Summary
Neural stem cell (NSC) proliferation and differentiation in the developing brain is a complex process precisely regulated by intrinsic and extrinsic signals. Although epigenetic modification has been reportedly involved in the regulation of the cerebral cortex, whether UTX, an H3K27me3 demethylase, regulates the development of cerebral cortex during the embryonic period is unclear. In this study, we demonstrate that Utx deficiency by knockdown and conditional knockout increases NSC proliferation and decreases terminal mitosis and neuronal differentiation. Furthermore, we find that impairment of cortical development caused by lack of Utx is less significant in males than in females. In addition, UTX demethylates H3K27me3 at the Pten promoter and promotes Pten expression. P-AKT and P-mTOR levels are increased in the Utx conditional knockout cortices. Finally, Utx or Pten overexpression can rescue the impairment of brain development caused by Utx loss. These findings may provide important clues toward deciphering brain diseases.
Keywords: neural stem cells, UTX, proliferation, differentiation, H3K27me3
Highlights
-
•
UTX is essential for cortical development and neuron production
-
•
Utx suppression impairs normal neocortical neurogenesis in a sex-specific manner
-
•
UTX affects the levels of H3K27 trimethylation at Pten promoters
In this article, Jiao and colleagues show that UTX is a critical regulator of neural progenitor cell proliferation and neurogenesis. When Utx is suppressed, the generation of neurons and terminal mitosis are affected. This study states that UTX controls the development of embryonic cortex through Pten in a sex-specific manner.
Introduction
The mammalian brain contains millions of neurons and glial cells. Normal cerebral cortical development plays an important role in controlling behavior, learning, and cognition. During neurogenesis, neural stem cells (NSCs) proliferate and differentiate into neurons of all cortical layers, which is precisely controlled by various intracellular and extracellular signaling pathways (Durak et al., 2016, Meissner, 2010, Yelagandula et al., 2014). During embryonic development, NSCs of the cortex mainly are PAX6-positive apical progenitor cells (also called radial glia [RG]) and TBR2-positive basal progenitor cells (also called intermediate progenitors) (Imayoshi and Kageyama, 2014, Pontious et al., 2008). Although more and more molecules are reportedly involved in the proliferation of progenitor cells, how neurogenesis is regulated by epigenetic factors during the embryonic development of the cerebral cortex remains to be studied.
During cortical development, histone methylases and demethylases are essential for the proliferation and self-renewal of NSCs (Imayoshi and Kageyama, 2014). Ubiquitously transcribed tetratricopeptide repeat (UTX) is a broadly expressed H3K27 demethylase belonging to a family of JmjC-domain-containing proteins (Hong et al., 2007). UTX contains a tetratricopeptide repeat (TRP) domain at the N terminus and a JMJC catalytic-active domain at the C terminus (Tie et al., 2012). UTX acts as a critical regulator to activate chromatin in cardiac genes during cardiac development by demethylating H3K27me3 (Lee et al., 2012, Northrup et al., 2017). Mutation of UTX on the JMJC domain (H1146A) abrogates the H3K27 demethylase activity. As a result of these mutations, the enzyme is non-functional. Most of the Utx gene mutations in humans disrupt its role in histone demethylation and even result in Kabuki syndrome, a pediatric congenital disorder with multiple congenital anomalies and intellectual disabilities (Kim and Lee, 2017, Morales Torres et al., 2013, Shpargel et al., 2012). Sometimes UTX exerts its function independent of its H3K27 demethylase activity (Lui et al., 2017, Shpargel et al., 2014, Yoo et al., 2016). Overall, UTX participates in the regulation of embryonic development both dependent on and independent of its demethylase activity.
The female mice have two X chromosomes, with one inactivated. The inactive X chromosome is silenced by its being packaged by heterochromatin and prevents females from containing twice as much X chromosome gene expression as males (Carrel and Willard, 2005, Lyon, 1972). While Utx escapes X chromosome inactivation in females, it is predicted to own a functionally similar Y-linked homolog. Interestingly, ubiquitously transcribed tetratricopeptide repeat gene on Y chromosome (Uty) is the homology of Utx located on the Y chromosome (Xu et al., 2002).
Here, we report that UTX could regulate proliferation of NSCs in a sex-specific manner. Deficiency of Utx resulted in a more increased population of PAX6-positive RG cells in ventricular zone (VZ)/subventricular zone (SVZ) and a more decreased population of neurons in cortical plates at embryonic day 16.5 (E16.5)/E17.5 in females than in males. Furthermore, we found that UTX could change the levels of H3K27 trimethylation at Pten promoters, and conditional knockout of Utx resulted in significant decrease of Pten expression in RNA and protein levels. Subsequently, the levels of phospho (P)-AKT and P-mTOR (mammalian target of rapamycin) were significantly increased. In addition, Utx or Pten overexpression rescued the impairment caused by Utx knockdown. Taken together, UTX regulates the development of embryonic cortex through Pten in a sex-specific manner.
Results
Utx Is Expressed in Neural Stem Cells of Developing Brain
To study the function of UTX in cortical development, we performed immunostaining of UTX in female cortices at E13 and E16. The results showed that UTX was expressed in the whole cortex both in the NSCs and in the neurons (Figure 1A). Immunostaining in embryonic neural stem cells (eNSCs) derived from E12 cortices also proved that UTX was highly expressed in eNSCs, which could be marked by SOX2, PAX6, or TBR2 (Figure 1B). Considering that UTX was encoded by the Utx gene on X chromosome and there were two X chromosomes in females and one in males, we tested the difference between female and male cortices at E13 and E15. The western blot results showed that Utx was more highly expressed in females than in males and that Utx expression was downregulated at E15 compared with that at E13 (Figures 1C and 1D). To further define the expression level of Utx and Uty, we carried out an absolute quantitative experiment. The results revealed that the copy number of Utx in females was about 1.4-fold that of Utx in males, the difference being statistically significant (Figure 1E).
Figure 1.
UTX and UTY Are Expressed in the Developing Brain, and Utx Knockdown Regulates Cell Position in the Embryonic Cortex
(A) UTX is expressed in the embryonic NSC in vivo. The E13 and E16 brain sections were immunostained with anti-UTX antibody. Scale bars, 50 μm.
(B) The eNSCs were immunostained with anti-SOX2/PAX6/TBR2 and anti-UTX antibody after 24 hr of culturing since separation from E12 brains. Scale bar, 10 μm.
(C) Western blot analysis of UTX expression at E13/E15 in female/male embryonic cortices.
(D) Quantification of UTX-expression level at E13/E15 in female/male embryonic cortices (normalized to β-ACTIN). Values are presented as mean ± SEM (n = 3 independent experiments; ∗p < 0.05, ∗∗p < 0.01).
(E) Absolute quantification analysis of Utx expression at E13 in female/male embryos and of Uty expression at E13 in male embryos by qRT-PCR. Values are presented as mean ± SEM (n = 3 independent experiments; ∗p < 0.05).
