Abstract
The utility of human induced pluripotent stem cells (iPSCs) is enhanced by an ability to precisely modify a chosen locus with minimal impact on the remaining genome. However, the derivation of gene-edited iPSCs typically involves multiple steps requiring lengthy culture periods and several clonal events. Here, we describe a one-step protocol for reliable generation of clonally derived gene-edited iPSC lines from human fibroblasts in the absence of drug selection or FACS enrichment. Using enhanced episomal-based reprogramming and CRISPR/Cas9 systems, gene-edited and passage-matched unmodified iPSC lines are obtained following a single electroporation of human fibroblasts. To minimize unwanted mutations within the target locus, we use a Cas9 variant that is associated with decreased nonhomologous end-joining (NHEJ) activity. This protocol outlines in detail how this streamlined approach can be used for both monoallelic and biallelic introduction of specific base changes or transgene cassettes in a manner that is efficient, rapid (~6–8 weeks), and cost-effective.
INTRODUCTION
Human iPSCs offer enormous potential for the development of autologous cell-based therapies, disease modeling, and drug discovery. However, the full potential of iPSC technology cannot be completely unleashed without an ability to genetically modify these cells in a precise and specific manner. Obviously, in the context of autologous cell therapy, in many instances it is desirable to correct an underlying disease-causing mutation before the use of patient cells in downstream transplantation applications. But even in the context of disease modeling, comparisons between gene-corrected and matched isogenic patient iPSC lines are critical to understanding the exact underlying molecular mechanisms governing disease, as any functional variation can be attributed solely to the patient-specific mutation rather than differences in genetic background. To date, numerous studies have demonstrated the utility of using such isogenic sets of iPSCs for parallel differentiation into specific disease-relevant cell types in an effort to directly assess the contribution of a specific mutation to cellular pathology1–3.
Owing to its simplicity, ease of manipulation, and high efficiency, the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system is quickly becoming the preferred method for the precise modification of human pluripotent stem cells (PSCs)4–12. Derived from the adaptive immune system of Streptococcus pyogenes13, the CRISPR/Cas9 system consists of the Cas9 nuclease and a small guide (g)RNA that directs Cas9 to the genetic locus of interest by RNA–DNA base pairing14. The specificity of CRISPR/Cas9 is mediated by a 20-nt sequence within the gRNA that is complementary to the target site (protospacer) in the host genome, with the additional requirement that this 20-bp target site lie immediately upstream of a 5′-NGG protospacer-adjacent motif (PAM)14. After binding to the specified target, the CRISPR/Cas9 complex induces a double-strand break (DSB) ~3 bp upstream of the PAM15. These DSBs are then repaired using error-prone NHEJ or the high-fidelity homology-directed repair (HDR) pathway. NHEJ is the predominate pathway in most cell types and involves the direct ligation of the break ends, leaving scars in the form of insertion/deletion (indel) mutations within the target locus8,9,16,17. Alternatively, the HDR pathway facilitates precise and defined modifications at a target locus in the presence of an exogenously introduced repair template. The repair template can either be in the form of double-stranded plasmid DNA, comprising homology arms flanking the insertion sequence, or single-stranded DNA oligonucleotides (ssODNs)10.
Generation of indel-free cell lines with Cas9-Gem
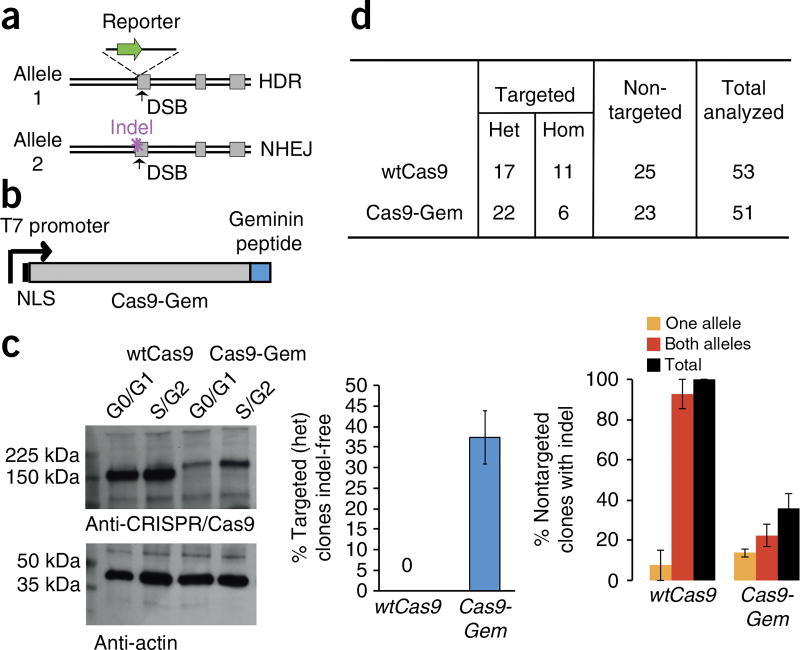
The CRISPR/Cas9 complex is extremely efficient and commonly results in biallelic DSBs at the specified target site18,19. However, typically, these DSBs are repaired by the NHEJ pathway, leading to the formation of nonspecific indels within the target locus8,9,16,17,20. Consequently, the few clones that have undergone HDR to successfully introduce a heterozygous modification will often carry disruptive secondary ‘on-target’ mutations within the other allele6,21 (Fig. 1a). We hypothesized that this issue could be addressed, at least in part, by controlling the presence of Cas9 protein during specific stages of the cell cycle19. Previous studies, including one in normal human fibroblasts, have demonstrated that HDR activity is virtually absent during the G1 phase, increases sharply in the S phase, and begins to decrease by G2/M, whereas NHEJ activity occurs throughout the cell cycle22,23. We have previously fused the Cas9 nuclease to a peptide derived from the human Geminin protein (Cas9-Gem) (Fig. 1b) to facilitate its degradation during the G1 phase of the cell cycle (Fig. 1c), when DNA repair by NHEJ predominates. In our study, we also used mRNA transfection to facilitate low and transient expression of Cas9-Gem. Although the frequency of HDR is not impaired by fusion of the Geminin peptide to Cas9 (ref. 17), we observed a two- to threefold decrease in NHEJ frequency associated with Cas9-Gem as compared with unmodified Cas9 (Fig. 1d). This bias toward the HDR pathway of Cas9-Gem allows efficient and reliable generation of both knock-in cell lines and gene-corrected patient-specific iPSC lines that are free of disruptive mutations within the untargeted allele.
Figure 1.
HDRCas9-Gem facilitates generation of indel-free gene-edited iPSCs. (a) Schematic diagram of ‘on-target’ indel mutations that are commonly observed in successfully targeted clones using CRISPR/Cas9. (b) Schematic diagram of the Cas9-Gem variant. An upstream T7 promoter is used to facilitate in vitro transcription. (c) Western blot analysis of Cas9-Gem and wtCas9 protein expression in sorted G0/G1 and S/G2 subpopulations shows enrichment of Cas9-Gem in the S/G2 subpopulation and lower levels in G1. Loading control is anti-actin. (d, Left) Proportion of heterozygous gene-targeted iPSC clones identified without disruption of the second allele following knock-in of an EGFP reporter within the DNMT3B locus using either wtCas9 (n = 17) or Cas9-Gem (n = 22). (Right) The proportion of nontargeted iPSC clones from the same experiment with one or both DNMT3B alleles disrupted by NHEJ is also shown for wtCas9 (n = 25) and Cas9-Gem (n = 23). Data represent mean ± s.e.m. from three independent experiments. b–d adapted from ref. 19, Howden et al. Het, heterozygous; Hom, homozygous; NLS, nuclear localization signal.
One-step reprogramming and gene editing of human fibroblasts
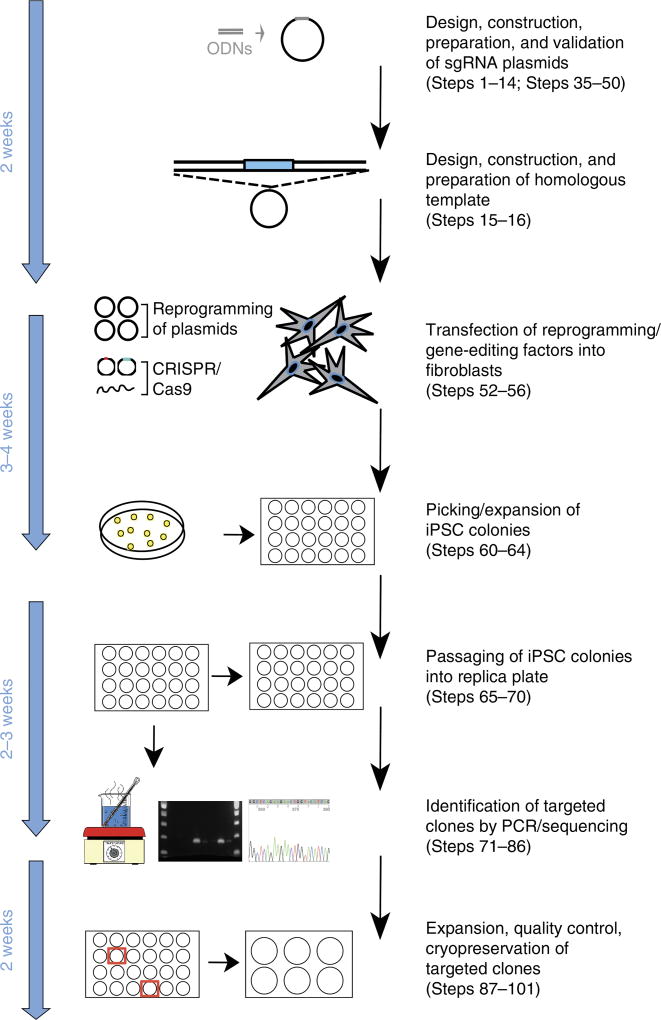
Here, we describe a simple protocol that enables simultaneous reprogramming and CRISPR/Cas9 gene editing, allowing rapid and clonal derivation of multiple gene-targeted iPSC lines from human fibroblasts (Fig. 2). This protocol uses an enhanced episomal-based reprogramming system6, which forces expression of the microRNA (miR) 302/367 cluster in addition to expression of the seven factors (OCT4, SOX2, NANOG, c-MYC, KLF4, LIN28, and SV40 large T antigen) encoded by three episomal-based vectors described previously24. The inclusion of this fourth miR302/367 vector results in substantial increases (> 100-fold) in reprogramming efficiency and is critical to obtaining large numbers of iPSC colonies, including potentially targeted lines. To minimize undesirable secondary mutations within the target locus, this protocol also uses Cas9-Gem19. This method can be used to faithfully insert large modifications (up to several kilobases in size) into a gene of interest, including fluorescent reporters, Cre drivers, or other cDNAs. Furthermore, because this procedure does not require the use of drug selection, gene knock-in can be accomplished with simple homologous templates that can be synthesized commercially in a rapid and cost-effective manner. Our one-step method also facilitates the generation of clones that contain ‘seamless’ single-base-pair changes, without leaving residual loxP or flippase recognition target (FRT) sites in the host genome. This is ideal for the repair of patient-specific mutations in fibroblasts expanded from patient biopsies or even for the introduction of heterozygous and homozygous disease-associated mutations into healthy cells. Multiple gene-edited and passage-matched unmodified (control) iPSC clones can be obtained from a single experiment; these can be used for downstream applications such as disease modeling and/or drug discovery assays.
Figure 2.
Time line and overview of the one-step reprogramming/gene-editing workflow.
