Summary
Histone H3 lysine 9 (H3K9) methylation is unevenly distributed in mammalian chromosomes. However, the molecular mechanism controlling the uneven distribution and its biological significance remain to be elucidated. Here, we show that JMJD1A and JMJD1B preferentially target H3K9 demethylation of gene-dense regions of chromosomes, thereby establishing an H3K9 hypomethylation state in euchromatin. JMJD1A/JMJD1B-deficient embryos died soon after implantation accompanying epiblast cell death. Furthermore, combined loss of JMJD1A and JMJD1B caused perturbed expression of metabolic genes and rapid cell death in embryonic stem cells (ESCs). These results indicate that JMJD1A/JMJD1B-meditated H3K9 demethylation has critical roles for early embryogenesis and ESC maintenance. Finally, genetic rescue experiments clarified that H3K9 overmethylation by G9A was the cause of the cell death and perturbed gene expression of JMJD1A/JMJD1B-depleted ESCs. We summarized that JMJD1A and JMJD1B, in combination, ensure early embryogenesis and ESC viability by establishing the correct H3K9 methylated epigenome.
Keywords: histone methylation, histone demethylation, transcription, embryonic stem cell
Highlights
-
•
JMJD1A/JMJD1B have redundant but essential roles for ESC survival
-
•
JMJD1A/JMJD1B target H3K9 demethylation at gene-dense euchromatin
-
•
JMJD1A/JMJD1B and G9A play collaborative roles in tuning H3K9me2 levels
Kuroki et al. showed that H3K9 demethylases JMJD1A and JMJD1B are redundantly but essentially required for ESC survival and early embryogenesis in mice. JMJD1A and JMJD1B ensure transcription accuracy by demethylating H3K9 at gene-dense euchromatin.
Introduction
Posttranslational modifications in the tail region of core histones are important epigenetic marks linked to various nuclear functions, including transcriptional activity control. The discovery of enzymes that add/remove methyl groups to/from histones suggests that histone methylation levels are not statically but dynamically controlled (Kooistra and Helin, 2012). So far, individual functions of histone methyltransferases or demethylases have been studied extensively, but the role sharing between methyltransferases and demethylases for the correct establishment of histone methylated epigenome is not yet fully understood.
Histone H3 lysine 9 (H3K9) methylation is considered as an epigenetic mark of transcriptionally silenced heterochromatin. The JMJD1 family of proteins, which includes JMJD1A and its isozyme JMJD1B, reportedly possesses intrinsic H3K9 demethylating activity (Kim et al., 2012, Kuroki et al., 2013a, Yamane et al., 2006). JMJD1A plays important roles in several biological processes such as spermiogenesis (Liu et al., 2010, Okada et al., 2007), metabolism (Inagaki et al., 2009, Tateishi et al., 2009), and sex determination (Kuroki et al., 2013b). Recently, JMJD1B was shown to be required for female fertility (Liu et al., 2015). In contrast to the individual function of JMJD1A and JMJD1B in postnatal mice, the role of JMJD1 proteins in early embryogenesis and their functional redundancy are still unknown.
In this study, we demonstrated that JMJD1A and JMJD1B are redundantly but essentially required for mouse development immediately after implantation. We also demonstrated that JMJD1A and JMJD1B exhibit pivotal roles in mouse embryonic stem cell (ESC) function through the correct establishment of H3K9 methylated epigenome. Not a single depletion but depletion of both JMJD1A and JMJD1B induced a massive increase in H3K9 methylation accompanied by rapid cell death and perturbed gene expression. In control ESCs, dimethylated H3K9 (H3K9me2) was abundant in the gene-poor regions and scarce in the gene-dense regions of the chromosomes. Intriguingly, JMJD1A/JMJD1B deficiency induced a remarkable increase of H3K9me2 in the gene-dense regions, causing aberrant H3K9me2 distribution; high levels of H3K9me2 decorate chromosomes throughout. This result implies that JMJD1A/JMJD1B preferentially target H3K9 demethylation at the gene-dense euchromatin. Finally, we found that the additional mutation for an H3K9 methyltransferase G9A in a JMJD1A/JMJD1B-deficient background restored not only H3K9 overmethylation but also rapid cell death and perturbed gene expression phenotypes, indicating collaborative roles of JMJD1A/JMJD1B and G9A not only for the tuning of the H3K9 methylation levels but also for the regulation of ESC function. Taking these results together, we propose that JMJD1A/JMJD1B ensures cellular viability and transcription accuracy through the establishment of the correct H3K9 methylated epigenome in early mouse development.
Results
JMJD1A and JMJD1B Are Essentially Required for Peri-implantation Development in Mice
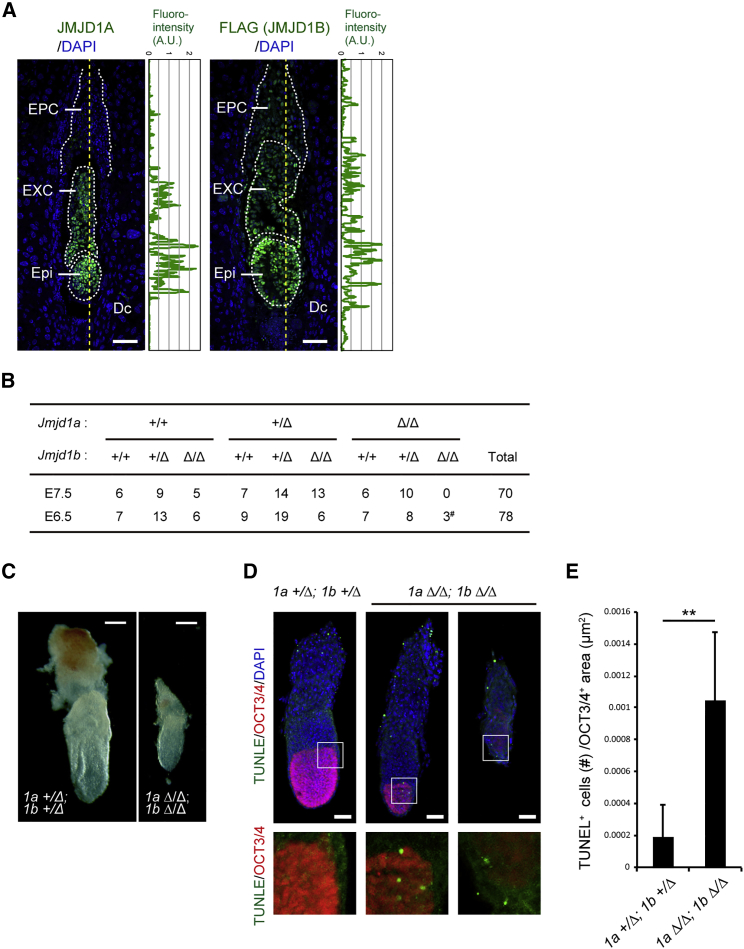
Previous studies had demonstrated that JMJD1A and JMJD1B possess intrinsic H3K9 demethylating activity (Kim et al., 2012, Kuroki et al., 2013a, Yamane et al., 2006), suggesting that JMJD1A and JMJD1B may function in a redundant manner. To evaluate the roles of JMJD1A and JMJD1B in peri-implantation development, we examined the expression profiles of JMJD1A and JMJD1B in developing embryos (Figure 1A). Immunofluorescence analysis with anti-JMJD1A antibodies revealed that JMJD1A is expressed in the epiblast and extraembryonic ectoderm at embryonic day (E) 6.5 (Figure 1A, left). To detect the endogenous JMJD1B protein with an anti-FLAG tag antibody, we established knockin mouse and ESC lines carrying modified Jmjd1b allele (referred as 1b hereafter). In the resultant lines, 1b+/Flag−KI allele produces the JMJD1B protein with a carboxy-terminal FLAG tag (Figure S1). Immunofluorescence analysis of 1b+/Flag−KI embryos with anti-FLAG tag antibody revealed that JMJD1B is expressed in the epiblast, extraembryonic ectoderm, and ectoplacental cone of E6.5 embryos (Figure 1A, right). We measured and plotted the signal intensities for JMJD1 proteins in the embryonic lineages (Figure 1A, plot analyses). Consequently, JMJD1A and JMJD1B were both found to be abundantly expressed in the epiblast of the developing embryos, indicating the potential roles of JMJD1A and JMJD1B in epiblast development.
