Abstract
We investigate the role of axonal transport in regulating neuronal mitochondrial density. We show that the density of mitochondria in the touch receptor neuron (TRN) of adult Caenorhabditis elegans is constant. Mitochondrial density and transport are controlled both by the Kinesin heavy chain and the Dynein-Dynactin complex. However, unlike in other models, the presence of mitochondria in C. elegans TRNs depends on a Kinesin light chain as well. Mutants in the three C. elegans miro genes do not alter mitochondrial density in the TRNs. Mutants in the Kinesin-1 associated proteins, UNC-16/JIP3 and UNC-76/FEZ1, show increased mitochondrial density and also have elevated levels of both the Kinesin Heavy and Light Chains in neurons. Genetic analyses suggest that, the increased mitochondrial density at the distal end of the neuronal process in unc-16 and unc-76 depends partly on Dynein. We observe a net anterograde bias in the ratio of anterograde to retrograde mitochondrial flux in the neuronal processes of unc-16 and unc-76, likely due to both increased Kinesin-1 and decreased Dynein in the neuronal processes. Our study shows that UNC-16 and UNC-76 indirectly limit mitochondrial density in the neuronal process by maintaining a balance in anterograde and retrograde mitochondrial axonal transport.
Introduction
Mitochondria are present along the entire length of the neuron and at greater density in regions with high energy needs such as at the Nodes of Ranvier and at synapses1,2. Additionally, neurons have defined mitochondrial density along neuronal processes that is altered in models of neurodegenerative disease3,4. Majority of the mitochondrial biogenesis in neurons is believed to occur in the cell body5,6. Therefore, transport is likely an important means to control both the presence and the density of mitochondria along the neuronal processes. Kinesin-1 and Dynein are the major anterograde and retrograde motors for mitochondrial transport7–10. Kinesin heavy chain in both Drosophila and vertebrates is recruited onto the mitochondrial surface via the adaptor Milton/TRAK, in addition, Miro, a Rho GTPase, is essential for mitochondrial transport11–14. Kinesin heavy chain (KHC), independent of Kinesin light chain (KLC), binds to Milton/TRAK11,14. Milton/TRAK in turn binds to Miro present on the mitochondrial surface and this binding facilitates mitochondrial transport11,14. Additionally, RIC-7, a C. elegans specific mitochondrial protein, has been shown to control the presence of mitochondria in the motor neuron process10, although its interaction with Kinesin-1 is not established.
Kinesin-1 has multiple interacting proteins, some of which are used to bind cargo1,9,11,12,15–20. UNC-16/JIP3/dSYD scaffolds the JNK pathway kinases and is a major Kinesin-1 adaptor15,16,21. JIP3/UNC-16 facilitates Kinesin-1 mediated transport of activated JNK Kinases22 and Dynein Light Intermediate Chain23. C. elegans JIP3/UNC-16 mutants also show an increase in levels of mitochondria, marked by TOMM20, in motor neuron axons24. JIP3/UNC-16 binds to both Kinesin heavy and light chains25,26 and increases motor velocity and run length25. Another protein, UNC-76/FEZ1 (fasciculation and elongation protein zeta 1), binds to the Kinesin heavy chain subunit and activates the motor27–29.
In this study, we use C. elegans touch receptor neuron (TRN) as a model to study mitochondrial transport in vivo. The TRN allows us to track mitochondria in a single isolated neuron where its distribution and transport can be easily observed. In wild type animals, mitochondrial density is constant in the TRN. This density is regulated by the Kinesin heavy chain, Kinesin light chains and the Dynein-Dynactin complex. Our data suggest that the adaptors of Kinesin-1 limit the density of mitochondria in the TRNs through regulation of the balance of anterograde and retrograde mitochondrial transport.
Results
Kinesin heavy chain, kinesin light chain and the dynein-dynactin complex maintain the density of mitochondria along the neuronal process
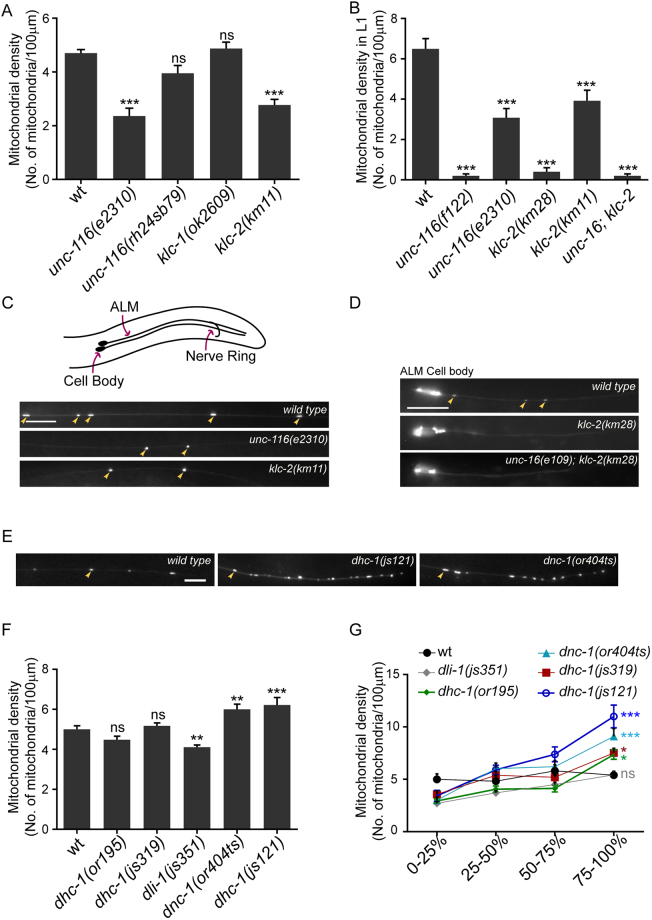
We visualized mitochondria in the anterior and posterior TRN processes using jsIs60930, a transgenic animal expressing matrix targeted GFP in the touch receptor neurons of C. elegans. The mitochondrial density in the anterior TRN (ALM anterior lateral mechanosensory) processes in 1-day adult animals is approximately 5 mitochondria/100 µm (Fig. 1A), similar to those reported earlier31.
Figure 1.
Kinesin-1 and Dynein regulate mitochondrial density in the anterior touch receptor neuronal process. (A) Mitochondrial densities in one day adult ALM (anterior lateral mechanosensory) TRNs (touch receptor neurons) of wild type (jsIs609) and Kinesin-1 motor subunit hypomorphic mutants. n = 16–20 worms. All comparisons made to wild type. (B) Average mitochondrial densities in ALM TRN processes of just hatched L1 animals of the following genotypes: Kinesin-1 subunit null alleles (f122, km28), Kinesin-1 subunit hypomorphic alleles (e2310, km11) and a double mutant unc-16(e109); klc-2(km28). n = 16–20 worms. All comparisons made to wild type. (C) Representative images of mitochondria in the ALM TRN processes of wild type (jsIs609), unc-116(e2310) and klc-2(km11) one day adults. Orientation of the anterior TRNs (here only ALM) and schematic representation of the worm is shown above fluorescent images. Scale bar = 10 μm. (D) Representative images of the ALM TRN processes of wild type (jsIs609), klc-2(km28) and unc-16(e109); klc-2(km28) L1 animals. Scale bar = 10 μm. (E) Representative images of the distal ALM TRN processes in wild type (jsIs609), dhc-1(js121) and dnc-1(or404ts) (grown at 22 °C). Scale bar = 10 μm (F) Average mitochondrial densities in the ALM TRN processes of Dynein-Dynactin complex mutants. n = 10–20 worms. All comparisons made to wild type (G) Mitochondrial density across different segments along the ALM TRN processes in wild type, Dynein mutants and Dynactin mutants. n = 10–16 worms. All comparisons made to wild type. Colours correspond to respective genotypes. Some mitochondria marked with yellow arrows in (C,D) and E. Data represented as Mean ± SEM. Statistical tests used for A,B and F: One-way ANOVA, for G: Two-way ANOVA. All tests use Bonferroni multiple comparisons correction, ns: not significant, p value * < 0.05, ** < 0.01, *** < 0.001.
