Summary
Background
Eosinophilic esophagitis (EoE) is understood in terms of quantifiable histologic, endoscopic, and molecular features. There are limited data on interrelations of these features and their potential to identify distinct disease endotypes.
Methods
In this cross-sectional study, esophageal biopsies (n=185) from pediatric and adult EoE subjects across 10 sites associated with the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR) were analyzed using the EoE Diagnostic Panel (EDP), a set of 95 informative transcripts. Histologic and endoscopic features were assessed by quantification of esophageal eosinophils, the EoE histology scoring system (HSS), and the EoE endoscopic reference score (EREFS). Associations among the various histologic, endoscopic, and molecular features were analyzed with Spearman correlation. Results were replicated in a biologically independent single center validation cohort of active EoE subjects (n=100).
Findings
The EDP showed intersite consistency, significant correlation with esophageal eosinophils (p<0·0001), and similar findings between pediatric and adult subjects. Of eight HSS domains, basal zone hyperplasia (BZH) correlated with the EDP (median Spearman ρ 0·47, interquartile range [IQR] 0·36–0·60). Of five EREFS features, furrows correlated with the EDP (median Spearman ρ 0·42, IQR 0·32–0·50). By analyzing active EoE, the EDP identified three clusters associated with distinct endotypes (termed EoEe1-3) despite similar eosinophil levels. EoEe1 was associated with a normal-appearing esophagus (risk ratio [RR] 3·27, 95% confidence intervals [CI] 1·04–10·27, p=0·0443) and showed relatively mild histologic, endoscopic and molecular changes. EoEe2 demonstrated an inflammatory and steroid-refractory phenotype (RR 2·77, 95% CI 1·11–6·95, p=0·0376), and showed the highest expression of cytokines and steroid-responding genes. EoEe3 was associated with a narrow-caliber esophagus (RR 7·98, 95% CI 1·84–34·64, p=0·0013), and showed the highest degree of endoscopic and histologic severity and the lowest expression of epithelial differentiation genes. These endotypes were replicated in a validation cohort by clustering and with an EoE endotype-prediction algorithm.
Interpretation
Differential molecular signatures established correlations among BZH, endoscopic furrows, and esophageal transcripts and identified three EoE endotypes, with likely clinical significance.
Funding
National Institutes of Health (In full).
Introduction
Eosinophilic esophagitis (EoE) is an emerging disease characterized by marked esophagus-specific eosinophilia that is typically driven by allergic sensitization to a variety of common foods.1,2 Diagnosis is dependent upon quantitative assessment of esophageal levels of eosinophils (i.e., a peak eosinophil count of ≥15 intraepithelial eosinophils in one high-power field [HPF]).3 Although the gold standard for diagnosing disease and monitoring disease activity is the esophageal eosinophil level, recent advances have identified the potential value of a deeper analysis based on a wide range of quantifiable molecular, endoscopic and histologic parameters.4,5 In particular, a unique EoE transcriptome, referred to as the EoE Diagnostic Panel (EDP), a set of 95 esophageal transcripts that distinguishes EoE from control individuals including those with gastroesophageal reflux disease, correlates with distinct disease features and can identify EoE amongst ambiguous cases.6 In addition, a deeper histologic assessment called the EoE histologic scoring system (HSS) has been described and takes into account disease stage and grade across eight different parameters beyond peak eosinophil levels.7 Finally, EoE endoscopic reference scoring (EREFS), which takes into account five endoscopic features (i.e., edema, concentric rings, white plaques/exudates, longitudinal furrows, and strictures), has significance in terms of understanding the clinical features and monitoring the effect of therapy in both children and adults.8,9
An outstanding need in the EoE field is to define the relationships among these various clinical, endoscopic, and histologic features (especially the gold standard of the disease, the esophageal eosinophil level) and the degree of patient heterogeneity, since current therapy is not governed by specific disease features.10 Although a fibrostenotic phenotype has been associated with a subset of subjects with EoE, its molecular features, particularly in comparison to a non-fibrostenotic phenotype, have not yet been determined. At present, EoE is treated by food elimination trials focused on the most highly allergenic foods, and topical glucocorticoid therapy, but only a minority achieved deep remission.11 It is not known whether these therapies, as well as emerging anti-IgE12 and anti-cytokine therapeutics (e.g., anti–IL-5, anti–IL-13, and anti–IL-4Rα), should be used in all subjects or only in subgroups of subjects.2 Understanding the relationships between the clinical and molecular features and their heterogeneity in patient subgroups has potential for implementation of personalized molecular medicine approaches.
In the case of respiratory allergic diseases, which often co-exist in subjects with EoE and share some common features including disease mechanisms, there is emerging evidence that distinct disease endotypes, i.e., disease subtypes defined by molecular and cellular markers involved in distinct mechanisms,13 have considerable significance in terms of prognosis and response to therapy, including recently approved biologic agents such as anti-eosinophil therapies.
Herein, we aimed to identify clinical-pathologic correlations between endoscopic and histologic disease parameters by transcription profiling of the esophagus of subjects with varying EoE severity and disease activity states. To approach this aim, we examined a cohort of EoE subjects across multiple sites associated with the Consortium of Eosinophilic Disease Researchers (CEGIR)14 by performing detailed analyses of the EDP, HSS, and EREFS, and their relationships.
Methods
Study design and participants
This study was conducted within the wider context of CEGIR, which is part of the Rare Diseases Clinical Research Network (RDCRN), an initiative of the Office of Rare Diseases Research (ORDR), National Center for Advancing Translational Sciences (NCATS).14 For the discovery cohort, between 19th November 2015 – 21th February 2017, children and adults with EoE (≥3 years of age) were enrolled in a multicenter prospective observational study associated with CEGIR. Data were entered and managed by the Data Management and Coordinating Center (DMCC) associated with the RDCRN. Subjects with EoE were defined as having symptomatic esophageal dysfunction and a peak count of 15 or more esophageal eosinophils/HPF.2,3 For the validation cohort, between 2nd February 2004 – 7th March 2016, children and adults with active EoE (≥3 years of age) presenting for standard care were enrolled in a biologically independent cohort at Cincinnati Children’s Hospital Medical Center with the same disease definition. Subjects in the validation cohort were not in the discovery cohort. This study was approved by the institutional review boards of the participating institutions via a central institutional review board at Cincinnati Children’s Hospital Medical Center.