Figure 1.
JMJD1A and JMJD1B Are Essential for Mouse Embryogenesis
(A) Immunofluorescence analysis of longitudinal sections of the E6.5 embryos with JMJD1A (left) and JMJD1B (right). E6.5 wild-type embryos were stained with anti-JMJD1A antibodies and DAPI (left). E6.5 Jmjd1b+/Flag−KI embryos were stained with anti-FLAG antibodies and DAPI (right). Fluorescence intensities along the dashed lines were quantified and plotted on the right side of the images. Scale bars, 100 μm. Epi, epiblast; EXC, extraembryonic ectoderm; EPC, ectoplacental cone; Dc, decidual cells. A.U., arbitrary unit.
(B) Jmjd1a+/Δ; Jmjd1a+/Δ males and females were crossed, and the resultant embryos were genotyped at the indicated embryonic periods. Jmjd1aΔ/Δ; Jmjd1aΔ/Δ embryos were found at E6.5, but not at E7.5. #, growth-retarded embryos.
(C) Gross appearances of Jmjd1a/Jmjd1b double-deficient embryos at E6.5 (left) when compared with a littermate control (right). 1a and 1b represent Jmjd1a and Jmjd1b, respectively. Scale bar, 100 μm.
(D) Whole-mount immunostaining analysis for the epiblast marker OCT3/4. Embryos were counterstained with DAPI and TUNEL to detect apoptotic cells. Scale bars, 100 μm.
(E) TUNEL-positive cells in the epiblast lineage were counted and summarized. Jmjd1a+/Δ; Jmjd1a+/Δ, n = 7; Jmjd1aΔ/Δ; Jmjd1aΔ/Δ, n = 3 different embryos (biological replicates). Data are presented as means ± SD. ∗∗p < 0.01 (Student's t test).
We established mutant mice bearing the 1bΔ allele, which lacks exons 20–21, corresponding with a portion of the catalytic JmjC domain (Figure S1). The 1bΔ/Δ mice were born at nearly the expected Mendelian ratio, but most of them died within a week after birth (Figure S2). Some of the 1bΔ/Δ mice survived and developed into adults; however, their body weights were almost half those of the controls (Figure S2). These phenotypes are in accordance with those reported in a recent study (Liu et al., 2015). To evaluate the effects of compound depletion of JMJD1A and JMJD1B on mouse development, we examined postnatal animals and embryos derived from the mating of 1a/1b double-heterozygous mutant mice. Among the 109 neonatal offspring, no JMJD1A/JMJD1B-deficient mice were found, strongly suggesting that JMJD1A/JMJD1B-deficient mice were embryonically lethal (Figure S2). Intriguingly, all of the mice carrying three mutant alleles of 1a or 1b were stillborn, indicating that the gene dosage of 1a/1b is critical for prenatal development (Figure S2). Embryos bearing the 1a/1b double-homozygous mutation were not found in 70 embryos at E7.5, whereas three embryos with this mutation were found in 78 embryos at E6.5 (Figure 1B). Notably, all JMJD1A/JMJD1B-deficient embryos were smaller than the controls at this stage (Figure 1C). These data suggest that JMJD1A/JMJD1B-deficient embryos display growth retardation and die around E6.5.
To examine the development of JMJD1A/JMJD1B-deficient embryos in more detail, we performed a whole-mount immunostaining analysis using antibodies against OCT3/4, which mark epiblast cells (Figure 1D). Apoptotic cells were detected by TUNEL labeling (Figure 1D). Strikingly, the mass size of OCT3/4-positive epiblasts in JMJD1A/JMJD1B-deficient embryos was smaller than those in the control embryos (Figure 1D, middle panels). We also found some JMJD1A/JMJD1B-deficient embryos without detectable epiblast cells (Figure 1D, right panels). TUNEL counterstaining analysis demonstrated a significant increase in the number of apoptotic cells in the epiblasts of JMJD1A/JMJD1B-deficient embryos (summarized in Figure 1E). These data indicate that growth retardation of JMJD1A/JMJD1B-deficient embryos can be attributed, in part, to the compromised development of the epiblast cells. We therefore conclude that JMJD1A and JMJD1B play redundant but essential roles for post-implantation development in mouse.
JMJD1A and JMJD1B Are Essentially Required for ESC Viability
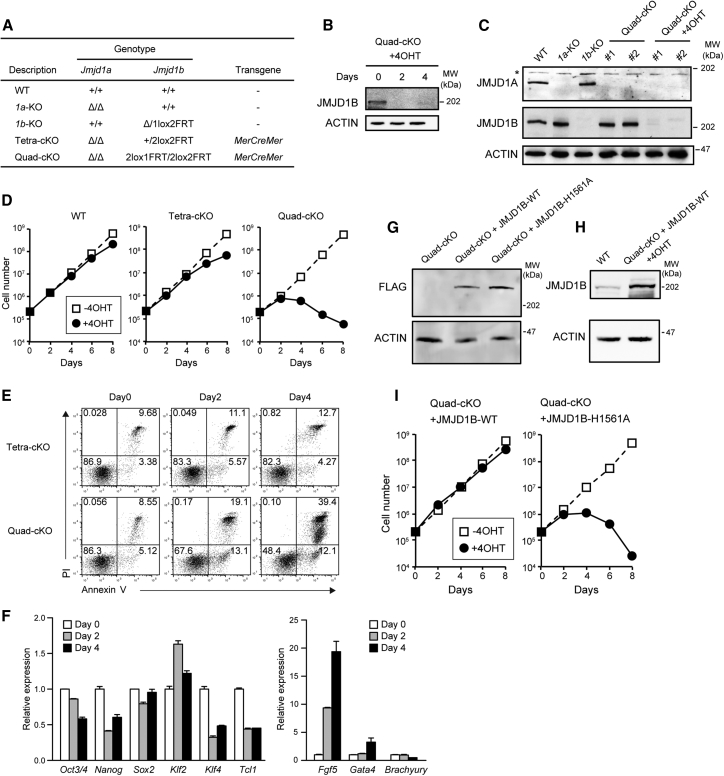
To further address the roles of JMJD1-mediated H3K9 demethylation in early embryogenesis, we used mouse ESCs, which provide a good tool for studying the developmental process of pre- and post-implantation embryos. Immunoblot analysis indicated that JMJD1A and JMJD1B were both expressed in ESCs (Figure 2). We previously generated ESCs lacking JMJD1A by a simple targeting method (Inagaki et al., 2009). Also, we have established ESCs lacking JMJD1B in this study (Figure S1), indicating that neither JMJD1A nor JMJD1B is essential for ESC survival. To address the impact of JMJD1A and JMJD1B double-deficiencies in ESC function, we tried to establish an ESC line with conditionally depleted JMJD1 proteins. The conditional targeting vector of Jmjd1b was constructed and then introduced into the JMJD1A-deficient ESC line (Figure S1). To convert functional 1b2lox alleles into defective 1b1lox alleles in a 4-hydroxy tamoxifen (4OHT)-dependent manner, the MerCreMer (Cre flanked by mutated estrogen receptor ligand-binding domains) expression plasmid was also introduced into the homologous recombinant lines. We consequently obtained 1a/1b-conditional knockout (KO) lines named Quad-cKO (Jmjd1aΔ/Δ; Jmjd1b2lox1FRT/2lox2FRT; Mer-Cre-Mer) (Figure 2A). Quad-cKO cell lines were cultured with 4OHT and then applied to immunoblot analysis (Figures 2B and 2C). Time-course experiments showed that JMJD1B signal disappeared after the 4OHT treatment for 2 days (Figure 2B). We confirmed the absence of JMJD1A signals, as well as JMJD1B signals, in the 4OHT-treated Quad-cKO cells (Figure 2C). We, thus, successfully generated ESC lines with conditionally depleted JMJD1 proteins.
Figure 2.
Depletion of JMJD1A and JMJD1B Induces Growth Arrest in ESCs
(A) List of the established ESC lines and their genotypes. MerCreMer, Cre flanked by mutated estrogen receptor ligand-binding domains.
