Abstract
Despite considerable recent attention to problems with reproducibility of scientific research, there is a striking lack of agreement about the definition of the term. That is a problem, because the lack of a consensus definition makes it difficult to compare studies of reproducibility, and thus to have even a broad overview of the state of the issue in natural language processing. This paper proposes an ontology of reproducibility in that field. Its goal is to enhance both future research and communication about the topic, and retrospective meta-analyses. We show that three dimensions of reproducibility, corresponding to three kinds of claims in natural language processing papers, can account for a variety of types of research reports. These dimensions are reproducibility of a conclusion, of a finding, and of a value. Three biomedical natural language processing papers by the authors of this paper are analyzed with respect to these dimensions.
Keywords: methodology, reproducibility, repeatability, replicability, replicatability
1. Introduction
The journal Language Resources and Evaluation recently published an editorial on reproducibility in language processing. The editorial announced a new topic for the journal, named the associate editors for the topic, called for papers on the topic, and defined a number of relevant terms (Branco et al., 2017).
Before the editorial had appeared, the authors (who include two of the authors of this paper) had already submitted a correction: on further assessment of the literature, they had realized that they should reverse the definitions of two crucial terms (viz. reproducibility and replicability)1. It was a textbook example of publication of an analysis that turned out not to hold, and of how such an analysis should be handled.
It is especially striking that the topic of the correction was the definition of two rather central terms—failures of reproducibility have been so present in both the global scientific and the public consciousness that one could reasonably expect that there would be at least a broad consensus about the terminology that is used to discuss the phenomenon. And yet: even a cursory (and certainly a careful) review of the literature shows that no such consensus exists. Indeed, the community is not even close to a consensus. In definitions in the literature (multiple examples omitted from the abstract due to space constraints), one observes at least the following three terms used frequently to refer to the same two concerns: replicability, repeatability, and reproducibility. Less frequently, one also sees the terms commensurate, valid, and validity, and all of these can be involved in the definition of rigor (Kilicoglu, 2017). It is not uncommon to see reproducibility and replicability or repeatability used interchangeably in the same paper.
As to the things that may or may not be replicated or reproduced, the literature includes the experiment itself; specific values, measured or calculated; findings; conclusions; and confirmation or non-confirmation of a hypothesis. These terms that are used to define reproducibility are explicitly defined less frequently than are reproducibility, replicability, etc., so in the end, one is often not sure what, exactly, would count as “reproducible” or not, even in the presence of a definition. The situation is further complicated by the introduction of modifiers, e.g. to refer to weak reproducibility. Biological research literature often involves additional occasional confusion with the noun replicate, a count noun (one can have two or more replicates, or none) that refers to copies of a macromolecule under study (Vaux et al., 2012).
This lack of consensus definitions related to reproducibility is a problem because without them, we cannot compare studies of reproducibility. A number of such studies have appeared very recently, and in general, the results have been depressing. Multiple studies over the course of the past two years have reported widespread failures of reproducibility (Collaboration and others, 2015; Collberg et al., 2015). They range from unusually large-scale studies in psychology (Collaboration and others, 2015), to surprisingly large ones in computer science (Collberg et al., 2015), to case studies in natural language processing (Schwartz, 2010; Borgholt et al., 2015; Cohen et al., 2016; Gomes et al., 2016; Névéol et al., 2016; Cassidy and Estival, 2017; Kilicoglu, 2017; Mieskes, 2017). Yet, it is still quite difficult to get even a rough sense of the actual scale of the problem in natural language processing, because the lack of agreement about what exactly is being assessed makes it difficult to compare findings across papers on reproducibility issues.
To address this problem of a lack of consensus definitions, this paper proposes a set of dimensions of reproducibility. Perhaps counter-intuitively, we first give the definition of replicability or repeatability that we assume in the paper:
Replicability or repeatability is a property of an experiment: the ability to repeat—or not—the experiment described in a study.
Thus, we reserve the terms replicability or repeatability for the ability to repeat an experiment’s methods (in the case of natural language processing, with the same data). As (Drummond, 2009) puts it, “replicability…means to exactly replicate the original experiment”—the experiment must be repeated, but whether or not the same values are obtained, the same findings emerge, or the same conclusion is reached is not relevant to the question of whether or not the experiment has been replicated. In the lexicon of (Goodman et al., 2016), which discusses reproducibility and replicability from the perspective of the broader field of both computational and non-computational science, this corresponds to methods reproducibility.
We differentiate between replicability or repeatability on the one hand, and reproducibility on the other. We propose the following:
Reproducibility is a property of the outcomes of an experiment: arriving—or not—at the same conclusions, findings, or values.
With the disjunction conclusions, findings, or values, we see a likely cause of differing assessments of whether or not a “paper” has been reproduced: a subsequent study could replicate or repeat the experiment described in that paper, or vary the methodology in some small but interesting way, and arrive at the same or different values, or findings, or conclusions. Our proposal of three separate dimensions of reproducibility begins with the hypothesis that the lack of consensus about definitions of reproducibility is directly related to the fact that there are these multiple ways in which a paper might, or might not, be supported by subsequent work. Problems then arise when a single one of these dimensions is isolated and labelled as reproducibility. The result is essentially one of polysemy—with its attendant ambiguity.
To address this issue, the paper proposes a set of three things that we will refer to as dimensions of reproducibility, using here the sense of the word dimension as one of a group of properties whose number is necessary and sufficient to determine uniquely each element of a system2. The proposed dimensions are then evaluated with an adequacy test—we ask the question of whether or not publications in natural language processing can be mapped to the proposed dimensions. If a dimension were found not to be relevant to any paper in natural language processing, that would constitute evidence that it is not a valid dimension, or at least not a very useful one. On the other hand, if an analysis of publications in natural language processing resulted in very disparate aspects of papers being lumped into the same dimension, that would be consistent with the hypothesis that the dimension in question needed to be split into finer-grained categories.
To avoid punishing our colleagues for graciously sharing their findings with the community, we do this analysis with our own papers. This has the methodological shortcoming that it almost certainly introduces an element of bias into the analysis. For example, it is almost certainly the case that we assume things to be obvious in those papers that probably are not obvious to very many people outside of our own laboratories. On the other hand, it has the methodological advantage that we are intimately familiar with the papers, and so failures to find the relevant aspects of the papers in question are very unlikely to be due to not being familiar with the topic areas, or the methodologies, or the rationales behind the analyses. If they were not our own papers, all of these factors would certainly be possible confounds. In any case, we return to this methodological shortcoming in the Discussion section, and discuss some ways that it might be avoided in future work.
