Abstract
Background: Whether load carriage (LC), an endurance exercise mode composed of the aerobic component of traditional endurance exercise [e.g., cycle ergometry (CE)] and contractile forces characteristic of resistive-type exercise, modulates acute mitochondrial adaptive responses to endurance exercise and supplemental nutrition [carbohydrate + essential amino acids (CHO+EAA)] is not known.
Objective: The aim of this study was to examine the effects of LC and CE, with or without CHO+EAA supplementation, on acute markers of mitochondrial biogenesis.
Methods: Twenty-five adults performed 90 min of metabolically matched LC (treadmill walking, wearing a vest equal to 30% of body mass) or CE exercise during which CHO+EAA (46 g carbohydrate and 10 g essential amino acids) or non-nutritive control (CON) drinks were consumed. Muscle biopsy samples were collected at rest (pre-exercise), post-exercise, and after 3 h of recovery to assess citrate synthase activity and the expression of mRNA (reverse transcriptase–quantitative polymerase chain reaction) and protein (Western blot).
Results: Citrate synthase and phosphorylated p38 mitogen-activated protein kinase (p38 MAPK)Thr180/Tyr182 were elevated postexercise compared with pre-exercise (time main effect, P < 0.05). Peroxisome proliferator-activated γ-receptor coactivator 1α (PGC-1α) expression was highest after recovery for CE compared with LC (exercise-by-time effect, P < 0.05). Sirtuin 1 (SIRT1) expression postexercise was higher for CON than for CHO+EAA treatments (drink-by-time, P < 0.05). Tumor suppressor p53 (p53), mitochondrial transcription factor A (TFAM), and cytochrome c oxidase subunit IV (COXIV) expression was greater for CON than for CHO+EAA treatments (drink main effect, P < 0.05). PGC-1α and p53 expressions were positively associated (P < 0.05) with TFAM (r = 0.629 and 0.736, respectively) and COXIV (r = 0.465 and 0.461, respectively) expressions.
Conclusions: Acute mitochondrial adaptive responses to endurance exercise appear to be largely driven by exogenous nutrition availability. Although CE upregulated PGC-1α expression to a greater extent than LC, downstream signaling was the same between modes, suggesting that LC, in large part, elicits the same acute mitochondrial response as traditional, non–weight-bearing endurance exercise. This trial was registered at clinicaltrials.gov as NCT01714479.
Keywords: PGC-1α, p53, SIRT1, TFAM, COXIV, concurrent exercise
Introduction
Skeletal muscle is a malleable tissue that responds to acute exercise and diet-induced alterations in molecular signaling, resulting in long-term aerobic adaptations with training (1, 2). The peroxisome proliferator-activated γ-receptor coactivator 1α (PGC-1α) is the key regulator of skeletal muscle adaptions to endurance-type exercise, responsive to exogenous and endogenous carbohydrate availability (3–5). Increased expression of PGC-1α has been linked to enhanced oxidative capacity by activating mitochondrial transcription factor A (TFAM) and upregulating mitochondrial biogenesis (6, 7). Exercise- and nutrition-induced mitochondrial biogenesis may also be stimulated by the tumor suppressor p53 (p53), which targets TFAM to stimulate mitochondrial DNA expression (8, 9). Multiple upstream targets, which include AMP-activated protein kinase (AMPK), p38 mitogen-activated protein kinase (p38 MAPK), and sirtuin 1 (SIRT1), govern the activation of PGC-1α and p53 and are responsive to muscle contractile activity and diminished intracellular energy availability (Figure 1) (9–14). Muscle aerobic capacity is enhanced with repeated stimulation of this pathway and subsequent increases in mitochondrial biogenesis with training, resulting in improved endurance exercise performance (15, 16).
FIGURE 1.
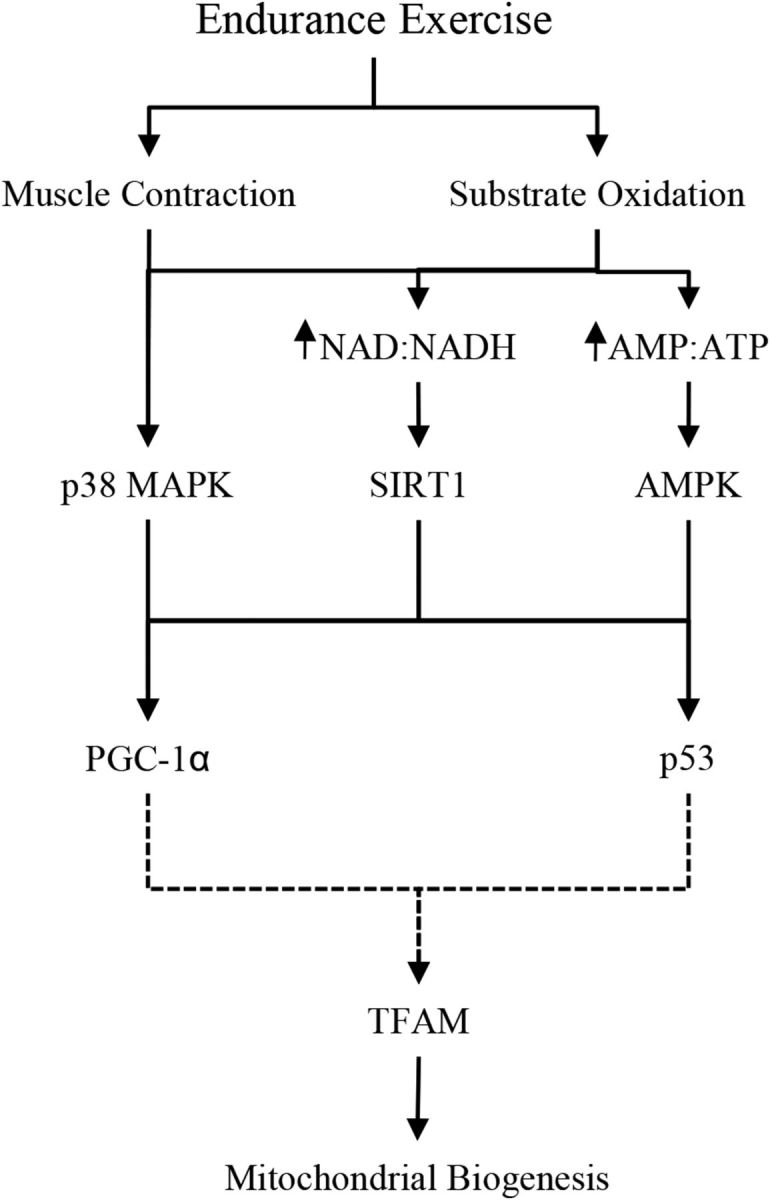