(B) Time-course analysis of 4OHT-dependent depletion of JMJD1B in Quad-cKO cells.
(C) Immunoblot analyses of JMJD1A/JMJD1B-depleted ESC lines. Whole extracts of the indicated ESC lines were fractionated using SDS-PAGE and then applied to immunoblot analysis with antibodies against JMJD1A and JMJD1B. JMJD1A and JMJD1B were depleted in the Quad-cKO cell lines cultured with 800 nM 4OHT for 4 days. Asterisk (∗) represents non-specific signals.
(D) The indicated ESC lines were cultured in the presence or absence of 4OHT. Cell numbers were determined every 2 days. Growth arrest became apparent when the Quad-cKO cell line was cultured in the presence of 4OHT for 4 days (right). In contrast, wild-type (left) and Tetra-cKO (middle) cells grew in the presence of OHT.
(E) Time-course analysis of JMJD1 depletion-induced cell death. The indicated ESC lines were cultured in the presence of 4OHT, following which the cells were stained with PI and annexin V and analyzed using flow cytometry.
(F) Expression levels of pluripotency-associated (left) and lineage-associated (right) genes in Quad-cKO cells treated with 4OHT were examined using RT-qPCR. We used Fgf5, Gata4, and Brachyury as the markers for primitive ectoderm, endoderm, and mesoderm, respectively. Representative data are presented from independent triplicate experiments. Error bars indicate means ± SD derived from technical replicates.
(G and H) Rescue of the growth arrest phenotype by exogenous introduction of JMJD1B into Quad-cKO cell line. (G) Expression vectors for FLAG-tagged wild-type JMJD1B or enzymatically inactive H1561A mutants of JMJD1B were individually and stably introduced into the Quad-cKO cell line. The expression levels of exogenously expressed proteins were compared by immunoblot analysis. (H) Comparison of protein expression levels of endogenously expressed JMJD1B and exogenously expressed JMJD1B using anti-JMJD1B antibody. JMJD1B expression levels were compared between wild-type ESCs and 4OHT-treated Quad-cKO cells expressing FLAG-JMJD1B-WT.
(I) Quad-cKO cell lines expressing wild-type JMJD1B (left) or the enzymatically inactive H1561A mutant of JMJD1B (right) were cultured in the presence of 4OHT. Exogenous expression of wild-type JMJD1B rescued the growth arrest phenotype of Quad-cKO cells in the presence of 4OHT, whereas the enzymatically inactive H1561A mutant did not.
Next, we examined the growth potential of Quad-cKO cell lines. Tetra-cKO (Jmjd1aΔ/Δ; Jmjd1b+/2lox2FRT; Mer-Cre-Mer) cells that carried two disrupted Jmjd1a alleles and single conditional allele of Jmjd1b were generated as controls (Figure 2A). The parental wild-type cells (TT2 line) and Tetra-cKO cells could grow exponentially in the presence of 4OHT (Figure 2D, left and middle panels, respectively). In contrast, when Quad-cKO cell lines were cultured in the presence of 4OHT, an increase in cell numbers was noted during the first 2 days, which was followed by a decrease in number (Figure 2D, right panel). Note that we could not establish cell lines lacking both JMJD1A and JMJD1B due to severe growth defect in the Quad-cKO cell lines when the two proteins were depleted. Taking these results together, we concluded that JMJD1A and JMJD1B are redundantly but essentially required for ESC survival.
To examine the cause of growth arrest in ESCs lacking JMJD1A and JMJD1B, we assessed the cell viability by staining with propidium iodide (PI) and annexin V (Figure 2E). The number of early apoptotic cells that stained positively with annexin V and negatively with PI increased when the Quad-cKO cell line was cultured in the presence of 4OHT for 2 days; remarkably, the number further increased with 4OHT treatment for another 2 days (Figure 2E, bottom panel). In addition, the number of dead cells that stained positively with both annexin V and PI simultaneously increased (Figure 2E, bottom panel). The progressive increase in the number of apoptotic cells might account for the growth defect of ESCs lacking JMJD1A and JMJD1B, at least in part. In contrast, 4OHT treatment had a moderate effect on the number of apoptotic and dead cells in the Tetra-cKO cell line (Figure 2E, top panel). We also conducted cell cycle analysis of Quad-cKO cells by measuring bromodeoxyuridine (BrdU) incorporation and the DNA contents (Figure S3). 4OHT treatment did not have a profound effect on cell cycle progression in both Quad-cKO and Tetra-cKO cell lines. Therefore, the growth defect phenotype in JMJD1A/JMJD1B-depleted ESCs partly results from an increase in the apoptotic cell death.
To investigate how JMJD1A/JMJD1B depletion influenced the pluripotency of ESCs, we examined mRNA expression levels of six pluripotency-associated genes (Oct3/4, Nanog, Sox2, Klf2, Klf4, and Tcl1) and three lineage-associated genes (Fgf5, Gata4, and Brachyury) in 4OHT-treated Quad-cKO cells (Figure 2F). The expression of four pluripotency-associated genes, Oct3/4, Nanog, Klf4, and Tcl1, was downregulated (Figure 2F, left), whereas that of the lineage-associated primitive endoderm marker gene Fgf5 was upregulated in 4OHT-treated Quad-cKO cells (Figure 2F, right). These results indicate that JMJD1A/JMJD1B depletion in ESCs might compromise the self-renewal property of pluripotent ESCs and/or prompt their differentiation.
It is important to elucidate whether the growth arrest of JMJD1A/JMJD1B-depleted cells was dependent on JMJD1-mediated enzymatic activity. To address this issue, we performed rescue experiments using Jmjd1b expression vectors. Expression vectors for FLAG-tagged wild-type JMJD1B and catalytically defective JMJD1BH1561A (Figure S3) were stably introduced into the Quad-cKO cells (Figure 2G). We confirmed that the protein expression levels of exogenously introduced JMJD1B were higher than that of endogenous JMJD1B (Figures 2G and 2H). As shown in Figure 2I, Quad-cKO cells carrying wild-type Jmjd1b transgene had a clearly restored growth potential in the presence of 4OHT. In contrast, the growth potential of Quad-cKO cells carrying Jmjd1bH1561A transgene was not restored, even though the expression level of exogenously introduced JMJD1BH1561A was higher than that of wild-type (Figure 2G). These findings indicate that the H3K9 demethylating activity of JMJD1 enzymes is absolutely required for ESC survival.
JMJD1A and JMJD1B Substantially Contribute to H3K9 Demethylation in ESCs
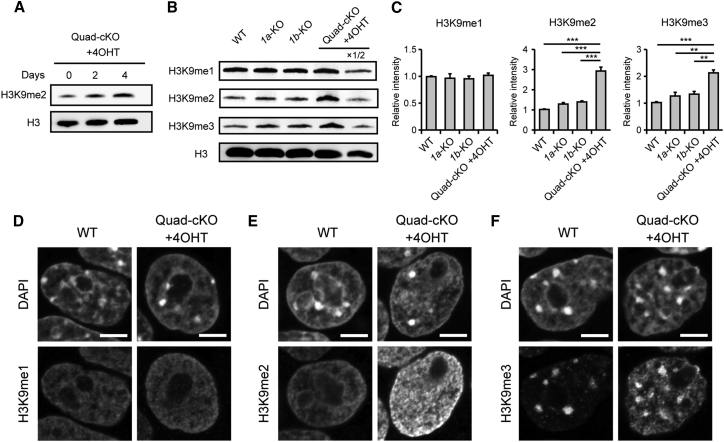
To address how JMJD1A and JMJD1B contribute to H3K9 methylation status of ESCs, we examined H3K9 methylation levels of mutant ESCs by immunoblot analysis (Figures 3A–3C). Dimethylated H3K9 (H3K9me2) was markedly increased in the Quad-cKO cell line when cultured in the presence of 4OHT for 2 days onward (Figure 3A). We next compared the H3K9 methylation levels among wild-type, JMJD1A-deficient, JMJD1B-deficient, and JMJD1A/JMJD1B-depleted ESCs using a panel of specific antibodies by immunoblot analysis (Figure 3B) (Kimura et al., 2008). H3K9me1 levels remained unchanged even after 1a/1b compound mutation (summarized in Figure 3C, left panel). In contrast, the compound mutation led to robust increases in the levels of H3K9me2 and me3 (summarized in Figure 3C, middle and right panels, respectively). Single mutations of 1a or 1b led to only moderate increases in the levels of H3K9me2 and me3, indicating that JMJD1A and JMJD1B redundantly catalyze H3K9 demethylation in ESCs.