To investigate the role of motors, we examined mitochondrial density in the loss of function mutants of the UNC-116/Kinesin Heavy Chain (KHC) and the two Kinesin Light Chain genes, klc-1 and klc-2. Partial loss of function alleles unc-116(e2310)32 and klc-2(km11)21 show significant reductions in their mitochondrial densities in the anterior TRN processes in both adult and L1 animals (Fig. 1A–C). unc-116(rh24sb79)33 has been shown to reduce mitochondrial density in motor neurons10 although we do not see a significant decrease in the anterior TRN (Fig. 1A). The null alleles unc-116(f122)34 and klc-2(km28)21 show a complete absence of mitochondria along the neuronal process in just hatched L1 animals compared to similarly staged wild type L1 (Fig. 1B,D). We could not examine the adult null mutants of the Kinesin-1 complex since they do not survive beyond the early L2 stage. The mitochondrial density in klc-1(ok2609)35 with a 1383 bp deletion in the gene was similar to that observed in wild type (Fig. 1A). Thus, both Kinesin-1 motor subunits UNC-116/KHC and KLC-2, but not KLC-1, play an essential role in maintaining mitochondrial density in the TRN process.
Dynein is known to transport mitochondria in the retrograde direction7. Thus, we assessed mitochondrial density in the mutants of the Dynein heavy chain (dhc-1), the Dynein light intermediate chain (dli-1) and the p150 Glued Dynactin subunit (dnc-1). We observed that a strong loss-of-function allele, dhc-1(js121)36, and dnc-1(or404ts)36 grown at the restrictive temperature, show a significant increase in overall mitochondrial density (Fig. 1F). All other dhc-1 mutant alleles examined show average overall densities of mitochondria, similar to wild type (Fig. 1F). However, all Dynein-Dynactin complex mutants, except dli-1(js351)36, show a significant increase in mitochondrial density at the distal quarter of the neuronal process (Fig. 1E,G). This phenotype is similar to that reported in the Dynein complex mutants, where Dynein-dependent cargo accumulate at the end of the anterior TRN process36. Additionally, dhc-1(js121), dhc-1(or195)37 and dnc-1(or404ts) all show a significant increase compared to wild type in the ratio of the density of mitochondria in the last quarter of the anterior TRN to the density in the first quarter (Figure S1A). Despite a reduction in average overall mitochondrial density (Fig. 1F), dli-1(js351), shows nearly a doubling of but not a statistically significant increase compared to wild type in the ratio of mitochondrial density in the distal TRN process to the proximal TRN process (Figure S1A). The fold change in mitochondrial density between the last quarter and first quarter of the neuronal process in dli-1(js351) is similar to that observed in the hypomorphic dhc-1(js319) allele (Figure S1A). dli-1(js351) animals are sterile and are the progeny of dli-1/+ hermaphrodites36. The weaker phenotype observed in dli-1(js351) animals thus may arise from the known maternal perdurance of wild type DLI-138.
Mitochondrial transport in both mammalian cells and Drosophila neurons has been shown to occur independent of the Kinesin Light Chain14. The dependence of mitochondrial density on KLC-2 suggests that C. elegans may have alternate and/or additional means to regulate mitochondrial transport in neurons, for instance, through RIC-710. The complete absence of mitochondria in the null mutants of klc-2 suggests that KLC-2 in conjunction with UNC-116 is an essential complex that mediates mitochondria entry into axons of C. elegans. The observed distal accumulation of mitochondria in Dynein and Dynactin mutants suggests a role for the Dynein-Dynactin complex in facilitating retrograde mitochondrial transport in C. elegans TRNs.
miro mutants do not significantly alter TRN mitochondrial density
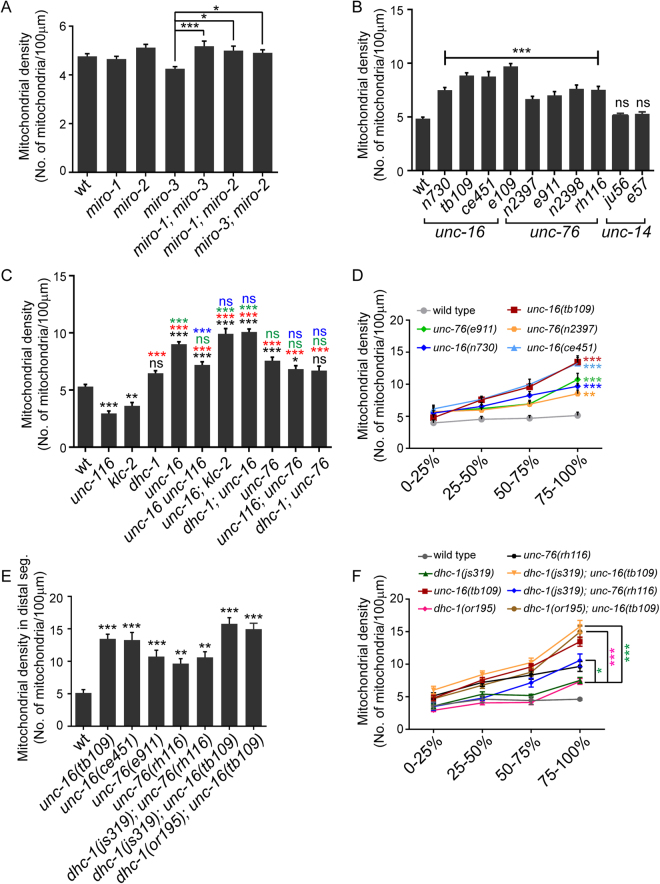
The Miro-Milton complex is the major mitochondrial adaptor for Kinesin-113,14. No Milton homologues have been found in the C. elegans genome39, but three Miro orthologues have been reported to be present40. Therefore, we investigated the role of all reported miro genes, whose mutants lack the mitochondria associated transmembrane domain, some of their EF hands and GTPase domains (Figure S2). C. elegans MIRO-1 shows ~60% protein sequence similarity with D. melanogaster Miro while C. elegans MIRO-2 and MIRO-3 show ~30% protein sequence similarities41. miro-1(tm1966), miro-2(tm2933) and miro-3(tm3150) single mutants do not alter mitochondrial density in the anterior TRN process (Fig. 2A). Double mutants [miro-1; miro-2], [miro-1; miro-3] and [miro-3; miro-2] do not show a significant change in mitochondrial density from wild type. Only miro-3 mutants show significantly lower density compared to all three double mutants [miro-1; miro-2], [miro-1; miro-3] and [miro-3; miro-2] in the neuronal process (Fig. 2A).
Figure 2.
