Abstract
Bacteria–plasmid associations can be mutualistic or antagonistic depending on the strength of positive selection for plasmid-encoded genes, with contrasting outcomes for plasmid stability. In mutualistic environments, plasmids are swept to high frequency by positive selection, increasing the likelihood of compensatory evolution to ameliorate the plasmid cost, which promotes long-term stability. In antagonistic environments, plasmids are purged by negative selection, reducing the probability of compensatory evolution and driving their extinction. Here we show, using experimental evolution of Pseudomonas fluorescens and the mercury-resistance plasmid, pQBR103, that migration promotes plasmid stability in spatially heterogeneous selection environments. Specifically, migration from mutualistic environments, by increasing both the frequency of the plasmid and the supply of compensatory mutations, stabilized plasmids in antagonistic environments where, without migration, they approached extinction. These data suggest that spatially heterogeneous positive selection, which is common in natural environments, coupled with migration helps to explain the stability of plasmids and the ecologically important genes that they encode.
Keywords: compensatory evolution, amelioration, species interactions, mobile genetic element, spatial heterogeneity, source-sink
1. Introduction
Conjugative plasmids are semi-autonomous mobile genetic elements that have control over their own replication and transmission, but rely on the bacterial cell for their propagation [1]. Because plasmids often carry accessory genes encoding ecologically important traits—such as toxin resistance, novel metabolic functions or virulence factors [2]—they play an important role in bacterial adaptation and genome evolution through horizontal gene transfer. However, the ubiquity of plasmids is difficult to explain. Plasmid acquisition is often costly for host cells, owing to the biosynthetic demand placed upon the cell and the disruption of cellular homeostasis [3,4]. The benefits of plasmid encoded traits meanwhile are often context dependent and only beneficial to the bacterial host under specific environmental conditions. Thus, interactions between plasmids and bacteria form a context-dependent parasitism–mutualism continuum [5,6]. In environments where the benefits conferred by plasmid-encoded traits outweigh the costs of plasmid carriage, the interaction is mutualistic [5,6]. Where these costs are not offset by the benefits of plasmid-encoded traits, plasmids are parasitic and the interaction is antagonistic [5,6]. The ecological population dynamics of plasmids are dependent on the balance of these costs and benefits: plasmids will be maintained at higher frequencies in mutualistic environments owing to positive selection. In antagonistic environments, plasmids which do not have sufficiently high rates of infectious transmission will be purged by purifying selection potentially leading to extinction of the plasmid and, concomitantly, reduced evolutionary potential for the bacterial community.
Compensatory evolution to ameliorate the cost of plasmid carriage can rescue plasmids from extinction by weakening purifying selection [5,7]. Compensatory evolution has been observed repeatedly in bacteria–plasmid co-culture studies and is therefore believed to be an important determinant of plasmid population dynamics [5,8–14]. Recent theory shows that compensatory evolution is more likely to occur in mutualistic environments because plasmids are at higher frequency for longer periods of time, increasing the probability that compensation mutations will arise [13,15]. We predicted, therefore, that under spatially heterogeneous positive selection, migration from mutualistic to antagonistic patches will stabilize plasmids across the entire landscape through an eco-evolutionary mechanism, whereby immigrants increase both the frequency of the plasmid and the supply of compensatory mutations ameliorating the plasmid cost.
The interaction between the bacterium Pseudomonas fluorescens SBW25 [16] and its conjugative plasmid, pQBR103 [17], forms a context-dependent parasitism–mutualism continuum. Plasmid carriage imposes a large fitness cost on the host cell, but this cost is progressively outweighed by the fitness benefit of plasmid-encoded mercury resistance at higher concentrations of toxic Hg(II), creating a fitness gradient from strongly negative to strongly positive selection [5,6]. We previously showed that P. fluorescens can ameliorate the cost of plasmid carriage through compensatory mutations targeting the GacA/GacS global regulatory system [5]. While parallel compensatory evolution was observed across the entire parasitism–mutualism continuum, it occurred with higher likelihood and at a faster rate in mutualistic environments. Since GacA/GacS positively regulates a well-characterized suite of secreted proteins we are able to track compensatory evolution dynamics through time using simple phenotypic assays for protease production [18]. Thus, we have developed a tractable experimental system that allows us to simultaneously follow the ecological dynamics of plasmid prevalence and the evolutionary dynamics of compensatory mutation in real-time. Here, we test how the spatial heterogeneity of positive selection and migration rate interact to determine plasmid stability through their joint effects on plasmid frequency and compensatory evolution dynamics.
