Abstract
Interleukin-10 (IL-10) polymorphisms have been shown to affect IL-10 production. This study investigated the influences of IL-10 polymorphisms on the susceptibility to chronic periodontitis (CP) and aggressive periodontitis (AP), and their possible role in the quantity of subgingival bacteria Aggregatibacter Actinomycetemcomitans and Porphyromonas gingivalis. 92 CP patients, 83 AP patients and 91 periodontal healthy controls were recruited. Serum IL-10 concentration was analyzed by enzyme-linked immunosorbent assay (ELISA). Gene polymorphisms were determined by multiplex SNaPshot technique. Bacteria were quantified by real-time polymerase chain reaction with TaqMan MGB probes. Taking into account age, gender and periodontal status, IL-10-592 AA, -819 TT and ATA/ATA genotype occurred more frequently in patients with CP than in healthy controls. In CP cases, higher quantity of subgingival A. actinomycetemcomitans and lower serum IL-10 levels could be detected in homozygous ATA/ATA carriers. These findings indicate that variants in IL-10 promoter gene were not only associated with predisposition to chronic periodontitis but also affected the subgingival number of A. Actinomycetemcomitans in a Chinese Han population.
Introduction
Periodontitis, as a multifactorial chronic inflammatory disease, affects an extremely large percentage of the human population and leads to tooth loss. Generally it emerges either with the chronic or the aggressive form1. Periodontitis is predominantly a bacterial infection, initiated by microorganisms which could cause the defense of host immune system in the subgingival plaque, and it is also influenced by both genetic and acquired risk factors2–4. In particular, growing evidence suggests that individual genetic factors may have influence on the host’s immune response to pathogens5. Reports have focused on the relationship between genetic variations and microbial colonization6,7. There are several factors contribute to the differences in subgingival microbial colonization among persons. The reason, to some extent, is that the genetic variants play an important role in deciding which species are able to colonize the host6. Nibali et al. elucidated part of the mechanism of periodontal infect genomics as follows: once bacteria have colonized the periodontal tissues, in susceptible individuals they can not only proliferate but also trigger the immune-pathological reactions which could lead to tissue destruction. Furthermore, increased inflammatory response to plaque accumulation in subjects carrying specific genes may enhance the chance of overgrowth of particular components of the opportunistic microbiota (such as A. actinomycetemcomitans)7. Understanding of the molecular basis for these various responses could improve our knowledge of pathogenesis of infectious diseases such as periodontitis.
Cytokines act as messengers to initiate, mediate and control immune and inflammatory responses8,9. It’s well known that the interplay of pro- and anti- inflammatory cytokines plays a crucial role in the progression of periodontitis10,11. Cytokine gene polymorphisms often affect cytokine expression profile, and then may regulate susceptibility to infection in part by affect the colonization of the periodontopathic bacteria12–22.
Interleukin-10 (IL-10) is the most potent anti-inflammatory cytokine as it induces T cell anergy and downregulates the production of some pro-inflammatory cytokines23,24. IL-10 promoter region is highly polymorphic, and three single nucleotide polymorphisms (SNPs) at positions -1082(rs1800896), -819(rs1800871), and -592 (rs1800872) have been reported to be tightly correlated with altered IL-10 production in vitro25–27. The three SNPs are in linkage disequilibrium, resulting in three preference haplotypes: ATA, ACC and GCC. IL-10 promoter haplotype ATA has been associated with low production of IL-10, while GCC and ACC have been identified as risk factors for high and intermediate IL-10 production respectively25,28. AndIL-10 polymorphisms may modulate the host’s risk for periodontitis through various IL-10 levels. This mechanism could be proved by the evidence that the mice lacking IL-10 gene were highly susceptible to alveolar bone loss in comparison with the normal controls29,30, which could attribute to the attenuation of anti-inflammatory and increase of pro-inflammatory mediators. To date, just a limited number of studies have been performed to assess the association between the above SNPs and either chronic periodontitis (CP) or aggressive periodontitis (AP)21,31–38, however, no clear consensus has been reached. Two meta-analysis taking ethnicity and sample size into consideration suggest that −819 and −592 polymorphisms seem to be genetic risk factors for CP39,40.
On the basis of these clinical data, IL-10 polymorphisms may influence the composition of the subgingival bacteria by regulating IL-10 levels, and then be responsible for susceptibility to periodontitis. The current clinical study was performed to investigate the distribution of IL-10 polymorphisms in Chinese patients suffering from CP and AP compared with the healthy controls using the multiple logistic regression analysis. A further aim was to examine whether the IL-10 genetic variants could influence the subgingival bacterial counts of two key periodontal pathogens Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis.
Subjects and Methods
Study population and clinical investigations
This study was approved by the Ethical Committee of Stomatological Hospital affiliated to Nanjing Medical University, Nanjing, China. The purposes and procedures of the study were explained and informed consents were obtained from all recruits. The study was also performed in accordance with the declaration of Helsinki.
Patients and controls were recruited from the Stomatological Hospital affiliated to Nanjing Medical University. All subjects were Han Chinese population. Inclusion criteria comprised partially or fully dentate patients (at least 14 natural teeth, including 10 posterior teeth, excluding third molars), systemically healthy with no evidence of known systemic modifiers of periodontitis such as rheumatoid arthritis, diabetes, and osteoporosis. Subjects who met the following criteria were excluded from the study: (1) systemic modifiers of periodontal disease, as described above; (2) current pregnancy or lactation; (3) administration of antibiotics or anti-inflammatory drugs in the past six months; (4) current and former smokers (a person who smoked at least one cigarette per day was considered as a smoker); (5) received periodontal therapy in the past six months.
After collection of medical and dental histories, clinical parameters were recorded. Probing depth (PD), clinical attachment loss (CAL) and bleeding on probing (BOP) were examined at six sites per tooth with Florida probe (Florida Probe Corporation, Gainesville, Florida, USA). All assessments were measured by only one experienced clinician; a randomly chosen sample of 53(20%) subjects was re-measured by the same examiner one day later in order to establish the intra-examiner variance. The intra-examiner reproducibility for PD and CAL was assessed by kappa statistic, and the score of kappa was 0.89 and 0.87 respectively.