Figure 3.
JMJD1A and JMJD1B Substantially Contribute to H3K9 Demethylation in ESCs
(A) Time-course analysis of JMJD1B-depletion-induced increase in H3K9me2 in 4OHT-treated Quad-cKO cells.
(B) Immunoblot analyses of H3K9 methylation levels in the indicated ESC lines.
(C) The methylation levels of H3K9me1 (left), H3K9me2 (middle), and H3K9me3 (right) in the indicated ESC lines were determined by immunoblot analysis. The intensities of H3K9me signals of wild-type cells were defined as 1. Data are presented as means ± SD (n = 3 independent experiments). ∗∗p < 0.01; ∗∗∗p < 0.001 (Student's t test).
(D–F) Nuclear distribution profiles of H3K9me1 (D), H3K9me2 (E), and H3K9me3 (F) in JMJD1A/JMJD1B-depleted ESCs compared with those in the wild-type ESCs. Scale bars, 5 μm.
We compared nuclear distribution profiles of H3K9 methylation between wild-type and JMJD1A/JMJD1B-depleted ESCs by immunocytochemistry. Neither distribution nor intensity of H3K9me1 signals was affected by JMJD1A/JMJD1B depletion (Figure 3D). H3K9me2 signals were detected mostly at the euchromatic regions in wild-type nuclei (Figure 3E, left panels). Distribution profiles of H3K9me2 signals seemed to be unaffected by JMJD1A/JMJD1B depletion; nevertheless, the signal intensities were dramatically increased (Figure 3E, right panels). H3K9me3 were detected mainly at the pericentric heterochromatin as dense signals in both wild-type and JMJD1A/JMJD1B-depleted cells (Figure 3F). Interestingly, we observed that JMJD1A/JMJD1B depletion led to a significant increase in H3K9me3 signals in the euchromatin (Figure 3F, right panels). These results strongly suggest that the increase in H3K9me2/3 in JMJD1A/JMJD1B-depleted ESCs is attributed to the increase in H3K9me2/3 in the euchromatin.
JMJD1A/JMJD1B-Mediated H3K9 Demethylation Targets Gene-Dense Regions of Chromosomes
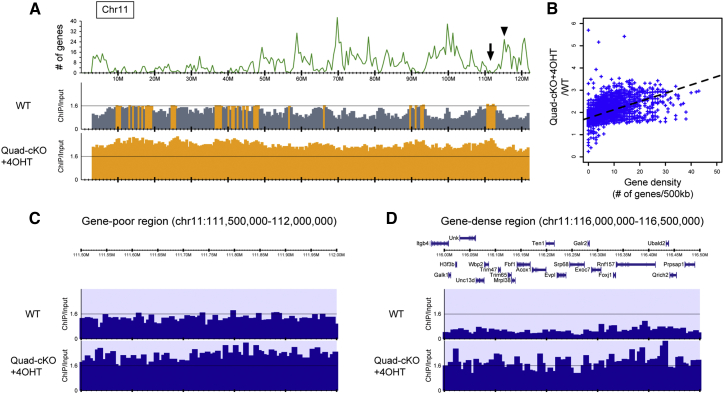
The next important question is which loci are targeted by JMJD1A/JMJD1B-mediated H3K9 demethylation in ESCs. To address this issue, we performed chromatin immunoprecipitation (ChIP) analysis using antibodies against H3K9me2. Mononucleosome was prepared by enzymatic digestion of ESC chromatin and then applied to ChIP sequencing (ChIP-seq) analysis (Figure 4). Importantly, approximately 1.7 times more DNA was immunoprecipitated with anti-H3K9me2 antibody from JMJD1A/JMJD1B-depleted nuclei than from wild-type nuclei, indicating that JMJD1A/JMJD1B depletion resulted in a genome-wide increase in H3K9me2 (Figure S4). The distribution profile of H3K9me2 on chromosome 11 is representatively shown in Figure 4A; the relatively gene-poor region spans 10–50 M and relatively gene-dense region spans 50–120 M (Figure 4A, upper panel). H3K9me2 was detected throughout chromosome 11 in wild-type cells, indicating high abundance of the H3K9me2 marks (Figure 4A, middle panel). It is intriguing that H3K9me2 distributes evenly with high levels of methylation in the gene-poor region, whereas H3K9me2 distributes unevenly with high and low levels of methylation in the gene-dense region of chromosome 11 in wild-type cells. In the gene-dense region, we observed a typical feature between the gene densities and enrichment of H3K9me2 in wild-type ESCs; loci with low gene densities were heavily methylated, while those with high gene densities were hypomethylated (Figure 4A, middle panel). JMJD1A/JMJD1B depletion resulted in increased levels of H3K9me2 throughout chromosome 11 (Figure 4A, bottom panel). These characteristics were common in the other chromosomes (Figure S4). Notably, the increase in H3K9me2 levels due to JMJD1A/JMJD1B depletion in the gene-dense region seemed to be higher than that in the gene-poor region. To address this quantitatively, the relationship between the increased H3K9me2 levels due to JMJD1A/JMJD1B depletion and gene densities of all chromosomes was statistically evaluated by calculating the average levels of H3K9me2 in comparison with the gene densities (number of genes per 500 kb genome). As shown in Figure 4B, there was a positive correlation between the increased H3K9me2 levels due to JMJD1A/JMJD1B depletion and gene densities, implying that JMJD1A/JMJD1B preferentially targets H3K9 demethylation in these regions (Figure 4B). The distribution profiles of H3K9me2 with narrower windows in the gene-poor and gene-dense regions of chromosome 11 are shown in Figures 4C and 4D, respectively. In the gene-poor region, H3K9me2 levels of JMJD1A/JMJD1B-depleted cells were approximately one and a half times higher than that of wild-type cells. In contrast, H3K9me2 levels of JMJD1A/JMJD1B-depleted cells were more than three times higher than that of wild-type cells, confirming that JMJD1A/JMJD1B preferentially targets H3K9 demethylation in gene-dense regions (Figures 4C and 4D).
Figure 4.
JMJD1A/JMJD1B-Mediated H3K9 Demethylation Targets Gene-Dense Euchromatin
(A) Distribution profiles of H3K9me2 along chromosome 11. The upper panel shows the number (#) of mm10 RefSeq genes smoothed with a width of 500 kb. The middle and lower panels represent the ratios of normalized read density between ChIP and whole-cell lysate (input) samples (ChIP/input) in wild-type and JMJD1A/JMJD1B-depleted cells (Quad-cKO+4OHT), respectively. The ratio of ChIP/input >1.6 is shown in orange.
(B) Correlation between gene density and increased H3K9me2 levels due to JMJD1A/JMJD1B depletion. The x and y axes indicate the number of mm10 RefSeq genes and the quotient of ChIP/input of Quad-cKO+4OHT divided by ChIP/input of wild-type (WT), respectively, smoothed with a 500 kb width. The dashed lines represent regression lines.
(C and D) Distribution profile of H3K9me2 in a gene-poor region (C), arrow showing the position in (A), or a gene-dense region (D), arrowhead showing the position in (A) within chromosome 11. The upper panels show the positions of the protein-coding genes, whereas lower tracks depict ChIP/input in the indicated samples.
