Abstract
The long-term chronic inflammation of cervical intraepithelial neoplasia (CIN) induces the initiation and progression of cervical cancer. Long non-coding RNAs (LncRNAs) are being identified to be involved into inflammation and carcinogenesis and could function as cancer biomarkers in clinical. However, the significance of inflammation-related LncRNA (e.g. LncRNA-IL7R) in cervical cancer is limited. We, here, investigated the clinical role of inflammation-related LncRNA-IL7R (Lnc-IL7R) in healthy cervical tissue (n=15), CIN 1/2/3 (n=35), cervical cancer (n=70), and clarified its function via knockdown in vitro and in vivo. The results showed that the expression of Lnc-IL7R was increased from normal tissues to neoplastic lesions and cervical cancer. Up-regulated Lnc-IL7R positively correlated to tumor size, International Federation of Gynaecology and Obstetrics (FIGO) stage, and lymph node metastasis (LNM). Patients with high expression of Lnc-IL7R had poor prognosis with short overall survival (OS) time, and Cox regression analysis revealed that Lnc-IL7R could be independent prognostic factor for cervical cancer. Moreover, knockdown of Lnc-IL7R by two different siRNAs in cervical cancer cell lines Hela and SiHa induced impaired cell vitality and caspase-3-dependent apoptosis in vitro. Furthermore, inhibition of Lnc-IL7R in vivo significantly restricted the tumor growth with decreased expressions of proliferation index Ki-67 and Lnc-IL7R. These data indicated that Lnc-IL7R predicts a poor clinical outcome of cervical cancer patients, and knockdown of Lnc-IL7R is amenable to the treatment of cervical cancer.
Keywords: cervical cancer, Inflammation, Lnc-IL7R, prognosis
Introduction
Cervical cancer is the major cause of death from gynecological cancers and is the third most common malignancy in women worldwide with a global incidence of 500000 and mortality of 250000. More than 85% of these cases and deaths occurred in developing countries, including China [1,2]. Persistent infection with oncogenic subtypes of the human papillomavirus (HPV) results in chronic inflammation, leading to the cervical intraepithelial neoplasia (CIN) and carcinogenesis of uterine cervix [3]. The signs and symptoms of cervical cancer often occur in the later stages of the infection (CIN 1, 2, and 3), thus, the detection of tumorigenesis at the microscopic level is inefficient in earliest stages of diagnosis [4]. Currently, some proteins and HPV DNA-based biomarkers are developed for the diagnosis of cervical cancer in clinical, such as SSC-Ag, CA-125, CEA, and Cytokeratins [4]. In addition, no single screening method exists that is highly sensitive, highly specific, affordable, and practical [5]. Therefore, it is still urgent to identify new and effective prognostic markers and therapeutic strategies to improve treatment of cervical cancer.
The majority of transcribed RNAs are non-coding in mammalian cells, which do not contain protein-coding sequences. These transcripts are eventually, on one hand, processed into small RNAs including miRNAs, Piwi-interacting RNAs (piRNAs), tRNA-derived stress-induced fragment RNAs, and small nucleolar RNAs (snoRNAs), and, on the other hand, processed into long non-coding RNAs (LncRNAs) [6,7]. The miRNAs and LncRNAs have demonstrated to be involved into carcinogenesis and functioned as diagnostic and prognostic biomarkers [8,9]. miR-138 expression in cervical cancer cells is significantly lower than that in normal tissues, which causes telomerase activation and carcinogenesis [10]. Let-7b, let-7c, miR-23b, miR-143, and miR-196b were down-regulated in cell lines and tumor tissue compared with normal tissue whereas miR-21 was up-regulated [10]. Up-regulated LncRNA HOTAIR in cervical cancer tissues and correlated with International Federation of Gynaecology and Obstetrics (FIGO) stage, lymphatic metastasis, size of tumor in cervical cancer progression and could be a potential target for diagnosis as well as an independent predictor for overall survival (OS) [11]. The tumor-suppressor LncRNA GAS5 was down-regulated in cervical cancer tissues, significantly correlated to advanced cancer progression, and identified as a biomarker for forecasting the clinical states of patients in cervical cancer [12]. Similarly, LncRNA MALAT1 and HOTAIR were reported to predict the poor clinical outcome of patients with cervical cancer [9]. Chronic inflammation is essential for the development of cervical cancer, but the role of inflammation-related LncRNA in cervical is unclear.