PGC-1α pathway schematic. AMPK, AMP-activated protein kinase; PGC-1α, peroxisome proliferator-activated γ-receptor coactivator 1α p38 MAPK, p38 mitogen-activated protein kinase; p53, tumor suppressor p53; RER, respiratory exchange ratio; SIRT1, sirtuin 1; TFAM, mitochondrial transcription factor A.
It is well accepted that mitochondrial biogenesis is stimulated by traditional endurance exercise (1), and acute responses appear to be sensitive to exogenous and endogenous carbohydrate availability and exercise mode. Maintaining endogenous carbohydrate availability (i.e., glycogen) by manipulating dietary carbohydrate intake, or by ingesting carbohydrate with exercise, is generally recommended to sustain or improve endurance exercise performance (17). However, exercising in a carbohydrate-restricted state (i.e., fasted with low muscle glycogen status) (18, 19) may produce greater acute mitochondrial adaptive stimulus because PGC-1α expression is upregulated to a greater extent than when exercise is performed in a carbohydrate-replete state (7, 9, 20). Interestingly, PGC-1α mRNA expression is upregulated to a greater extent after endurance exercise when resistance exercise is performed concurrently (21), suggesting the contractile forces of traditional resistance exercise may modulate mitochondrial biogenesis signaling (22). Studies have also reported that performing resistance exercise in a carbohydrate-restricted state potentiates resistance exercise–induced changes in PGC-1α transcription (23). These studies suggest that combining the mechanical strain of traditional resistance exercise and the metabolic stress of traditional endurance exercise, particularly when carbohydrate availability is limited, may enhance activation of mitochondrial biogenesis-related signaling.
Load carriage (LC), an endurance exercise mode commonly performed by military personnel (24), elicits mechanical forces that exceed traditional, non–weight-bearing endurance exercise. We recently reported greater myofibrillar muscle protein synthetic responses to LC than to cycle ergometry (CE) (25). This suggests that acute mitochondrial biogenesis-related signaling may be more pronounced for LC than for CE, particularly when carbohydrate availability is limited, although to our knowledge this has never been studied. This study compared molecular markers of mitochondrial biogenesis in response to LC and CE, with or without consuming a combined carbohydrate and essential amino acids (CHO+EAA) drink during exercise (i.e., reflecting restricted and adequate carbohydrate availability). We hypothesized that LC would stimulate a greater upregulation in the expression of various markers of mitochondrial biogenesis than CE, and that this response would be greatest when exogenous energy and carbohydrate were not consumed during the exercise bout.
Methods
Experimental design
After providing informed consent, 25 adults (23 men and 2 women; mean ± SD age, 22 ± 1 y; height, 177 ± 2 cm; weight, 82 ± 2 kg; VO2peak, 50 ± 1 mL ⋅ kg−1 ⋅ min−1) participated in this randomized, double-blind, placebo-controlled study (25). The data presented in this study were derived from a larger investigation; however, the sample size is smaller due to limited tissue availability. Volunteers were randomly assigned to 1 of 2 exercise experimental groups. One group performed CE and the other performed LC (weighted vest equal to 30% of body mass) on a treadmill. Within each exercise group, participants consumed either combined CHO+EAA [223 kcal, 46 g carbohydrate, and 10 g essential amino acids (EAA; 0.7 g histidine, 0.7 g isoleucine, 3.6 g leucine, 1.2 g lysine, 0.3 g methionine, 1.4 g phenylalanine, 1.0 g threonine, and 1 g valine)] or flavor-matched, non-nutritive control (CON; 22 kcal, 5 g carbohydrate) drinks at 0, 30, 60, and 90 min during the exercise session (LC-CHO+EAA: n = 6; CE-CHO+EAA: n = 7; LC-CON: n = 5; CE-CON: n = 7).