2. The proposed dimensions
We begin by establishing and constraining the scope of these dimensions.
We exclude issues related to the ability to repeat the experiments reported in a paper. We define that above as replicability or repeatability.
-
We take the unit of analysis as a paper. This could include conference papers, journal articles, or chapters in an edited volume. We exclude longer works, such as books, as well as shorter ones, such as published abstracts.
There is an impressive amount of research going back to at least 1993 (Yentis et al., 1993) on the topic of subsequent publication of work that was originally presented at medical conferences in abstract form. It is clear from these publications that there are things related to reproducibility to be investigated in abstracts, as well (Yentis et al., 1993; Scherer et al., 1994; Marx et al., 1999; Sanders et al., 2001; Bydder et al., 2004; Byerly et al., 2000; Jackson et al., 2000; Oliver et al., 2003; Herbison, 2004; Ng et al., 2004; Autorino et al., 2006; Balasubramanian et al., 2006; Peng et al., 2006; Rao et al., 2006; Smollin and Nelson, 2006; Scherer et al., 2007; Dahllof¨ et al., 2008; Harel et al., 2011; Varghese et al., 2011). However, since they are not a common publication type in natural language processing and this is a paper about reproducibility in natural language processing, we do not have the requisite data to establish whether or not these dimensions apply to them.
Within this scope, then, we propose the following three dimensions of reproducibility:
Reproducibility of a conclusion.
Reproducibility of a finding.
Reproducibility of a value.
We now expand on those dimensions.
2.1. Reproducibility of a conclusion
By conclusion, we mean a broad induction that is made based on the results of the reported research. Some examples of conclusions from our papers include:
The abstracts of scientific journal articles and the bodies of scientific journal articles have meaningfully different structural and linguistic characteristics. This conclusion in our paper (Cohen et al., 2010), which was quite clearly stated—it actually formed the title of the paper—was important at the time (and presumably now) because it demonstrated the importance of what has since become a major theme in biomedical natural language processing. Prior to this, the majority of biomedical natural language processing papers treated only the abstracts of scientific journal articles; full text was becoming widely available, and this paper’s conclusion supported the idea that there would be a crucial need for a very new research direction.
-
Clinical documents and scientific literature have very different distributional characteristics at multiple levels of negation. This conclusion in the paper (Cohen et al., 2017a) appeared in the context of a recent paper by Wu et al. (Wu et al., 2014) that had concluded that negation was very much not a solved problem in natural language processing—despite the fact that many papers had suggested that it is—and that in order for it to become a solved problem in natural language processing, the way forward was not to annotate more data from the same genres in which it was already available, but to annotate data with different distributional characteristics from the data that was already available. Wu et al.’s conclusion was well-reasoned, but at the time it was difficult to act on it, as there wasn’t actually much published data on what those distributions actually were. Our paper then showed some of the relevant characteristics of the distributions. In that context, our paper presented methodologies for doing apples-to-apples comparisons of the distributions of negation at both the phrasal and the sub-word (affixal) levels, as well as showing that those can vary completely independently.
One might ask whether a “conclusion” is even capable of being reproduced. Data and objects can be reproduced, but to the extent that conclusions happen in people’s minds, it is difficult to claim that they can be demonstrated to be the same.
So, it is important to specify that by “conclusion,” we mean an explicit statement in a paper. The fact that scientific papers often include a section labelled Conclusions should give even the most stalwart logical positivist (for a classic example in linguistics, consider Leonard Bloomfield (Bloomfield, 1936)) some confidence that such things exist.
2.2. Reproducibility of a finding
By finding, we mean a relationship between the values for some reported figure of merit with respect to two or more dependent variables. Two values could be equal—or not. These may be direct measurements (e.g. counts of true and false positives) or calculated numbers (e.g. a p-value less than some value for alpha, or not), but there must be a comparison involved. Findings of specific relationships between values—an F-measure higher with one classification algorithm than another, a strength of lexical association that is stronger in one genre than another—are at the heart of applications of natural language processing in the digital humanities (Moretti et al., 2008) and the essential starting point for natural-language-processing-based approaches to social science (Chateauraynaud, 2003; Née, 2017). More generally, they lie at the very heart of evaluation in natural language processing, where the most common trope is to compare the performance of one system as measured by some figure of merit to that of another (Resnik and Lin, 2010).
In contrast with a conclusion, a finding is a repeatable discovery, whereas a conclusion is not—it is instead a broader statement inferred (justifiably or not) from one or more findings. A finding deals with computable properties of some entity; a conclusion does not, but rather makes a statement that the findings support or lead to. Two papers could have the same findings but reach different conclusions based on those findings because the conclusions of a paper are based on an interpretation of its findings—two researchers might interpret a given set of findings quite differently. Some examples of findings from our papers include:
Explicit phrasal negation is normally distributed in the abstracts of scientific journal articles and in the bodies of scientific journal articles. This finding, reported in (Cohen et al., 2017a), was important in the context of that paper because it constrained a central aspect of the methodology of the work—the statistical hypothesis tests that could be applied to the raw data.
Negation is normally distributed in scientific journal articles and in clinical documents. The finding was derived from a statistical hypothesis test that showed that in the cases of both document types, the p-value of a Shapiro-Wilk test was less than 0.05. This finding in our paper is notable in this context because it is a clear example of a finding in the previous paper not being reproduced. The finding was even more significant in this paper than in the previous one, due to the motivation that was described above for this specific paper—the need to know the distributional characteristics of negation in a variety of types of biomedical text—in contrast to the previous paper, which studied textual characteristics of the genre more broadly.
In the lexicon of (Goodman et al., 2016), the dimension of reproducibility of a finding corresponds to results reproducibility.
2.3. Reproducibility of a value
By value, we mean a number, whether measured (e.g. a count of false positives) or calculated (e.g. a standard deviation). Actual values are important in finding constants, e.g. the coefficient of a long-tail distribution (see the extensive discussion of relevant topics for language in (Muller, 1977; Tweedie and Baayen, 1998; Baayen, 2001)), or the best smoothing value when calculating relative frequencies (Kilgarriff, 2012).)
