Abstract
Breeding for drought tolerance is a challenging task that requires costly, extensive, and precise phenotyping. Genomic selection (GS) can be used to maximize selection efficiency and the genetic gains in maize (Zea mays L.) breeding programs for drought tolerance. Here, we evaluated the accuracy of genomic selection (GS) using additive (A) and additive + dominance (AD) models to predict the performance of untested maize single-cross hybrids for drought tolerance in multi-environment trials. Phenotypic data of five drought tolerance traits were measured in 308 hybrids along eight trials under water-stressed (WS) and well-watered (WW) conditions over two years and two locations in Brazil. Hybrids’ genotypes were inferred based on their parents’ genotypes (inbred lines) using single-nucleotide polymorphism markers obtained via genotyping-by-sequencing. GS analyses were performed using genomic best linear unbiased prediction by fitting a factor analytic (FA) multiplicative mixed model. Two cross-validation (CV) schemes were tested: CV1 and CV2. The FA framework allowed for investigating the stability of additive and dominance effects across environments, as well as the additive-by-environment and the dominance-by-environment interactions, with interesting applications for parental and hybrid selection. Results showed differences in the predictive accuracy between A and AD models, using both CV1 and CV2, for the five traits in both water conditions. For grain yield (GY) under WS and using CV1, the AD model doubled the predictive accuracy in comparison to the A model. Through CV2, GS models benefit from borrowing information of correlated trials, resulting in an increase of 40% and 9% in the predictive accuracy of GY under WS for A and AD models, respectively. These results highlight the importance of multi-environment trial analyses using GS models that incorporate additive and dominance effects for genomic predictions of GY under drought in maize single-cross hybrids.
Introduction
Accurate prediction of the performance of untested genotypes in one or more environments is essential to maximize genetic gains in breeding programs (Bernardo 1994, 1996). Pedigree-based analyses have been widely used to evaluate field experiments, estimate genetic parameters, and predict breeding values (Piepho et al. 2008). However, due to the decreasing genotyping costs of thousands or millions of markers, and to the increasing phenotyping costs (Krchov and Bernardo 2015), genomic selection (GS; Meuwissen et al. 2001) is emerging as an alternative genome-wide marker-based method to predict yet-to-be seen genetic responses. Appropriate GS methods provide accurate predictions even for untested genotypes, resulting in a considerable progress for breeding programs, reducing the number of field-tested genotypes, with a consequent reduction in the phenotyping costs (Krchov and Bernardo 2015). The benefits of GS are more evident when traits are difficult, time-consuming, and/or expensive to measure, or when several environments need to be evaluated.
In breeding programs for drought tolerance, genotypes are evaluated in water-managed environments, such as well-watered (WW) and water-stressed (WS) conditions, in which an effective phenotypic screening for several traits is often laborious and time-consuming. Thus, the release of new cultivars with yield stability for areas that are prone to water limitations is considered a critical and challenging task. Due to the impact of climate changes and the limitation of water resources to irrigation, yield stability obtained via improved drought tolerance will be highly desirable in the future (Cooper et al. 2014). In maize, a major effect of drought stress is to increase the anthesis-sliking interval (due to a delay in silking), resulting in yield losses (Ribaut et al. 2009; Maazou et al. 2016). In addition, there are also other important traits related to drought tolerance, such as those described in Ribaut et al. (2009) and Tuberosa (2012). Most of the drought tolerance-related traits are controlled by many genes of small effects and strongly influenced by the environment (Ribaut et al. 2009; Zhang et al. 2015). Therefore, GS is expected to increase genetic gains, because its capacity to accurately predict the performance of untested genotypes based on markers distributed throughout the genome. Ziyomo and Bernardo (2013) compared GS to phenotypic selection based on grain yield (GY) and on secondary traits, and showed the advantage of GS to increase genetic gains for drought tolerance in maize. Beyene et al. (2015) and Zhang et al. (2015) also reported superior results with GS in comparison to phenotypic selection for drought tolerance in maize.
Although the estimation of both additive and non-additive (dominance and epistasis) effects helps to improve the understanding of the genetic architecture of target traits and to define optimal breeding strategies, most genetic analyses focus only on the estimation of additive or total genetic effects. The estimation of these effects and their corresponding variance components are often difficult, requiring appropriate mating designs and a large number of observations, given the lack of orthogonality in the estimation process. Some studies have shown that the use of molecular-based relationship matrices greatly improves orthogonality and predictability of both additive and non-additive effects (Vitezica et al. 2013; Muñoz et al. 2014; Nazarian and Gezan 2016b). Furthermore, the inclusion of dominance effects in the GS models is essential for the accurate prediction of untested genotypes in species with some level of heterosis, such as single-cross hybrids in maize (Technow et al. 2014; Almeida Filho et al. 2016; Santos et al. 2016).
Models that evaluate genotype-by-environment (GxE) interaction are essential to any plant breeding program regardless of the method used for genetic prediction. Understanding GxE provides valuable information for breeders, including: (i) evaluation of stability of the genotypes’ response across environments; (ii) selection of genotypes to specific environments; (iii) evaluation/definition of breeding zones; (iv) definition of target environments; and (v) definition of strategies to maximize genetic gain. Recent studies have shown the advantages of GS models that incorporate GxE to predict untested genotypes for quantitative traits (Burgueño et al. 2012; Heslot et al. 2014; Oakey et al. 2016). In maize, prediction of single-cross hybrids has been done with high level of predictive accuracy using GxE models (Technow et al. 2014; Kadam et al. 2016). In the case of drought tolerance in maize, Zhang et al. (2015) showed the advantage of modeling GxE effects for the prediction of untested genotypes.
Several modeling approaches exist to explore GxE. The most interesting ones consider modeling the genetic variance-covariance (VCOV) matrix across environments. Following this matrix structure, it is possible to improve the understanding of GxE and of the genetic architecture of breeding traits, together with the estimation of all environment-to-environment genetic correlations. One parsimonious way to model this genetic VCOV matrix is by using a factor analytic (FA) structure (Piepho 1997, 1998; Smith et al. 2001). The FA structure approximates the unstructured (UN) matrix but with a reduction in the number of parameters to be estimated, which is particularly relevant when the number of environments is large, and also in early generation trials of breeding programs (Kelly et al. 2007). Many studies have shown that FA models are good approximations of the UN models and that they can be implemented in most breeding programs (Burgueño et al. 2008; Cullis et al. 2014; Smith et al. 2015). Another advantage of the FA model is that it can be extended to estimate additive and non-additive effects simultaneously (Kelly et al. 2009).
Few studies have simultaneously incorporated additive, dominance (Azevedo et al. 2015; Bouvet et al. 2015; Santos et al. 2016), and GxE interaction effects (Burgueño et al. 2012; Lopez-cruz et al. 2015; Oakey et al. 2016) into GS models. Reports on the inclusion of additive and dominance effects into multi-environment trials genomic selection (MET-GS) models is limited (Kumar et al. 2015; Kadam et al. 2016). Additionally, some of these studies have not taken full advantage of linear mixed models, such as FA models that can fit different genetic and residual variance components for each environment, and genetic and residual correlations between pairs of environments using additive and dominance effects simultaneously. To our knowledge, latent regression, including additive and dominance effects, has not yet been used to understand GxE in multi-environment trial (MET) analyses. Therefore, the goals of this study were: (i) to evaluate the accuracy of GS to predict the performance of untested maize single-cross hybrids for five traits under two different water conditions using a high-density single-nucleotide polymorphism (SNP) marker panel and multi-environmental trials analyses; (ii) to compare the predictive accuracy achieved by models that account only for additive effects (A model) against models with additive and dominance (AD model) effects; and (iii) to explore the stability of hybrids via latent regression plots using AD models.