Exercise intensity (2.2 ± 0.6 L/min) and energy expenditure (1000 ± 26 kcal/90 min) were matched between modes as described elsewhere (25). Muscle biopsies of the vastus lateralis were performed at rest (pre-exercise), postexercise, and after 3 h of recovery and analyzed for markers of mitochondrial biogenesis-related signaling and activity. Dietary intake and physical activity were controlled as previously described (25). This study was approved by the Institutional Review Board at the US Army Research Institute of Environmental Medicine and registered at www.clinicaltrials.gov as NCT01714479.
Substrate oxidation
Respiratory exchange ratio (RER; 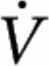 CO2/
CO2/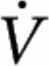 O2) was determined by using indirect calorimetry (ParvoMedics) every 30 min during exercise. Nonprotein substrate oxidation was calculated from
O2) was determined by using indirect calorimetry (ParvoMedics) every 30 min during exercise. Nonprotein substrate oxidation was calculated from 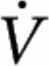 CO2 and
CO2 and 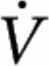 O2 measurements during exercise (ParvoMedics) (26, 27), as follows:
O2 measurements during exercise (ParvoMedics) (26, 27), as follows:
 |
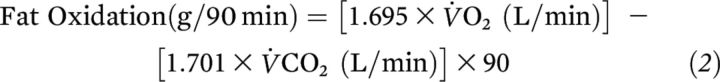 |
Citrate synthase activity assay
Citrate synthase activity was assessed to determine mitochondrial activity by using pre-exercise and postexercise time points due to limited sample availability. The analysis was performed by using whole-cell lysate from ∼30 mg muscle homogenized in ice-cold RIPA (radio-immunoprecipitation assay) (ThermoFisher) homogenization buffer (1:10 wt:vol) containing 1 mM DTT, phosphatase (PhosSTOP; Roche), and protease (cOmplete, ULTA Tablet; Roche) inhibitors. Enzyme activity was determined by using a colorimetric assay by combining 10 μL diluted (1:10; 0.1 M Tris HCl, pH 8.1) sample to 150 μL reaction master mix (1 mL DNTB [5,5-dithio-bis-(2-nitrobenzoic acid)], 3 mg acetyl-CoA, and 8 mL 0.1 M Tris HCl, pH 8.1) and 10 μL 10 mM oxaloacetate. Samples were analyzed at 412 nm on a SpectraMax 3 microplate reader (Molecular Devices). Data were normalized to protein content.
Western blotting
Western blot analysis was only performed for pre- and postexercise time periods due to limited sample availability. Homogenates used for the citrate synthase activity assay were centrifuged for 15 min at 10,000 × g at 4°C, the supernatant (lysate) was collected, and protein content was analyzed (ThermoFisher). Equal amounts of total protein (15 µg) were used in precast 4–20% Mini-PROTEAN TGX gels (Bio-Rad Laboratories). Proteins were transferred to polyvinylidene fluoride membranes and exposed to phosphorylated p38 MAPKThr180/Tyr182 and phosphorylated AMPKThr172 (Cell Signaling Technology) primary antibodies at 4°C overnight. Labeling was performed by using secondary antibody (anti-rabbit IgG conjugate with HRP; Cell Signaling Technology) and chemiluminescent reagents (Super Signal, West Pico Kit; Pierce Biotechnology). Blots were quantified by using a phosphoimager (ChemiDoc XRS; Bio-Rad) and Image Lab software (Bio-Rad). GAPDH was used to confirm equal protein loading per well. All data are presented as fold changes relative to the pre-exercise time period.
mRNA expression
Total RNA was isolated from ∼20 mg muscle (pre-exercise, postexercise, and recovery period) by using a miRNeasy Mini kit (Qiagen) and assessed for quantity and quality with the use of a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies). Equal amounts of RNA were reverse transcribed by using a TaqMan cDNA with RNase Inhibitor Reverse Transcription kit (Applied Biosystems). Real-time PCR with the use of individual TaqMan Gene Expression Assays (PGC-1α, SIRT1, p53, and TFAM; Applied Biosystems) were performed in triplicate. Data were normalized to the geometric mean of 18S and RNU48. Fold changes for mRNA were calculated by using the ∆∆CT method (28) and expressed relative to the pre-exercise time period.
Statistical analyses
A univariate ANOVA was used to determine the main effect of exercise mode (LC compared with CE) and drink (CHO+EAA compared with CON) on RER. Mixed-model repeated-measures ANOVA was used to determine main effects and interactions of exercise mode, drink, and time (pre-exercise, postexercise, and recovery period) for mRNA expression. Mixed-model repeated-measures ANOVA was also used to determine main effects and interactions for Western blotting and citrate synthase activity; however, due to the limited sample availability, analyses were only included at pre- and postexercise time periods. Bonferroni adjustments were used for post hoc comparisons if interactions were observed. Correlations between mRNA data were assessed by using Pearson correlation coefficients. Data were log transformed (log2) for correlations to maintain equal scale between positive and negative values. Significance was set at P < 0.05. Data were analyzed by using SPSS (version 21.0, 2010; SPSS, Inc.) and expressed as means ± SEMs.
Results
Independent of drink, RER was higher (P < 0.05) during CE (0.06 ± 0.02 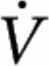 CO2/
CO2/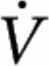 O2) than during LC (Table 1). Carbohydrate oxidation was higher (P < 0.05) during CE (64 ± 16 g/90 min) than during LC, regardless of drink. Fat oxidation was lower (P < 0.05) during CE (22 ± 6 g/90 min) than during LC, independent of drink.
