Abstract
With ongoing global change, shifts in the ranges of non-native species and resulting novel communities can modify biotic interactions and ecosystem processes. We hypothesized that traits and not biogeographic origin of novel plant communities will determine community structure of organisms that depend on plants for habitat or as a food resource. We tested the functional redundancy of novel tree communities by verifying if six pairs of congeneric European and North American tree species bearing similar leaf litter traits resulted in similar ecological filters influencing the assembly of springtail (Collembola) communities at two sites. Litter biogeographic origin (native versus non-native) did not influence springtail community structure, but litter genus, which generally reflected trait differences, did. Our empirical evidence suggests that a functional trait approach may be indeed as relevant as, and complementary to, studying biogeographic origin to understand the ecological consequences of non-native tree species in soils of novel forest ecosystems.
Keywords: biodiversity, exotic species, functional diversity, IDENT, mesofauna, Collembola
1. Introduction
With global change due to anthropogenic activities, historical distributions of species are changing faster than ever through range shifts and new introductions [1]. Deliberate introductions such as forest plantations, but also unintentional ones, can reshape species combinations and ecosystem properties leading to novel ecosystems [2]. The novel ecosystem concept suggests that management decisions should be based on functions conferred by species in altered ecosystems, regardless of their biogeographic origin [3]. This concept has been hotly debated in environmental conservation, partly because critics and proponents disagree on strategies regarding non-native species management (e.g. [4,5]). There is however convergence that biogeographic origin should not be the only criterion for management options of non-native species, because the relevance of their impact depends on the stage along the introduction–naturalization–invasion–impact continuum [6]. However surprisingly, few empirical studies so far have attempted to dissociate origin effects from other criteria, despite the actual debate.
Studies focusing on non-native species generally contain multiple confounding factors, making it challenging to isolate the sole influence of biogeographic origin on ecosystems [7]. The differential performance of introduced species may, in fact, depend on characteristics of the source and/or recipient environment and on traits of the introduced and/or recipient community species [7]. One way to isolate these factors would be to compare the influence of congeneric species with similar traits, but with different biogeographic origin, in the same recipient environment [8]. Experimental approaches with congeneric pairs have been used in invasion ecology, but functional trait effects are rarely controlled, despite being key to understanding non-native species invasiveness (e.g. [9]). Functional traits have been assumed to be similar within a genus (e.g. [10]), although trait variation within genera and even within species can be extensive for plants [11] and driven by the environmental context [12]. Studies using a functional approach are also generally limited to a single trophic level (i.e. plants), strongly limiting our understanding of emerging properties of novel ecosystems. Some experimental approaches have gone beyond taxonomically controlled experiments to study origin effects within a multi-trophic context (e.g. leaf herbivore consumption on tree congeneric species with similar functional traits [13]). However, empirical evidence derived from such designs remain rare and little is known about how non-native plant communities might influence other trophic levels from a bottom-up perspective, particularly within soils of novel forest ecosystems.
Forest soils, including the leaf litter layer, provide habitat to a diverse community of fauna that play an important role in litter decomposition, soil structuration and biogeochemical cycling [14–16]. Springtails (Collembola) are small and particularly abundant invertebrates that respond quickly to environmental change, including forest management, making them ideal study organisms [14]. Leaf litter can influence springtail community structure through abiotic filters (by influencing their physical habitat [17]) and resource-driven effects [14]. Such effects can be related to particular leaf litter traits such as C/N ratio and leaf thickness [18,19]. To date, studies on how springtail communities respond to non-native tree litters have focused primarily on understanding whether litter from Eucalyptus plantations were changing springtail communities compared to native Quercus-dominated forests [20] or grasslands [21]. Other studies have rather focused on how novel tree species might influence litter decomposition [22,23]. For example, physico-chemical litter traits of native (Alnus sp. and Populus sp.) contrasted to non-native tree species (Acacia sp. and Eucalyptus sp.) had a stronger effect on springtail-mediated decomposition than biogeographic origin of species [22]. However, litters of non-native and native tree species were functionally different (e.g. polyphenol content) making it impossible to isolate origin effects. Similarly, Makkonen et al. [23] demonstrated in a global reciprocal transplant experiment that litter decomposition by mesofauna was quality- and not origin-driven across four different biomes (subarctic, temperate, Mediterranean or tropical). Yet again, it was difficult to dissociate origin from biome-specific traits.
The aim of our study was to isolate origin effects by providing a first test of whether tree biogeographic origin influenced springtail community structure when comparing functionally redundant congeneric tree species (i.e. bearing similar litter functional traits) in a common recipient environment that was fully replicated at two distinct sites. We hypothesized that litter genus will influence springtail community structure more than tree biogeographic origin, as traits of litters will more likely influence key springtail distribution factors (e.g. resource quality and microhabitat) than biogeographic origin on its own.