Cui et al. [13] found an inflammation-regulated LncRNA-IL7R (Lnc-IL7R) was capable of diminishing the LPS-induced inflammatory response, inhibiting the expressions of LPS-induced E-selectin, VCAM-1, IL-6, and, IL-8. Ding et al. [14] reported that Lnc-IL7R was also induced in response to TLR3 stimulation and negatively regulated the TNF-α and IL-8. Lnc-IL7R has been found to participate in multiple sclerosis, chemotherapy, and acute respiratory distress syndrome (ARDS) [14–16]. The Lnc-IL7R levels were correlated with the severity of ARDS and could predict 28-day mortality in the patient’s cohort [15]. In the present study, we investigated the potential clinical role of Lnc-IL7R in cervical cancer. The expression of Lnc-IL7R in normal, CIN, and cervical cancer samples, and its correlation to clinical characteristics were analyzed. The functions of Lnc-IL7R in vitro and in vivo were also assessed in cervical cancer cell lines Hela and SiHa.
Materials and methods
Patients and tissue specimens
Healthy cervical tissue (n=15), CIN 1/2/3 (n=35), cervical cancer (n=70) were collected to determine the expressions of Lnc-IL7R and TNF-α, and median age was 35, 37, and 51 years, respectively. The sample collection in the present study was approved by Yan’an People’s Hospital and all patients completed informed consent forms and the healthy individuals’ recruitment were also obtained from Yan’an People’s Hospital. All these retrospective specimens were handled and anonymized according to ethical and legal standards. The tissue samples were isolated from surgical removal and then stored at −80°C until use.
The patients with primary cervical cancer were diagnosed by Hematoxylin and Eosin staining by experienced pathologists from Department of Pathology at Yan’an People’s Hospital. None of the patients underwent preoperative chemotherapy and/or radiotherapy. Patients with other kinds of cancer or some autoimmune disease (e.g. rheumatoid arthritis, systemic lupus erythematous, diabetes etc.) were absolutely excluded. Besides, pregnant and lactating individuals were also excluded from the present study. The 70 patients were followed up until 1 October 2017. The high or low expressions of Lnc-IL7R were defined by the median of the expression. The three grades of Lnc-IL7R expression were defined as Grade 1 (25th percentile), Grade 2 (25–50th percentile), and Grade 3 (>50th percentile).
RNA isolation and quantitative real-time RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) according to standard RNA isolation protocol. The conditions of reverse-transcription for cDNA with Reverse Transcription System were 42°C for 5 min, 99°C for 20 min, 4°C for 5 min. The SYBR Green PCR Master Mix (Applied Biosystems), according to the manufacturer’s instructions, was performed for quantitative real-time RT-PCR (qRT-PCR) and 2−ΔΔCT method was used to estimate relative expression changes in genes including Lnc-IL7R and TNF-α. The expression levels were normalized to GAPDH for gene expression.
Cell lines and reagents
The cervical cancer cell lines Hela and SiHa were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS (Life Technologies, U.S.A.), ampicillin and streptomycin at 37°C, 5% CO2 conditions. siRNA-Lnc-IL7R or negative control was purchased from RiboBio (Guangzhou, China). Anti-GAPDH, Ki-67, caspase-3, and Bcl-2 antibodies were obtained from Cell Signaling Technology (Denver, MA) and Abcam (U.S.A.).
Cell transfection
The Hela and SiHa cell lines were cultured to ~80% confluence in 12/96-well plates and then, using Lipofectamine 2000 (Invitrogen, U.S.A.), the cells were transfected with indicated agents according to the manufacturer’s instructions. After transfection for the indicated time, the cells were harvested for further experiments.
CCK-8 assay
The Hela and SiHa cell lines were transfected with siRNAs and then were harvested to wash with PBS, and then cell counting kit-8 (Kumamoto, Japan) mixed with DMEM was used for cell viability assay, and the absorbance was measured at 450 nm by a microplate reader.
Hoechst staining assay
The conditional Hela and SiHa cell lines were harvested and incubated with Hoechst 33342 (5 μg/ml, Sigma, U.S.A.) for 10 min at room temperature. Following washing with 0.5% Triton X-100 in PBS, the changes in nuclear morphology were observed under a fluorescence microscope (Olympus, Tokyo, Japan). Each experiment was performed in triplicate and repeated three times.