2. Material and methods
Experimental populations were established using isogenic strains of the bacteria P. fluorescens SBW25 with and without the mercury resistance plasmid, pQBR103. Strain SBW25-Gm carries a gentamicin resistance marker and strain SBW25-Sm-lacZ carries both a streptomycin resistance marker and the lacZ gene. Antibiotic markers were used to introduce the plasmid by conjugation [19] and the lacZ gene was used to distinguish between strains when spread on to media containing X-gal. Populations were grown in 30 ml glass vials in 6 ml Kings B broth shaking at 28°C.
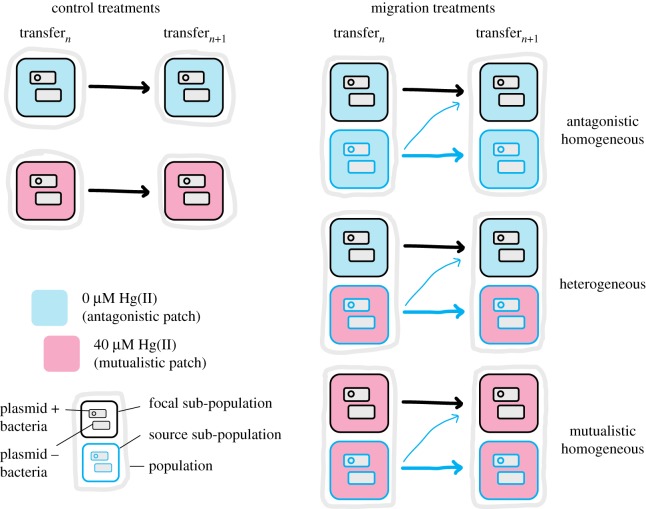
Six replicate populations were established for each treatment. Experimental treatments consisted of three mercury selection ‘landscapes’ and three immigration rates in a factorial design, with the addition of two control treatments which experienced no immigration (figure 1). Experimental landscapes consisted of a focal ‘patch’, which was represented by a 6 ml sub-population initiated with 50 : 50 plasmid-containing and plasmid-free SBW25-Gm, and a source patch, represented by a 6 ml sub-population of 50 : 50 plasmid-containing and plasmid-free SBW25-Sm-lacZ. The three mercury selection landscapes consisted of a heterogeneous landscape with a mutualistic (40 µM HgCl2) source patch and an antagonistic (0 µM HgCl2) focal patch, and two homogeneous landscapes: purely antagonistic (0 µM HgCl2 in both patches) and purely mutualistic (40 µM HgCl2 in both patches). All populations were propagated by serial transfer every two days. For each replicate population, 60 µl of the source sub-population was transferred directly to a fresh microcosm while focal sub-populations were first mixed with bacteria from their source sub-population at three rates of immigration (0.1, 1 and 10%) and then transferred (figure 1). Carry-over of HgCl2 from mutualistic source patches owing to migration is expected to be negligible. The mer operon provides resistance through detoxification of Hg(II) into the less toxic Hg(0) which evaporates. After approximately 6 h the supernatant of plasmid-containing cultures is non-toxic to plasmid-free sensitive cells, suggesting that the concentration of Hg(II) is already substantially reduced (electronic supplementary material, figure S1). Control populations, with no immigration, were established at either 0 µM or 40 µM HgCl2.
Figure 1.
Transfer strategy for selection experiment. Bacterial populations were propagated by serial transfer of 1% of the population to fresh media (represented by arrows) every 48 h. Figure shows the strategy for a single bacterial transfer step for the two control treatments and three migration treatments. Control populations were propagated by simple transfer of bacteria from one population to a fresh environment. Populations in the migration treatments consisted of two paired sub-populations. At each transfer bacteria from the source (blue line) sub-population were transferred as normal and bacteria from the focal (black line) sub-population were first mixed with bacteria from the source (blue line) sub-population at three migration rates (0.1, 1 and 10%) before being transferred.
