Soc. 2007;129:11177–11184. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]
- 17.Kurosaka A, Yano A, Itoh N, Kuroda Y, Nakagawa T, Kawasaki T. J Biol Chem. 1991;266:4168–4172. [PubMed] [Google Scholar]; Yang BY, Gray JSS, Montgomery R. Carbohyd Res. 1996;287:203–212. doi: 10.1016/0008-6215(96)00073-0. [DOI] [PubMed] [Google Scholar]
- 18.Viricel W, Mbarek A, Leblond J. Angew Chemie Int Edit. 2015;54:12743–12747. doi: 10.1002/anie.201504661. [DOI] [PubMed] [Google Scholar]
- 19.Gong D, Smith MD, Manna D, Bostic HE, Cho W, Best MD. Bioconjugate Chem. 2009;20:310–316. doi: 10.1021/bc8004107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Losey EA, Smith MD, Meng M, Best MD. Bioconjugate Chem. 2009;20:376–383. doi: 10.1021/bc800414k. [DOI] [PubMed] [Google Scholar]; Rowland MM, Bostic HE, Gong D, Lucas N, Cho W, Best MD. Chem Phys Lipids. 2012;165:207–215. doi: 10.1016/j.chemphyslip.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang WQ, Gao SH, Gao XM, Karnati VVR, Ni WJ, Wang BH, Hooks WB, Carson J, Weston B. Bioorg Med Chem Lett. 2002;12:2175–2177. doi: 10.1016/s0960-894x(02)00339-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