Shannon’s early work on the entropy of written English text provides an example of a language-related value that stimulated an enormous amount of academic work, some of which has been evaluated with respect to the extent to which it does or does not reproduce the values reported in (Shannon, 1951). For example, (Cover and King, 1978) used a very different method from Shannon’s original one and found a value of 1.3 bits for the entropy of written English. The paper explicitly states that this value “agrees well with Shannon’s estimate,” suggesting that the authors considered their value to have reproduced Shannon’s original value in (Shannon, 1951)3. In a very different tone, (Brown et al., 1992) reported an upper bound of exactly 1.75 bits, but did not explicitly compare that to previous findings, although it is clear from the paper that they considered it different from—and better than—previously reported values. As the authors put it:
We see this paper as a gauntlet thrown down before the computational linguistics community.
A relevant value from our papers that was not reproduced is the mean value for the frequency of negation. We reported this in our papers (Cohen et al., 2010) and (Cohen et al., 2017a). They were different by roughly a factor of 2, even though we used the same corpus in both cases.
This is especially notable because the second of these papers is completely replicable, and yet we were later unable to reproduce our initial value. There was a doubly non-reproducible value here—the value in (Cohen et al., 2010) was not reproduced in (Cohen et al., 2017a), and that value in turn was not reproduced when we later repeated the experiment—all in our own papers.
The dimension of reproducibility of a value does not have an equivalent in the lexicon of (Goodman et al., 2016), perhaps because that paper points out a number of problems in determining whether or not studies have the “same” values, even when we can avail ourselves of statistical hypothesis tests.
3. Meta-analyses of some papers in natural language processing
3.1. Case study: A paper that was replicable but only partly reproducible
The motivation for this work came from a paper by Wu et al. (Wu et al., 2014) that discussed the potentially misleading nature of much recent work on negation in natural language processing of biomedical text. The contention of that paper was that in order to achieve reportable results that give a better estimate of the reality of performance on negation, one needs data with different distributional properties than the data that has been used in previous research on the topic. To address this, our paper undertook a study of two kinds of negation in two kinds of biomedical texts. We studied explicit phrasal negation (e.g. is not involved in) and sub-word or affixal negation (e.g. unknown) in biomedical journal articles and in physician progress notes on Intensive Care Unit patients from the MIMIC II database (Saeed et al., 2002). This was a quantitative study that did hypothesis tests on the rates of the two kinds of negation, finding that phrasal negation was more common in clinical texts than in scientific journal articles, while affixal negation was more common in scientific journal articles than in clinical texts—a surprising finding, given the relative amounts of research on negation in the two genres (much more on clinical text than on scientific text).
3.2. Why this example?
We select this study to illustrate the application of the proposed dimensions of reproducibility because it is the most heavily evaluated work, with respect to both replicability (the ability to repeat the experiments) and reproducibility (the outcomes of that experiment), that we have ever done. The reasons that we say that:
First, we made a very deliberate effort to archive all data and all code for this study on GitHub4.
Second, we then had two trainees repeat the experiments, during the course of which one of the students found a bug in the analysis code, suggesting that they examined it quite closely.
In addition to the great effort that was made to ensure the replicability of this project—and we note that it appears to have been a very strong effort, as indicated by the fact that the student was able to repeat the experiment closely enough to locate a bug in the code—we had the opportunity to do a fortuitous assessment of the reproducibility of the work, because the night before giving a talk on this work, we found another bug in the code. That then gave us an opportunity to see whether or not the original conclusion, findings, and values would be reproduced after we fixed the bug.
So, this was a rather unusual piece of research, both in terms of the documented efforts that went into ensuring its replicability, and the unexpected opportunity to assess its reproducibility.
Additionally, there’s this: the first author of the work is an associate editor for reproducibility issues in natural language processing. Three of the four authors of the paper are actively involved in research on reproducibility. It is difficult to say that they were not aware of how difficult of a problem this is, and it is clear from the GitHub repository (see Footnote 1) and from the replicability check that they had a student do that they were making a concentrated effort to ensure the reproducibility of the work.
3.3. What happened
The research was carried out, and the paper written, with no more stress or problems than one would expect. The analyses were all done in R markdown, as is often recommended in order to ensure the replicability of an analysis. (Gandrud, 2013; Leeper, 2014; Wickham and Grolemund, 2016). All data, code, and outputs of analyses were put in a publicly accessible GitHub repository, as is also often recommended in order to ensure replicability (Pedersen, 2008). The paper was written and submitted to a large conference on biomedical informatics.
While the paper was under review, two trainees (one doctoral student and one post-doctoral fellow) were asked to check out the GitHub repository and repeat the work. This immediately led to two observations:
We had forgotten to upload two data files.
Replicating the analysis required editing some paths in the R code.
The two forgotten data files were then uploaded, and the fact that some editing of paths in the R code was required was duly noted. (The email chain that documents the chain of events here has been uploaded to the GitHub repository as a series of screen shots.) With that done, the trainees were able to replicate the analysis. (Note that we use the term replicate here because the analysis technique is a part of the experimental method, rather than one of the outcomes of the experiment.)
At that point, one of the trainees attempting to replicate the work noticed a bug in the R markdown file for calculating the inter-annotator agreement: two file names had been reversed. The code was fixed and the analysis was rerun. The calculated value for inter-annotator agreement changed, but the overall finding of relative incidence of negation still held, so neither this finding, nor the overall conclusion of the paper, was affected.
The paper was accepted for publication in the conference proceedings. The value for inter-annotator agreement in the paper that had been based on the incorrect file names was replaced with the correct value; again, neither the finding nor the conclusion was affected.
The night before giving the talk, one of the authors was finalizing the slides. Figuring that the most explicit way to demonstrate what had been done in counting the phrasal explicit negatives (e.g. no and not) would be to show the regular expression that had been used to detect them, he looked at the code in order to copy that regular expression into the slides. A sinking feeling ensued: he had written that code, he knew what he had intended for it to do, and it was not at all likely that the code in question had done it. The regular expression would work fine on the clinical data, which had been converted to one token per line in a preprocessing step. However, the scientific journal articles had not undergone this preprocessing, and the regular expression would need to have a global switch (which directs the regular expression engine to match as many times as the pattern occurs in the input, rather than just once) in order to do the count properly. Without that global switch, the code would only find at most one explicit phrasal negative in a line; since the corpus contained one paragraph per line, that meant that the code would find at most one explicit phrasal negative per paragraph.