Western blot
According to the manufacturer’s protocol, cells for Western blot were collected and total protein was isolated from the cell samples. Detailed procedures for immunoblotting are described elsewhere [17]. GAPDH was used as the loading control in the Western blotting.
Immunohistochemistry
To determine the expression of Ki-67 in tissues, 2-μm thick, formalin-fixed, and paraffin-embedded specimen sections were used. After the slides were incubated in xylene for 5 min, 100% ethanol was used for 10 min, 95% ethanol for 10 min. Antigen unmasking was performed and then the slides were blocked with 3% hydrogen peroxide for 30 min at room temperature. Then the primary antibody for Ki-67 was incubated the FFPE specimen sections at 4°C overnight, then the biotinylated horse secondary antibody and streptavidin-horseradish peroxidase (Zymed Laboratories Inc.) were used for the detection of Ki-67. After that, the EnVision Detection System kit (DAKO, Denmark) was used for the DAB chromogen followed by nuclear staining using Hematoxylin.
Tumor model
To investigate the role of Lnc-IL7R in vivo, Hela cells were transfected with lentivirus vector of siRNA-Lnc-IL7R or negative control, 2 × 106 Hela cells were subcutaneously injected in rear flank of nude mice (five per group). The tumor sizes were measured 3 days apart and the tumor volumes were calculated: V (cm3) = width2 (cm2) * length (cm)/2. On day 2, the mice were killed.
Statistical analyses
The results were analyzed by the Statistical Package for Social Sciences version 16.0 (SPSS 16.0, SPSS Inc., Chicago, IL, U.S.A.) and the Prism statistical software package (version 5.0, GraphPad Software Inc.). Kolmogorov–Smirnov and Shapiro–Wilk tests showed that the expression of Lnc-IL7R in each group did not follow a normal distribution. The Mann–Whitney U-test was used to compare the two groups (e.g. normal compared with CIN) and the differences between more than two groups (e.g. CIN1/2/3) were analyzed by the Kruskal–Wallis test. Kaplan–Meier survival curves and the log-rank statistic were used to analyze the prognostic significance of Lnc-IL7R. Correlations between the expression of Lnc-IL7R and TNF-α were analyzed by Spearman’s Rho analysis. Correlations of expression of Lnc-IL7R and clinicopathological characters were analyzed by Pearson chi-square. Cox proportional hazards regression was used for univariate and multivariate analysis of OS according to Lnc-IL7R expression. P<0.05 was considered statistically significant. All experiments were performed at least three times.
Results
The expression of Lnc-IL7R is increased during the development of cervical cancer
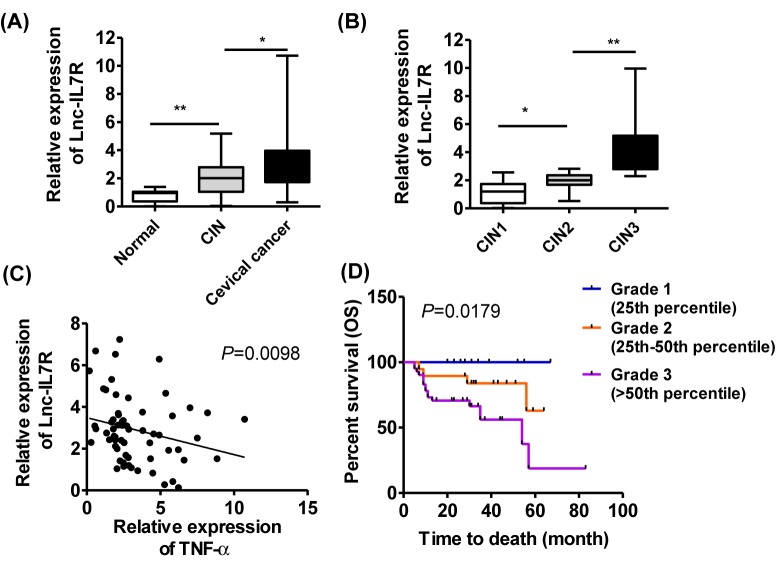
To investigate the expression pattern of Lnc-IL7R in cervical cancer, normal cervix (n=15), (CIN1/2/3) (n=35), and cervical cancer samples (n=70) were collected and the results of Q-PCR indicated that the expression of Lnc-IL7R was increased during the development of cervical cancer. The cancer tissues harbored the highest Lnc-IL7R level (Figure 1A). We further analyzed the expression of Lnc-IL7R in different groups, 12 in 15 (80%) of normal cervix showed the low expression of Lnc-IL7R and only 3 in 15 (20%) showed the high expression of Lnc-IL7R. But in the CIN samples, the high expression of Lnc-IL7R accounted for (57%) 20 in 35, which was further increased to (59%) 41 in 70 in cervical cancer (Table 1).