Procedures
Distal esophageal biopsies were used throughout the study, since this is typically obtained during endoscopy, represents the conventional location of biopsies and allows ready comparison to the previous transcriptomic studies, which were generally limited to this region. Considering the potential patchiness, 2–3 biopsies were taken from the distal esophagus (2–4 cm above the lower esophageal sphincter). Transcriptomic signatures in distal esophageal biopsy samples were obtained using an EDP as previously reported.6 Histologic and endoscopic features were assessed by peak eosinophil counts, the EoE HSS7, and EREFS.8 Clinical features of subjects were captured across sites by the CEGIR questionnaires, which include self-reported demographic, race/ethnicity, and clinical variables. Clinical phenotypes were defined using previously reported metrics.15–17 Steroid-sensitivity/-refractoriness was determined using a positive/negative response, respectively, to whether swallowed topical steroids have been effective on the basis of the pathology (see the REGID [Registry for Eosinophilic Gastrointestinal Disease] questionnaire in the appendix p 30).
We evaluated for associations between peak eosinophil counts and disease parameters (EDP, HSS, and EREFS). Furthermore, the EDP (either as a whole or individual genes) was examined for associations with the HSS and EREFS features. Spearman correlation analysis was performed between the gene expression levels on the EDP and the HSS and EREFS features.
EDP data from subjects with active EoE were further examined by unbiased/unsupervised clustering. Consensus clustering was performed by the partition-around-medoids (PAM) algorithm using the Euclidean distance.18 Bootstrapping was performed by randomly removing 10% of the data and repeating the clustering a total of 1,000 times. To determine the optimal number of clusters (k), stability was assessed using the cumulative distribution function (CDF),19 cluster-consensus (CLC) values19 and silhouette width analysis.20 To identify specific clinical associations for each cluster, strength of associations were measured. Multiple correspondence analysis (MCA) was also performed to present the pattern of relationships among the clusters and several phenotypes. To develop the algorithm for identifying EoE endotypes, stepwise linear discriminant analysis was performed with a stepwise selection method. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were measured to evaluate the diagnostic accuracy of the algorithm. Results were replicated in the validation cohort. See the appendix (pp 3–8) for further description of the methodology and analyses.
Outcomes
The primary objectives of this study were to identify relationships between the various endoscopic, histologic, and molecular features and to determine whether EoE endotypes exist and their significance in terms of histologic, endoscopic, and clinical features.
Statistical analysis
Data are n (%) or median (interquartile range [IQR]) unless otherwise stated. Missing data were excluded from all formal statistical analyses. Sample size required for endotype analysis was estimated by establishing subjects greater than five times the number of independent variables measured (appendix p 3), and was not pre-planned. Statistical analyses were performed using the JMP v13·0 (SAS Institute, Cary, NC), GeneSpring GX 12·6 (Agilent Technologies, Santa Clara, CA), GraphPad Prism 7 (GraphPad Software, Inc., San Diego, CA), and the R statistical computing environment (version 3·1·2). Correlation analysis was performed using Spearman’s rank correlation coefficient followed by Bonferroni adjustment. Risk ratios (RR) and 95% confidence intervals (CI) were calculated for each endotype with reference to all other endotypes. To compare differences between endotypes, the Kruskal-Wallis test and Dunn’s multiple comparison test were used for nonparametric continuous variables, and chi-square tests for categorical variables. A significant p value was defined as less than 0·05 (further statistical description is in the appendix pp 3–8).
Role of the funding source
The funders of the study, the National Institutes of Health (NIH), were involved in the study design, data collection, data analysis, data interpretation, and writing of the report. The authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Gene expression profiles from a total of 285 esophageal biopsy samples were analyzed, comprised of the discovery cohort of 185 individual subjects from 10 clinical sites and the validation cohort of 100 individual subjects from a single site (appendix p 22). The basic demographic characteristics are detailed in Table 1 and the appendix (p 15).
Table 1.
Basic characteristics of subjects in the discovery and validation cohorts
| Discovery cohort (N=185) | Validation cohort (N=100) | ||
|---|---|---|---|
| Demographics | |||
| Age at biopsy | 18·3 (8·8–37·1) | 10·2 (6·3–15·2) | |
| Gender | Male | 125 (67·6%) | 80 (80·0%) |
| Race | White | 170 (91·9%) | 95 (95·0%) |
| History of eosinophilic gastrointestinal diseases | |||
| History of EoE | 185 (100%) | 100 (100%) | |
| History of eosinophilic gastritis | 4 (2·2%) | 6 (6·0%) | |
| History of eosinophilic colitis | 4 (2·2%) | 0 (0%) | |
| Treatment at biopsy | |||
| Current PPI treatment | 62 (33·5%) | 84 (84·0%) | |
| Current topical steroid treatment | 95 (51·4%) | 58 (58·0%) | |
| Ongoing diet therapy | 97 (52·4%) | 63 (63·0%) | |
| Disease parameters at biopsy | |||
| Peak eosinophil counts | 15 (1–46) | 68 (34·3–143·8) | |
| EoE score from EDP | 242·2 (67·1–352·0) | 125·9 (67·4–248·9) | |
| HSS total score | 0·5 (0·2–0·9) | 0·8 (0·5–1·0) | |
| EREFS total Score | 2·0 (0–6·0) | 1·0 (1·0–2·0)* | |
Data are n (%) or median (IQR).
Simplified endoscopic severity score (ESS) was used.
EDP=EoE diagnostic panel. EoE=eosinophilic esophagitis. HSS=EoE histology scoring system. EREFS=EoE endoscopic reference score. PPI=Proton pump inhibitor.