Because all of the data and code was available on the GitHub site, repeating the counts and the subsequent analysis was literally a matter of about two minutes’ work. When this was done, the following emerged:
The counts of explicit phrasal negatives in the clinical documents had not changed; the counts of explicit phrasal negatives in the scientific journal articles had changed quite a bit. The mean frequency of explicit phrasal negation in the scientific journal articles was now much higher than it had been.
Contrary to what we had published in the paper, the distribution of frequencies in the scientific journal articles was not normal. This meant that the t-test that had been used for hypothesis testing in the published version of the paper should not be used—rather, a non-parametric hypothesis test should have been used. Happily, when a Wilcoxon signed rank test was then done, the means were still significantly different, p = 2.138e-15.
3.4. Analysis in terms of the three proposed dimensions of reproducibility
Working upwards from the most granular dimension to the most general one, we find:
Values One of the values was not reproduced. The mean for the scientific journal articles was much higher after the bug fix than before it. In contrast, the value for the clinical documents was reproduced.
Findings The finding that the frequencies of explicit phrasal negation in the scientific journal articles were normally distributed was not reproduced. In contrast, the finding that the mean of the frequencies of explicit phrasal negation in the scientific journal articles was statistically significantly lower than the mean in the clinical documents was reproduced.
Conclusion Since of those two findings, only the finding that the mean of the frequencies of explicit negation in the scientific journal articles was lower than in the clinical documents was used to support the conclusion of the paper—that the distribution of explicit phrasal negation is different in the two genres—the conclusion was reproduced.
To summarize: the conclusion of the paper and the finding that was used to support that conclusion were both reproduced. The other finding, and the value that led to that finding, were not reproduced.
3.5. Case study: A paper on reproducibility whose conclusion is not reproducible
Cohen et al. (Cohen et al., 2016) published a case study on reproducibility that involved evaluating two R libraries for biomedical text mining. Both of those libraries provided connections to a web-based service. They concluded that reproducing the original work was difficult, but not impossible. Even before the paper went to press, that had ceased to be the case. As the authors put it:
[T]wo hours after we submitted this paper for review, one of the libraries stopped working completely……the behavior [of the library] has not been the same since…and so far, we have been unable to reproduce our previous results. The research that relied on the library is at a standstill.
3.6. Case study: An attempt to reproduce an influential paper that was unable to reproduce its findings
Gomes et al. (Gomes et al., 2016) described a paper on their results when they replicated an influential approach to domain adaption. As the work is described, they were able to replicate the methodology. However, they were not able to reproduce the findings: the performance of their machine translation system did improve in one translation direction, but not in the other. In contrast, the original paper had shown improved performance in all cases that it examined. As the authors put it:
While we were able to improve the Portuguese-to-English translation of in-domain texts using the…technique, the [method] did not outperform the in-domain trained baseline in the English-to-Portuguese direction.
This is an example of the dimension of reproducibility of a finding because it consists of a failure to reproduce the relative values of the system under test with respect to the baseline system. The dimension of reproducibility of a value is not relevant here because values were not being directly compared. Neither is the dimension of reproducibility of a conclusion relevant, since the paper does not make one beyond the statement about the findings.
4. Discussion and Conclusions
4.1. Is there really a problem here?
This paper is motivated by the claim of the existence of a problem of lack of consensus on terminology. Is there really such a problem? Related literature is consistent with the claim that there is. For example, a 2016 paper from one of the scientists most responsible for the recent notion of a “reproducibility crisis” in science notes that
The language and conceptual framework of “research reproducibility” are nonstandard and unsettled across the sciences…As the movement to examine and enhance the reliability of research expands, it is important to note that some of its basic terms—reproducibility, replicability, reliability, robustness, and generalizability—are not standardized. (Goodman et al., 2016)
Is that “problem in theory” really a “problem in practice”? Goodman et al. suggest that it is:
This diverse nomenclature has led to confusion …about what kind of confirmation is needed to trust a given scientific result. (Goodman et al., 2016)
In the field of natural language processing, work on the topic has concluded that such problems exist, as well. Ten years ago, (Pedersen, 2008) discussed the extent to which replicability and reproducibility issues go right to the heart of our field’s claim to being an empirical science, and 9 years later, Olorisade et al. showed that the problem is still quite widespread (Olorisade et al., 2017). Fokken et al. showed that it is a difficult problem to address—the repercussions are grave (Fokkens et al., 2013).
As Goodman et al. point out, not knowing what kind of confirmation is needed to trust a given “scientific fact”— presumably, what we report in computational linguistics meetings and journals—has a very practical consequence. Not knowing what kind of confirmation is needed prevents us from operationalizing the solutions to the problems pointed out by writers on the topic of replicability and repeatability problems in natural language processing— we have no “clear operational criteria for what constitutes successful replication or reproduction” (Goodman et al., 2016). As we show in this paper in our case study on negation, the things that we often think are sufficient for ensuring replicability (e.g. shared data, the use of publicly available repositories to share our code, and markdown languages) are clearly not in and of themselves sufficient.
4.2. What is the origin of the problem?
Where does that lack of consensus come from, and does the source of the consensus tell us anything about the possible success (or lack thereof) of any proposal to address it? On some level, we can trace the lack of consensus to a case of synonymy: the words reproducibility and repeatability are close enough to synonymous in general English that they often appear in each other’s definitions. For example, in monolingual English dictionaries, we see:
replicate, sense 3: to repeat, duplicate, or reproduce, esp. for experimental purposes. (Random House Unabridged 1999).
This is reflected in the very ways that scientists themselves define the terms when they write about the topics. For example:
…reproducibility means that the process of establishing a fact, or the conditions under which the same fact can be observed, is repeatable.
(Teten, 2016), cited in (Atmanspacher et al., 2014)—our emphasis.
Previous work has established three things about reproducibility in natural language processing: it is important (Pedersen, 2008; Schwartz, 2010; Branco et al., 2017), it can be quite difficult to achieve (Fokkens et al., 2013; Névéol et al., 2016), and the causes of reproducibility problems can be well-hidden—see (Johnson et al., 2007; Cohen et al., 2017b), as well as the bug that we report in this paper.