2. Material and methods
(a). Experimental site and design
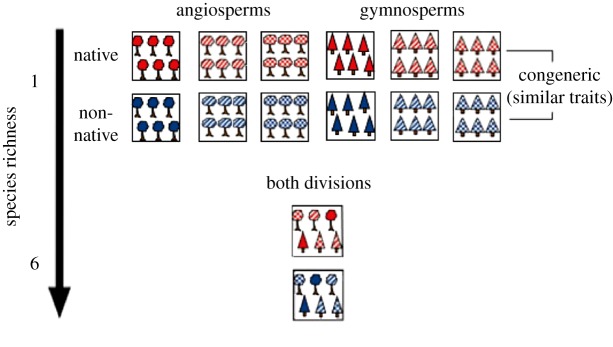
The study was conducted in the AuCl experiment of the International Diversity Experiment Network with Trees (IDENT), a network of Biodiversity – Ecosystem Functioning experiments in North America and Europe [8]. The two study sites, which were planted at the same time with trees grown from the same nurseries, were located in Auclair (Au), Québec, Canada (47.7° N, 68.6° W) and Cloquet (Cl), Minnesota, USA (46.7° N, 92.5° W), respectively at an altitude of 333 m and 383 m. These high-density tree experiments were established in 2010 on 0.34 ha low-input abandoned pasture with loam soil (Au) or a previously forested site with sandy-loam soil (Cl) [24]. Further site description including climate and soil variables on the AuCl experiment are provided in appendix S1 in the electronic supplementary material. With the aim of allowing for multi-site studies, both sites were identically designed with one, two, and six-species treatments for a total of 48 different tree communities randomized in four blocks. Plots were distanced from each other by 1 m and consisted of 49 trees (7 × 7 trees with 40 cm intervals). Species pool of the AuCl experiment was composed of 6 North American temperate tree species (Acer saccharum, Betula papyrifera, Larix laricina, Quercus rubra, Picea glauca, Pinus strobus) and 6 congeneric European temperate tree species (A. platanoides, B. pendula, L. decidua, Q. robur, P. abies, P. sylvestris). Here, North American species are considered as native and European species as non-native. Based on wood density, seed mass and leaf N content, native and non-native congeneric species were functionally more similar than non-congeneric species, thereby allowing the use of tree genus as a proxy for tree functional identity [8]. For this study, we subsampled a subset of plots including all 12 single-species treatments of native and non-native trees (figure 1). Additionally, the design allowed us to explore diversity effects by sampling two six-species mixtures, one composed of native species and the other one of non-native species (figure 1). These 14 treatments were replicated four times at each site for a total of 112 sampled plots.
Figure 1.
Schematic representation of selected treatments in Auclair and Cloquet (AuCl) experimental design (adapted from Tobner et al. [8] with permission). Shading pattern corresponds to functional traits and colour shows biogeographic origin.
(b). Litter sampling and litter trait measurements
Considering that the original experimental design was based on traits of the living trees, functional traits were measured on freshly senesced leaf litter to verify that congeneric litters were indeed more similar than non-congeneric litters. In October 2015, freshly senesced leaves were collected on single-species plots at both sites and on at least five random trees per plot after gentle shaking of the trunks. Senesced evergreen needles were collected throughout the field season in June, July, August and October 2015. All leaves per species per site were then pooled together and mixed homogeneously before trait measurements: leaf water saturation capacity (% H2O dry matter), leaf thickness (mm), leaf resistance to fracture (g mm−2), N concentration (% d.m.), cellulose, hemicellulose and lignin concentrations (% organic matter), C/N and lignin/N ratios. For microbial basal respiration rate (μg CO2-C g−1 h−1), litter was manually collected on the forest floor at five random positions per plot and air-dried until analysed in a microplate-based respiration system (MicroResp) following the colorimetric detection protocol [25]. Here, we consider the mean litter microbial basal respiration rate per species (across blocks) as a litter species functional trait. Details about trait measurements are provided in appendix S2.1 in the electronic supplementary material.
(c). Environmental variable measurements
Soil moisture measurements were taken at each plot centre during springtail sampling with a FieldScout TDR 300 moisture meter (n = 3); additional measurements for single-species plots at both sites were taken in June 2015 and October 2015. Litter layer mean height (to the nearest mm) was measured prior to springtail sampling with a ruler from the top of the soil to the top of the litter layer for each subsample. In order to assess if there was any difference in soil microbial resource abundance across treatments within an experimental site, soil microbial biomass (μg microbial-C g−1) was determined for Auclair plots. During springtail sampling, a superficial layer of soil (0–2 cm) was collected in a sterile tube at three random positions per plot and then frozen at −20°C. Soils were adjusted to 45% of their water holding capacity and pre-incubated for 7 days at 25°C before MicroResp analyses with glucose addition [25]. Resulting glucose-induced respiration rates were converted into microbial biomasses according to Anderson & Domsch [26].