Material and methods
Phenotypic data
Field data comprised of 308 single-cross maize hybrids evaluated under WW and WS conditions at two locations in Brazil (Janaúba–Minas Gerais state, and Teresina–Piauí state) over 2 years (2010 and 2011) in a total of eight trials/environments. Hybrids were obtained from single crosses between two testers and 188 inbred lines, representing dent (85 lines) and flint (86 lines) heterotic groups, and also another group–here called group C (17 lines) which is unrelated to both dent and flint sources. The two testers were a flint (L3) and a dent (L228-3) inbred lines. Fifty-two inbred lines were crossed with L228-3 only, and 16 with L3 only, whereas 120 lines were crossed with both testers. Each trial comprised of the 308 maize single-cross hybrids randomly split into six sets i.e., sets 1–3 for L3 crosses, with 61, 61, 14 hybrids each, and sets 4–6 for L228-3 crosses, with 80, 77, and 15 hybrids each. In the field, each set was augmented with four common checks (commercial maize cultivars), and arranged as a randomized complete block design. Although the hybrids within each set were kept the same across trials, hybrids and checks were randomly allocated to groups of plots within each set, and this allocation was different between replicates of sets and between trials. The WS trials had three replicates, except for the sets of 15 hybrids and the trials evaluated in 2010 that had two replicates. All WW trials had two replicates, except the trials in 2011, when both locations had a single replicate. In Janaúba, plots had a 3.6-m row, and in Teresina each plot consisted of a 4-m row. In all trials, the distance between rows was 0.8 m with a density of four plants per meter.
The experiments were performed in a dark red latosol in Janaúba, and in a red-yellow argisol in Teresina. In the WS experiments, the water supply was interrupted before flowering, and the drought stress was imposed during flowering and grain filling. WW conditions were ensured by compensating evapotranspiration losses, based on local climatic data obtained from an automatic weather station; i.e., the frequency and the amount of water supplied by irrigation in the WW experiments were based on the daily crop evapotranspiration index, obtained using the reference evapotranspiration calculated from the Penman–Monteith equation and the crop coefficient per phase, ensuring the occurrence of no stress until the stage of physiological maturity. The water content in the soil was monitored up to a depth of 0.70 m using a DIVINER 2000® probe (Sentek Sensor Technologies, Australia). Other agronomical practices were performed as recommended for maize crops.
Five drought tolerance traits were evaluated: GY (ton/ha), determined by weighing all the grains in each plot, adjusted to 13% of grain moisture and converted to tons per hectare (t/ha), considering the differences in the plot sizes across trials; number of ears per plot (EPP); female and male flowering times (FFT and MFT), measured as the number of days from sowing until silks have emerged on 50% of the plants, and 50% of the plants have begun to shed pollen, respectively; and anthesis-silking interval (ASI, days), which corresponds to the time between FFT and MFT. For the traits GY and EPP, the number of plants per plot was used as a covariate. A summary of phenotypic distribution and phenotypic correlations for all evaluated traits under WW and WS are presented in Fig. S1.
Genotypic data
Genomic DNA was extracted from young leaves of inbred lines based on the cetyl trimethylammonium bromide method (Saghai-Maroof et al. 1984). DNA samples were quantified using the Fluorometer Qubit® 2.0, following the manufacturer’s instructions (Life TechnologiesTM, USA). Samples were also evaluated on 1% agarose gel in Tris-acetate-EDTA buffer, stained with GelRedTM (Biotium, USA) and recorded under UV light in the Imager Gel Doc L-PIX (Loccus Biotecnologia, Brazil). Genotyping-by-sequencing (GBS) was carried out by the Genomic Diversity Facility at Cornell University (Ithaca, NY, USA) using the GBS standard protocol (Elshire et al. 2011) with the restriction enzyme ApeKI and 96 samples per sequencing lane. Tags were aligned to the B73 reference genome (AGPv3) using the Burrows–Wheeler alignment tool (Li and Durbin 2009). Then, SNPs were called using the GBS pipeline, available in the software TASSEL v. 5 (Glaubitz et al. 2014). SNPs were obtained for the 188 inbred lines and the two testers used as parents of the 308 maize hybrids (relationship between lines within and across heterotic group are shown in Fig. S2). SNPs were discarded if: the minor allele frequency was smaller than 5%, more than 20% of missing genotypes were found, and/or there were more than 5% of heterozygous genotypes. After filtering, missing data were imputed using NPUTE (Roberts et al. 2007). Then, for each SNP, the genotypes of the hybrids were inferred based on the genotype of their parents (inbred line and tester). The number of SNPs per chromosome ranged from 3121 (chromosome 10) to 7705 (chromosome 1), totalizing 47,127 markers.
Genomic relationship matrices
Genetic relationships between hybrids were constructed based on the SNP information. Additive (Ag) and dominance (Dg) genomic relationship matrices were calculated following the methods described by Yang et al. (2010) and Vitezica et al. (2013), respectively. Both methods consider two alleles (A and a) for a given marker locus. The Ag matrix was calculated as:
Here, m is the total number of markers, pk is the observed allele frequency of the kth SNP, gki and gkj represent the number of copies of a given allele A for individuals i and j at SNP k, assuming values 2, 1, and 0 for the genotypes AA, Aa, and aa, respectively. The allele A was considered as the most frequent one. The Dg matrix was calculated as:
with
Additive and dominance genomic relationship matrices were obtained using the software GenoMatrix (Nazarian and Gezan 2016a). If Ag and Dg were not positive definite, their inverse were obtained by iterative bending methods as described by Nazarian and Gezan (2016a).
Dependence between additive and dominance variances were evaluated as described by Muñoz et al. (2014). For this, the portions of asymptotic VCOV matrices attributed to additive and dominance components were used to evaluate the non-independence among variance components by calculating and plotting the eigenvalues of the corresponding correlation matrix.
Statistical analyses
Genomic best linear unbiased predictions (GBLUP) were performed for models with additive and dominance effects in single-environment and MET analyses. For MET analyses, different groups of trials were considered: (i) the four trials under WW conditions; (ii) the four trials under WS conditions; and (iii) all eight trials under both WW and WS conditions. However, note that MET analysis considering all the eight trials was used only to investigate additive-by-environment and dominance-by-environment interactions, and to obtain the latent regression plots. The following generic linear mixed model with genomic relationship matrices was fitted for each trait and group:
| 1 |
where y (n × 1) is the vector of phenotypes for q sets, j replicates, m hybrids, and s trials, , where ni is the number of plots in trial s; µ is the overall mean; s (s × 1) is the vector of fixed effects of trials; b.s (qs × 1) is the vector of fixed effects of sets within trials; r.s (jqs × 1) is the vector of fixed effects of replicates within sets within trials; a.s (ms × 1) is the vector of random additive effects of hybrids within trials, with ; d.s (ms × 1) is the vector of random dominance effects of hybrids within trials, with ; and e (n × 1) is the vector of residuals, with . , , , , and represent incidence matrices for their respective effects, 1 is a (n x 1) vector of ones, and is an identity matrix of its corresponding order. R is a diagonal VCOV matrix, in which each trial has a different and independent variance component for the residuals. and are VCOV matrices for the additive and dominance genetic effects of hybrids across trials, with dimensions of 4 × 4 for the group of trials under WW or WS conditions, and 8 × 8 when considering all trials. A FA model of order k (FAk) was considered for both ΣA and ΣD, in which k is the number of multiplicative components (for more details, see Piepho 1998 and Smith et al. 2001). The genomic relationship matrices Ag and Dg were previously specified, and the kronecker product is denoted by ⊗.
The model presented in Eq. (1) corresponds to the AD model, which contains both additive and dominance effects. An alternative model including only additive effects (A model) was also fitted by dropping the term d.s. Narrow-sense heritability (h2) and the proportion of the variance explained by the dominance effects (d2) were estimated for each trait using the following expressions: and , where the bars over the variance components represent the average variance (i.e., diagonal terms) across trials within each water condition. Broad-sense heritability was estimated as .
The FA structures used in the MET analyses were fitted as described in Oakey et al. (2007) and Kelly et al. (2009), in which the vector of genetic effects (ug), that includes both additive and dominance effects, is defined as:
| 2 |
where uA and uD are random vectors of additive and dominance effects within s trials, respectively. These vectors are assumed to be independent with a multivariate normal distribution with zero mean and VCOV matrices and . Here, ΣA and ΣD are s × s matrices for the additive and dominance genetic effects, respectively. The structure of ΣA and ΣD matrices were defined based on a FAk (i.e., of order k) model as:
| 3 |
| 4 |
where and are s × k matrices of factor loadings (common factors) for the additive and dominance effects, respectively, for s trials; and and (diagonal matrices of dimension s × s) are specific factors for the additive and dominance effects, respectively. Common and specific factors are assumed to be independent and normally distributed. Hence, under the above definitions the VCOV matrix of the genetic effects, i.e. cov(ug), can be derived as:
| 5 |
where all terms were previously defined. Note, the use of these VCOV matrices allowed to estimate site-to-site additive, dominance, and additive + dominance genetic correlations.