Patients were clinically diagnosed as generalized CP or generalized AP in accordance with the criteria established in 1999 at the World Workshop for a classification of Periodontal Diseases and Conditions, which was based on clinical, radiographic and historical findings1. CP patients were selected if they showed an attachment loss in at least 30% of the teeth with a minimum PD of 4 mm and lesions distributed on more than two teeth in each quadrant, CP often occurs in adults (but it may also affect younger patients), in addition, periodontal destruction is consistent with the amount of plaque present and other local factors. AP patients were included if they meet the following criteria: the disease occurred often before the age of 35 years (but it may also affect older people); history of rapid attachment loss and bone destruction as well as familial aggregation (if available); the presence of more than eight teeth with CAL ≥ 5 mm and PD ≥ 6 mm, and more than three affected teeth that were not first molars or incisors; the amounts of microbial deposits or subgingival calculus less than what would be expected for the amounts of periodontal destruction. No case that produced doubt in classification was included in the study. Periodontal healthy controls (PH) were included if they did not show any attachment loss, PD ≤ 3 mm and no history of periodontal disease. It is worth mentioning that CAL ≥ 3 mm as a result of traumatic tooth brushing, overhanging dental fillings, etc., was not considered as a case of periodontitis.
Microbiological assessment of subgingival plaque samples
The subgingival plaque samples were obtained from the deepest site (excluding teeth with hopeless prognosis) in each quadrant before subgingival scaling and root planning were done, in addition, the CAL and PD of sampled sites were recorded, that is, the average of data in deepest site of each quadrant. The subgingival plaque samples were collected by inserting a sterile paper points for 30 s into the pocket after carefully removing the supragingival plaque, and then the samples were pooled from 4 sites per individual. Preparation of bacterial DNA was carried out using the QIAamp DNA Mini kit (Qiagen, Germany) according to the manufacturer’s instructions.
A real-time PCR assay was applied to achieve quantification of the pathogens, and the methods were described previously41,42. The oligonucleotide primers and probes, designed by Primer Express (version 2.0) software, are listed in Table 1. The primers and probes were based on A. actinomycetemcomitans- and P. gingivalis- specific conserved regions from lktA43 and 16S rRNA genes, respectively. Identification of conserved regions was done by multiple sequence alignment with ClustalW software based on the published sequences. Primers and probes were checked for possible cross-hybridization with bacterial genes using the database similarity search program BLAST. The fluorescent TaqManMGB probes were dually labeled with a reporter dye FAM attached to the 5′ end and a quencher dye MGB attached to the 3′ end. The primers used in real-time PCR were also used in conventional PCR, which were performed to confirm the specificities of the primers as well as to obtain the species-specific gene products. Consequently, the primers for A. actinomycetemcomitans and P. gingivalis demonstrated specific amplification products of each bacterial species and did not amplify DNA from other species containing P. intermedia and T. forsythia.
Table 1.
Oligonucleotide primers and probes for real-time PCR.
| Primers and probes | Sequence (5′–3′) | Production (bp) | Target |
|---|---|---|---|
| A. actinomycetemcomins | |||
| Forward | TTGATCGTGCGAGAATGCTT | ||
| Reverse | ATCGCCGTTATAACCAAATTTCTT | ||
| Probe | FAM-AGGAATACTCGAAACGC-MGB | 65 | lktA |
| P. gingivalis | |||
| Forward | TACCCATCGTCGCCTTGGT | ||
| Reverse | CGGACTAAAACCGCATACACTTG | ||
| Probe | FAM-ATTTATAGCTGTAAGATAGGC-MGB | 126 | 16S rRNA |
Quantitative PCR was carried out in duplicates in ABI PRISM 7300 sequence detection system (Applied Biosystems, USA) with the following cycle profile: 95 °C for 30 s followed by 40 cycles of 95 °C for 5 s and 60 °C for 31 s.
Serum IL-10 level estimation
Venous blood samples were centrifuged at 1500 rpm for 10 min, and serum was then collected and kept in −70 °C conditions until tested. The level of IL-10 in the serum was measured by using ELISA kits (Invitrogen, USA) according to manufacturer’s instructions. The IL-10 level was obtained by comparison with the standard curve prepared. The sensitivity for IL-10 ELISA’s was 1 pg/mL, IL-10 level below the limit of the assay’s detectability was scored as 0.
Genetic studies
For genetic investigations, fresh blood samples were collected in ethylenediaminetetraacetic acid (EDTA)–treated tubes and stored at −70 °C. Preparation of genomic DNA was carried out using a QIAamp DNA blood Mini kit (Qiagen, Germany) in accordance with the manufacturer’s manual.
Genomic regions containing the IL-10-592, -819 and -1082 SNPs were amplified by PCR using the following primers: -592, 5′-AAGAGGTGGAAACATGTGCC-3′ (forward) and 5′-TACCCAAGACTTCTCCTTGC-3′ (reverse); -819, 5′-ATGGTGTACAGTAGGGTGAG-3′ (forward) and 5′-TTTCCACCTCTTCAGCTGTC-3′ (reverse); -1082, 5′-AGAAGTCCTGATGTCACTGC-3′ (forward) and 5′-AAGTCAGGATTCCATGGAGG-3′ (reverse). The investigated SNPs were genotyped by a single-base primer extension assay using the SNaPshot™ Multiplex kit (Applied Biosystems, USA), according to the manufacturer’s instructions. The following primers were used: -592, 5′-TTTTTTCACATCCTGTGACCCCGCCTGT-3′; -819, 5′-TTACCCTTGTACAGGTGATGTAA-3′; -1082, 5′-CACTACTAAGGCTTCTTTGGGA-3′ (Table 1).
Statistical Analysis
The SPSS 19.0 package was used for statistical analysis, values of p < 0.05 were considered significant. Continuous, normally distributed variables were reported as means ± standard deviations (SD). The genotype distributions in all groups were found to be in Hardy–Weinberg equilibrium. Association between IL-10 polymorphisms and CP/AP were analyzed by multiple logistic regression analysis. Age, gender, BOP, PD and CAL were entered in the analyses as covariates. We used Kruskal-Wallis ANOVA or a Mann-Whitney U test to analyze the impact of genetic variants on the number of subgingival bacteria and IL-10 serum levels, and a Dunn-Bonferroni test for post hoc comparisons (for values were not distributed normally), as appropriate, and by multiple linear regression analysis adjusted for age, gender, BOP, PD and CAL.
Results
Patient demographics and clinical characteristics
Demographic information and periodontal parameters of participants and sampled sites were listed in Table 2.
Table 2.
