Abstract
The ancestral tracks in admixed genomes are valuable for population history inference. While a few methods have been developed to infer admixture history based on ancestral tracks, these methods suffer the same flaw: only population admixture history under some specific models can be inferred. In addition, the inference of history might be biased or even unreliable if the specific model deviates from the real situation. To address this problem, we firstly proposed a general discrete admixture model to describe the admixture history with multiple ancestral populations and multiple-wave admixtures. We next deduced the length distribution of ancestral tracks under the general discrete admixture model. We further developed a new method, MultiWaver, to explore multiple-wave admixture histories. Our method could automatically determine an optimal admixture model based on the length distribution of ancestral tracks, and estimate the corresponding parameters under this optimal model. Specifically, we used a likelihood ratio test (LRT) to determine the number of admixture waves, and implemented an expectation–maximization (EM) algorithm to estimate parameters. We used simulation studies to validate the reliability and effectiveness of our method. Finally, good performance was observed when our method was applied to real data sets of African Americans and Mexicans, and new insights were gained into the admixture history of Uyghurs and Hazaras.
Introduction
Admixture among previously isolated populations has been a common phenomenon throughout the evolution of modern humans (Li et al., 2008; Reich et al., 2009; Wall et al., 2009). The history of population admixture has a strong influence on the landscape of genetic variation in individuals from admixed populations. Therefore, the population history of admixed populations can be reconstructed by utilizing genetic variation information (Xu et al., 2008; Pool and Nielsen, 2009; Price et al., 2009; Moorjani et al., 2011; Pugach et al., 2011; Gravel, 2012; Jin et al., 2012; Loh et al., 2013; Hellenthal et al., 2014; Jin et al., 2014; Pickrell et al., 2014; Ni et al., 2016; Pugach et al., 2016).
A few methods have been developed to infer admixture history based on ancestral tracks information (Pool and Nielsen, 2009; Pugach et al., 2011; Gravel, 2012; Jin et al., 2012; Jin et al., 2014; Ni et al., 2016; Pugach et al., 2016). Pool and Nielsen firstly used the length of ancestral tracks to infer population history (Pool and Nielsen, 2009). They introduced a theoretical framework describing the length distribution of ancestral tracts and proposed a likelihood inference method to estimate the parameters related to historical change in migration rates. In addition, Pugach et al. performed wavelet transforms on the ancestral tracks in an admixed population to obtain the dominant frequency of ancestral tracks to estimate the admixture time (Pugach et al., 2011; Pugach et al., 2016). Jin et al. further explored admixture dynamics by comparing the empirical and simulated distribution of ancestral tracks under three typical two-way admixtures models: the hybrid isolation (HI) model, the gradual admixture (GA) model, and the continuous gene flow (CGF) model (Jin et al., 2012). They later deduced the theoretical distributions of ancestral tracks under HI and GA models (Jin et al., 2014). Gravel extended these studies to multiple ancestral populations and discrete migrations, and provided a numerical estimation of tract length distribution (Gravel, 2012).
However, all of the aforementioned methods have a significant shortcoming. Before estimating the parameters of admixture history, a prior admixture model was required. The method by Pool and Nielsen considered the model in which a target population received migrants from a source population (Pool and Nielsen, 2009). Pugach et al.’s method involved an HI model, and Jin et al.’s methods involved HI, GA, and CGF models (Pugach et al., 2011; Jin et al., 2012; Jin et al., 2014). While Gravel considered models of multiple ancestral populations and discrete migrations, a prior admixture model was also required when dealing with the problem of admixture history inference (Gravel, 2012). However, in real data analysis, we always have little information of admixture history, and the admixture model is often uncertain for some complex admixed populations (Xu et al., 2008; Xu and Jin, 2008; Lipson et al., 2014; Bryc et al., 2015). Therefore, when the prior model deviates from real history, these methods might be unreliable.
In our previous work (Ni et al., 2016), we proposed some general principles in parameter estimation and model selection with the length distribution of ancestral tracks under a general model. However, with the increase of the number of parameters, it is complex and time-consuming to find the optimal solution, and too many parameters can lead to over-fitting. Thus, we only developed a method to infer admixture history under three typical two-way admixtures models.
In this work, we introduced a new method to select the optimal admixture model and estimate the corresponding parameters under a general model. Firstly, we proposed a general discrete admixture model with an arbitrary number of ancestral populations and an arbitrary number of admixture events. This was similar to the general model in our previous work (Ni et al., 2016). Then, we deduced the theoretical distribution of ancestral tracks with some reasonable approximations under the general discrete admixture model. We selected an optimal admixture model based on the length distribution of ancestral tracks. Specifically, we used a likelihood ratio test (LRT) (Wilks, 1938) to determine the number of admixture waves, and employed a method of exhaustion to determine the order of admixtures. We then applied an expectation–maximization (EM) algorithm (Dempster et al., 1977) to estimate the parameters under the optimal model. In our method, no prior knowledge about the admixture history is required, and the admixture model and its corresponding parameters could both be inferred by ancestral tracks. Finally, we conducted simulation studies to demonstrate the effectiveness of our method, and then applied our method to African Americans and Mexicans from the HapMap project phase III data set (International HapMap et al. 2010), and Uyghurs and Hazaras from the Human Genome Diversity Project (HGDP) data set (Li et al., 2008).
Materials and methods
General discrete admixture model
In our previous study (Ni et al., 2016), we modeled admixture history generation by generation and proposed a general admixture model. The model was determined by a K × T admixture proportion matrix M = {mi(t)}1 ≤ i ≤ K,1 ≤ t ≤ T, where K is the number of ancestral populations, T is the time the admixed population arose, and mi(t) is the ancestry contribution of ith ancestral population at time t. If the admixed population did not receive any gene flows of ith ancestral population at time t, we set mi(t) as 0. This general model covers all of the scenarios of an admixed population with an arbitrary number of ancestral populations and an arbitrary number of admixture events. However, the parameters for this general model are redundant, and will lead to over-fitting in most cases. For example, if we consider an HI model of two ancestral populations and the admixed population that arose T. generations ago, the number of parameters is 2T. However, 2(T − 1) parameters should be equal to 0, since ancestral populations contribute nothing after the first admixture; in fact, only two parameters must be estimated. Thus, to reduce redundancy and maintain the universality of our model, we proposed a general discrete admixture model that only records the information of effective admixture events (Fig. 1).