In a FAk structure, adaptability and stability of genetic effects can be easily assessed using latent regression plots (Cullis et al. 2014; Smith et al. 2015). These plots show the genetic responses to each environment—i.e., the predicted breeding values from marginal prediction as shown in Cullis et al. (2014)—considering the genetic effects as dependent variables against the independent variables—i.e., the rotated estimated factor loadings . Here, the regression plots proposed for additive effects (Cullis et al. 2014) were extended to dominance effects as well, based on the MET-GS model jointly fitted for all eight trials (WS and WW conditions). Plots for the first and second factors were obtained for WW and WS trials together using: (i) plot FA1: against ; and (ii) plot FA2: against , where is the vector of breeding values or dominance deviations of hybrid i within trial s, f is a vector of factors scores, and (*) denotes the vectors after rotation. The important difference here is that plots FA1 and FA2 were constructed for additive and dominance effects separately. Therefore, patterns of additive and dominance effects in terms of adaptability and stability can be easily identified. The rotation of factor loading and factor scores using Varimax rotation was performed to facilitate and simplify the results of FA models.
A and AD models were fitted for all five traits using the library ASReml-R v. 3 (Butler et al. 2009), within the statistical package R v. 3.2.5 (R Core Team 2016) that estimates variance components using restricted maximum likelihood (REML) through the average information algorithm (Gilmour et al. 1995), followed by estimation of fixed and random effects by solving the mixed model equations. Diagnostic plots were used to verify the outliers and normality of the residuals. Akaike information criterion (AIC, Akaike 1974) was used for model comparisons. The standard errors of h2, d2, and H2 were estimated through the Delta method as implemented in the library nadiv (Wolak 2012) available in R v. 3.2.5 (R Core Team 2016), which estimates approximated standard errors of the variance components. It is worth mentioning that preliminary analyses were performed by fitting single-environment models to each trial.
Cross-validation scheme
In order to compare the advantage of models that borrow information between correlated trials, two cross-validation schemes, CV1 and CV2, were performed for MET data analyses as proposed by Burgueño et al. (2012). CV1 is the most traditional cross-validation scheme, in which new hybrids have not been evaluated in any trial/environment. In this case, prediction of the performance of an untested hybrid does not borrow information from any trial for this particular hybrid, but it borrows information from related hybrids; hence, predictions derived from CV1 are based on phenotypic and genotypic information of other hybrids. The CV2 scheme speculates a situation where hybrids are evaluated in some trials but not measured in others. In this case, predictions of an untested hybrid borrow some information from other trials for this particular hybrid, in addition to the information of related hybrids; hence, it takes some advantage of information from correlated trials.
A five-fold cross-validation procedure was implemented for CV1 and CV2 by splitting the total set of hybrids through the stratified sampling method, in a way that both training (80% of the hybrids) and validation (20% of the hybrids) sets contained proportionally all genetic backgrounds (Dent × Dent, Dent × Flint, Flint × Flint, C × Dent, C × Flint) for each fold. Untested hybrids (i.e., hybrids in the validation set) were predicted for each fold of the cross-validation procedure. Then, the prediction accuracy was estimated as the Pearson’s correlation between the observed genotypic values— i.e., obtained from single-environment trial analyses considering hybrids as fixed effects— and the genomic predicted genotypic values. The entire cross-validation process was repeated 10 times for both CV1 and CV2, and it was performed separately for each trait and water condition (WW or WS) using the MET-GS model [1] for additive and additive + dominance effects.
In the CV1 scheme, hybrids in the validation set were considered as not measured in any trial, and in the CV2 hybrids were considered as not evaluated in two trials. For the latter, two trials were randomly selected, e.g. trials 1 and 3, and missing values were set in these trials for the hybrids in the validation set. Latter, for the other two trials, e.g. trials 2 and 4, the same validation set was used to assign missing values for the hybrids in these two trials, and the phenotypic values of trials 1 and 3 were considered as observed for the genomic predictions in trials 2 and 4. This process was repeated for each fold in CV2.
Results
Estimates of genetic parameters using high-density SNP markers
Broad-sense and narrow-sense heritabilities (H2 and h2, respectively) varied considerably between WW and WS conditions in the MET-GS analyses (Table 1). For GY, ASI, and FFT, the heritabilities were lower in WS than under WW conditions. The highest H2 and h2 were found for ASI under WW and for MFT under WS respectively. Based on the AD model, ASI showed h2 in WS conditions 51% smaller than in WW conditions. Similar trends were observed for ASI broad-sense heritabilities, with values of 0.35 and 0.53 in WS and WW conditions, respectively. AD models exhibited a considerable decrease in the additive variance component, and consequently in h2, for GY in both water conditions and for EPP, FFT, and MFT in WS conditions. For example, GY showed a decrease of 50% in h2 in WS conditions when the dominance effects were incorporated in the GS model. However, for ASI and EPP, h2 values were almost the same for A and AD models in WW conditions. A similar pattern was also observed in the single-environment trial analyses (Supplementary Tables S1–S5).
Table 1.
Estimates of the genetic parameters and the goodness-of-fit-measures obtained via MET analyses (model [1]) for WW and WS conditions, including additive (model A) and additive-dominance (model AD) effects
| WS | WW | ||||
|---|---|---|---|---|---|
| A | AD | A | AD | ||
| GY | h 2 | 0.40 (0.02) | 0.20 (0.03) | 0.45 (0.03) | 0.28 (0.03) |
| d 2 | – | 0.14 (0.02) | – | 0.20 (0.02) | |
| H 2 | – | 0.34 (0.03) | – | 0.48 (0.02) | |
| LogL | −2332.79 | −2237.95 | −1661.97 | −1611.48 | |
| AIC | 4697.58 | 4531.90 | 3355.94 | 3278.95 | |
| EPP | h 2 | 0.33 (0.02) | 0.24 (0.02) | 0.23 (0.02) | 0.22 (0.03) |
| d 2 | – | 0.12 (0.01) | – | 0.07 (0.03) | |
| H 2 | – | 0.35 (0.02) | – | 0.29 (0.03) | |
| LogL | −5082.60 | −5042.37 | −2718.68 | −2707.91 | |
| AIC | 10197.22 | 10140.73 | 5469.37 | 5471.82 | |
| ASI | h 2 | 0.30 (0.06) | 0.25 (0.06) | 0.50 (0.03) | 0.49 (0.03) |
| d 2 | – | 0.10 (0.02) | – | 0.04 (0.02) | |
| H 2 | – | 0.35 (0.05) | – | 0.53 (0.03) | |
| LogL | −3455.46 | −3428.31 | −678.13 | −669.34 | |
| AIC | 6942.87 | 6912.62 | 1388.26 | 1394.69 | |
| FFT | h 2 | 0.44 (0.02) | 0.31 (0.03) | 0.49 (0.03) | 0.43 (0.03) |
| d 2 | – | 0.09 (0.04) | – | 0.08 (0.02) | |
| H 2 | – | 0.40 (0.04) | – | 0.51 (0.03) | |
| LogL | −4824.27 | −4775.49 | −2298.95 | −2277.53 | |
| AIC | 9681.13 | 9606.98 | 4629.91 | 4611.06 | |
| MFT | h 2 | 0.58 (0.02) | 0.44 (0.03) | 0.48 (0.03) | 0.41 (0.03) |
| d 2 | – | 0.07 (0.01) | – | 0.08 (0.02) | |
| H 2 | – | 0.51 (0.02) | – | 0.49 (0.03) | |
| LogL | −3567.26 | −3538.55 | −2112.20 | −2089.74 | |
| AIC | 7166.52 | 7133.11 | 4256.40 | 4235.47 | |
h2 narrow-sense heritability, d2 proportion of the total genetic variance explained by the dominance effects, H2 broad-sense heritability, LogL REML log-likelihood of the fitted model. Traits correspond to grain yield (GY, t/ha), number of ears per plot (EPP), anthesis-silking interval (ASI, days), female flowering time (FFT, days), and male flowering time (MFT, days)
Values within parentheses are approximated standard errors
For almost all traits, except GY and MFT, the proportion of the total genetic variance explained by the dominance effects (d2) was smaller in WW than in WS conditions (Table 1). For example, ASI showed d2 more than twice larger in WS than in WW conditions. In general, the ratio h2/d2 ranged from 1.43 (GY) to 6.28 (MFT) in WS, and from 1.40 (GY) to 12.25 (ASI) in WW conditions. Moreover, the AIC criterion showed that, in general, the inclusion of dominance effects improved the model fitness compared with the A model, except for ASI and EPP under WW. The dependency between additive and dominance genomic relationship matrices is presented in Supplementary Fig. S3. A small dependency between Ag and Dg was observed when compared with a hypothetical situation of orthogonality (diagonal solid line, Fig. S3). However, in 2011 for the WW condition of Janaúba and Teresina, where unreplicated trials were used to evaluate hybrids, the dependency between Ag and Dg was higher for the secondary traits (ASI, EPP, FFT and MFT).