Populations were evolved for 24 transfers (approx. 180 bacterial generations). Every transfer for the first 12 transfers, and thereafter every two transfers, samples of the focal sub-populations were spread on to skimmed milk agar (10% milk powder in KB agar) containing 20 mg µl−1 X-gal with and without 20 µM HgCl2. Skimmed milk agar was used to identify the spontaneous appearance of GacA/S compensatory mutations, as the GacA/S regulator controls the production of exoprotease [20]. Colonies positive for GacA/S function can be distinguished by a zone of clearing around the colony. X-gal was used to distinguish immigrant (blue) and resident (white) bacteria. Milk plates supplemented with X-gal therefore allowed us to estimate the total population density, the frequency of immigrants and residents and their GacA/S status, while milk plates supplemented with X-gal and mercury allowed us to estimate the proportions of these genotypes which contained the plasmid.
All analyses were conducted in the R statistical package (R Foundation for Statistical Computing) using endpoint data (from transfer 24) unless specified. Data were analysed with ANOVA and further interrogated using planned contrasts, defined using the ‘contrasts’ package, allowing specific comparisons between treatments. Where used, specific contrasts are specified in lowercase capital letters.
3. Results
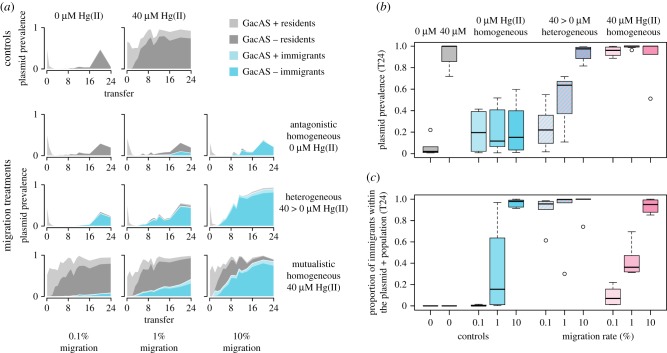
To determine the effects of spatial heterogeneity of positive selection and migration rate on plasmid population dynamics we tracked plasmid frequencies in focal patches over time. In control populations that were propagated without immigration, plasmids persisted in all populations but rapidly declined to very low frequencies in antagonistic patches, consistent with the high cost of plasmid carriage (figure 2a). With migration, we observed interactive effects of selection landscape and migration rate on plasmid frequency in focal patches (figure 2a,b; landscape × migration rate: F2,48 = 14.81, p < 0.001). This was driven by variation in response to migration between the heterogeneous treatment and the two homogeneous treatments (landscape[homogeneous(i.e. mutualistic + antagonistic) vs heterogeneous] × migration rate: t = 5.405, p < 0.001). In homogeneous landscapes, plasmid frequencies in focal patches varied according to the strength of positive selection, such that they were higher in mutualistic compared to antagonistic landscapes (figure 2b; landscape[mutualistic vs antagonistic]: t = 9.89, p < 0.001), but did not vary with migration rate (landscape[mutualistic vs antagonistic] × migration rate: t = −0.65, p = 0.52). In the heterogeneous landscape, however, plasmid frequency increased in antagonistic focal patches with increasing migration rate (figure 2b). While at the lowest migration rate, the plasmid frequency was similar to those observed in antagonistic focal patches within homogeneous landscapes (landscape[migration = 0.1%; heterogeneous vs antagonistic homogeneous]: F = 0.114, p = 0.742), at higher migration rates, the plasmid frequencies in focal patches of heterogeneous landscapes exceeded those observed in homogeneous landscapes (landscape[migration > 0.1%; heterogeneous vs antagonistic homogeneous]: F = 26.71 p < 0.0001). This suggests that plasmid stability was enhanced by higher migration rates under spatially heterogeneous positive selection, whereas migration had no effect on plasmid frequency in spatially homogeneous selection environments.
Figure 2.
The impacts of migration across treatments. (a) Population dynamics within plasmid-containing individuals over the course of the selection experiment. The total shaded area shows the proportion of plasmid containing individuals in the focal subpopulations, averaged across six replicate populations. Shading is broken down by genotype, showing the relative proportion of resident (grey) and immigrant (blue) bacteria which were positive (light) or negative (dark) for the GacA/S phenotype (i.e. dark areas indicate compensatory mutations). (b) Summary of endpoint (transfer 24) mean plasmid prevalence (n = 6). (c) Mean proportion of plasmid containing individuals that are from the source population (lacZ+) at the final time point (n = 6).