The 185 subjects in the discovery cohort had an age range of 3·5 to 69·6 years, and consisted of 88 children and 97 adults. The pediatric and adult groups both exhibited a male predominance. Although there were different proportions of pediatric-onset versus adult-onset EoE, the length of time from the initial EoE diagnosis to the biopsy sample collection was similar in the pediatric and adult individuals. Peak eosinophil counts ranged from 0 to 174 eosinophils/HPF. Approximately half of the subjects (46·5%, n=86) had active EoE (≥15 eosinophils/ HPF) and 20·5% of the subjects (n=38) had biopsy specimens without eosinophils. There were no significant differences in peak eosinophil counts between the pediatric and adult individuals with active EoE; however, some clinical and endoscopic findings differed by age. Among the subjects with active EoE, nearly all subjects (91·9%, n=79/86) were from PPI-resistant EoE. A larger proportion of the pediatric subjects had an inflammatory phenotype at endoscopy than did the adults (p=0·0144), and significantly more adults had a fibrostenotic phenotype than did pediatric individuals (p<0·0001): the phenotype definitions are defined in the appendix (p 13). There was no significant difference in the EDP score or HSS score between the pediatric and adult individuals with active EoE, whereas the EREFS total score was significantly higher in adults than children (p<0·0001). The distributions of the different types of therapy were similar between the pediatric and adult populations. Among the subjects with a history of swallowed steroid treatment (n=91), there were fewer subjects in the category of steroid-refractory (n=25/91, 27·5%) than steroid-sensitive (n=66/91, 72·5%), and this distribution was similar in the pediatric and adult individuals (p=0·8155).
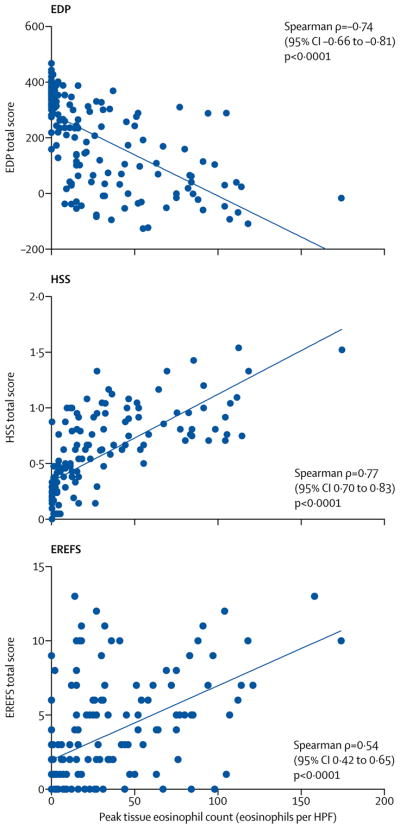
The EDP showed consistency across sites (Spearman ρ= −0·73 to −0·80, p<0·0001) and had similar values across pediatric and adult subjects with EoE (p=0·1125) (appendix p 24). To define the relationships among various clinical, endoscopic, and histologic features in relation to the accepted gold standard of assessing disease activity, i.e., the esophageal eosinophil level, we evaluated the associations between the peak eosinophil counts and the disease parameters (EDP, HSS, and EREFS). Using total scores, which represent the overall values of each platform, we found significant correlations between the peak eosinophil counts and each platform (EoE EDP total score: Spearman ρ= −0·74 (95% CI −0·66 – −0·81), p=1·0E-29; HSS total score: Spearman ρ=0·77 (95% CI 0·70 – 0·83), p=2·0E-33; and EREFS total score: Spearman ρ=0·54 (95% CI 0·42 – 0·65), p=3·0E-13, respectively) (Figure 1, appendix p 25). We subsequently focused on the individual components of the HSS and EREFS and their relationships with the individual expression level of the 95 EDP genes (overall EDP). There were associations between the overall EDP and several HSS domains (absolute median Spearman ρ=0·30 [IQR, 0·20–0·40]) (Figure 2). In particular, the basal zone hyperplasia (BZH) domain from the distal esophagus exhibited the highest magnitude of correlation with the overall EDP (absolute median Spearman ρ=0·47 [IQR, 0·36–0·60]). There were associations between several EREFS domains and the overall EDP (absolute median Spearman ρ=0·25; [IQR, 0·12–0·38]) (Figure 2). In particular, distal furrows, as a single endoscopic feature, exhibited the highest magnitude of correlation with the overall EDP (absolute median Spearman ρ=0·42 [IQR: 0·32–0·50]). A clustering tree based on the Spearman correlations showed their hierarchic relationships, supporting that the HSS and EREFS features aligned with the biological features associated with EoE (appendix p 26).
Figure 1.
Associations between the peak esophageal eosinophil counts in EoE and diagnostic platforms. A linear correlation is seen between the peak esophageal eosinophils/HPF and the total score from the EDP (left), HSS (middle) and EREFS (right), with Spearman ρ, 95% confidence intervals (CI), and p-values shown. EDP=EoE diagnostic panel. EoE=eosinophilic esophagitis. HSS=EoE histology scoring system. EREFS=EoE endoscopic reference score. HPF=high-power field.
Figure 2.
Associations between the EDP and the HSS domains and EREFS features. Spearman correlation analysis between the gene levels on the EDP and HSS domains (left) and the EREFS features (right), using the absolute value to account for differences in the direction of the effect across genes. Statistical significance was calculated with the Kruskal-Wallis test and Dunn’s post-hoc test. *p<0·0001 vs EA, ESL, DEC, DIS, SEA and LPF. †p<0·05 vs edema, exudates, rings and stricture. EDP=EoE diagnostic panel. EoE=eosinophilic esophagitis. HSS=EoE histology scoring system. EREFS=EoE endoscopic reference score. EI=eosinophilic inflammation. BZH=basal zone hyperplasia. EA=eosinophilic abscess. ESL=eosinophilic surface layering. DIS=dilated intercellular spaces. SEA=surface epithelial alteration. DEC=dyskeratotic epithelial cells. LPF=lamina propria fibers.