Average additive and dominance genetic correlations, estimated through the FA models between pairs of trials within and across water conditions, are shown in Fig. 1. Additive genetic correlations ranged from 0.39 (WS_GY) to 0.81 (WS_MFT), and dominance genetic correlations ranged from 0.003 (WS_ASI) to 0.72 (WS_MFT) within water conditions. Based on these correlations for each water condition, it was noted that the dominance effects exhibited, in general, lower correlation between trials (higher crossover interaction) than the additive effects, except for WS_EPP and WS_GY. The additive effects showed low interaction with environments (exhibit smaller crossover interaction) for the traits MFT and FFT in both conditions, with correlations greater than 0.70. In contrast, all the dominance correlations were smaller than 0.60, except for MFT in WS. Similar trends were observed across water conditions, with dominance correlation lower than additive correlation for all traits. Further information showing the correlations for additive and dominance effects for each pair of trials is presented in supplementary heatmaps (Figs. S4, S5). Likelihood ratio test for the additive and dominance by environment interaction are shown in Table S6.
Fig. 1.
Average additive (A) and dominance (D) genetic correlations, estimated via additive + dominance (AD) model (Eq. (1)) between pairs of trials within each water regime—i.e., well-watered (WW) and water-stressed (WS) conditions, and across water regimes (WW/WS). Traits correspond to grain yield (GY, t/ha), number of ears per plot (EPP), anthesis-silking interval (ASI, days), female flowering time (FFT, days), and male flowering time (MFT, days)
Accuracy of MET-GS models for drought tolerance-related traits
Predictive accuracy varied considerably across WW and WS conditions (Fig. 2). In general, considering only CV1, the predictive accuracy was higher in WW conditions, ranging from 0.36 (GY) to 0.57 (FFT) for the A model, and from 0.43 (EPP) to 0.59 (MFT and FFT) for the AD model. For GY, when the dominance effects were included in the model, the predictive accuracy was approximately 28% and 100% higher in WW and WS conditions, respectively, than using the A model. EPP, FFT, and MFT in WS showed an increase of 26, 17, and 16% in the predictive accuracy, when the dominance effects were considered in the MET-GS model, respectively. However, ASI in WS conditions, and ASI, EPP, FFT, and MFT in WW conditions, did not exhibit considerable increase in the predictive accuracy (lower than 10%) when dominance effects were included in the MET-GS model.
Fig. 2.
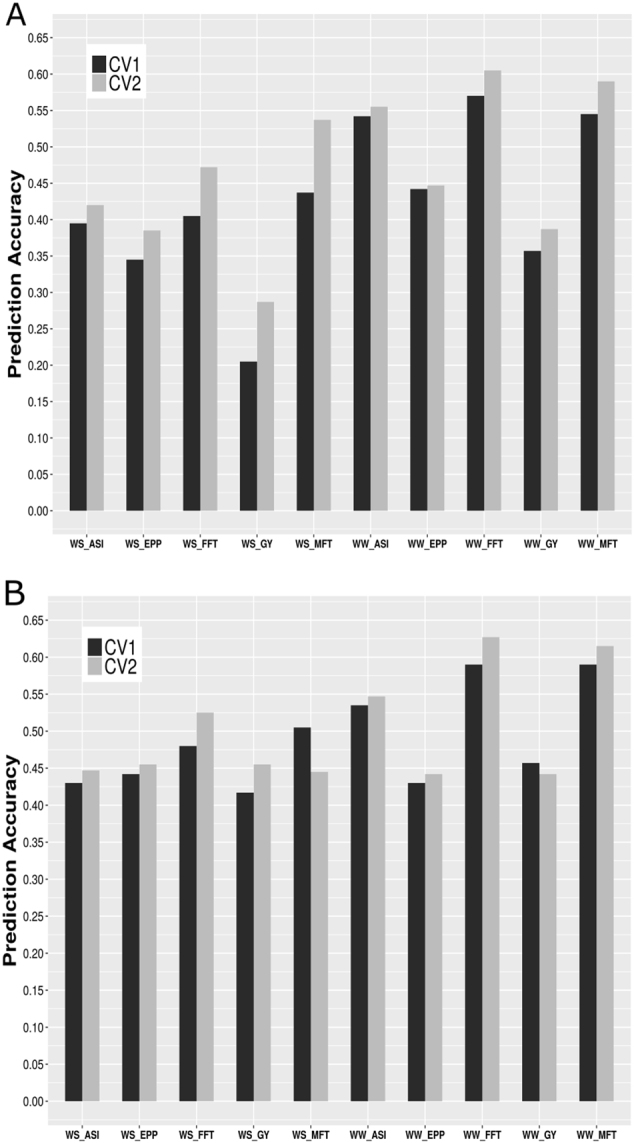
Average predictive accuracy (across four trials) for the additive (a) and for the additive + dominance (b) models under well-watered (WW) and water-stressed (WS) conditions. Traits correspond to grain yield (GY, t/ha), number of ears per plot (EPP), anthesis-silking interval (ASI, days), female flowering time (FFT, days), and male flowering time (MFT, days)
Important differences in terms of predictive accuracies were observed between GY and the secondary traits for the A model in both CV1 and CV2 (Fig. 2). The A model in WW conditions resulted in a predictive accuracy higher than 0.44 for all secondary traits, which was at least 22% higher than the predictive accuracy observed for GY in CV1. For the A model in WS conditions, such differences were more evident, where the predictive accuracies for ASI, FFT, and MFT were almost twice larger than the one observed for GY in CV1. For the AD models, the differences between the average predictive accuracies obtained for the secondary traits and GY were less evident in both conditions and cross-validation schemes compared to the A model (Fig. 2, Tables 2 and 3).
Table 2.
Predictive accuracy based on the CV1 scheme for five traits evaluated in WW and WS trials using additive (A) and additive + dominance (AD) models [1]
| Model | Trial | GY | EPP | ASI | FFT | MFT |
|---|---|---|---|---|---|---|
| WS | ||||||
| A | WSJ10 | 0.28 (0.01) | 0.21 (0.04) | 0.69 (0.01) | 0.67 (0.01) | 0.56 (0.03) |
| WSJ11 | 0.03 (0.02) | 0.20 (0.01) | 0.43 (0.03) | 0.53 (0.01) | 0.57 (0.02) | |
| WST10 | 0.34 (0.02) | 0.61 (0.01) | 0.09 (0.03) | 0.11 (0.04) | 0.29 (0.03) | |
| WST11 | 0.17 (0.03) | 0.36 (0.03) | 0.37 (0.02) | 0.31 (0.02) | 0.33 (0.02) | |
| AD | WSJ10 | 0.36 (0.02) | 0.27 (0.03) | 0.70 (0.01) | 0.71 (0.02) | 0.63 (0.02) |
| WSJ11 | 0.49 (0.02) | 0.44 (0.02) | 0.51 (0.02) | 0.68 (0.02) | 0.64 (0.01) | |
| WST10 | 0.31 (0.02) | 0.61 (0.01) | 0.11 (0.04) | 0.09 (0.03) | 0.32 (0.03) | |
| WST11 | 0.51 (0.02) | 0.45 (0.01) | 0.40 (0.02) | 0.44 (0.02) | 0.43 (0.01) | |
| WW | ||||||
| A | WWJ10 | 0.37 (0.02) | 0.40 (0.03) | 0.52 (0.02) | 0.60 (0.01) | 0.56 (0.02) |
| WWJ11 | 0.33 (0.01) | 0.44 (0.01) | 0.61 (0.02) | 0.44 (0.01) | 0.49 (0.02) | |
| WWT10 | 0.42 (0.02) | 0.53 (0.01) | 0.54 (0.01) | 0.69 (0.01) | 0.62 (0.01) | |
| WWT11 | 0.31 (0.02) | 0.40 (0.02) | 0.50 (0.02) | 0.55 (0.02) | 0.51 (0.01) | |
| AD | WWJ10 | 0.58 (0.02) | 0.39 (0.02) | 0.52 (0.02) | 0.65 (0.02) | 0.65 (0.02) |
| WWJ11 | 0.40 (0.01) | 0.40 (0.02) | 0.60 (0.01) | 0.46 (0.02) | 0.51 (0.03) | |
| WWT10 | 0.52 (0.01) | 0.56 (0.01) | 0.54 (0.02) | 0.72 (0.01) | 0.62 (0.02) | |
| WWT11 | 0.33 (0.02) | 0.37 (0.02) | 0.48 (0.01) | 0.61 (0.01) | 0.58 (0.02) | |
Traits correspond to grain yield (GY, t/ha), number of ears per plot (EPP), anthesis-silking interval (ASI, days), female flowering time (FFT, days), and male flowering time (MFT, days)
Values within parentheses are standard deviations
Table 3.