O2) than during LC (Table 1). Carbohydrate oxidation was higher (P < 0.05) during CE (64 ± 16 g/90 min) than during LC, regardless of drink. Fat oxidation was lower (P < 0.05) during CE (22 ± 6 g/90 min) than during LC, independent of drink.
TABLE 1.
Substrate oxidation1
| CHO+EAA | CON | |||
|---|---|---|---|---|
| Load carriage | Cycle ergometry | Load carriage | Cycle ergometry | |
RER, 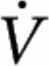 CO2/ CO2/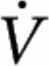 O2 O2
|
0.85 ± 0.02 | 0.91 ± 0.02‡ | 0.87 ± 0.02 | 0.93 ± 0.02‡ |
| Carbohydrate oxidation, g/90 min | 123 ± 16 | 196 ± 16‡ | 140 ± 15 | 195 ± 15‡ |
| Fat oxidation, g/90 min | 52 ± 7 | 30 ± 7‡ | 45 ± 6 | 24 ± 6‡ |
Values are means ± SEMs. ‡Different from load carriage, P < 0.05. CHO+EAA, carbohydrate (46 g) plus essential amino acids (10 g); CON, control (non-nutritive); RER, respiratory exchange ratio; 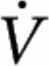 CO2, volume of carbon dioxide produced;
CO2, volume of carbon dioxide produced; 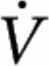 O2, volume of oxygen consumed.
O2, volume of oxygen consumed.
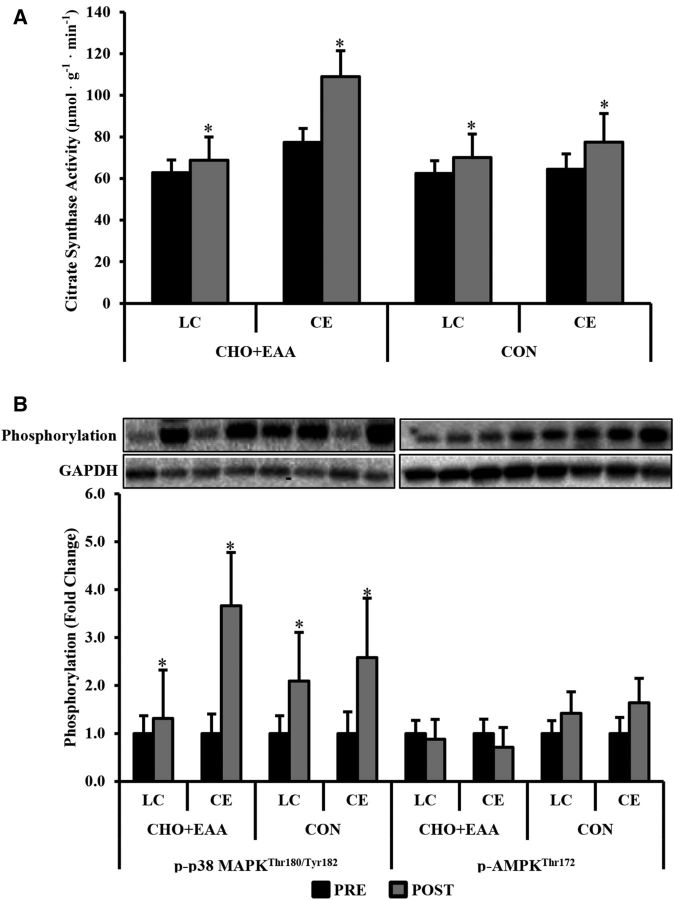
Citrate synthase activity was higher postexercise than pre-exercise (14.6 ± 5.4 µmol ⋅ g−1 ⋅ min−1; P < 0.05), regardless of exercise mode (Figure 2A). Postexercise phosphorylated p38 MAPKThr180/Tyr182 was 2.41 ± 0.55-fold higher than at pre-exercise (P < 0.05), independent of exercise mode and drink (Figure 2B). No changes were observed in phosphorylated AMPKThr172.
FIGURE 2.
Citrate synthase activity (A) and p-p38 MAPKThr180/Tyr182 and p-AMPKThr172 (B). Representative bands correspond to x axis labels below in panel B. Phosphorylation status was normalized to GAPDH, with data presented as fold changes relative to the pre-exercise time period for each group. Values are means ± SEMs. *Different from pre-exercise, P < 0.05. CE, cycle ergometry; CHO+EAA, carbohydrate plus essential amino acids; CON, control; LC, load carriage; p-AMPK, phosphorylated AMP-activated protein kinase; p-p38 MAPK, phosphorylated p38 mitogen-activated protein kinase; POST, postexercise; PRE, pre-exercise.