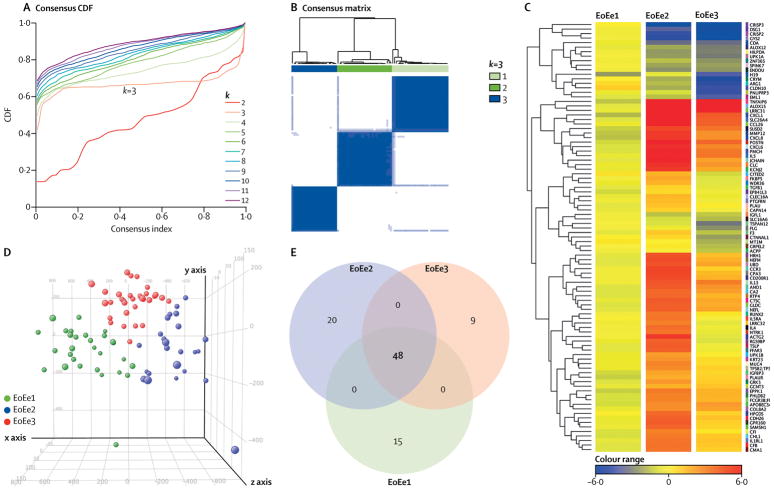
To determine if EoE demonstrates heterogeneous molecular profiling, we focused on analyzing the subjects with active EoE (n=86, we did not include samples from subjects with inactive EoE). Consensus clustering based solely on the EDP was examined to assess stability for a number of potential cluster numbers varying from 2 to 12. This established 3 stable groups (i.e., endotypes, referred to as EoEe1-3) after resampling, as defined by a flat middle part of the consensus CDF (Figure 3A) and well-defined squares within the consensus matrix (Figure 3B), in addition to the CLC values and silhouette widths (appendix p 27). Although a few clinical sites enrolled a larger number of subjects, there was no significant difference in the distribution of the endotypes at any given site (appendix p 16). On the heat map and 3-dimensional plot by principal component analysis (PCA), these endotypes were well separated from each other (Figures 3C and D). As a control, multiple biopsies obtained from the same endoscopy maintained each separate endotype (n=4, each endotype) (appendix p 28). Differentially expressed genes between each endotype were identified using a Benjamini-Hochberg false discovery rate of less than 0·05 and greater than 2-fold change. Using this threshold, differentially expressed genes totaled 15 in EoEe1, 20 in EoEe2, and 9 in EoEe3 (Figure 3E, Table 2).
Figure 3.
Clustering analysis of the active EoE group in the discovery cohort. (A) Consensus cumulative distribution function (CDF) with increasing number of clusters (k2 to k12). (B) Unsupervised consensus clustering of the active EoE showed optimal partitioning to 3 clusters (endotypes). (C) Comparison of esophageal transcriptomes by endotype. Heat maps were generated on the basis of the 95 EDP genes. (D) A 3-dimensional plot containing sample points from the 3 endotypes was derived from principal component analysis (PCA) of the entities demonstrated in the heat map to visualize the geometric distance between any given samples. (E) Venn diagrams comparing the number of genes identified as differentially expressed genes (adjusted p<0·05 and 2-fold change) that characterize the 3 endotypes. EDP=EoE diagnostic panel. EoE=eosinophilic esophagitis. EoEe=eosinophilic esophagitis endotype.
Table 2.
Differentially expressed genes between each endotype (≥ 2-fold change and p < 0·05)
| Gene symbol | EoEe1 | Adjusted p-value | Gene symbol | EoEe2 | Adjusted p-value | Gene symbol | EoEe3 | Adjusted p-value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||
| FC (EoEe1 vs. EoEe2) | FC (EoEe1 vs. EoEe3) | FC (EoEe2 vs. EoEe1) | FC (EoEe2 vs. EoEe3) | FC (EoEe3 vs. EoEe1) | FC (EoEe3 vs. EoEe2) | ||||||
| ALOX12 | 3·3 | 6·3 | 6·0E-06 | ACTG2 | 28·9 | 44·2 | 2·0E-07 | ACPP | −2·8 | −3·1 | 9·2E-09 |
| ALOX15 | −263·1 | −176·4 | 3·3E-11 | CCR3 | 24·0 | 12·9 | 8·7E-11 | CITED2 | −2·9 | −5·7 | 7·6E-08 |
| APOBEC3A | −11·0 | −8·3 | 6·8E-08 | CLEC16A | 3·3 | 3·0 | 1·5E-07 | CTNNAL1 | −5·9 | −8·0 | 2·9E-10 |
| CDA | 10·4 | 10·1 | 7·0E-09 | FFAR3 | 18·0 | 14·5 | 1·6E-06 | EML1 | −10·7 | −5·6 | 1·6E-06 |
| COL8A2 | −6·3 | −4·3 | 2·8E-04 | FKBP5 | 3·2 | 5·1 | 1·0E-05 | FLG | −6·4 | −5·5 | 1·3E-07 |
| CRISP3 | 307·6 | 540·6 | 6·0E-11 | HPGDS | 9·1 | 6·1 | 1·8E-07 | GRPEL2 | −3·4 | −4·0 | 2·7E-09 |
| ENDOU | 6·1 | 5·6 | 1·0E-08 | IL4 | 9·9 | 10·8 | 1·0E-08 | MT1M | −4·9 | −4·5 | 1·2E-05 |
| EPB41L3 | −3·3 | −2.0 | 1·5E-02 | IL5RA | 7·3 | 7·8 | 1·1E-06 | PNLIPRP3 | −21·8 | −12·8 | 2·4E-09 |
| GCNT3 | −3·6 | −2·7 | 8·0E-07 | KRT23 | 5·4 | 3·1 | 1·2E-05 | TSPAN12 | −10·1 | −5·3 | 2·0E-07 |
| HILPDA | 5·3 | 7·5 | 3·9E-09 | LRRC32 | 7·1 | 9·2 | 1·4E-07 | ||||
| IGFL1 | 2·9 | 2·7 | 1·4E-03 | MUC4 | 7·1 | 4·2 | 2·3E-08 | ||||
| PLAUR | −7·3 | −4·4 | 9·0E-07 | NTRK1 | 8·4 | 10·6 | 2·3E-08 | ||||
| SPINK7 | 4·9 | 5·3 | 1·9E-05 | PTGFRN | 2·5 | 2·4 | 1·2E-07 | ||||
| UPK1A | 5·4 | 9·1 | 4·3E-06 | RGS9BP | 14·8 | 21·1 | 3·4E-06 | ||||
| ZNF365 | 2·8 | 5·5 | 3·5E-06 | RUNX2 | 7·9 | 6·1 | 2·7E-08 | ||||
| SAMSN1 | 13·4 | 7·2 | 1·0E-08 | ||||||||
| TGFB1 | 2·5 | 2·2 | 2·3E-05 | ||||||||
| TSLP | 20·8 | 14·6 | 1·6E-06 | ||||||||
| UPK1B | 4·6 | 3·3 | 3·1E-04 | ||||||||
| WDR36 | 3·2 | 4·1 | 8·1E-09 | ||||||||
EoE=eosinophilic esophagitis. EoEe=eosinophilic esophagitis endotype. FC=fold change.