(d). Springtail sampling
Springtails were sampled in late July (Au) and early August (Cl) 2015. For each plot, two subsamples were randomly collected at its centre to minimize any effect of neighbouring plots, yielding a total of 224 subsamples. At each subsample site, litter was collected manually in a 15 × 15 cm quadrat. At the centre of the quadrat, soil was extracted with a split soil core sampler (diameter 5.08 cm × depth 5 cm). Both litter and soil were transferred in hermetic containers and kept at 4°C for transport to the laboratory for a maximum of 48 h. Litter and soil subsamples were then pooled together prior to Tullgren (Au) or Kempson (Cl) extraction [27], during which the temperature was gradually increased during 7 days (20°C to 50°C) and fauna collected in 70% ethanol. Springtails were cleared in lactic acid at 60°C to better see structures needed for identification (e.g. chaetotaxy and post-antennal organ). Patterns and colouration were carefully noted before this step. Identification to the species level was done with a Leica DM1000 LED phase contrast microscope (800×) by using the keys of Christiansen & Bellinger [28] and Hopkin [29]. Bellinger et al. [30] was used as complementary information to these keys. Cleared specimens were all slide-mounted in Hoyer's medium (50 ml distilled water—30 g gum arabic—200 g chloral hydrate—20 ml glycerol). Damaged specimens were identified only to the family or genus level (less than 3%) and excluded in further analyses. Exclusions included six samples for which springtail total abundance equalled zero: B. pendula (block A), Q. robur (block B), P. glauca (block B) and a non-native six-species mixture (block D) in Auclair, and A. saccharum (block A) and P. abies (block D) in Cloquet. Given the small size of Megalothorax minimus (less than 0.4 mm), this species was also excluded from the analyses due to possible sampling bias as some individuals may not have been detected.
(e). Statistical analyses
Variation of environmental variables depending on study site, litter genus and litter origin was analysed with ANOVAs and post hoc Tukey Honest Significant Difference (HSD) test (normality confirmed with Shapiro–Wilk test). The variation of litter traits and springtail species among and across congeneric tree species was visualized with principal component analysis ordinations (PCA) after trait data standardization and species abundance Hellinger transformation [31]. Prior to variance analyses, springtail species abundances were transformed in Hellinger distance [32]. PERMANOVAs nested by blocks were then used to test the effect of litter genus, litter biogeographic origin, study site and their interactions on Hellinger distance of springtail species abundances by site and across sites (9999 permutations) [33]. We ensured that homogeneity of variance was respected with a multivariate analogue of Levene's test. To explore association between litter traits and springtail species matrices, procrustes analyses were used [34]. As we did not have all trait measurements for six-species mixtures (e.g. microbial biomass), procrustes analyses included monocultures only. Considering the low number of shared species and high intraspecific trait variation of litters between sites, we decided to execute procrustes analyses on each site separately.
All statistical data analyses were performed in R (v. 3.2.1) (R Development Core Team, Vienna, Austria) with RStudio environment (v. 0.99.903) (RStudio Inc., Boston, USA). Vegan package (v. 2.4-0) by Oksanen et al. [35] was used for Hellinger transformation/distance (decostand/vegdist), litter trait standardization (decostand), PCAs (rda), homogeneity of variance test (betadisper), normality test (shapiro.test), ANOVAs (aov/TukeyHSD), PERMANOVA model (adonis) and procrustes analyses (protest).
3. Results
(a). Litter trait variation of congeneric tree species
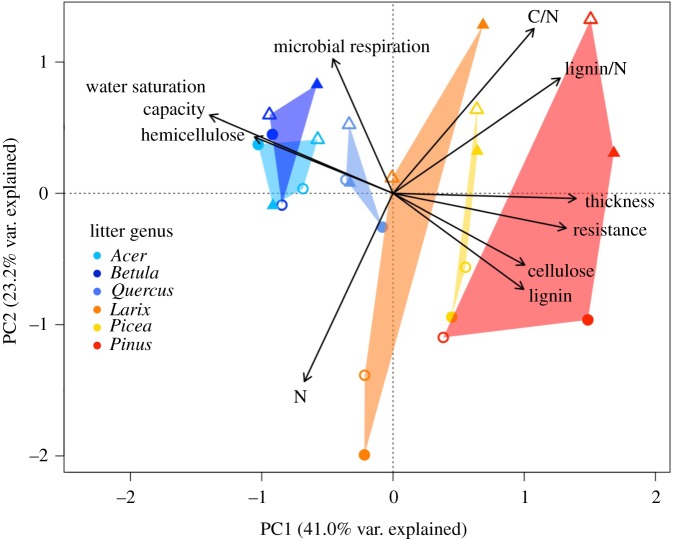
Congeneric species generally had similar leaf litter traits, but also showed substantial intraspecific variation between sites with the first two PCA axes explaining 63.2% litter trait score variation (figure 2). The first axis (41.0%) explained the trait variation due to genera along a deciduous-evergreen gradient, while the second axis explained intraspecific trait variation between sites (23.2%; figure 2, appendix S2.2 in the electronic supplementary material for all specific trait values). For example, mean N concentration was higher at the Auclair (1.25 ± 0.24% d.m.) than Cloquet site (0.82 ± 0.17% d.m.), and conversely, mean microbial respiration was lower at the Auclair (27.43 ± 5.24 µg CO2-C g−1 h−1) than Cloquet site (61.26 ± 7.88 µg CO2-C g−1 h−1).