(F) Coronal sections of E13.5–E16.5 female/male mouse brains electroporated at E13.5 with GFP plus the control (Ctrl; top) or Utx shRNA2 (bottom) constructs. GFP-positive cells are derived from transfected cortical progenitor cells. Sections were stained with DAPI (blue). Scale bar, 100 μm.
(G) Analysis of the distribution of GFP-positive cells in electroporated cortices of different sexes. Values are presented as mean ± SEM (n ≥ 3 individual brains for all constructs, one-way ANOVA with Turkey's test for post hoc multiple comparisons; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). CP, cortical plate; IZ, intermediate zone; SVZ, subventricular zone; VZ, ventricular zone.
See also Figure S1.
To study the function of UTX in the embryonic cortical development, we constructed two short hairpin RNA (shRNA) plasmids targeting Utx mRNA. Western blotting showed that both Utx-shRNA1 and Utx-shRNA2 significantly decreased UTX expression (Figures S1A and S1B). To detect the function of UTX in embryonic neurogenesis, we electroporated control or Utx shRNA plasmids with GFP-expressing vector into cerebral cortices via in utero electroporation (IUE) at E13.5 and harvested them at E16.5. Compared with control, fewer GFP-positive cells were located in the cortical plate (CP), while the percentage of GFP-positive cells in the VZ and SVZ was significantly increased (Figures 1F and 1G).
Considering that UTY exists in males but not in females, we further constructed a knockdown plasmid targeting Uty (shUTY). RT-PCR with eNSCs showed that shUTY downregulated the mRNA level of Uty significantly (Figure S1C). Next, shUTY and Utx shRNA2 were simultaneously electroporated into cerebral cortices at E13.5. Brain section analysis demonstrated that downregulating Uty in male Utx-depleted cortices further increased the impairments on GFP-positive cell distribution (Figures S1D and S1E).
Knockdown of Utx Results in Increased Proliferation in Neural Progenitors
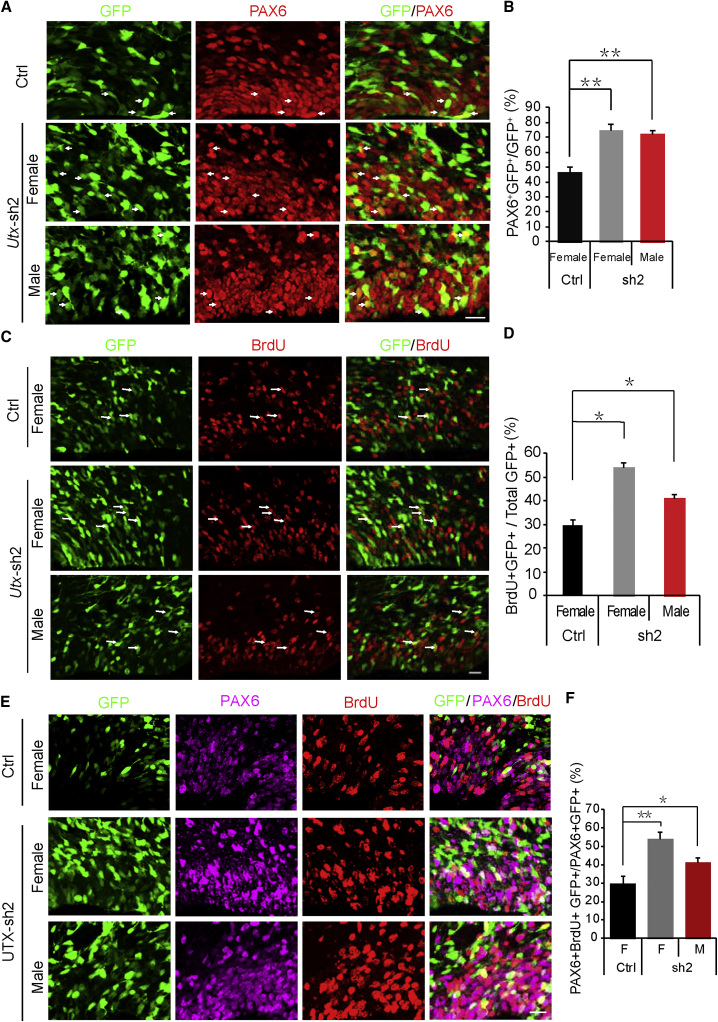
Considering that more GFP-positive cells were located in the VZ/SVZ, and that PAX6-positive neuronal progenitor cells (RG) were located in the VZ and the TBR2-positive intermediate progenitor cells were mostly located in the SVZ, immunostaining for PAX6 or TBR2 was performed. The results indicated that Utx depletion significantly increased GFP and PAX6 double-positive progenitor cells in the VZ (Figures 2A and 2B). However, Utx depletion led no significant increase on GFP and TBR2 double-positive intermediate progenitors in the SVZ (Figures S2A and S2B).
Figure 2.
Utx Knockdown Increases Proliferation of Pax6-Positive Radial Progenitor Cells
(A) Immunostaining of PAX6 3 days after electroporation with control or Utx shRNA constructs into female or male embryonic brains at E13.5. The white arrows indicate GFP and Pax6 double-positive cells. Scale bar, 20 μm.
(B) Bar graph shows the percentage of PAX6 and GFP double-positive cells relative to the total GFP-positive cells in the VZ/SVZ. Values are presented as mean ± SEM (n = 3 individual brains for all constructs; ∗∗p < 0.01).
(C) BrdU incorporation is increased in Utx shRNA plasmid-electroporated brains. The brains of pregnant mice electroporated at E13.5 were injected with BrdU (100 mg/kg, intraperitoneally) 2 hr prior to euthanasia at E16.5. The white arrows indicate GFP and BrdU double-positive cells. Scale bar, 20 μm.
(D) Bar graph shows the percentage of BrdU and GFP double-positive cells relative to the total GFP-positive cells in the VZ/SVZ. Values are presented as mean ± SEM (n = 3 individual brains for all constructs; ∗p < 0.05).
(E) Co-immunostaining of PAX6 (red) and BrdU (pink) of brain sections electroporated with control or Utx shRNA constructs at E13.5. BrdU was injected into the electroporated pregnant mouse 2 hr before euthanasia. Scale bar, 20 μm.
(F) Bar graph shows the percentage of GFP/PAX6/BrdU triple-positive cells relative to the total GFP/PAX6 double-positive cells in the VZ/SVZ. Values are presented as mean ± SEM (n = 3 individual brains for all constructs; ∗p < 0.05, ∗∗p < 0.01).
See also Figures S2 and S3.