Comparisons with other methods
Previous methods for the generation of gene-edited iPSC lines are based upon a sequential approach, whereby iPSC lines are first derived from somatic cells (such as human fibroblasts) and gene editing is performed in the resulting iPSCs12. This requires cells to be in culture for extensive periods, as well as involving multiple clonal events, and entails considerable amounts of time and resources. Furthermore, when gene editing is performed in established iPSCs, isolation of a pure population of correctly targeted cells is often cumbersome and laborious, particularly in the absence of drug selection. This is largely because human PSCs do not survive single-cell dissociation without the addition of a rho-associated coiled-coil kinase inhibitor25 and must form contacts with neighboring cells. As a result, gene-edited iPSCs are often associated with unmodified cells, and successive steps are usually required to ensure clonality of a gene-edited line. The purification of transfected cells by FACS, followed by single-cell plating into a 96-well plate to generate clonal lines is associated with poor cell viability and often necessitates low-oxygen incubators and/or optimization of flow cytometry parameters26. Moreover, single-cell passaging of human iPSCs has also been shown to increase genomic abnormalities27.
Using the direct approach detailed here, multiple gene-edited iPSC clones can be generated following a single electroporation of human fibroblasts. Colonies can be obtained in as little as 2 weeks, requiring considerably less time and resources, as compared with conventional multistep protocols, which typically take several months to complete. In addition, our one-step procedure does not require the use of drug selection or FACS purification for the generation of clonal populations of knock-in or gene-corrected iPSC lines. This greatly minimizes the complications and costs associated with single-cell dissociation, FACS, drug selection, and extended culture of human iPSCs.
Limitations of simultaneous reprogramming/gene editing
Although gene-editing frequencies can vary substantially from experiment to experiment, using the one-step protocol described here, successfully modified iPSCs typically comprise 2–20% of the total iPSC population. This is notably higher than the gene-targeting frequencies associated with multistep protocols that use iPSCs as the starting material, for which correctly targeted cells typically comprise < 1% of the total population in the absence of drug selection6. Because our protocol involves screening 20–50 iPSC colonies to identify targeted clones, actively dividing fibroblast cultures amenable to reprogramming are most suited to this method. Fibroblasts derived from younger donors (< 20 years) are generally associated with higher reprogramming efficiencies than those derived from older donors28. Furthermore, variations in the manner in which primary fibroblasts are isolated from human biopsies and the number of passages the cells undergo before reprogramming may also substantially impact reprogramming efficiency.
A number of studies have also demonstrated that iPSC lines derived from skin biopsies often harbor a unique subset of de novo genetic abnormalities, either in the form of copy-number variation (CNV) mutations or single base-pair changes29–31, and that a positive correlation exists between donor age and mutational burden32. Furthermore, iPSC lines derived from the same parental cell population can vary substantially with respect to whole-genome gene expression in the differentiated state33. Nonetheless, it is reasonable to expect that any confounding effects that may arise from these variations can be minimized by evaluating multiple clones. We routinely observe targeting efficiencies > 5%, enabling the generation and evaluation of multiple reporter iPSC lines or multiple gene-corrected and ‘matched’ uncorrected clones from a single experiment.
Experimental design
Single-guide RNA selection
The identification, ranking, and selection of single-guide RNA (sgRNA) sequences for a given locus can be determined using several online tools (e.g., http://www.rgenome.net/cas-designer and http://crispr.mit.edu) and have been described previously10. Importantly, these tools can aid in the identification of sgRNAs that are associated with low predicted off-target activity (Step 1). We recommend selecting sgRNAs that bind close (within 20 bp) to the intended modification. If performing gene correction of a heterozygous mutation, sgRNAs that overlap the patient-specific mutation can be used in order to minimize Cas9-induced cleavage of the wild-type allele. Occasionally, a given sgRNA may exhibit little or no activity for reasons that remain largely unknown and may not be predictable34. We therefore recommend designing and evaluating the efficiencies of at least two sgRNAs for each locus of interest before use in downstream gene-editing/reprogramming experiments. We also use the U6 promoter to drive sgRNA expression, which works most effectively when a guanine (G) is used as the first base at the site of transcription initiation35. For this reason, the first base of a 20-nt sgRNA is replaced with a G when the sequence does not inherently begin with a G.
Source of human fibroblasts
Normal and disease-specific human fibroblasts are available from cell banks, such as ATCC (https://www.atcc.org) and Coriell Cell Repositories (https://catalog.coriell.org). Alternatively, a skin punch biopsy obtained by a trained physician under the approval of the relevant authorities with informed patient consent can be used to isolate primary fibroblasts as previously described36. For transfection experiments, we recommend working with cells that are low in passage number (preferably < 10), as primary fibroblasts may senesce quickly. Proper cell growth is critical to adequate transfection, reprogramming, and gene-targeting efficiencies. Fibroblasts isolated from younger donors tend to be associated with higher reprogramming efficiency and are more likely to yield successful results. Newly derived or acquired fibroblast lines should also be tested before transfection to ensure the cells are not contaminated with mycoplasma. Because cell viability and growth rates may vary among different fibroblast lines following transfection, we recommend plating cells at different densities across a six-well plate immediately following electroporation, especially when using a new line for the first time. When using a new line for the first time, we also recommend performing a transfection control experiment using a simple fluorescent reporter plasmid (e.g., pCMV-tdTomato) to monitor transfection efficiency. The percentage of fluorescent cells in the transfection control can be estimated by using a fluorescence microscope or flow cytometry. Typically, a good transfection efficiency is > 50% when plasmid size is < 5 kb.
Repair template design
Plasmid-based repair templates that contain homology arms flanking the site of alteration have traditionally been used for targeted DNA modification37,38. Although the homology arms can vary in length, 400–1,000 bp is generally sufficient. More recently, ssODNs (100–200 bp) have been used for short modifications, such as correction or introduction of a specific mutation within a defined locus39. The main advantage of ssODNs over plasmid-based repair templates is that they can be readily purchased from a DNA synthesis service (e.g., Integrated DNA Technologies, https://www.idtdna.com) and do not require additional cloning steps. ssODNs are also less likely to integrate randomly into the host genome. However, errors during the DNA synthesis process can lead to the introduction of unwanted mutations6, and it is therefore highly recommended that ssODNs be HPLC- or PAGE-purified. If possible, include a consecutive 3-bp synonymous change to facilitate the identification of successfully edited clones by allele-specific PCR (Step 77B). This synonymous change can also double as a CRISPR/Cas-blocking mutation to prevent CRISPR/Cas9 recutting and induction of NHEJ mutations following successful HDR with the repair template11,40.
The incorporation of larger modifications, including the insertion of reporter genes such as fluorescent proteins, antibiotic resistance markers, or other cDNA sequences, requires the design and construction of plasmid constructs (Step 15B). The location of the reporter should be based upon known information for the gene of interest. Insertion of a reporter gene and downstream poly(A) signal at the endogenous start codon of the target gene may best recapitulate the gene expression pattern of the endogenous target gene. However, this approach creates a null allele for the targeted gene and should, therefore, be used only when knockout of one allele is tolerated and not associated with a haploinsufficient phenotype that may impact downstream applications. If knockout of one allele is not desirable, a self-cleaving 2A peptide (e.g., T2A) can be inserted between the reporter and the gene of interest to preserve expression of the targeted gene. Although we, and others, have used self-cleaving 2A peptide to successfully drive reporter genes from various genomic loci, this approach may occasionally result in incomplete cleavage, which may in turn lead to lower levels of gene expression or protein that is incorrectly localized41.
Picking and expansion of iPSC colonies
In a typical experiment with the enhanced episomal reprogramming system used here, the first iPSC colonies begin to appear within ~2 weeks post transfection, with the bulk of these colonies ready for picking 2.5–4 weeks post transfection. Although reprogramming efficiencies can vary, the three fibroblast lines (ATCC ID: CRL-2429, ATCC ID: PCS-201-010, and RG_120.153 (patient-specific fibroblasts from a 20-year-old donor with kidney disease)) used in this study resulted in > 50 independent iPSC colonies per electroporation. We suggest picking 30–50 individual iPSC colonies and placing each into a single well of a Matrigel-coated 24-well cell culture plate (Steps 60–64). Avoid picking colonies that are growing in very close proximity to one another and that are likely to be mixed. Replica splitting should be performed after 2–5 d (depending on colony size) by transferring a small fraction (25–30%) of the cells in each well to a new 24-well plate (Steps 65–70). The remaining cells should be cultured for an additional 2–5 d until confluent and then harvested for subsequent genomic analysis. The replica plate should be maintained (daily media changes and passaging as necessary) until genomic analysis is complete and successfully modified clones are selected for further expansion, karyotype analysis, and cryopreservation. We recommend culturing iPSCs in feeder-free conditions using Matrigel-coated plates, Essential 8 medium, and EDTA, and passaging as previously described42. Prewarm the required volume of Essential 8 medium at room temperature (20–25 °C) until it is no longer cool to the touch.
Identification of targeted clones
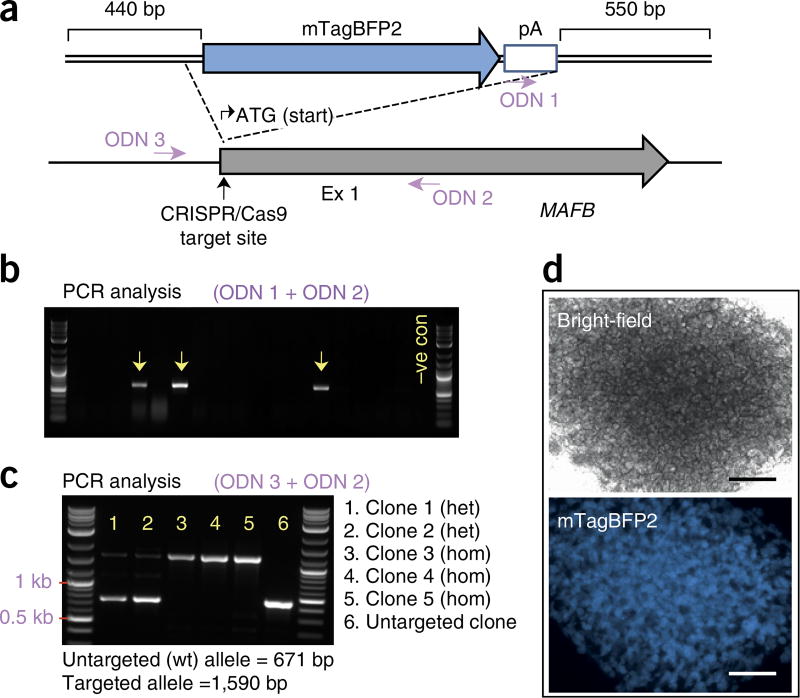
In this protocol, we describe a simple, rapid, and cost-effective PCR-based genotyping protocol for identifying successfully modified clones without the need for genomic DNA extraction/purification (Steps 71–76). Before screening, iPSCs are dissociated, resuspended in 50–100 µl of Tris–EDTA (TE) buffer, and placed in a boiling water bath for 5–10 min. After cooling to room temperature, the samples are ready to be used for subsequent PCR analysis. For large modifications, such as knock-in of a reporter gene, HDR can be detected using primers that flank the recombination junctions by using one primer that binds within the inserted sequence and a second primer that binds just outside the region of homology (Fig. 3a). The second primer should anneal outside the region spanned by the homology arms to avoid false detection of residual repair template. A second PCR analysis should also be performed on successfully targeted clones, using primers that flank the insertion, to distinguish monoallelic from biallelic modifications (heterozygous versus homozygous clones) (Fig. 3). For heterozygous clones, this PCR amplicon should also be Sanger-sequenced to ensure the untargeted allele is free of disruptive indels resulting from NHEJ. For the detection of NHEJ-induced mutations, it is important to design primers that bind 200–1,000 bp from the Cas9 target site to allow for the detection of longer indels.
Figure 3.