Predictive accuracy based on CV2 scheme for five traits evaluated in WW and WS trials using additive (A) and additive + dominance (AD) models [1]
| Model | Trial | GY | EPP | ASI | FFT | MFT |
|---|---|---|---|---|---|---|
| WS | ||||||
| A | WSJ10 | 0.31 (0.04) | 0.26 (0.04) | 0.70 (0.01) | 0.71 (0.02) | 0.65 (0.03) |
| WSJ11 | 0.18 (0.07) | 0.28 (0.05) | 0.47 (0.04) | 0.62 (0.04) | 0.64 (0.03) | |
| WST10 | 0.37 (0.02) | 0.61 (0.01) | 0.12 (0.02) | 0.17 (0.05) | 0.40 (0.04) | |
| WST11 | 0.29 (0.06) | 0.39 (0.03) | 0.39 (0.02) | 0.40 (0.06) | 0.46 (0.06) | |
| AD | WSJ10 | 0.38 (0.03) | 0.27 (0.02) | 0.70 (0.01) | 0.73 (0.02) | 0.59 (0.02) |
| WSJ11 | 0.54 (0.02) | 0.48 (0.03) | 0.55 (0.02) | 0.70 (0.02) | 0.57 (0.04) | |
| WST10 | 0.34 (0.04) | 0.60 (0.01) | 0.13 (0.02) | 0.16 (0.02) | 0.16 (0.02) | |
| WST11 | 0.56 (0.02) | 0.47 (0.01) | 0.41 (0.02) | 0.51 (0.02) | 0.46 (0.05) | |
| WW | ||||||
| A | WWJ10 | 0.42 (0.03) | 0.42 (0.03) | 0.54 (0.02) | 0.65 (0.03) | 0.62 (0.03) |
| WWJ11 | 0.37 (0.03) | 0.44 (0.01) | 0.64 (0.01) | 0.50 (0.03) | 0.55 (0.04) | |
| WWT10 | 0.46 (0.02) | 0.55 (0.01) | 0.55 (0.01) | 0.70 (0.01) | 0.62 (0.01) | |
| WWT11 | 0.30 (0.02) | 0.38 (0.02) | 0.49 (0.02) | 0.57 (0.02) | 0.57 (0.04) | |
| AD | WWJ10 | 0.52 (0.02) | 0.41 (0.02) | 0.55 (0.01) | 0.66 (0.01) | 0.67 (0.01) |
| WWJ11 | 0.40 (0.02) | 0.42 (0.02) | 0.62 (0.02) | 0.51 (0.04) | 0.56 (0.02) | |
| WWT10 | 0.53 (0.02) | 0.57 (0.01) | 0.54 (0.01) | 0.71 (0.01) | 0.62 (0.02) | |
| WWT11 | 0.32 (0.03) | 0.37 (0.03) | 0.48 (0.02) | 0.63 (0.02) | 0.61 (0.01) | |
Traits correspond to grain yield (GY, t/ha), number of ears per plot (EPP), anthesis-silking interval (ASI, days), female flowering time (FFT, days), and male flowering time (MFT, days)
Values within parentheses are standard deviations
GxE interaction model for additive and additive + dominance effects
For the A model, in general, and as expected, predictive accuracies were higher in CV2 than in CV1; however, for the AD model, the differences between CV2 and CV1 were smaller (Tables 2 and 3). Average predictive accuracy (across four trials under each water regime) for CV1 and CV2 are given in Fig. 2. In the CV2 scheme the benefits of borrowing information from correlated trials with the same hybrid were more evident under WS conditions, with values ranging from 0.29 (GY) to 0.54 (MFT) for the A model, and from 0.45 (ASI) to 0.53 (FFT) for the AD model. For GY under WS, the predictive accuracy in CV2 increased 40% for the A model, and 9% for the AD model when compared to CV1, highlighting the advantage of CV2 for MET genomic predictions.
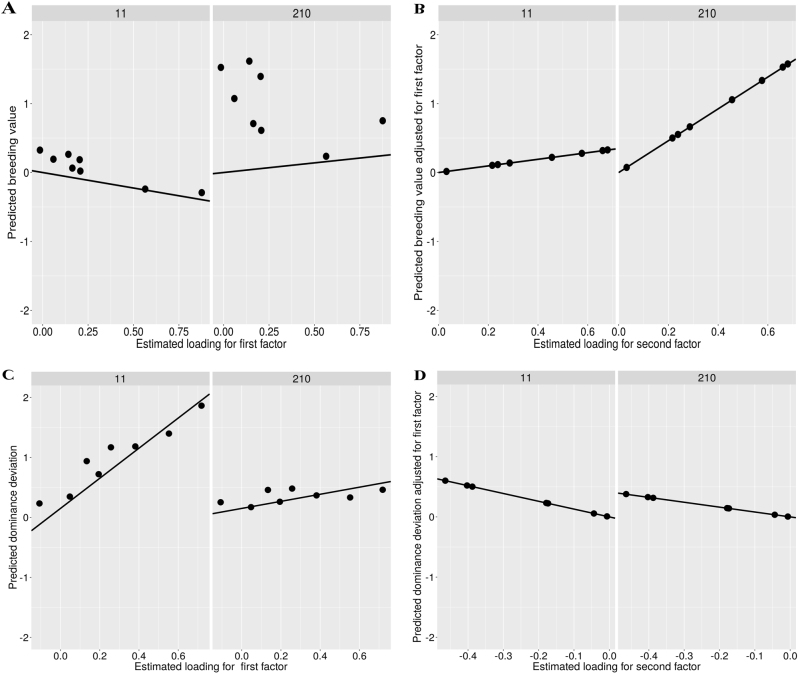
Factor scores of the additive and the dominance effects for the 15 top hybrids for GY are shown in Table 4. Among them, 13 hybrids represent crosses between flint and dent heterotic groups. Here, additive and dominance effects with the first and the second factor scores close to the origin — i.e., close to (0, 0) — have stable performance across all trials, and, therefore, across WW and/or WS conditions. Four hybrids (35, 47, 258, and 265) showed stability (i.e., all factor scores lower than 0.4) for additive and dominance effects in both water conditions for GY (data not shown). However, these hybrids had poor performance for GY. Among the best hybrids, some exhibited only stable additive effects, whereas others had only stable dominance effects. For example, hybrid 11 showed only stable additive effects, whereas hybrid 210 exhibited only stable dominance effects (Table 4). Latent regression plots for the first and the second factors of additive and dominance effects of hybrids 11 and 210 are shown in Fig. 3. Note that the additive effects of hybrid 210 increased in trials with higher estimated loadings, as suggested by the latent regression (Fig. 3a, b). On the other hand, for hybrid 11, the dominance effects increased in the trials with higher estimated loadings (Fig. 3c, d).
Table 4.