As most of the cells in the zones were NSCs with highly proliferative activity, we presumed that UTX might affect the proliferation of NSCs. To verify this speculation, we injected 5-bromo-2′-deoxyuridine (BrdU) into the electroporated pregnant mice 2 hr before they were harvested at E16.5. The knockdown of Utx led to an increase in GFP and BrdU double-positive neural progenitor cells in the VZ/SVZ (Figures 2C and 2D). The knockdown of Utx led to a larger increase in GFP and BrdU double-positive neural progenitor cells in the VZ/SVZ in females than in males (Figures 2C and 2D). Based on the fact that RG cells only come from its proliferation, we further detected the RG proliferation by co-immunostaining PAX6 and BrdU. The results showed that Utx knockdown indeed increased the proliferation of RG (Figures 2E and 2F).
To confirm the effect of UTX on proliferation, we cultured primary progenitors derived from E12 cortices and infected them with recombinant lentivirus expressing control or Utx shRNA. Subsequently, eNSCs were subjected to Ki67 staining. Compared with control, more Ki67-positive cells were detected in Utx knockdown cells (Figures S3A and S3B). We also performed western blot and RT-PCR experiments to check the protein and mRNA levels of proliferation-related genes. The results show that Ki67 and proliferating cell nuclear antigen (PCNA) were both upregulated in Utx knockdown cells (Figures S3C and S3D). These results signify that UTX inhibits the proliferation of progenitor cells.
Utx Knockdown Inhibits Cell Cycle Exit of Neural Progenitors and Neuronal Differentiation
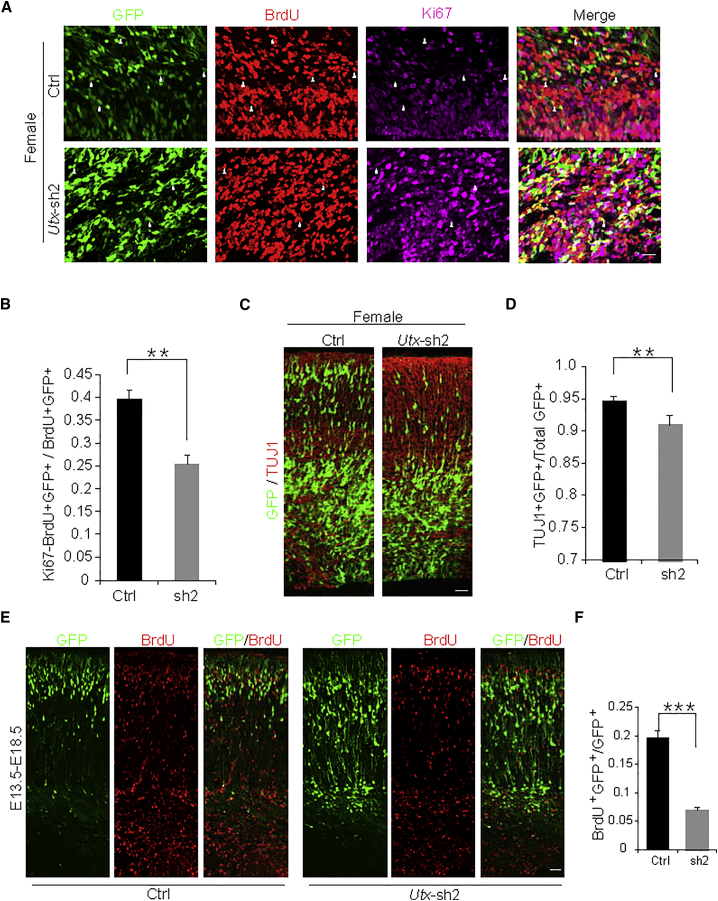
Neuronal progenitor cells exit the cell cycle into the differentiation phase during neurogenesis. Our observations that Utx knockdown increased the number of GFP-positive cells in the VZ/SVZ and of neural progenitor proliferation suggest that the differentiation of eNSCs into neurons may be affected. To test the role of UTX in cell cycle exiting, we injected BrdU into the electroporated pregnant mice at E15.5, 24 hr before they were harvested, and immunostained the sections for BrdU and Ki67. The analysis showed that Utx knockdown caused a significant reduction in the percentage of GFP+BrdU+Ki67− cells relative to GFP and BrdU double-positive cells in the VZ/SVZ (Figures 3A and 3B), suggesting that Utx knockdown resulted in a decreased number of neural progenitor cells that exit the cell cycle.
Figure 3.
Utx Knockdown Inhibits Neural Differentiation In Vivo
(A) Knockdown of Utx in females decreases premature cell cycle exit. Embryonic mouse brains were electroporated at E13.5 with GFP plus the control or Utx shRNA2 construct; BrdU (100 mg/kg) was injected at E15.5. The embryonic brains were harvested and analyzed 24 hr after BrdU injection at E16.5. GFP+BrdU+Ki67− cells are indicated by white arrowheads. Scale bar, 20 μm.
(B) Bar graph shows the percentage of premature cell cycle exit (GFP+BrdU+Ki67− cells/GFP+BrdU+ cells in VZ/SVZ). Values are presented as mean ± SEM (n = 3 individual brains for all constructs; ∗∗p < 0.01).
(C) Utx knockdown decreases neuronal differentiation in female in vivo. E16.5 brain sections electroporated at E13.5 were immunostained for TUJ1. Scale bar, 50 μm.
(D) Bar graph shows the percentage of GFP and TUJ1 double-positive cells relative to the total GFP-positive cells. Values are presented as mean ± SEM (n = 3 individual brains for all constructs; ∗∗p < 0.01).
(E) Control or Utx shRNA constructs were electroporated into embryonic mouse cortices at E13.5, and BrdU was injected 24 hr later. The female electroporated embryonic brains were then collected for immunostaining with anti-BrdU antibody at E18.5. Scale bar, 25 μm.
(F) Bar graph shows the percentage of GFP and BrdU double-positive cells relative to the total number of GFP-positive cells in the CP. Values are presented as mean ± SEM (n = 3 individual brains for all constructs; ∗∗∗p < 0.001).
See also Figure S4.
To investigate whether UTX affects the differentiation of progenitor cells, we immunostained female sections with an anti-neuronal class III β-tubulin (TUJ1) antibody. The statistics of GFP and TUJ1 double-positive cells revealed that Utx knockdown caused a significant decrease in the percentage of GFP and TUJ1 double-positive cells (Figures 3C and 3D). We also carried out experiments on the differentiation of NSCs in vitro. eNSCs were infected with control or Utx shRNA lentivirus and cultured for 3 days in differentiating conditions. Immunostaining with anti-TUJ1 antibodies was performed, and the quantification of GFP and TUJ1 double-positive cells revealed that the knockdown of Utx in vitro decreased neuronal differentiation compared with the control treatment (Figures S4A and S4B). RT-PCR for Tuj1 also showed that the expression of Tuj1 was obviously decreased when Utx was knocked down in eNSCs (Figure S4C).