Generation of MAFP:mTagBFP2 reporter iPSC lines following one-step gene editing/reprogramming of healthy fibroblasts. (a) Schematic diagram of the MAFB locus and the homologous template used for reporter gene knock-in. The primers (ODNs) used for identifying targeted clones by PCR are indicated. (b) Representative gel following PCR analysis of 17 iPSC clones using primers ODN 1 and ODN 2, which flank the 3′ recombination junction. ODN 1 binds within the reporter cassette, and ODN 2 binds outside the region of homology. Arrows indicate PCR products amplified from correctly targeted clones. (c) PCR analysis of correctly targeted clones using primers ODN 3 and ODN 2, which flank the mTagBFP2 reporter cassette. These primers preferentially amplify the untargeted allele in heterozygous clones but will amplify only the knock-in alleles in homozygous clones. (d) Generation of kidney organoids from MAFB:mTagBFP2 iPSCs reveals appropriate mTagBFP2 reporter gene expression that is localized and restricted to the podocyte population in developing glomeruli. Scale bars, 500 µm. Ex, exon; pA, polyA signal.
For short modifications, such as correction or introduction of a point mutation, targeted clones can be detected by allele-specific PCR by using one primer that overlaps the synonymous change incorporated into the repair template (see Repair template design section above) and a second primer that binds outside the region of homology. The synonymous change should be at the very 3′ end of the primer. In our experience, a 3-bp mismatch is extremely effective for distinguishing wild-type and successfully modified alleles. Potentially targeted clones identified using this method should be subjected to a second round of PCR using primers that flank the target site and are capable of amplifying both the modified and wild-type alleles. Again, primers should bind at least 200 bp either side of the Cas9 target. These amplicons should be Sanger-sequenced to confirm successful gene editing, to distinguish heterozygous from homozygous modifications, and to ensure the untargeted allele is intact and free of NHEJ mutations (in heterozygous clones). We recommend extracting genomic DNA from successfully targeted clones and repeating PCR/sequencing screens for further confirmation of clone identity before use in downstream applications.
Quality control of gene-edited iPSC lines
Off-target analysis: online tools are available to determine the top predicted off-target sites of a given sgRNA (e.g., http://crispr.mit.edu, http://www.rgenome.net/cas-offinder)10,43. PCR and Sanger sequencing can be performed to confirm that no additional indels have occurred within the top predicted off-target sites, particularly those that lie within coding regions. More comprehensive approaches for identifying off-target sites have also been described and include whole-genome sequencing44, BLESS (breaks labeling, enrichment on streptavidin, and sequencing)45, Guide-seq46, and Digenome-seq47. Although a more comprehensive analysis of off-target mutations is highly recommended for cells that are intended for therapeutic use, we believe this is less critical for lines used for research purposes.
Loss of reprogramming vectors
PCR analysis can be performed on genomic DNA isolated from individual iPSC lines using primers specific to the ampicillin resistance gene (which is present on all four reprogramming vectors) to ensure lines are free of the reprogramming transgenes. If a band is detected, the PCR analysis should be repeated after another four to five passages, as the episomes should eventually be lost in rapidly dividing cells (such as iPSCs). In the rare case in which reprogramming vectors are detectable after more than ten passages, an additional subcloning step may be necessary, and PCR analysis should be repeated following the expansion of five to ten subclones.
Assessment for random integration of targeting constructs
We use supercoiled plasmid DNA or ssODNs for gene-editing experiments, as this is much less likely to integrate randomly into the host genome than linear dsDNA. However, a simple PCR analysis can be performed to assess for additional randomly integrated repair plasmids in successfully edited iPSC clones. For this analysis, we suggest using primers specific to sequences on the plasmid backbone, such as the bacterial origin of replication or antibiotic resistance gene. To minimize potential amplification of residual reprogramming vectors, it is preferable to use one primer that binds within the genomic insert and a second primer in the reverse orientation that binds within the plasmid backbone.
Genomic integrity
We recommend performing either a karyotype analysis by standard G-banding or single-nucleotide polymorphism (SNP) microarray (Illumina) to assess the genomic integrity of gene-edited iPSC lines (and uncorrected iPSC lines, if applicable) soon after derivation and before use in downstream experiments. Although SNP microarrays will not detect balanced translocations, this type of analysis can identify CNV mutations down to 0.5 MB, a resolution that is much higher than that of standard G-band karyotyping. The genomic integrity of iPSC lines should also be assessed following extensive culture periods, as this has been associated with the accumulation of karyotypic abnormalities48–50.
Pluripotency markers
Pluripotency of newly derived gene-edited iPSC lines can be confirmed by standard immunofluorescence assays using antibodies to common pluripotency markers such as OCT4, SSEA4, TRA-1-60, NANOG, and/or alkaline phosphatase51,52. Teratoma assays in immunocompromised mice and/or in vitro differentiation assays can also be performed to confirm pluripotency52.
MATERIALS
REAGENTS
sgRNA and homologous template preparation
pSMART-sgRNA(Sp) plasmid (Addgene, plasmid ID 80427)
Oligonucleotides (ODNs) for sgRNA construction (Integrated DNA Technologies; see Table 1 for design guidelines and Supplementary Table 1 for sgRNA sequences used in our experiments)
ODNs and/or gBlocks for construction of homologous recombination templates (Integrated DNA Technologies)
pSMART-HCKan Blunt Cloning Kit (Lucigen, cat. no. 40704-2)
Nuclease-free dH2O (Thermo Fisher Scientific, cat. no. AM9932)
1 M Tris-HCl (pH 8; Sigma-Aldrich, cat. no. T2788)
0.5 M EDTA (Sigma-Aldrich, cat. no. 03690)
5 M NaCl (Sigma-Aldrich, cat. no. S6546)
BbsI restriction endonuclease (NEB, cat. no. R0539S)
PCR and gel extraction kit (Bioline, cat. no. BIO-52060)
6× DNA loading dye (NEB, cat. no. B7024S)
TAE buffer (10×; Sigma-Aldrich, cat. no. T9650)
Agarose (Bioline, cat. no. BIO-41025)
-
Ethidium bromide solution (Sigma-Aldrich, cat. no. E1510)
! CAUTION Ethidium bromide is toxic and carcinogenic; handle it with nitrile gloves at all times.
Scalpel (BD, cat. no. 371611)
T4 DNA ligase (NEB, cat. no. M0202S)
DH5-alpha chemically competent cells (Bioline, cat. no. BIO-85047)
SOC (super optimal broth with catabolite repression) medium (NEB, cat. no. B9020S)
LB medium (Sigma-Aldrich, cat. no. L3022)
LB agar medium (Sigma-Aldrich, cat. no. L2897)
Kanamycin (50 mg/ml, sterile-filtered; Sigma-Aldrich, cat. no. K0254)
Ampicillin (100 mg/ml, sterile-filtered; Sigma-Aldrich, cat. no. A5354)
QIAprep Spin Miniprep Kit (Qiagen, cat. no. 27104)
Plasmid Maxi Kit (Qiagen, cat. no. 12163)
TE buffer (Thermo Fisher Scientific, cat. no. 12090015)
100% (vol/vol) Ethanol (Sigma-Aldrich, cat. no. E7023)
2-Propanol (Sigma-Aldrich, cat. no. 278475)
MycoAlert mycoplasma detection kit (Lonza, cat. no. LT07-118)
Table 1.
ODNs for sgRNA plasmid construction and sequence validation.
| ODN | Sequence | Purpose |
|---|---|---|
| sgRNA-top | CACCGNNNNNNNNNNNNNNNNNNN | Insertion of gene-specific (sense) sgRNA sequence into pSMART-sgRNA |
| sgRNA-bottom | AAACNNNNNNNNNNNNNNNNNNNC | Insertion of gene-specific (antisense) sgRNA sequence into pSMART-sgRNA |
| pSMARTseqF | CAGTCCAGTTACGCTGGAGTC | Sanger sequence verification of cloned sgRNA ODNs |
The G residue in bold indicates the transcription start site.
Fibroblast culture
! CAUTION Experiments must conform to all relevant governmental and institutional regulations relating to human ethics, biosafety, and genetic modification. It is recommended that the cells be checked to ensure that they are chromosomally stable and that they are not infected with mycoplasma.
DMEM, high glucose (Thermo Fisher Scientific, cat. no. 11995-073)
Dulbecco’s PBS (DPBS; Thermo Fisher Scientific, cat. no. 14190-144)
FBS (Interpath Services, cat. no. SFBSF)
GlutaMAX Supplement (Thermo Fisher Scientific, cat. no. 35050-061)
Non-essential amino acids (Thermo Fisher Scientific, cat. no. 11140-050)
TrypLE Select (Thermo Fisher Scientific, cat. no. 12563-029)
pCMV-tdTomato vector (Clontech, cat. no. 632534)
Reprogramming
Plasmids: pEP4 E02S ET2K (Addgene, plasmid ID 20927), pEP4 E02S EN2L (Addgene, plasmid ID 20922), pEP4 E02S EM2K (Addgene, plasmid ID 20923), and pSimple-miR302/367 (Addgene, plasmid ID 98748)
TesR-E7 reprogramming medium (Stem Cell Technologies, cat. no. 05910)
Sodium butyrate (Sigma-Aldrich, cat. no. B5887)
In vitro transcription of EBNA1, Cas9, and Cas9-Gem mRNA
Plasmids: pSP6-EBNA2A + DBD (Addgene, plasmid ID 98749), pDNR-SpCas9-Gem (Addgene, plasmid ID 80424), and pDNR-SpCas9 (Addgene, plasmid ID 80425)
PmeI restriction endonuclease (NEB, cat. no. R0560S)
EcoRI-HF restriction endonuclease (NEB, cat. no. R3101S)
mMessage mMachine SP6 Transcription Kit (Thermo Fisher Scientific, cat. no. AM1340)
mMessage mMachine T7 Ultra Transcription Kit (Thermo Fisher Scientific, cat. no. AM1345)
RNA ladder (Promega, cat. no. G3191)
Pluripotent stem cell culture
hESC-qualified Matrigel (LDEV (lactose dehydrogenase-elevating virus)-free; Corning, cat. no. 354277)
DMEM/F-12 (Thermo Fisher Scientific, cat. no. 11320-082)
Essential 8 medium (Thermo Fisher Scientific, cat. no. A1517001)
DMSO (Sigma-Aldrich, cat. no. D5879)
Genotyping analysis
Custom-designed ODNs to generate ~200- to 800-bp amplicons around the sgRNA target site and identify targeted clones (Integrated DNA Technologies; see Supplementary Table 1 for primer sequences used in our experiments)
GoTaq Green Master Mix (Promega, cat. no. M7123)
Exonuclease I (NEB, cat. no. M0293S)
rAPid alkaline phosphatase (Sigma-Aldrich, cat. no. 4898133001)
BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, cat. no. 4337455)
DNeasy Blood & Tissue Kit (Qiagen, cat. no. 69506)
EQUIPMENT
Sequence analysis software (e.g., Snapgene (http://www.snapgene.com), Vector NTI (https://www.thermofisher.com/us/en/home/life-science/cloning/vector-nti-software.html), and DNAstar (https://www.dnastar.com))
Sterile pipette tips with filters (Fisher Scientific, cat. nos. 14-222-765, 14-222-777 and 14-222-781)
Disposable serological pipettes (Fisher Scientific, cat. nos. 13-675-20 and 13-675-22)
Standard microcentrifuge tubes (1.5 ml; Eppendorf, cat. no. 0030 125.150)
Sealing film for PCR plates (VWR, cat. no. UC-500)
PCR plates (96 well; VWR, cat. no. PCR-96M2-HSC)
PCR tubes (individual; VWR, cat. no. PCR-02-C)
Falcon tubes (polypropylene, 15 ml; BD Falcon, cat. no. 352097)
Falcon tubes (polypropylene, 50 ml; BD Falcon, cat. no. 352070)
Petri dishes (60 × 15 mm; BD Biosciences, cat. no. 351007)
Cryovials (Thermo Fisher Scientific, cat. no. 4000198)
Tissue culture plate (24 wells; Thermo Fisher Scientific, cat. no. 142475)
Tissue culture plate (six wells; Thermo Fisher Scientific, cat. no. 140675)
Tissue culture dish (10 cm; Thermo Fisher Scientific, cat. no. 150288)
175-cm2 Tissue culture flask (Nunc, cat. no. 159910)
75-cm2 Tissue culture flask (Nunc, cat. no. 156472)
25-cm2 Tissue culture flask (Nunc, cat. no. 156367)
Hemocytometer (Fisher Scientific)
Mr. Frosty freezing container (Thermo Fisher Scientific, cat. no. 5100-0001)
Gradient thermocycler (96 well; Veriti; Applied Biosystems, cat. no. 4375786)
Microcentrifuge (e.g., Eppendorf, cat. no. 5424)
Centrifuge for 50-ml and 15-ml tubes (e.g., Eppendorf, cat. no. 5702)
Gel electrophoresis system (PowerPac power supply; Bio-Rad, cat. no. 164-5050) and Sub-Cell GT System gel tray (Bio-Rad, cat. no. 170-4401)
Digital gel-imaging system (GelDoc EZ; Bio-Rad, cat. no. 170-8270) and blue sample tray (Bio-Rad, cat. no. 170-8273)
UV-light transilluminator (Thermo Fisher) ! CAUTION UV light is harmful to eyes and skin. Wear gloves, lab coat, and face shield.