Figure 2.
Principal component analysis (PCA) ordination of centred and standardized litter traits (arrows) arranged by study site (circle: Auclair/triangle: Cloquet), litter genus (coloured polygon) and origin (open: native/closed: non-native).
(b). Environmental variables
Soil moisture was almost double at the Auclair (19.6 ± 1.7% H2O d.m.) compared with the Cloquet (10.4 ± 1.5% H2O d.m.) site (p < 0.01; see appendix S1 in the electronic supplementary material), which was consistent with respective differences in soil texture (loam versus sandy-loam). At the time of sampling, trees at Auclair were bigger than at Cloquet, despite their identical age (field observations). Some treatments also had a particularly thin litter layer (mean height < 1 cm) (Auclair: Betula sp./Cloquet: Acer sp. and Quercus sp.; see appendix S1.2 in the electronic supplementary material). Soil microbial biomass, measured only in Auclair, did not vary by litter genus, origin or origin × genus interaction.
(c). Springtail communities and tree treatment effects
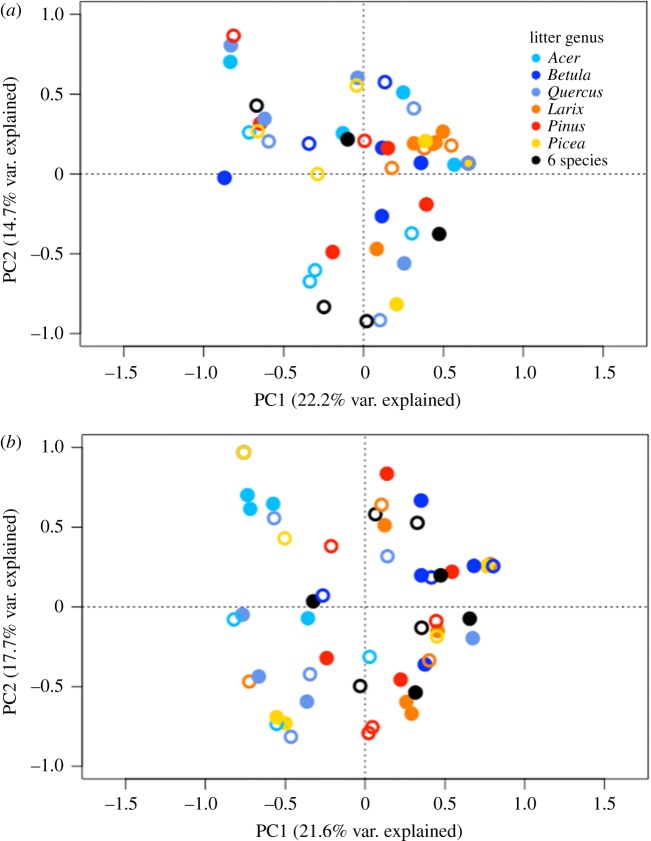
Total richness of springtail communities across sites was 57 species, with 34 species sampled at Cloquet (n = 758) and 37 species sampled at Auclair (n = 1073; see appendix S3 in the electronic supplementary material). Litter biogeographic origin had no influence on springtail community structure (no significant litter origin × genus interaction; table 1, figure 3). Study site had the strongest influence on springtail community structure (R2 = 0.21), followed by litter genus (R2 = 0.07) (table 1). Additionally, these factors interacted significantly with each other (R2 = 0.07) indicating that the influence of litter genus on springtail community structure was different at both sites (table 1 and figure 3). This observation was concordant with the intraspecific trait variation observed between sites (figure 2). Indeed, distinct PERMANOVAs for each site on Hellinger distance matrices (9999 permutations) showed a stronger litter genus influence in Cloquet (R2 = 0.19, p < 0.001) than in Auclair (R2 = 0.15, p < 0.05). When testing the direct relationship between litter traits and springtail community structure, the procrustes analyses demonstrated a significant 0.4 correlation at Cloquet (p < 0.01; see appendix S4 in the electronic supplementary material). This relationship was, however, not significant at Auclair (0.32 correlation, p = 0.34). These observations highlighted similar patterns as those observed when testing treatment effects with our PERMANOVA models: tree genus influenced springtail community structure at both sites, but not necessarily through litter traits. Springtail communities of single- and six-species treatments answered similarly to the factors included in the model (see black versus coloured dots in figure 3).
Table 1.