To determine the dynamics of compensatory evolution in focal patches, we tracked the frequency of the protease negative phenotype associated with mutated gacA/gacS loci of P. fluorescens SBW25. Protease negative phenotypes appeared rapidly in all populations regardless of treatment (figure 2a; landscape: F2,50 = 0.36, p = 0.702, migration rate: F1,50 = 2.66, p = 0.110), and swept to high frequency among plasmid-bearers (68–100% of mercury resistant colonies were protease negative at transfer-24; figure 2a), indicating that compensatory evolution played a key role in the survival of the plasmid in our experiment. We next estimated the proportion of immigrant genotypes among the plasmid-bearers in focal patches. Immigrant and resident genotypes were distinguished using the lacZ marker. Although the lacZ marked strain appears to have had a slight fitness advantage over the unlabelled strain, the response to migration rate differed significantly between homogeneous and heterogeneous treatments (landscape[homogeneous vs heterogeneous] × migration rate: t = 5.41, p < 0.001). In both types of homogeneous landscape, the proportion of immigrant plasmid-bearers in focal patches increased with the rate of migration (figure 2c; migration rate[homogeneous only]: F = 111.883, p < 0.001) with no significant difference between treatments (landscape[mutualistic vs antagonistic]: t = 1.151, p = 0.256). By contrast, in the heterogeneous landscape, immigrant plasmid-bearers comprised of more than 90% of the plasmid-bearing population regardless of the migration rate (migration rate[heterogeneous only]: F = 0.517, p = 0.482). Taken together, these data suggest that plasmid stability in antagonistic focal patches under spatially heterogeneous positive selection required the immigration from mutualistic patches of plasmid-bearing genotypes that had acquired compensatory mutations.
4. Discussion
Using a tractable bacteria–plasmid model system, where the ecological plasmid population dynamics and the compensatory evolution dynamics can be jointly tracked in real-time, we show that migration stabilized plasmids under spatially heterogeneous positive selection by simultaneously increasing both the plasmid frequency and the supply of compensatory mutations. This adds to our understanding of the key role for compensatory evolution in plasmid stability, illustrating how ecological context can enhance this evolutionary process within heterogeneous environments. The likelihood of compensatory evolution, and thus plasmid survival, increases with the strength and frequency of positive selection [15], and, as shown here, with the rate of immigration from subpopulations experiencing positive selection. Spatial heterogeneity is widely thought to be a common feature of the environments bacterial communities inhabit across a wide range of ecological scales. Spatially structured environments, such as soils, are likely to contain heterogeneous microenvironments with localized patches of positive selection [21,22]. Indeed, positive selection for plasmid-encoded traits can vary at the µm scale, creating microscale population structure [23] that may be overlooked by less sensitive measurement approaches. Even low rates of migration in spatially heterogeneous selection landscapes can spread beneficial mutations from localized hotspots of positive selection to facilitate adaptation across the entire landscape [24–26]. At larger spatial scales, antibiotic use in hospitals and farms will create hotspots of positive selection for resistance plasmids, leading to higher plasmid frequencies and higher rates of compensatory evolution. Emigration of compensated plasmid-bearers from these environments, e.g. via waste-water systems [27], spreads not just the antibiotic resistance genes carried by the plasmid, but also bacterial lineages able to maintain plasmids in the absence of antibiotics with minimal fitness cost. By acting as plasmid ‘sources’ in their new communities, these lineages could maintain community-wide access to the mobile gene pool [28]. Thus, the joint eco-evolutionary effects of migration on plasmid frequency and compensatory evolution could help to explain why resistance plasmids are so commonly isolated from uncontaminated environments [29].
Our work has shown that compensatory mutations arise rapidly and have the potential to spread widely. We have previously shown that compensatory evolution is more likely to evolve in environments where plasmids are under positive selection [15]. Here we extend this to show that the invasion of compensatory evolution need not be limited by the prevailing local environment if migration increases the supply of compensatory mutations. Within our experimental system compensatory mutations occur at relatively high frequency as the gacA/gacS loci are known to have an elevated mutation rate [30]. Thus, because compensatory mutations arose in all focal patches, an effect of migration on plasmid frequency could not be detected at the lowest migration rate. The frequency of compensated plasmid-carrying genotypes was, however, significantly increased by higher migration rates in environments with heterogeneous positive selection. For alternative mechanisms of compensation with lower mutation rates we would expect even low rates of migration to enhance the spread of compensatory mutations.