UV-filter face mask (Thermo Fisher)
UV spectrophotometer (Thermo Fisher Scientific, model no. NanoDrop 2000c)
Inverted contrasting tissue culture microscope (Nikon, model no. TS100)
Cell-culture-hood-embedded dissection microscope
Biological safety cabinet (LAF Technologies, model no. TOP-SAFE 1.2)
CO2 incubators (Thermo Scientific, model no. Heracell 150i)
Neon Transfection System (Thermo Fisher Scientific, cat. no. MPK5000)
Neon transfection kit (Thermo Fisher Scientific, cat. no. MPK10096)
Hot plate (e.g., Fisher Scientific, cat. no. S504631H)
Glass beaker (2-liter capacity, e.g., Fisher Scientific, cat. no. 02-540R)
Floating tube rack (e.g., VWR, cat. no. 82017-634)
Pipetboy (Integra Biosciences, PIPETBOY pro model)
Pipettes (Pipetman Classic; Gilson, model nos. P10, P200, and P1000)
Serological pipettes (5 and 10 ml; Corning, cat. no. 357551)
Syringe-driven (0.22-µm filter unit; Merck Millipore, cat. no. SLMP025SS)
REAGENT SETUP
ODN annealing buffer
ODN annealing buffer is 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 1 mM EDTA. Prepare 10 ml of ODN annealing buffer by combining 9.3 ml of nuclease-free H2O, 500 µl of 1 M Tris-HCl (pH 8), 200 µl of 5 M NaCl, and 20 µl of 0.5 M EDTA (pH 8). Store at room temperature for up to 12 months.
TAE electrophoresis solution
Dilute 10× TAE buffer stock in dH2O to a 1× working solution. Store at room temperature for up to 6 months.
Fibroblast medium
Prepare fibroblast medium by supplementing DMEM with GlutaMAX (1×), nonessential amino acid solution (1×), and 15% (vol/vol) FBS. Store at 4 °C for up to 1 month.
Matrigel aliquots
Place a 5-ml Matrigel bottle on ice and thaw overnight in a 4 °C refrigerator or cold room. Prechill 1.5-ml tubes and pipette tips at −20 °C and divide into aliquots of the desired volume according to Matrigel lot specifications (typically 300–500 µl each for later dilution to 50 ml). Freeze at −20 or −80 °C for up to 6 months.
Matrigel-coated culture plates
Thaw one aliquot of Matrigel on ice. Add the aliquot of Matrigel to 50 ml of cold DMEM/F12 and add 1 ml of this solution per well of a six-well plate or 300 µl per well of a 24-well plate. Incubate the cultureware in a 37 °C incubator for at least 30 min before use. Coated plates can be stored in a 37 °C incubator or at 2–8 °C for up to 2 weeks. Before use, remove residual DMEM/F12 medium and replace with the appropriate culture medium.
Essential 8 medium
Thaw a 10-ml vial of supplement (provided in the kit) overnight at 4 °C and combine with 500 ml of Essential 8 base medium. Store at 4 °C for up to 2 weeks.
0.5 mM EDTA
Combine 500 µl of 0.5 M EDTA and 500 ml of DPBS. Store at room temperature for up to 12 months.
Sodium butyrate solution (100 mM)
Dissolve 110 mg of sodium butyrate in 10 ml of dH2O. Filter-sterilize through a syringe-driven 0.22-µm filter unit and store at −20 °C in 500-µl aliquots for up to 12 months.
Reprogramming medium
Prepare TeSR-E7 reprogramming medium by combining base medium with the supplied supplement solution. Add 500 µl of sodium butyrate solution to 500 ml of TeSR-E7 reprogramming medium (to achieve a final concentration of 100 µM). Complete medium can be stored at 4 °C for up to 2 weeks.
Freezing medium
On the day of freezing, supplement Essential 8 medium with 20% (vol/vol) sterile DMSO.
PROCEDURE
CRISPR selection and design ● TIMING 1 h
-
1|
Input of target genomic DNA sequence. Use an online design tool (e.g., http://crispr.mit.edu, http://www.rgenome.net/cas-designer) to identify and rank potential targets from an input sequence as previously described10,53 (e.g., from a 200-bp genomic fragment spanning the region of interest). Alternatively, select a CRISPR guide sequence manually by identifying the 20-nt sequence directly upstream of any 5′-NGG.
-
2|
Order sgRNA-top (sense) and sgRNA-bottom (complementary) ODNs required for sgRNA assembly from Integrated DNA Technologies (https://www.idtdna.com), with the added overhangs indicated in Table 1.
Δ CRITICAL STEP For efficient transcription from the U6 promoter, the first base of a 20-nt sgRNA is replaced with a G if it does not inherently begin with a G.
Cloning of sgRNA ODNs into the pSMART-sg RNA vector ● TIMING 3–4 d
-
3|Digestion of pSMART-sgRNA expression vector. Set up a restriction endonuclease digestion reaction as described below. Incubate at 37 °C for at least 1 h.
Component Amount Final concentration pSMART-sgRNA vector 10 µg 100 ng/µl 10× Buffer 2.1 10 µl 1× BbsI (10 U/µl) 5 µl 0.5 U/µl Nuclease-free H2O Make up to 100 µl -
4|
Purify the digestion reaction using a PCR purification kit according to the manufacturer’s directions. Elute the DNA in 50 µl of elution buffer.
-
5|
Gel extraction of BbsI-digested pSMART-sgRNA vector. Load the entire sample mixed with 1× DNA loading dye on a 1–2% (wt/vol) agarose gel containing 0.5 µg/ml ethidium bromide. Separate the bands of DNA by electrophoresis in 1× TAE buffer at 100 V for 1–2 h and check the bands using a gel imager. Use a scalpel to extract the ~2.2-kb DNA band visualized on a UV gel imaging box.
! CAUTION UV exposure to skin and eyes is harmful; wear full laboratory protective equipment, including a UV-filtering facemask.
-
6|
Purification of BbsI-digested pSMART-sgRNA vector. Purify gel-extracted pSMART-sgRNA vector using a gel extraction kit according to the manufacturer’s directions.
■ PAUSE POINT Purified BbsI-digested pSMART-sgRNA vector can be stored at −20 °C for several months.
-
7|
Measure the concentration of the BbsI-digested pSMART-sgRNA vector using a NanoDrop spectrophotometer and adjust the concentration to 5–20 ng/µl.
-
8|
Preparation of sgRNA ODN inserts. Dissolve ODNs in annealing buffer to a final concentration of 100 µM. Combine 5 µl of each ODN in a separate PCR tube and anneal in a thermocycler using the following parameters: 95 °C for 2 min; 52 °C for 10 min; and hold at 4 °C.
-
9|Cloning of annealed sgRNA ODNs into pSMART-sgRNA. Set up the ligation reaction as shown below and incubate at room temperature for 1 h.
Component Amount Final concentration BbsI-digested pSMART-sgRNA vector from Step 6 20 ng 1 ng/µl Annealed sgRNA ODNs from Step 8 (100 µM) 1 µl 5 µM 10× Ligation buffer 2 µl 1× T4 DNA ligase (400 U/µl) 1 µl 20 U/µl Nuclease-free H2O Make up to 20 µl ■ PAUSE POINT After incubation, the ligation reaction can be stored at −20 °C for several weeks.
-
10|
Transformation. Transform the ligated plasmid into a competent Escherichia coli strain according to the manufacturer’s instructions. Briefly, add 3–5 µl of the ligation mixture from Step 9 to 30–50 µl of freshly thawed DH5-alpha cells and incubate the mixture on ice for 10–30 min. Heat-shock the cells at 42 °C for 30 s, and then cool on ice for 2–5 min. Add 950 µl of SOC medium and incubate with agitation for 60 min at 37 °C. Spread 100–200 µl of the culture onto an LB plate containing 50 µg/ml kanamycin and incubate overnight upside down at 37 °C.
-
11|
Cloning day 2. Inspect the plates for colony growth. Typically, there are tens to hundreds of colonies.
? TROUBLESHOOTING
■ PAUSE POINT Colonies on agar plates can be left on the bench at room temperature for up to 1 week.
-
12|
Pick four to six individual colonies using a sterile nonfilter pipette tip, and place each colony into a 15-ml tube containing 2 ml of LB medium with 50 µg/ml kanamycin. Incubate overnight at 37 °C with vigorous shaking.
-
13|
Cloning day 3. Isolate the plasmid DNA from cultures by using the QIAprep Spin Miniprep Kit according to the manufacturer’s instructions. Elute with 50 µl of buffer EB.
-
14|
Confirmation of sgRNA sequences. Verify the insertion and integrity of sgRNA ODNs by Sanger sequencing of the purified plasmid DNA using the pSMARTseqF ODN (binds just upstream of the U6 promoter). Map and sequence information for the pSMART-sgRNA vector can be found on the Addgene website (https://www.addgene.org).
? TROUBLESHOOTING
HDR template design and construction ● 1–2 weeks
-
15|Design plasmid templates for either gene knock-in (option A) or gene repair (option B). An example for each is shown in Figures 3 and 4. Alternatively, 100- to 200-bp ssODNs can also be used for gene repair or knock-in of point mutations with the inclusion of synonymous changes as detailed in the Experimental design section. An example is also shown in Figure 5.
- Design and construction of a plasmid template for reporter knock-in
- Design a repair template by following the specifications detailed in the Experimental design section.
- Generate a map of the targeting vector using sequence analysis software (e.g., Snapgene, VectorNTI, and DNAStar).
- Order a phosphorylated gBlock (double-stranded DNA fragments) encoding the designed template from Integrated DNA Technologies (https://www.idtdna.com).
-
Reconstitute the gBlock in 50 µl of TE buffer and ligate into a minimal plasmid vector, e.g., pSMART-HCKan, according to the manufacturer’s instructions. If the template is too long to be encoded by a single gBlock, two gBlocks, each encoding half of the template, can be inserted simultaneously into a plasmid vector. A unique restriction site can be included at each end of the repair template to enable ligation/cloning into a suitable plasmid vector.Δ CRITICAL STEP Some sequences, such as those with extremely low or high GC content, homopolymeric runs, or other structural motifs such as repeats or hairpins, may not be able to be synthesized using gBlock technology.See the TROUBLESHOOTING section for further advice.? TROUBLESHOOTING
- Design and construction of a plasmid template for repair or introduction of a point mutation
- Design a repair template by following the specifications detailed in the Experimental design section.
- Order a phosphorylated gBlock encoding the designed repair template from Integrated DNA Technologies (http://www.idtdna.com).