To identify whether UTX controls the differentiation process through regulating premature neural progenitor cell (NPC) terminal mitosis during early cortical development, we performed a BrdU birth dating study (Duque and Rakic, 2011). BrdU injection was injected to pregnant mice at E14.5 after electroporation, and the electroporated brains were collected at E18.5. The percentage of BrdU and GFP double-labeled cells in all cortical layers showed a significant decrease of newborn neurons in Utx knockdown brains compared with control (Figures 3E and 3F). In addition, Utx loss also had no obvious effects on migration during the period of E15 to postnatal day 3 (Figures S4D and S4E). These results indicate that Utx knockdown represses premature NPCs terminal mitosis and then decreases neural differentiation.
Utx Knockout Results in Increased Cell Proliferation and Changes in Gene Expression Profile
To further investigate the function of UTX in whole cortical development, we bred the Utxfl/fl mice with Nestin-Cre mice to generate Utx conditional knockout mice (Utx cKO). To verify the function of UTX in neurogenesis, we performed IUE with GFP-expressing vector on the embryonic brains of Utx fl/fl mice and Utx cKO mice at E14.5, then harvested the brains at E17.5. Compared with Utx fl/fl mice, Utx cKO mice showed abnormal distribution of GFP-positive cells (Figures 4A and 4B). Further knockdown of Uty in male Utx cKO cortices resulted in almost the same distribution of GFP-positive cells (Figures S5A and S5B). As in the Utx knockdown cortices, we also detected PAX6 and TBR2 changes in the VZ/SVZ. The results showed that PAX6-positive RG cells (Figures 4C and 4D), but not the TBR2-positive cells, were significantly increased (Figures S5C and S5D). Consistently, immunostaining of BrdU on cortical sections showed that more BrdU-positive cells were detected in Utx cKO embryos, indicating that proliferation of progenitor cells was affected when Utx were deleted (Figures 4C and 4D). The knockdown and cKO experiments demonstrate that the loss of Utx resulted in an increase in proliferation of progenitors. However, there was no significant difference in the levels of apoptosis between control and Utx cKO embryonic cortices (Figure S5E), confirming that apoptosis is not the major reason for the reduced number of neurons.
Figure 4.
Loss of Utx Affects Neurogenesis and Proliferation of Neural Progenitors
(A) Coronal sections of E14.5–E17.5 female/male mouse brains electroporated at E14.5 with GFP plasmid. GFP+ cells are derived from transfected cortical progenitor cells. Sections were stained with DAPI (blue). Scale bar, 100 μm.
(B) Analysis of the distribution of GFP+ cells in electroporated cortices of different sexes. Values are presented as mean ± SEM (n = 3 individual brains for all constructs; ∗∗p < 0.01, ∗∗∗p < 0.001).
(C) Immunostaining of PAX6 in female wild-type or Utx cKO cortices at E17.5. Scale bar, 50 μm.
(D) Bar graph shows the average number of PAX6-positive cells per 1,000 μm2 in the VZ/SVZ. Values are presented as mean ± SEM (n = 3 individual brains; ∗∗p < 0.01).
(E) BrdU incorporation is increased in Utx cKO female brains. The brains of pregnant mice were injected with BrdU (100 mg/kg, intraperitoneally) 2 hr prior to euthanasia at E18. Female control or Utx cKO brains were immunostained. Scale bar, 50 μm.
(F) Bar graph shows the average number of BrdU-positive cells per 1,000 μm2 in the VZ/SVZ. Values are presented as mean ± SEM (n = 3 individual brains for all constructs; ∗∗p < 0.01).
See also Figure S5.
To examine the gene expression changes in Utx conditional knockout cortices, we conducted RNA sequencing (RNA-seq) to examine the genome-wide changes by Utx depletion. The cortices were obtained from embryos of control or Utx cKO at E14.5, and RNA was extracted for RNA-seq. The heatmap (Figure 5A) and the volcano map (Figure 5B) showed that the gene expression profile was changed between Utx cKO and control. The different expression analysis revealed that 1,331 genes were downregulated or upregulated significantly between the Utx cKO and the control. Gene ontology (GO) term analysis of downregulated genes in the samples showed a significant enrichment of biological processes related to NSC differentiation (for example, negative regulation of glomerular mesangial cell proliferation, spinal cord association neuron differentiation) (Figure 5C). These results indicate that UTX is involved in a wider range of neural developmental and embryonic development processes. We also performed RT-PCR to detect expression levels of several genes that are reported to be associated with proliferation. The result showed that when Utx was deleted, the expression of Pten was also decreased, which was identical with the volcano plot analysis (Figure 5D). These results prompt us to speculate that the impact of UTX on cortical development is likely associated with the Pten pathway.
Figure 5.
Loss of Utx Affects Gene Expression Profile
(A) Cluster analysis of control and UTX conditional knockout (cKO) cortices. The Utxfl/fl (Ctrl) or Utx cKO cortices were isolated at E14 and total RNA was isolated and used for RNA-seq.
(B) GO analysis of control and Utx cKO cortices.
(C) Volcano plot of control and Utx cKO cortices. Pcna, a marker of proliferation, was upregulated and Pten significantly downregulated in Utx cKO cortices. The genes that were non-significantly regulated are marked with gray points; genes significantly downregulated are marked with green points; genes significantly upregulated are marked with red points.
(D) RT-PCR analysis of eNSCs infected with control or Utx shRNA constructs. The cells were collected 3 days after infection and total RNA isolated by TRIzol was used for the RT-PCR. Values are presented as mean ± SEM (n = 3 independent experiments; ∗∗p < 0.01, ∗∗∗p < 0.001).