Figure 1. Up-regulated Lnc-IL7R predicts poor clinical outcome in cervical cancer.
(A,B) The expression of Lnc-IL7R in normal cervix (n=15), (CIN 1/2/3) (n=35), and cervical cancer samples (n=70) tissue samples were determined by Q-PCR. (C) The correlation between Lnc-IL7R and TNF-α was analyzed. (D) The OS time was analyzed by Kaplan–Meier survival curves according to the three degrees of Lnc-IL7R. *P<0.05, **P<0.01, data represent the means ± S.D.
Table 1. Lnc-IL7R expression in patients with cervical cancer.
| Types | n | Lnc-IL7R expression | χ2 | P | |
|---|---|---|---|---|---|
| Low (%) | High (%) | ||||
| Normal cervix | 15 | 12 (80.0) | 3 (20.0) | 7.672 | 0.02* |
| CIN | 35 | 15 (42.8) | 20 (57.2) | ||
| Cervical cancer | 70 | 29 (41.4) | 41 (58.6) | ||
*P<0.05, statistically significant, χ2 represents Pearson chi-square value.
Moreover, the Lnc-IL7R levels in different stages of CIN were determined. The results indicated the similar trend that high stage of CIN correlated to high Lnc-IL7R level in tissue samples (Figure 1B) and 83% CIN3 tissues harbor high expression of Lnc-IL7R (Table 2).
Table 2. The Lnc-IL7R expression in patients with CIN.
| CIN speicemen | n | Lnc-IL7R expression | χ2 | P | |
|---|---|---|---|---|---|
| Low (%) | High (%) | ||||
| CIN1 | 13 | 9 (69.2) | 4 (30.8) | 7.087 | 0.029* |
| CIN2 | 10 | 4 (40) | 6 (60) | ||
| CIN3 | 12 | 2 (16.7) | 10 (83.3) | ||
*P<0.05, statistically significant, χ2 represents Pearson chi-square value.
Because TNF-α as an antitumor factor is negatively regulated by Lnc-IL7R [14], thus, we investigated its correlation in cervical cancer. We found that higher Lnc-IL7R level corresponded to lower TNF-α level in tumor tissues (Figure 1C). The Lnc-IL7R-induced decreased expression of TNF-α might be conducive to the progression of cervical cancer.
Up-regulated expression of Lnc-IL7R correlates to poor clinical outcome in cervical cancer
Since the Lnc-IL7R increased with the progression of cervical cancer, we next estimated the correlation between the expression of Lnc-IL7R and the clinicopathological characteristics of cervical patients. As shown in Table 3, the expression of Lnc-IL7R had no association with age, histology, differentiation, vascular invasion, and the HPV status. But patients with higher tumor size, FIGO stage, and lymph node metastasis (LNM) have more high-expressed Lnc-IL7R. These data implicated that, in patients with cervical cancer, Lnc-IL7R could predict a poor clinical outcome including tumor size, FIGO, and LNM.
Table 3. Relationships between Lnc-IL7R expression and clinicalpathologic characteristics in cervical cancer patients.
| Characteristics | n (=70) | Lnc-IL7R expression | P | |
|---|---|---|---|---|
| Low (n/%) | High (n/%) | |||
| Age | ||||
| <50 | 31 | 14 (45.2) | 17 (54.8) | 0.572 |
| ≥50 | 39 | 15 (37.1) | 24 (62.9) | |
| Tumor size (cm) | ||||
| ≤4 | 45 | 23 (51.1) | 22 (48.9) | 0.027* |
| >4 | 25 | 6 (24) | 19 (76) | |
| Histology | ||||
| Squamous | 52 | 24 (46.2) | 28 (53.8) | 0.173 |
| Adenocarcinoma | 18 | 5 (27.8) | 13 (72.2) | |
| FIGO stage | ||||
| I–II | 39 | 21 (53.8) | 18 (46.2) | 0.018* |
| III–IV | 31 | 8 (25.8) | 23 (74.2) | |
| Differentiation | ||||
| Well | 26 | 12 (46.2) | 14 (53.8) | 0.537 |
| Moderate to poor | 44 | 17 (38.6) | 27 (61.4) | |
| LNM | ||||
| No | 49 | 25 (51) | 24 (49) | 0.013* |
| Yes | 21 | 4 (19) | 17 (81) | |
| Vascular invasion | ||||
| No | 46 | 20 (43.5) | 26 (56.5) | 0.63 |
| Yes | 24 | 9 (37.5) | 15 (62.5) | |
| HPV | ||||
| Negative | 30 | 14 (46.7) | 16 (43.3) | 0.441 |
| Positive | 40 | 15 (37.5) | 25 (62.5) | |
| Total | 70 | 29 (100) | 41 (100) | |
*P<0.05, statistically significant.
