Abstract
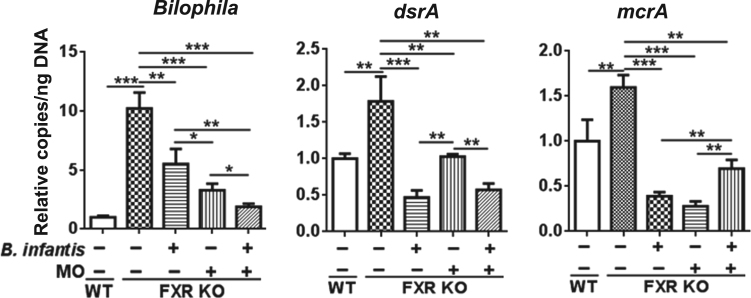
Probiotic Bifidobacterium longum subspecies infantis (Bifidobacterium infantis) consumes human milk oligosaccharides (MO) and protects intestinal permeability thereby having anti-inflammatory effects (Underwood et al., 2015; Bode, 2006; Asakuma et al., 2011) [1–3]. Via the gut-liver axis, gut barrier disruption and dysbiosis lead to hepatic inflammation (Sheng et al., 2017; Jena et al., 2017) [4,5,6]. Our published data revealed that butyrate, as well as synbiotics of B. infantis in combination with MO, had protective effects against cancer-prone non-alcoholic steatohepatitis (NASH) mouse models, i.e., Western diet (WD)-fed bile acid receptor FXR (farnesoid x receptor) knockout (KO) mice (Jena et al., 2018) [6,7]. In addition, MO was particularly effective in increasing the blooming of butyrate-generating bacteria (Jena et al., 2018) [7]. In the present study, we further showed that the reduced ileal short chain fatty acid (SCFA) signaling found in WD-fed FXR KO mice could be reversed by B. infantis and/or MO treatment. Moreover, ileal mRNA levels of SCFA receptors i.e. Gpr41 (Ffar3), Gpr109 (Hcar2), and Gpr43 (Ffar2) were increased in B. infantis and/or MO-treated mice suggesting increased SCFA signaling (Fig. 1). Further, nuclear magnetic resonance (NMR) data revealed that MO and B. Infantis plus MO increased intestinal acetate, propionate, butyrate, and valerate levels (Fig. 2). In addition, B. infantis and/or MO reduced the abundance of genus Bilophila and the relative copy number of bacterial genes including dissimilatory sulfite reductase (dsrA) and methyl coenzyme M reductase A (mcrA), which were all increased in cancer-prone FXR KO mice (Fig. 3).
Keywords: Probiotics, Western diet, Short chain fatty acid, Inflammation
Specifications Table
| Subject area | Biology, Microbiology, Molecular Biology |
| More specific subject area | Synbiotics |
| Type of data | Table, figure, graph |
| How data was acquired | Real time PCR, NMR |
| Data format | Analyzed |
| Experimental factors | C57BL/6 wild type and FXR KO male mice were fed with Western diet after weaning. When mice were 3-month old, they were given B. infantis (109cfu per mouse, orally, once a week) in saline, bovine MO (7% in a diet that lacks 7% cellulose), or a combination of B. infantis plus MO while mice continued a WD for 7 months and euthanized when they were 10-month old. |
| Experimental features | Gene expression in ileum and SCFA concentration in cecal content were measured by real time PCR and NMR, respectively. |
| Data source location | Sacramento, California, United States of America |
| Data accessibility | Data with this article |
Value of the data
-
–
In our earlier report, B. infantis and MO treatment reduced hepatic and ileal inflammation [7]. It has been shown that B. infantis and MO protects from gut barrier function and inflammation [1], [2], [3]. In addition, the abundance of bacterial butyrate-generating genes bcoA (butyryl-CoA: acetate CoA-transferase) and buk (butyrate kinase) in the cecum was increased with MO and B. infantis plus MO treatment. Data in this study described the mRNA expression of SCFA receptors regulated by B. infantis and MO treatment. Our data showed that increased SCFA signaling with B. infantis and MO treatment was associated with reduced inflammation.