However, the success of plasmid-bearing emigrants in new environments may be limited by context-dependent effects of the compensatory mutations themselves. For example, compensatory mutations targeting the GacA/S global regulatory system prevent expression of a large set of bacterial secreted proteins which are important for competitive interactions with other microbes [31,32], protection from predators [33] and virulence against eukaryotic hosts [34]. In extreme cases, compensatory mutations can be costly in the absence of the plasmid even in the environment where they evolved [35]. Similarly, some compensatory mutations are beneficial only in the absence of positive selection [36,37], for example where the cost of the plasmid is linked to the expression of its beneficial trait [38]; under this scenario the effects of migration on the spread of compensatory mutations may be limited. Thus, pleiotropic effects of compensatory mutations may lead to compensated emigrants being at a disadvantage in their new environment, limiting their dissemination. Additionally, compensatory evolution could effectively ‘lock’ bacteria—by reducing the strength of purifying selection—into associations with plasmids that are not beneficial under local environmental conditions, a scenario akin to symbiont addiction [39]. This could be detrimental to the lineage's long-term evolvability, because it would prevent acquisition of alternative plasmids from the same incompatibility group [40], limiting access to the mobile gene pool.
Plasmids are the principal mobile genetic elements driving horizontal gene transfer in bacterial communities and, thus, plasmid stability is an important determinant of bacterial evolution. Environments without positive selection for plasmid-encoded functions have a greater degree of plasmid horizontal transmission [41] and of interspecific gene mobilization [42]. Thus, by boosting plasmid residence times in these environments through jointly increasing both the frequency of plasmids and the supply of compensatory mutations, migration could enhance rates of horizontal gene transfer in bacterial communities.
Supplementary Material
Supplementary Material
Data accessibility
The data supporting this article are available in the electronic supplementary material, table S1.
Authors' contributions
E.H., J.P.J.H. and M.A.B. conceived the experiment. E.H. conducted the experiment, analysis and wrote the first draft. E.H., J.P.J.H. and M.A.B. contributed substantially to the discussion, writing and revisions of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by funding from the European Research Council awarded to M.A.B. (StG-2012-311490-COEVOCON), a Leverhulme Prize from the Leverhulme Trust awarded to M.A.B. (PLP-2014-242) and a NERC Research Fellowship awarded to E.H. (NE/P017584/1).
References
- 1.Norman A, Hansen LH, Sørensen SJ. 2009. Conjugative plasmids: vessels of the communal gene pool. Phil. Trans. R. Soc. B 364, 2275–2289. ( 10.1098/rstb.2009.0037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304. ( 10.1038/35012500) [DOI] [PubMed] [Google Scholar]
- 3.Baltrus DA. 2013. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489–495. ( 10.1016/j.tree.2013.04.002) [DOI] [PubMed] [Google Scholar]
- 4.San Millan A, MacLean RC. 2017. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol Spectr. 5, MTBP 0016-2017 ( 10.1128/microbiolspec.MTBP-0016-2017) [DOI] [PubMed] [Google Scholar]
- 5.Harrison E, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. 2015. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 25, 2034–2039. ( 10.1016/j.cub.2015.06.024) [DOI] [PubMed] [Google Scholar]
- 6.Hall JPJ, Harrison E, Lilley AK, Paterson S, Spiers AJ, Brockhurst MA. 2015. Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects. Environ. Microbiol. 17, 5008–5022. ( 10.1111/1462-2920.12901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison E, Brockhurst MA. 2012. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 20, 262–267. ( 10.1016/j.tim.2012.04.003) [DOI] [PubMed] [Google Scholar]