Profound Effect of JMJD1A/JMJD1B Depletion on Gene Expression
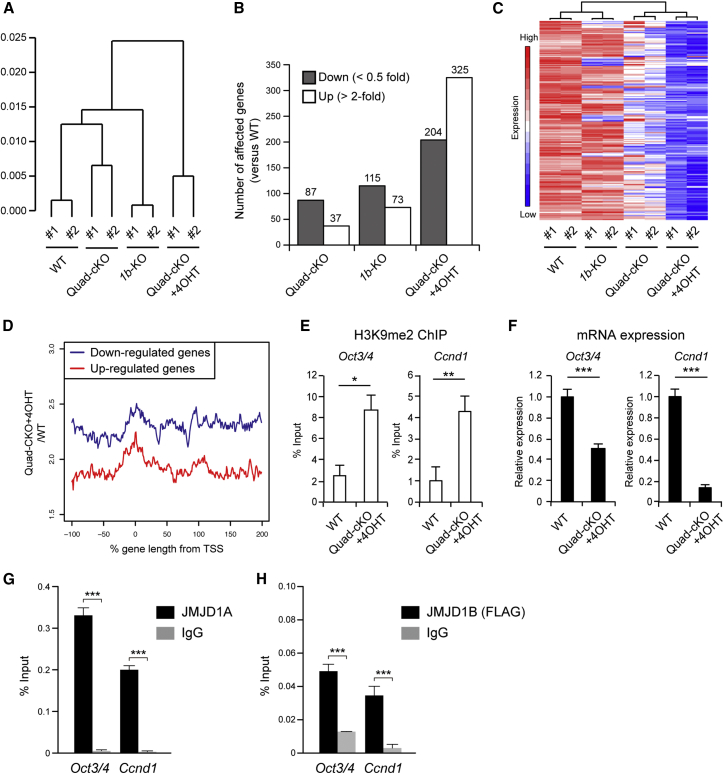
As the enrichment of H3K9 methylation marks is relevant to transcriptionally silenced heterochromatin, the distorted distribution of H3K9me2 might induce the alteration of gene expression in JMJD1-depleted cells. To address this, we performed a microarray analysis using RNAs from wild-type, JMJD1B-deficient, JMJD1A-deficient (Quad-cKO cell lines), and JMJD1A/JMJD1B-depleted cells (Quad-cKO cell lines cultured in the presence of 4OHT). Hierarchical clustering analysis revealed that each of the genotype-identical sets forms the smallest clusters, indicating that it is not the clonal but the genetic difference that influences gene expression (Figure 5A). The cluster of JMJD1A/JMJD1B-depleted cells was the farthest from those of the other cells, indicating that compound depletion of JMJD1A and JMJD1B has the most prominent effect on transcription. Based on this notion, the numbers of genes up- or downregulated by JMJD1A/JMJD1B double depletion were found to be larger than the numbers affected by single mutation (Figure 5B, genes are listed in Table S2).
Figure 5.
Depletion of JMJD1A and JMJD1B Alters Gene Expression Profile in ESCs
RNAs were prepared from ESCs and subjected to a microarray analysis using an Affymetrix mouse genome 430 2.0 array.
(A) Hierarchical clustering analysis of gene expression profiles in wild-type cells (TT2 lines), JMJD1A-deficient cells (Quad-cKO cell lines), JMJD1B-deficient cells, and JMJD1A/JMJD1B-double-depleted cells (Quad-cKO cell lines cultured with 4OHT). The farthest distance was observed between the cluster of JMJD1A/JMJD1B-double-depleted cells and the other cell clusters, indicating that the compound depletion of JMJD1A and JMJD1B has the most prominent effect on transcription.
(B) Numbers of genes affected by the JMJD1A and/or JMJD1B depletion in ESCs. Downregulated genes (<0.5-fold) and upregulated genes (>2.0-fold) due to each mutation are represented as dark and white bars, respectively.
(C) Comparison of mRNA levels in the 204 genes between the indicated samples, which were downregulated by JMJD1A/JMJD1B double depletion. Heatmap analysis demonstrated that single depletion had a moderate effect on the transcription of these genes compared with double depletion.
(D) Quantitative analysis of H3K9me2 levels of the genes regulated by JMJD1A and JMJD1B. Averaged increase in H3K9me2 around the gene bodies down- (blue) and upregulated (red) due to JMJD1A/JMJD1B depletion was plotted. The x axes indicate % gene length; 0% and 100% represent transcription start site and transcription end site, respectively. The y axes indicate the average of the quotient of ChIP/input of Quad-cKO+4OHT divided by ChIP/input of wild-type.
(E) H3K9 methylation levels of the promoter regions of Oct3/4 (left) and Ccnd1 (right) of the indicated ESCs.
(F) mRNA expression levels of Oct3/4 (left) and Ccnd1 (right) of the indicated ESCs.
(G) ChIP analysis for JMJD1A at the promoter regions of Oct3/4 (left) and Ccnd1 (right) with anti-JMJD1A antibodies.
(H) ChIP analysis of JMJD1B at the promoter regions of Oct3/4 (left) and Ccnd1 (right) using anti-FLAG antibody. To detect endogenous JMJD1B with anti-FLAG antibody, we established knockin ESC line carrying1b+/Flag−KI allele, in which the modified 1b allele produces JMJD1B protein with a FLAG tag at its carboxy terminus (Figure S1).
For (E–H), data are presented as means ± SD from n = 3 independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 (Student's t test).
Next, we examined the individual roles of JMJD1A and JMJD1B on the transcriptional regulation of the 204 genes that were downregulated by JMJD1A/JMJD1B double depletion. A heatmap analysis revealed that JMJD1A or JMJD1B deficiency alone had only moderate effects on the downregulation of those genes, indicating that JMJD1A and JMJD1B upregulate these genes in a redundant manner (Figure 5C). Among the 204 genes, 134 were downregulated only when both JMJD1A and JMJD1B were depleted (Figure S5). Gene ontology (GO) analyses demonstrated that metabolic processes such as one-carbon metabolism and amino acid biosynthesis were significantly enriched among the 134 genes (Figure S5). On the other hand, the genes of many pathways, such as reproductive process, inflammatory pathway, and cellular differentiation, were significantly enriched among the genes upregulated only when both JMJD1A and JMJD1B were depleted (Figure S5). To elucidate the cause of distorted gene expression in JMJD1A/JMJD1B-depleted cells, we examined H3K9 methylation levels in the genomic portion surrounding the down- and upregulated genes (Figure 5D). We found that JMJD1A/JMJD1B depletion increased H3K9 methylation levels of the downregulated genes more profoundly than those of the upregulated genes (Figure 5D). Therefore, it is conceivable that these genes are the direct targets of JMJD1A and JMJD1B.
Because previous studies have demonstrated that H3K9 methylation negatively controls the transcription of genes including Oct3/4 (Feldman et al., 2006) and a D-type cyclin Ccnd1 (Shirato et al., 2009), we hypothesized that JMJD1A/JMJD1B-mediated H3K9 demethylation will upregulate the transcription of Oct3/4 and Ccnd1. To evaluate this hypothesis, we examined the H3K9 methylation levels of Oct3/4 and Ccnd1 promoters and the mRNA expression levels of Oct3/4 and Ccnd1 in JMJD1A/JMJD1B-depleted ESCs. As shown in Figures 5E and 5F, JMJD1A/JMJD1B depletion increased the levels of H3K9me2 at Oct3/4 and Ccnd1 and decreased their transcription. To further confirm the effect of JMJD1-mediated H3K9 demethylation on Oct3/4 and Ccnd1 regulation, we examined the enrichment of JMJD1A and JMJD1B at Oct3/4 and Ccnd1 loci. ChIP analysis revealed that JMJD1A was significantly enriched at the promoter regions of Oct3/4 and Ccnd1 in ESCs (Figure 5G). ChIP analysis of 1b+/Flag−KI ESCs demonstrated that endogenous JMJD1B was significantly enriched at the promoter regions of Oct3/4 and Ccnd1 (Figure 5H). Taking these results together, we conclude that JMJD1A and JMJD1B catalyze H3K9 demethylation of Oct3/4 and Ccnd1 gene promoters, thereby activating their transcription.