Kinesin-1 associated proteins- UNC-16/JIP3 and UNC-76/FEZ1- regulate mitochondrial density in the anterior touch neuronal process. (A) Mitochondrial density in the ALM (anterior lateral mechanosensory) touch neuronal processes of one day adult wild type (jsIs609), miro-1(tm1966), miro-2(tm2933), miro-3(tm3150) and double mutants [miro-1(tm1966); miro-3(tm3150)], [miro-1(tm1966); miro-2(tm2933)] and [miro-3(tm3150); miro-2(tm2933)]. n ≥ 22 worms. Only significant comparisons have been marked. (B) Mitochondrial density in one day adult ALM neuronal processes of mutants in other Kinesin-1 associated proteins-unc-16(n730), unc-16(tb109), unc-16(ce451), unc-16(e109), unc-76(n2397), unc-76(e911), unc-76(n2398), unc-76(rh116), unc-14(ju56) and unc-14(e57). n ≥ 10 worms. All comparisons made to wild type (jsIs609). (C) Mitochondrial densities in one day adult wild type (jsIs609), unc-116(e2310), klc-2(km11), dhc-1(js319), unc-16(tb109), unc-76(rh116) and double mutants between Kinesin-1 adaptors and motor hypomorphs [(tb109 e2281), (tb109; km11), (js319; tb109), (e2310; rh116) and (js319; rh116)]. e2310 and e2281 are the same allele, see Table S1. n ≥ 18 worms. Black * or ns (not significant) are comparisons to wild type. Green * or ns are comparisons of adaptor single mutants and double mutants with adaptors to dhc-1 single mutant. Red * or ns are comparisons of dhc-1, adaptor single mutants and double mutants with adaptors to both unc-116 and klc-2. Blue * or ns are comparisons of motor subunit; adaptor double mutants to respective adaptor single mutants. (D) Mitochondrial density across different segments along the neuronal process in one day adult wild type (jsIs609) and mutants in Kinesin-1 associated proteins. n ≥ 18 worms. All comparisons made to wild type. Colours correspond to respective genotypes. (E) Mitochondrial density in the distal segment (last quarter) in wild type (jsIs609), unc-16 alleles, unc-76 alleles, [dhc-1(js319); unc-76(rh116)], [dhc-1(js319); unc-16(tb109)] and [dhc-1(or195); unc-16(tb109)]. n ≥ 10 worms. All comparisons made to wild type. (F) Mitochondrial densities in one day adults of wild type (jsIs609), Dynein mutants, unc-16, unc-76 and double mutants between Kinesin-1 binding proteins and Dynein mutants across different segments along the neuronal process. n ≥ 10 worms. All comparisons made between double mutants and the corresponding dynein complex single mutant. Data represented as Mean ± SEM. Statistical tests one-way ANOVA A,B,C,E) and two-way ANOVA (D,F) with Bonferroni multiple comparisons correction, ns: not significant, p value * < 0.05, ** < 0.01, *** < 0.001.
miro-1 and miro-2 RNAi has been shown to reduce mitochondrial density and over-expression of MIRO-1 increases mitochondrial density in C. elegans GABA neurons42. Additionally, Miro-1 mutants show a reduction in the amount of mitochondria in the hypodermis43. Our results do not rule out a role for Miro in mitochondrial movement along the touch neuronal process but suggest that the Kinesin Heavy Chain-Miro complex may not be critical for mitochondrial exit from the cell body of touch neurons.
Kinesin-1 associated proteins UNC-16/JIP3 and UNC-76/FEZ1 regulate mitochondrial density
Since the miro genes did not have a significant effect on mitochondrial density, we investigated mutants in other Kinesin-1 associated proteins on mitochondrial density. Three proteins, UNC-16/JIP3, UNC-76/FEZ1 and UNC-14 are known to associate with the Kinesin-1 complex and play multiple roles in transport of cargo15,16,21,25,28,29. Multiple alleles of unc-16 (e10944, tb10945, n73044 and ce45124) show a significant increase in mitochondrial density along the anterior TRN process compared to wild type (Fig. 2B). unc-76 alleles (n239727, n239827, e91127,46 and rh11627) show a significant but less pronounced increase in mitochondrial density than unc-16 when compared to wild type (Fig. 2B). However, unc-14 (e5747 and ju5621) alleles do not show a significant change in mitochondrial density compared to wild type in the TRN processes (Fig. 2B). A significant increase in mitochondrial density is observed in anterior TRNs of L1 larval stage unc-16(tb109) and unc-76(n2397) animals (Figure S1B).
Since UNC-16 and UNC-76 affect anterograde axonal transport through their interaction with Kinesin-121,28, we examined whether the increased mitochondrial density regulation in unc-16 and unc-76 depends on the presence of Kinesin-1. Double mutants between unc-16(e109) and the null allele klc-2(km28) result in an absence of mitochondria from the neuronal process identical to klc-2(km28) alone (Fig. 1B,D). We were unable to build recombinants between three different unc-16 alleles and the unc-116(f122) null allele perhaps due to a combination of the lethality of f122 allele and that the two genes are about 0.2 map units apart. Using double mutants with hypomorphic alleles in the Kinesin-1 subunits, we observe that [unc-16 unc-116], [unc-16; klc-2] and [unc-116; unc-76] show significantly increased mitochondrial density compared to both wild type (Fig. 2C, black*) and the respective Kinesin-1 subunit single mutant alleles (Fig. 2C, red*). unc-16 unc-116 animals show a significant reduction in mitochondrial density compared to unc-16 alone (Fig. 2C, blue*). [unc-16; klc-2] and [unc-116; unc-76] animals do not show a significant reduction in mitochondrial density compared respectively to unc-16 and unc-76 alone (Fig. 2C, blue ns). In conjunction with unc-16; klc-2(km28) (Fig. 1B), we conclude that, Kinesin-1 appears necessary for increased mitochondrial density at least in unc-16.
Both hypomorphic alleles unc-116(e2310) and klc-2(km11) produce lower levels of full length UNC-116 and KLC-2 respectively48. One possible explanation to account for the elevated mitochondrial density in double mutants between the adaptors and hypomorphic alleles of Kinesin-1 is that, presence of even low levels of the Kinesin-1 motor maybe sufficient to cause increased mitochondrial density in unc-16 and unc-76 mutants.
unc-16 and unc-76 animals have elevated levels of Kinesin-1 in the neuronal process
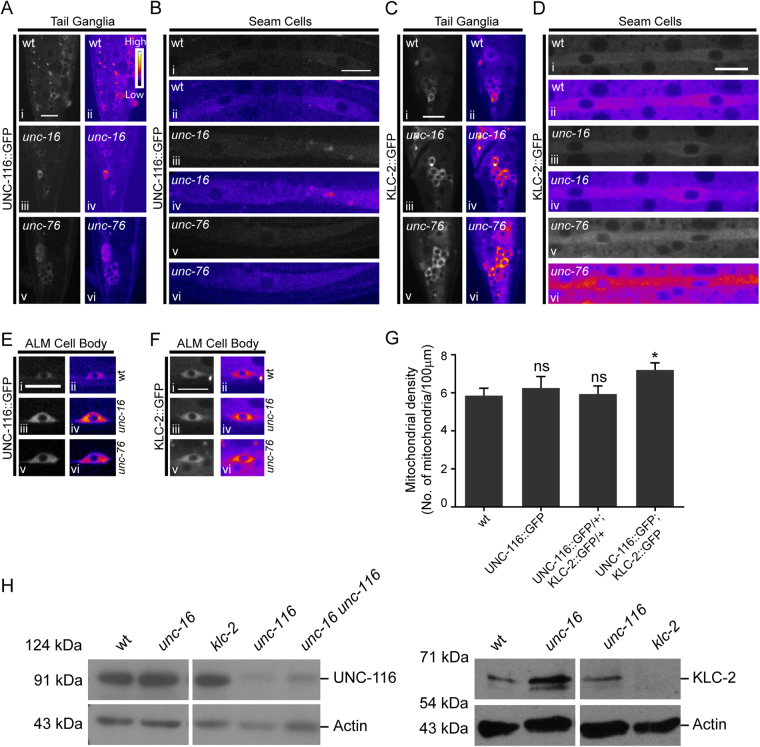
Hypomorphic mutations in either Kinesin heavy chain or Kinesin light chain genes do not substantially reduce the increased mitochondrial density observed in unc-16 and unc-76 mutant animals. Therefore, we sought to determine if Kinesin-1 levels were altered in unc-16 and unc-76 mutants. Using transgenic UNC-116::GFP and KLC-2::GFP integrated lines, driven by their endogenous promoters, that rescue their respective null alleles48, we observe an increase in levels of UNC-116::GFP and KLC-2::GFP in neurons of both unc-16 and unc-76 animals (Fig. 3A,C,E,F). This increase is seen early in development in L1 animals as well (Figure S3A, B). This observation is consistent with a significant overall increase in endogenous protein levels of ~30% in UNC-116 and ~300% in KLC-2 in unc-16(tb109) mutants (Figs 3H, S3C, S3D). We do not see an increase in Kinesin-1::GFP levels in non-neuronal cells like the seam cells of unc-16 animals (Fig. 3B,D) suggesting that the overall increase may arise largely from an increase of motor levels in neurons. KLC-2::GFP but not UNC-116::GFP levels increase in the seam cells and perhaps other non-neuronal cells of unc-76 animals (Figs 3B,D, S3A,B). Levels of unc-116 and klc-2 mRNA in two alleles each of unc-16 and unc-76, using primers described in Table 1, do not show any change. This suggests that the increase in the Kinesin-1 motor levels in these mutants may occur through a post-translational step (Figure S3E). Additionally, although statistically not significant (p = 0.06), unc-16 unc-116 animals show a 40% increase in overall UNC-116 protein levels (Figs 3H and S3D) compared to unc-116. This small increase may partially contribute to the elevated mitochondria density seen in unc-16 unc-116 animals (Fig. 2C).