-
–
In our earlier report, the concentration of butyrate was reduced in FXR KO mice [6]. Data in this study described that B. infantis and MO increased SCFAs, i.e., acetate, propionate, butyrate, and valerate. Additionally, such increases were accompanied by reduced inflammation in FXR KO mice. It has been shown that dysregulated bile acid and gut dysbiosis causes hepatic inflammation [4], [5], [6], [7]. Moreover, the abundance of genus Bilophila and dsrA gene, which are involved in the generation of hydrogen sulfide, was increased by FXR inactivation and B. infantis and MO treatment reversed those changes.
-
–
These data suggested that the anti-inflammatory effect of B. infantis and MO may due to increased SCFAs and reduced hydrogen sulfide and methane.
1. Data
The data showed the mRNA level of ileal SCFA receptors i.e. Gpr41, Gpr109a, and Gpr43 in WD-fed wild type (WT) mice and WD-fed FXR KO mice supplemented with or without Bifidobacterium infantis and/or MO (Fig. 1). NMR data generated from cecal content showed the concentration of acetate, propionate, butyrate, and valerate (Fig. 2). Moreover, bacteria and their functional genes were determined by real-time quantitative PCR (Fig. 3).
Fig. 1.
The effects of B. infantis and/or MO on SCFA receptor signaling. Ileal mRNA level of indicated genes in WD-fed WT mice and WD-fed FXR KO mice supplemented with and without B. infantis and/or MO for 7 months. Data expressed as mean ± SD. n ≥ 6 per group. *p< 0.05, **p< 0.01. WT mice compared with FXR KO mice, and untreated FXR KO mice compared with treated FXR KO mice on the same diet.
Fig. 2.
NMR analysis of SCFA concentration in cecal content of WD-fed WT mice and WD-fed FXR KO mice treated with and without B. infantis and/or MO. Data expressed as mean±SD. n ≥ 6 per group. *p< 0.05. WT mice compared with FXR KO mice, and untreated FXR KO mice compared with treated FXR KO mice.
Fig. 3.
Targeted functional quantitative PCR analyses of microbial genes in WD-fed WT mice and WD-fed FXR KO mice supplemented with and without B. infantis and/or MO. Data expressed as mean±SD. n ≥ 6 per group. *p< 0.05, **p< 0.01, ***p< 0.001, WT mice compared with FXR KO mice, and untreated FXR KO mice compared with treated FXR KO mice.
2. Experimental design
2.1. Mice
Specific pathogen-free male C57BL/6 WT mice (Jackson Laboratory, Sacramento, CA, USA) and FXR KO mice were housed in steel microisolator cages at 22 °C with a 12-h light/dark cycle. They were given a WD containing 21.2% fat, 34% sucrose, and 0.2% cholesterol (Envigo, Indianapolis, IN, USA) right after weaning (3 weeks, 6–10 mice per group). When mice were 3-month old, they were given B. infantis (109 cfu per mouse, orally, once a week) in saline, bovine MO (7% in a diet that lacks 7% cellulose), or a combination of B. infantis plus MO while mice continued a WD until they were 10-month old. Experiments were conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals under protocols approved by the Institutional Animal Care and Use Committee of the University of California, Davis.
2.2. Gene expression and quantification of bacterial genes
RNA was isolated from mouse tissues using TRIzol (Invitrogen, Carlsbad, CA, USA) and reverse transcribed into cDNA. qRT-PCR was performed on an ABI 7900HT Fast real-time PCR system using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The mRNA levels were normalized to the level of Gapdh mRNA. The primer sets used are described in Table 1.
Table 1.
Primers used for qPCR.
| Gene | Forward (5’ – 3’) | Reverse (5’ – 3’) | |
|---|---|---|---|
| qPCR primers used to quantify gene expression in mice | |||
| Gpr41 | GTGACCATGGGGACAAGCTTC | CCCTGGCTGTAGGTTGCATT | |
| Gpr109a | ATGGCGAGGCATATCTGTGTAGCA | TCCTGCCTGAGCAGAACAAGATGA | |
| Gpr43 | GGCTTCTACAGCAGCATCTA | AAGCACACCAGGAAATTAAG | |
| qPCR primers used to quantify bacterial genes | |||
| Bilophila (tpA) | CGGTATCGAAATCGTGAAGG | CAGAGGGTCAGGGTGTTGTT | |
| dsrA | GCCGTTACTGTGACCAGCC | GGTGGAGCCGTGCATGTT | |
| mcrA | TTCGGTGGATCDCARAGRGC | GBARGTCGWAWCCGTAGAATCC | |
For bacterial gene quantification, cecal (0.05 g) DNA was extracted using ZR Fecal DNA MiniPrep Kit (Zymo Research, Irvine, CA, USA), quantified by NanoDrop (Thermo Scientific, Wilmington, DE, USA), and amplified using primers listed in Table 1. A dissociation step was included to analyze the melting profile of amplified products. In parallel, qPCR was performed using ten-fold serial diluted synthetic DNA fragments (Integrative DNA technologies, Redwood city, CA, USA) of a bacterial gene with known concentrations. Bacterial DNA concentration was calculated using standard curves of diluted synthetic DNA fragment based on method described elsewhere [8].