Utx Regulates Embryonic Neurogenesis via the PTEN-AKT-mTOR Pathway
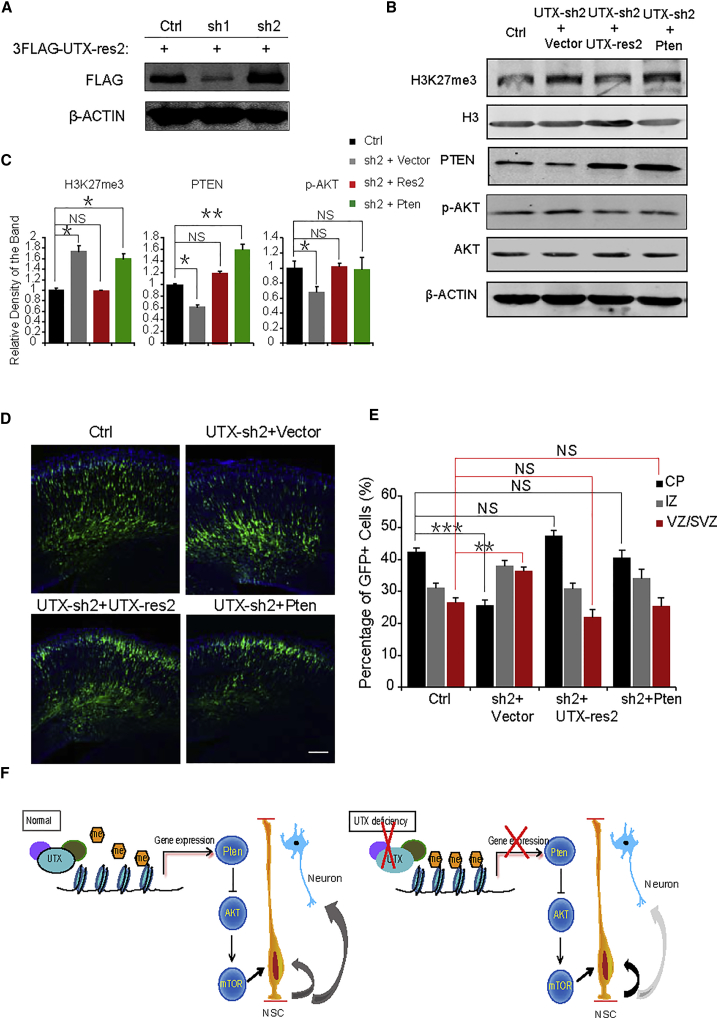
To test the hypothesis on the mechanism, we checked the H3K27me3 level through western blot and found that with the condition of Utx knockout, the total H3K27me3 level was increased (Figures 6A and 6B). These results demonstrate that UTX acts as an epigenetic regulator. In addition, the PTEN (phosphatase and tensin homolog) protein level was significantly decreased in the cortices after Utx conditional knockout. Simultaneously, P-AKT and P-mTOR were decreased in the UTX deficiency group, with no change of the total AKT and mTOR expression level (Figures 6A and 6B). To further test the hypothesis, we constructed the shRNA plasmid targeting Pten (Pten-sh1) (Figures 6C and 6D) and performed IUE at E13.5. The proportion of GFP-positive cells in each zone showed a phenotype similar to that of Utx shRNA (Figures 6E and 6F). The immunostaining in eNSCs infected with control or Utx shRNA lentivirus showed that Utx deletion downregulated the PTEN level and upregulated the P-AKT level compared with the control group (Figure 6G). These results indicate that UTX is involved in the AKT-mTOR signaling pathway via Pten regulation.
Figure 6.
UTX Regulates PTEN-AKT-mTOR Pathway through Demethylating H3K27me3
(A) Western blot analysis of UTX, H3K27me3, PTEN, P-AKT, P-mTOR of Utxfl/fl (Ctrl) or Utx cKO female/male cortices at E16.5. β-ACTIN was the normal reference.
(B) Quantification of changes of protein level detected by western blot (UTX and PTEN were normalized to β-ACTIN and H3K27me3/P-AKT/P-mTOR were respectively normalized to H3/AKT/mTOR). Values are presented as mean ± SEM (n = 3 independent experiments; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(C) Western blot analysis revealing the efficiency of Pten knockdown by Pten-sh1 in vitro. A notable variation of Pten protein expression was observed in N2a cells.
(D) Quantification of Pten-sh1 knockdown efficiency (normalized to β-ACTIN). Values are presented as mean ± SEM (n = 3 independent experiments; ∗∗∗p < 0.001).
(E) Coronal sections of E13.5–E16.5 female mouse brains electroporated at E13.5 with GFP plus the control (left) or Pten-sh1 constructs (right). GFP+ cells are derived from transfected cortical progenitor cells. Sections were stained with DAPI (blue). Scale bar, 100 μm.
(F) Analysis of the distribution of GFP-positive cells in electroporated cortices. Values are presented as mean ± SEM (n ≥ 3 individual brains for all constructs; ∗p < 0.05, ∗∗∗p < 0.001).
(G) Immunostaining of UTX, PTEN, and P-AKT in the eNSCs infected with Control and Utx shRNA lentivirus 3days after infection. The white arrows and circles indicate the immunostaining of UTX, PTEN, or P-AKT in a single cell. Scale bar, 10 μm.
(H) Chromatin immunoprecipitation analysis showed that UTX promoted Pten expression by promoting H3K27me3 demethylation on the Pten promoter. The eNSCs infected with control, Utx shRNA, and Utx overexpression lentivirus were harvested. The DNA fragments were quantified using real-time PCR with primers for the Pten promoter and for the Pten coding sequences. Values are presented as mean ± SEM (n = 3 independent experiments; ∗p < 0.05, ∗∗p < 0.01).
(I) Western blot analysis of UTX, H3K27me3, PTEN, P-AKT, and P-mTOR of eNSCs infected with control and Utx overexpression lentivirus. β-ACTIN was the normal reference. H3, AKT, or mTOR was internal reference, respectively, for H3K27me3, P-AKT, and P-mTOR.
(J) Quantification of changes of protein level detected by western blot (UTX and PTEN were normalized to β-ACTIN and H3K27me3/P-AKT/P-mTOR were respectively normalized to H3/AKT/mTOR). Values are presented as mean ± SEM (n = 3 independent experiments; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
See also Figure S6.
Next, we needed to prove that UTX controlled Pten expression by regulating the H3K27me3 level at the Pten gene sites. Chromatin immunoprecipitation experiments were performed, and the RT-PCR results showed that the H3K27me3 level at the Pten promoter was upregulated after Utx knockdown (Figure 5H).
To further verify the roles of UTX in embryonic neurogenesis, we constructed Utx-overexpressed (OE) plasmid (Figures S6A and S6B). UTX-OE plasmids were electroporated into the brain of E13.5 embryos and also harvested at E16.5. Consistent with the results of Utx knockdown, fewer GFP-positive cells were observed in the VZ/SVZ while more GFP-positive cells were observed in the CP (Figures S6C and S6D). Moreover, immunostaining revealed that the proportion of GFP and BrdU double-positive cells decreased when UTX was overexpressed (Figures S6E and S6F), and the protein level of PTEN/P-AKT/P-mTOR was correspondingly changed (Figures 6I and 6J). Therefore, UTX is necessary for normal cell proliferation in the developing neocortex. Taken together, these results suggest that UTX regulates eNSC proliferation through the PTEN-AKT-mTOR signaling pathway.
Overexpression of PTEN Rescues the Deficiency Resulting from Utx Depression
To eliminate the impact of Utx shRNA off target, we constructed a UTX-resistant overexpression plasmid that could resist the Utx knockdown caused by Utx shRNA2 (UTX-res2). Western blot analysis revealed that UTX-res2 could overexpress UTX protein despite the presence of Utx shRNA2 but not UTX-shRNA1 (Figure 7A). As expected, western blot revealed that co-expression of UTX-res2 or PTEN with Utx shRNA2 could rescue PTEN and P-AKT level to the control level (Figures 7B and 7C). In addition, UTX-res2 or PTEN rescued the abnormal distribution of GFP-positive cells resulting from Utx shRNA2 (Figures 7D and 7E) and the change in the percentage of BrdU and GFP double-positive cells in the VZ/SVZ (Figures S7A and S7B). We also constructed a catalytic-inactive plasmid of UTX (UTX-mu), which lost the catalytic activity of demethylating H3K27me3/me2. Unlike the UTX, UTX-mu resulted in no significant changes in the H3K27me3 and PTEN levels (Figure S7C). Moreover, UTX-mu could not rescue the abnormal distribution of GFP-positive cells resulting from Utx shRNA2 (Figures S7D and S7E).