-
Reconstitute the gBlock in 50 µl of TE buffer and ligate into a minimal plasmid vector, e.g., pSMART-HCKan, according to the manufacturer’s instructions.Δ CRITICAL STEP Some sequences, such as those with extremely low or high GC content, homopolymeric runs, and other structural motifs such as repeats or hairpins, may not be able to be synthesized using gBlock technology.See the TROUBLESHOOTING section for further advice.? TROUBLESHOOTING
-
16|
Preparation of transfection-grade plasmids. Sequence-verified sgRNA (Step 14) and HDR template plasmid DNA (Step 15) can be prepared using the Qiagen Plasmid Maxi Kit according to the manufacturer’s instructions. Plasmid concentration should be ≥1 µg/µl.
■ PAUSE POINT Plasmid DNA can be stored for > 12 months at −20 °C.
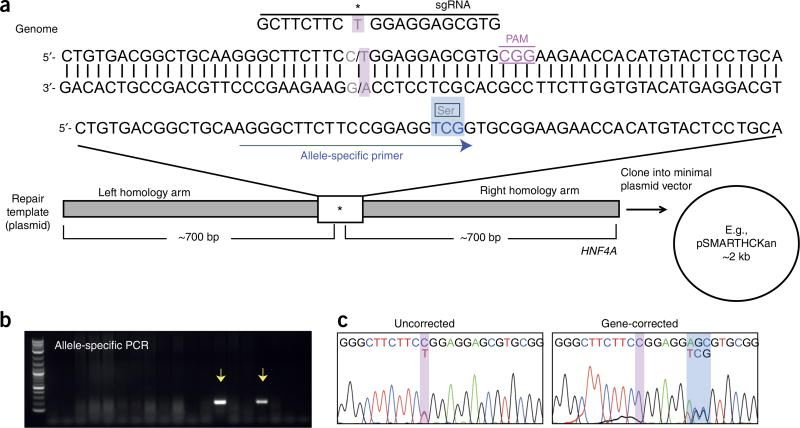
Figure 4.
Simultaneous reprogramming and genetic correction of fibroblasts from a kidney disease patient with an autosomal dominant mutation in HNF4A. (a) Schematic diagram of the HNF4A gene and the homologous template used for gene repair. The patient mutation (c.187C>T) (pink box), sgRNA, and 3-bp synonymous change (blue box) incorporated into the repair template and allele-specific primer used for identification of gene-corrected clones are shown. (b) Representative gel following PCR analysis of 18 iPSC clones using primers that specifically amplify the successfully gene-edited allele. Arrows indicate PCR products amplified from correctly edited clones. (c) Sanger sequencing analysis of the HNF4A target region of an uncorrected and a gene-corrected iPSC clone. *, patient-specific mutation; Ser, serine residue.
Figure 5.
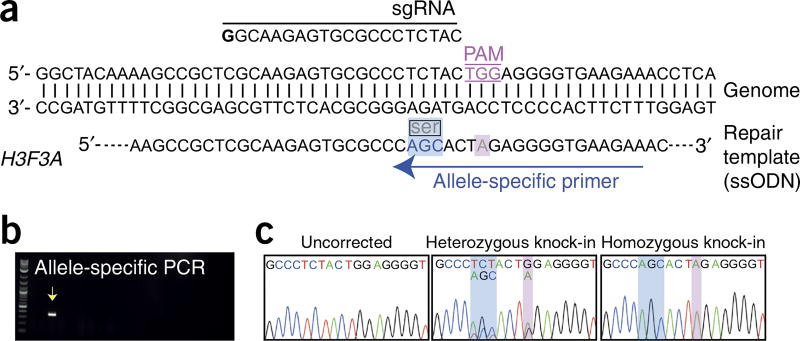
Generation of iPSC lines with a point mutation in H3F3A following one-step gene editing/reprogramming of healthy fibroblasts. (a) Schematic diagram of the H3F3A gene and the ssODN used for gene repair. The sgRNA, 3-bp synonymous change (blue box), and G > A mutation (pink box) incorporated into the ssODN are shown. The allele-specific primer used for identification of gene-corrected clones is also shown. (b) Representative gel following PCR analysis of 18 iPSC clones using primers that specifically amplify the successfully gene-edited allele. The arrow indicates PCR product amplified from a correctly edited clone. (c) Sequencing analysis of the H3F3A target region of a normal and two gene-edited (one heterozygous and one homozygous) iPSC clones. The 3-bp synonymous change introduced by the repair template is indicated.
In vitro transcription of Cas9 and Cas9-Gem m RNA ● TIMING 1–2 d
Δ CRITICAL Extra precautions should be taken to minimize RNase contamination. Use tubes, tips, and reagents that are guaranteed to be RNase-free. Note that DNA from some miniprep procedures may be contaminated with residual RNase A and that some reagents may occasionally introduce RNase or other inhibitors of transcription.
-
17|Linearize the pDNR-SpCas9 or pDNR-SpCas9-Gem plasmid DNA by setting up the following digestion reaction. Incubate at 37 °C for at least 1 h.
Component Amount Final concentration pDNR-SpCas9-Gem OR pDNR-Spcas9 10 µg 100 ng/ µl 10× CutSmart buffer 10 µl 1× PmeI (10 U/µl) 5 µl 0.5 U/µl Nuclease-free H2O Make up to 100 µl -
18|Terminate the reaction by adding the reagents in the following table. Mix well and incubate at −20 °C for 15 min.
Component Amount Final concentration 0.5 M EDTA 5 µl 6.8 mM 5 M Sodium acetate (supplied in mMessage mMachine Kit) 10 µl 137 mM Ethanol 250 µl -
19|
To pellet the DNA, centrifuge for 15 min in a microcentrifuge at 17,000g at room temperature. Remove the supernatant, respin the tube (17,000g, room temperature, 10 s), and remove the residual fluid with a fine-tipped pipette. Resuspend in 10 µl of TE buffer to achieve a concentration of 0.3–1 µg/µl.
-
20|Assemble the transcription reaction using the T7 mMessage mMachine Ultra Kit as described below. Mix thoroughly and incubate at 37 °C for 3–4 h.
Component Amount Final concentration 2× NTP/CAP (15 mM) 30 µl 7.5 mM GTP (20 mM) 1.5 µl 0.5 mM 10× Reaction buffer 6 µl 1× Linearized pDNR-SpCas9-Gem OR pDNR-Spcas9 template from Step 19 3 µg 50 ng/µl Enzyme mix 6 µl 0.1 µl/µl Nuclease-free H2O To 60 µl -
21|
Add 3 µl of Turbo DNase, mix well by flicking the tube, and incubate at 37 °C for 15 min.
■ PAUSE POINT After the incubation, the reactions can be stored at −20 °C for several days.
-
22|Add the poly(A) tailing reagents (supplied in the T7 mMessage mMachine Ultra Kit) to the reaction in the following order.
Component Amount Final concentration mRNA reaction from Step 21 63 µl Nuclease-free water 108 µl 5× E-PAP buffer 60 µl 1× 25 mM MnCl2 30 µl 2.5 mM ATP solution (10 mM) 30 µl 1 mM -
23|
Remove 2 µl from the reaction mixture before adding the E-PAP enzyme in Step 24; this minus-enzyme control can be run on a gel next to the tailed RNA at the end of the experiment. Store at −20 °C for up to several days.
-
24|
Add 12 µl of E-PAP enzyme, mix gently, and incubate at 37 °C for 45 min.
-
25|
Stop the reaction and precipitate the mRNA by adding 300 µl of LiCl precipitation solution (provided in the kit). Mix thoroughly and chill for 2–24 h at −20 °C.
■ PAUSE POINT The precipitation reaction can be left at −20 for 24 h.
-
26|
Pellet the mRNA in a microcentrifuge at 17,000g for 15 min at 4 °C. Remove the supernatant and wash the pellet with ~1 ml of 70% (vol/vol) ethanol and recentrifuge (17,000g, 15 min, 4 °C). Remove the supernatant and resuspend the pellet in 80 µl of nuclease-free water to achieve a final mRNA concentration of 1–2 µg/µl.
■ PAUSE POINT In vitro-transcribed mRNA can be stored for > 12 months at −20 or −80 °C.
In vitro transcription of EBNA1 m RNA ● TIMING 1–2 d
-
27|Linearize the pSP6-EBNA2A + DBD plasmid DNA by assembling the following digestion reaction. Incubate at 37 °C for 1–3 h.
Component Amount Final concentration pSP6-EBNA2A + DBD 10 µg 100 ng/µl 10× CutSmart buffer 10 µl 1× EcoRI-HF (10 U/µl) 5 µl 0.5 U/µl Nuclease-free H2O Make up to 100 µl -
28|
Repeat Steps 18 and 19 to terminate the reaction.
-
29|Assemble the transcription reaction using the mMessage mMachine SP6 Kit as described below. Mix thoroughly and incubate at 37 °C for 3–4 h.
Component Amount Final concentration 2× NTP/CAP (15 mM) 30 µl 7.5 mM GTP (20 mM) 1.5 µl 0.5 mM 10× Reaction buffer 6 µl 1× Linearized pSP6-EBNA2A + DBD template from Step 28 3 µg 50 ng/µl Enzyme mix 6 µl 0.1 µl/µl Nuclease-free H2O To 60 µl -
30|
Add 3 µl of Turbo DNase, mix well, and incubate at 37 °C for 15 min.
-
31|
Stop the reaction and precipitate the mRNA by adding 90 µl of nuclease-free H2O and 90 µl of LiCl precipitation solution. Mix thoroughly and chill for 2–24 h at −20 °C.
■ PAUSE POINT The precipitation reaction can be left at −20 °C for 24 h.
-
32|
Pellet the mRNA in a microcentrifuge at 17,000g for 15 min at 4 °C. Remove the supernatant and wash the pellet with ~1 ml of 70% (vol/vol) ethanol and recentrifuge as above. Remove the supernatant and resuspend the pellet in 100 µl of nuclease-free water to achieve a final mRNA concentration of 1–2 µg/µl.
■ PAUSE POINT In vitro-transcribed mRNA can be stored for > 12 months at −20 or −80 °C.
Gel analysis of in vitro-transcribed mRNA ● TIMING 3 h
-
33|
Add 2 µl of each mRNA sample to 2 µl of formaldehyde loading dye (supplied in the kit). In a separate tube, add 4 µl of the RNA ladder to 4 µl of formaldehyde loading dye. Incubate the samples at 85–90 °C for 5 min and load on a 1–2% (wt/vol) agarose gel containing 1 µg/ml ethidium bromide. Separate the bands by electrophoresis in 1× TAE buffer at 100 V for 1 h.
-
34|Check the bands using a gel imager. Although some smearing below the expected band size is typical, a strong band at the expected size should be clearly evident if the transcription reaction was successful (see the table below for expected band sizes). The Cas9 and Cas9-Gem mRNA samples post polyA tailing should also run more slowly than the sample taken before the tailing reaction.
mRNA Size (bp) Cas9 (before polyA tailing) ~4.2 kb Cas9 (after polyA tailing) ~4.8 kb Cas9-Gem (before polyA tailing) ~4.5 kb Cas9-Gem (after polyA tailing) ~5 kb EBNA1 ~1.5 kb Δ CRITICAL STEP To minimize degradation of the RNA during electrophoresis, we recommend using autoclaved 1× TAE buffer. Gel trays, combs, and tanks should also be rinsed with autoclaved H2O before use.
? TROUBLESHOOTING
Functional validation of sgRNA: human fibroblast culture and transfection ● TIMING 1 week
Δ CRITICAL Below, we detail transfection conditions for primary human fibroblasts (e.g., foreskin fibroblasts from ATCC ID CRL-2429) using the Neon Transfection System. Alternatively, sgRNA constructs can be evaluated in commonly used human cell lines such as HEK293; however, transfection conditions may vary across different cell types and should be optimized accordingly. Tips for optimizing transfection conditions using the Neon Transfection System can be found on the Thermo Fisher website (https://www.thermofisher.com/au/en/home/life-science/cell-culture/transfection/transfection---selection-misc/neon-transfection-system/neon-protocols-cell-line-data.html).