4.3. Definitions of dimensions of reproducibility in the larger context of natural language processing
The bigger picture in which this work is situated is that of a lack of a fully developed epistemology of computational linguistics and natural language processing. Enormous advancements in this area have come from the shared task model of evaluation (Hirschman, 1990; Hirschman, 1994; Jones and Galliers, 1995; Resnik and Lin, 2010; Hirschman, 1998; Chapman et al., 2011; Huang and Lu, 2015), from the development of a science of evaluation in our field (Daelemans and Hoste, 2002; Voorhees et al., 2005; Buckley and Voorhees, 2017), and from the development of a science of annotation (Palmer et al., 2005; Ide, 2007; Wilcock, 2009; Pustejovsky and Stubbs, 2012; Stubbs, 2012; Styler IV et al., 2014; Bonial et al., 2017; Green et al., 2017; Ide and Pustejovsky, 2017; Savova et al., 2017). But, large holes remain in our development of an epistomology of computational linguistics and natural language processing that integrates these strengths of our field and also explores the relationships between natural language processing; computational and corpus linguistics; artificial intelligence, theoretical linguistics, and cognitive science (Cori et al., 2002). (See also (Cori and Léon, 2002) for a discussion of how issues of definition of what our field is affect that epistemology and (Bès, 2002; Habert and Zweigenbaum, 2002; Amblard, 2016) for how taxonomization of methodologies in natural language processing, computational linguistics, and engineering interact with it).
4.4. Novel observations
The work reported here allows some observations that to our knowledge have not been made before. First: reproducibility is not a binary, you-are-or-you-aren’t condition— it is more nuanced than that, as can be seen from the examples of the dimensions, as well as from the extended case study.
Second: despite suggestions to the contrary, there are no “silver bullets” where reproducibility or replicability is concerned. The work that is described in the case study made heavy use of the most-commonly-advocated architectures for enhancing both replicability and reproducibility— and yet, we were initially not able to replicate the experiments. Once we could, we found that although the conclusion was reproducible, a crucial value and a key finding were not. The distinction between replicability and reproducibility that we make from the outset of this paper, along with the three dimensions of reproducibility that this paper proposes, allowed us to make these distinctions. A failure to distinguish between the ability to replicate an experiment and to reproduce its outcomes would not allow for a description of these circumstances, and a binary reproduced/not reproduced distinction would not allow us to do so, either.
We also note that trying to replicate the work was very productive—it led not only to discovering that some files were absent from the repository (which directly affects the repeatability of the work), but it led directly to the finding of the first bug. This might be surprising in the context of (Drummond, 2009)’s strong stance against the very notion that replicability is valuable. He makes the same distinction between replicability (the ability to repeat an experiment’s methods) and reproducibility, and says the following about replicability in a paper titled Replicability is not Reproducibility—Nor is it Good Science:
It would cause a great deal of wasted effort by members of our community. …I am also far from convinced that it will deliver the benefits that many think it will. I suspect that, at best, it would serve as little more than a policing tool, preventing outright fraud.
The analysis of the case of our study on negation is a clear example of the success of what (Goodman et al., 2016) call “a proof-of-principle study…sufficient to show that [a phenomenon is] possible:” pace Drummond, our attempt to replicate an experiment improved our science.
4.5. Conclusions
We have shown examples from the natural language processing and computational linguistics literature of all three of the proposed dimensions of reproducibility—cases where conclusions, findings, and values were reproduced, and cases where they were not. We have also shown that the value for one dimension is not dependent on the others. For example, in the extended analysis of (Cohen et al., 2017a), we showed a case where a value was not reproduced and a finding was not reproduced, but the conclusion was. In the discussion of (Gomes et al., 2016), we showed a case where a finding was not reproduced, but neither values nor conclusions were relevant to asking whether or not the earlier paper as a whole had been reproduced. In (Cohen et al., 2016), we see a paper whose conclusion is not reproducible, independent of specific values or findings. Taken together, these suggest that the proposed dimensions of reproducibility are, indeed, applicable to research in natural language processing. We have also shown how they map to definitions of the relevant phenomena in other work on reproducibility in science more broadly.
Moving forward, what can be done with the dimensions proposed in this paper that could not be done before? With this more nuanced set of definitions of reproducibility, we can better understand the state of the science in our field. Once we know what that state is, then we can build on the suggestions of papers like (Pedersen, 2008; Fokkens et al., 2013; Olorisade et al., 2017) that make concrete recommendations about dealing with issues of reproducibility in natural language processing—and make it better.
While we do this, we should be charitable to each other other, recognizing that failures of reproducibility can occur even in spite of the best intentions of the researchers. Facing our reproducibility problems will probably be painful for the field, but in the end, it will be of benefit to all of us, and to our science.
Figure 1.
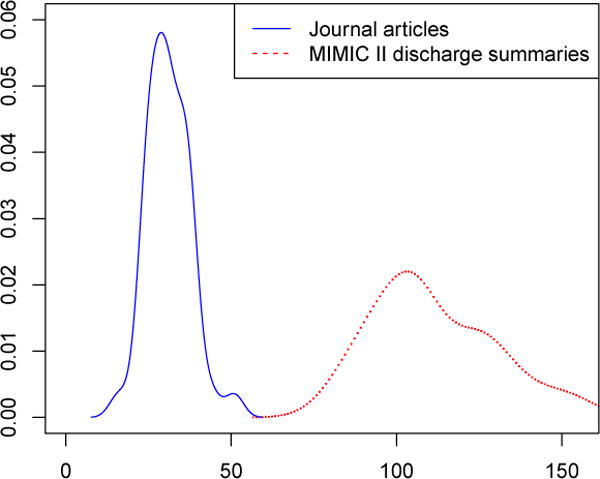
MIMIC II progress notes mean = 111 per 10,000-word sample, CRAFT corpus mean = 31 per 10,000-word sample. Welch 2-sample t-test: t = −27.092, df = 53.822, p-value < 2.2e-16.
Figure 2.
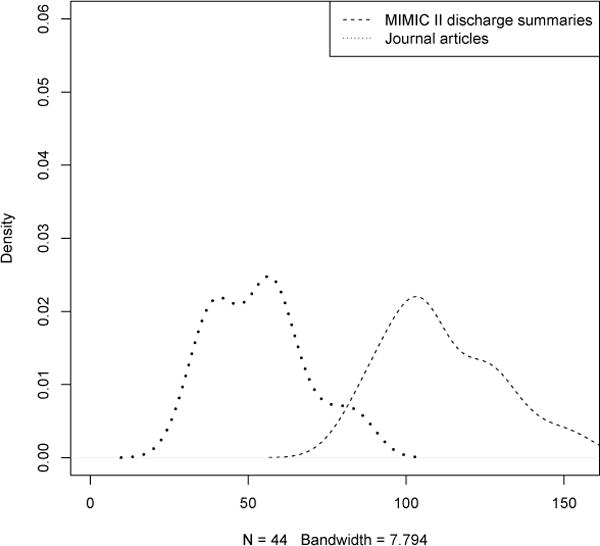
MIMIC II progress notes mean = 111 per 10,000-word sample, CRAFT corpus mean = 53 per 10,000-word sample. Wilcoxon (Mann-Whitney U) W = 5.5, p-value = 2.138e-15.