PERMANOVA of the Hellinger distance for AuCl springtail community composition in relation to study site (Site), litter origin (Origin), litter genus (Genus) and their interactions (9999 permutations, nested by block). d.f., degrees of freedom; SS, sum of squares; MS, mean sum of squares; F, F value by permutation. Treatments included in analyses were single- and six-species mixtures. ***p < 0.001, **p < 0.01, *p < 0.05, n.s., not significant.
| factor | d.f. | SS | MS | F | R2 | p |
|---|---|---|---|---|---|---|
| Genus | 6 | 5.54 | 0.92 | 1.55 | 0.066 | ** |
| Origin | 1 | 0.44 | 0.44 | 0.74 | 0.005 | n.s. |
| Site | 1 | 17.44 | 17.44 | 29.31 | 0.209 | *** |
| Site × Genus | 6 | 5.74 | 0.96 | 1.61 | 0.069 | ** |
| Origin × Genus | 6 | 3.89 | 0.65 | 1.09 | 0.047 | n.s. |
| Origin × Site | 1 | 0.53 | 0.53 | 0.89 | 0.006 | n.s. |
| Origin × Site × Genus | 6 | 3.65 | 0.61 | 1.02 | 0.044 | n.s. |
| Residuals | 78 | 46.42 | 0.60 | 0.555 |
Figure 3.
Principal component analysis (PCA) ordination of springtail species abundances of (a) Auclair and (b) Cloquet arranged by litter genus (colour) and origin (open: native/closed: non-native). Hellinger transformation was applied to data before analyses.
4. Discussion
Our study explores a new dimension of novel forest ecosystems by looking at how tree litter might modify the community structure of a dominant soil invertebrate taxa. Across all 14 tested tree community types, as hypothesized, biogeographic origin of the leaf litter did not significantly influence springtail community structure, in contrast to litter genus (table 1 and figure 3), which we considered here as a proxy for leaf litter traits (figure 2). While congeneric tree species generally had more similar litter traits, we acknowledge that trait variation was particularly large for two of our 12 congeneric pairs (Larix in Cloquet and Pinus in Auclair; figure 2). Such intra-genus trait variation further emphasizes the necessity to go beyond strict taxonomically controlled experiments to isolate origin effects. Interestingly, the similarity of how springtail communities of one- and six-species mixtures varied according to the studied factors (table 1 and figure 3), suggests additive effects of the litter species when comparing genus and biogeographic origin effects. Despite a lower number of six-species replicates in the experiment, this observation suggests that one could expect similar results for single-species plantations and multi-species forests, reinforcing the above conclusions for real-life scenarios.
Several previous studies on invertebrate herbivores [13,36,37] or on fauna-mediated litter decomposition [38] have shown similar results to those reported here. Species or genus effects were more important than biogeographic origin to predict feeding of invertebrate herbivores [13,37]. Similarly, Connor et al. [36] demonstrated that leaf-mining insects can switch resources if an introduced plant species is closely related to its native resource (e.g. congeners), regardless of biogeographic origin. While these studies demonstrated that resource consumption is not affected by leaf origin, our results show that even potential consumer community structure (i.e. springtails) is not influenced by litter biogeographic origin. We recognize that the arrival of an invasive, non-native tree species in a novel forest ecosystem may change this result. However, any potential effect of a non-native tree species (invasive or not) on this soil invertebrate community will likely be mediated by traits of the tree, and not be a consequence of simply biogeographic origin.
In addition to observing indirect litter trait effects through similar congeneric species, we also examined the direct correlation of litter traits with springtail community structure (appendix S4 in the electronic supplementary material). Our results showed that despite a significant litter genus effect at both sites, leaf litter traits covaried significantly with springtail community structure only at the Cloquet, but not at the Auclair site. This contrasting result might be the consequence of different resource utilization due to site-related soil conditions [39]. For example, springtail diet at the Auclair site might have been, contrary to the traditional perception, more root- than litter-driven [39,40]. In contrast, the less favourable environment in Cloquet, with drier and sandier soil, might have strengthened the influence of litter resources on its springtail communities. If resource filters were indeed different at both sites, the site-dependent genus effect (significant site × genus interaction) may reflect the reality of root traits not necessarily being coordinated with leaf traits [41]. Moreover, by using traits of decomposed litter, covariation with springtails could have been stronger than with freshly senescent litter, as they are secondary decomposers [14]. Nonetheless, we acknowledge that this tree genus influence could either be associated with abiotic- and/or resource-driven effects as the experimental design did not allow us to distinguish their respective contributions.
Despite the identical experimental design at both sites including source trees from common tree nurseries, site-specific effects may have been due to intraspecific variation in some litter traits (N and microbial respiration; figure 2), different environmental conditions (appendix S1 in the electronic supplementary material) and/or different local species pools (appendix S3 in the electronic supplementary material) influencing springtail community structure. Considering that these communities are shaped from local species pools, each with numerous abiotic and biotic filters, the low proportion of variance explained by litter genus (thus traits) in comparison to study site (table 1) is therefore not surprising. Interestingly, the variance explained by litter genus within each site was more than two times higher than across site which highlights the importance of context dependency [42]. While local species pools varied, we observed a consistent response of springtail communities to litter treatments across sites reaffirming the strength of ‘filtering effects’ despite the context dependency. Within each local species pool, the proportion of springtails with differing vertical habitat preference (epigeic, hemiedaphic or eudaphic; [14]) may vary possibly influencing the correlation between litter and springtail community structure. However, the most abundant species at both our sites were different, yet within the Lepidocyrtus genus (which generally regroups drought-tolerant epigeic species; [43]), suggesting that it is unlikely to have contributed to the observed site-specific effect. One final source of variation between sites may have been methodological, as we were constrained by the use of slightly different extraction methods for Auclair (Tullgren) and Cloquet (Kempson) that might have influenced the number of total individuals collected in each site. However, given that the most efficient extraction method (i.e. Kempson, due to its capacity to maintain better gradients of humidity and temperature through the sample; [27]) resulted in lower total abundance at Cloquet, it is unlikely that this contributed to much variation.