Collaborative Roles of JMJD1A/JMJD1B Demethylases and G9A Methyltransferase for Tuning the H3K9 Methylation Levels and Regulation of ESC Function
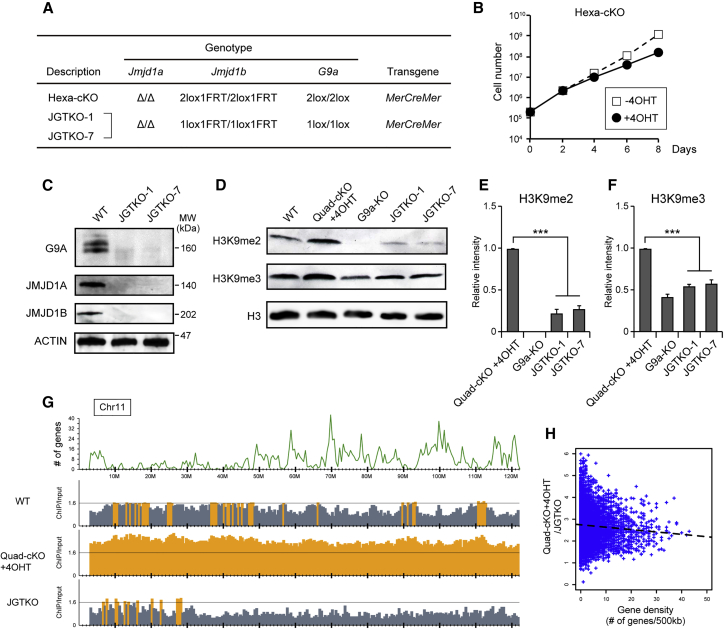
The data presented here suggest that the excess accumulation of H3K9 methylation might cause rapid cell death and perturbed gene expression in JMJD1A/JMJD1B-depleted cells. To verify this possibility, we tried to identify the enzymes responsible for H3K9 overmethylation in JMJD1A/JMJD1B-depleted cells. We have previously demonstrated that GLP/G9A methyltransferase complex catalyzes H3K9 methylation in the euchromatin (Tachibana et al., 2002, Tachibana et al., 2005). Assuming the strong candidate of this complex for euchromatic H3K9 overmethylation in the JMJD1A/JMJD1B-depleted cells, we introduced G9A-conditional alleles into the Quad-cKO cell line. Consequently, Hexa-cKO cells, in which JMJD1A, JMJD1B, and G9A could be inducibly depleted, were obtained (Figure 6A). Surprisingly, the Hexa-cKO cells demonstrated normal growth when they were cultured in the presence of 4OHT (Figure 6B), suggesting that G9A depletion restored the growth potential of JMJD1A/JMJD1B-depleted ESCs. As expected by this finding, ESC lines lacking JMJD1A, JMJD1B, and G9A (named as JGTKO) were easily established from a pool of Hexa-cKO cells cultured with 4OHT, confirming that the triple-KO ESCs have normal growth potential (Figures 6A and 6C). Next, H3K9 methylation levels of the JGTKO lines were assessed by immunoblot analysis (Figures 6D–6F). The additional G9a mutation induced drastic reductions in H3K9me2 and H3K9me3 levels in ESCs with JMJD1A/JMJD1B-deficient background (Figures 6E and 6F). Taken together, we conclude that G9A is a bona fide enzyme responsible for the H3K9 overmethylation in JMJD1A/JMJD1B-depleted ESCs and that G9A-mediated H3K9 overmethylation is the cause of cell death in JMJD1A/JMJD1B-depleted ESCs.
Figure 6.
Collaborative Roles of JMJD1A/JMJD1B Demethylases and G9A Methyltransferase for Tuning the H3K9 Methylation Levels and Regulating ESC Function
(A) Genotypes of the established ESC line. Hexa-cKO cell line carries alleles for the null mutation of Jmjd1a, the conditional mutation of Jmjd1b, and the conditional mutation of G9a. ESC lines lacking JMJD1A, JMJD1B, and G9A (named JGTKO-1 and JGTKO-7) were established by cloning from a pool of Hexa-cKO cells cultured in the presence of 4OHT.
(B) Growth curve of Hexa-cKO cell line in the presence or absence of 4OHT.
(C) JMJD1A, JMJD1B, and G9A proteins were absent in JGTKO-1 and JGTKO-7 ESC lines. Whole-cell extracts were fractionated by SDS-PAGE, and then subjected to immunoblot analysis with the indicated antibodies.
(D–F) G9a mutation rescues the H3K9 overmethylation phenotype of JMJD1A/JMJD1B-double-depleted cells. H3K9me2 and H3K9me3 levels in the indicated cell lines were examined by immunoblot analysis (D) and summarized (E and F, respectively). The intensities of H3K9me signals of Quad-cKO + 4OHT were defined as 1. Data are presented as means ± SD from n = 3 independent experiments. ∗∗∗p < 0.001 (Student's t test).
(G) Distribution profiles of H3K9me2 of JGTKO cells along chromosome 11. The top panel shows the number of mm10 RefSeq genes smoothed over 500 kb. The middle panels represent the ratios of normalized ChIP/input of ESCs of the indicated genotypes, including wild-type and JMJD1A/JMJD1B-depleted cells. The ratio of ChIP/input >1.6 is shown in orange.
(H) Correlation between gene density and decreased H3K9me2 levels due to G9a mutation in JMJD1A/JMJD1B-depleted cells. The x and y axes indicate the number of mm10 RefSeq genes and the quotient of ChIP/input of Quad-cKO+4OHT divided by ChIP/input of JGTKO cells, respectively, smoothed over 500 kb. Dashed lines represent regression curves.
To investigate how G9a mutation altered the genome-wide distribution of H3K9 methylation, we examined the H3K9me2 distribution profile of JGTKO cells by ChIP-seq analysis (Figure 6G). Notably, less than half the amount of input DNA was immunoprecipitated with anti-H3K9me2 antibody from JGTKO cell nuclei than from JMJD1A/JMJD1B-depleted cell nuclei, indicating that G9A might actually contribute to H3K9 di-methylation in JMJD1A/JMJD1B-depleted cells (Figure S4). The distribution profile of H3K9me2 on chromosome 11 of JGTKO cells is shown in Figure 6G. We found that G9a mutation resulted in decreased H3K9me2 levels throughout chromosome 11, confirming that G9A substantially contributes to H3K9 di-methylation in JMJD1A/JMJD1B-depleted cells. To address the target preference of G9A-mediated H3K9 methylation, the relationship between the decreased H3K9me2 levels due to G9a mutation in JMJD1A/JMJD1-depleted cells and gene densities of all chromosomes was statistically evaluated as in Figure 4B. No significant differences were detected in G9A-mediated H3K9 methylation levels between the gene-poor and gene-dense regions (Figure 6H). These data suggest that the target preference of G9A-mediated H3K9 methylation is less strict than that of JMJD1A and JMJD1B-mediated H3K9 demethylation, which preferentially targets gene-dense regions (Figure 4B).