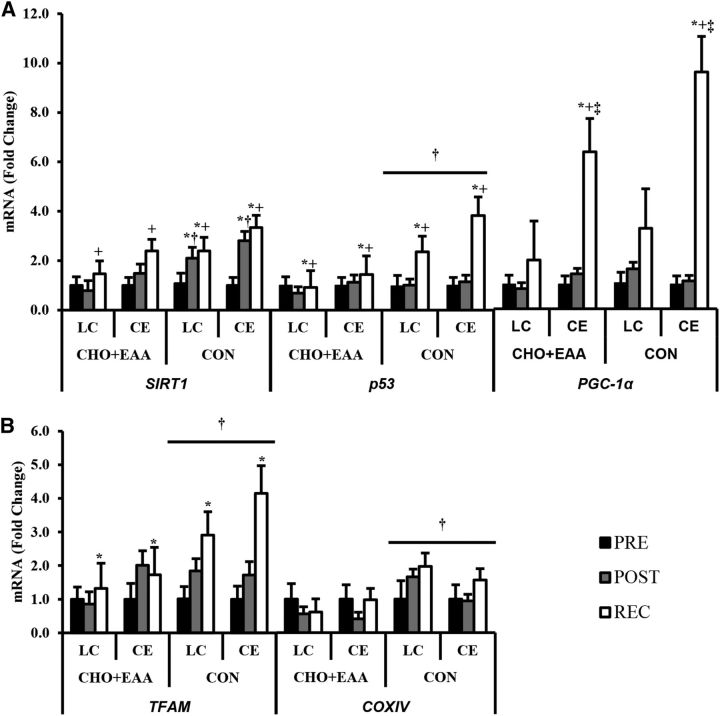
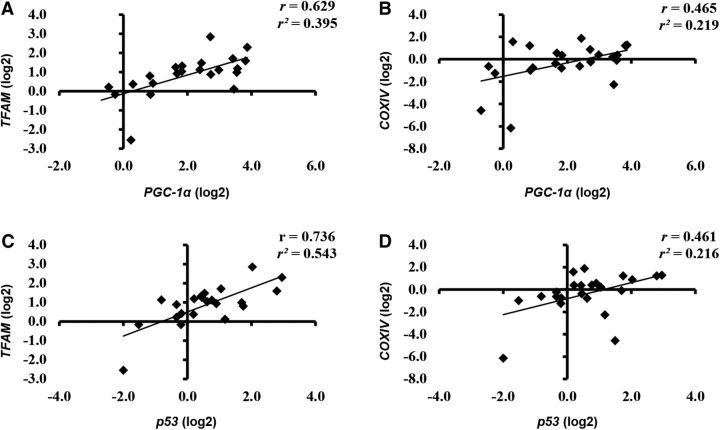
PGC-1α expression was highest with CE compared with LC during the recovery period (exercise-by-time interaction; P < 0.05; Figure 3A). SIRT1 expression was elevated in CON to a greater extent than CHO+EAA at postexercise (drink-by-time interaction; P < 0.05). The expression of p53 and TFAM was higher during the recovery period than at pre-exercise (time main effect; P < 0.05). p53, TFAM, and cytochrome c oxidase subunit IV (COXIV) expression was higher with CON than with CHO+EAA (drink main effect; P < 0.05; Figure 3B). No other main effects of interactions were observed. PGC-1α and p53 expressions were positively associated (P < 0.05) with TFAM (r = 0.629 and 0.736, respectively) and COXIV (r = 0.465 and 0.461, respectively) expressions during the recovery period (Figure 4).
FIGURE 3.
mRNA expressions of SIRT1, PGC-1α, and p53 (A) and TFAM and COXIV (B). Data are presented as fold changes relative to mean pre-exercise values for each group. Values are means ± SEMs. *Different from pre-exercise, P < 0.05. +Different from postexercise, P < 0.05. ‡Different from LC, P < 0.05. †Different from CHO+EAA, P < 0.05. CE, cycle ergometry; CHO+EAA, carbohydrate plus essential amino acids; CON, control; COXIV, cytochrome c oxidase subunit IV; LC, load carriage; PGC-1α, peroxisome proliferator-activated γ-receptor coactivator 1α POST, postexercise; PRE, pre-exercise; p53, tumor suppressor p53; REC, recovery; SIRT1, sirtuin 1; TFAM, mitochondrial transcription factor A.
FIGURE 4.
Correlation of PGC-1α to TFAM (A) and COXIV (B) expression and of p53 to TFAM (C) and COXIV (D) expression. Data are reported as log2 values for correlations to maintain equal scale between positive and negative values. COXIV, cytochrome c oxidase subunit IV; PGC-1α, peroxisome proliferator-activated γ-receptor coactivator 1α p53, tumor suppressor p53; TFAM, mitochondrial transcription factor A.
Discussion
The primary outcome of this study was that mRNA expression of PGC-1α was markedly greater after a bout of CE than after LC. However, despite the marked differences in PGC-1α expression between LC and CE, SIRT1, p53, TFAM, and COXIV transcription was not affected by exercise mode. SIRT1, p53, TFAM, and COXIV expressions were, however, upregulated to a greater extent when participants ingested the non-nutritive CON drink than after the CHO+EAA drink. These findings suggest that the expression of metabolic genes associated with regulating mitochondrial biogenesis is largely driven by substrate availability and only modestly affected by mode.
Contrary to our hypothesis, PGC-1α mRNA expression during recovery was upregulated to a greater extent for CE than for LC. One explanation is that the localized contractile forces and metabolic stress imposed on the quadriceps biopsied may have differed between LC and CE. We previously reported that the muscle protein synthetic response to LC exceeded the response to CE, which suggests that, in the current analysis, the metabolic stress with CE exceeded LC. This theory is supported, in part, by RER, which was greater for CE, despite matching the exercise modes for oxygen consumption (2.2 L/min) and energy expenditure (1000 kcal/90 min). Elevated RER resulted in higher rates of carbohydrate oxidation and lower rates of fat oxidation for CE than for LC. Similarly, others have reported lower fat oxidation with CE than with a more whole-body endurance exercise mode (e.g., running) when exercise was matched at 60% VO2max (29). In the study that showed that PGC-1α expression was higher after performing concurrent CE (65% VO2max) and resistance exercise compared with CE alone, the participants performed 6 sets of leg presses (70–80% 1-repetition maximum) isolating the quadriceps, which likely generated contractile forces different from LC (21). Taken together, the difference between the metabolic and contractile stressors of LC (i.e., whole body) and the more localized stress of CE likely explains the observed differences in PGC-1α.