Predicted factor scores for the additive and the dominance effects of the top 15 hybrids based on best linear unbiased predictions (BLUP) for GY (t/ha) from an MET analysis with additive and dominance effects (model AD)
| Hybrid | Background | BLUPa | Additive effect | Dominance effect | |||
|---|---|---|---|---|---|---|---|
| Additive | Dominance | Factor 1 | Factor 2 | Factor 1 | Factor 2 | ||
| 42 | D_F | 0.81 | 0.68 | 0.53 | 1.55 | 1.17 | −1.18 |
| 97 | D_F | 0.85 | 0.63 | 0.66 | 1.58 | 2.10 | −0.05 |
| 44 | D_F | 0.81 | 0.63 | 0.39 | 1.55 | 1.45 | −0.52 |
| 283 | F_F | 0.91 | 0.51 | −0.22 | 1.51 | 0.95 | −1.59 |
| 210 | C_F | 1.21 | 0.20 | 0.28 | 2.31 | 0.59 | −0.81 |
| 132 | D_F | 0.77 | 0.64 | 0.40 | 1.49 | 1.97 | −0.48 |
| 120 | D_F | 0.84 | 0.55 | −0.21 | 1.33 | 1.17 | −0.36 |
| 185 | D_F | 0.57 | 0.77 | −0.31 | 0.92 | 2.12 | −0.83 |
| 11 | D_F | 0.27 | 1.03 | −0.45 | 0.48 | 2.51 | −1.29 |
| 90 | D_F | 0.87 | 0.42 | 0.11 | 1.47 | 0.82 | −0.70 |
| 239 | D_F | 0.83 | 0.44 | 1.16 | 1.50 | 0.72 | −0.94 |
| 181 | D_F | 0.66 | 0.60 | 0.69 | 1.16 | 1.55 | −0.79 |
| 291 | D_F | 0.78 | 0.48 | 0.44 | 1.58 | 0.46 | −1.39 |
| 48 | D_F | 0.65 | 0.60 | −0.72 | 1.08 | 1.31 | −1.48 |
| 138 | D_F | 0.60 | 0.65 | 0.22 | 1.18 | 1.46 | −0.75 |
For all trials, factor scores are rotated with the Varimax rotation. Background is D_F (dent × flint), C_F (c × flint), F_F (flint × flint)
aEstimated genotypic values (additive plus dominance effects) from AD models considering all trials
Fig. 3.
Latent regression plots for the first (a) and the second (b) factors of additive effects, and latent regression plots for the first (c) and the second (d) factors of dominance effects for hybrids 11 and 210
Discussion
Improving accuracy of genomic prediction for untested hybrids in maize is a recurrent challenge for the successful application of GS in breeding programs. This requires models that account for multi-environment trials data, as well as for additive and non-additive effects. Our results, based on tropical maize germplasm cultivated in Brazil, show that it is possible to achieve high levels of predictive accuracy of untested hybrids for drought tolerance-related traits by including GxE, additive, and dominance effects simultaneously into a MET model that incorporates genomic relationship matrices.
Partition of the genetic variance through the incorporation of SNP markers
Orthogonal partitioning of the genetic variance into additive and dominance effects was performed based on the use of genomic relationship matrices. This partitioning contributes to a better understanding of the genetic architecture of target traits. In maize, for instance, this knowledge assists breeders to decide if target traits should be better evaluated in inbred lines or hybrids. The genomic relationship matrices used in this study were constructed based on the quantitative genetics theory, under the assumption that the covariance between additive and dominance effects is zero (Vitezica et al. 2013). This parameterization has been previously used in plants (Muñoz et al. 2014; Bouvet et al. 2015), humans (Zhu et al. 2015), and in simulation studies (Almeida Filho et al. 2016; Nazarian and Gezan 2016b; Santos et al. 2016), and showed reasonable orthogonal partitioning of additive and non-additive effects. These studies also highlighted the importance of a better partitioning of the genetic components whenever genomic relationship matrices are incorporated.
In the present study, narrow-sense heritabilities decreased when the dominance effects were included in the MET-GS models for GY (Table 1), probably due to the distribution of allele frequencies at causal loci (Hill et al. 2008). In this case, depending on the allele frequencies, part of these effects can be estimated as additive variance, even in the presence of non-additive effects (Hill et al. 2008). Our findings, showing that h2 values decreased when dominance effects were included in the models, agree with results published by Muñoz et al. (2014) and Bouvet et al. (2015) in pine and eucalyptus, respectively. However, to our knowledge, ours is the first study to incorporate GxE, additive, and dominance effects into the genomic prediction framework of single-cross hybrids for drought tolerance-related traits in maize. Therefore, the presence of dominance in the genetic models is important for obtaining a realistic and more accurate partitioning of the genetic variance. As seen in Table 1, narrow-sense heritabilities and the genetic gains may be overestimated when only an additive model is considered.
Accuracy of METs GS models
The differences in predictive accuracy between A and AD models found in this study were more evident in WS than in WW conditions (Fig. 2). Small differences were observed between the predictive accuracies of the A and AD models for the secondary traits — i.e., EPP and ASI (under WS) and ASI, EPP, and FFT (under WW). In contrast, dominance effects had an important contribution for the genomic predictions of GY. One possible explanation for this is that traits with small differences in the predictive accuracies between A and AD models are expected to exhibit smaller contribution of dominance effects in the total genetic variance (Table 1). Thus, the results of this study suggest that the genetic architecture of a drought tolerance trait affects the prediction accuracy of A and AD model.
Secondary traits, such as ASI, FFT, and MFT, had similar or higher prediction accuracies than GY in WW and WS conditions for both CV schemes. These differences were less evident in CV2, where models borrowed information from correlated trials. These results are in good agreement with the results of other GS studies for drought tolerance in maize (Ziyomo and Bernardo 2013; Zhang et al. 2015). Additionally, comparisons between GS and phenotypic selection for drought tolerance in biparental maize populations showed that, after three cycles of selection, GS achieved larger genetic gains (Beyene et al. 2015). The values of predictive accuracies found in this study, which are similar or higher than the ones reported in other GS studies, suggested that including a dominance term in the model can improve considerably selection of untested hybrids for drought tolerance.
In general, prediction accuracy within D_D (dent × dent) or F_F (flint × flint) groups were higher than in the D_F (dent × flint) group for GY (data not shown). However, other relevant studies performed in maize, which have often considered larger population sizes, have suggested that GS models, including specific marker effects for each heterotic group, had little or no advantage over models that assume consistent marker effects across heterotic groups (Technow et al. 2012; Technow et al. 2014). Thus, future studies in tropical maize with a larger population size, and considering additional testers, are required to generalize this conclusion.
In recent years, many statistical models have been proposed for the application of GS in plant and animal breeding programs. Although comparisons between different statistical models for GS are important, GBLUP has shown high levels of predictive accuracy, often ranking among the best predictive approaches (for further details, see Heslot et al. 2012; Resende Jr. et al. 2012). Azevedo et al. (2015) compared different models for GS that incorporate additive and dominance effects, and concluded that GBLUP was among the best methods and provided an accurate prediction of total genotypic values, as well as the additive and dominance effects. In addition, GBLUP often reduces computing resources and allows for fitting complex linear mixed models such as single-step models applied to METs and multi-trait analyses (Meyer 2009; Cullis et al. 2014; Oakey et al. 2016). Here, we extended the GBLUP method to account for additive and dominance effects in the context of MET data using FA structures, which has been widely recommended as an efficient VCOV model to perform MET analysis.
GxE interaction for additive and dominance effects
Estimation of GxE effects in the MET analysis provides valuable information about the stability of hybrids in both WW and WS conditions. However, an appropriate statistical model that accounts for genetic and residual correlations across trials and deals with unbalanced data is required. GxE explicit models that have a main genetic effect and a GxE interaction effect provide the same results as GxE implicit models where there is a single term for genotype effects within environments, with a VCOV matrix based on the compound symmetry structure (Smith et al. 2001; Cullis et al. 2014). This means that the GxE explicit models assume the same genetic variance and covariance (and hence correlation) across all trials and pairs of trials, respectively (Smith et al. 2001, 2015). Therefore, the GxE implicit models in comparison to the explicit models have the advantage of evaluating different VCOV matrix structures, such as UN or FA, allowing to fit more realistic models. Hence, the presence of high GxE for most of the traits analyzed by breeders highlights the importance of using models that can deal with MET data and can properly model GxE interactions.
Modeling complex genetic VCOV matrices did not improve the predictive accuracy in CV1 (see Table S7). For example, genomic predictions of GY under WS using additive effects, considering even an identity, a diagonal, or a FA2 VCOV structure for the genetic effects, and a residual diagonal matrix, had similar values of predictive accuracy. Further, advantages of CV2 were observed for a FA2 model (see Table S8). As also indicated by Burgueño et al. (2011, 2012), the CV1 scheme provides results similar as the ones provided by fitting a GBLUP model for each trial. In other words, modeling the genetic covariances in CV2 allows to borrow the information of hybrids across trials, whereas CV1 assumes that the hybrids in the validation set have not been evaluated in any environment.