Fig. 1.
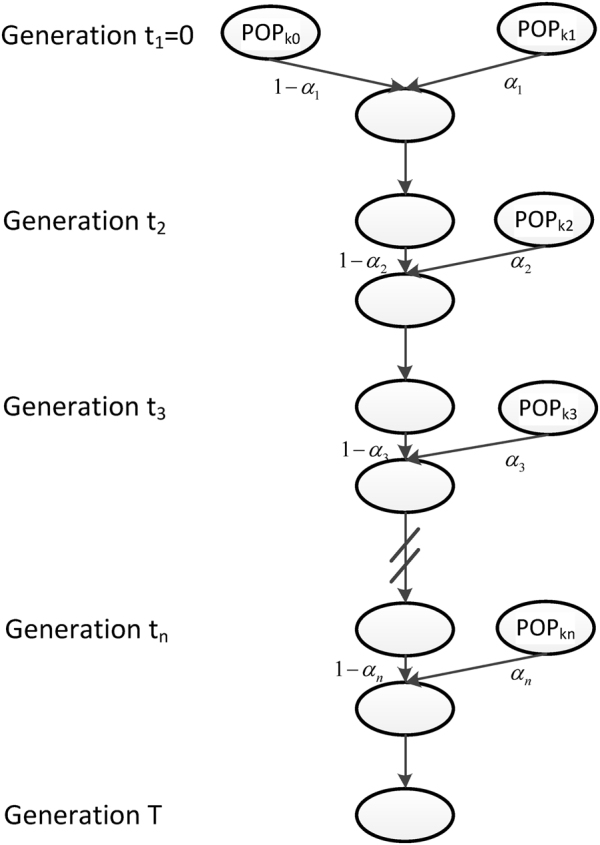
The general discrete admixture model. Here, we illustrate an admixed population with K ancestral populations and n-wave discrete admixtures, which started to admix T generations ago. POPki is the ancestral population of the ith admixture, αi is the admixture proportion of the ith admixture, and ti is the admixture time of the ith admixture
We considered an admixed population with K ancestral populations and n-wave discrete admixtures. Here, the time of the admixture in generations increases over time, with T being the present time. For the first wave admixture (i = 1), there are two ancestral populations. We denote one ancestral population as population k0 and the other as population k1. When i ≥ 2, we denote ki as the ancestral population of ith admixture. Then, we denote a vector O = (k0, k1, …, kn). as the admixture order of ancestral populations. Let αi. be the admixture proportion of the ith admixture and ti be the admixture time of the ith admixture. We note that 0 ≤ αi ≤ 1 for 1 ≤ i ≤ n, and t1 ≤ t2 ≤ ‧‧‧ ≤ tn ≤ tn+1 = :T. For convenience in our later description, we denote the admixture event from population k0 as the 0th admixture, which means that the ancestral population k0 is regarded as an admixed population before the first wave admixture. Thus, we set the corresponding admixture proportion α0 = 1 and admixture time t0 = t1. With this definition, each wave (ith wave) of admixture can be determined by three parameters: ki, αi, and ti.
Now, denote Ik = {i:ki = k}, then, Ik(j) represents the wave ordinal of the jth admixture from ancestral population k. Let nk denote the number of admixture waves from ancestral population k, and thus we have nk = |Ik| and 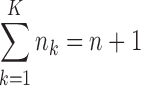 , where n is the total number of admixture waves. The general discrete admixture model is determined by the admixture order O = (k0, k1,…, kn), the admixture proportion {αi}0 ≤ i ≤ n, and the admixture time {ti}0 ≤ i ≤ n+1. If we set
, where n is the total number of admixture waves. The general discrete admixture model is determined by the admixture order O = (k0, k1,…, kn), the admixture proportion {αi}0 ≤ i ≤ n, and the admixture time {ti}0 ≤ i ≤ n+1. If we set
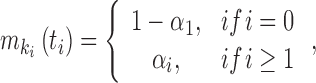 |
and set the rest of the elements of the admixture proportion matrix {mi(t)}1 ≤ i ≤ K,1 ≤ t ≤ T as 0, we can get the admixture proportion matrix of the previous general model (Ni et al., 2016). This shows that our new model is similar to the previous model. Furthermore, this new model can also cover all of the scenarios of an admixed population with an arbitrary number of ancestral populations and an arbitrary number of admixture events.
Length distribution of ancestral tracks
Next, we deduced the length distribution of the ancestral tracks from ancestral population k. The deduction process is similar to that of our previous work (Ni et al., 2016). The wave ordinals of the admixture from ancestral population k are Ik(1), Ik(2), …, Ik(nk), respectively. Define si as the survival proportion of the ancestral tracks from the ith admixture at generation T. Then,
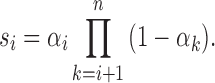 |
1 |
Denote Hk(t) as the total ancestry proportion of the kth ancestral population in the admixed population at t generation, and we have
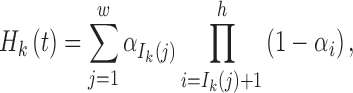 |
2 |
where 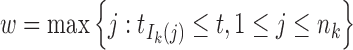 and
and 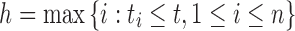 .
.