Figure 7.
Utx-res2 and Pten Rescue the Deficiency Caused by Utx shRNA
(A) Western blot analysis revealing the efficiency of Utx-res2 overexpression in vitro and its resistance over the Utx shRNA2 but not the Utx shRNA1.
(B) Western blot analysis of UTX, H3K27me3, PTEN, and P-AKT with Utx-res2/Pten co-expressed with Utx shRNA in the eNSCs derived from cortices at E12. β-ACTIN was the normal reference.
(C) Quantification of changes of protein level detected by western blot (PTEN was normalized to β-ACTIN and H3K27me3/P-AKT were respectively normalized to H3/AKT). Values are presented as mean ± SEM (n = 3 independent experiments; ∗p < 0.05, ∗∗p < 0.01; NS, not significant).
(D) Coronal sections of E13.5–E16.5 female mouse brains electroporated at E13.5 with GFP plus the control, Utx shRNA2, or Utx shRNA2 together with Utx-res2 or Pten plasmids. GFP+ cells are derived from transfected cortical progenitor cells. Sections were stained with DAPI (blue). Scale bar, 100 μm.
(E) Analysis of the distribution of GFP-positive cells in electroporated cortices. Values are presented as mean ± SEM (n ≥ 3 individual brains for all constructs; ∗∗p < 0.01, ∗∗∗p < 0.001).
(F) Model for the role of UTX in regulating embryonic neurogenesis.
See also Figure S7.
Discussion
UTX gene mutations in humans result in Kabuki syndrome, a pediatric congenital disorder with multiple congenital anomalies and intellectual disabilities (Kim and Lee, 2017, Miyake et al., 2013a, Miyake et al., 2013b). However, there is no direct evidence regarding whether or how UTX regulates brain development and neurogenesis. Neurogenesis in the developing brain is precisely controlled by various intracellular and extracellular signaling pathways. Here, we provide several lines of evidence demonstrating that UTX is essential for cerebral cortical neurogenesis. These results indicate a role of UTX in the embryonic developing mammalian cortex. Our data may provide an understanding of the epigenetic mechanism underlying brain pathology.
During the embryonic development, NSCs are located in the VZ and SVZ of the cortex, and the NSCs mainly include two groups: RG, which are Pax6 positive, and intermediate progenitor (IP) cells, which are TBR2 positive. During the process of early brain development, PAX6-positive RG cells can divide symmetrically to proliferate or divide asymmetrically to self-renew and generate one TBR2-positive IP cell or one neuron. IP cells divide symmetrically to produce two IP cells or two neurons. The symmetrical or asymmetrical division are dependent on the changes in the mitotic spindle positioning and angle of cleavage plane during cell division (Andrews et al., 2017, Arbeille et al., 2015, LaMonica et al., 2013, Yingling et al., 2008). In our study, while PAX6-positive RG cells were significantly increased after UTX knockdown, TBR2-positive IP cells slightly decreased but not significantly. Meanwhile, Utx knockdown inhibits the premature NPC terminal mitosis. UTX is a widely existing demethylase, and the RNA-seq results showed that most genes were downregulated. There is a possibility that UTX may regulate the symmetrical or asymmetrical division through the genes that influence the mitotic spindle positioning and angle of cleavage plane.
Utx is one of several X-linked genes that escape X inactivation in both humans and mice. In this study, we found Utx to be expressed at a slightly higher RNA and protein level in female than male mice in the embryonic cortex. Utx and Uty were expressed at a similar RNA level in male mouse cortices. Unfortunately, we failed in testing the UTY expression at protein level because the anti-UTY antibodies also recognized UTX protein (data not shown), and knockdown of Utx slightly increased Uty expression at the RNA level (∼1.3-fold). UTX participates in the regulation of embryonic development both dependent on and independent of its demethylase activity in H3K27. We found that Utx knockdown increased the eNSC proliferation and inhibited neuron differentiation. UTX-mu could not rescue the defects and PTEN partially rescued the defects. The data imply that there is also a mechanism whereby UTX is involved in neurogenesis regulation independent of its catalytic activity.
In summary, we demonstrate that UTX functions as a regulator of neurogenesis. We identify UTX as the promotional transcription factor of Pten through demethylating H3K27me3 at the Pten promoter to regulate neurogenesis. UTX-mu could not rescue the defects while PTEN partially rescued the defects.
Experimental Procedures
Animals
Pregnant ICR female mice were purchased from Vital River Laboratories. Utx flox/flox (Utx fl/fl) mice were purchased from The Jackson Laboratory (stock number 021926). The Nestin-Cre mice (catalog #003771: B6.Cg-Tg(Nes-cre)1Kln/J) were obtained from The Jackson Laboratory. Utx fl/fl mice were mated with Nestin-Cre mice to generate Utx conditional knockout mice. The Utx fl/fl mice were identified by PCR genotyping using the primers in Table S1. Sex identification of fetal mice after E16.5 is identified by gonad. To determine sexual identify of mouse fetuses before E15, we synthesized IL3 primers and male-specific SRY primers (Thermo Fisher Scientific). The primer sequences are listed in Table S1 (Lambert et al., 2000).
All the mice involved in procedures were in line with the Guide for the Care and Use of Laboratory Animals. Animal experiments and protocols were approved by the Animal Committee of Institute of Zoology, Chinese Academy of Sciences.
Reagents
The following primary antibodies and dilutions were used for immunohistochemistry (IHC) staining and western blotting: rabbit polyclonal anti-UTX (1:1,000; Millipore, #ABE409); mouse monoclonal anti-SOX2 (1:500; R & D, #MAB2018); rabbit anti-PAX6 (1:1,000; Millipore, #AB2237); rabbit anti-TBR2 (1:1,000; Abcam, #AB23345); mouse monoclonal anti-β-ACTIN (1:20,000; Proteintech, #60008-1-Ig); rat monoclonal anti-BrdU (1:1,000; Abcam, #AB6362); rabbit monoclonal anti-Ki67 (1:1,000; Abcam, #AB15580); rabbit anti-PCNA (1:500; Santa Cruz Biotechnology, #SC7907), rabbit anti-TUJ1 (1:1,000; Sigma, #T2200); rabbit monoclonal anti-PTEN (1:1,000; Cell Signaling Technology, #9188); rabbit monoclonal anti-AKT (1:1,000; Cell Signaling, #4685); rabbit monoclonal anti-phospho-AKT (1:1,000; Cell Signaling, #3787); rabbit monoclonal anti-mTOR (1:1,000; Cell Signaling, #2983); rabbit monoclonal anti-phospho-mTOR (1:1,000; Cell Signaling, #5536); rabbit monoclonal anti-H3 (1:4,000; Cell Signaling, #4499S); rabbit polyclonal anti-trimethyl-histone H3 (Lys27) (1:2,000; Cell Signaling, #3377S); and rabbit anti-FLAG (1:1,000; Sigma, #7425).