Figure 3.
unc-16 and unc-76 mutants have elevated levels of Kinesin-1 subunits in neurons. (A) Representative images and corresponding heat maps of UNC-116::GFP in wild type, unc-16(e109) and unc-76(n2398) tail ganglia of L4 animals. (B) Representative images and corresponding heat maps of UNC-116::GFP in wild type, unc-16(e109) and unc-76(n2398) in seam cells of L4 animals. (C) Representative images and corresponding heat maps of KLC-2::GFP in wild type, unc-16(e109) and unc-76(rh116) tail ganglia of L4 animals. (D) Representative images and corresponding heat maps of KLC-2::GFP in L4 wild type, unc-16(e109) and unc-76(rh116) seam cells. (E,F) Representative images and corresponding heat maps of UNC-116::GFP (E) and KLC-2::GFP (F) in ALM TRN cell bodies of wild type, unc-16(e109) and unc-76(n2398) L4 animals. (G) Average mitochondrial density in the PLM (posterior lateral mechanosensory) touch neuronal process of L1 when Kinesin-1 subunits are over-expressed. Data represented as Mean ± SEM, n ≥ 8 worms. Mann Whitney test, p value * < 0.05. (H) Western Blots showing levels of Kinesin-1 protein subunits in whole worm lysates prepared from wild type, unc-16(tb109) and unc-16(tb109) unc-116(e2281) animals. Lysates prepared from unc-116(e2310) and klc-2(km11) were used as controls respectively for the UNC-116 and KLC-2 antibodies. e2310 and e2281 are the same allele, see Table S1. The experiment was performed at least three independent times in duplicates, quantitation shown in supplementary Figure 3D. Scale bar:10 μm.
Table 1.
Primers sequences used.
| Gene | Primer (Forward, Reverse) | Size of product |
|---|---|---|
| unc-116 | TT36, 5′CGGAAAAACACATACAATGGAGGGAGTAATCG TT37, 5′CTCGGGGTCTAATAAATCTCGAATCTTCTCG |
(TT36 spans exon 2 & 3), 181 bp |
| klc-2 | TT461, 5′AGGAGGCTGAGGAGTTGTACAAGC TT462, 5′GCCCATCCAGCTTGTTCGTTGG |
Exon 4, 163 bp |
| dhc-1 | TT46, 5′CTCTCGGACAAGTTTGGGATGC TT47, 5′CATCCCATCCTTTGATCAAACGTGTC |
(TT46 spans exon 4 & 5), 186 bp |
| act-3 | TT40, 5′GTAGACAATGGATCCGGAATGTGCAAGG TT41, 5′GGTACTTGAGGGTAAGGATACCTCTCTTGG |
(TT41 spans exon 1 & 2) 181 bp |
| miro-1 | TT408, 5′AATCTTCTACTACGCCCA TT409, 5′TTCCCAATTTTACTCCTCC |
800 bp |
| miro-2 | TT193, 5′GGCTGCAGGTCAATTGACTC TT194, 5′CGAGTATCATGGACTTATCG |
1482 bp |
| miro-3 | TT195, 5′TTGTACGAGTATCGTGGACT TT196, 5′GTATTCCCAGCACGACCTGA |
935 bp |
The increase in Kinesin-1 motor levels in unc-16 and unc-76 mutant animals may at least partially account for the increase in mitochondrial density in these animals. To test whether an elevation in Kinesin-1 levels alone is sufficient to result in an increase in mitochondrial density in the touch receptor neurons, we measured the density of mitochondria after over-expression of UNC-116::GFP and KLC-2::GFP in wild type animals using jsIs1073, with tagRFP targeted to the mitochondrial matrix49. Animals over-expressing Kinesin heavy chain alone or wild type animals heterozygous for both Kinesin heavy and light chain-2 expressing transgenes do not show an increase in mitochondrial density (Fig. 3G). Homozygous animals over-expressing both UNC-116::GFP and KLC-2::GFP encoding transgenes, show a significant increase in mitochondrial density in the TRN process (Fig. 3G). This suggests that elevation of just the Kinesin heavy chain or lower levels of increase in Kinesin-1 may be insufficient to elevate mitochondrial density. The smaller increased mitochondrial density in animals homozygous for the Kinesin heavy and light chain transgenes compared to unc-16 and unc-76 may arise from the relatively modest increase in levels of Kinesin-1 in a wild type background compared to the increase in motor levels seen in unc-16 and unc-76 animals (Fig. 3E,F). Additionally, the elevation of just the anterograde motor levels in unc-16 and unc-76 may be insufficient to account for the observed large increase in density of mitochondria in the TRNs of these mutants.
UNC-16 regulates axonal mitochondrial density in part through Dynein
UNC-16 has a role in retrograde transport as an adaptor of Kinesin-1, by facilitating binding of KLC-2 to Dynein23. Additionally, UNC-76 associates with the Kinesin heavy chain and activates Kinesin-1, thus facilitating the transport of Kinesin-1 cargo, one of which is Dynein23,29,50,51. Thus both mutants are likely to reduce Dynein mediated transport through different mechanisms.
We first examined whether unc-16 and unc-76 mutants shared mitochondrial distribution phenotypes with mutants in the Dynein complex. unc-16 but not unc-76 has an overall mitochondrial density significantly above that of dhc-1 (Fig. 2C, green*). However, similar to Dynein-Dynactin mutants, both unc-16 and unc-76 animals also show a significant increase in mitochondrial density in the distal quarter of the neuronal process (Figs 1G and 2D). In all alleles of both mutants examined, the density of mitochondria in the last quarter of the TRN process is significantly higher than that observed in wild type (Fig. 2D,E). The distal accumulation phenotype is more severe in the strong loss of function unc-16 alleles24,45 (ce451, tb109) compared to the strong loss of function unc-76 alleles27 (rh116, n2397, e911) (Fig. 2D,E). The phenotypes where mitochondria accumulate towards the end of the neuronal process are consistent with the known accumulation of Dynein-dependent cargo at the distal end of the anterior TRN36. The distal mitochondrial accumulation phenotypes in unc-16 and unc-76 may be accounted for by mitochondria moving to the distal part of the neuronal process using anterograde motors and accumulating towards the end of the neuronal process due to their inability to return to the cell body due to insufficient functional Dynein activity. We observe that two alleles of both unc-16 and unc-76 have dhc-1 mRNA levels, using primers described in Table 1, similar to wild type (Figure S3E). We were unable to effectively image DHC-1 in neurons as the dhc-1 promoter driven DHC-1::GFP line was very faint52.