G9a Mutation Rescues JMJD1A/JMJD1B-Depletion-Induced Transcriptional Downregulation
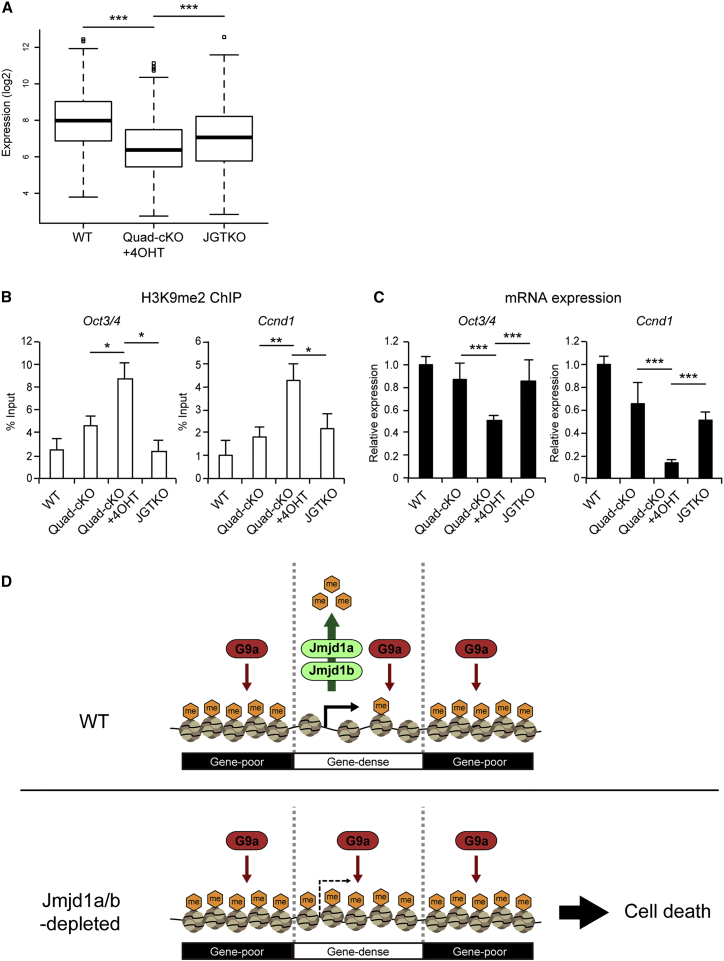
To address the question of whether G9a mutation also restores gene expression profiles in JMJD1A/JMJD1B-depleted cells, we examined the expression levels of 204 genes in JGTKO cells that were downregulated due to JMJD1A/JMJD1B depletion. As shown in Figure 7A, G9a mutation remarkably restored the expression levels of those genes. To investigate the roles of JMJD1A/JMJD1B-mediated H3K9 demethylation and G9A-mediated H3K9 methylation in transcription regulation at single-gene level, we examined H3K9 methylation and mRNA expression of Oct3/4 and Ccnd1 in the serial mutant ESC lines (Figures 7B and 7C, respectively). ChIP analyses demonstrated that the elevated H3K9me2 levels were rescued by the introduction of G9a mutation (Figure 7B), strongly suggesting that H3K9me2 levels of Oct3/4 and Ccnd1 were regulated by both JMJD1A/JMJD1B-mediated H3K9 demethylation and G9A-mediated H3K9 methylation. mRNA analyses indicated typical inverse correlations between the H3K9me2 levels and the expression levels of Oct3/4 and Ccnd1; the decreased mRNA expression levels of Oct3/4 and Ccnd1 were rescued by G9a mutation (Figures 7B and 7C). Thus, we conclude that the antagonistic activities between JMJD1A/JMJD1B and G9A collaboratively contribute to the tuning of H3K9 methylation levels in Oct3/4 and Ccnd1. We summarize the function of JMJD1A/JMJD1B in the maintenance of ESCs as follows: H3K9 demethylases JMJD1A and JMJD1B ensure cell viability and transcriptional accuracy by removing the H3K9 methylation marks deposited by G9A in the gene-dense euchromatin (Figure 7D).
Figure 7.
G9a Mutation Rescues JMJD1A/JMJD1B-Depletion-Induced Transcriptional Downregulation
(A) Comparison of average expression levels of 204 genes that were downregulated by JMJD1A/JMJD1B depletion between the mutant ESCs. Introduction of G9a mutation in JMJD1A/JMJD1B-deficient background significantly restored the expression levels of those genes. ∗∗∗p < 0.001 (Student's t test).
(B and C) JMJD1A/JMJD1B and G9A antagonistically tune the H3K9 methylation levels of Oct3/4 and Ccnd1 to ensure accurate transcription. (B) The H3K9me2 levels in the promoter regions of Oct3/4 (left) and Ccnd1 (right) were examined by performing ChIP-qPCR analyses. Increased H3K9me2 levels in these genes in JMJD1A/JMJD1B-double-depleted cells were rescued by G9A depletion. (C) The expression levels of Oct3/4 (left) and Ccnd1 (right) were examined by RT-qPCR analyses. Reduced expression levels of these genes in JMJD1A/JMJD1B-double-depleted cells were rescued by G9A depletion. mRNA expression levels of wild-type cells were defined as 1. For (B) and (C), data are presented as means ± SD. n = 3 independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 (Student's t test).
(D) Schematic representation of the roles of JMJD1A/JMJD1B in ESCs. JMJD1A/JMJD1B ensure cell viability and transcriptional accuracy by antagonizing G9a-mediated H3K9 overmethylation in gene-rich euchromatin in ESCs. In wild-type ESCs, JMJD1A/JMJD1B preferentially remove H3K9 methylation marks from gene-rich euchromatin. The compound loss of JMJD1A/JMJD1B results in G9a-mediated H3K9 overmethylation in euchromatin, thereby inducing cell death and impaired gene expression.
As shown in Figure 6B, JMJD1A/JMJD1B- and G9A-triple-KO cells clearly re-acquired growth potential, indicating that ESCs can survive in the absence of JMJD1A/JMJD1B- and G9A-mediated H3K9 methylation tuning. To address the possible importance of JMJD1A/JMJD1B- and G9A-mediated H3K9 methylation tuning for differentiation of ESCs, we cultured the JMJD1A/JMJD1B/G9A-triple-deficient JGTKO-1 line under differentiating condition without leukemia inhibitory factor. As shown in Figure S6, the JGTKO cell line immediately lost growth potential in the differentiating condition. We further examined the kinetics of mRNA expression of several pluripotency-associated genes (Figure S6). In wild-type cells, pluripotency-associated genes were downregulated concomitantly with differentiation. In contrast, some pluripotency-associated genes were not downregulated in the JGTKO line in the differentiating condition (Figure S6). These results indicate that collaborative H3K9 methylation tuning by JMJD1A/JMJD1B and G9A plays an indispensable role in the suppression of pluripotency-associated genes when ESCs undergo differentiation.
Discussion
We previously demonstrated that JMJD1A and JMJD1B have intrinsic enzymatic activities for H3K9 demethylation (Kuroki et al., 2013a). Here, we report the functional overlap of JMJD1A and JMJD1B in early embryogenesis and ESC maintenance in mice. As expected, JMJD1A and JMJD1B are both expressed in developing embryos and ESCs (Figures 1 and 2, respectively). On the other hand, Jmjd1a single-mutant mice display unique phenotypes, such as defects of spermiogenesis (Liu et al., 2010, Okada et al., 2007), metabolism (Inagaki et al., 2009, Tateishi et al., 2009), and sex determination (Kuroki et al., 2013b). We speculate that Jmjd1a is highly expressed in these tissues and therefore plays a dominant role in H3K9 demethylation. According to this notion, protein expression profiles of JMJD1A and JMJD1B are distinctive in the human tissues (http://www.humanproteomemap.org/).
We demonstrated that the combined depletion of JMJD1A and JMJD1B has a synergistic effect on gene expression (Figure 5). It is plausible that the growth defect phenotype of JMJD1A/JMJD1B-depleted ESCs is attributed to distorted gene expression, at least partly. Note that the GO terms for the multiple metabolic pathways downregulated by JMJD1A/JMJD1B deficiency include the one-carbon and amino acid biosynthetic pathways (Figure S5). It is feasible that disorders in multiple metabolic pathways might induce the loss of ESC viability.
Mammalian cells have multiple H3K9 methyltransferases and demethylases with different product/substrate specificities and target specificities (Greer and Shi, 2012, Kooistra and Helin, 2012). The antagonistic activities of H3K9 methyltransferase and demethylase are considered to contribute to the tuning of the H3K9 methylation levels, but how and which combination of methyltransferase/demethylase contributes to this regulation were still unresolved. Here, we demonstrated that JMJD1A/JMJD1B demethylases and G9A methyltransferase are involved in the tuning of the H3K9 methylation levels (summarized in Figure 7D). To the best of our knowledge, this is the first study revealing the role sharing between demethylase and methyltransferase for the correct establishment of histone methylated epigenome. In this context, G9A-mediated H3K9 methylation seems to exhibit less specificity for the target region of chromosomes (Figure 6H). According to this notion, a previous ChIP-seq analysis demonstrated that G9A is located in almost all chromosomal regions (Mozzetta et al., 2014). On the other hand, ChIP-seq analysis in our present study revealed that JMJD1A/JMJD1B-mediated H3K9 demethylation may preferentially target gene-dense euchromatin (Figure 4B). How JMJD1A/JMJD1B exerts target specificity toward euchromatin needs to be ascertained. Like most of the histone modification enzymes, JMJD1A and JMJD1B do not contain typical DNA recognition motifs, strongly suggesting that the target specificity of these proteins is dependent on the interaction of other proteins. JMJD1A and JMJD1B bear the signature LXXLL motif indicative of a protein-protein interaction with the nuclear receptors. It was reported that JMJD1A is recruited to androgen-receptor target genes in a hormone-dependent manner (Yamane et al., 2006). A recent study revealed that phosphorylation of JMJD1A strengthens the binding capacity to binding partner proteins, such as PPARγ (Abe et al., 2015). However, as shown in Figure 4D, it seems likely that JMJD1A/JMJD1B-mediated H3K9 demethylation can target megabase ranges of the chromosomal region. Thus, it is also conceivable that JMJD1A/JMJD1B forms a complex with a nuclear protein, which can recognize the global chromatin structure rather than specific DNA sequences.