Performing LC and CE in a fasted state upregulated SIRT1, p53, TFAM, and COXIV expressions to a greater extent than when the CHO+EAA drinks were consumed. These findings are consistent with past studies that showed that the provision of exogenous energy and carbohydrate during endurance exercise dampens transcriptional activation regulating mitochondrial biogenesis (5, 9, 20). Although we were unable to detect diet differences in PGC-1α expression between the CON and CHO+EAA treatments, there was a trend toward significance (P = 0.09) and PGC-1α and p53 were strongly associated with downstream targets, TFAM and COXIV, during the recovery period. These correlation data, together with previous reports (30, 31), provide further evidence that after endurance exercise, PGC-1α and p53 likely stimulate the transcription of TFAM to upregulate mitochondrial DNA expression (6, 7, 32), as well as COXIV, an indicator of mitochondrial count (30, 33). The lack of an effect on transcription of TFAM and COXIV suggests that LC likely does not inhibit mitochondrial biogenesis compared with CE.
It is unlikely that the addition of EAA to the carbohydrate altered the effects of carbohydrate availability on mitochondrial biogenesis-related gene expression. In vitro analysis has suggested that the EAA leucine may increase mitochondrial size in skeletal muscle through the activation of SIRT1 and subsequent stimulation of PGC-1α (34, 35). However, previous in vivo human investigations have reported no difference in mitochondrial protein synthesis with the consumption of a combined carbohydrate and protein supplement compared with a non-nutritive control (36) or carbohydrate alone (37) after a bout of endurance exercise. Similarly, consumption of dietary protein (casein hydrolysate or whey) alone during or immediately after endurance (38) or resistance (39) exercise does not elevate PGC-1α mRNA expression to a greater extent than does a non-nutritive control. Together, these studies suggest that the inclusion of protein will not alter the downregulation of mitochondrial biogenesis markers when carbohydrate is consumed during exercise.
Despite endurance exercise mode and nutrient supplementation effects on the transcription of PGC-1α, SIRT1, p53, TFAM, and COXIV during the recovery period, no treatment effects were observed for phosphorylation status of AMPK and p38 MAPK, or citrate synthase activity. Previous work has shown that the activation of AMPK and p38 MAPK (9, 10, 23) and citrate synthase (40, 41) is sensitive to exercise and carbohydrate availability. The lack of an effect between treatment groups in the present study may potentially be due to our limited sample size per group. In addition, the lack of a recovery muscle sample to assess signaling and citrate synthase activity may have also contributed to no treatment effects being observed. Because transcriptional modifications of the mitochondrial markers were most pronounced at recovery, treatment effects on signaling and citrate synthase may have been missed due to a lack of tissue to sample at this time point. Furthermore, reliance on citrate synthase to assess mitochondrial activity may have limited our ability to distinguish differences between exercise mode and dietary supplementation. Additional assessments of isocitrate dehydrogenase and α-ketoglutarate dehydrogenase, rate-limiting enzymes of the TCA cycle, may have provided further insight into the impact of our intervention on mitochondrial activity. However, the current study was limited by sample availability.
In conclusion, results from this investigation further suggest that performing endurance exercise when energy and carbohydrate availability is low stimulates the expression of metabolic genes associated with mitochondrial biogenesis. Although LC resulted in a lower expression of PGC-1α than CE, the change in PGC-1α expression was proportional to change in expression of its downstream targets TFAM and COXIV, which were not negatively affected by exercise mode. This finding indicates that LC may not inhibit mitochondrial biogenesis. Future investigation should focus on the long-term training impact of LC compared with CE on mitochondrial biogenesis and adaptations to training.
Acknowledgments
We acknowledge Gregory Lin and Patrick Radcliffe for their significant contributions to the project. The authors' responsibilities were as follows—LMM, HLM, and SMP: designed the research (project conception, development of overall research plan, and study oversight); LMM, NEM, CTC, and SMP: analyzed the data, interpreted the results of experiments, prepared the figures, and drafted the manuscript; and all authors: conducted the research (hands-on conduct of the experiments and data collection) and revised the manuscript, and read and approved the final manuscript.
Abbreviations
- AMPK
AMP-activated protein kinase
- CE
cycle ergometry
- CHO+EAA
carbohydrate plus essential amino acids
- CON
control
- COXIV
cytochrome c oxidase subunit IV
- EAA
essential amino acids
- LC
load carriage
- PGC-1α
peroxisome proliferator-activated γ-receptor coactivator 1α
- p38 MAPK
p38 mitogen-activated protein kinase
- p53
tumor suppressor p53
- RER
respiratory exchange ratio
- SIRT1
sirtuin 1
- TFAM
mitochondrial transcription factor A
-
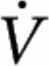 CO2
CO2
volume of carbon dioxide produced
-
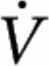 O2
O2
volume of oxygen consumed
-
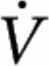 O2max
O2max maximal oxygen uptake
-
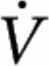 O2peak
O2peak peak volume of oxygen consumed
Footnotes
The investigators adhered to the policies for protection of human subjects as prescribed in Army Regulation 70-25, and the research was conducted in adherence with the provisions of 32 CFR part 219. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Army or the Department of Defense. Any citations of commercial organizations and trade names in this report do not constitute an official Department of the Army endorsement of approval of the products or services of these organizations. This study was approved by the Institutional Review Board at the US Army Research Institute of Environmental Medicine.