Demographic and clinical characteristics of three groups.
| Variable | Chronic periodontitis (n = 92) | Aggressive periodontitis (n = 83) | Periodontal healthy controls (n = 91) |
|---|---|---|---|
| Average age at diagnosis (years) | 42 ± 7.8 | 28 ± 6.1 | 45 ± 9.8 |
| Female (%) | 56.6 | 41.3 | 52.8 |
| Early tooth loss due to periodontitis among relatives (%) | 41.3* | 50.6* | 17.6 |
| No. of lost teeth (n) | 2.0 ± 1.4* | 0.9 ± 1.4* | 0.1 ± 0.3 |
| Bleeding on probing (BOP, %) | 78.4 ± 9.0* | 81.2 ± 12.0* | 12.1 ± 6.3 |
| Probing depth (PD, mm) | 5.7 ± 0.9* | 6.0 ± 0.9* | 1.7 ± 0.2 |
| Clinical attachment loss (CAL, mm) | 6.4 ± 1.0* | 6.7 ± 0.9* | 0.2 ± 0.3 |
| Teeth with CAL ≥ 6 mm (%) | 60.3* | 57.6* | 0 |
| PD (sampled site, mm) | 6.8 ± 0.5* | 7.2 ± 0.5* | 1.9 ± 0.4 |
| CAL (sampled site, mm) | 7.0 ± 0.5* | 7.3 ± 0.6* | 0 |
*There were significant differences between chronic periodontitis or aggressive periodontitis and periodontal healthy controls (P < 0.05), but there was no significant difference between chronic periodontitis and aggressive periodontitis.
In comparison with the PH group, no statistically significant difference in gender could be detected. In accordance with the inclusion criteria, the AP patients are younger than CP and PH individuals, but there were no significant differences. Among their relatives, both patient groups reported significantly more often early tooth loss as a consequence of periodontitis (p < 0.001). Mean values of periodontal status, such as BOP (%), PD (mm) and CAL (mm) in patient groups were significantly higher than those in PH group. Severe generalized attachment loss for both patient groups was indicated via the results that the mean values for both PD (mm) and CAL (mm) were ≥ 5 mm, and the mean percentage of teeth with CAL ≥ 6 mm was more than 50%.
Quantitative Detection of Periodontal pathogens
Sensitivity of the real-time PCR assay was evaluated using 100 to 108 plasmid copies of each pathogen. Limit for minimum detection was 102 cells, and the sample was recognized as negative if its initial target genes were less than 102.
When the bacterial counts were compared, there was a great variability between subjects (Fig. 1). Evaluated from the mean value, no significant difference was detected in the number of A. actinomycetemcomitans and P. gingivalis between CP and AP groups. The difference of the two bacterial load was analyzed within each group, and the amount of P. gingivalis was much higher in comparison with that of A. actinomycetemcomitans (p < 0.001). Further, periodontal patients showed a higher number of the tested pathogens, compared to healthy controls (p < 0.001).
Figure 1.
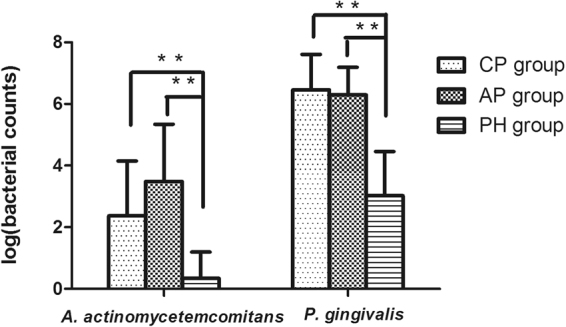
Bacterial counts (through logarithmic transformation) of A. actinomycetemcomitans and P. gingivalis. No significance (NS) for both pathogens by Mann-Whitney U test between CP and AP groups. Patients from CP and AP groups showed significant higher number of both pathogens compared with PH controls (P < 0.001 by Mann-Whitney U test). **Significant (p < 0.001 by Mann-Whitney U test) PH, periodontal healthy; CP, chronic periodontitis; AP, aggressive periodontitis.
Allele and genotype distribution of three IL-10 SNPs
Association between IL-10 polymorphisms and CP/AP were analyzed by multiple logistic regression analysis. Age, gender, BOP, PD and CAL were entered in the analysis as covariates.
The A allele and AA genotype at position -592 (p = 0.03, OR = 2.32, 95%CI = 1.45–3.41; p = 0.023, OR = 4.23, 95%CI = 1.26–6.78), and the T allele and TT genotype at position -819 (p = 0.034, OR = 1.88, 95%CI = 1.32–2.59; p = 0.021, OR = 3.59, 95%CI = 1.45–7.06) occurred more frequently in patients with CP than in healthy controls. Related with AP, no association between IL-10 polymorphisms and AP was found (Table 3). According to the haplotype analysis, the dominant haplotype was ATA (-1082-819-592) in three groups (57.7% in PH group, 70.1% in CP group, and 64.5% in AP group). There was a trend that the haplotype ATA was expressed more in patients with CP (p = 0.076, OR = 1.72, 95%CI = 0.93–2.65) (Table 4). In patients with CP, the frequency of the combination ATA/ATA was increased compared with healthy controls (p = 0.017, OR = 2.43, 95%CI = 1.21–3.83) (Table 4).
Table 3.
Genotype and allele frequencies of SNPs in IL-10 promoter gene of three groups and results of logistic regression analyses.
| Single nucleotide | PH | CP | AP | CP vs.PH | AP vs.PH | ||
|---|---|---|---|---|---|---|---|
| polymorphism | n (%) | n (%) | n (%) | p | OR (95%CI) | p | OR (95%CI) |
| IL-10-592 Genotype | n = 91 | n = 92 | n = 83 | ||||
| CC | 17 (18.7) | 7 (7.6) | 10 (12) | 1 | 1 | ||
| AC | 42 (46.2) | 39 (42.4) | 36 (43.4) | 0.146 | 2.45 (0.93–5.44) | 0.341 | 1.69 (0.44–3.52) |
| AA | 32 (35.2) | 46 (50) | 37 (44.6) | 0.023 | 4.23 (1.26–6.78) | 0.163 | 2.14 (0.71–5.63) |
| A carrier | 106 (58.2) | 131 (71.2) | 110 (66.3) | 0.03 | 2.32 (1.45–3.41) | 0.198 | 1.12 (0.73–1.85) |
| IL-10–819 Genotype | n = 91 | n = 92 | n = 83 | ||||
| CC | 17 (18.7) | 7 (7.6) | 10 (12) | 1 | 1 | ||
| TC | 39 (42.9) | 40 (43.5) | 38 (45.8) | 0.205 | 1.25 (0.67–2.34) | 0.361 | 1.52 (0.47–2.65) |
| TT | 35 (38.5) | 45 (48.9) | 35 (42.2) | 0.021 | 3.59 (1.45–7.06) | 0.322 | 1.96 (0.45–5.67) |
| T carrier | 109 (59.9) | 130 (70.7) | 108 (65.1) | 0.034 | 1.88 (1.32–2.59) | 0.184 | 1.36 (0.79–2.13) |
| IL-10-1082 Genotype | n = 91 | n = 92 | n = 83 | ||||
| GG | 2 (2.2) | 1 (1.1) | 1 (1.2) | 1 | 1 | ||
| AG | 16 (17.6) | 9 (9.8) | 10 (12.0) | 0.879 | 1.02 (0.83–11.79) | 0.933 | 1.12 (0.57–12.47) |
| AA | 73 (80.2) | 82 (89.1) | 72 (86.7) | 0.654 | 2.67 (0.65–24.29) | 0.704 | 2.87 (0.2–24.24) |
| A carrier | 162 (89.0) | 173 (94.0) | 154 (92.8) | 0.584 | 2.31 (0.75–7.63) | 0.653 | 1.92 (0.55–5.28) |
PH, periodontal healthy subjects; CP, chronic periodontitis subjects; AP, aggressive periodontitis subjects.