Since, both UNC-16 and UNC-76 contribute to the anterograde transport of Dynein, we investigated the genetic interactions between mutants in both genes with a Dynein complex mutant. We examined whether a further decrease in Dynein function exacerbated the distal TRN mitochondria accumulation phenotypes of unc-16 and unc-76.
[dhc-1; unc-16] and [dhc-1; unc-76] do not show any significant increase in overall mitochondrial density compared respectively to unc-16 and unc-76 alone (Fig. 2C, blue ns). Further the overall mitochondria density in dhc-1; unc-16 but not in dhc-1; unc-76 is significantly higher than the dhc-1 single mutant (Fig. 2C, green* & ns). Double mutants [dhc-1(js319); unc-16(tb109)], [dhc-1(or195); unc-16(tb109)] and [dhc-1(js319); unc-76(rh116)] all continue to show greater numbers of distal mitochondria compared to wild type (Fig. 2E). The distal mitochondrial density in all three double mutants listed above are significantly higher compared to the respective dynein heavy chain single mutants (Fig. 2F). [dhc-1(js319); unc-16(tb109)], [dhc-1(or195); unc-16(tb109)] and [dhc-1(js319); unc-76(rh116)] show a small but a statistically not significant increase in the distal mitochondrial density (17%, 11% and 10% respectively) compared to the distal mitochondria accumulation seen in the corresponding the unc-16 or unc-76 single mutant alleles in the same region (Fig. 2F). The lack of a bigger increase could arise from the combination of alleles used i.e. weak loss of function mutants in the Dynein complex built with strong loss of function alleles of unc-16 and unc-76.
We think that reduced Dynein transport could contribute to the increased density of mitochondria at the distal end of the neuron in unc-16 and unc-76 animals. In unc-16 animals, due to the absence of the adaptor, anterograde transport of Dynein and retrograde transport of Dynein-dependent vesicles has been shown to be reduced23, this may account for the more prominent mitochondrial distal accumulation in this adaptor mutant. The absence of UNC-76/FEZ1 may lead to weaker activation of the Kinesin-1 motor, also potentially reducing the available Dynein at the end of the neuronal process. However, other molecules such as Kinesin light chain are also known to activate Kinesin-153,54. The redundancy in Kinesin-1 activation along with elevated levels of Kinesin-1 seen in unc-76 may lead to greater levels of Dynein in the end of the neuron in unc-76 compared to unc-16; perhaps accounting for the less severe distal mitochondrial accumulation phenotype in unc-76 compared to unc-16.
unc-16 and unc-76 show increased anterograde flux when Kinesin-1 levels are limiting
Our genetic data suggests that the altered mitochondrial density in these adaptor mutants may arise indirectly due to an altered ratio of anterograde and retrograde motors leading to changes in transport. Thus, we examined mitochondrial transport in unc-16 and unc-76 mutants.
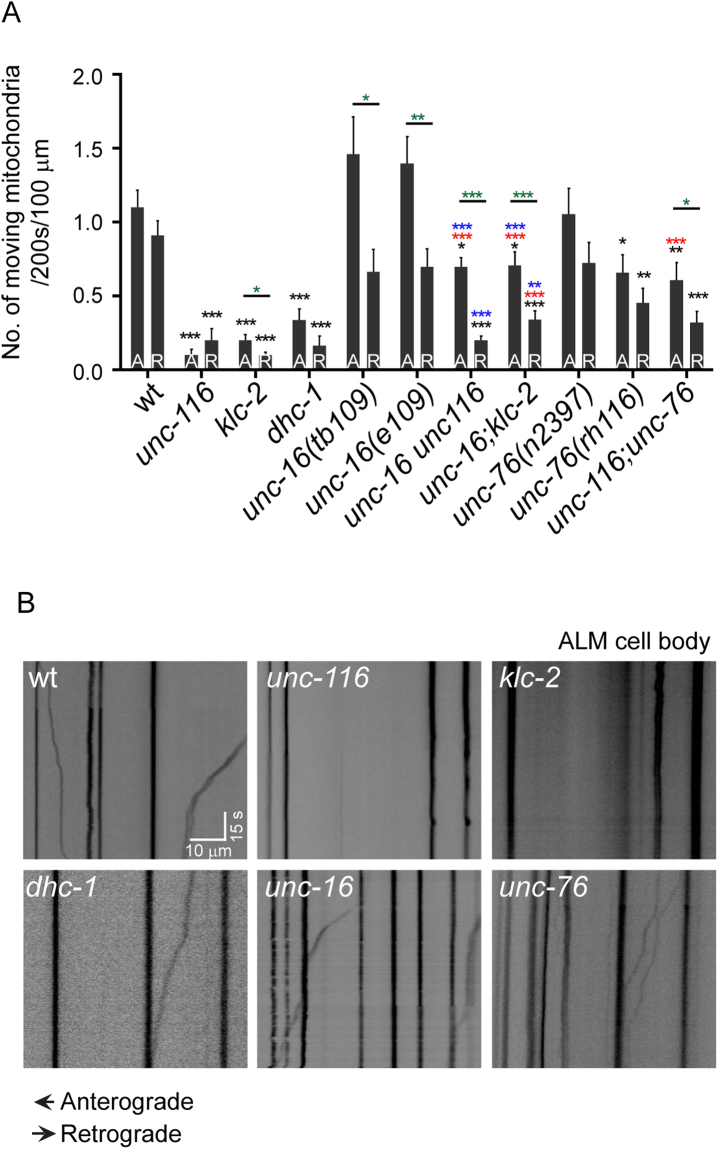
Our data show that anterograde mitochondrial movement is dependent on the Kinesin-1 complex since mutants in both unc-116(e2310) and klc-2(km11) have significantly reduced anterograde flux compared to wild type animals (Fig. 4A, black*). Surprisingly, dhc-1 mutants also show a significant (~68%) decrease in anterograde transport compared to wild type (Fig. 4A, black*). Despite an increase in Kinesin-1 motor levels both alleles of unc-16 and unc-76(n2397) do not show a significant increase in anterograde transport compared to wild type (Fig. 4A). Puzzlingly, unc-76(rh116), shows a significant reduction in the anterograde flux compared to wild type, contrary to the expectation that increased Kinesin-1 motors should lead to increased anterograde flux (Figs 3D,F, 4A). However, the anterograde mitochondrial flux in unc-16 unc-116 is significantly increased compared to unc-116 (red*) perhaps arising from statistically not significant (p = 0.06) but a 40% elevation in anterograde motor levels seen in the double mutant compared to unc-116 alone (Figs 3H, S3D). unc-16 unc-116 has significantly lower anterograde flux compared both to wild type (black*) and the unc-16 single mutant (blue*) (Fig. 4A). This might arise due to the reduction in UNC-116 levels in unc-16 unc-116 compared to wild type and unc-16 alone (Fig. 3H). Double mutants [unc-16 unc-116], [unc-16; klc-2] and [unc-116; unc-76] show at least a 250% increase in anterograde mitochondrial flux, compared to the corresponding Kinesin-1 single mutants alone (Fig. 4A, red*). This suggests a recovery of anterograde mitochondrial flux may occur in part due to an increase in Kinesin-1 levels in these double mutants. These data support the hypothesis that the lack of UNC-16 and UNC-76 can limit anterograde transport of mitochondria in the neuronal processes at least when Kinesin-1 levels are significantly reduced. However a simple explanation that there is an increase in Kinesin-1-dependent anterograde transport probably mediated through the elevated levels of UNC-116 and KLC-2, thereby contributing to increased mitochondrial density in unc-16 and unc-76, may be insufficient.
Figure 4.