For simplicity, we assumed that the chromosome length was infinite and there was no genetic drift. In addition, we defined the recombination among tracks from different ancestral populations as effective recombination because we only observed these recombination events among different ancestries. The length of the ancestral tracks was changed by these recombination events. For the tracks from ancestral population k, the effective recombination rate is 1 − Hk(t) at t generation. Let uk(j) be the total effective recombination rate for the ancestral tracks from the jth admixture of ancestral population k. Then, we have
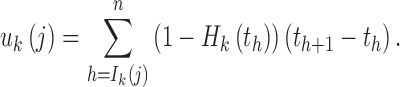 |
3 |
The length distribution of the ancestral tracks from the jth admixture of ancestral population k is an exponential distribution with a rate of uk(j) (Pool and Nielsen, 2009; Gravel, 2012; Ni et al., 2016). A chromosome from the jth admixture of ancestral population k is expected to be split into uk(j) pieces per unit length (units in Morgan). Thus, for the admixed population at T generation, the number of ancestral tracks from the jth admixture of ancestral population k is proportional to 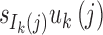 . Let Xk be the length of the ancestral tracks from ancestral population k at generation T, and fk(x) is the probability density of Xk. Then,
. Let Xk be the length of the ancestral tracks from ancestral population k at generation T, and fk(x) is the probability density of Xk. Then,
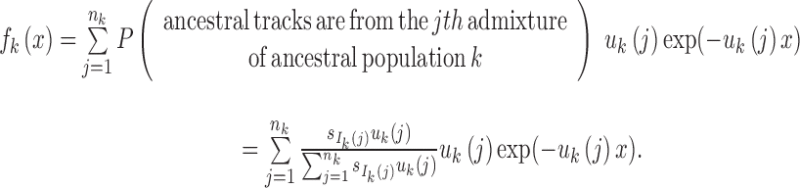 |
4 |
The length distribution of the ancestral tracks was a mixed exponential distribution, and consisted with the results from our previous study (Ni et al., 2016).We deduced the length distribution of the ancestral tracks without drift and assumed infinite chromosome length. These assumptions had little influence on the length distribution. In our previous study, simulations had demonstrated that these assumptions were reasonable and accurate (Ni et al., 2016).
Model selection and parameter estimation
If the admixture model is determined, the length distribution of the ancestral tracks can be written as Formula (4), which is a mixed exponential distribution. The EM algorithm (Dempster et al., 1977) can be used to estimate the parameters in this distribution. However, the admixture model is often unclear in real situations, which means that the number of admixture waves (nk) and the order of admixtures (O) are unknown. Thus, we must first determine nk and O. Here we used LRT (Wilks, 1938) to select the optimal nk. (Details of the LRT procedures are in Supplementary Text S1) After that, we used the method of exhaustion to validate the accuracy of O. For any order of admixtures, we estimated the admixture proportion {αi}0 ≤ i ≤ n and admixture time {ti}0 ≤ i ≤ n+1 using the EM algorithm. However, these parameter estimations must satisfy the following constraint conditions:
(a) 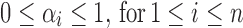 ; and
; and
(b) 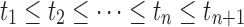 .
.
If the estimations do not satisfy these conditions, the order is incorrect. After traversing all of the admixture orders, we could determine the correct ones.
The detailed procedures are as follows:
Step 1: Estimate the total admixture proportion mk of ancestral population k. With the inferred ancestral tracks, divide the total length of tracks from population k by the total length of tracks in the admixed population, then we get the estimation of total admixture proportion mk.
Step 2: Determine the number of admixture waves (nk) for each ancestral population and estimate the parameters of the mixed exponential distribution. For ancestral population k, use LRT to select the optimal number of admixture waves nk and then estimate the parameters 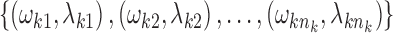 of the mixed exponential distribution using the EM algorithm, where
of the mixed exponential distribution using the EM algorithm, where 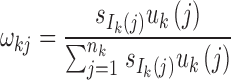 and λkj = uk(j). Details of the EM algorithm and LRT procedures are in Supplementary Text S1.
and λkj = uk(j). Details of the EM algorithm and LRT procedures are in Supplementary Text S1.
Step 3: Select admixture order O without a replacement from the set 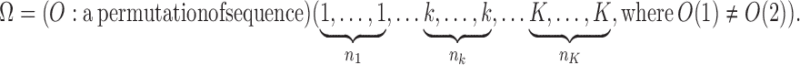 Get Ik for each k base on the selected O.
Get Ik for each k base on the selected O.
Step 4: Determine {si}0 ≤ i ≤ n and {uk(j),1 ≤ j ≤ nk,1 ≤ k ≤ K} from the following equations:
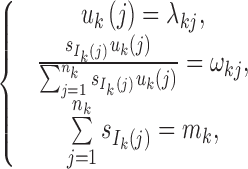 |
where 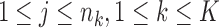 .
.
Step 5: Determine {αi}0 ≤ i ≤ n from the following equations:
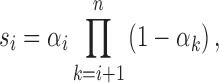 |
where 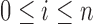 .
.
Step 6: Determine {ti}0 ≤ i ≤ n+1 from the following equations:
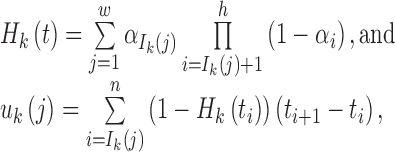 |
where 1 ≤ j ≤ nk, 1 ≤ k ≤ K
Step 7: Judge whether {αi}0≤i≤n and {ti}0 ≤ i ≤ n+1 satisfy the following conditions:
(a) 0 ≤ αi ≤ 1, for 1 ≤ i ≤ n; and
(b) 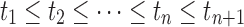 .
.
If these conditions are satisfied, record the corresponding admixture order O, admixture proportion {αi}0 ≤ i ≤ n, and admixture time {ti}0 ≤ i ≤ n+1. Then return to Step 3 until all of the possible admixture orders are checked.
Through these above procedures, we obtained all of the reasonable admixture orders O, the corresponding estimators of admixture proportion {αi}0 ≤ i ≤ n, and admixture time {ti}0 ≤ i ≤ n+1. Based on the estimations of these parameters, we could recover the history of the admixed population.
However, due to a lack of accuracy in local ancestry inference, only these relatively long tracks are reliable (Pool and Nielsen, 2009; Gravel, 2012). Therefore, we are interested in the conditional length distribution of ancestral tracks longer than specific threshold C. As we know, the length distribution of ancestral tracks from each ancestral population is a mixed exponential distribution. When we consider only tracks larger than C, the length distribution from ancestral population k becomes:
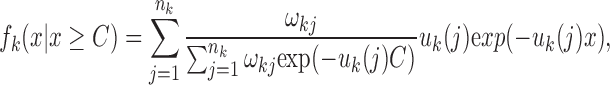 |
where 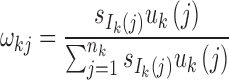 . However, since this condition distribution is not a mixed exponential distribution, we cannot use the EM algorithm to estimate the parameters. Fortunately, when we consider the random variable Yk = Xk − C, we find that the distribution of Yk is a mixed exponential distribution, which can be written as follows:
. However, since this condition distribution is not a mixed exponential distribution, we cannot use the EM algorithm to estimate the parameters. Fortunately, when we consider the random variable Yk = Xk − C, we find that the distribution of Yk is a mixed exponential distribution, which can be written as follows:
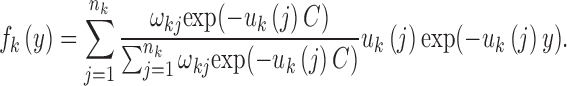 |
To take the threshold C into consideration, we must change the procedures of aforementioned Step 2. We can easily obtain samples of Yk from samples of Xk. Then, we can use the EM algorithm and LRT to obtain the distribution parameters of Yk. Furthermore, by the relationship between fk(x) and fk(y), we can obtain the parameters of the mixed exponential distribution of Xk. Then, the following steps are the same as those aforementioned in Steps 3–7. These procedures were all implemented in our MultiWaver, which can be downloaded at http://www.picb.ac.cn/PGG/resource.php.