Table 1.
Mean and distribution of explicit phrasal negation in scientific journal articles before and after fixing the bug. The Before values are the values published in (Cohen et al., 2017a). The After values are the values after we fixed the bug.
| Metric | Before | After |
|---|---|---|
| Mean | 31/10K words | 53/10K words |
| Distribution | normal | bimodal |
Acknowledgments
The work reported here was supported by NIH grants LM008111 and LM009254 to Lawrence E. Hunter. Cohen’s work was supported by Hunter’s grants, by grant AHRQ R21HS024541-01 to Foster Goss (as was Goss’s), and by generous funding from Labex DigiCosme (project ANR11LABEX0045 DIGICOSME), operated by ANR as part of the program Investissement d’Avenir Idex Paris-Saclay (ANR11 IDEX000302), as well as by a Jean d’Alembert fellowship from the Fondation Campus Paris-Saclay as part of the Investissement d’Avenir program operated by ANR. TJC’s work was supported by a supplement to NIH grant LM009254 to Hunter and Callahan. PZ and AN received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant 676207. The authors thank:
Nicoletta Calzolari
The participants in the workshop on The role of text mining and image processing in fostering responsible research practices at the 5th World Conference on Research Integrity
… for their insightful comments.
Footnotes
We do not evaluate that claim ourselves because Shannon’s paper actually reports a range of values; it is clear from the quotation that (Cover and King, 1978) felt that they reproduced Shannon’s value, but it is not clear to us exactly how they came to that conclusion from the range of values in Shannon’s 1951 paper.
Bibliographical References
- Amblard M. Pour un TAL responsable. Traitement Automatique des Langues. 2016;57(2):21–45. [Google Scholar]
- Atmanspacher H, Lambert LB, Folkers G, Schubiger PA. Relevance relations for the concept of reproducibility. Journal of the Royal Society Interface. 2014;11(94):20131030. doi: 10.1098/rsif.2013.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autorino R, Quarto G, Sio MD, Lima E, Quarto E, Damiano R, Oliviero R, Osorio L, Marcelo F, D’Armiento M. Fate of abstracts presented at the world congress of endourology: are they followed by publication in peer-reviewed journals? Journal of endourology. 2006;20(12):996–1001. doi: 10.1089/end.2006.20.996. [DOI] [PubMed] [Google Scholar]
- Baayen RH. Word frequency distributions. Vol. 18. Springer Science & Business Media; 2001. [Google Scholar]
- Balasubramanian S, Kumar I, Wyld L, Reed M. Publication of surgical abstracts in full text: a retrospective cohort study. The Annals of The Royal College of Surgeons of England. 2006;88(1):57–61. doi: 10.1308/003588406X82961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bès GG. La linguistique entre science et ingénierie. Traitement Automatique des Langues. 2002;43(2):57–81. [Google Scholar]
- Bloomfield L. Language or ideas? Language. 1936:89–95. [Google Scholar]
- Bonial C, Conger K, Hwang JD, Mansouri A, Aseri Y, Bonn J, O’Gorman T, Palmer M. Handbook of Linguistic Annotation. Springer; 2017. Current directions in English and Arabic PropBank; pp. 737–769. [Google Scholar]
- Borgholt L, Simonsen P, Hovy D. EMNLP. Lisbon, Portugal: 2015. The rating game: Sentiment rating reproducibility from text; pp. 2527–2532. [Google Scholar]
- Branco A, Cohen KB, Vossen P, Ide N, Calzolari N. Replicability and reproducibility of research results for human language technology: introducing an LRE special section 2017 [Google Scholar]
- Brown PF, Pietra VJD, Mercer RL, Pietra SAD, Lai JC. An estimate of an upper bound for the entropy of english. Computational Linguistics. 1992;18(1):31–40. [Google Scholar]
- Buckley C, Voorhees EM. ACM SIGIR Forum. Vol. 51. ACM; 2017. Evaluating evaluation measure stability; pp. 235–242. [Google Scholar]
- Bydder SA, Joseph DJ, Spry NA. Publication rates of abstracts presented at annual scientific meetings: how does the Royal Australian and New Zealand College of Radiologists compare? Journal of Medical Imaging and Radiation Oncology. 2004;48(1):25–28. doi: 10.1111/j.1440-1673.2004.01243.x. [DOI] [PubMed] [Google Scholar]
- Byerly WG, Rheney CC, Connelly JF, Verzino KC. Publication rates of abstracts from two pharmacy meetings. Annals of Pharmacotherapy. 2000;34(10):1123–1127. doi: 10.1345/aph.19375. [DOI] [PubMed] [Google Scholar]
- Cassidy S, Estival D. Supporting accessibility and reproducibility in language research in the Alveo virtual laboratory. Computer Speech & Language. 2017;45:375–391. [Google Scholar]
- Chapman WW, Nadkarni PM, Hirschman L, D’Avolio LW, Savova GK, Uzuner O. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. 2011 doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateauraynaud F. Prospéro. Une technologie littéraire pour les sciences humaines. CNRS; 2003. [Google Scholar]
- Cohen KB, Johnson HL, Verspoor K, Roeder C, Hunter LE. The structural and content aspects of abstracts versus bodies of full text journal articles are different. BMC bioinformatics. 2010;11(1):492. doi: 10.1186/1471-2105-11-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen KB, Xia J, Roeder C, Hunter L. Reproducibility in natural language processing: A case study of two R libraries for mining PubMed/MEDLINE. Workshop on Research Results Reproducibility and Resources Citation in Science and Technology of Language; Portoroz, Slovenia. 2016. pp. 6–12. [PMC free article] [PubMed] [Google Scholar]
- Cohen KB, Goss F, Zweigenbaum P, Hunter LE. Translational morphosyntax: Distribution of negation in clinical records and biomedical journal articles. MEDINFO; Hangzhou, China: 2017a. [PubMed] [Google Scholar]
- Cohen K, Névéol A, Xia J, Hailu N, Hunter L, Zweigenbaum P. Reproducibility in biomedical natural language processing. Proc AMIA Annu Symp 2017b [Google Scholar]