Constructs
The sequences for shRNAs targeting Utx are as follows: Utx shRNA-1 (sh1): 5′-AGT TAG CAG TGG AAC GTT ATG-3′; Utx shRNA-2 (sh2): 5′-GCT ACG AAT CTC TAA TCT TAA-3′. Utx sh2 recognize mouse and human Utx. The sequence for mouse Uty is 5′-TCA TTG TTA GCT TCT ACC A-3′. Utx and Uty shRNA oligonucleotides were cloned into the pSicoR vector.
Alternatively, we created pCDH-3xFLAG vector (pCDH-FLAG). For Utx-expression plasmid construction, the transcript variant 5 sequence of Utx was cloned by RT-PCR using RNA isolated from the brain of E12 mice as the template and inserted into PCDH-FLAG to express FLAG-tagged UTX (3FLAG-UTX-OE). FLAG-tagged enzyme-dead (H1152A and E1154A) UTX (3FLAG-UTX-mu) and UTX expression against sh2 (the sequence at which nt 242–255 were mutated from GCT ACG AAT CTC TA to GTT ATG AGT CAC TG) (3FLAG-UTX-res2) were generated.
For Pten-expression plasmid construction, the full-length coding sequence of mouse Pten was cloned by RT-PCR using RNA isolated from the brain of E12 mice as the template and inserted into pCDH-CMV-EF1-copGFP to express PTEN (pCDH-PTEN).
In Utero Electroporation
ICR pregnant mice were deeply anesthetized with pentobarbital and their embryos surgically manipulated. The effector plasmids (shRNA and mouse Utx or Pten-expression plasmids) and one-third volume of GFP expression vector were mixed and introduced into the VZ of E13.5 embryos by in utero electroporation. For rescue experiments, Utx shRNA and half-volume of Utx or Pten-expression plasmids were co-electroporated. Embryos were euthanized on E16.5 and the brains were dissected, fixed for 24 hr in 4% paraformaldehyde (PFA) at 4°C, and dehydrated for 48 hr in 30% sucrose/1× PBS at 4°C for cryoprotection. The brains were then frozen in OCT compound and sectioned at 15 μm thickness.
Western Blotting
Cells or brain tissues were extracted by laying cells in RIPA buffer (Solarbio) (10 mM PMSF and 10 mM cocktail added). The concentration of protein was measured by a Pierce BCA Protein Assay (Thermo Fisher). Ten to thirty micrograms of protein (in 4× loading buffer) was added into SDS-PAGE gels and transferred onto nitrocellulose or polyvinylidene fluoride membranes. The membranes were incubated with primary antibodies, and the primary antibody was visualized using IRDye 800CW or 680CW (LI-COR) donkey anti-mouse or anti-rabbit secondary antibodies (1:10,000, Odyssey).
Immunostaining
Cortical sections (15 μm) were harvested using a freezing microtome (Leica CM1950). During immunohistochemistry, the brain sections were washed with 1% PBST (1% Triton X-100 in 1 M PBS [pH 7.4]), fixed for 30 min by 4% PFA, and washed by PBST for 15 min three times. The slices were incubated with blocking buffer (5% BSA in 1% PBST) for 1 hr at room temperature. The slices were incubated in primary antibodies at 4°C overnight and then washed with PBST for 15 min three times. The secondary antibodies conjugated with Alexa Fluor dyes (1:1,000 dilution; Jackson ImmunoResearch) were added for 1 hr at room temperature. The brain sections were washed three times for 15 min in PBST, incubated with 2 mg/mL DAPI (Sigma, D9542) for 2 min, and washed three times for 5 min with PBST. When IHC was required for BrdU, the slices were incubated in ice-cold 1 M HCl for 10 min, 2 M HCl for 10 min at room temperature, and 2 M HCl for 20 min at 37°C, then washed with PBST three times before blocking and anti-BrdU antibody incubation.
Immunostaining for cultured cells was performed according to the following procedure: the cells were washed with PBST (0.1% Triton X-100 in PBS), fixed in 4% PFA, blocked by 5% BSA, incubated with primary antibodies overnight at 4°C, and visualized using fluorescence-labeling secondary antibodies.
RNA Isolation and qRT-PCR
Total RNA was extracted with TRIzol (Invitrogen) and then inverse transcribed into cDNA using the FastQuant RT Kit (Tiangen Biotech). Real-time qPCR was performed according to the Super Real PreMix Plus (SYBR Green I) (Tiangen Biotech) in a 20-μL system on an ABI PRISM 7500 sequence detector system (Applied Biosystems). The primers are listed in Table S1. The relative expression level of each mRNA was analyzed by the 2-ΔΔCt method. All experiments were repeated three times.
TUNEL Assay
Mouse cortices of control and Utx cKO were harvested at E18. The cryopreserved cortical sections were determined by the In Situ Cell Death Detection Kit, TMR red (Roche). In brief, the sections were fixed with 4% PFA in PBS, washed 30 min with PBS, and permeablized for 2 min on ice with freshly prepared 0.1% Triton X-100 in 0.1% sodium citrate. After rinsing slides twice with PBS, TUNEL reaction mixture was added and the slices were incubated in a humidified condition for 60 min at 37°C. The slides were rinsed three times with PBS, labeled with DAPI, and mounted. The labeled samples were directly analyzed under a fluorescence microscope.
For two negative controls, the slides were incubated in 50 μL/sample label solution instead of TUNEL reaction mixture. For a positive control, the slide was incubated with 3 U/mL DNase I recombinant for 10 min at room temperature to induce DNA strand breaks, prior to labeling procedures.
RNA Sequencing Analysis
Total RNA was extracted with TRIzol (Invitrogen) from E14 telencephalic tissue of wild-type and UTX cKO mice. Total RNA was then quality controlled and quantified using an Agilent 2100 Bioanalyzer. After converting to cDNA and building the library, high-throughput sequencing was performed using the Illumina HiSeq 2500 platform in Annoroad Genomics.
Chromatin Immunoprecipitation
The NSCs derived from E12.5 cortices were infected with lentivirus for 8 hr with polybrene. The cells were continuously cultured with proliferation medium for 3 days. Thereafter, the cells were treated with 1% formaldehyde for 15 min at room temperature to crosslink the chromosome and protein, and 2.5 M glycine was added to stop the reaction. Cells were washed three times with cold 1 M PBS and collected in lysis buffer 1 (140 mM NaCl, 1 mM EDTA, 50 mM HEPES-KOH [pH 7.5], 0.25% Triton X-100, 10% glycerol, 0.5% NP-40, and protease inhibitor). Then sample was resuspended in lysis buffer (10 mM Tris-HCl [pH 8.0], 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and protease inhibitor). After centrifugation, the samples were resuspended and sonicated in lysis buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, and protease inhibitor). Each sample was then incubated with anti-H3K27me3 magnetic beads at 4°C overnight. After washing six times with washing buffer, the beads-antibody-DNA complex was incubated at 65°C to reverse the covalent histone-DNA bonds. The DNA was extracted and analyzed by real-time PCR. The primers are listed in Table S1. The data were analyzed as previously described (Zhang et al., 2014).