UNC-16 and UNC-76 change mitochondrial flux in anterior TRNs. Mitochondrial anterograde and retrograde flux in the distal ALM processes in L3 animals of (A) wild type (jsIs609), unc-116(e2310), klc-2(km11), dhc-1(js319), unc-16(tb109), unc-16(e109), unc-76(n2397), unc-76(rh116) and double mutants [unc-16(e109) unc-116(e2310)], [unc-16(e109); klc-2(km11)] and [unc-116(e2310); unc-76(rh116)]. Data represented as Mean ± SEM. A: Anterograde; R: Retrograde. n ≥ 34 worms. Statistical tests used: The above multiple comparisons were done using multiple t-test with Sidak-Bonferroni correction. Black * are comparisons to respective wild type direction of movement. Red * are comparisons to respective directions of movement between motor; adaptor double mutants and corresponding motor subunit single mutants. Blue * are comparisons to respective directions of movement between motor; adaptor double mutants and corresponding adaptor single mutants. Green * are comparisons between anterograde and retrograde movements in the same genotype. p value * < 0.05, ** < 0.01, *** < 0.001. Comparisons that are not marked eg retrograde transport in tb109 to retrograde transport in wild type, are not statistically significant. (B) Representative kymographs of L3 wild type, unc-116(e2310), klc-2(km11), dhc-1(js319), unc-16(tb109) and unc-76(n2397) animals. Scale bars: x axis 10 μm, y axis 15 seconds.
unc-16 and unc-76 mutants show an altered ratio of anterograde to retrograde transport
We examined if changes in retrograde transport could account for the elevated mitochondrial density in unc-16 and unc-76. We observed significantly reduced retrograde flux of mitochondria in unc-76(rh116), dhc-1, klc-2 and unc-116 animals compared to wild type (Fig. 4A black*, B). Retrograde mitochondrial flux is reduced by ~82% in dhc-1 animals compared to wild type. The reduced retrograde flux in unc-116(e2310) and klc-2(km11) likely arises from both the fewer mitochondria in the neuronal process available for retrograde transport and the known role of these motors in transporting Dynein in the axon23. However neither allele of unc-16 nor unc-76(n2397) show significant changes in retrograde mitochondrial flux compared to wild type (Fig. 4A), despite a demonstrated reduction in Dynein transport at least in unc-1623. The double mutants [unc-116 unc-16], [unc-16; klc-2] and [unc-116; unc-76] show at least a 60% significant reduction in retrograde mitochondrial flux, compared to wild type (Fig. 4A, black*). The double mutants [unc-116 unc-16], [unc-16; klc-2] have significantly lower retrograde transport compared to unc-16 alone, consistent with a reduced amount of Dynein transport (Fig. 4A, blue*). unc-116; unc-76 does not show a significant reduction in retrograde mitochondrial transport compared to unc-76 alone, perhaps as there may not be as strong an effect of lack of UNC-76 on Dynein transport. The changes in retrograde transport are complex and not easily accounted for by a simple model where all Dynein is transported with the help of UNC-16 and UNC-76 regulated pathways. Mere changes in retrograde transport cannot readily account for the observed increase in mitochondrial density in unc-16 and unc-76.
The net mitochondrial density probably arises due to a balance in transport of anterograde and retrograde flux, thus we decided to examine the ratios of mitochondrial transport in the different mutants. Although there is little statistical change in the anterograde flux of unc-16(tb109) and unc-16(e109) animals, there is an increase of > 30% compared to wild type (Fig. 4A). unc-76(n2397) animals show a 10% increase and unc-76(rh116) animals show a 38% decrease in anterograde flux respectively compared to wild type. unc-16(tb109) and unc-16(e109) animals respectively show a 13% and 22% reduction in retrograde flux compared to wild type. unc-76(n2397) and unc-76(rh116) animals respectively show a 16% and 50% decrease in retrograde mitochondrial flux when compared to wild type. We thus examined whether the difference in anterograde and retrograde transport or its ratio might account for the observed overall mitochondrial density increase observed in adaptor mutants and adaptor; motor subunit double mutants. Anterograde and retrograde flux of mitochondria do not differ from each other in wild type but significantly differ from each other in both unc-16 mutants and all adaptor; Kinesin-1 subunit double mutant combinations examined (Fig. 4A, green*). These genetic backgrounds all significantly elevate mitochondrial density. Notably, while the anterograde and retrograde flux of mitochondria balance each other in wild type animals, mutants in unc-16(tb109), unc-16(e109), unc-76(n2397), unc-76(rh116) and dhc-1(js319) have higher anterograde/retrograde flux ratios respectively of 1.9, 2, 1.6, 1.5 and 2 compared to the wild type ratio of ~1. The double mutants [unc-116 unc-16], [unc-16; klc-2] and [unc-116; unc-76] have an anterograde to retrograde flux ratio of 3.5, 2.1 and 1.9 respectively. All genotypes that show an anterograde/retrograde flux ratio much greater than 1 also have significantly increased mitochondrial density in the neuron compared to wild type (flux ratio of ~1). These data suggest that, UNC-16 and UNC-76 may act to maintain the balance between anterograde and retrograde flux of mitochondria in the neuronal processes. This anterograde bias in the ratio of transport in unc-16 and unc-76 may contribute in part to the increased mitochondrial density in the TRN processes of these genotypes.
Discussion
The results described in this study suggest that, the Kinesin-1 mediated regulation of mitochondrial transport by UNC-16 and UNC-76 could occur at two levels. (i) The increase in Kinesin-1 levels in adaptor mutants. This increase coupled with the inability of the increased Kinesin-1 to recruit UNC-16 and UNC-76 dependent cargo may lead to more Kinesin available for mitochondrial anterograde transport. This is consistent with the striking observation where double mutants between Kinesin-1 and the adaptor mutants show higher levels of anterograde mitochondrial transport compared to motor mutants alone. (ii) In addition to the anterograde transport effects, we also see reduced retrograde transport. This may be due to reduced availability of dynein complex subunits at the distal end of the neuron as has been reported23. Additionally, UNC-16 is also independently known to interact with the retrograde transport complex55. These dual effects may both contribute to the increased density of mitochondria in the neuronal process with the greatest density at the distal end of the process.
Mitochondrial density regulation in neurons is a complex process
Cultured neurons from rats and zebrafish sensory neurons in vivo show a defined density of mitochondria56,57. This mitochondrial density changes, both, during development57 and when neurons express human disease causing variants of TDP-43 and SOD156. We show that wild type C. elegans TRNs also have a defined density of mitochondria. Localization of mitochondria in specific regions in the neuronal process and their overall density in the axon are likely dependent on multiple factors. In our study, we find that changes in axonal transport act as one factor in regulating density of mitochondria. Specifically, our data suggest that cargo competition for the Kinesin-1 motor through the availability of adaptors may contribute to this organelle’s density. However, there are likely to be additional factors. This is illustrated by the vast reduction in anterograde transport in a weaker loss-of-function allele dhc-1 mutant, perhaps as a compensatory mechanism to maintain overall density of mitochondria in the neuronal process. Thus, we think that the relationship between motor levels, mitochondrial flux and average axonal mitochondrial density is likely to be complex.
Materials and Methods
Worm maintenance and imaging
Worms were grown using standard protocols58, mitochondrial density was assessed in worms grown without any contamination using 30 mM sodium azide or 10 mM sodium azide as an anesthetic. Each animal was imaged within 10 minutes of anesthetisation. Overlapping images were used to reconstruct the entire neuronal process. The neuronal length was measured using the background fluoresecence in the TRN and the total number of mitochondria were counted. Mitochondrial density was calculated as number of mitochondria/100 micron of the neuronal process using 20–25 animals. L3 animals were anaesthetized in 3 mM Levamisole solution and time lapse images were acquired at 2fps for 200s in the distal regions of the ALM (Anterior Lateral Mechanosensory neuron listed as anterior TRN in the text). Kymographs were made using the ImageJ Kymograph plugin59 and anterograde and retrograde movements were determined using a displacement of 2 µm as the minimum cut-off. Each mitochondrion was counted only once. The strains used in this study have been mentioned in Table S1.