- 8.Dahlberg C, Chao L. 2003. Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics 165, 1641–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Gelder L, Williams JJ, Ponciano JM, Sota M, Top EM. 2008. Adaptive plasmid evolution results in host-range expansion of a broad-host-range plasmid. Genetics 178, 2179–2190. ( 10.1534/genetics.107.084475) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dionisio F, Conceição IC, Marques ACR, Fernandes L, Gordo I. 2005. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 1, 250–252. ( 10.1098/rsbl.2004.0275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heuer H, Fox RE, Top EM. 2006. Frequent conjugative transfer accelerates adaptation of a broad-host-range plasmid to an unfavorable Pseudomonas putida host: adaptation of a plasmid to an unfavorable host. FEMS Microbiol. Ecol. 59, 738–748. ( 10.1111/j.1574-6941.2006.00223.x) [DOI] [PubMed] [Google Scholar]
- 12.Porse A, Schønning K, Munck C, Sommer MOA. 2016. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol. Biol. Evol. 33, 2860–2873. ( 10.1093/molbev/msw163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.San Millan A, Peña-Miller R, Toll-Riera M, Halbert ZV, McLean AR, Cooper BS, MacLean RC. 2014. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 5, 5208 ( 10.1038/ncomms6208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yano H, Wegrzyn K, Loftie-Eaton W, Johnson J, Deckert GE, Rogers LM, Konieczny I, Top EM. 2016. Evolved plasmid-host interactions reduce plasmid interference cost. Mol. Microbiol. 101, 743–756. ( 10.1111/mmi.13407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison E, Dytham C, Hall JPJ, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. 2016. Rapid compensatory evolution promotes the survival of conjugative plasmids. Mob. Genet. Elements 6, e1179074 ( 10.1080/2159256X.2016.1179074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey MJ, Lilley AK, Thompson IP, Rainey PB, Ellis RJ. 1995. Site directed chromosomal marking of a fluorescent pseudomonad isolated from the phytosphere of sugar beet; stability and potential for marker gene transfer. Mol. Ecol. 4, 755–763. ( 10.1111/j.1365-294X.1995.tb00276.x) [DOI] [PubMed] [Google Scholar]
- 17.Tett A, et al. 2007. Sequence-based analysis of pQBR103; a representative of a unique, transfer-proficient mega plasmid resident in the microbial community of sugar beet. ISME J. 1, 331–340. ( 10.1038/ismej.2007.47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevenson C, Hall JPJ, Brockhurst MA, Harrison E. 2018. Plasmid stability is enhanced by higher-frequency pulses of positive selection. Proc. R. Soc. B 285, 20172497 ( 10.1098/rspb.2017.2497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simonsen L, Gordon DM, Stewart FM, Levin BR. 1990. Estimating the rate of plasmid transfer: an end-point method. J. Gen. Microbiol. 136, 2319–2325. ( 10.1099/00221287-136-11-2319) [DOI] [PubMed] [Google Scholar]
- 20.Cheng X, de Bruijn I, van der Voort M, Loper JE, Raaijmakers JM. 2013. The Gac regulon of Pseudomonas fluorescens SBW25. Environ. Microbiol. Rep. 5, 608–619. ( 10.1111/1758-2229.12061) [DOI] [PubMed] [Google Scholar]
- 21.Vos M, Wolf AB, Jennings SJ, Kowalchuk GA. 2013. Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol. Rev. 37, 936–954. ( 10.1111/1574-6976.12023) [DOI] [PubMed] [Google Scholar]
- 22.Becker JM, Parkin T, Nakatsu CH, Wilbur JD, Konopka A. 2006. Bacterial activity, community structure, and centimeter-scale spatial heterogeneity in contaminated soil. Microb. Ecol. 51, 220–231. ( 10.1007/s00248-005-0002-9) [DOI] [PubMed] [Google Scholar]
- 23.Slater FR, Bruce KD, Ellis RJ, Lilley AK, Turner SL. 2010. Determining the effects of a spatially heterogeneous selection pressure on bacterial population structure at the sub-millimetre scale. Microb. Ecol. 60, 873–884. ( 10.1007/s00248-010-9687-5) [DOI] [PubMed] [Google Scholar]
- 24.Bell T. 2010. Experimental tests of the bacterial distance–decay relationship. ISME J. 4, 1357–1365. ( 10.1038/ismej.2010.77) [DOI] [PubMed] [Google Scholar]
- 25.Franklin RB, Mills AL. 2003. Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol. Ecol. 44, 335–346. ( 10.1016/S0168-6496(03)00074-6) [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung C-K, Pourmand N, Austin RH. 2011. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333, 1764–1767. ( 10.1126/science.1208747) [DOI] [PubMed] [Google Scholar]