Lnc-IL7R is an independent factor for cervical cancer
The prognostic role of Lnc-IL7R was also investigated in the present study. The 70 patients were followed up until 1 October 2017. The OS time was analyzed according to the expression of Lnc-IL7R, which indicated that patients with highly expressed Lnc-IL7R had shorter OS than those with lowly expressed Lnc-IL7R (chi square = 5.605 by log-rank test) (Figure 1D).
Univariate and multivariate analyses were performed, the results revealed that age, histology, FIGO, LNM, vascular invasion, and differentiation were not independent prognostic indicators for OS, but the tumor size, the HPV status, and the expression of Lnc-IL7R were the independent prognostic factors for the OS of patients with cervical cancer (Table 4).
Table 4. Prognostic factors in the Cox proportional hazards model.
| Variables | OS | |||||
|---|---|---|---|---|---|---|
| HR | Univariate 95% CI | Sig. | HR | Multivariate 95% CI | Sig. | |
| Age | ||||||
| <50 compared with ≥50 | 1.125 | 0.827–2.491 | 0.736 | |||
| Tumor size (cm) | ||||||
| <4 compared with ≥4 | 2.435 | 1.730–3.440 | 0.008* | 3.837 | 2.312–6.384 | 0.0001* |
| Histology | ||||||
| Squamous compared with adenocarcinoma | 0.837 | 0.842–2.391 | 0.721 | |||
| FIGO stage | ||||||
| I–II compared with III–IV | 1.923 | 1.723–3.342 | 0.023* | 3.128 | 3.023–4.298 | 0.127 |
| Differentiation | ||||||
| Low compared with moderate-high | 1.182 | 0.728–1.942 | 0.732 | |||
| LNM | ||||||
| – compared with + | 2.442 | 1.401–3.894 | 0.013* | 4.104 | 1.202–3.390 | 0.073 |
| Vascular invasion | ||||||
| No compared with high | 1.879 | 1.237–2.823 | 0.026* | 2.923 | 1.949–5.127 | 0.533 |
| HPV | ||||||
| Negative compared with positive | 0.394 | 0.122–0.739 | 0.011* | 0.418 | 0.232–0.823 | 0.027* |
| Lnc-IL7R expression | ||||||
| Low compared with high | 3.392 | 1.750–5.979 | 0.008* | 5.826 | 2.213–7.129 | 0.0001* |
Abbreviations: CI, confidence interval; HR, hazard ratio; Sig, significance. *P<0.05, statistically significant.
Knockdown of Lnc-IL7R induces caspase-3 dependent apoptosis in vitro
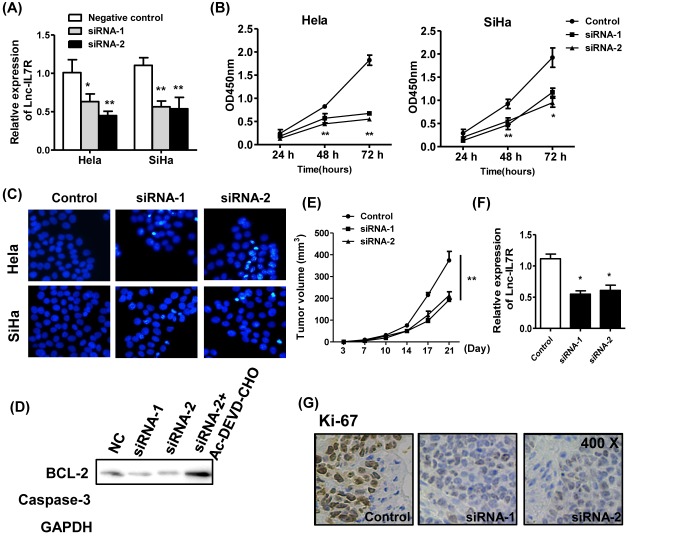
To provide the potential mechanism of the tumorigenic role of Lnc-IL7R, two cervical cancer cell lines Hela and SiHa were used. Knockdown of Lnc-IL7R by two independent siRNA in two cell lines effectively decreased the Lnc-IL7R level (Figure 2A). The cell vitalities were analyzed and found that knockdown of Lnc-IL7R significantly inhibited the cell vitalities of both the cell lines (Figure 2B). The apoptosis of two cell lines were also found to be induced by the siRNA- Lnc-IL7R (Figure 2C).