2.3. 1H NMR spectroscopy for metabolic profiling
Cecal samples (50−60 mg) were prepared and analyzed by 1H NMR based on published method [6], [9]. Each cecal sample was homogenized and mixed with sodium-potassium phosphate buffer and centrifuged at 14,000g for 20 min. All 1H NMR spectra were collected using a Bruker advance 600 NMR spectrometer (Bruker; Rheinstetten, Germany). The samples were pre-cooled to 277 K before being loaded into the magnet, and were warmed to 298 K and equilibrated for 5 min before data acquisition. For each sample, 64 scans were collected into 64 K data points over a spectral width of 20 ppm with a relaxation delay of 2 s. Identification and quantification of metabolites were accomplished using Chenomx NMR Suite 7.6 (Chenomx, Inc., Edmonton, Alberta, Canada).
2.4. Statistical analysis
Data are presented as the mean ± SD. Statistical analysis was performed by unpaired Student׳s t-test or one-way analysis of variance by using GraphPad Prism 6.0 (GraphPad, La Jolla, CA, USA). P < 0.05 was considered statistically significant.
Acknowledgements
This study is supported by grants funded by National Institutes of Health U01CA179582 and R01CA 222490.
Footnotes
Transparency document associated with this article can be found in the online version at 10.1016/j.dib.2018.05.127.
Transparency document. Supporting information
Supporting information
References
- 1.Underwood M.A., German J.B., Lebrilla C.B., Mills D.A. Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr. Res. 2015;77:229–235. doi: 10.1038/pr.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bode L. Recent advances on structure, metabolism, and function of human milk oligosaccharides. J. Nutr. 2006;136:2127–2130. doi: 10.1093/jn/136.8.2127. [DOI] [PubMed] [Google Scholar]
- 3.Asakuma S., Hatakeyama E., Urashima T., Yoshida E., Katayama T., Yamamoto K. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 2011;286:34583–34592. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheng L., Jena P.K., Liu H.X., Kalanetra K.M., Gonzalez F.J., French S.W. Gender differences in bile acids and microbiota in relationship with gender dissimilarity in steatosis induced by diet and FXR inactivation. Sci. Rep. 2017;7:1748. doi: 10.1038/s41598-017-01576-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jena P.K., Sheng L., Liu H.-X., Kalanetra K.M., Mirsoian A., Murphy W.J. Western diet–induced dysbiosis in farnesoid X receptor knockout mice causes persistent hepatic inflammation after antibiotic treatment. Am. J. Pathol. 2017;187:1800–1813. doi: 10.1016/j.ajpath.2017.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng LJ P.K., Hu Y., Liu H.X., Nagar N., Kalanetra K.M., French S.W., French S.W., Mills D.A., Wan Y.Y. Hepatic inflammation caused by dysregulated bile acid synthesis is reversible by butyrate supplementation. J. Pathol. 2017;243:431–441. doi: 10.1002/path.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jena P.K., Sheng L., Nagar N., Wu C., Barile D., Mills D.A., Wan Y., Y. Synbiotics Bifidobacterium infantis and milk oligosaccharides are effective in reversing cancer-prone non-alcoholic steatohepatitis using western diet-fed FXR knockout mouse models. J. Nutr. Biochem. 2018 doi: 10.1016/j.jnutbio.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ou J., Carbonero F., Zoetendal E.G., DeLany J.P., Wang M., Newton K. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 2013;98:111–120. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian Y., Zhang L., Wang Y., Tang H. Age-related topographical metabolic signatures for the rat gastrointestinal contents. J. Proteom. Res. 2012;11:1397–1411. doi: 10.1021/pr2011507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information