The clinical and demographic characteristics (which were not included in the consensus clustering) for each EoE endotype are described below and detailed (appendix pp 17–18). The 3 endotypes did not differ significantly by their peak eosinophil level, age at time of biopsy collection, gender, race, or length of time from diagnosis of EoE to biopsy collection. To address whether the identified endotypes have histologic and endoscopic distinctions, we evaluated the association of each endotype with several disease parameters (peak eosinophil count, EDP, HSS, and EREFS). Notably, the EDP, HSS, and EREFS parameters were associated with the endotype classification (Figure 4A). Among the HSS domains, the BZH and surface epithelial alteration (SEA) domains showed the most significant association with endotype (Figure 4B). BZH was significantly higher in EoEe2 compared to EoEe1 (p=0·0004), whereas SEA was significantly higher in EoEe3 compared to EoEe1 (p=0·0178). Among the EREFS features, edema, exudates and furrows showed significant association with endotype (Figure 4B). Endoscopic edema was significantly higher in EoEe2 and EoEe3 compared to EoEe1 (p=0·0096 and p=0·0017, respectively). Occurrence of exudates was significantly higher in EoEe2 compared to EoEe1 (p=0·0317), whereas occurrence of furrows was significantly higher in EoEe3 compared to EoEe1 (p=0·0163).
Figure 4.
The clinical features of each EoE endotype. (A) Comparison of each EoE endotype by diagnostic platform. Peak esophageal eosinophil counts (upper left), EoE score from EDP (upper right), HSS scores (lower left) and EREFS score (lower right) in each EoE endotype. Data are medians (IQR). Each dot represents an individual subject. Statistical significance was calculated with the Kruskal-Wallis test and Dunn’s post-hoc test. * p<0·05, **p<0·01, ***p<0·001, ****p<0·0001, vs EoEe1. (B) Comparison of each HSS domain in each EoE endotype (upper) and each EREFS feature in each EoE endotype (lower). Data are means ± SEM. (C) Summary of significant associations for each endotype. The dashed line in the forest plots indicates a risk ratio of 1. (D) Multiple correspondence analyses plot of the relationships between clinical phenotypes and endotypes. Distance between variables (phenotype and endotype) indicates the approximate relationship between variables. The distance between variables is inversely proportional to the strength of the relationship. EDP=EoE diagnostic panel. EoE=eosinophilic esophagitis. EoEe=eosinophilic esophagitis endotype. HSS=EoE histology scoring system, EREFS=EoE endoscopic reference score. HPF=high-power field. RR=risk ratio. CI= confidence interval.
To determine which clinical characteristics were specific to each endotype, strength of associations were measured (appendix p 19). EoEe1 was associated with a normal-appearing esophagus (RR=3·27, 95%CI 1·04–10·27, p=0·0443) and inversely associated with a history of esophageal dilation (RR=0·27, 95% CI 0·09–0·82, p=0·0105). EoEe2 was associated with steroid refractoriness (RR=2·77, 95% CI 1·11–6·95, p=0·0376). EoEe3 was associated with the presence of a narrow-caliber esophagus (RR=7·98, 95% CI 1·84–34·64, p=0·0013) and adult-onset of disease (RR=2·22, 95% CI 1·19–4·12, p=0·0155). (Figure 4C). Furthermore, MCA was performed to demonstrate the pattern of the endotypes with regard to the clinical features (Figure 4D). EoEe1 was situated near atopic, steroid-sensitivity, and normal endoscopic appearance. EoEe2 was situated close to pediatric-onset, inflammatory, and steroid-refractory phenotypes, whereas EoEe3 was located near non-atopic, adult-onset, and fibrostenotic phenotypes.
To facilitate potential translation to clinical practice, a clinically reproducible method to identify endotypes was developed (appendix p 8). Stepwise linear discriminant analysis using the same 95 EDP genes for active EoE (including EoEe1-3) identified the 8 strongest discriminatory genes (Figure 5A). Using these 8 genes, 84 (97·8%) subjects in the discovery cohort were assigned to the appropriate endotype. The three endotypes were well discriminated from each other with excellent diagnostic accuracy (Figure 5B).
Figure 5.
EoE endotype prediction based on machine learning with high accuracy. (A) Stepwise discriminant analysis shows the 15 strongest discriminatory genes for cluster assignment. (B) Canonical plot in which subjects are plotted in a 2-dimensional space. Each dot represents an individual subject. A 95% confidence level ellipse (inner) and an ellipse denoting a 50% contour (outer) are plotted for each group. The flow of the analysis was graphed in the lower table. The diagnostic accuracy was summarized in the right table. EDP=EoE diagnostic panel. EoE=eosinophilic esophagitis. EoEe=eosinophilic esophagitis endotype. PPV=positive predictive value. NPV=negative predictive value.
To further validate the EoE endotype findings, the same analysis was performed on the validation cohort (Table 1, appendix pp 8,22). Two separate strategies (consensus clustering based solely on the EDP, and endotype prediction based on the 8 genes) were used for assignment of endotype, and then the validation cohort (n=100) was compared to the discovery cohort. For validation of endotypes by clustering, similar to the discovery cohort, subjects with active EoE (n=60) in the validation cohort segregated into the three endotypes with optimal quality and stability (appendix p 27). Aside from the clustering, subjects with active EoE (n=40) in the validation cohort were assigned to the three endotypes on the basis of the results of the endotype-prediction algorithm developed with the discovery cohort (appendix p 29). Notably, the endotypes generated from the validation cohort were similar to the discovery cohort in that the gene expression relationships among the endotypes were maintained. Furthermore, differences in the EoE scores and several gene expression levels among the endotypes were similar between the validation and discovery cohorts. The three endotypes in the validation cohort had peak eosinophil counts that were not statistically different. Also, consistent with the discovery cohort, the three endotypes showed similar differential trends in the clinical and endoscopic findings (appendix p 29).