In the MultiWaver software, two estimations of admixture time for the first wave were output. One was an estimation of t0, while the other was an estimation of t1. In theory, t1 is equal to t0, but in real data analysis the estimations may be not equal because of random errors and tracks inference errors. Thus, we presented two estimations of admixture time for the first admixture wave in our results. When t1 is proximate or equal to t0, we regard t0 or t1 as a successful estimation of time of the first wave of admixture.
Simulation studies
We conducted simulations to evaluate the performance of MultiWaver. The simulation data were generated by the forward-time simulator AdmixSim (Yang et al., 2016) (AdmixSim can be downloaded at http://www.picb.ac.cn/PGG/resource.php). General settings of our simulation were the same as those in our previous study (Ni et al., 2016).
Here we divided multiple-wave admixture models into two different types of models. We denoted the model as a simple model if each ancestral population could contribute only once to the admixed population. The others were denoted as a complex model. In the complex model, at least one ancestral population donates admixture more than once. It is important to note that when we infer the admixture history under the complex model, it is very challenging to distinguish the different admixture waves from the same ancestral population.
We focused on evaluating the performance of MultiWaver under these two types of models. For the simple model, we considered a scenario of three ancestral populations (Supplementary Figure S1, Scenario (I)) and a scenario of five ancestral populations (Supplementary Figure S1, Scenario (II)). For the complex model, we considered a scenario of two ancestral populations with two-wave admixtures (Supplementary Figure S1, Scenario (III)). We evaluated the performance of MultiWaver with different admixture times and admixture proportions. For simplicity, we supposed the admixture proportions (αi,1 ≤ i ≤ n) were equal. We set three different values of admixture proportions for each scenario: 0.1, 0.3, and 0.5. For Scenario (I), the admixture time was set as two different cases: (a) t2 = 20, T = 40, and (b) t2 = 40, T = 60. For Scenario (II), the admixture time was also set as two different cases: (a) t2 = 20, t3 = 40, t4 = 60, T = 80, and (b) t2 = 40, t3 = 80, t4 = 120, T = 140. For Scenario (III), the admixture time was set as four different cases: (a) t2 = 20, T = 40, (b) t2 = 40, T = 60, (c) t2 = 60, T = 80, and (d) t2 = 80, T = 100. Each case was repeated 10 times for a total of 240 simulations across the three scenarios. MultiWaver was applied to the simulated data with the default settings; the results were recorded and summarized.
In real situations, due to the limitations of local ancestry inference, only the ancestral tracks longer than a special threshold can be accurately inferred. Thus, to make our method more available to real situations, we chose the thresholds ranging from 0 cM to 2 cM in steps of 0.25 cM, and then evaluated the robustness of our method under the different thresholds.
We evaluated the performance of MultiWaver with different sample sizes. We considered Scenario (I) and Scenario (III) in which the admixture proportion was set as 0.3 and admixture time was set as t2 = 40, T = 60. At the end of the simulation, we sampled 20, 40, 100, 200, 300 and 400 “individuals”, corresponding to 2, 4, 10, 20, 30, and 40 human samples.
Application to real data sets
First, we applied our method to some real data sets of African Americans and Mexicans. These two populations are typical admixed populations and their histories are relatively clear. Therefore, they could be used to test the performance of our method for real data. We obtained the data sets of African Americans (ASW), Mexicans (MEX), and the reference populations African (YRI) and European (CEU) from the HapMap Project Phase III data set (International HapMap et al., 2010). Meanwhile, Maya and Pima populations represented American Indian ancestry, which were obtained from the HGDP data set (Li et al., 2008). According to prior studies, African Americans and Mexicans have more than two ancestries (Kidd et al., 2012; Jin et al., 2014). However, the proportion of Native American ancestry of African Americans is <5% (Baharian et al., 2016), and thus, we only considered two dominant ancestries (African and European ancestry) of African Americans. For Mexicans, we considered three ancestries: African, European, and American Indian ancestry (Moreno-Estrada et al., 2013).
Then, our method was used to reconstruct the population history of Uyghurs and Hazaras. The histories of these two populations are more complex. Uyghurs and Hazaras populations were obtained from the HGDP data set. Previous studies have shown that Uyghurs and Hazaras had admixed ancestries mainly from East and West (Li et al., 2008; Hellenthal et al., 2014). Here we used Han and French as the proxies of Eastern ancestry and Western ancestry, respectively (Loh et al., 2013). These reference populations were also obtained from the HGDP data set.
To enhance the reliability of our analysis, HAPMIX (Price et al., 2009) was selected as the local ancestry inference method since it shows good performance in admixture break points inference (Hinch et al., 2011). However, HAPMIX can only be used to detect ancestral tracks for two-way admixtures, and thus it might not be proper for the Mexican population. PCAdmix (Brisbin et al., 2012) has shown great power in inferring the local ancestry of Mexican populations (Moreno-Estrada et al., 2013), and thus we used PCAdmix in this study. The generations pre-set in HAPMIX inference were 10 for African Americans and 80 for Uyghurs and Hazaras. The window size set in PCAdmix was the default. Since phasing data were required for both HAPMIX and PCAdmix, SHAPEIT 2 (Delaneau et al., 2012) was used to infer the haplotype phase. Finally, MultiWaver was used to determine the optimal model and estimate the admixture time accordingly with tracks longer than 1 cM.