Table 4.
Distribution of IL-10 haplotypes (arranged as allele frequencies) and IL-10 combinations (arranged as genotype frequencies) of three groups and results of logistic regression analyses.
| Haplotype | PH (n = 182) | CP (n = 184) | AP (n = 166) | CP vs.PH | AP vs.PH | ||
|---|---|---|---|---|---|---|---|
| -1082-819-592 | n (%) | n (%) | n (%) | p | OR (95%CI) | p | OR (95%CI) |
| ATA | 105 (57.7) | 129 (70.1) | 107 (64.5) | 0.076 | 1.72 (0.93–2.65) | 0.105 | 1.27 (0.78–2.31) |
| Others | 77 (42.3) | 55 (29.9) | 59 (35.5) | ||||
| ACC | 52 (28.6) | 41 (22.3) | 43 (25.9) | 0.745 | 0.84 (0.57–1.42) | 0.886 | 1.04 (0.58–1.80) |
| Others | 130 (71.4) | 143 (77.7) | 123 (74.1) | ||||
| Genotype | PH (n = 91) | CP (n = 92) | AP (n = 83) | CP vs.PH | AP vs.PH | ||
| -1082-819-592/-1082-819-592 | n (%) | n (%) | n (%) | p | OR (95%CI) | p | OR (95%CI) |
| ATA/ATA | 31 (34.1) | 44 (47.8) | 34 (41.0) | 0.017 | 2.43 (1.21–3.83) | 0.149 | 1.57 (0.84–2.60) |
| others | 60 (65.9) | 48 (52.2) | 49 (59.0) | ||||
| ATA/ACC | 28 (30.8) | 30 (32.6) | 31 (37.3) | 0.553 | 1.32 (0.42–2.75) | 0.447 | 1.61 (0.77–2.87) |
| others | 63 (69.2) | 62 (67.4) | 52 (62.7) | ||||
PH, periodontal healthy subjects; CP, chronic periodontitis subjects; AP, aggressive periodontitis subjects.
The impact of genetic variants on the number of subgingival bacteria and the serum IL-10 level
The amount of A. actinomycetemcomitans was statistically different among -592 AA, AC and CC groups in patients with CP (χ2 = 9.29 p = 0.028), and AA individuals showed higher bacterial counts of A. actinomycetemcomitans than AC subjects in the following all pairwise multiple comparison test (p = 0.035). The amount of A. actinomycetemcomitans also differed among -819 TT, TC and CC groups in patients with CP (χ2 = 10.13 p = 0.017), and TT individuals showed higher bacterial counts of A. actinomycetemcomitans than TC subjects in post hoc comparisons (p = 0.023) (Fig. 2). Moreover, the amount of subgingival A. actinomycetemcomitans was significantly increased in ATA/ATA-positive individuals of CP subjects (p = 0.009) (Fig. 3). It seemed that IL-10 polymorphisms may not influence the amount of A. actinomycetemcomitans in AP cases and the healthy controls (data were not shown). Moreover, no significant association between IL-10 polymorphisms and the bacterial load of P. gingivalis was observed in any group (data were not shown). Further multiple regression analysis using the stepwise method was carried out, ATA/ATA carriers were associated with increased bacterial counts of A. actinomycetemcomitans in CP group after adjustment for age, gender, BOP, PD and CAL (p = 0.013) (Table 6).
Figure 2.
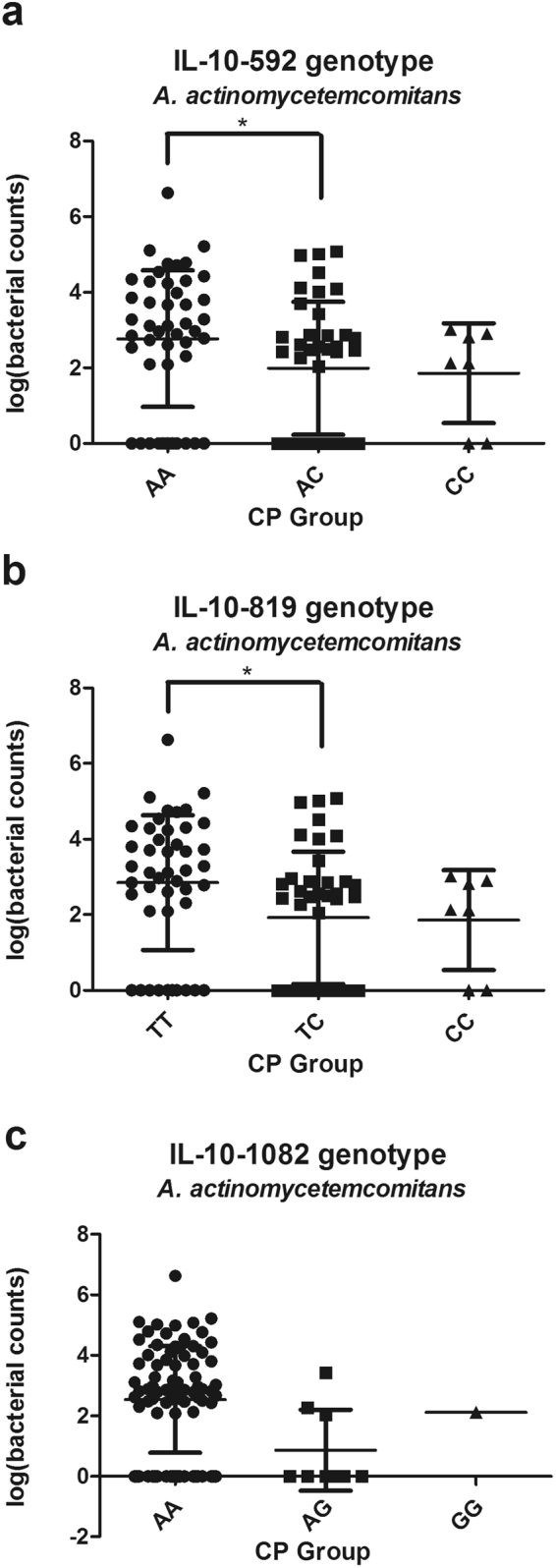
Association between SNPs in IL-10 promoter gene (IL-10-592, -819 and -1082 genotype) and the subgingival number of A. actinomycetemcomitans in CP group. *Significant (p < 0.05 by Kruskal-Wallis ANOVA test followed by Dunn-Bonferroni test) CP, chronic periodontitis.