References
- 1. Hood DA, Irrcher I, Ljubicic V, Joseph AM.. Coordination of metabolic plasticity in skeletal muscle. J Exp Biol 2006;209:2265–75. [DOI] [PubMed] [Google Scholar]
- 2. Margolis LM, Pasiakos SM.. Optimizing intramuscular adaptations to aerobic exercise: effects of carbohydrate restriction and protein supplementation on mitochondrial biogenesis. Adv Nutr 2013;4:657–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO.. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J 2002;16:1879–86. [DOI] [PubMed] [Google Scholar]
- 4. Baar K.. Nutrition and the adaptation to endurance training. Sports Med 2014;44(Suppl 1):S5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pilegaard H, Osada T, Andersen LT, Helge JW, Saltin B, Neufer PD.. Substrate availability and transcriptional regulation of metabolic genes in human skeletal muscle during recovery from exercise. Metabolism 2005;54:1048–55. [DOI] [PubMed] [Google Scholar]
- 6. Safdar A, Little JP, Stokl AJ, Hettinga BP, Akhtar M, Tarnopolsky MA.. Exercise increases mitochondrial PGC-1alpha content and promotes nuclear-mitochondrial cross-talk to coordinate mitochondrial biogenesis. J Biol Chem 2011;286:10605–17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7. Irrcher I, Adhihetty PJ, Sheehan T, Joseph AM, Hood DA.. PPARgamma coactivator-1alpha expression during thyroid hormone- and contractile activity-induced mitochondrial adaptations. Am J Physiol Cell Physiol 2003;284:C1669–77. [DOI] [PubMed] [Google Scholar]
- 8. Saleem A, Hood DA.. Acute exercise induces tumour suppressor protein p53 translocation to the mitochondria and promotes a p53-Tfam-mitochondrial DNA complex in skeletal muscle. J Physiol 2013;591:3625–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bartlett JD, Louhelainen J, Iqbal Z, Cochran AJ, Gibala MJ, Gregson W, Close GL, Drust B, Morton JP.. Reduced carbohydrate availability enhances exercise-induced p53 signaling in human skeletal muscle: implications for mitochondrial biogenesis. Am J Physiol Regul Integr Comp Physiol 2013;304:R450–8. [DOI] [PubMed] [Google Scholar]
- 10. Bartlett JD, Hwa Joo C, Jeong TS, Louhelainen J, Cochran AJ, Gibala MJ, Gregson W, Close GL, Drust B, Morton JP.. Matched work high-intensity interval and continuous running induce similar increases in PGC-1alpha mRNA, AMPK, p38, and p53 phosphorylation in human skeletal muscle. J Appl Physiol (1985) 2012;112:1135–43. [DOI] [PubMed] [Google Scholar]
- 11. Gibala MJ, McGee SL, Garnham AP, Howlett KF, Snow RJ, Hargreaves M.. Brief intense interval exercise activates AMPK and p38 MAPK signaling and increases the expression of PGC-1alpha in human skeletal muscle. J Appl Physiol (1985) 2009;106:929–34. [DOI] [PubMed] [Google Scholar]
- 12. Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P.. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J 2007;26:1913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, Spiegelman BM.. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol Cell 2001;8:971–82. [DOI] [PubMed] [Google Scholar]
- 14. Philp A, Schenk S.. Unraveling the complexities of SIRT1-mediated mitochondrial regulation in skeletal muscle. Exerc Sport Sci Rev 2013;41:174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tadaishi M, Miura S, Kai Y, Kano Y, Oishi Y, Ezaki O.. Skeletal muscle-specific expression of PGC-1alpha-b, an exercise-responsive isoform, increases exercise capacity and peak oxygen uptake. PLoS One 2011;6:e28290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Calvo JA, Daniels TG, Wang X, Paul A, Lin J, Spiegelman BM, Stevenson SC, Rangwala SM.. Muscle-specific expression of PPARgamma coactivator-1alpha improves exercise performance and increases peak oxygen uptake. J Appl Physiol (1985) 2008;104:1304–12. [DOI] [PubMed] [Google Scholar]
- 17. Cermak NM, van Loon LJ.. The use of carbohydrates during exercise as an ergogenic aid. Sports Med 2013;43:1139–55. [DOI] [PubMed] [Google Scholar]
- 18. Hansen AK, Fischer CP, Plomgaard P, Andersen JL, Saltin B, Pedersen BK.. Skeletal muscle adaptation: training twice every second day vs. training once daily. J Appl Physiol (1985) 2005;98:93–9. [DOI] [PubMed] [Google Scholar]
- 19. Yeo WK, Paton CD, Garnham AP, Burke LM, Carey AL, Hawley JA.. Skeletal muscle adaptation and performance responses to once a day versus twice every second day endurance training regimens. J Appl Physiol (1985) 2008;105:1462–70. [DOI] [PubMed] [Google Scholar]
- 20. Psilander N, Frank P, Flockhart M, Sahlin K.. Exercise with low glycogen increases PGC-1alpha gene expression in human skeletal muscle. Eur J Appl Physiol 2013;113:951–63. [DOI] [PubMed] [Google Scholar]