-
35|
Fibroblast cell culture. Maintain cells in fibroblast medium at 37 °C and 5% CO2. To passage, remove the medium and rinse the cells once by adding DPBS to the side of the vessel, so as not to dislodge the cells. Remove DPBS and add 8 ml of TrypLE to a T175 flask. Incubate the cells for 5 min at 37 °C. Add 10 ml of warm fibroblast medium and dissociate the cells by pipetting them up and down gently. Reseed them into new flasks at a split ratio of 1:3–1:5.
Δ CRITICAL STEP Growth rates among different primary fibroblast cultures vary. An actively expanding fibroblast culture with an acceptable growth rate generally reaches confluency and requires passaging every 3–5 d after a split ratio of 1:3–1:5. Cells should be discarded when a slower growth rate becomes apparent.
-
36|
Preparation of cells for transfection (Day 1). Passage fibroblasts (1:3–1:5) when confluent. One T175 flask is usually enough for two to three transfections.
-
37|
Day 3. Detach cells with TryPLE (as in Step 35) 2 d after passage, when cells are ~50–70% confluent. Add 7 ml of fibroblast medium to the cells after detaching and transfer them to a 15-ml Falcon tube. Centrifuge at 300g for 3 min at room temperature to pellet the cells.
-
38|
Aspirate and discard the medium, and resuspend the cells in 10 ml of DPBS; take an aliquot of the cell suspension and count the cells using a hemocytometer to determine the cell density. Recentrifuge at 300g for 3 min at room temperature to pellet the cells.
-
39|
Aspirate and discard the PBS, and resuspend the cell pellet in Resuspension Buffer R (provided in the Neon Transfection Kit) at a final density of 1.0 × 107 cells per ml. Gently pipette the cells to obtain a single-cell suspension.
-
40|
Transfect sgRNA plasmids. Place a Neon tube with 3 ml of Electrolytic Buffer E2 into the Neon pipette station and set the following pulse conditions: 1,400 V; 20 ms; two pulses. Transfer 100 µl of the cell suspension from Step 39 to a tube containing 2 µg of sgRNA plasmid DNA from Step 16 and 5 µl of Cas9 mRNA from Step 26. Mix by gently flicking the tube. Aspirate the mixture into a 100-µl Neon tip and perform electroporation according to the manufacturer’s instructions.
Δ CRITICAL STEP Avoid air bubbles during pipetting, as air bubbles cause arcing during electroporation, leading to lowered or failed transfection. If you notice air bubbles in the Neon tip, dispel the sample and carefully aspirate sample into the tip again until air bubbles are no longer apparent.
-
41|
Immediately transfer the electroporated cells to a T25 flask containing 15 ml of prewarmed fibroblast medium. Maintain the culture at 37 °C and 5% CO2.
Δ CRITICAL STEP We recommend including a transfection control when electroporating a new fibroblast line for the first time (e.g., pCMV-tdTomato vector) to monitor transfection efficiency. Transfect 2.5 µg of reporter plasmid using the conditions outlined in Step 40 and assess the number of fluorescent cells 2 d post transfection by using a fluorescence microscope or flow cytometry.
-
42|
Harvesting of cells for genomic DNA extraction. At 48 h post transfection, detach cells with TryPLE (as in Step 35) and extract genomic DNA from each sample using the Qiagen DNeasy Kit according to the manufacturer’s instructions. Genomic DNA should also be extracted from an equal number of untransfected fibroblasts as a control.
■ PAUSE POINT Extracted genomic DNA can be stored at −20 °C for several months.
-
43|PCR amplification of the targeted genomic locus. Use primers that flank the sgRNA target site to amplify ~500 bp of genomic sequence (each primer should bind an approximately equal distance upstream or downstream of the target site). Prepare the reaction as follows. Be sure to include genomic DNA from the untransfected control sample.
Component Amount Final concentration GoTaq Green Master Mix (2×) 12.5 µl 1× Forward primer (10 µM) 1 µl 400 nM Reverse primer (10 µM) 1 µl 400 nM Genomic DNA 50–200 ng 2–8 ng/µl Nuclease-free H2O To 25 µl -
44|Perform PCR using the following cycling conditions.
Cycle number Denature Anneal Extend 1 95 °C, 3 min 2–32 95 °C, 20 s 57 °C, 20 s 72 °C, 60 s per kb 33 72 °C, 5 min Δ CRITICAL STEP Although these PCR conditions are designed to work with most primers, some primers may need additional optimization, e.g., adjustment of the template concentration, MgCl2 concentration, and/or the annealing temperature.
-
45|
Run 5 µl of the PCR product on a 1% (wt/vol) agarose gel containing 0.5 µg/ml ethidium bromide. Separate the bands by electrophoresis in 1× TAE buffer at 100 V for 1–2 h and check for the band of the expected size using a gel imager.
? TROUBLESHOOTING
-
46|
Add 1 µl of exonuclease I and 1 µl of rAPid Alkaline Phosphatase to the remaining PCR product generated from Step 44. Incubate the samples at 37 °C for 30 min, and then at 80 °C for 10 min to heat inactivate.
-
47|Sanger sequencing of PCR product. Set up a BigDye sequencing reaction using a nested primer that binds just downstream of the forward primer (or just upstream of the reverse primer) used to amplify the genomic region of interest in Step 43. In addition, set up a reaction for the PCR product from the untransfected control sample.
Component Amount Final concentration BigDye buffer (5×) 2 µl 1× Sequencing primer (1 µM) 2 µl 200 nM Treated PCR product from Step 46 5 µl BigDye Terminator 1 µl -
48|Place in a thermocycler and run the following program:
Cycle number Denature Anneal Extend 1 95 °C, 30 s 2–32 95 °C, 10 s 50 °C, 10 s 60 °C, 4 min -
49|
Submit the samples to a Sanger sequencing service. Make sure to check with the facility for the requirements of the submitted DNA.
-
50|
Indel analysis using the TIDE algorithm. To estimate sgRNA efficacy, upload the sequence traces to the TIDE web tool (http://tide-calculator.nki.nl) as previously described54. Only sgRNAs that exhibit nuclease activity as determined in this step should be used for editing in subsequent steps. In our experience, sgRNAs that cause an indel rate of > 10% perform well when used for genome editing in subsequent steps.
Δ CRITICAL STEP Chromatograms should be inspected by eye to ensure they are of sufficient quality before uploading into the TIDE algorithm.
? TROUBLESHOOTING
Preparation of episomal reprogramming vectors ● TIMING 1–2 d
-
51|
We prepare large stocks of transfection-grade plasmid DNA by cesium chloride density gradient centrifugation55. Alternatively, prepare episomal reprogramming vectors using the Qiagen Plasmid Maxi Kit according to the manufacturer’s instructions. Ensure that the plasmid concentration is > 1 µg/µl.
■ PAUSE POINT Plasmid DNA can be stored for > 12 months at −20 °C.
Introduction of reprogramming/gene-editing factors into human fibroblasts ● TIMING 3 d
-
52|
Prepare fibroblasts as described in Steps 36–39.
-
53|Electroporation setup. Place a Neon tube with 3 ml of Electrolytic Buffer E2 into the Neon pipette station and set the following pulse conditions: 1,400 V; 20 ms; two pulses. Transfer 100 µl of the cell suspension from Step 52 to a tube containing the following:
Component Amount pEP4 E02S ET2K 2.5 µg pEP4 E02S EN2L 2.5 µg pEP4 E02S EM2K 2.5 µg pSimple-miR302/367 2.5 µg HDR template Plasmid OR 5 µg ssODN 0.3 nmol sgRNA plasmid 2 µg EBNA1 mRNA 5 µg Cas9-Gem mRNA 5 µg Δ CRITICAL STEP We recommend adding mRNA immediately before electroporation to minimize potential degradation by any contaminating RNases.
-
54|
Mix by gently flicking the tube. Aspirate the mixture into a 100-µl Neon tip and perform electroporation according to the manufacturer’s instructions
-
55|
Immediately transfer the electroporated cells at varying densities across a six-well Matrigel-coated plate containing 2 ml of prewarmed fibroblast medium. Maintain the culture at 37 °C and 5% CO2.
-
56|
Inspect the cells the next day and perform medium change if a noticeable amount of cell death or debris is observed.
Induction of iPSC colonies ● TIMING 3–4 weeks
-
57|
On day 3 or 4 post transfection, remove fibroblast medium from the culture and replace it with 10 ml of reprogramming medium (TesR-E7 supplemented with 100 µM sodium butyrate).
-
58|
Replace the medium every 2 d until iPSC colonies become evident. Cultures should be monitored daily by standard light microscopy for morphological changes and the emergence of iPSC colonies. Although vast morphological changes should become apparent within the first week post transfection, the first iPSC colonies are usually not evident until ~2 weeks after electroporation.
-
59|
Once the first iPSC colonies emerge (~2 weeks post transfection), replace the reprogramming medium with Essential 8 medium. Perform daily medium changes after this point.
? TROUBLESHOOTING
Picking of iPSC colonies ● TIMING 3–5 d
Δ CRITICAL Colonies should be ~1 mm in diameter before picking. This generally corresponds to 2.5–4 weeks after electroporation. Avoid letting the colonies become too big (> 2–3 mm), as they tend to spontaneously differentiate.
-
60|
Prepare a 24-well Matrigel-coated plate by adding 0.5 ml of Essential 8 medium to each well. Prewarm the plate to 37 °C.
-
61|
Replace the medium in the 10-cm dish containing the iPSC colonies. Place the dish on the stage of a dissecting stereomicroscope fitted within a biosafety cabinet.
-
62|
Detach single iPSC colonies with a sterile 200-µl filter tip. Slowly aspirate the colony into the tip and transfer the pieces from a single colony to one well of the 24-well plate prepared in Step 60.
Δ CRITICAL STEP Make sure not to pipette multiple colonies into the same well and avoid picking colonies growing in very close proximity to one another that may be mixed. Take care to transfer or remove every part of a detached colony, to prevent them from mixing with other colonies on the plate.
-
63|
If possible, pick 30–50 colonies (obviously this depends on reprogramming efficiency). Colonies will be of varying sizes and will most likely require picking over the course of several days. Be sure to replace culture medium before and after each session.
-
64|
The next day, use a standard light microscope to check attachment, viability, and morphology of the picked iPSC colonies in the 24-well plate. Perform daily medium changes (Essential 8 medium) until cells are ready for replica splitting, when individual colonies are typically 1–2 mm in diameter.
Δ CRITICAL STEP Avoid letting the colonies become too big (> 2–3 mm), as they tend to spontaneously differentiate.
Replica splitting of iPSC clones ● TIMING 1–2 weeks
-
65|
EDTA passaging of iPSC cultures. Remove the culture medium from the well and wash the cells once with 0.5 ml of 0.5 mM EDTA solution.
-
66|
Remove the wash, add 0.5 ml of fresh 0.5 mM EDTA solution, and incubate the cells at room temperature for 2–5 min until the cells start to separate and round up, and the colonies appear to have holes in them when viewed under a microscope.
Δ CRITICAL STEP EDTA incubation times can vary depending on cell line and colony size, so it is important to continue to monitor them under a microscope.
-
67|
Remove the EDTA solution, leaving the iPSC colonies still loosely attached to the plate.
Δ CRITICAL STEP If iPSCs start to dislodge before removal of EDTA solution, add 1 ml of Essential 8 medium directly to the well without removal of the EDTA. Transfer the cells to a 1.5-ml or 15-ml tube and centrifuge at 300g for 3 min at room temperature to pellet the cells. Remove the supernatant and resuspend the cells in 1 ml of Essential 8 medium, and divide the cells across two wells of the 24-well plate. Then proceed to Step 69.
-
68|
Remove the cells from the well(s) by gently squirting Essential 8 medium directly onto the cells. Transfer a small fraction (25–30%) of the cells in each well to a new 24-well plate.