Figure 3.
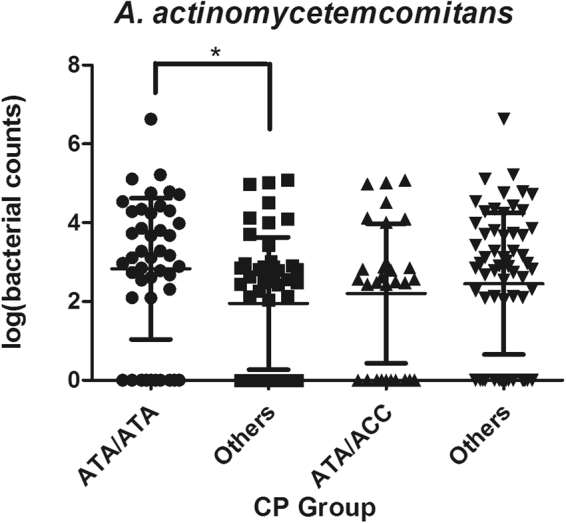
Association between the IL-10 haplotype and the subgingival number of A. actinomycetemcomitans in CP group. *Significant (p < 0.05 by Mann-Whitney U test) CP, chronic periodontitis.
Table 6.
Results of multiple regression analyses for the impact of genetic variants on the number of subgingival bacteria and the serum IL-10 level.
| Unstandardized coefficients | Standardized coefficients | T | P | ||
|---|---|---|---|---|---|
| B | Standard error | β | |||
| Subgingival counts of Aa | |||||
| ATA/ATA | 0.79 | 0.46 | 0.25 | 2.43 | 0.013 |
| Serum level of IL-10 | |||||
| ATA/ATA | −2.03 | 1.58 | −0.96 | −3.21 | 0.021 |
Aa, A. actinomycetemcomitans.
Serum IL-10 levels differed among -592 AA, AC and CC groups in CP subjects (χ2 = 8.74 p = 0.028), lower IL-10 levels were significantly detected in subjects carrying the AA genotype compared with AC genotype (p = 0.036). Serum IL-10 levels were also statistically different among -819 TT, TC and CC groups in CP subjects (χ2 = 8.32 p = 0.031), and TT subjects presented significant lower IL-10 levels than TC subjects in post hoc comparisons (p = 0.04). Serum IL-10 concentrations was significantly lower in ATA/ATA-positive individuals of CP subjects (p = 0.033) (Table 5). However, any of the IL-10 haplotypes had no significant influence on the serum IL-10 levels in the healthy group, the AP group and the whole diseased group (data were not presented). Moreover, multiple linear regression analysis by stepwise method, adjusted for age, gender, BOP, PD and CAL, was used to investigate the impact of the above IL-10 haplotypes in subjects with CP. The analysis revealed that ATA/ATA-positive individuals were still related to lower serum IL-10 levels among CP patients (p = 0.021) (Table 6).
Table 5.
Serum level (mean ± SD) of IL-10 in relation to genetic polymorphisms in CP group.
| polymorphism | genotype | Serum IL-10 (pg/ml) |
|---|---|---|
| IL-10-592 | CC (n = 7) | 6.2 ± 5.5 |
| AC (n = 39) | 7.6 ± 7.4 | |
| AA (n = 46) | 4.8 ± 4.1* | |
| IL-10-819 | CC (n = 7) | 6.2 ± 5.5 |
| TC (n = 40) | 7.6 ± 7.3 | |
| TT (n = 45) | 4.8 ± 4.1* | |
| 1082-819-592/1082-819-592 | ATA/ATA (n = 44) | 4.8 ± 4.0§ |
| Others (n = 48) | 7.2 ± 5.8 |
CP, chronic periodontitis.
*Significant (p < 0.05 by Kruskal-Wallis ANOVA test followed by Dunn-Bonferroni test) IL-10-592 AA subjects vs. AC subjects; IL-10-819 TT subjects vs. TC subjects.
§Significant (p < 0.05 by Mann-Whitney U test).
Discussion
The present study analyzed the distribution of three SNPs of IL-10 promoter gene among patients with CP, patients with AP and periodontal healthy controls in Chinese Han population. Furthermore, possible impact of IL-10 genetic variants on the quantity of subgingival bacteria A. actinomycetemcomitans and P. gingivalis were investigated.
The microbial analysis revealed that the two bacteria investigated were also found in the healthy controls, however, with significantly lower number compared to the patient groups (Fig. 1). It suggests that besides periodontal pathogens, the host immune response and its genetic regulation and control may be crucial to the development of periodontitis. Moreover, the amount of A. actinomycetemcomitans cannot be used to distinguish patients with AP and patients with CP due to its great variability between individuals within groups (Fig. 1), which was in accordance with some recent studies44,45.
All the periodontitis patients involved expressed severe generalized attachment loss, hence, they were suitable for the identification of genetic risk factors. Multiple logistic regressions revealed that that IL-10-592 AA, -819 TT and ATA/ATA genotype may confer a slight increase in the risk for CP after adjustment for age, gender and periodontal status (Tables 3, 4). With regard to the -592 SNP, the related studies most presented the similar trends in their populations, including two meta-analysis32,33,35,39,40,46. In agreement with our results concerning the -819 SNP, only one study suggested that -819 CC genotype may decrease the risk for CP33, while the others yielded negative associations. As to the -1082 loci, negative result was in accordance with previous studies. It was showed that in Brazilian female cases, there was a trend with the predominance of haplotype ATA in CP group (p = 0.061)33. In German subjects the haplotype ATA could increase the odds ratio for AP21. In Taiwanese population the ATA/ATA haplotype could increase the risk of AP and individuals with the ATA/ACC genotype were less susceptible to CP35. A possible reason for these conflicting results might be based on ethnic differences in the distribution of IL-10 polymorphisms. The genotype frequencies of IL-10 SNPs in Chinese subjects vary from those of Caucasians47, but they are racially close to those of Japanese48 and Taiwanese individuals35. The ATA haplotype was dominant in Asian population, and the frequency of it was 0.64 in our study. GCC and ACC haplotypes were in the majority among Caucasians whereas ATA just accounted for 0.21. In addition, other factors, such as the sample size, oral hygiene and smoking can have impact on the inconsistent genetic results.