UNC-116::GFP and KLC-2::GFP imaging
L1 and L4 animals were anaesthetized with 30 mM Sodium azide and imaged using 60×/100× objective on a Olympus IX83 microscope with spinning disc (Perkin Elmer Ultraview, Waltham, MA, USA) with a Hamamatsu (SZK, Japan) monochrome EMCCD camera. Images for a specific cell type were collected from all genotypes grown identically, at the same time, using identical imaging conditions. Imaging conditions varied for different cell types.
Western blotting
Protein extraction using Homogenization buffer (HEPES pH7.6–15 mM; KCl-10 mM; MgCl2-1.5 mM; EDTA-0.1 mM; EGTA-0.5 mM; Sucrose-44 mM in water) with Protease Inhibitor Cocktail (Complete Mini EDTA-free Protease Inhibitor Cocktail Tablets; Roche; Catalog No. 04 693 159 001) was carried out from a growing plate. Western blot analysis was carried out as described in60. Polyclonal rabbit antibodies against UNC-116 and KLC-2 were raised by Bioklone, Chennai using HIS tagged proteins made from constructs as described earlier33. We validated the antibodies on Western blot using protein extracts from Ex[punc-116::unc-116::gfp]; unc-116(f122) and Ex[Pklc-2::klc-2::gfp]; klc-2(km28) worms (data not shown). Antibody dilutions used were Rabbit α-UNC-116 (1:40000), Rabbit α-KLC-2 (1:40000), mouse α-Actin (1:1500) (NeoLabMarkers)]. Secondary antibodies dilutions were anti-Rabbit-HRP (1:10000), anti-Mouse-HRP (1:6000) (ABclonal). Three or more independent biological repeats were carried out in duplicates.
Quantitation of levels of UNC-116 RNA
RNA was extracted from whole worm lysate from a just starved plate using Sigma TRI reagent. cDNA was synthesized using Superscript IV Reverse Transcriptase (ThermoFisher Scientific) using random hexamers. qPCR was done with Kapa SYBR FAST universal qPCR mastermix on Roche LightCycler- LC96. For purposes of quantitation, the samples were normalized to wild type. At least three independent biological repeats were carried out with at least two technical repeats. No genomic DNA contamination was observed when using actin primers. Quantitation was carried out as described in61.
Generation of domain structures for Miro
The domain structure has been generated using ScanProsite62 and MyDomains63. The genomic DNA sequences were sourced from the NCBI Database.
Data availability
All data and reagents are available upon request.
Electronic supplementary material
Acknowledgements
This paper is dedicated to the memory of GPRS who passed away in April 2013 in a motor cycle accident. jsIs609 was made by SPK in Dr. M. Nonet’s laboratory. We thank Drs. Yishi Jin, Francis McNally and Kenneth Miller for reagents. We thank Drs. Krishnamurthy, Manoj Matthew at CIFF, NCBS supported by the DST – Centre for Nanotechnology (No. SR/55/NM-36-2005). We thank Parul Sood and Neena Ratnakaran for confocal images. We thank Souvik Modi for discussions and help during analyses of miro domains. We thank Shubha Shanbhag for integrating UNC-116::GFP and KLC-2::GFP transgenic lines. We thank Salik Ansari for help in building strains. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). miro-1(tm1966), miro-2(tm2933) and miro-3(tm3150) was provided by the Mitani Lab through the National Bio-Resource Project of the MEXT, Japan. This study was supported by grants to SPK from DBT, DBT overseas associateship, HHMI and TIFR. Salary support comes from TIFR and HHMI (NM), TIFR (AC, NM), CSIR (GPRS), UGC (SD), BITS (AA) and DST (SM). The spinning disc confocal microscope was supported by HHMI and DAE Project 12-R&D-IMS-5.02-0202. We thank members of the SPK lab for their critical input on the manuscript.
Author Contributions
Designed research: Sandhya P. Koushika (S.P.K.), Guruprasada Reddy Sure (G.P.R.S.), Anusheela Chatterjee (A.C.), Nikhil Mishra (N.M.), Vidur Sabharwal (V.S.). Conducted research: All authors. Interpreted data: S.P.K., G.P.R.S., A.C., N.M., V.S. Wrote the paper: S.P.K., A.C., N.M.
Competing Interests
The authors declare no competing interests.
Footnotes
Guruprasada Reddy Sure and Anusheela Chatterjee contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-27211-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cai Q, Gerwin C, Sheng Z-H. Syntabulin-mediated anterograde transport of mitochondria along neuronal processes. The Journal of cell biology. 2005;170:959–969. doi: 10.1083/jcb.200506042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frederick RL, Shaw JM. Moving mitochondria: establishing distribution of an essential organelle. Traffic. 2007;8:1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunawardena S, Goldstein LS. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron. 2001;32:389–401. doi: 10.1016/S0896-6273(01)00496-2. [DOI] [PubMed] [Google Scholar]
- 4.Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144:1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amiri M, Hollenbeck PJ. Mitochondrial biogenesis in the axons of vertebrate peripheral neurons. Developmental neurobiology. 2008;68:1348–1361. doi: 10.1002/dneu.20668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis AF, Clayton DA. In situ localization of mitochondrial DNA replication in intact mammalian cells. The Journal of Cell Biology. 1996;135:883–893. doi: 10.1083/jcb.135.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Molecular biology of the cell. 2006;17:2057–2068. doi: 10.1091/mbc.e05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxton WM, Hollenbeck PJ. The axonal transport of mitochondria. J Cell Sci. 2012;125:2095–2104. doi: 10.1242/jcs.053850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka Y, et al. Targeted disruption of mouse conventional kinesin heavy chain kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/S0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- 10.Rawson RL, et al. Axons degenerate in the absence of mitochondria in C. elegans. Current Biology. 2014;24:760–765. doi: 10.1016/j.cub.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo X, et al. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Stowers RS, Megeath LJ, Górska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/S0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- 13.van Spronsen M, et al. TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron. 2013;77:485–502. doi: 10.1016/j.neuron.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 14.Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. The Journal of cell biology. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowman AB, et al. Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell. 2000;103:583–594. doi: 10.1016/S0092-8674(00)00162-8. [DOI] [PubMed] [Google Scholar]
- 16.Byrd DT, et al. UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans. Neuron. 2001;32:787–800. doi: 10.1016/S0896-6273(01)00532-3. [DOI] [PubMed] [Google Scholar]
- 17.Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 18.Kamal A, Almenar-Queralt A, LeBlanc JF, Roberts EA, Goldstein LS. Kinesin-mediated axonal transport of a membrane compartment containing β-secretase and presenilin-1 requires APP. Nature. 2001;414:643–648. doi: 10.1038/414643a. [DOI] [PubMed] [Google Scholar]
- 19.Kamal A, Stokin GB, Yang Z, Xia C-H, Goldstein LS. Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron. 2000;28:449–459. doi: 10.1016/S0896-6273(00)00124-0. [DOI] [PubMed] [Google Scholar]