Figure 2. Knockdown of Lnc-IL7R inhibits the tumor growth of cervical cancer.
(A) The efficiency of knockdown in two cervical cancer cell lines Hela and SiHa was determined. (B,C) The cell vitalities and apoptosis of two cell lines were estimated by CCK-8 and Hoechst. (D) The expressions of BCL-2 and caspase-3 were assessed by western blot. (E) The mean tumor size (mm3) was analyzed. (F,G) The expressions of Lnc-IL7R and Ki-67 proliferation index were estimated by Q-PCR and immunohistochemistry. *P<0.05,**P<0.01.
In addition, the apoptosis-related molecules were determined by WB. The anti-apoptosis factor BCL-2 was down-regulated by the knockdown of Lnc-IL7R and the caspase-3 was also cleaved in response to the knockdown of Lnc-IL7R. This process could be restored by the treatment of caspase-3 inhibitor Ac-DEVD-CHO (25 μmol/l) (Figure 2D).
Lnc-IL7R inhibition in vivo restricts tumor growth
Given the tumorigenic role of Lnc-IL7R in vitro, we next estimated the efficiency of Lnc-IL7R knockdown for tumor progression by lentivirus vector of siRNA-Lnc-IL7R in vivo. The xenograft model of human HeLa was established. The results showed that the inhibition of Lnc-IL7R could effectively inhibit tumor growth (Figure 2E), which might be related to decreased expression of Lnc-IL7R (Figure 2F). The expression of tumor proliferation indication Ki-67 was also inhibited by knockdown of Lnc-IL7R in vivo (Figure 2G).
Discussion
In some low-income countries, cervical cancer is the most common cancer in women. Therefore, cervical cancer is a public health problem worldwide [5]. The treatment strategy for cervical cancer depends on the clinical stage, which is defined by the FIGO staging system. Traditional clinicopathological characteristics are not sufficiently reliable for predicting clinical outcomes or for guiding optimal treatment strategies [18]. In the present study, an inflammation-related LncRNA, Lnc-IL7R, was up-regulated with the initiation and development of cervical cancer, which positively correlated to the tumor size, FIGO stage, and LNM, and could predict the poor prognosis of patients with cervical cancer.
The non-coding RNAs (ncRNAs) participate in the post-transcription of gene expressions or interact with proteins to regulate the target mRNAs and proteins that were involved into carcinogenesis [19]. Amounting evidences identified the clinical diagnostic and prognostic role of ncRNAs. Using an established PCR-based miRNA assay to analyze 102 cervical cancer samples, Hu et al. [20] identified miR-200a and miR-9 that could predict poor patient survival and patients with high miR-200a and miR-9 had shorter survival time. Another study in small cell carcinoma of the cervix (SCCC) demonstrates that down-regulation of seven miRNAs (e.g. let-7c, miR-100, miR-125b) associated with advanced-stage SCCC patients (FIGO IB2-IV) compared with early-stage SCCC patients (FIGO IB1), six miRNAs with metastasis, and two with poor prognosis (e.g. miR-100, miR-125b) [21]. Yang et al. [22] reported that the expression of oncogene LncRNA-MALAT1 was significantly increased in cervical cancer than in normal tissues and correlated with tumor size, FIGO stage, vascular invasion, and LNM and is an independent predictor for OS of cervical cancer. LncRNA CCAT2 was also found to be up-regulated in cervical squamous cell cancer tissues, patients with high expression of lncRNA CCAT2 had poor OS and PFS rates and was an independent poor prognostic factor for cervical cancer patients [23]. In the present study, we found the clinical significance of Lnc-IL7R in cervical cancer, the expression of Lnc-IL7R was elevated in CIN tissues and cervical cancers, which predicted the poor clinical outcome of patients and could be an independent factor for cervical cancer. This finding suggested that the inflammation-related Lnc-IL7R functioned as an oncogene in cervical cancer.