5. Conclusion
Our study on springtail communities in soils of dense, young plantations of native and non-native tree communities showed that community assembly was not driven by biogeographic origin of litter resources, but partially driven by genus-related leaf litter traits. Such empirical evidence studied here in novel forest ecosystems emphasizes the importance of focusing on the function of species, instead of origin only for management decisions. Still, considering the scarce number of studies on non-native species trying to correct biases related to species traits and environment, further efforts to understand empirically the full range of biogeographic origin effects are needed. Such efforts might include a similar experimental design but in an array of different scenarios (e.g. longer time sequence study, influence on vertebrates, specialist versus generalist consumers, top-down versus bottom-up effects of non-native species, different mixed tree species combinations, etc.). Further efforts in multi-trophic functional studies to better understand trait-matching between resource (e.g. litter and roots) and consumers (e.g. mouthparts) will also be important to better predict responses of soil communities and their role in key ecosystem functions as plant communities change. If strict taxonomy (as highlighted by Agrawal et al. [9]) and origin are indeed poor predictors to determine the influence of novel species on ecosystems, we encourage researchers to consider potential trait effects when comparing the influence of native and non-native species on ecosystem functioning and/or community assembly. The management of non-native species remains a sensitive subject that extends beyond pure biological reasoning, but nonetheless, we need collectively to advance our empirical understanding of potential ecological impacts of novel species and how such impacts may depend on environmental context. To this extent, multi-site experimental biodiversity studies such as IDENT allow not only to isolate origin effects, but to identify context-dependency, that may induce modification of community assembly filters and intraspecific variation. Such variation will undoubtedly allow us to better understand how changing biodiversity, including non-native species, will affect novel ecosystem functioning in a range of real-world scenarios.
Supplementary Material
Supplementary Material
Acknowledgements
We are indebted to our colleagues in the IDENT network, particularly Alain Paquette and Christian Messier who established and coordinated the original experiments and have facilitated interactions among IDENT members since. We thank Artur Stefanski for field support and providing environmental data for the Cloquet site. We also thank Dominique Bélanger (C and N concentration analyses), Jonathan Brassard (pH), Peter Kennedy (pH), Laura Willliams (soil moisture) for site description data and the multiple undergraduate students who provided assistance with data collection. Alain Paquette and Jérôme Cortet provided thoughtful comments on an earlier version of the manuscript.
Data accessibility
Litter trait values and springtail abundances used in this study can be found in supplementary support (see appendices S2.2 and S5 in the electronic supplementary material).
Authors' contributions
L.J.R.-L. and I.T.H. conceived the study, developed the methodology, interpreted results and led the writing of the manuscript. L.J.R.-L. collected the data and performed analyses. D.G. and P.B.R. are local principal investigators, respectively, of the Auclair and Cloquet IDENT sites. They facilitated data acquisition logistics and contributed through critical review of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by NSERC Discovery and Canadian Foundation for Innovation Leader grants to I.T.H. and NSERC Collaborative Research and Training Experience program in Forest Complexity Modeling, NSERC Alexander Graham Bell and FQRNT graduate scholarships to L.J.R.-L.