Recently, Lopez-cruz et al. (2015) showed in three wheat data sets that GS models that account for GxE resulted, as expected, in larger prediction accuracies than models without GxE modeling. In the case of drought tolerance in biparental maize populations, the prediction accuracy was also increased by fitting a model that incorporated GxE for GY (Zhang et al. 2015). Our findings confirm the results of Burgueño et al. (2012), Lopez-cruz et al. (2015), and Zhang et al. (2015), which showed that predicting performance of new genotypes is more challenging than predicting genotypes that have been evaluated in some correlated trials. If WW and WS information were combined, based on CV2 scheme, then prediction accuracy would likely increase, since the additive and the dominance genetic correlations were modeled across water conditions.
In the present study, we have used latent regression plots based on a FA VCOV structure to visualize additive-by-environment and dominance-by-environment interaction effects (Fig. 3). These plots are useful to infer about the stability of additive and dominance effects across environments for a given hybrid. Based on these plots, it is possible to select, among all high-performance hybrids, those with stable additive effects across environments to inter-mate their parents (inbred lines) in the subsequent breeding cycles. Then, optimized crosses through the selection of the best and stable parents can be performed aiming to improve hybrids stability and expected genetic gains (Toro and Varona 2010). This approach can be easily extended to other crops, such as sugarcane, eucalyptus, and pine, in which the evaluated individual is used as parent to generate the next breeding cycle. In maize breeding, testcrosses are commonly used to evaluate the performance of new inbred lines using elite inbred testers from complementary heterotic groups. Thus, in this case, the new inbred lines identified as parents of high-performance testcross hybrids, with stable additive effects across environments, can be selected to be used for inbred recycling and/or inter-population selection. Moreover, high-performance testcross hybrids exhibiting stable additive and dominance effects across environments can be directly indicated as a potential hybrid cultivar for a given mega environment.
Implementation of GS in a maize breeding program for drought tolerance
There are at least two scenarios in which GS can be applied in a maize breeding program for drought tolerance. First, focusing on additive effects, GS can be used for parental selection and for successive cycles of intermating and selection within breeding populations (usually biparental populations). In this case, a GS model can be used to perform more than one breeding cycle per year, increasing the genetic gains per unit of time. Applying a similar approach, Beyene et al. (2015) showed the advantage of this GS strategy over phenotypic selection to increase genetic gains in drought tolerance breeding programs in maize. In another scenario, that focuses on additive and dominance effects, GS can be employed to predict untested hybrids, which has an important role in maize breeding programs, allowing for the reduction in the number of tested (phenotyped) hybrids. In both scenarios, there are time and financial resources benefits. Recently, Krchov and Bernardo (2015) reported that some financial resources can be saved when using GS in a breeding program, given that the genotyping costs are currently decreasing. These authors, also emphasized the importance of GS to predict the performance of double haploid (DH) lines that do not have enough seeds for testcrossing and phenotyping.
Our findings suggest that GBLUP models that account simultaneously for GxE, additive, and dominance effects should be routinely used in any MET-GS analysis to predict the performance of untested hybrids for drought tolerance in maize breeding programs. These MET models that incorporate genomic relationship matrices can be easily extended to other crops, such as outcrossing species, in which non-additive effects are important. However, in advanced stages of a breeding program, one-stage analysis using FA models, as used in this study, may be challenging to fit, since hybrids are usually evaluated across many locations and years. One option is to perform a two-stage analysis that first fits every single environment and then combines the estimated means into a second and larger analysis (for further details, see Mohring and Piepho 2009). By fitting MET linear mixed models that included additive and dominance effects through a FA structure, it was possible to investigate the stability of these effects across environments, as well as additive-by-environment and dominance-by-environment interactions, with interesting applications for parental and hybrid selection in maize. Moreover, our results contributed to a better understanding of the genetic architecture of important traits related to drought tolerance in WW and WS conditions and highlighted the importance of MET-GS that account for dominance effects for the prediction of maize single-cross hybrids.
Data archiving
Data available in the Dryad Digital Repository: 10.5061/dryad.ps22r.
Electronic supplementary material
Acknowledgements
This research was supported by FAPEMIG (Fundação de Amparo à Pesquisa de Minas Gerais), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, program PREMIO 2045/2014, grant 23038.007195/2012-39), and Embrapa (Brazilian Agricultural Research Corporation). KOGD was also supported by FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo, grant 2016/12977-7). The authors thank the anonymous reviewers for their suggestions, and Jhonathan Pedroso Rigal dos Santos, Samuel Bonfim Fernandes, and Matheus Dalsente Krause to carefully read and give suggestions for the final English version of the manuscript. The authors also thank all the research and field assistants who helped to conduct field experiments at Embrapa Maize and Sorghum and Embrapa Mid-North in Brazil.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41437-018-0053-6) contains supplementary material, which is available to authorized users.
Contributor Information
Lauro José Moreira Guimarães, Email: lauro.guimaraes@embrapa.br.
Maria Marta Pastina, Email: marta.pastina@embrapa.br.
References
- Akaike H. New look at statistical-model identification. Trans Autom Control. 1974;19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- Almeida Filho JE, Guimarães JFR, Silva FF, Resende MDV, Muñoz P, Kirst M, et al. The contribution of dominance to phenotype prediction in a pine breeding and simulated population. Heredity. 2016;117:33–41. doi: 10.1038/hdy.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo CF, Resende MDV, Silva FF, Viana JMS, Valente MSF, Resende JRMFR, Muñoz P. Ridge, Lasso and Bayesian additive dominance genomic models. BMC Genet. 2015;16:1–13. doi: 10.1186/s12863-015-0264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo R. Prediction of maize single-cross performance using RFLPs and information from related hybrids. Crop Sci. 1994;34:20–25. doi: 10.2135/cropsci1994.0011183X003400010003x. [DOI] [Google Scholar]
- Bernardo R. Testcross additive and dominance effects in best linear unbiased prediction of maize single-cross performance. Crop Sci. 1996;93:1098–1102. doi: 10.1007/BF00230131. [DOI] [PubMed] [Google Scholar]
- Beyene Y, Semagn K, Mugo S, Tarekegne A, Babu R, Meisel B, et al. Genetic gains in grain yield through genomic selection in eight bi-parental maize populations under drought stress. Crop Sci. 2015;55:154–163. doi: 10.2135/cropsci2014.07.0460. [DOI] [Google Scholar]
- Bouvet JM, Makouanzi G, Cros D, Vigneron PH. Modeling additive and non-additive effects in a hybrid population using genome-wide genotyping: prediction accuracy implications. Heredity. 2015;115:146–157. doi: 10.1038/hdy.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgueño J, Crossa J, Cornelius PL, Yang RC. Using factor analytic models for joining environments and genotypes without crossover genotype x environment interaction. Crop Sci. 2008;48:1291–1305. doi: 10.2135/cropsci2007.11.0632. [DOI] [Google Scholar]
- Burgueño J, Crossa J, Cotes JM, Vicente FS, Biswanath D. Prediction assessment of linear mixed models for multi-environment trials. Crop Sci. 2011;51:944–954. doi: 10.2135/cropsci2010.07.0403. [DOI] [Google Scholar]
- Burgueño J, De Los Campos G, Weigel K, Crossa J. Genomic prediction of breeding values when modeling genotype x environment interaction using pedigree and dense molecular markers. Crop Sci. 2012;52:707–719. doi: 10.2135/cropsci2011.06.0299. [DOI] [Google Scholar]
- Butler DG, Cullis BR, Gilmour AR, Gogel BJ (2009) ASReml-R Reference Manual. Release 3. Technical Report, Queensland Department of Primary Industries, 160 pp
- Cooper M, Gho C, Leafgren R, Tang T, Messina C. Breeding drought tolerant maize hybrids for the US corn-belt: discovery to product. J Exp Bot. 2014;65:1–14. doi: 10.1093/jxb/eru064. [DOI] [PubMed] [Google Scholar]