Δ CRITICAL STEP Do not scrape the cells from the dish or pipette too vigorously. Very little pipetting is required to break up cell clumps, and one to two triturations should be sufficient. Ensure that the position of the clones is identical for the two plates and that both plates are labeled the same way to minimize potential for mix-ups.
-
69|
The replica plate from Step 68 should be maintained (daily medium changes and passaging every 3–4 d as necessary) until genomic analysis is complete and successfully modified clones are selected for further expansion, analysis, and cryopreservation.
-
70|
The cells remaining in the original 24-well plate should be cultured for an additional 2–5 d with daily medium changes until > 50% confluent before harvesting for subsequent genomic analysis.
Preparation of iPSCs for PCR analysis ● TIMING 1–2 h
-
71|
Remove the medium from each well of the plate from Step 70 marked for harvesting and add 0.2 ml of TrypLE. Incubate the cells for 2–3 min at room temperature until cells begin to separate but are still loosely attached.
-
72|
Remove the TrypLE, leaving the iPSC colonies still loosely attached to the plate.
-
73|
Add 50–100 µl of TE buffer to each well and pipette vigorously to remove as many cells as possible from the well. Transfer the cells from each well to a correspondingly labeled 1.5-ml tube.
-
74|
Place the samples into a floating foam tube rack and incubate the cells for 5–10 min in a beaker of boiling water.
-
75|
Carefully remove the samples and allow them to cool to room temperature.
-
76|
Centrifuge the samples in a benchtop microcentrifuge at 17,000g for 5 min at room temperature to pellet any cell debris. It is not necessary to transfer the supernatant to a new tube.
Δ CRITICAL STEP Samples prepared in this way are suitable for PCR amplification of fragments up to 1 kb in size. For longer amplicons, we recommend extracting genomic DNA from iPSCs using the Qiagen DNeasy Blood & Tissue Kit.
■ PAUSE POINT Samples can be stored for several weeks at 4 °C but should be recentrifuged (17,000g, room temperature, 5 min) before re-use.
Genotyping of iPSC clones by PCR amplification ● TIMING 1 d
-
77|Identification of gene-edited clones. For identification of large knock-in modifications, follow option A; for analysis of point-mutation correction or introduction, follow option B.
- Screening for large knock-in modifications (e.g., reporter lines)
- Use primers that flank the recombination junctions; one primer should bind within the inserted sequence (e.g., the reporter cassette) and a second primer should bind just outside the region of homology (see Fig. 3a, for example). The second primer should anneal outside the region spanned by the homology arms, to avoid false detection of residual repair template.
- Screening for correction or introduction of a point mutation
-
If a synonymous change (of at least 2 bp) has been included in the repair template, detect the targeted clones by allele-specific PCR using a primer that overlaps the synonymous change, which should be at the very 3′ end of the primer. The second primer should bind outside the region of homology.Δ CRITICAL STEP We recommend performing a control PCR reaction using primers that amplify an irrelevant sequence (e.g., ~500 bp of the GAPDH gene) to ensure samples are adequate for PCR amplification.
-
-
78|Set up the following reaction for each sample from Step 76 and transfer 20 µl to a well of a 96-well PCR plate.
Component Amount Final concentration GoTaq Green Master Mix (2×) 12.5 µl 1× Forward primer (10 µM) 1 µl 400 nM Reverse primer (10 µM) 1 µl 400 nM Nuclease-free H2O To 20 µl -
79|Add 5 µl of each sample from Step 76 to the corresponding wells containing PCR mix in the 96-well plate. Include a sample from untransfected fibroblasts or iPSCs as a negative control. Place the plate in a thermocycler and run the following program:
Cycle number Denature Anneal Extend 1 95 °C, 3 min 2–32 95 °C, 20 s 57 °C, 20 s 72 °C, 60 s per kb 33 72 °C, 5 min Δ CRITICAL STEP Although these PCR conditions are designed to work with most primers, some primers may need additional optimization, e.g., adjustment of the template concentration, MgCl2 concentration, and/or the annealing temperature.
-
80|
Run 10 µl of the PCR product on a 1% (wt/vol) agarose gel to check for potentially targeted clones that have a DNA band of the expected size. Ensure that this band is absent for negative-control sample.
? TROUBLESHOOTING
Sequencing analysis of targeted iPSC clones ● TIMING 2–3 d
-
81|Use primers from Step 43 that flank the sgRNA target site and amplify ~250 bp of genomic sequence on either side of the Cas9 target site. Prepare the following reaction for each potentially targeted clone identified in Step 80. Add 20 µl to an individual PCR tube.
Component Amount Final concentration GoTaq Green Master Mix (2×) 12.5 µl 1× Forward primer (10 µM) 1 µl 400 nM Reverse primer (10 µM) 1 µl 400 nM Nuclease-free H2O To 20 µl -
82|Add 5 µl of sample for each potentially targeted clone identified in Step 80 to the reaction prepared in Step 81. Include a sample from untransfected fibroblasts or iPSCs as a negative control. Place PCR tubes in a thermocycler and perform PCR using the following cycling conditions (or those previously optimized in Step 44).
Cycle number Denature Anneal Extend 1 95 °C, 3 min 2–32 95 °C, 20 s 57 °C, 20 s 72 °C, 60 s per kb 33 72 °C, 5 min -
83|
Run 10 µl of PCR product on a 1% (wt/vol) agarose gel to check for a DNA band of the expected size.
? TROUBLESHOOTING
-
84|
Add 1 µl of exonuclease I and 1 µl of rAPid Alkaline Phosphatase to the PCR product generated from Step 81. Incubate the samples at 37 °C for 30 min, and then at 80 °C for 10 min to heat inactivate.
-
85|
Repeat Steps 47–49.
-
86|
Analysis of sequencing chromatograms. Reference the sequencing results against the expected genomic sequence to check for the presence of Cas9-induced NHEJ or HDR modifications.
? TROUBLESHOOTING
Cryopreservation of iPSCs ● TIMING 1 h
-
87|
Harvest iPSCs cells by EDTA dissociation (Steps 65–68) and transfer them to a 15-ml tube. Ensure that the cells are maintained as clumps.
-
88|
Prepare the freezing medium by mixing 100 µl of DMSO and 400 µl of Essential 8 medium (for one vial).
-
89|
Centrifuge the cells from Step 87 for 3 min at 300g at room temperature, and then discard the supernatant. Add 500 µl of fresh Essential 8 medium to the cell pellet and gently flick the tube to aid resuspension.
-
90|
Dropwise, add the freezing medium and transfer the cell clumps to a cryotube (final volume of 1 ml per vial).
-
91|
Place the cryotube into a Mr. Frosty freezing container containing 200 ml of 2-propanol and place the container into a 80 °C freezer overnight to achieve a −1 °C per min freezing rate.
-
92|
The next day, transfer the cryotube to a liquid nitrogen tank for long-term storage (indefinitely).
iPSC quality control ● TIMING 2–3 weeks
-
93|
Confirmation of pluripotency. Confirm pluripotency markers in gene-edited iPSC lines (and uncorrected control lines, if applicable) by performing immunofluorescence for common pluripotency markers (e.g., OCT4, NANOG, and SSEA-4), in vitro differentiation, or in vivo teratoma assays as previously described51,52.
-
94|
Karyotyping. CRISPR/Cas9 editing, reprogramming, and extensive culture periods can make cells susceptible to chromosomal re-arrangements and other chromosomal aberrations. Confirm iPSC lines used have a normal karyotype by standard G-banding or SNP microarray. Many companies perform karyotyping analysis.
-
95|
Mycoplasma testing. Test for mycoplasma. Several companies provide mycoplasma testing services. Alternatively, confirm the absence of mycoplasma using the MycoAlert Kit according to the manufacturer’s instructions.
-
96|Confirmation of loss of episomal vectors. Perform PCR analysis of genomic DNA extracted from iPSC lines using primers specific to the ampicillin resistance gene encoded by all four reprogramming vectors and/or the EBNA1 gene, which is carried by three of the reprogramming vectors. Prepare the following reaction for each iPSC clone identified in Step 80. Place each reaction into an individual PCR tube.
Component Amount Final concentration GoTaq Green Master Mix (2×) 12.5 µl 1× Forward primer (10 µM) 1 µl 400 nM Reverse primer (10 µM) 1 µl 400 nM Genomic DNA 50–200 ng 2–8 ng/µl Nuclease-free H2O To 25 µl Δ CRITICAL STEP In addition, set up a negative-control reaction containing genomic DNA from untransfected cells and a positive-control sample that contains 1 ng of reprogramming plasmid.
-
97|Place the PCR tubes in a thermocycler and perform PCR using the following cycling conditions.
Cycle number Denature Anneal Extend 1 95 °C, 3 min 2–32 95 °C, 20 s 57 °C, 20 s 72 °C, 60 s per kb 33 72 °C, 5 min -
98|
Run 10 µl of PCR product on a 1% (wt/vol) agarose gel to check for the presence of a DNA band of the expected size in positive-control reaction and the absence of this band in reactions containing iPSC genomic DNA and the negative-control reaction.
? TROUBLESHOOTING
-
99|Assessment of random integration of the targeting template. Perform PCR analysis using primers specific to sequences on the plasmid backbone of the targeting vector, such as the bacterial origin of replication or antibiotic resistance gene. Prepare the following reaction for each iPSC clone identified in Step 80. Place each reaction into an individual PCR tube.
Component Amount Final concentration GoTaq Green Master Mix (2×) 12.5 µl 1× Forward primer (10 µM) 1 µl 400 nM Reverse primer (10 µM) 1 µl 400 nM Genomic DNA 50–200 ng 2–8 ng/µl Nuclease-free H2O To 25 µl ΔCRITICAL STEP In addition, set up a negative-control reaction containing genomic DNA from untransfected cells and a positive-control sample that contains 1 ng of targeting vector.
-
100|Place the PCR tubes in a thermocycler and perform PCR using the following cycling conditions.