- Collaboration, O. S. et al. Estimating the reproducibility of psychological science. Science. 2015;349(6251):aac4716. doi: 10.1126/science.aac4716. [DOI] [PubMed] [Google Scholar]
- Collberg C, Proebsting T, Warren AM. Repeatability and benefaction in computer systems research. University of Arizona TR. 2015;14:4. [Google Scholar]
- Cori M, Léon J. La constitution du TAL. Traitement Automatique des Langues. 2002;43(3):21–55. [Google Scholar]
- Cori M, David S, Léon J. Pour un travail épistémologique sur le TAL. Traitement Automatique des Langues. 2002;43(3) [Google Scholar]
- Cover T, King R. A convergent gambling estimate of the entropy of english. IEEE Transactions on Information Theory. 1978;24(4):413–421. [Google Scholar]
- Daelemans W, Hoste V. Evaluation of machine learning methods for natural language processing tasks. 3rd International conference on Language Resources and Evaluation (LREC 2002) European Language Resources Association 2002 [Google Scholar]
- Dahllöf G, Wondimu B, Maniere MC. Subsequent publication of abstracts presented at the international association of paediatric dentistry meetings. International journal of paediatric dentistry. 2008;18(2):91–97. doi: 10.1111/j.1365-263X.2007.00898.x. [DOI] [PubMed] [Google Scholar]
- Drummond C. Replicability is not reproducibility: nor is it good science 2009 [Google Scholar]
- Fokkens A, Van Erp M, Postma M, Pedersen T, Vossen P, Freire N. Offspring from reproduction problems: What replication failure teaches us. ACL (1) 2013:1691–1701. [Google Scholar]
- Gandrud C. Reproducible research with R and R studio. CRC Press; 2013. [Google Scholar]
- Gomes L, van Noord G, Branco A, Neale S. Seeking to reproduce “Easy Domain Adaptation”. Workshop on Research Results Reproducibility and Resources Citation in Science and Technology of Language; Portoroz, Slovenia. 2016. p. 1. [Google Scholar]
- Goodman SN, Fanelli D, Ioannidis JP. What does research reproducibility mean? Science translational medicine. 2016;8(341):341ps12–341ps12. doi: 10.1126/scitranslmed.aaf5027. [DOI] [PubMed] [Google Scholar]
- Green M, Hargraves O, Bonial C, Chen J, Clark L, Palmer M. Handbook of Linguistic Annotation. Springer; 2017. Verbnet/ontonotes-based sense annotation; pp. 719–735. [Google Scholar]
- Habert B, Zweigenbaum P. Régler les règles. Traitement Automatique des Langues. 2002;43(3):83–105. [Google Scholar]
- Harel Z, Wald R, Juda A, Bell CM. Frequency and factors influencing publication of abstracts presented at three major nephrology meetings. International archives of medicine. 2011;4(1):40. doi: 10.1186/1755-7682-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbison P. Full publication of abstracts of randomised controlled trials published at international continence society meetings’. Neurourology and urodynamics. 2004;23(2):101–103. doi: 10.1002/nau.20003. [DOI] [PubMed] [Google Scholar]
- Hirschman L. Speech and Natural Language. Hidden Valley; Pennsylvania: 1990. Natural language evaluation. [Google Scholar]
- Hirschman L. Human Language Technology. Hidden Valley; Pennsylvania: 1994. Human language evaluation. [Google Scholar]
- Hirschman L. The evolution of evaluation: Lessons from the message understanding conferences. Computer Speech & Language. 1998;12(4):281–305. [Google Scholar]
- Huang CC, Lu Z. Community challenges in biomedical text mining over 10 years: success, failure and the future. Briefings in bioinformatics. 2015;17(1):132–144. doi: 10.1093/bib/bbv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide N, Pustejovsky J. Handbook of Linguistic Annotation. Springer; 2017. [Google Scholar]
- Ide N. Annotation science from theory to practice and use 2007 [Google Scholar]
- Jackson KR, Daluiski A, Kay RM. Publication of abstracts submitted to the annual meeting of the Pediatric Orthopaedic Society of North America. Journal of Pediatric Orthopaedics. 2000;20(1):2. [PubMed] [Google Scholar]
- Johnson HL, Cohen KB, Hunter L. Pacific Symposium on Biocomputing. Maui, Hawaii: NIH Public Access; 2007. A fault model for ontology mapping, alignment, and linking systems; p. 233. [PMC free article] [PubMed] [Google Scholar]
- Jones KS, Galliers JR. Evaluating natural language processing systems: An analysis and review. Vol. 1083. Springer Science & Business Media; 1995. [Google Scholar]
- Kilgarriff A. International conference on text, speech and dialogue. Springer; 2012. Getting to know your corpus; pp. 3–15. [Google Scholar]
- Kilicoglu H. Biomedical text mining for research rigor and integrity: tasks, challenges, directions. Briefings in Bioinformatics. 2017 doi: 10.1093/bib/bbx057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeper TJ. Archiving reproducible research with r and dataverse. R Journal. 2014;6(1) [Google Scholar]
- Marx WF, Cloft HJ, Do HM, Kallmes DF. The fate of neuroradiologic abstracts presented at national meetings in 1993: rate of subsequent publication in peer-reviewed, indexed journals. American journal of neuroradiology. 1999;20(6):1173–1177. [PMC free article] [PubMed] [Google Scholar]
- Mieskes M. A quantitative study of data in the NLP community. EACL 2017. 2017:1. [Google Scholar]
- Moretti F, Dobenesque E, Jeanpierre L. Graphes, cartes et arbres: modèles abstraits pour une autre histoire de la littérature. Les prairies ordinaires 2008 [Google Scholar]
- Muller C. Principes et méthodes de statistique lexicale. Classiques Hachette; 1977. [Google Scholar]
- Née E. Méthodes et outils informatiques pour l’analyse des discours. Presses universitaires de Rennes; 2017. [Google Scholar]
- Névéol A, Cohen K, Grouin C, Robert A. Replicability of research in biomedical natural language processing: a pilot evaluation for a coding task. Proceedings of the LOUHI Seventh International Workshop on Health Text Mining and Information Analysis; Austin, Texas. 2016. pp. 78–84. [Google Scholar]