The positive associations between IL-10 certain genotypes and CP might be explained by the following reasons. IL-10 promoter haplotype ATA has been related to low production of IL-1025. After the initial colonization of the pathogens in the periodontal region of IL-10 hypo-producers, reduced IL-10 levels might modulate the host immune response, with attenuation of anti-inflammatory and increase of pro-inflammatory mediators49. For instance, it might result in increased production of tumor necrosis factor- α, which was known for its important role in alveolar bone loss50. Moreover, the absence of IL-10 could result in accelerated alveolar bone loss29,30. Obviously, individuals with certain genotypes were more likely to develop CP characterized by alveolar bone loss.
In the present study, we obtained a positive association between IL-10 ATA/ATA genotype and lower serum IL-10 levels, in addition, a positive relation to increased subgingival bacterial counts of A. actinomycetemcomitans by multiple regression analysis in Chinese patients with chronic periodontitis for the first time (Table 6). The findings indicated the possible effect of IL-10 polymorphisms on the periodontopathic bacteria. This study supported the hypothesis that the host genotype can influence the composition of the subgingival microbiota7. However, limiting the microbial analysis to two periodontal pathogens may overlook the possible influence on subgingival microbiota, so comprehensive microbiological assessment should be adopted in the following study. Moreover, oral hygiene may also influence the subgingival colonization51,52, based on this point, plaque index should also be listed as a covariate in the multiple regression analysis, but we lack these data, so more comprehensive data should be collected when the participants are recruited in further study. Hyper-inflammatory genotypes (IL-10 ATA/ATA) cluster presented higher subgingival counts of A. actinomycetemcomitans and higher risk for chronic periodontitis which revealed that complex interactions between the host genetic variants and the subgingival microbiota are at the basis of predisposition to periodontitis. The reasons for the above associations are not entirely clear, we hypothesize a possible explanation for the findings. We assume that ATA/ATA carriers (IL-10 hypo-producers) may be prone to the growth of bacteria, such as A. actinomycetemcomitans. After the initial colonization of the pathogens in the periodontal region of IL-10 hypo-producers, they may benefit from the hyperactivation of host cells, the stimulation of the inflammatory cascade and the increased production of multiple inflammatory cytokines49,53 due to the effect of IL-10 genetic variants. In a word, the pathogens could gain more favorable environment for their survival and well overgrowth in inflamed areas in subjects with specific genotype. And increased pathogens in turn may affect the local tissues which would lead to enhanced inflammation and periodontal pocket formation. In addition, the absence of IL-10 could result in accelerated alveolar bone loss29,30. Increased alveolar bone loss and deepened periodontal pockets would further favor the growth of A. actinomycetemcomitans due to the anaerobic environment. The study revealed that IL-10 ATA/ATA individuals (with lower serum IL-10 levels) were more susceptible to CP, and it may be partly caused by higher subgingival counts of A. actinomycetemcomitans which increased the risk for characteristic periodontal tissue destruction. In the present study, there was no association between IL-10 polymorphisms and AP. Moreover, any of the IL-10 haplotypes had no significant influence on the serum IL-10 levels and the amount of A. actinomycetemcomitans and/or P. gingivalis in the AP group. The above results may be related to the sample size, the exclusion of current and former smokers, and the lack of plaque index. However, the results may indicate that the susceptibility to AP might be not closely tied with the change of A. actinomycetemcomitans and/or P. gingivalis caused by IL-10 genetic variants to some extent.
To date several clinical studies have been conducted to assess the influence of some cytokine genes polymorphisms on the subgingival microbiota. Among these, IL-612,13,15 and IL-116–18,22 polymorphisms have received most attention, and it has been proved that they were associated with subgingival detection of A. actinomycetemcomitans and P. gingivalis. IFN-γ-AA carriers had a lower odds ratio for the presence of A. actinomycetemcomitans54. IL-2-330,166 TT: TT combination seemed to affect the occurrence of P. gingivalis and bacteria of the ‘red’ complex19. The Q551R IL-4R polymorphism was associated with the presence of T. forsythia20. In addition, a study obtained IL-10 ATA haplotype was associated with increased odds ratio for AP among Caucasians population and the presence of P. intermedia was found to be decreased in ATA- positive individuals21. The results are contradictory to ours, and the possible reasons for these conflicting results might be based on ethnic differences in the distribution of IL-10 polymorphisms, sample size, or sensitivity of molecular assessment for periodontopathic bacteria.
In conclusion, despite of the limitations of sample size, the results of this study suggest that the possible influence of IL-10 polymorphisms on the susceptibility to chronic periodontitis. In addition, IL-10 ATA/ATA genotype is associated with the subgingival quantity of A. actinomycetemcomitansi in subjects with chronic periodontitis. This study supports the hypothesis that complex interactions between the host genetic variants and the subgingival microbiota are at the basis of susceptibility to periodontitis.
Acknowledgements
This study is supported by grants from the National Natural Science Foundation of China (Grants No. 81470749&81771074), the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, 2014-37) and the Major Project of Natural Science Research in Colleges and Universities of Jiangsu Province (Grant No.16KJA320001).