Results
MultiWaver performed well under simple and complex models
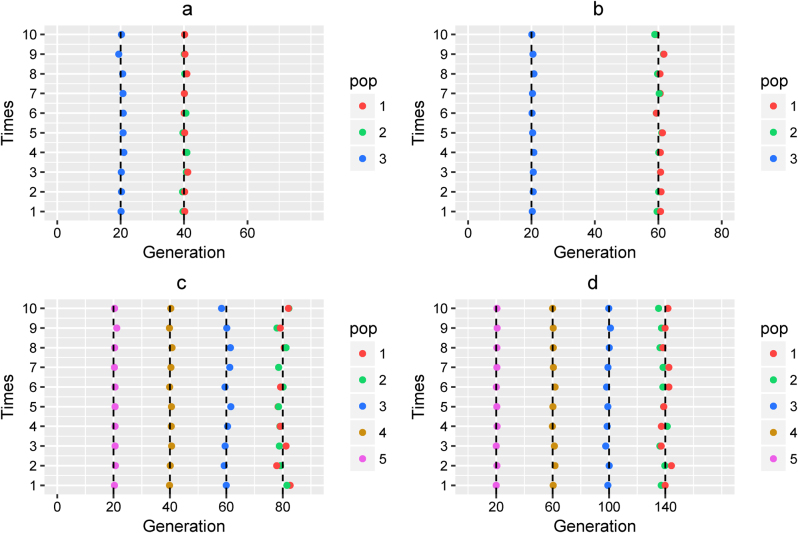
We compared the admixture histories inferred by MultiWaver with the histories set in simulations, and then evaluated the performance of our method in model selection and parameters estimation. For Scenario (I) and (II), the results showed that the estimations of admixture time were highly consistent with the time simulated if we pre-set the admixture model as the simple model (-s option in MultiWaver) (Fig. 2). Our method also performed well when we did not pre-set this option (-s) (Supplementary Figure S2). The model was correctly selected in 85% of the simulations, and only in a few simulations the incorrect model was selected. When the model was correctly selected, the admixture time estimated was consistent with the simulated time.
Fig. 2.
Admixture time estimated under a simple model. Admixture time estimated under Scenario (I) for (a) α1 = α2 = 0.3 and t2 = 20, T = 40; and (b) α1 = α2 = 0.3 and t2 = 40, T = 60. Admixture time estimated under Scenario (II) for (c) α1 = α2 = α3 = α4 = 0.3 and t2 = 20, t3 = 40, t4 = 60, T = 80; and (d) α1 = α2 = α3 = α4 = 0.3 and t2 = 40, t3 = 80, t4 = 120, T = 140. X-coordinate is the admixture time in generations ago, with 0 being the present time. Each case was repeated 10 times, and Y = i means the ith simulation. The points in the line (Y = i) represent the admixture time estimated from the ith simulation, and the color of the points indicates the ancestral population. The dashed lines represent the simulated admixture time. For the first wave admixture, two estimations of admixture time are presented
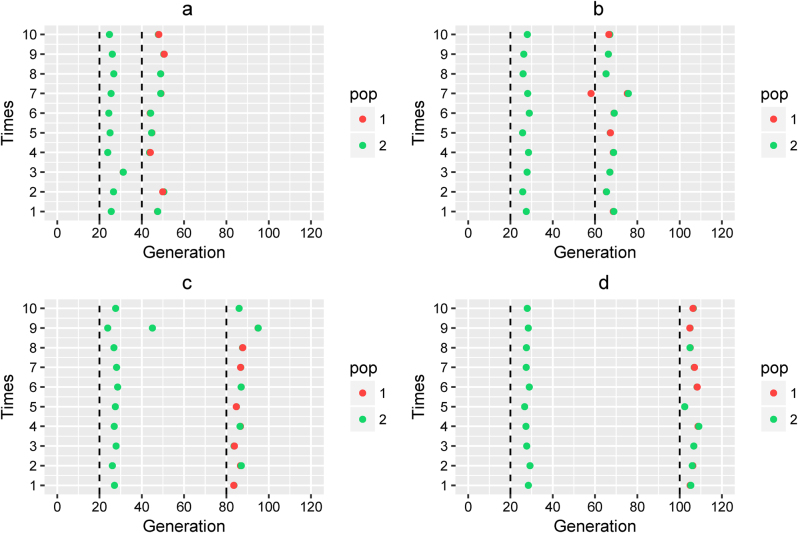
For the complex model, we found that our method could select the right model with high accuracy (Fig. 3). Model selection was incorrect for only 3 in 40 simulations. In these three cases, the number of admixture waves was wrongly estimated, which led to the inaccurate estimation of admixture time. Thus, selecting a correct model is of crucial importance for admixture history inference. When the admixture model could be correctly selected, only a slight overestimation occurred for the admixture time.
Fig. 3.
Admixture time estimated under a complex model. Admixture time estimated under Scenario (III) for (a) α1 = α2 = 0.3 and t2 = 20, T = 40; (b) α1 = α2 = 0.3 and t2 = 40, T = 60; (c) α1 = α2 = 0.3 and t2 = 60, T = 80; and (d) α1 = α2 = 0.3 and t2 = 80, T = 100. X-coordinate is the admixture time in generations ago, with 0 being the present time. Each case was repeated 10 times, and Y = i means the ith simulation. The points in the line (Y = i) represent the admixture time estimated from the ith simulation, and the color of the points indicates the ancestral population. The dashed lines represent the simulated admixture time. For the first wave admixture, two estimations of admixture time are presented
We also evaluated the performance of MultiWaver with different admixture proportions. We found that the overestimation of admixture time in the complex model was related to the admixture proportions (Supplementary Figure S3). When the proportions of each admixture wave became smaller, the estimation error decreased. However, for the simple model, our method performed well for all of the situations.
In conclusion, regardless of which type of admixture model, our method performed well for model selection. Furthermore, the admixture time was estimated well for the simple model, and with only slight overestimation for the complex model.
Robustness for different thresholds of track length
We tested the robustness of our method for different thresholds of track length. The results showed that our method was robust to thresholds for both the simple model and complex model (Fig. 4). Due to the limitations of our method, the local ancestry inference was not so accurate for short ancestral tracks. Thus, in real data analysis, we had to discard tracks smaller than a threshold. However, short ancestral tracks contain ancient admixture information, and if the threshold was too large, lots of information would be lost. Therefore, we had to balance the trade-off between information and accuracy. In our real data analysis, we set the threshold as 1 cM.