- Cullis B, Jefferson P, Thompson R, Smith AB. Factor analytic and reduced animal models for the investigation of additive genotype-by-environment interaction in outcrossing plant species with application to a Pinus radiata breeding program. Theor Appl Genet. 2014;127:2193–2210. doi: 10.1007/s00122-014-2373-0. [DOI] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler E, et al. A robust simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6:1–10. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour AR, Thompson R, Cullis BR. AI, an efficient algorithm for REML estimation in linear mixed models. Biometrics. 1995;51:1440–1450. doi: 10.2307/2533274. [DOI] [Google Scholar]
- Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, et al. A high capacity genotyping by sequencing analysis pipeline. PLoS ONE. 2014;9:903–916. doi: 10.1371/journal.pone.0090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslot N, Yang HP, Sorrels ME, Jannink JL. Genomic selection in plant breeding: a comparison of models. Crop Sci. 2012;52:146–160. doi: 10.2135/cropsci2011.06.0297. [DOI] [Google Scholar]
- Heslot N, Akdemir D, Sorrels ME, Jannink JL. Integrating environmental covariates and crop modeling into the genomic selection framework to predict genotype by environment interactions. Theor Appl Genet. 2014;127:463–489. doi: 10.1007/s00122-013-2231-5. [DOI] [PubMed] [Google Scholar]
- Hill W, Goddard M, Visscher P. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4:1–10. doi: 10.1371/journal.pgen.0040001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-cruz M, Crossa J, Bonnett D, Dreisigacker S, Poland J, Jannink JL, et al. Increased prediction accuracy in wheat breeding trials using a marker × environment interaction genomic selection model. G3: Genes Genom Genet. 2015;5:569–582. doi: 10.1534/g3.114.016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam DC, Potts SM, Bohn MO, Lipka AE, Lorenz AJ. Genomic prediction of single crosses in the early stages of a maize hybrid breeding pipeline. G3: Genes Genom Genet. 2016;6:3443–3453. doi: 10.1534/g3.116.031286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AM, Cullis BR, Gilmour AR, Eccleston AE, Thompson R. Estimation in a multiplicative mixed model involving a genetic relationship matrix. Genet Sel Evol. 2009;41:1–9. doi: 10.1186/1297-9686-41-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AM, Smith AB, Eccleston JA, Cullis BR. The accuracy of varietal selection using factor analytic models for multi-environment plant breeding trials. Crop Sci. 2007;47:1063–1070. doi: 10.2135/cropsci2006.08.0540. [DOI] [Google Scholar]
- Krchov LM, Bernardo R. Relative efficiency of genome wide selection for testcross performance of doubled haploid lines in a maize breeding program. Crop Sci. 2015;55:2091–2099. doi: 10.2135/cropsci2015.01.0064. [DOI] [Google Scholar]
- Kumar S, Molloy C, Muñoz P, Daetwyler H, Chagné D, Volz R. Genome-enabled estimates of additive and nonadditive genetic variances and prediction of apple phenotypes across environments. G3: Genes Genom Genet. 2015;5:2711–2718. doi: 10.1534/g3.115.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maazou ARS, Tu J, Ju Q, Liu Z. Breeding for drought tolerance in maize (Zea mays L.) Am J Plant Sci. 2016;7:1858–1870. doi: 10.4236/ajps.2016.714172. [DOI] [Google Scholar]
- Meyer K. Factor-analytic models for genotype × environment type problems and structured covariance matrices. Genet Sel Evol. 2009;41:1–11. doi: 10.1186/1297-9686-41-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829 [DOI] [PMC free article] [PubMed]
- Mohring J, Piepho HP. Comparison of weighting in two-stage analysis of plant breeding trials. Crop Sci. 2009;49:1977–1988. doi: 10.2135/cropsci2009.02.0083. [DOI] [Google Scholar]
- Muñoz PR, Resende JRMFR, Gezan SA, Resende MDV, De Los Campos G, Kirst M, et al. Unraveling additive from nonadditive effects using genomic relatinship matrices. Genetics. 2014;198:1759–1768. doi: 10.1534/genetics.114.171322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian A, Gezan SA. GenoMatrix: a software package for pedigree-based and genomic prediction analyses on complex traits. J Hered. 2016;107:372–379. doi: 10.1093/jhered/esw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian A, Gezan SA. Integrating non-additive genomic relationship matrices into the study of genetic architecture of complex traits. J Hered. 2016;107:153–162. doi: 10.1093/jhered/esv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakey H, Cullis B, Thompson R, Comadran J, Halpin C, Waugh R. Genomic selection in multi-environment crop trials. G3: Genes Genom Genet. 2016;6:1313–1326. doi: 10.1534/g3.116.027524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakey H, Verbyla A, Cullis B, Wei X, Pitchford W. Joint modelling of additive and no-additve (genetic line) effects in mult-environment trials. Theor Appl Genet. 2007;114:1319–1332. doi: 10.1007/s00122-007-0515-3. [DOI] [PubMed] [Google Scholar]
- Piepho HP. Analyzing genotype-environment data by mixed models with multiplicative terms. Biometrics. 1997;53:761–767. doi: 10.2307/2533976. [DOI] [Google Scholar]
- Piepho HP. Empirical best linear unbiased prediction in cultivar trials using factor analytic variance-covariance structures. Theor Appl Genet. 1998;97:195–201. doi: 10.1007/s001220050885. [DOI] [Google Scholar]
- Piepho HP, Mohring J, Melchinger AE, Buchse BLUP for phenotypic selection in plant breeding and variety testing. Euphytica. 2008;161:209–228. doi: 10.1007/s10681-007-9449-8. [DOI] [Google Scholar]
- R Core Team (2016) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
- Resende MF, Jr, Muñoz P, Garrick DJ, Fernardo RL, Davis JM, Jokela EJ, et al. Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.) Genetics. 2012;190:1503–1510. doi: 10.1534/genetics.111.137026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribaut JM, Betran J, Monneveux P, Setter T (2009) Drought tolerance in maize. In: Bennetzen JL, Hake SC (eds) Handbook of maize: its biology. Springer, New York, pp 311–344
- Roberts A, Mcmillan L, Wang W, Parker J, Rusyn I, Threadgill D. Inferring missing genotypes in large SNP panels using fast nearest-neighbor searches over sliding windows. Bioinformatics. 2007;23:401–407. doi: 10.1093/bioinformatics/btm220. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JPR, Vasconcelhos RCC, Pires LPM, Balestre M, Von Pinho RG. Inclusion of dominance effects in the multivariate GBLUP model. PLoS ONE. 2016;11:1–21. doi: 10.1371/journal.pone.0152045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Cullis BR, Thompson R. Analysing variety by environment data using multiplicative mixed models and adjustment for spatial field trend. Biometrics. 2001;57:1138–1147. doi: 10.1111/j.0006-341X.2001.01138.x. [DOI] [PubMed] [Google Scholar]
- Smith A, Ganesalingam A, Kuchel H, Cullis BR. Factor analytic mixed model for the provision of grower information from national crop variety testing programs. Theor Appl Genet. 2015;128:55–72. doi: 10.1007/s00122-014-2412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technow F, Riedelsheimer C, Schrag TA, Melchinger AE. Genomic prediction of hybrid performance in maize with models incorporating dominance and population specific marker effects. Theor Appl Genet. 2012;125:1181–1194. doi: 10.1007/s00122-012-1905-8. [DOI] [PubMed] [Google Scholar]
- Technow F, Schrag TA, Schipprack W, Bauer E, Simianer H, Melchinger AE. Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics. 2014;197:1343–1355. doi: 10.1534/genetics.114.165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro MA, Varona L. A note on mate allocation for dominance handling in genomic selection. Genet Sel Evol. 2010;42:1–9. doi: 10.1186/1297-9686-42-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanraden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Vitezica ZG, Varona L, Legarra L. On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics. 2013;195:1223–1230. doi: 10.1534/genetics.113.155176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolak EM. Nadiv: an R package to create relatedness matrices for estimating non-additive genetic variances in animal models. Methods Ecol Evol. 2012;3:792–796. doi: 10.1111/j.2041-210X.2012.00213.x. [DOI] [Google Scholar]
- Yang J, Benyamin B, Mcevoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziyomo C, Bernardo R. Drought tolerance in maize—indirect selection through secondary traits versus genome wide selection. Crop Sci. 2013;52:1269–1275. doi: 10.2135/cropsci2012.11.0651. [DOI] [Google Scholar]
- Zhang X, Pérez-Rodríguez P, Semagn K, Beyene Y, Babu R, López-Cruz MA, et al. Genomic prediction in biparental tropical maize populations in water-stressed and well-watered environments using low-density and GBS SNPs. Heredity. 2015;114:291–299. doi: 10.1038/hdy.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Bakshi A, Vinkhuyzen AAE, Hemani G, Lee SH, Nolte IM, et al. Dominance genetic variation contributes little to the missing heritability for human complex traits. Am J Hum Genet. 2015;96:1–9. doi: 10.1016/j.ajhg.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuberosa, R (2012) Phenotyping for drought tolerance of crops in the genomics era. Frontiers in Physiology 3: 1-26 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.