- 27.Kotay S, Chai W, Guilford W, Barry K, Mathers AJ. 2017. Spread from the sink to the patient: in situ study using green fluorescent protein (GFP)-expressing Escherichia coli to model bacterial dispersion from hand-washing sink-trap reservoirs. Appl. Environ. Microbiol. 83, e03327-16 ( 10.1128/AEM.03327-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall JPJ, Wood AJ, Harrison E, Brockhurst MA. 2016. Source-sink plasmid transfer dynamics maintain gene mobility in soil bacterial communities. Proc. Natl Acad. Sci. USA 113, 8260–8265. ( 10.1073/pnas.1600974113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lilley AK, Bailey MJ, Day MJ, Fry JC. 1996. Diversity of mercury resistance plasmids obtained by exogenous isolation from the bacteria of sugar beet in three successive years. FEMS Microbiol. Ecol. 20, 211–227. ( 10.1111/j.1574-6941.1996.tb00320.x) [DOI] [Google Scholar]
- 30.van den Broek D, Chin-A-Woeng TFC, Bloemberg GV, Lugtenberg BJJ. 2005. Molecular nature of spontaneous modifications in gacS which cause colony phase variation in Pseudomonas sp. strain PCL1171. J. Bacteriol. 187, 593–600. ( 10.1128/JB.187.2.593-600.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berry CL, Nandi M, Manuel J, Brassinga AKC, Fernando WGD, Loewen PC, de Kievit TR. 2014. Characterization of the Pseudomonas sp. DF41 quorum sensing locus and its role in fungal antagonism. Biol. Control 69, 82–89. ( 10.1016/j.biocontrol.2013.11.005) [DOI] [Google Scholar]
- 32.Koch B, Nielsen TH, Sørensen D, Andersen JB, Christophersen C, Molin S, Givskov M, Sørensen J, Nybroe O. 2002. Lipopeptide production in Pseudomonas sp. strain DSS73 is regulated by components of sugar beet seed exudate via the Gac two-component regulatory system. Appl. Environ. Microbiol. 68, 4509–4516. ( 10.1128/AEM.68.9.4509-4516.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bjørnlund L, Rønn R, Péchy-Tarr M, Maurhofer M, Keel C, Nybroe O. 2009. Functional GacS in Pseudomonas DSS73 prevents digestion by Caenorhabditis elegans and protects the nematode from killer flagellates. ISME J. 3, 770–779. ( 10.1038/ismej.2009.28) [DOI] [PubMed] [Google Scholar]
- 34.Cha JY, Lee DG, Lee JS, Oh J-I, Baik HS. 2012. GacA directly regulates expression of several virulence genes in Pseudomonas syringae pv. tabaci 11528. Biochem. Biophys. Res. Commun. 417, 665–672. ( 10.1016/j.bbrc.2011.11.124) [DOI] [PubMed] [Google Scholar]
- 35.Bouma JE, Lenski RE. 1988. Evolution of a bacteria plamid association. Nature 335, 351–352. ( 10.1038/335351a0) [DOI] [PubMed] [Google Scholar]
- 36.Paulander W, Maisnier-Patin S, Andersson DI. 2007. Multiple mechanisms to ameliorate the fitness burden of mupirocin resistance in Salmonella typhimurium. Mol. Microbiol. 64, 1038–1048. ( 10.1111/j.1365-2958.2007.05713.x) [DOI] [PubMed] [Google Scholar]
- 37.Schulz zur Wiesch P, Engelstadter J, Bonhoeffer S. 2010. Compensation of fitness costs and reversibility of antibiotic resistance mutations. Antimicrob. Agents Chemother. 54, 2085–2095. ( 10.1128/AAC.01460-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bottery MJ, Wood AJ, Brockhurst MA. 2017. Adaptive modulation of antibiotic resistance through intragenomic coevolution. Nat. Ecol. Evol. 1, 1364–1369. ( 10.1038/s41559-017-0242-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aanen DK, Hoekstra RF. 2007. The evolution of obligate mutualism: if you can't beat 'em, join 'em. Trends Ecol. Evol. 22, 506–509. ( 10.1016/j.tree.2007.08.007) [DOI] [PubMed] [Google Scholar]
- 40.Novick RP. 1987. Plasmid incompatibility. Microbiol. Rev. 51, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevenson C, Hall JP, Harrison E, Wood AJ, Brockhurst MA. 2017. Gene mobility promotes the spread of resistance in bacterial populations. ISME J. 11, 1930–1932. ( 10.1038/ismej.2017.42) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall JPJ, Williams D, Paterson S, Harrison E, Brockhurst MA. 2017. Positive selection inhibits gene mobilisation and transfer in soil bacterial communities. Nat. Ecol. Evol. 1, 1348–1353. ( 10.1038/s41559-017-0250-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this article are available in the electronic supplementary material, table S1.