- 21. Wang L, Mascher H, Psilander N, Blomstrand E, Sahlin K.. Resistance exercise enhances the molecular signaling of mitochondrial biogenesis induced by endurance exercise in human skeletal muscle. J Appl Physiol (1985) 2011;111:1335–44. [DOI] [PubMed] [Google Scholar]
- 22. Burd NA, Andrews RJ, West DW, Little JP, Cochran AJ, Hector AJ, Cashaback JG, Gibala MJ, Potvin JR, Baker SK, et al. . Muscle time under tension during resistance exercise stimulates differential muscle protein sub-fractional synthetic responses in men. J Physiol 2012;590:351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Camera DM, Hawley JA, Coffey VG.. Resistance exercise with low glycogen increases p53 phosphorylation and PGC-1alpha mRNA in skeletal muscle. Eur J Appl Physiol 2015;115:1185–94. [DOI] [PubMed] [Google Scholar]
- 24. Pasiakos SM, Margolis LM, Murphy NE, McClung HL, Martini S, Gundersen Y, Castellani JW, Karl JP, Teien HK, Madslien EH, et al. . Effects of exercise mode, energy, and macronutrient interventions on inflammation during military training. Physiol Rep 2016;4 pii: e12820. doi: 10.14814/phy2.12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pasiakos SM, McClung HL, Margolis LM, Murphy NE, Lin GG, Hydren JR, Young AJ.. Human muscle protein synthetic responses during weight-bearing and non-weight-bearing exercise: a comparative study of exercise modes and recovery nutrition. PLoS One 2015;10:e0140863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jeukendrup AE, Wallis GA.. Measurement of substrate oxidation during exercise by means of gas exchange measurements. Int J Sports Med 2005;26(Suppl 1):S28–37. [DOI] [PubMed] [Google Scholar]
- 27. Péronnet F, Massicotte D.. Table of nonprotein respiratory quotient: an update. Can J Sport Sci 1991;16:23–9. [PubMed] [Google Scholar]
- 28. Pfaffl MW.. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pfeiffer B, Stellingwerff T, Zaltas E, Hodgson AB, Jeukendrup AE.. Carbohydrate oxidation from a drink during running compared with cycling exercise. Med Sci Sports Exerc 2011;43:327–34. [DOI] [PubMed] [Google Scholar]
- 30. Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM.. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998;92:829–39. [DOI] [PubMed] [Google Scholar]
- 31. Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, et al. . Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999;98:115–24. [DOI] [PubMed] [Google Scholar]
- 32. Campbell CT, Kolesar JE, Kaufman BA.. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta 2012;1819:921–9. [DOI] [PubMed] [Google Scholar]
- 33. Uguccioni G, Hood DA.. The importance of PGC-1alpha in contractile activity-induced mitochondrial adaptations. Am J Physiol Endocrinol Metab 2011;300:E361–71. [DOI] [PubMed] [Google Scholar]
- 34. Liang C, Curry BJ, Brown PL, Zemel MB.. Leucine modulates mitochondrial biogenesis and SIRT1-AMPK signaling in C2C12 myotubes. J Nutr Metab 2014;2014:239750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun X, Zemel MB.. Leucine modulation of mitochondrial mass and oxygen consumption in skeletal muscle cells and adipocytes. Nutr Metab (Lond) 2009;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coffey VG, Moore DR, Burd NA, Rerecich T, Stellingwerff T, Garnham AP, Phillips SM, Hawley JA.. Nutrient provision increases signalling and protein synthesis in human skeletal muscle after repeated sprints. Eur J Appl Physiol 2011;111:1473–83. [DOI] [PubMed] [Google Scholar]
- 37. Breen L, Philp A, Witard OC, Jackman SR, Selby A, Smith K, Baar K, Tipton KD.. The influence of carbohydrate-protein co-ingestion following endurance exercise on myofibrillar and mitochondrial protein synthesis. J Physiol 2011;589:4011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taylor C, Bartlett JD, van de Graaf CS, Louhelainen J, Coyne V, Iqbal Z, Maclaren DP, Gregson W, Close GL, Morton JP.. Protein ingestion does not impair exercise-induced AMPK signalling when in a glycogen-depleted state: implications for train-low compete-high. Eur J Appl Physiol 2013;113:1457–68. [DOI] [PubMed] [Google Scholar]
- 39. Smiles WJ, Areta JL, Coffey VG, Phillips SM, Moore DR, Stellingwerff T, Burke LM, Hawley JA, Camera DM.. Modulation of autophagy signaling with resistance exercise and protein ingestion following short-term energy deficit. Am J Physiol Regul Integr Comp Physiol 2015;309:R603–12. [DOI] [PubMed] [Google Scholar]
- 40. Philp A, Chen A, Lan D, Meyer GA, Murphy AN, Knapp AE, Olfert IM, McCurdy CE, Marcotte GR, Hogan MC, et al. . Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) deacetylation following endurance exercise. J Biol Chem 2011;286:30561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Philp A, Hargreaves M, Baar K.. More than a store: regulatory roles for glycogen in skeletal muscle adaptation to exercise. Am J Physiol Endocrinol Metab 2012;302:E1343–51. [DOI] [PubMed] [Google Scholar]