Discussion
Herein, we dissected EoE disease molecular heterogeneity via the EDP, across a multi-site cohort of subjects associated with CEGIR, and assessed its relevance using a combination of standardized histologic, endoscopic, and clinical platforms. First, we demonstrate that the EDP showed consistency across sites, had similar findings between pediatric and adult EoE subjects, and correlated with esophageal eosinophil levels. Second, we report the existence of three disease endotypes and present evidence for their clinical, histologic, and endoscopic significance. Notably, these disease endotypes were shown to be stable using distinct statistical methodology including unsupervised clustering, 3-dimensional PCA, and cumulative distribution functionality. Third, disease endotypes occurred independent of peak eosinophil counts, underscoring that these findings surpass the information provided by eosinophil counts alone, consistent with prior findings that disease severity and clinical symptoms do not simply reflect eosinophil levels.21 Fourth, focusing on the unique features of the disease endotypes, we report that EoEe1 has the mildest phenotype, most closely resembling findings seen in normal biopsies; EoEe2 is characterized by substantial inflammatory changes, type 2 immune responses, and evidence of refractoriness to steroids; and EoEe3 is associated with the presence of a narrow-caliber esophagus, the highest degree of endoscopic and histologic severity, and the lowest expression of epithelial differentiation genes. Fifth, we demonstrate that machine learning can be used to reproducibly separate these disease endotypes. Six, we have uncovered associations between eosinophilic inflammation, BZH and endoscopic furrowing. Lastly, we have identified genes that are modulated within each of the endotypes, establishing insight into distinct disease mechanisms. Collectively, this new disease classification stratifies subjects with EoE into subgroups having potential clinical and therapeutic significance, and it provides a framework for a precision-medicine approach to EoE. For example, given that fibrostenotic EoE is often steroid resistant,22 EoEe2 and EoEe3 likely represent more complex or severe phenotypes and may well require novel therapies in addition to—or instead of—inflammatory control. By uncovering three distinct disease endotypes, each associated with different clinical features and, importantly, molecular pathways, our findings provide a potential framework for distinct prognostic medicine and future therapeutic interruption strategies for specific patient populations.
In the present study, we highlighted the associations between the EDP, HSS and EREFS, which are important assessments of disease severity. The overall EDP exhibited the strongest association with the BZH domain. Although eosinophil levels in the esophageal epithelium are the hallmark of EoE, recent work supports a substantial role of the basal epithelium, particularly related to loss of cellular differentiation.23 We also found that the endoscopic finding of furrowing stands out as a unique feature, related to transcript changes, particularly those involved in inflammatory responses. Of note, this association was consistent across age groups, even though it is well recognized that clinical and endoscopic features differ between children and adults.
Our study is the first to characterize endotypes in EoE. EoEe1, representing 35% of the subjects, had relatively small changes in epithelial differentiation genes, a pauci-inflammatory state and a greater proportion of normal-appearing esophagus by endoscopy. EoEe2, representing 29% of subjects, had particularly high type 2 immune response mechanisms and a steroid-refractory phenotype. EoEe3, representing 36% of subjects, had particularly low expression of epithelial differentiation genes and a greater frequency of subjects with a narrow-caliber esophagus. Interestingly, we found that endotypes were associated with distinct clinical features, including pediatric-onset versus adult-onset EoE (EoEe1 and EoEe2 vs. EoEe3), atopic versus non-atopic (EoEe1 and EoEe2 vs. EoEe3), normal versus inflammatory versus fibrostenotic endoscopic appearance (EoEe1 vs. EoEe2 vs. EoEe3), and steroid-sensitive versus steroid-refractory (EoEe1vs EoEe2 and EoEe3). Our findings showed a consistent association of EoEe3 with a presence of narrow-caliber esophagus, which is recently recognized as a subgroup of EoE.22
EoEe1 is characterized by markedly low expression of ALOX15, suggesting that this is associated with a more mild phenotype24 and that perhaps suppression of this gene and/or the metabolic products of 15-lipoxygenase may be therapeutic, particularly in subjects with EoEe2 or EoEe3. EoEe2 is characterized by a marked inflammatory response, observed by endoscopy and molecular transcript profiling. EoEe2 transcript profiles are derived from genes that encode for a variety of pro-inflammatory cytokines, especially those characterized by type 2 immune responses (e.g., IL-4 and TSLP). It is notable that the highest relative expression is seen in the ACTG2 gene, encoding for the actin gamma smooth muscle 2 protein. This protein has been shown to be involved in epithelial cell responses including mesenchymal transition,25 which is observed in EoE.26 EoEe3 is enriched for epithelial genes that lose expression, particularly ACPP, CITED2, CTNNAL1, EML1, FLG, GRPEL2, MT1M, PNLIPPR3 and TSPAN12. This is the first molecular analysis of the fibrostenotic disease group and provides pathogenic insight and potential points of therapeutic intervention for this difficult-to-treat EoE group. For example, TSPAN12 is a tetraspan protein involved in epithelial cell contact, proliferation and migration27 and could be a therapeutic point of intervention. Collectively, these molecular-pathogenic connections begin to open novel ways to both understand and potentially treat EoE, including patient subgroups. It is tempting to speculate that EoE endotypes may represent a temporal transition from EoEe1 to EoEe2 to EoEe3, which merits further investigation. Additionally, our findings suggest distinct therapeutic strategies, with EoEe2 being more amenable to specific anti-type 2 immune therapy such as anti–IL-4Rα rather than anti–IL-13 (which shows less differentiation between the three endotypes). Interestingly, recent work also suggested the presence of a subgroup that has a Th2-type inflammatory profile with high expression of TSLP.28 Thus, subjects in EoEe2 could represent those subjects whose EoE would better respond to anti-TSLP biologics currently being tested.29 Additionally, our findings imply that pediatric and adult EoE have comparable pathogenesis and are likely amenable to similar therapeutic interventions. Furthermore, eosinophil levels may not be the primary determinant of disease severity or the clinically relevant subgroup; accordingly, we propose the potential value of endotyping for disease classification.