References
- 1.Ricciardi A. 2007. Are modern biological invasions an unprecedented form of global change? Conserv. Biol. 21, 329–336. ( 10.1111/j.1523-1739.2006.00615.x) [DOI] [PubMed] [Google Scholar]
- 2.Hobbs RJ, et al. 2006. Novel ecosystems: theoretical and management aspects of the new ecological world order. Glob. Ecol. Biogeogr. 15, 1–7. ( 10.1111/j.1466-822X.2006.00212.x) [DOI] [Google Scholar]
- 3.Miller JR, Bestelmeyer BT. 2016. What's wrong with novel ecosystems, really? Restor. Ecol. 24, 577–582. ( 10.1111/rec.12378) [DOI] [Google Scholar]
- 4.Davis MA, et al. 2011. Don't judge species on their origins. Nature 474, 153–154. ( 10.1038/474153a) [DOI] [PubMed] [Google Scholar]
- 5.Simberloff D. 2011. Non-natives: 141 scientists object. Nature 475, 36 ( 10.1038/475036a) [DOI] [PubMed] [Google Scholar]
- 6.Larson BM, van der Wal R, Fischer A, Selge S. 2016. Origin might matter; people matter, too (a response to the comment by Rejmánek and Simberloff (2016)). Environ. Conserv. 44, 100–101. ( 10.1017/S0376892916000503) [DOI] [Google Scholar]
- 7.Buckley YM, Catford J. 2016. Does the biogeographic origin of species matter? Ecological effects of native and non-native species and the use of origin to guide management. J. Ecol. 104, 4–17. ( 10.1111/1365-2745.12501) [DOI] [Google Scholar]
- 8.Tobner CM, Paquette A, Reich PB, Gravel D, Messier C. 2014. Advancing biodiversity–ecosystem functioning science using high-density tree-based experiments over functional diversity gradients. Oecologia 174, 609–621. ( 10.1007/s00442-013-2815-4) [DOI] [PubMed] [Google Scholar]
- 9.Agrawal AA, Kotanen PM, Mitchell CE, Power AG, Godsoe W, Klironomos J. 2005. Enemy release? An experiment with congeneric plant pairs and diverse above- and belowground enemies. Ecology 86, 2979–2989. ( 10.1890/05-0219) [DOI] [Google Scholar]
- 10.Agrawal AA, Kotanen PM. 2003. Herbivores and the success of exotic plants: a phylogenetically controlled experiment. Ecol. Lett. 6, 712–715. ( 10.1046/j.1461-0248.2003.00498.x) [DOI] [Google Scholar]
- 11.Siefert A, et al. 2015. A global meta-analysis of the relative extent of intraspecific trait variation in plant communities. Ecol. Lett. 18, 1406–1419. ( 10.1111/ele.12508) [DOI] [PubMed] [Google Scholar]
- 12.Van Zandt PA. 2007. Plant defense, growth, and habitat: a comparative assessment of constitutive and induced resistance. Ecology 88, 1984–1993. ( 10.1890/06-1329.1) [DOI] [PubMed] [Google Scholar]
- 13.Wein A, Bauhus J, Bilodeau-Gauthier S, Scherer-Lorenzen M, Nock C, Staab M. 2016. Tree species richness promotes invertebrate herbivory on congeneric native and exotic tree saplings in a young diversity experiment. PLoS ONE 11, e0168751 ( 10.1371/journal.pone.0168751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopkin SP. 1997. Biology of the springtails (Insecta: Collembola). Oxford, UK: University Press. [Google Scholar]
- 15.Bardgett RD, van der Putten WH. 2014. Belowground biodiversity and ecosystem functioning. Nature 515, 505–511. ( 10.1038/nature13855) [DOI] [PubMed] [Google Scholar]
- 16.Handa IT, et al. 2014. Consequences of biodiversity loss for litter decomposition across biomes. Nature 509, 218–221. ( 10.1038/nature13247) [DOI] [PubMed] [Google Scholar]
- 17.Ponge JF, Arpin P, Vannier G. 1993. Collembolan response to experimental perturbations of litter supply in a temperate forest ecosystem. Eur. J. Soil Biol. 29, 141–153. [Google Scholar]
- 18.Ilieva-Makulec K, Olejniczak I, Szanser M. 2006. Response of soil micro-and mesofauna to diversity and quality of plant litter. Eur. J. Soil Biol. 42, S244–S249. ( 10.1016/j.ejsobi.2006.07.030) [DOI] [Google Scholar]
- 19.Santorufo L, Cortet J, Arena C, Goudon R, Rakoto A, Morel JL, Maisto F. 2014. An assessment of the influence of the urban environment on collembolan communities in soils using taxonomy- and trait-based approaches. Appl. Soil Ecol. 78, 48–56. ( 10.1016/j.apsoil.2014.02.008) [DOI] [Google Scholar]
- 20.Sousa JP, da Gama MM, Ferreira C, Barrocas H. 2000. Effect of eucalyptus plantations on Collembola communities in Portugal: a review. Belg. J. Zool. 2, 187–201. [Google Scholar]
- 21.Rieff GG, Natal-da-Luz T, Sousa JP, Wallau MO, Hahn L, Saccol de Sá EL. 2016. Collembolans and mites communities as a tool for assessing soil quality: effect of eucalyptus plantations on soil mesofauna biodiversity. Curr. Sci. 110, 713–719. ( 10.18520/cs/v110/i4/713-719) [DOI] [Google Scholar]
- 22.Pereira AP, Graca MA, Molles M. 1998. Leaf litter decomposition in relation to litter physico-chemical properties, fungal biomass, arthropod colonization, and geographical origin of plant species. Pedobiologia 42, 316–327. [Google Scholar]
- 23.Makkonen M, Berg MP, Handa IT, Hättenschwiler S, Ruijven J, Bodegom PM, Aerts R. 2011. Highly consistent effects of plant litter identity and functional traits on decomposition across a latitudinal gradient. Ecol. Lett. 15, 1033–1041. ( 10.1111/j.1461-0248.2012.01826.x) [DOI] [PubMed] [Google Scholar]
- 24.TreeDivNet. 2018. IDENT (Canada, USA, Germany, Italy). See http://www.treedivnet.ugent.be/ExpIDENT.html (accessed 21 February 2018).