Fig. 4.
Admixture time estimated with different thresholds. (a) Admixture time estimated under Scenario (I), where α1 = α2 = 0.3 and t2 = 40, T = 60; and (b) admixture time estimated under Scenario (III), where α1 = α2 = 0.3 and t2 = 40, T = 60. X-coordinate is the admixture time in generations ago, with 0 being the present time. Y-coordinate represents the thresholds, and the color of the points indicates the ancestral population. The dashed lines represent the simulated admixture time. For the first wave admixture, two estimations of admixture time are presented
Good performance with different sample sizes
The results showed that MultiWaver performed well with different sample sizes. For both scenarios, even with 20 “individuals” (corresponding to two human samples), it could select the right model and estimate the admixture time with high accuracy (Supplementary Figure S4). Therefore, our method was insensitive to sample sizes.
Bootstrapping results with simulation data
To assess uncertainty in the optimal model and parameter estimation, we performed an analysis of the simulation data with bootstrapping sampling, which provided a supporting rate of the optimal model and 95% confidence interval (CI) of the estimations of admixture time (Table S1). We found that our inferred model was consistent with the optimal model based on bootstrapping analysis, giving the highest supporting rate and indicating high accuracy of our method as well as reliability in model selection and parameter estimation. Details of bootstrapping procedures are described in Supplementary Text 1(d).
Real data analysis
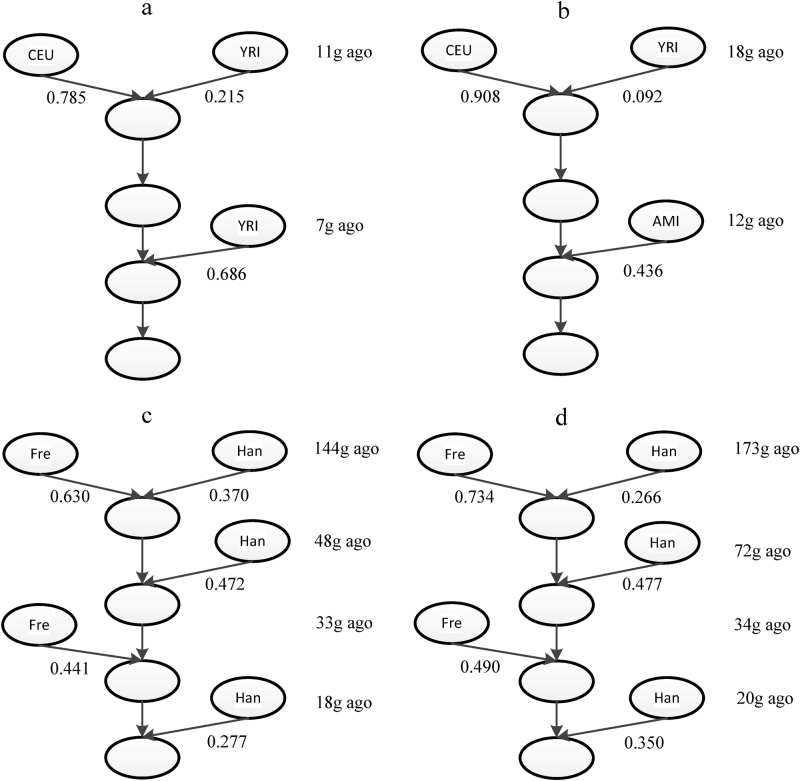
We applied our method to infer the admixture histories of some real data sets. For African Americans, HAPMIX was used to infer the ancestries with Africans (YRI) and European (CEU) as the two ancestral populations. The admixture model was inferred as two ancestral populations with a two-wave admixtures model (Fig. 5a). The African population (YRI) contributed 2 wave admixtures, and the admixture time was 11 generations ago and 7 generations ago, respectively. The time of the first admixture was about the 17th century, which was consistent with the time that most African ancestors arrived in America via slave trading. This inferred time was close to previous findings (Gravel, 2012; Jin et al., 2012; Kidd et al., 2012; Baharian et al., 2016; Ni et al., 2016). After slave trading, many African people settled down in America. The second wave admixture might have been caused by these people or by recent migrations from Africa to America. The admixture model inferred by our method suggested that the admixture history of African Americans was not one pulse of admixture, which was also reported in previous studies (Jin et al., 2012; Kidd et al., 2012; Baharian et al., 2016).
Fig. 5.
Inferred admixture history of real data sets. Inferred admixture history of (a) African Americans, (b) Mexicans, (c) Uyghurs, and (d) Hazaras. The time of the first wave admixture was the average of estimations for time t0 and t1. AMI denotes the combined data set of populations Maya and Pima, which represent American Indian ancestry, Han denotes Han population, which represent Asian ancestry, Fre denotes French population, which represent European ancestry
For Mexicans, we used PCAdmix to infer the local ancestries, and a two-wave admixtures model was inferred (Fig. 5b). Each ancestral population contributed once to the admixed population. The time of the first admixture wave was about 18 generations ago, which was close to previous findings (Price et al., 2007; Tian et al., 2007; Wang et al., 2008; Kidd et al., 2012; Moreno-Estrada et al., 2013). The time of the second admixture was 12 generations ago. This time period (12–18 generations ago) was consistent with the time of the exploration of the new world. For our analysis of African Americans and Mexicans, the admixture histories inferred by our method were consistent with recorded histories, thus showing the power of our method in real data analysis.
Finally, we applied our method to reconstruct the admixture histories of Uyghurs and Hazaras (Fig. 5c–d, Table S2). The results showed that these two populations shared a similar admixture model. The earliest admixture event of Uyghurs occurred about 144 (127–182, 95%CI) generations ago, with subsequent admixture waves from both ancestries 20–50 generations ago. The earliest admixture event of Hazaras occurred around 173 (147–198, 95%CI) generations ago, with following gene flows occurred 20–70 generations ago. Although Hazaras might be more likely derived their ancestry from an ancient admixture compared to Uyghurs, the 95% CI overlaps with the earliest admixture event of these two populations and there is no significant difference of admixture time between Hazaras and Uyghurs. Compared with the results obtained by the admixture history inference method ALDER (Loh et al., 2013), our method found an additional ancient admixture event in Uyghurs and Hazaras. To explain the discrepancies in theory, ALDER considers only the decay curve of weighted linkage disequilibrium (LD) between the pairs of sites whose genetic distances were longer than 0.5 cM (Loh et al., 2013), and thus the information of ancient signals within shorter loci pairs would be lost. Meanwhile, our method saved some of these ancient signals by deducing the conditional length distribution of ancestral tracks even if we discarded ancestral tracks shorter than 1 cM.