Reassessment of clinical trial designs to include biomarkers reflecting the status of the host response is supported by a growing body of evidence.30 The molecular endotypes described here show that EoE is heterogeneous, with distinct pathophysiologic profiles that are not distinguishable by esophageal eosinophil counts alone. This has potential value for future clinical trials that could stratify subjects prospectively or retrospectively to examine subgroups with distinct responsiveness. In addition, by deriving the machine-learning prediction for each endotype, we provide evidence that the technology such as an automated medical algorithm exists and that molecular subtyping of subjects with EoE is feasible in clinical settings.
Our study has several notable strengths and limitations. As for the strengths, first, we analyzed samples from multiple sites across the US, which increases the generalizability of the results. Second, the subjects were evaluated with several validated diagnostic platforms prospectively, allowing us to examine the associations between endotypes and clinical phenotypes. Third, the differences that we observed derived from exclusively examining samples from the distal esophagus. We allowed an inclusion of EoE subjects that had extra-esophageal involvement (e.g. eosinophilic gastritis) as the esophagus of these subjects expresses similar EDP-based gene transcripts compared with isolated EoE.6 Fourth, we validated gene expression differences between endotypes in an independent, local cohort. As for the limitations, clustering analysis of this study was restricted to the 95 genes included in the EDP. Genome-wide approaches would likely reveal additional subgroups of subjects and provide more insight into genetic associations with clinical characteristics. Second, detailed clinical characterization was not available. Therefore, the absence of an association between the endotypes and other clinical characteristics might be due to missing data or a consequence of the unbalanced presence of these clinical characteristics across the EoE endotypes. Finally, we did not analyze the data prospectively or longitudinally to assess the day-to-day variability or the long-term usefulness of these endotypes. The data presented are limited by the cross-sectional approach, highlighting the importance of additional replication, particularly in prospective and/or longitudinal studies.
In conclusion, we have established that esophageal eosinophilia correlates with distinct features of molecular transcripts, histology, and endoscopy. In particular, we identified a relationship between transcript changes and endoscopic furrowing and basal zone hyperplasia. Furthermore, we determined that EoE exists in at least three disease endotypes, each of which bears unique molecular, histologic, endoscopic, and clinical features. These findings transcend the current gold standard of relying strictly on esophageal eosinophil levels. Therefore, we have provided deep insight into the disease classes and pathogenesis of EoE.
Supplementary Material
Acknowledgments
CEGIR (U54 AI117804) is part of the Rare Disease Clinical Research Network (RDCRN), an initiative of the Office of Rare Diseases Research (ORDR), National Center for Advancing Translational Sciences (NCATS), and is co-funded by National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), and NCATS. CEGIR is also supported by patient advocacy groups including the American Partnership for Eosinophilic Disorders (APFED), Campaign Urging Research for Eosinophilic Disease (CURED), and Eosinophilic Family Coalition (EFC). Dr. Jeffrey Krischer is supported by NIH U54TR001263. The authors are also grateful to Dr. Amanda Rudman-Spergel for her guidance and input and Shawna Hottinger for editorial assistance. CEGIR investigators are grateful to their colleagues and clinical support staff for procuring biopsies and clinical data.
Footnotes
Conflicts of Interest. M.E.R. is a consultant for Pulm One, Spoon Guru, Celgene, Shire, Astra Zeneca, Glaxosmithkline, Allakos, Adare, Regeneron and Novartis and has an equity interest in Pulm One, Spoon Guru, Celgene and Immune Pharmaceuticals and royalties from reslizumab (Teva Pharmaceuticals). M.E.R. is an inventor of patents, owned by Cincinnati Children’s. G.W.F. has received research support from Celgene/Receptos, Regeneron, Shire and Adare. M.H.C. is a consultant for Shire, Regeneron, Receptos and Adare and has received research funding from Shire, Regeneron and Receptos. S.K.G. is a consultant for Abbott, Allakos, QOL, Meritage, and Receptos and receives research support from Shire. V.A.M. is a consultant for Shire and has received research funding from Shire. N.G. is a consultant for Allakos. E.S.D. is a consultant for Adare, Alivio, Allakos, Banner, Enumeral, GSK, Receptos/Celegene, Regeneron and Shire, has received research funding from Adare, Meritage, Miraca, Nutricia, Receptos/Celgene and Shire, and has received educational grants from Banner and Holoclara. S.S.A. is a consultant for Regeneron, is an inventor of oral viscous budesonide, patented by UCSD and licensed by Shire, and has research funding from Ferring Research Institute. J.M.S. is a consultant for Regeneron and DBV Technology, and his research is supported by NIH, EATS foundation, AImmune Therapeutics, FARE and DBV Technology. I.H. is a consultant for Regeneron, Receptos, Shire, Allakos and Adare and has received research funding from Regeneron, Receptos, Shire and Adare. G.T.F. is a consultant for Shire and a co-founder of EnteroTrack. All other authors declare they have no competing interests.
Contributors
TS, TW, and MER conceived the study and design. TS performed the laboratory work. TS contributed to the statistical analysis. TW and MPT provided analytical and bioinformatic support. MHC, KEC, NZ and GY performed the pathological assessments. SSA, PJA, DA, PAB, JMC, CLC, ESD, NG, SKG, GWF, IK, JTK, JPK, JL, VAM, JMS, and GTF provided administrative, clinical, or material support through CEGIR. TS and MER drafted the paper. MER obtained funding and led the study. All of the authors discussed the results and commented on the manuscript.