The potential mechanisms of LncRNA in cervical cancer had been reported. Kim et al. [11] found that knockdown of HOTAIR in cervical cancer cell lines inhibited cell proliferation, migration, and invasion via the regulation of epithelial-to-mesenchymal transition (EMT)-related genes. We here found that knockdown of Lnc-IL7R significantly impaired the cell vitalities of two cervical cancer cell lines HeLa and SiHa. The apoptosis was also induced by the Lnc-IL7R inhibition by reduced expressions of BCL-2 and caspase-3, which could be restored by the caspase-3 inhibitor. CCHE1 overexpression promoted the proliferation of cervical cancer cell. RNA pull-down assays confirmed that CCHE1 physically associated with proliferating cell nuclear antigen (PCNA) to enhance the expression of PCNA [24]. The tumorigenic role of LncRNA in cervical cancer in vivo was limited. Using the siHOXA11-AS-transfected HeLa cells revealed that HOXA11-AS strongly induced tumor growth in xenograft experiments with the decreased cancer stemness and triggered the EMT program [25]. We found that the administration of siRNA for Lnc-IL7R could inhibit the tumor growth in vivo and decrease the expression of Ki67.
Conclusion
We reported that an oncogene Lnc-IL7R is increased during the development of cervical cancer, and could be an independent factor for the patients with cervical cancer. Knockdown of Lnc-IL7R inhibited the tumor growth in vivo, which could be a potential treatment for cervical cancer.
Abbreviations
- ARDS
acute respiratory distress syndrome
- CIN
cervical intraepithelial neoplasia
- DMEM
Dulbecco’s modified Eagle’s medium
- EMT
epithelial-to-mesenchymal transition
- FIGO
International Federation of Gynaecology and Obstetrics
- HPV
human papillomavirus
- LncRNA
long non-coding RNA
- LNM
lymph node metastasis
- OS
overall survival
- PCNA
proliferating cell nuclear antigen
- SCCC
small cell carcinoma of the cervix
Funding
The authors declare that there are no sources of funding to be acknowledged.
Author contribution
Y.F. and Y.N. were responsible for the protocol development and data collection or management. J.H. was responsible for the data analyses and manuscript writing/editing. H.Z. and W.Z. were responsible for the project development and manuscript editing.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Siegel R.L., Miller K.D. and Jemal A. (2017) Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Obel J., Souares Y., Hoy D. et al. (2014) A systematic review of cervical cancer incidence and mortality in the pacific region. Asian Pac. J. Cancer Prev. 15, 9433–9437 10.7314/APJCP.2014.15.21.9433 [DOI] [PubMed] [Google Scholar]
- 3.Tewari K.S. and Monk B.J. (2014) New strategies in advanced cervical cancer: from angiogenesis blockade to immunotherapy. Clin. Cancer Res. 20, 5349–5358 10.1158/1078-0432.CCR-14-1099 [DOI] [PubMed] [Google Scholar]
- 4.Dasari S., Wudayagiri R. and Valluru L. (2015) Cervical cancer: biomarkers for diagnosis and treatment. Clin. Chim. Acta 445, 7–11 10.1016/j.cca.2015.03.005 [DOI] [PubMed] [Google Scholar]
- 5.Maza M., Schocken C.M., Bergman K.L., Randall T.C. and Cremer M.L. (2017) Cervical precancer treatment in low- and middle-income countries: a technology overview. J. Glob. Oncol. 3, 400–408 10.1200/JGO.2016.003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayraktar R., Van Roosbroeck K. and Calin G.A. (2017) Cell to cell communication: microRNAs as hormones. Mol. Oncol. 10.1002/1878-0261.12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun W., Yang Y., Xu C. and Guo J. (2017) Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 216–217, 105–110 10.1016/j.cancergen.2017.06.003 [DOI] [PubMed] [Google Scholar]
- 8.Li J., Liu Q., Clark L.H., Qiu H., Bae-Jump V.L. and Zhou C. (2017) Deregulated miRNAs in human cervical cancer: functional importance and potential clinical use. Future Oncol. 13, 743–753 10.2217/fon-2016-0328 [DOI] [PubMed] [Google Scholar]