Conclusively, Uyghurs and Hazaras had a similar admixture history. The ancient admixture might have been caused by the migrations of Indo-Aryan speaking people into the Indian subcontinent (1500 BC). Uyghurs mainly settled in West China, and Hazaras mainly settled in Afghanistan and Pakistan. The residences of these two populations were all near the Silk Road, and thus we thought the recent multiple admixtures might have been caused by the trades or migrations along the Silk Road. In fact, the real history of Uyghurs and Hazaras might be more complex than inferred. However, our method could detect some admixture events and provide some useful information to understand the origin and development of these complex populations.
Discussion
Compared to previous methods, our method showed superiority in two aspects. First, our method explicitly models more complex admixture scenarios than currently available techniques, jointly inferring >2 episodes of admixture at different times and/or with changing ancestral source populations. Second, while previous methods rely on assuming some special admixture models when trying to infer admixture history, no prior admixture model was required in our method. Therefore, the inferred history might be biased or even unreliable if the provided model deviates from real history. However, our method avoided this problem by selecting an optimal admixture model based on ancestral tracks. Some other methods, such as MALDER (Patterson et al., 2012; Pickrell et al., 2014), GLOBETROTTER (Hellenthal et al., 2014), can also deal with multi-wave admixtures. Compared with these methods, GLOBETROTTER can only analyze populations with signals consistent with at most three-way admixture and infer at most two-wave admixture. MALDER tests all of the possible pairs of reference populations and each test was assumed to be independent, which ignored the effects of many other populations when a certain pair of reference populations were selected. Our method can handle populations with more than three-way admixture and takes into account the effects of different reference populations as well as different admixture events. However, there is also space to improve in our method. In contrast to some available techniques for detecting admixture, such as GLOBETROTTER, ROLLOFF (Moorjani et al., 2011), ALDER, and MALDER, our method requires assigning ancestry to local tracts of the genome, and the efficiency of our method relies on the validity of this inference. In addition, the LTR method might tend to select a multi-wave admixture model rather than a continuous admixture model due to the reduction of the number of parameters. Therefore, the multiple waves admixture inferred from the data could be also an alternative scenario of continuous migration.
Our method introduced a practical solution to the complex admixture history inference. However, some problems still exist. When inferring admixture history under the complex model, overestimation occurred for the admixture time. In our method, we assumed chromosome length was infinite and there was no genetic drift, and then we found that the length distribution was a mixed exponential distribution. However, Liang and Nielsen pointed out that the length distribution did not follow an exponential distribution when the admixture time was too small or too large (Liang and Nielsen, 2014). In the complex model, the non-exponential property would be accumulated, which might be the reason behind the overestimation we observed with our method. We also found that the overestimation was related to the admixture time and the admixture proportion of each admixture wave. We performed simple linear regression analysis on the errors for admixture time estimations and admixture proportion (Supplementary Figure S5). We found that the overestimation increased when the estimated admixture proportion increased, and that the magnitude of overestimation also elevated as the true time increased.
In our method, it is possible that more than one optimal admixture model satisfied the constraint conditions and should be recorded. However, for all of the simulations we conducted, this phenomenon did not appear. This was reasonable because the admixture history had a one-to-one correspondence with the length distribution of ancestral tracks. If one situation had more than one optimal model, it implied the ancestral tracks were not accurately inferred. To assess uncertainties in the optimal model and parameter estimation, we applied a bootstrapping technique to provide a supporting rate of the chosen model and the CI of estimations of admixture time.
The efficiency of our method was also influenced by the validity of the local ancestry inference. We tested the performance of our method with the inferred ancestral tracks (Supplementary Text S1). We found that MultiWaver tended to overestimate the number of waves, and thus led to overestimating the admixture time with the ancestral tracks inferred by HAPMIX (Supplementary Figure S6 (a)). For multiple-way admixtures, the inaccuracy of ancestral tracks inferred by PCAdmix led to underestimating the time of the first admixture wave (Supplementary Figure S6 (b)). It was very difficult to obtain relatively accurate ancestral tracks with a small length for all of the local ancestry inference methods. To improve the effectiveness of the inference, we suggest using the ancestral tracks longer than a certain threshold C in our method. However, when the threshold became large, ancient admixture information would be lost rapidly. The development of sequencing technology and computational methods is expected to improve the detection of short ancestral tracks in the near future. Then, our method would be promising in recovering even more ancient admixture history, such as the admixture between modern humans and ancient humans (Prüfer et al., 2014; Sankararaman et al., 2014).
Conclusions
Complex admixture history inference has long been a challenging problem in population genetics. In this work, we proposed a general discrete admixture model to describe admixture history with multiple ancestral populations and multiple-wave admixtures. We deduced that the length distribution of ancestral tracks was a mixed exponential distribution. On the basis of this distribution, we developed a new method, MultiWaver, to infer the multiple-wave admixture histories. We used LRT to select the number of admixture waves, and implemented a method of exhaustion to determine the order of admixtures. When the admixture model was determined, we applied the EM algorithm to estimate the parameters. Simulations and real data analysis showed that MultiWaver was precise and efficient in inferring admixture history.
Data archiving
MultiWaver can be downloaded at http://www.picb.ac.cn/PGG/resource.php.
AdmixSim can be downloaded at http://www.picb.ac.cn/PGG/resource.php.
All of the data used to perform the application described in this paper are available for free. The data of African Americans and Mexicans were obtained from the HapMap project phase III data set; and the data of Uyghurs and Hazaras were obtained from the Human Genome Diversity Project (HGDP) data set.