References
- 1.Simon D, Cianferoni A, Spergel JM, et al. Eosinophilic esophagitis is characterized by a non-IgE-mediated food hypersensitivity. Allergy. 2016;71(5):611–20. doi: 10.1111/all.12846. [DOI] [PubMed] [Google Scholar]
- 2.Lucendo AJ, Molina-Infante J, Arias A, et al. Guidelines on eosinophilic esophagitis: evidence-based statements and recommendations for diagnosis and management in children and adults. United European Gastroenterol J. 2017;5(3):335–58. doi: 10.1177/2050640616689525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dellon ES, Gonsalves N, Hirano I, et al. ACG clinical guideline: Evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE) Am J Gastroenterol. 2013;108(5):679–92. doi: 10.1038/ajg.2013.71. [DOI] [PubMed] [Google Scholar]
- 4.Wen T, Rothenberg ME. Clinical Applications of the Eosinophilic Esophagitis Diagnostic Panel. Front Med. 2017;4:108. doi: 10.3389/fmed.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warners MJ, Hindryckx P, Levesque BG, et al. Systematic Review: Disease Activity Indices in Eosinophilic Esophagitis. Am J Gastroenterol. 2017;112(11):1658–69. doi: 10.1038/ajg.2017.363. [DOI] [PubMed] [Google Scholar]
- 6.Wen T, Stucke EM, Grotjan TM, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145(6):1289–99. doi: 10.1053/j.gastro.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins MH, Martin LJ, Alexander ES, et al. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring. Dis Esophagus. 2017;30(3):1–8. doi: 10.1111/dote.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirano I, Moy N, Heckman MG, Thomas CS, Gonsalves N, Achem SR. Endoscopic assessment of the oesophageal features of eosinophilic oesophagitis: validation of a novel classification and grading system. Gut. 2013;62(4):489–95. doi: 10.1136/gutjnl-2011-301817. [DOI] [PubMed] [Google Scholar]
- 9.Wechsler JB, Bolton S, Amsden K, Wershil BK, Hirano I, Kagalwalla AF. Eosinophilic Esophagitis Reference Score Accurately Identifies Disease Activity and Treatment Effects in Children. Clin Gastroenterol Hepatol. 2017 doi: 10.1016/j.cgh.2017.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atkins D, Furuta GT, Liacouras CA, Spergel JM. Eosinophilic esophagitis phenotypes: Ready for prime time? Pediatr Allergy Immunol. 2017;28(4):312–9. doi: 10.1111/pai.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greuter T, Bussmann C, Safroneeva E, et al. Long-Term Treatment of Eosinophilic Esophagitis With Swallowed Topical Corticosteroids: Development and Evaluation of a Therapeutic Concept. Am J Gastroenterol. 2017;112(10):1527–35. doi: 10.1038/ajg.2017.202. [DOI] [PubMed] [Google Scholar]
- 12.Andorf S, Purington N, Block WM, et al. Anti-IgE treatment with oral immunotherapy in multifood allergic participants: a double-blind, randomised, controlled trial. The lancet Gastroenterology & hepatology. 2018;3(2):85–94. doi: 10.1016/S2468-1253(17)30392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lotvall J, Akdis CA, Bacharier LB, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127(2):355–60. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 14.Cheng K, Gupta SK, Kantor S, et al. Creating a multi-center rare disease consortium - the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR) Transl Sci Rare Dis. 2017;2(3–4):141–55. doi: 10.3233/TRD-170016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoepfer AM, Safroneeva E, Bussmann C, et al. Delay in diagnosis of eosinophilic esophagitis increases risk for stricture formation in a time-dependent manner. Gastroenterology. 2013;145(6):1230–6. e1–2. doi: 10.1053/j.gastro.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Dellon ES, Kim HP, Sperry SL, Rybnicek DA, Woosley JT, Shaheen NJ. A phenotypic analysis shows that eosinophilic esophagitis is a progressive fibrostenotic disease. Gastrointest Endosc. 2014;79(4):577–85. e4. doi: 10.1016/j.gie.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singla MB, Chehade M, Brizuela D, et al. Early Comparison of Inflammatory vs. Fibrostenotic Phenotype in Eosinophilic Esophagitis in a Multicenter Longitudinal Study. Clin Transl Gastroenterol. 2015;6:e132. doi: 10.1038/ctg.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn. 2003;52(1–2):91–118. [Google Scholar]
- 19.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–3. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rousseeuw PJ. Silhouettes - a Graphical Aid to the Interpretation and Validation of Cluster-Analysis. J Comput Appl Math. 1987;20:53–65. [Google Scholar]
- 21.Rothenberg ME, Wen T, Greenberg A, et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. 2015;135(2):500–7. doi: 10.1016/j.jaci.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 22.Eluri S, Runge TM, Cotton CC, et al. The extremely narrow-caliber esophagus is a treatment-resistant subphenotype of eosinophilic esophagitis. Gastrointest Endosc. 2016;83(6):1142–8. doi: 10.1016/j.gie.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rochman M, Travers J, Miracle CE, et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2017;140(3):738–49. e3. doi: 10.1016/j.jaci.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siddique AS, Corney DC, Mangray S, et al. Clinicopathologic and gene expression analysis of initial biopsies from patients with eosinophilic esophagitis refractory to therapy. Hum Pathol. 2017;68:79–86. doi: 10.1016/j.humpath.2017.08.027. [DOI] [PubMed] [Google Scholar]
- 25.Benzoubir N, Mussini C, Lejamtel C, et al. Gamma-smooth muscle actin expression is associated with epithelial-mesenchymal transition and stem-like properties in hepatocellular carcinoma. PLoS One. 2015;10(6):e0130559. doi: 10.1371/journal.pone.0130559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kagalwalla AF, Akhtar N, Woodruff SA, et al. Eosinophilic esophagitis: epithelial mesenchymal transition contributes to esophageal remodeling and reverses with treatment. J Allergy Clin Immunol. 2012;129(5):1387–96. e7. doi: 10.1016/j.jaci.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otomo R, Otsubo C, Matsushima-Hibiya Y, et al. TSPAN12 is a critical factor for cancer-fibroblast cell contact-mediated cancer invasion. Proc Natl Acad Sci U S A. 2014;111(52):18691–6. doi: 10.1073/pnas.1412062112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lexmond WS, Pardo M, Rooney K, et al. Elevated levels of leukotriene C4 synthase mRNA distinguish a subpopulation of eosinophilic oesophagitis patients. Clin Exp Allergy. 2013;43(8):902–13. doi: 10.1111/cea.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gauvreau GM, O’Byrne PM, Boulet LP, et al. Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med. 2014;370(22):2102–10. doi: 10.1056/NEJMoa1402895. [DOI] [PubMed] [Google Scholar]
- 30.Fiorentino R, Liu G, Pariser AR, Mulberg AE. Cross-sector sponsorship of research in eosinophilic esophagitis: a collaborative model for rational drug development in rare diseases. J Allergy Clin Immunol. 2012;130(3):613–f6. doi: 10.1016/j.jaci.2012.07.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.