- 20.Muresan Z, Muresan V. Coordinated transport of phosphorylated amyloid-β precursor protein and c-Jun NH2-terminal kinase–interacting protein-1. The Journal of cell biology. 2005;171:615–625. doi: 10.1083/jcb.200502043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakamoto R, et al. The Caenorhabditis elegans UNC-14 RUN domain protein binds to the kinesin-1 and UNC-16 complex and regulates synaptic vesicle localization. Molecular biology of the cell. 2005;16:483–496. doi: 10.1091/mbc.e04-07-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drerup CM, Nechiporuk AV. JNK-interacting protein 3 mediates the retrograde transport of activated c-Jun N-terminal kinase and lysosomes. PLoS Genet. 2013;9:e1003303. doi: 10.1371/journal.pgen.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arimoto M, et al. The Caenorhabditis elegans JIP3 protein UNC-16 functions as an adaptor to link kinesin-1 with cytoplasmic dynein. Journal of Neuroscience. 2011;31:2216–2224. doi: 10.1523/JNEUROSCI.2653-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards SL, et al. An organelle gatekeeper function for Caenorhabditis elegans UNC-16 (JIP3) at the axon initial segment. Genetics. 2013;194:143–161. doi: 10.1534/genetics.112.147348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun F, Zhu C, Dixit R, Cavalli V. Sunday Driver/JIP3 binds kinesin heavy chain directly and enhances its motility. The EMBO journal. 2011;30:3416–3429. doi: 10.1038/emboj.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verhey KJ, et al. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. The Journal of cell biology. 2001;152:959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloom L, Horvitz HR. The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proceedings of the National Academy of Sciences. 1997;94:3414–3419. doi: 10.1073/pnas.94.7.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gindhart JG, et al. The kinesin-associated protein UNC-76 is required for axonal transport in the Drosophila nervous system. Molecular biology of the cell. 2003;14:3356–3365. doi: 10.1091/mbc.e02-12-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blasius TL, Cai D, Jih GT, Toret CP, Verhey KJ. Two binding partners cooperate to activate the molecular motor Kinesin-1. The Journal of cell biology. 2007;176:11–17. doi: 10.1083/jcb.200605099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fatouros C, et al. Inhibition of tau aggregation in a novel Caenorhabditis elegans model of tauopathy mitigates proteotoxicity. Human molecular genetics. 2012;21:3587–3603. doi: 10.1093/hmg/dds190. [DOI] [PubMed] [Google Scholar]
- 31.Morsci NS, Hall DH, Driscoll M, Sheng Z-H. Age-related phasic patterns of mitochondrial maintenance in adult caenorhabditis elegans neurons. Journal of Neuroscience. 2016;36:1373–1385. doi: 10.1523/JNEUROSCI.2799-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel N, Thierry-Mieg D, Mancillas JR. Cloning by insertional mutagenesis of a cDNA encoding Caenorhabditis elegans kinesin heavy chain. Proceedings of the National Academy of Sciences. 1993;90:9181–9185. doi: 10.1073/pnas.90.19.9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H-y, Mains PE, McNally FJ. Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. J Cell Biol. 2005;169:447–457. doi: 10.1083/jcb.200411132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNally KL, Martin JL, Ellefson M, McNally FJ. Kinesin-dependent transport results in polarized migration of the nucleus in oocytes and inward movement of yolk granules in meiotic embryos. Developmental biology. 2010;339:126–140. doi: 10.1016/j.ydbio.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consortium CeDM. Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3: Genes| Genomes| Genetics. 2012;2:1415–1425. doi: 10.1534/g3.112.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koushika SP, et al. Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. Journal of Neuroscience. 2004;24:3907–3916. doi: 10.1523/JNEUROSCI.5039-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Developmental cell. 2002;3:673–684. doi: 10.1016/S1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 38.Yoder JH, Han M. Cytoplasmic dynein light intermediate chain is required for discrete aspects of mitosis in Caenorhabditis elegans. Molecular Biology of the Cell. 2001;12:2921–2933. doi: 10.1091/mbc.12.10.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mercer KB, et al. A C. elegans homolog of huntingtin-associated protein 1 is expressed in chemosensory neurons and in a number of other somatic cell types. Journal of Molecular Neuroscience. 2009;37:37–49. doi: 10.1007/s12031-008-9109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu S, Wang Z, Kim KW, Jin Y, Chisholm AD. Targeted Mutagenesis of Duplicated Genes in Caenorhabditis elegans Using CRISPR-Cas9. Journal of Genetics and Genomics. 2016;2:103–106. doi: 10.1016/j.jgg.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McWilliam H, et al. Analysis tool web services from the EMBL-EBI. Nucleic acids research. 2013;41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han SM, Baig HS, Hammarlund M. Mitochondria localize to injured axons to support regeneration. Neuron. 2016;92:1308–1323. doi: 10.1016/j.neuron.2016.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen Y, et al. C. elegans miro-1 Mutation Reduces the Amount of Mitochondria and Extends Life Span. PloS one. 2016;11:e0153233. doi: 10.1371/journal.pone.0153233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwasaki K, Liu D, Thomas JH. Genes that control a temperature-compensated ultradian clock in Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 1995;92:10317–10321. doi: 10.1073/pnas.92.22.10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choudhary B, et al. UNC-16/JIP3 regulates early events in synaptic vesicle protein trafficking via LRK-1/LRRK2 and AP complexes. PLoS Genet. 2017;13:e1007100. doi: 10.1371/journal.pgen.1007100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bloom GS, Wagner MC, Pfister KK, Brady ST. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry. 1988;27:3409–3416. doi: 10.1021/bi00409a043. [DOI] [PubMed] [Google Scholar]
- 47.Ogura K, Shirakawa M, Barnes TM, Hekimi S, Ohshima Y. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes Dev. 1997;11:1801–1811. doi: 10.1101/gad.11.14.1801. [DOI] [PubMed] [Google Scholar]
- 48.Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 2009;136:2725–2733. doi: 10.1242/dev.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng Q, et al. The vesicle protein SAM-4 regulates the processivity of synaptic vesicle transport. PLoS Genet. 2014;10:e1004644. doi: 10.1371/journal.pgen.1004644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada M, et al. LIS1 and NDEL1 coordinate the plus‐end‐directed transport of cytoplasmic dynein. The EMBO journal. 2008;27:2471–2483. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamada M, et al. mNUDC is required for plus‐end‐directed transport of cytoplasmic dynein and dynactins by kinesin‐1. The EMBO journal. 2010;29:517–531. doi: 10.1038/emboj.2009.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Rourke SM, Christensen SN, Bowerman B. Caenorhabditis elegans EFA-6 limits microtubule growth at the cell cortex. Nat Cell Biol. 2010;12:1235–1241. doi: 10.1038/ncb2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stenoien DL, Brady ST. Immunochemical analysis of kinesin light chain function. Molecular biology of the cell. 1997;8:675–689. doi: 10.1091/mbc.8.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morfini G, Szebenyi G, Elluru R, Ratner N, Brady ST. Glycogen synthase kinase 3 phosphorylates kinesin light chains and negatively regulates kinesin‐based motility. The EMBO journal. 2002;21:281–293. doi: 10.1093/emboj/21.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavalli V, Kujala P, Klumperman J, Goldstein LS. Sunday Driver links axonal transport to damage signaling. The Journal of cell biology. 2005;168:775–787. doi: 10.1083/jcb.200410136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Magrané J, Sahawneh MA, Przedborski S, Estévez ÁG, Manfredi G. Mitochondrial dynamics and bioenergetic dysfunction is associated with synaptic alterations in mutant SOD1 motor neurons. Journal of Neuroscience. 2012;32:229–242. doi: 10.1523/JNEUROSCI.1233-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Donnell KC, Vargas ME, Sagasti A. WldS and PGC-1α regulate mitochondrial transport and oxidation state after axonal injury. Journal of Neuroscience. 2013;33:14778–14790. doi: 10.1523/JNEUROSCI.1331-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics international. 2004;11:36–42. [Google Scholar]
- 60.Kumar J, et al. The Caenorhabditis elegans Kinesin-3 motor UNC-104/KIF1A is degraded upon loss of specific binding to cargo. PLoS Genet. 2010;6:e1001200. doi: 10.1371/journal.pgen.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 62.De Castro E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic acids research. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sigrist CJ, et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic acids research. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and reagents are available upon request.