Unexpectedly, our results indicate that JMJD1A/JMJD1B- and G9A-mediated H3K9 methylation tuning is dispensable for the maintenance of ESCs (Figure 6B). On the other hand, this tuning machinery seems to play an indispensable role when ESCs undergo differentiation because the downregulation of pluripotency-associated genes was compromised in the JGTKO line when cultured in the differentiating condition (Figure S6). A previous study demonstrated that the forced expression of protein kinase A in ESCs induces cellular differentiation concomitantly with G9A-mediated H3K9 methylation of the Oct3/4 locus (Yamamizu et al., 2012). We demonstrated that the Oct3/4 locus undergoes not only G9A-mediated H3K9 methylation but also JMJD1A/JMJD1B-mediated H3K9 demethylation to ensure correct expression levels (Figures 7B and 7C). It is plausible that the collaboration of JMJD1A/JMJD1B-mediated H3K9 demethylation and G9A-mediated H3K9 methylation may be essentially required for the rearrangement of H3K9 methylated epigenome upon differentiation. Based on the result that G9a mutation rescued compromised gene expression due to JMJD1A/JMJD1B depletion (Figure 7A), we speculate that the fine-tuning of H3K9 methylation by JMJD1 and G9A may contribute to transcriptional regulation of not only Oct3/4 but also other genes important for ESC function.
It has been demonstrated that lysine methyltransferases and demethylases regulate methylation dynamics not only for histones but also for non-histone proteins (Hamamoto et al., 2015). Most recently, it was demonstrated that the GLP/G9A complex methylates lysine residues within the TARK motif of DNA Ligase 1, which is similar to the H3K9 methylation within the TARK motif of histone H3 (Ferry et al., 2017). To the best of our knowledge, the contribution of JMJD1A and JMJD1B toward the demethylation of non-histone protein substrates has not been reported. The possibility that JMJD1A/JMJD1B and G9A regulate methylation of non-histone proteins warrants further investigation.
Note that the levels of H3K9me2 and H3K9me3 in the triple-KO cells were slightly but significantly higher than those in the G9a-deficient cells (Figures 6D–6F). These data indicate that JMJD1A and JMJD1B demethylate the H3K9me2 and H3K9me3 marks deposited by H3K9 methyltransferase(s) other than G9A. We propose that the balance in H3K9 methylation levels is maintained in a highly orchestrated manner, governed by multiple enzymes for methylation and demethylation.
Experimental Procedures
Generation of Mutant Mice and ESCs
Serial knockin targeting vectors were constructed by the bacterial artificial chromosome recombineering technique (Copeland et al., 2001) and then introduced into the ESC line TT2. Detailed information for the generation of mutant mice and ESCs is described in Supplemental Information. All animal experiments were performed under the animal ethical guidelines of Tokushima University (experiment number 14,108, approved by The Ethics Committee of Tokushima University for Animal Research) and Kyoto University (experiment number A12-6-2, approved by Animal Experimentation Committee of Kyoto University).
Author Contributions
S. Kuroki and M.T. designed experiments. S. Kuroki, Y.N., R.M., N.O., M.A., Y.Y., S. Kitano, H.M., R.N., K.I., K.S., and M.T. performed experiments and analyzed data. H.K. and Y.S. provided resources. S. Kuroki and M.T. wrote the paper.
Acknowledgments
We are grateful to Hiroyuki Sasaki (Kyushu University) and Naoko Yokota (Tokyo University) for support during the in silico analysis. We also thank Enago for the English language review. We are especially grateful to Toru Nakano for critical reading of the manuscript. We thank all members of the Tachibana laboratory. This work was supported by JSPS KAKENHI grant numbers JP26250037 (M.T.), JP16H01218 (M.T.), JP16H01409 (M.T.), and JP16K21196 (S. Kuroki); the Funding Program for Next Generation World-Leading Researchers (M.T.); the Takeda Science Foundation (M.T. and S. Kuroki); the Suntory Foundation for Life Sciences (S. Kuroki); and a Promotion of Science Cooperative Research Grant of the Institute for Enzyme Research, Joint Usage/Research Center, Tokushima University (H.M.).
Published: March 8, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.02.002.
Accession Numbers
The accession numbers for the microarray data and the ChIP-seq data reported in this paper are GEO: GSE98761 and DDBJ: DRA006496, respectively.
Supplemental Information
References
- Abe Y., Rozqie R., Matsumura Y., Kawamura T., Nakaki R., Tsurutani Y., Tanimura-Inagaki K., Shiono A., Magoori K., Nakamura K. JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat. Commun. 2015;6:7052. doi: 10.1038/ncomms8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland N.G., Jenkins N.A., Court D.L. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- Feldman N., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Ferry L., Fournier A., Tsusaka T., Adelmant G., Shimazu T., Matano S., Kirsh O., Amouroux R., Dohmae N., Suzuki T. Methylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol. Cell. 2017;67:550–565.e5. doi: 10.1016/j.molcel.2017.07.012. [DOI] [PubMed] [Google Scholar]
- Greer E.L., Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto R., Saloura V., Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat. Rev. Cancer. 2015;15:110–124. doi: 10.1038/nrc3884. [DOI] [PubMed] [Google Scholar]
- Inagaki T., Tachibana M., Magoori K., Kudo H., Tanaka T., Okamura M., Naito M., Kodama T., Shinkai Y., Sakai J. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells. 2009;14:991–1001. doi: 10.1111/j.1365-2443.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Kim K.B., Eom G.H., Choe N., Kee H.J., Son H.J., Oh S.T., Kim D.W., Pak J.H., Baek H.J. KDM3B is the H3K9 demethylase involved in transcriptional activation of lmo2 in leukemia. Mol. Cell Biol. 2012;32:2917–2933. doi: 10.1128/MCB.00133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Hayashi-Takanaka Y., Goto Y., Takizawa N., Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- Kooistra S.M., Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Kuroki S., Akiyoshi M., Tokura M., Miyachi H., Nakai Y., Kimura H., Shinkai Y., Tachibana M. JMJD1C, a JmjC domain-containing protein, is required for long-term maintenance of male germ cells in mice. Biol. Reprod. 2013;89:93. doi: 10.1095/biolreprod.113.108597. [DOI] [PubMed] [Google Scholar]
- Kuroki S., Matoba S., Akiyoshi M., Matsumura Y., Miyachi H., Mise N., Abe K., Ogura A., Wilhelm D., Koopman P. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science. 2013;341:1106–1109. doi: 10.1126/science.1239864. [DOI] [PubMed] [Google Scholar]
- Liu Z., Chen X., Zhou S., Liao L., Jiang R., Xu J. The histone H3K9 demethylase Kdm3b is required for somatic growth and female reproductive function. Int. J. Biol. Sci. 2015;11:494–507. doi: 10.7150/ijbs.11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhou S., Liao L., Chen X., Meistrich M., Xu J. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J. Biol. Chem. 2010;285:2758–2770. doi: 10.1074/jbc.M109.066845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzetta C., Pontis J., Fritsch L., Robin P., Portoso M., Proux C., Margueron R., Ait-Si-Ali S. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol. Cell. 2014;53:277–289. doi: 10.1016/j.molcel.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Okada Y., Scott G., Ray M.K., Mishina Y., Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- Shirato H., Ogawa S., Nakajima K., Inagawa M., Kojima M., Tachibana M., Shinkai Y., Takeuchi T. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J. Biol. Chem. 2009;284:733–739. doi: 10.1074/jbc.M804994200. [DOI] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K., Okada Y., Kallin E.M., Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamizu K., Fujihara M., Tachibana M., Katayama S., Takahashi A., Hara E., Imai H., Shinkai Y., Yamashita J.K. Protein kinase A determines timing of early differentiation through epigenetic regulation with G9a. Cell Stem Cell. 2012;10:759–770. doi: 10.1016/j.stem.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.