- 9.Peng L., Yuan X., Jiang B., Tang Z. and Li G.C. (2016) LncRNAs: key players and novel insights into cervical cancer. Tumour Biol. 37, 2779–2788 10.1007/s13277-015-4663-9 [DOI] [PubMed] [Google Scholar]
- 10.Zhou N., Fei D., Zong S., Zhang M. and Yue Y. (2016) MicroRNA-138 inhibits proliferation, migration and invasion through targeting hTERT in cervical cancer. Oncol. Lett. 12, 3633–3639 10.3892/ol.2016.5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H.J., Lee D.W., Yim G.W. et al. (2015) Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int. J. Oncol. 46, 521–530 10.3892/ijo.2014.2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao S., Liu W., Li F., Zhao W. and Qin C. (2014) Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int. J. Clin. Exp. Pathol. 7, 6776–6783 [PMC free article] [PubMed] [Google Scholar]
- 13.Cui H., Xie N., Tan Z. et al. (2014) The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur. J. Immunol. 44, 2085–2095 10.1002/eji.201344126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L., Ren J., Zhang D. et al. (2017) The TLR3 agonist inhibit drug efflux and sequentially consolidates low-dose cisplatin-based chemoimmunotherapy while reducing side effects. Mol. Cancer Ther. 16, 1068–1079 10.1158/1535-7163.MCT-16-0454 [DOI] [PubMed] [Google Scholar]
- 15.Wan B., Xu W.J., Xu W.N. et al. (2017) Plasma long noncoding RNA IL-7R as a prognostic biomarker for clinical outcomes in patients with acute respiratory distress syndrome. Clin. Respir. J. 12, 1607–1614, 10.1111/crj.12717 [DOI] [PubMed] [Google Scholar]
- 16.Bina P., Pahlevan Kakhki M., Sahraian M.A. and Behmanesh M. (2017) The expression of lnc-IL-7R long non-coding RNA dramatically correlated with soluble and membrane-bound isoforms of IL-7Ra gene in multiple sclerosis patients. Neurosci. Lett. 642, 174–178 10.1016/j.neulet.2017.01.068 [DOI] [PubMed] [Google Scholar]
- 17.Liang D., Xiao-Feng H., Guan-Jun D. et al. (2015) Activated STING enhances Tregs infiltration in the HPV-related carcinogenesis of tongue squamous cells via the c-jun/CCL22 signal. Biochim. Biophys. Acta 1852, 2494–2503 10.1016/j.bbadis.2015.08.011 [DOI] [PubMed] [Google Scholar]
- 18.Bast R.C. Jr, Lilja H., Urban N. et al. (2005) Translational crossroads for biomarkers. Clin. Cancer Res. 11, 6103–6108 10.1158/1078-0432.CCR-04-2213 [DOI] [PubMed] [Google Scholar]
- 19.Bergmann J.H. and Spector D.L. (2014) Long non-coding RNAs: modulators of nuclear structure and function. Curr. Opin. Cell Biol. 26, 10–18 10.1016/j.ceb.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu X., Schwarz J.K., Lewis J.S. Jr et al. (2010) A microRNA expression signature for cervical cancer prognosis. Cancer Res. 70, 1441–1448 10.1158/0008-5472.CAN-09-3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang L., Lin J.X., Yu Y.H., Zhang M.Y., Wang H.Y. and Zheng M. (2012) Downregulation of six microRNAs is associated with advanced stage, lymph node metastasis and poor prognosis in small cell carcinoma of the cervix. PLoS ONE 7, e33762 10.1371/journal.pone.0033762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L., Bai H.S., Deng Y. and Fan L. (2015) High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur. Rev. Med. Pharmacol. Sci. 19, 3187–3193 [PubMed] [Google Scholar]
- 23.Chen X., Liu L. and Zhu W. (2015) Up-regulation of long non-coding RNA CCAT2 correlates with tumor metastasis and poor prognosis in cervical squamous cell cancer patients. Int. J. Clin. Exp. Pathol. 8, 13261–13266 [PMC free article] [PubMed] [Google Scholar]
- 24.Yang M., Zhai X., Xia B., Wang Y. and Lou G. (2015) Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumour Biol. 36, 7615–7622 10.1007/s13277-015-3465-4 [DOI] [PubMed] [Google Scholar]
- 25.Kim H.J., Eoh K.J., Kim L.K. et al. (2016) The long noncoding RNA HOXA11 antisense induces tumor progression and stemness maintenance in cervical cancer. Oncotarget 7, 83001–83016 10.18632/oncotarget.12863 [DOI] [PMC free article] [PubMed] [Google Scholar]