Confocal Imaging and Statistical Analysis
All images were captured with a Zeiss 780 laser scanning confocal microscope. Statistical analyses were performed using a one-way ANOVA or t test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 in figures). All bar graphs show the mean ± SEM. For analysis of all western blotting and RT-PCR experiments, at least three individual experiments were performed.
Author Contributions
X.L. designed the research, performed the research, analyzed the data, and wrote the manuscript; J.J. supervised the project and designed the research.
Acknowledgments
This work was supported by grants from the National Science Foundation of China (31730033 and 31621004), the National Key Basic Research Program of China (2015CB964500 and 2014CB964903), the Strategic Priority Research Program (XDA16020602), and the K.C. Wong Education Foundation.
Published: March 15, 2018
Footnotes
Supplemental Information includes seven figures and one table and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.02.008.
Accession Numbers
The RNA-seq raw data in female E14 wild-type (control) and Utx cKO forebrain have been deposited in NCBI's GEO and are accessible through series accession number GEO: GSE110392.
Supplemental Information
References
- Andrews W.D., Barber M., Nemitz M., Memi F., Parnavelas J.G. Semaphorin3A-neuropilin1 signalling is involved in the generation of cortical interneurons. Brain Struct. Funct. 2017;222:2217–2233. doi: 10.1007/s00429-016-1337-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeille E., Reynaud F., Sanyas I., Bozon M., Kindbeiter K., Causeret F., Pierani A., Falk J., Moret F., Castellani V. Cerebrospinal fluid-derived Semaphorin3B orients neuroepithelial cell divisions in the apicobasal axis. Nat. Commun. 2015;6:6366. doi: 10.1038/ncomms7366. [DOI] [PubMed] [Google Scholar]
- Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Duque A., Rakic P. Different effects of bromodeoxyuridine and [3H]thymidine incorporation into DNA on cell proliferation, position, and fate. J. Neurosci. 2011;31:15205–15217. doi: 10.1523/JNEUROSCI.3092-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durak O., Gao F., Kaeser-Woo Y.J., Rueda R., Martorell A.J., Nott A., Liu C.Y., Watson L.A., Tsai L.H. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat. Neurosci. 2016;19:1477–1488. doi: 10.1038/nn.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Cho Y.W., Yu L.R., Yu H., Veenstra T.D., Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I., Kageyama R. bHLH factors in self-renewal, multipotency, and fate choice of neural progenitor cells. Neuron. 2014;82:9–23. doi: 10.1016/j.neuron.2014.03.018. [DOI] [PubMed] [Google Scholar]
- Kim J., Lee C.G. Coinheritance of novel mutations in SCN1A causing GEFS+ and in KDM6A causing Kabuki syndrome in a family. Ann. Clin. Lab. Sci. 2017;47:229–235. [PubMed] [Google Scholar]
- Lambert J.F., Benoit B.O., Colvin G.A., Carlson J., Delville Y., Quesenberry P.J. Quick sex determination of mouse fetuses. J. Neurosci. Methods. 2000;95:127–132. doi: 10.1016/s0165-0270(99)00157-0. [DOI] [PubMed] [Google Scholar]
- LaMonica B.E., Lui J.H., Hansen D.V., Kriegstein A.R. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nat. Commun. 2013;4:1665. doi: 10.1038/ncomms2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee J.W., Lee S.K. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev. Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui N.C., Tam W.Y., Gao C., Huang J.D., Wang C.C., Jiang L., Yung W.H., Kwan K.M. Lhx1/5 control dendritogenesis and spine morphogenesis of Purkinje cells via regulation of Espin. Nat. Commun. 2017;8:15079. doi: 10.1038/ncomms15079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M.F. X-chromosome inactivation and developmental patterns in mammals. Biol. Rev. Camb. Philos. Soc. 1972;47:1–35. doi: 10.1111/j.1469-185x.1972.tb00969.x. [DOI] [PubMed] [Google Scholar]
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- Miyake N., Koshimizu E., Okamoto N., Mizuno S., Ogata T., Nagai T., Kosho T., Ohashi H., Kato M., Sasaki G. MLL2 and KDM6A mutations in patients with Kabuki syndrome. Am. J. Med. Genet. A. 2013;161A:2234–2243. doi: 10.1002/ajmg.a.36072. [DOI] [PubMed] [Google Scholar]
- Miyake N., Mizuno S., Okamoto N., Ohashi H., Shiina M., Ogata K., Tsurusaki Y., Nakashima M., Saitsu H., Niikawa N. KDM6A point mutations cause Kabuki syndrome. Hum. Mutat. 2013;34:108–110. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- Morales Torres C., Laugesen A., Helin K. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS One. 2013;8:e60020. doi: 10.1371/journal.pone.0060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrup D., Yagi R., Cui K., Proctor W.R., Wang C., Placek K., Pohl L.R., Wang R., Ge K., Zhu J. Histone demethylases UTX and JMJD3 are required for NKT cell development in mice. Cell Biosci. 2017;7:25. doi: 10.1186/s13578-017-0152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontious A., Kowalczyk T., Englund C., Hevner R.F. Role of intermediate progenitor cells in cerebral cortex development. Dev. Neurosci. 2008;30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- Shpargel K.B., Sengoku T., Yokoyama S., Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel K.B., Starmer J., Yee D., Pohlers M., Magnuson T. KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet. 2014;10:e1004507. doi: 10.1371/journal.pgen.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F., Banerjee R., Conrad P.A., Scacheri P.C., Harte P.J. Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol. Cell. Biol. 2012;32:2323–2334. doi: 10.1128/MCB.06392-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Burgoyne P.S., Arnold A.P. Sex differences in sex chromosome gene expression in mouse brain. Hum. Mol. Genet. 2002;11:1409–1419. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- Yelagandula R., Stroud H., Holec S., Zhou K., Feng S., Zhong X., Muthurajan U.M., Nie X., Kawashima T., Groth M. The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell. 2014;158:98–109. doi: 10.1016/j.cell.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J., Youn Y.H., Darling D., Toyo-Oka K., Pramparo T., Hirotsune S., Wynshaw-Boris A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo K.H., Oh S., Kang K., Wang C., Robinson G.W., Ge K., Hennighausen L. Histone demethylase KDM6A controls the mammary luminal lineage through enzyme-independent mechanisms. Mol. Cell. Biol. 2016;36:2108–2120. doi: 10.1128/MCB.00089-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ji F., Liu Y., Lei X., Li H., Ji G., Yuan Z., Jiao J. Ezh2 regulates adult hippocampal neurogenesis and memory. J. Neurosci. 2014;34:5184–5199. doi: 10.1523/JNEUROSCI.4129-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.