Author Contributions
Ying Geng and Lu Li conducted the experiments and analysed the results. Xiaoqian Wang and Mifang Yang participated in its design and helped to draft the manuscript. Fanzhen He and Yi Zhou helped to conduct the experiments. Yan Xu conceived the experiments and guided the whole research. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Ying Geng and Lu Li contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Armitage GC. Development of a classification system for periodontal diseases and conditions. Annals of periodontology. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Michalowicz BS. Genetic and heritable risk factors in periodontal disease. Journal of periodontology. 1994;65:479–488. doi: 10.1902/jop.1994.65.5s.479. [DOI] [PubMed] [Google Scholar]
- 3.Hart TC, Kornman KS. Genetic factors in the pathogenesis of periodontitis. Periodontology 2000. 1997;14:202–215. doi: 10.1111/j.1600-0757.1997.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 4.Genco RJ, S.Borgnakke W. Risk factors for periodontal disease. Periodontology 2000. 2013;62:59–94. doi: 10.1111/j.1600-0757.2012.00457.x. [DOI] [PubMed] [Google Scholar]
- 5.Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nature reviews. Genetics. 2012;13:175–188. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- 6.Kellam P, Weiss RA. Infectogenomics: insights from the host genome into infectious diseases. Cell. 2006;124:695–697. doi: 10.1016/j.cell.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nibali L, Donos N, Henderson B. Periodontal infectogenomics. J Med Microbiol. 2009;58:1269–1274. doi: 10.1099/jmm.0.012021-0. [DOI] [PubMed] [Google Scholar]
- 8.Elenkov I, Iezzoni D, Daly A, Harris A, Chrousos G. Cytokine dysregulation, inflammation and well-being. Neuroimmunomodulation. 2005;12:255–269. doi: 10.1159/000087104. [DOI] [PubMed] [Google Scholar]
- 9.Loo WT, et al. Gene polymorphism and protein of human pro- and anti-inflammatory cytokines in Chinese healthy subjects and chronic periodontitis patients. J Transl Med. 2012;10(Suppl 1):S8. doi: 10.1186/1479-5876-10-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y-CG, Lerner UH, Teng Y-TA. Cytokine responses against periodontal infection. Periodontology 2000. 2010;52:163–206. doi: 10.1111/j.1600-0757.2009.00321.x. [DOI] [PubMed] [Google Scholar]
- 11.Preshaw PM, Taylor JJ. How has research into cytokine interactions and their role in driving immune responses impacted our understanding of periodontitis? Journal of clinical periodontology. 2011;38(Suppl 11):60–84. doi: 10.1111/j.1600-051X.2010.01671.x. [DOI] [PubMed] [Google Scholar]
- 12.Nibali L, et al. Gene Polymorphisms and the Prevalence of Key Periodontal Pathogens. Journal of dental research. 2007;86:416–420. doi: 10.1177/154405910708600505. [DOI] [PubMed] [Google Scholar]
- 13.Nibali L, et al. Interleukin-6 polymorphisms are associated with pathogenic bacteria in subjects with periodontitis. Journal of periodontology. 2008;79:677–683. doi: 10.1902/jop.2008.070453. [DOI] [PubMed] [Google Scholar]
- 14.Nibali L, et al. Association between interleukin-6 -174 polymorphism and Aggregatibacter actinomycetemcomitans in chronic periodontitis. Journal of periodontology. 2010;81:1814–1819. doi: 10.1902/jop.2010.100084. [DOI] [PubMed] [Google Scholar]
- 15.Nibali L, et al. IL6 -174 genotype associated with Aggregatibacter actinomycetemcomitans in Indians. Oral Dis. 2011;17:232–237. doi: 10.1111/j.1601-0825.2010.01731.x. [DOI] [PubMed] [Google Scholar]
- 16.Agerbaek MR, Lang NP, Persson GR. Microbiological composition associated with interleukin-1 gene polymorphism in subjects undergoing supportive periodontal therapy. Journal of periodontology. 2006;77:1397–1402. doi: 10.1902/jop.2006.050212. [DOI] [PubMed] [Google Scholar]
- 17.Ferreira SB, et al. An interleukin-1beta (IL-1beta) single-nucleotide polymorphism at position 3954 and red complex periodontopathogens independently and additively modulate the levels of IL-1beta in diseased periodontal tissues. Infect Immun. 2008;76:3725–3734. doi: 10.1128/IAI.00546-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulz S, et al. Single nucleotide polymorphisms in interleukin-1gene cluster and subgingival colonization with Aggregatibacter actinomycetemcomitans in patients with aggressive periodontitis. Hum Immunol. 2011;72:940–946. doi: 10.1016/j.humimm.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Reichert S, et al. Interleukin-2 -330 and 166 gene polymorphisms in relation to aggressive or chronic periodontitis and the presence of periodontopathic bacteria. J Periodontal Res. 2009;44:628–635. doi: 10.1111/j.1600-0765.2008.01173.x. [DOI] [PubMed] [Google Scholar]
- 20.Reichert S, et al. The genetic impact of the Q551R interleukin-4 receptor alpha polymorphism for aggressive or chronic periodontitis and the occurrence of periodontopathic bacteria. Arch Oral Biol. 2011;56:1485–1493. doi: 10.1016/j.archoralbio.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Reichert S, et al. The interleukin-10 promoter haplotype ATA is a putative risk factor for aggressive periodontitis. J Periodontal Res. 2008;43:40–47. doi: 10.1111/j.1600-0765.2007.00992.x. [DOI] [PubMed] [Google Scholar]
- 22.Socransky SS, Haffajee AD, Smith C, Duff GW. Microbiological parameters associated with IL-1 gene polymorphisms in periodontitis patients. Journal of clinical periodontology. 2000;27:810–818. doi: 10.1034/j.1600-051x.2000.027011810.x. [DOI] [PubMed] [Google Scholar]
- 23.Moore KW, de W Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 24.de Waal Malefyt R, Abrams J, Bennett B, Figdor CG, de Vries JE. Interleukin 10 (IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. The Journal of experimental medicine. 1991;174:1209–1220. doi: 10.1084/jem.174.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner DM, et al. An investigation of polymorphism in the interleukin-10 gene promoter. European journal of immunogenetics: official journal of the British Society for Histocompatibility and Immunogenetics. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 26.Gibson AW, et al. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. The journal of Immunology. 2001;166:3915–3922. doi: 10.4049/jimmunol.166.6.3915. [DOI] [PubMed] [Google Scholar]
- 27.Reuss E, et al. Differential regulation of interleukin-10 production by genetic and environmental factors–a twin study. Genes and immunity. 2002;3:407–413. doi: 10.1038/sj.gene.6363920. [DOI] [PubMed] [Google Scholar]