- Ng L, Hersey K, Fleshner N. Publication rate of abstracts presented at the annual meeting of the American Urological Association. BJU international. 2004;94(1):79–81. doi: 10.1111/j.1464-410X.2004.04905.x. [DOI] [PubMed] [Google Scholar]
- Oliver D, Whitaker I, Chohan D. Publication rates for abstracts presented at the British Association of Plastic Surgeons meetings: how do we compare with other specialties? British journal of plastic surgery. 2003;56(2):158–160. doi: 10.1016/s0007-1226(03)00037-7. [DOI] [PubMed] [Google Scholar]
- Olorisade BK, Brereton P, Andras P. Reproducibility of studies on text mining for citation screening in systematic reviews: Evaluation and checklist. Journal of biomedical informatics. 2017;73:1–13. doi: 10.1016/j.jbi.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Palmer M, Gildea D, Kingsbury P. The proposition bank: An annotated corpus of semantic roles. Computational linguistics. 2005;31(1):71–106. [Google Scholar]
- Pedersen T. Empiricism is not a matter of faith. Computational Linguistics. 2008;34(3):465–470. [Google Scholar]
- Peng PH, Wasserman JM, Rosenfeld RM. Factors influencing publication of abstracts presented at the AAO-HNS Annual Meeting. Otolaryngology-Head and Neck Surgery. 2006;135(2):197–203. doi: 10.1016/j.otohns.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Pustejovsky J, Stubbs A. Natural Language Annotation for Machine Learning: A guide to corpus-building for applications. O’Reilly Media, Inc; 2012. [Google Scholar]
- Rao AR, Beatty JD, Laniado M, Motiwala HG, KARIM O. Publication rate of abstracts presented at the British Association of Urological Surgeons Annual Meeting. BJU international. 2006;97(2):306–309. doi: 10.1111/j.1464-410X.2006.05863.x. [DOI] [PubMed] [Google Scholar]
- Resnik P, Lin J. Evaluation of NLP systems. The handbook of computational linguistics and natural language processing. 2010;57:271–295. [Google Scholar]
- Saeed M, Lieu C, Raber G, Mark RG. Computers in Cardiology, 2002. IEEE; 2002. MIMIC II: a massive temporal ICU patient database to support research in intelligent patient monitoring; pp. 641–644. [PubMed] [Google Scholar]
- Sanders D, Carter M, Hurlstone D, Lobo A, Hoggard N. Research outcomes in British gastroenterology: an audit of the subsequent full publication of abstracts presented at the British Society of Gastroenterology. Gut. 2001;49(1):154–155. doi: 10.1136/gut.49.1.154a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savova G, Pradhan S, Palmer M, Styler W, Chapman W, Elhadad N. Handbook of Linguistic Annotation. Springer; 2017. Annotating the clinical text–MiPACQ, ShARe, SHARPn and THYME corpora; pp. 1357–1378. [Google Scholar]
- Scherer RW, Dickersin K, Langenberg P. Full publication of results initially presented in abstracts: a meta-analysis. Jama. 1994;272(2):158–162. [PubMed] [Google Scholar]
- Scherer RW, Langenberg P, Von Elm E, et al. Full publication of results initially presented in abstracts. Cochrane Database Syst Rev. 2007;2(2):MR000005. doi: 10.1002/14651858.MR000005.pub3. [DOI] [PubMed] [Google Scholar]
- Schwartz L. Proceedings of the Joint Fifth Workshop on Statistical Machine Translation and MetricsMATR. Uppsala, Sweden: Association for Computational Linguistics; 2010. Reproducible results in parsing-based machine translation: the JHU shared task submission; pp. 177–182. [Google Scholar]
- Shannon CE. Prediction and entropy of printed english. Bell Labs Technical Journal. 1951;30(1):50–64. [Google Scholar]
- Smollin CG, Nelson LS. Publication of abstracts presented at 2001 NACCT. Journal of Medical Toxicology. 2006;2(3):97–100. doi: 10.1007/BF03161017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs A. Developing specifications for light annotation tasks in the biomedical domain. Third Workshop on Building and Evaluating Resources for Biomedical Text Mining; Istanbul, Turkey. 2012. p. 71. [Google Scholar]
- Styler WF, IV, Bethard S, Finan S, Palmer M, Pradhan S, de Groen PC, Erickson B, Miller T, Lin C, Savova G, et al. Temporal annotation in the clinical domain. Transactions of the Association for Computational Linguistics. 2014;2:143. [PMC free article] [PubMed] [Google Scholar]
- Teten H. Enzyklopädie Philosophie und Wissenschaftstheorie: Bd 3: G–Inn. Springer-Verlag; 2016. [Google Scholar]
- Tweedie FJ, Baayen RH. How variable may a constant be? measures of lexical richness in perspective. Computers and the Humanities. 1998;32(5):323–352. [Google Scholar]
- Varghese RA, Chang J, Miyanji F, Reilly CW, Mulpuri K. Publication of abstracts submitted to the annual meeting of the pediatric orthopaedic society of north america: is there a difference between accepted versus rejected abstracts? Journal of Pediatric Orthopaedics. 2011;31(3):334–340. doi: 10.1097/BPO.0b013e31820f8783. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Fidler F, Cumming G. Replicates and repeats—what is the difference and is it significant?: A brief discussion of statistics and experimental design. EMBO reports. 2012;13(4):291–296. doi: 10.1038/embor.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees EM, Harman DK, et al. TREC: Experiment and evaluation in information retrieval. Vol. 1. MIT press; Cambridge: 2005. [Google Scholar]
- Wickham H, Grolemund G. R for data science: import, tidy, transform, visualize, and model data. O’Reilly Media, Inc; 2016. [Google Scholar]
- Wilcock G. Introduction to linguistic annotation and text analytics. Synthesis Lectures on Human Language Technologies. 2009;2(1):1–159. [Google Scholar]
- Wu S, Miller T, Masanz J, Coarr M, Halgrim S, Carrell D, Clark C. Negation’s not solved: generalizability versus optimizability in clinical natural language processing. PloS ONE. 2014;9(11):e112774. doi: 10.1371/journal.pone.0112774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yentis S, Campbell F, Lerman J. Publication of abstracts presented at anaesthesia meetings. Canadian journal of anaesthesia. 1993;40(7):632–634. doi: 10.1007/BF03009700. [DOI] [PubMed] [Google Scholar]