Electronic supplementary material
Acknowledgements
This work was supported by the Strategic Priority Research Program (XDB13040100) and Key Research Program of Frontier Sciences (QYZDJ-SSW-SYS009) of the Chinese Academy of Sciences (CAS), the National Natural Science Foundation of China (NSFC) (grants nos. 91331204, 91731303, 31771388, 11426237 and 31711530221), the National Science Fund for Distinguished Young Scholars (31525014), the Program of Shanghai Academic Research Leader (16XD1404700), the 973 Project (2011CB808000), the Fundamental Research Funds for the Central Universities (2017JBM071 and 2015IBM099), the National Excellent Doctoral Dissertation Foundation of PR China (201213), the National Center for Mathematics and Interdisciplinary Sciences of CAS, and the Key Laboratory of Random Complex Structures and Data Science, CAS (2008DP173182). S.X. also gratefully acknowledges the support of the National Program for Top-Notch Young Innovative Talents of the “Wanren Jihua” Project. We thank LetPub (www.LetPub.com) for its linguistic assistance during the preparation of this manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Xumin Ni, Kai Yuan and Xiong Yang contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1038/s41437-017-0041-2) contains supplementary material, which is available to authorized users.
Contributor Information
Zhiming Ma, Email: mazm@amt.ac.cn.
Shuhua Xu, Email: xushua@picb.ac.cn.
References
- Baharian S, Barakatt M, Gignoux CR, Shringarpure S, Errington J, Blot WJ, et al. The great migration and African-American genomic diversity. PLoS Genet. 2016;12:e1006059. doi: 10.1371/journal.pgen.1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisbin A, Bryc K, Byrnes J, Zakharia F, Omberg L, Degenhardt J, et al. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum Biol. 2012;84:343–64. doi: 10.3378/027.084.0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet. 2015;96:37–53. doi: 10.1016/j.ajhg.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc B. 1977;39:1–38. [Google Scholar]
- Gravel S. Population genetics models of local ancestry. Genetics. 2012;191:607–19. doi: 10.1534/genetics.112.139808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellenthal G, Busby GB, Band G, Wilson JF, Capelli C, Falush D, et al. A genetic atlas of human admixture history. Science. 2014;343:747–51. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinch AG, Tandon A, Patterson N, Song Y, Rohland N, Palmer CD, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–75. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Li R, Zhou Y, Xu S. Distribution of ancestral chromosomal segments in admixed genomes and its implications for inferring population history and admixture mapping. Eur J Hum Genet. 2014;22:930–7. doi: 10.1038/ejhg.2013.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Wang S, Wang H, Jin L, Xu S. Exploring population admixture dynamics via empirical and simulated genome-wide distribution of ancestral chromosomal segments. Am J Hum Genet. 2012;91:849–62. doi: 10.1016/j.ajhg.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Gravel S, Byrnes J, Moreno-Estrada A, Musharoff S, Bryc K, et al. Population genetic inference from personal genome data: impact of ancestry and admixture on human genomic variation. Am J Hum Genet. 2012;91:660–71. doi: 10.1016/j.ajhg.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–4. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Liang M, Nielsen R. The lengths of admixture tracts. Genetics. 2014;197:953–67. doi: 10.1534/genetics.114.162362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson M, Loh PR, Patterson N, Moorjani P, Ko YC, Stoneking M, et al. Reconstructing Austronesian population history in Island Southeast Asia. Nat Commun. 2014;5:4689. doi: 10.1038/ncomms5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Lipson M, Patterson N, Moorjani P, Pickrell JK, Reich D, et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–54. doi: 10.1534/genetics.112.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorjani P, Patterson N, Hirschhorn JN, Keinan A, Hao L, Atzmon G, et al. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet. 2011;7:e1001373. doi: 10.1371/journal.pgen.1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Estrada A, Gravel S, Zakharia F, McCauley JL, Byrnes JK, Gignoux CR, et al. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9:e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X, Yang X, Guo W, Yuan K, Zhou Y, Ma Z, et al. Length distribution of ancestral tracks under a general admixture model and its applications in population history inference. Sci Rep. 2016;6:20048. doi: 10.1038/srep20048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, et al. Ancient admixture in human history. Genetics. 2012;192:1065–93. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Patterson N, Loh PR, Lipson M, Berger B, Stoneking M, et al. Ancient west Eurasian ancestry in southern and eastern Africa. Proc Natl Acad Sci USA. 2014;111:2632–7. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Nielsen R. Inference of historical changes in migration rate from the lengths of migrant tracts. Genetics. 2009;181:711–9. doi: 10.1534/genetics.108.098095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, et al. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80:1024–36. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–9. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach I, Matveev R, Spitsyn V, Makarov S, Novgorodov I, Osakovsky V, et al. The Complex Admixture History and Recent Southern Origins of Siberian Populations. Mol Biol Evol. 2016;33:1777–95. doi: 10.1093/molbev/msw055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach I, Matveyev R, Wollstein A, Kayser M, Stoneking M. Dating the age of admixture via wavelet transform analysis of genome-wide data. Genome Biol. 2011;12:R19. doi: 10.1186/gb-2011-12-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–94. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Dannemann M, Prufer K, Kelso J, Paabo S, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–7. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Hinds DA, Shigeta R, Adler SG, Lee A, Pahl MV, et al. A genomewide single-nucleotide-polymorphism panel for Mexican American admixture mapping. Am J Hum Genet. 2007;80:1014–23. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Lohmueller KE, Plagnol V. Detecting ancient admixture and estimating demographic parameters in multiple human populations. Mol Biol Evol. 2009;26:1823–7. doi: 10.1093/molbev/msp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ray N, Rojas W, Parra MV, Bedoya G, Gallo C, et al. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet. 2008;4:e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks SS. The large-sample distribution of the likelihood ratio for testing composite hypotheses. Ann Math Stat. 1938;9:60–2. doi: 10.1214/aoms/1177732360. [DOI] [Google Scholar]
- Xu S, Huang W, Qian J, Jin L. Analysis of genomic admixture in Uyghur and its implication in mapping strategy. Am J Hum Genet. 2008;82:883–94. doi: 10.1016/j.ajhg.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Jin L. A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery. Am J Hum Genet. 2008;83:322–36. doi: 10.1016/j.ajhg.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ni X, Zhou Y, Guo W, Yuan K and Xu S (2016) AdmixSim: a forward-time simulator for various and complex scenarios of population admixture. bioRxiv 037135 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.