- 28.Schippers EF, et al. IL-10 and toll-like receptor-4 polymorphisms and the in vivo and ex vivo response to endotoxin. Cytokine. 2005;29:215–228. doi: 10.1016/j.cyto.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Al-Rasheed A, Scheerens H, Rennick DM, Fletcher HM, Tatakis DN. Accelerated alveolar bone loss in mice lacking interleukin-10. Journal of dental research. 2003;82:632–635. doi: 10.1177/154405910308200812. [DOI] [PubMed] [Google Scholar]
- 30.Claudino M, et al. Down-regulation of expression of osteoblast and osteocyte markers in periodontal tissues associated with the spontaneous alveolar bone loss of interleukin-10 knockout mice. European journal of oral sciences. 2010;118:19–28. doi: 10.1111/j.1600-0722.2009.00706.x. [DOI] [PubMed] [Google Scholar]
- 31.Berglundh T, Donati M, Hahn-Zoric M, Hanson L-A, Padyukov L. Association of the -1087 IL 10 gene polymorphism with severe chronic periodontitis in Swedish Caucasians. Journal of clinical periodontology. 2003;30:249–254. doi: 10.1034/j.1600-051X.2003.10274.x. [DOI] [PubMed] [Google Scholar]
- 32.Sumer AP, et al. Association of interleukin-10 gene polymorphisms with severe generalized chronic periodontitis. Journal of periodontology. 2007;78:493–497. doi: 10.1902/jop.2007.060309. [DOI] [PubMed] [Google Scholar]
- 33.Scarel-Caminaga RM, et al. Interleukin 10 gene promoter polymorphisms are associated with chronic periodontitis. Journal of clinical periodontology. 2004;31:443–448. doi: 10.1111/j.1600-051X.2004.00500.x. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi T, et al. Cytokine gene polymorphisms associated with rheumatoid arthritis and periodontitis in Japanese adults. Journal of periodontology. 2009;80:792–799. doi: 10.1902/jop.2009.080573. [DOI] [PubMed] [Google Scholar]
- 35.Hu KF, et al. Interleukin-10 (-592 C/A) and interleukin-12B (+16974 A/C) gene polymorphisms and the interleukin-10 ATA haplotype are associated with periodontitis in a Taiwanese population. J Periodontal Res. 2009;44:378–385. doi: 10.1111/j.1600-0765.2008.01116.x. [DOI] [PubMed] [Google Scholar]
- 36.Brett PM, et al. Functional gene polymorphisms in aggressive and chronic periodontitis. Journal of dental research. 2005;84:1149–1153. doi: 10.1177/154405910508401211. [DOI] [PubMed] [Google Scholar]
- 37.Mellati E, Arab HR, Tavakkol-Afshari J, Ebadian AR, Radvar M. Analysis of -1082 IL-10 gene polymorphism in Iranian patients with generalized aggressive periodontitis. Med Sci Monit. 2007;13:CR510–514. [PubMed] [Google Scholar]
- 38.Tervonen T, Raunio T, Knuuttila M, Karttunen R. Polymorphisms in the CD14 and IL-6 genes associated with periodontal disease. Journal of clinical periodontology. 2007;34:377–383. doi: 10.1111/j.1600-051X.2007.01067.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhong Q, Ding C, Wang M, Sun Y, Xu Y. Interleukin-10 gene polymorphisms and chronic/aggressive periodontitis susceptibility: a meta-analysis based on 14 case-control studies. Cytokine. 2012;60:47–54. doi: 10.1016/j.cyto.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Albuquerque CM, et al. Association of the IL-10 polymorphisms and periodontitis a meta analysis. Molecular Biology Reports. 2012;39:9319–9329. doi: 10.1007/s11033-012-1738-1. [DOI] [PubMed] [Google Scholar]
- 41.Kuboniwa M, et al. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral microbiology and immunology. 2004;19:168–176. doi: 10.1111/j.0902-0055.2004.00135.x. [DOI] [PubMed] [Google Scholar]
- 42.Nonnenmacher C, et al. RealTime Polymerase Chain Reaction for Detection and Quantification of Bacteria in Periodontal Patients. Journal of periodontology. 2005;76:1542–1549. doi: 10.1902/jop.2005.76.9.1542. [DOI] [PubMed] [Google Scholar]
- 43.Goncharoff P, Figurski DH, Stevens RH, Fine DH. Identification of Actinobacillus actinomycetemcomitans: polymerase chain reaction amplification of lktA-specific sequences. Oral microbiology and immunology. 1993;8:105–110. doi: 10.1111/j.1399-302X.1993.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 44.Tomita S, et al. Prevalence of Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythia in Japanese patients with generalized chronic and aggressive periodontitis. Microb Pathog. 2013;61-62:11–15. doi: 10.1016/j.micpath.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Heller D, Silva-Boghossian CM, do Souto RM, Colombo AP. Subgingival microbial profiles of generalized aggressive and chronic periodontal diseases. Arch Oral Biol. 2012;57:973–980. doi: 10.1016/j.archoralbio.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 46.Claudino M, et al. The broad effects of the functional IL-10 promoter-592 polymorphism: modulation of IL-10, TIMP-3, and OPG expression and their association with periodontal disease outcome. J Leukoc Biol. 2008;84:1565–1573. doi: 10.1189/jlb.0308184. [DOI] [PubMed] [Google Scholar]
- 47.Turner D, et al. An investigation of polymorphism in the interleukin‐10 gene promoter. European Journal of Immunogenetics. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 48.Yamazaki, K. et al. Interleukin-10 gene promoter polymorphism in Japanese patients with adult and early-onset periodontitis. Journal of clinical periodontology28, 828–832 (2001). [DOI] [PubMed]
- 49.Cavalcante LB, et al. Expression of the interleukin-10 signaling pathway genes in individuals with Down syndrome and periodontitis. Journal of periodontology. 2012;83:926–935. doi: 10.1902/jop.2011.110056. [DOI] [PubMed] [Google Scholar]
- 50.Garlet GP, et al. The dual role of p55 tumour necrosis factor-alpha receptor in Actinobacillus actinomycetemcomitans-induced experimental periodontitis: host protection and tissue destruction. Clinical and experimental immunology. 2007;147:128–138. doi: 10.1111/j.1365-2249.2006.03260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hellstrom MK, Ramberg P, Krok L, Lindhe J. The effect of supragingival plaque control on the subgingival microflora in human periodontitis. Journal of clinical periodontology. 1996;23:934–940. doi: 10.1111/j.1600-051X.1996.tb00514.x. [DOI] [PubMed] [Google Scholar]
- 52.Haffajee AD, et al. Efficacy of manual and powered toothbrushes (II). Effect on microbiological parameters. Journal of clinical periodontology. 2001;28:947–954. doi: 10.1034/j.1600-051x.2001.028010947.x. [DOI] [PubMed] [Google Scholar]
- 53.Belibasakis GN, et al. Cytokine responses of human gingival fibroblasts to Actinobacillus actinomycetemcomitans cytolethal distending toxin. Cytokine. 2005;30:56–63. doi: 10.1016/j.cyto.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 54.Reichert S, et al. Interferon-gamma and interleukin-12 gene polymorphisms and their relation to aggressive and chronic periodontitis and key periodontal pathogens. Journal of periodontology. 2008;79:1434–1443. doi: 10.1902/jop.2008.070637. [DOI] [PubMed] [Google Scholar]