- 25.Campbell CD, Chapman SJ, Cameron CM, Davidson MS, Potts JM. 2003. A rapid microtiter plate method to measure carbon dioxide evolved from carbon substrate amendments so as to determine the physiological profiles of soil microbial communities by using whole soil. Appl. Environ. Microbiol. 69, 3593–3599. ( 10.1128/AEM.69.6.3593-3599.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson JPE, Domsch KH. 1978. A physiological method for the quantitative measurement of microbial biomass in soils. Soil Biol. Biochem. 10, 215–221. ( 10.1016/0038-0717(78)90099-8) [DOI] [Google Scholar]
- 27.Edwards CA. 1991. The assessment of populations of soil-inhabiting invertebrates. Agric. Ecosyst. Environ. 34, 145–176. ( 10.1016/0167-8809(91)90102-4) [DOI] [Google Scholar]
- 28.Christiansen KA, Bellinger P. 1998. The Collembola of North America North of the Rio Grande, 2nd edn Grinnell, IA: Grinnell College. [Google Scholar]
- 29.Hopkin SP. 2007. A key to the Collembola (springtails) of Britain and Ireland. Telford, UK: FSC Publications. [Google Scholar]
- 30.Bellinger PF, Christiansen KA, Janssens F. 1996–2016 Checklist of the Collembola of the World. See http://www.collembola.org. (accessed 21 February 2018).
- 31.Legendre P, Legendre LF. 2012. Numerical ecology, 3rd edn Oxford, UK: Elsevier. [Google Scholar]
- 32.Legendre P, Gallagher ED. 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129, 271–280. ( 10.1007/s004420100716) [DOI] [PubMed] [Google Scholar]
- 33.Anderson MJ, Walsh DC. 2013. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol. Monogr. 83, 557–574. ( 10.1890/12-2010.1) [DOI] [Google Scholar]
- 34.Peres-Neto PR, Jackson DA. 2001. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 129, 169–178. ( 10.1007/s004420100720) [DOI] [PubMed] [Google Scholar]
- 35.Oksanen J, et al. 2018. vegan : Community Ecology Package. R package 2.4-6 version. See https://cran.r-project.org/web/packages/vegan/index.html. (accessed 21 February 2018).
- 36.Connor EF, Faeth SH, Simberloff D, Opler PA. 1980. Taxonomic isolation and the accumulation of herbivorous insects: a comparison of introduced and native trees. Ecol. Entomol. 5, 205–211. ( 10.1111/j.1365-2311.1980.tb01143.x) [DOI] [Google Scholar]
- 37.Engelkes T, Meisner A, Morriën E, Kostenko O, Van der Putten WH, Macel M. 2016. Herbivory and dominance shifts among exotic and congeneric native plant species during plant community establishment. Oecologia 180, 507–517. ( 10.1007/s00442-015-3472-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finerty GE, de Bello F, Bílá K, Berg MP, Dias ATC, Pezzatti GB, Moretti M. 2016. Exotic or not, leaf trait dissimilarity modulates the effect of dominant species on mixed litter decomposition. J. Ecol. 104, 1400–1409. ( 10.1111/1365-2745.12602) [DOI] [Google Scholar]
- 39.Endlweber K, Ruess L, Scheu S. 2009. Collembola switch diet in presence of plant roots thereby functioning as herbivores. Soil Biol. Biochem. 41, 1151–1154. ( 10.1016/j.soilbio.2009.02.022) [DOI] [Google Scholar]
- 40.Fujii S, Mori AS, Kominami Y, Tawa Y, Inagaki Y, Takanashi S, Takeda H. 2016. Differential utilization of root-derived carbon among collembolan species. Pedobiologia 59, 225–227. ( 10.1016/j.pedobi.2016.05.001) [DOI] [Google Scholar]
- 41.Weemstra M, Mommer L, Visser EJ, Ruijven J, Kuyper TW, Mohren GM, Sterck FJ. 2016. Towards a multidimensional root trait framework: a tree root review. New Phytol. 211, 1159–1169. ( 10.1111/nph.14003) [DOI] [PubMed] [Google Scholar]
- 42.Wardle DA. 2016. Do experiments exploring plant diversity–ecosystem functioning relationships inform how biodiversity loss impacts natural ecosystems? J. Veg. Sci. 27, 646–653. ( 10.1111/jvs.12399) [DOI] [Google Scholar]
- 43.Salmon S, Ponge J-F, Gachet S, Deharveng L., Lefebvre N, Delabrosse F. 2014. Linking species, traits and habitat characteristics of Collembola at European scale. Soil Biol. Biochem. 75, 73–85. ( 10.1016/j.soilbio.2014.04.002) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Litter trait values and springtail abundances used in this study can be found in supplementary support (see appendices S2.2 and S5 in the electronic supplementary material).