Cycle number Denature Anneal Extend 1 95 °C, 3 min 2–32 95 °C, 20 s 57 °C, 20 s 72 °C, 60 s per kb 33 72 °C, 5 min -
101|
Run 10 µl of PCR product on a 1% (wt/vol) agarose gel to check for the presence of a DNA band of the expected size in the positive-control reaction and the absence of this band in reactions containing iPSC genomic DNA and the negative-control reaction.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
Table 2.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 11 | No colonies on the plate | Inefficient ligation/transformation | Repeat ODN annealing and ligation/transformation steps |
| Incorrect ODN sequences | Check ODN sequence and re-order if necessary | ||
| 14 | No sgRNA sequence; wrong sgRNA sequence | Incomplete digestion of cloning plasmid; insertion of incorrectly synthesized ODN; ligation failure | Screen additional colonies; re-anneal sgRNA ODNs; titrate sgRNA oligo concentration during ligation; redigest pSMART-sgRNA vector |
| 15A(iv), 15B(iii) | Repair template unable to be synthesized as gBlock | Low or high GC content; homopolymeric runs of ten or more As and Ts, or six or more Gs and Cs; repetitive sequences | Adjust the length of the repair template; add a synonymous change within the sequence that is problematic; use an alternative strategy to generate part or the entire repair template, such as PCR amplification from genomic DNA or a BAC clone |
| 34 | No visible band; smeared bands; multiple bands | RNA degraded due to RNase contamination; incomplete linearization of template DNA | If RNA ladder is also degraded thoroughly, rinse gel trays, combs, and tanks with autoclaved H2O; if RNA ladder is intact but the sample is degraded, add 1–2 µl of RNAse inhibitor to the DNA template sample and repeat transcription reaction; repeat template linearization step and analyze by gel electrophoresis to ensure complete digestion |
| 45 | Multiple bands or no amplification visible on the gel | Inefficient or nonspecific priming; incorrect template concentration | Titrate MgCl2 (0–1 mM final concentration); normalize and titrate template concentration (50–200 ng total), titrate annealing temperature (55–62 °C); redesign primers to minimize primer dimer formation or nonspecific amplification |
| 50 | Indel frequency low or absent | sgRNAs exhibit no or very low activity; low transfection efficiency | Redesign sgRNAs; add EGFP transfection control; reseed cells evenly at recommended density; prepare new DNA/mRNA and repeat transfection; repeat transfection in a different cell line |
| 59 | No iPSC colony formation or morphological changes observed | Electroporation failure; poor-quality fibroblasts; poor-quality plasmid DNA | Repeat electroporation, ensuring the tip is free of air bubbles; perform a control experiment using a well-established cell line (such as neonatal fibroblasts from ATCC, e.g., CRL-2429); check plasmid DNA quality and quantity by spectrophotometry and gel electrophoresis |
| 80 | No bands; multiple nonspecific bands; strong band in negative control | No gene-edited iPSC clones; samples not adequate for PCR amplification; inefficient or nonspecific priming | Screen more iPSC clones; run a control PCR with another primer set (e.g., for GADPH) to check sample quality; adjust PCR conditions (see above); design a new primer pair |
| 83 | Incorrectly sized band | Indel mutations present in one or both alleles; larger than expected band may be indicative of a homozygous clone if performing knock-in (e.g., of a reporter gene) | If homozygous clone is suspected, confirm by Sanger sequencing; discard clones that harbor indel mutations |
| 86 | No gene-edited clones; gene-edited clones with additional indels | Ineffective sgRNA, sgRNA, and/or Cas9-Gem dosage too high | Design/construct/validate new sgRNA plasmids; repeat electroporation with lower sgRNA plasmid and/or Cas9-Gem mRNA concentrations |
| 98 | Bands present in reactions containing genomic DNA from iPSC clones | Persistence of reprogramming vectors | Repeat PCR analysis after another five to ten passages. Subcloning can also be performed to ensure iPSC lines are free of reprogramming transgenes |
● TIMING
Steps 1 and 2, CRISPR selection and design: 1 h
Steps 3–14, cloning of sgRNA ODNs into the pSMART-sgRNA vector: 3–4 d
Steps 15 and 16, HDR template design and construction: 1–2 weeks
Steps 17–26, in vitro transcription of Cas9 and Cas9-Gem mRNA: 1–2 d
Steps 27–32, in vitro transcription of EBNA1 mRNA: 1–2 d
Steps 33 and 34, gel analysis of in vitro-transcribed mRNA: 3 h
Steps 35–50, functional validation of sgRNA: 1 week
Step 51, preparation of episomal reprogramming vectors: 1–2 d
Steps 52–56, introduction of reprogramming/gene-editing factors into human fibroblasts: 3 d
Steps 57–59, induction of iPSC colonies: 3–4 weeks
Steps 60–64, picking of iPSC colonies: 3–5 d
Steps 65–70, replica splitting of iPSC clones: 1–2 weeks
Steps 71–76, preparation of iPSCs for PCR analysis: 1–2 h
Steps 77–80, genotyping of iPSC clones by PCR amplification: 1 d
Steps 81–86, sequencing analysis of targeted iPSC clones: 2–3 d
Steps 87–92, cryopreservation of iPSCs: 1 h
Steps 93–101, iPSC quality control: 2–3 weeks
ANTICIPATED RESULTS
We have rigorously validated our streamlined one-step approach in numerous human fibroblast lines, with successfully edited iPSCs typically comprising 5–20% of the total iPSC population6,19. Here, we describe the use of this protocol for knock-in of a reporter gene into the MAFB locus using commercially available fibroblasts, repair of an autosomal dominant mutation within the HNF4A locus in patient fibroblasts, and introduction of heterozygous and homozygous mutations in H3F3A, again using commercially available fibroblasts. In all cases, cells were tested and confirmed to be free of mycoplasma contamination before and after reprogramming.
To generate podocyte-specific reporter lines, we chose to knock-in the mTagBFP2 gene56 into the endogenous MAFB locus. We used a functionally validated sgRNA associated with low off-target activity (determined in silico) that overlaps the MAFB start codon and is unlikely to induce cleavage within the MAFB:mTagBFP2 repair template. The repair template consisted of the mTagBFP2 reporter and BGH-poly(A) signal flanked by 440-bp and 550-bp homology arms specific to sequences directly upstream and downstream of the MAFB start codon, respectively (Fig. 3a). Two fragments encoding each end of the repair template were cloned into the AatII and EcoRI sites of the commercially available pDNR-Dual plasmid vector (from Clontech). The repair template, MAFB-specific sgRNA plasmid, and mRNA encoding Cas9-Gem were co-transfected, along with the episomal reprogramming plasmids, into commercially available neonatal fibroblasts (ATCC, ID CRL-2429). Of 40 iPSC colonies screened (using primers that flank the recombination junctions), we identified five clones that were successfully targeted (Fig. 3b). Using primers that flank the mTagBFP2 reporter cassette (that bind within the MAFB locus), we were able to determine that three of these clones were homozygous for the mTagBFP2 insertion (Fig. 3c). The untargeted allele in the remaining two heterozygous clones was sequence-verified, which confirmed lack of any disruptive indel mutations. Using the human kidney organoid differentiation protocol described previously57,58, appropriate reporter gene expression was also confirmed, with mTagBFP2 expression localized and restricted to the podocyte population within developing glomeruli (Fig. 3d).
We have also used simultaneous gene editing and reprogramming for the generation of gene-corrected and uncorrected iPSC lines from skin fibroblasts derived from an adult kidney disease patient. The disease-causing mutation is an autosomal dominant missense mutation in exon 2 of the HNF4A gene (c.187C > T), which we confirmed by PCR/Sanger sequencing before reprogramming/gene correction. To minimize Cas9-mediated cleavage of the wild-type allele, we used an sgRNA that overlaps the patient-specific mutation (Fig. 4a). We also used a plasmid-based repair template with ~0.7-kb homology arms flanking the target site. A 3-bp synonymous change (c.193_195AGC > TCG; p.Ser65Ser) was also incorporated within the sgRNA target region of the repair template to act as a Cas9-blocking mutation and to facilitate the identification of gene-corrected iPSC clones by allele-specific PCR (Fig. 4b). We identified two gene-corrected clones out of a total of 44 screened. Both clones were sequence-verified, which revealed repair of the patient-specific mutation, gain of the 3-bp synonymous change, and lack of indel formation on the other allele (Fig. 4c).
We also demonstrated the use of our one-step protocol for the generation of iPSCs from a second commercially available fibroblast line from ATCC (ID PCS-201-010), into which we have inserted a specific point mutation in the H3F3A gene. Here, we used an ssODN repair template to introduce a G > A transition in exon 2 of H3F3A (p.Gly34Arg). A 3-bp synonymous change (c.94_96TCT > AGC; p.Ser32Ser) was also incorporated within the sgRNA target region of the repair template to act as a Cas9-blocking mutation and to facilitate the identification of correctly modified iPSC clones by allele-specific PCR (Fig. 5b). In this experiment, we screened a total of 32 clones, and identified a single heterozygous clone and a single homozygous clone, which were verified by Sanger sequencing of the H3F3A target site (Fig. 5c).
Supplementary Material
Acknowledgments
We acknowledge A. Mallett, Royal Brisbane and Women’s Hospital; C. Patel, Genetic Health Queensland; C. Simons and J. Crawford, University of Queensland; B. Bennetts, G. Ho, and K. Holman, Childrens Hospital Westmead; and A. Nandini and team, Pathology Queensland, acting as part of the KidGen Collaborative; for clinical and genetic evaluation, mutation identification, and recruitment of fibroblasts from RG_0120.153 (patient with HNF4A mutation). This work was supported by the National Institutes of Health (DK107344-01) and the National Health and Medical Research Council (NHMRC; GNT1098654 and GNT1100970). M.H.L. is a Senior Principal Research Fellow of the NHMRC (GNT1042093). The Murdoch Children’s Research Institute is supported by the Victorian Government’s Operational Infrastructure Support Program.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHORCONTRIBUTIONS S.E.H. and J.A.T. conceived and developed the protocol. S.E.H. prepared the manuscript. J.A.T. and M.H.L. provided supervision and assisted in manuscript preparation.
COMPETING INTERESTS
The authors declare no competing interests.
References
- 1.Chung CY, et al. Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013;342:983–987. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan SD, et al. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 2013;155:1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, et al. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J. Am. Coll. Cardiol. 2014;64:451–459. doi: 10.1016/j.jacc.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, et al. Engineering human stem cell lines with inducible gene knockout using CRISPR/Cas9. Cell Stem Cell. 2015;17:233–244. doi: 10.1016/j.stem.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez F, et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howden SE, et al. Simultaneous reprogramming and gene correction of patient fibroblasts. Stem Cell Reports. 2015;5:1109–1118. doi: 10.1016/j.stemcr.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou Z, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwart D, Paquet D, Teo S, Tessier-Lavigne M. Precise and efficient scarless genome editing in stem cells using CORRECT. Nat. Protoc. 2017;12:329–354. doi: 10.1038/nprot.2016.171. [DOI] [PubMed] [Google Scholar]
- 12.Hockemeyer D, Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell. 2016;18:573–586. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 14.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. Structural biology. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- 16.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canver MC, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 2017;292:2556. doi: 10.1074/jbc.A114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howden SE, et al. A Cas9 variant for efficient generation of indel-free knockin or gene-corrected human pluripotent stem cells. Stem Cell Reports. 2016;7:508–517. doi: 10.1016/j.stemcr.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 21.Merkle FT, et al. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep. 2015;11:875–883. doi: 10.1016/j.celrep.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 26.Forsyth NR, et al. Physiologic oxygen enhances human embryonic stem cell clonal recovery and reduces chromosomal abnormalities. Cloning Stem Cells. 2006;8:16–23. doi: 10.1089/clo.2006.8.16. [DOI] [PubMed] [Google Scholar]
- 27.Bai Q, et al. Temporal analysis of genome alterations induced by single-cell passaging in human embryonic stem cells. Stem Cells Dev. 2015;24:653–662. doi: 10.1089/scd.2014.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahmoudi S, Brunet A. Aging and reprogramming: a two-way street. Curr. Opin. Cell Biol. 2012;24:744–756. doi: 10.1016/j.ceb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abyzov A, et al. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gore A, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howden SE, et al. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc. Natl. Acad. Sci. USA. 2011;108:6537–6542. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lo Sardo V, et al. Influence of donor age on induced pluripotent stem cells. Nat. Biotechnol. 2017;35:69–74. doi: 10.1038/nbt.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinhardt P, et al. Genetic correction of a LRRK2 mutation in human iPSCs links Parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12:354–367. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guschin DY, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 36.Villegas J, McPhaul M. Establishment and culture of human skin fibroblasts. Curr. Protoc. Mol. Biol. 2005 doi: 10.1002/0471142727.mb2803s71. Unit 28.23. [DOI] [PubMed] [Google Scholar]
- 37.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 38.Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 39.Chen F, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paquet D, et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 41.Kim JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen G, et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bae S, Park J, Kim J-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith C, et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crosetto N, et al. Nucleotide-resolution DNA double-strand breaks mapping by next-generation sequencing. Nat. Methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 48.Draper JS, et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat. Biotechnol. 2004;22:53–54. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- 49.Cowan CA, et al. Derivation of embryonic stem-cell lines from human blastocysts. New Eng. J. Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 50.Baker DE, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 52.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 53.Park J, Bae S, Kim J-S. Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics. 2015;31:4014–4016. doi: 10.1093/bioinformatics/btv537. [DOI] [PubMed] [Google Scholar]
- 54.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radloff R, Bauer W, Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc. Natl. Acad. Sci. USA. 1967;57:1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One. 2011;6:e28674. doi: 10.1371/journal.pone.0028674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takasato M, Er PX, Chiu HS, Little MH. Generation of kidney organoids from human pluripotent stem cells. Nat. Protoc. 2016;11:1681–1692. doi: 10.1038/nprot.2016.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takasato M, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.