Abstract
This article provides details about the various cancer types recorded in Northeastern states of Nigeria currently being affected by insurgency in Nigeria. The dataset was described and chi-square test was used to determine the dependency of the variables under consideration on each other. Also, linear, logarithmic, inverse, quadratic, cubic, power, growth, exponential and logistic regression models were fitted to the dataset to show the relationship between them.
Keywords: Cancer, Chi-square test of independence, Insurgency, Nigeria, Regression model, Statistics
Specifications Table
| Subject area | Medicine |
| More specific subject area | Oncology, Public health, Biostatistics |
| Type of data | Table and text file |
| How data was acquired | Secondary data from University of Maiduguri Teaching Hospital. |
| Data format | Raw and partially analyzed (Descriptive and Inferential) |
| Experimental factors | Analysis of cancer incidences |
| Experimental features | Observations on the age, gender and the topographical location of cancer on the body of affected patients |
| Data source location | University of Maiduguri Teaching hospital, Maiduguri, Borno state, Northeast Nigeria. |
| Data accessibility | All the data are available this article |
Value of the data
-
•
The data is useful in the study of epidemiology of cancer in the affected areas.
-
•
The data is an indication of the public health crisis in insurgency affected region in Nigeria.
-
•
The data can be useful in cancer awareness, management and treatment.
-
•
The data could be used in oncologic studies.
-
•
The data can be used to test the performance of statistical models.
1. Data
The data set represents the age, gender and topological (Top) location of cancer on the body of cancer patients in the University of Maiduguri Teaching hospital located in Maiduguri, the capital of Borno state, Nigeria.
The teaching hospital is the only tertiary health care facility in the state and often serves the other northeast states like Yobe, Taraba, Adamawa, Bauchi and Gombe.
A total of 1671 patients were considered between the period of study and SPSS version 20 was used to perform the analysis. The dataset is available as Supplementary data while a brief summary of the data is presented in Table 1.
Table 1.
Brief summary of the data.
|
Statistics | ||||
|---|---|---|---|---|
| Gender | Age | Top | ||
| N | Valid | 1670 | 1671 | 1671 |
| Missing | 1 | 0 | 0 | |
| Mean | 1.53 | 50.06 | 37.59 | |
| Mode | 2 | 60 | 5 | |
| Variance | 0.249 | 281.086 | 816.431 | |
| Skewness | −0.115 | −0.258 | 0.241 | |
| Std. Error of Skewness | 0.060 | 0.060 | 0.060 | |
| Kurtosis | −1.989 | −0.220 | −1.149 | |
| Std. Error of Kurtosis | 0.120 | 0.120 | 0.120 | |
| Minimum | 1 | 3 | 1 | |
| Maximum | 2 | 95 | 117 | |
| Sum | 2553 | 83,658 | 62,806 | |
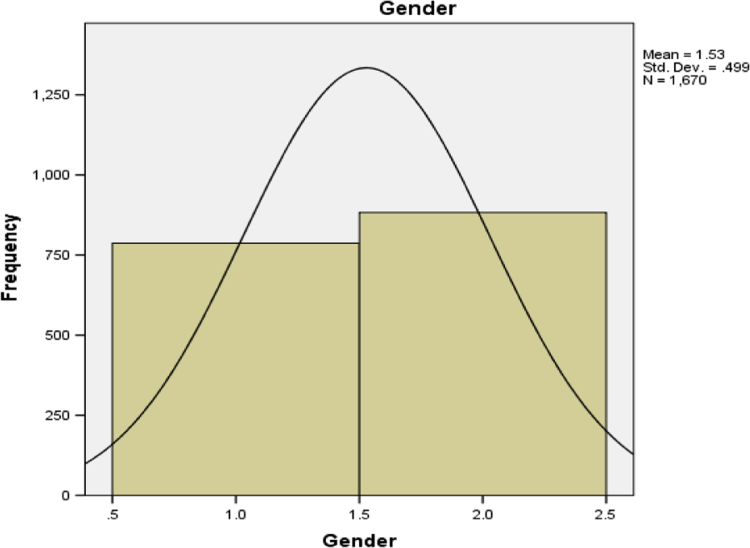
It was observed from Table 1 that information about the gender of a patient was not available, hence the missing data of 1.
The frequency distribution of the gender of the patients is presented in Table 2.
Table 2.
Frequency distribution of the patients’ gender.
| Gender | Frequency | Percent | Cumulative Percent | |
|---|---|---|---|---|
| Valid | Male | 787 | 47.1 | 47.1 |
| Female | 883 | 52.8 | 100.0 | |
| Total | 1670 | 99.9 | ||
| Missing | System | 1 | .1 | |
| Total | 1671 | 100.0 | ||
The frequency distribution of the patients’ age is presented in Table 3.
Table 3.
Frequency distribution of the patient's age.
| Age (years) | Frequency | Percent | Cumulative Percent |
|---|---|---|---|
| 3 | 6 | 0.4 | 0.4 |
| 4 | 5 | 0.3 | 0.7 |
| 5 | 1 | 0.1 | 0.7 |
| 6 | 5 | 0.3 | 1.0 |
| 7 | 5 | 0.3 | 1.3 |
| 8 | 2 | 0.1 | 1.4 |
| 9 | 1 | 0.1 | 1.5 |
| 10 | 2 | 0.1 | 1.6 |
| 12 | 4 | 0.2 | 1.9 |
| 14 | 4 | 0.2 | 2.1 |
| 15 | 8 | 0.5 | 2.6 |
| 16 | 6 | 0.4 | 2.9 |
| 17 | 4 | 0.2 | 3.2 |
| 18 | 9 | 0.5 | 3.7 |
| 19 | 6 | 0.4 | 4.1 |
| 20 | 15 | 0.9 | 5.0 |
| 22 | 9 | 0.5 | 5.5 |
| 23 | 12 | 0.7 | 6.2 |
| 24 | 11 | 0.7 | 6.9 |
| 25 | 17 | 1.0 | 7.9 |
| 26 | 11 | 0.7 | 8.6 |
| 27 | 15 | 0.9 | 9.5 |
| 28 | 19 | 1.1 | 10.6 |
| 29 | 7 | 0.4 | 11.0 |
| 30 | 51 | 3.1 | 14.1 |
| 31 | 6 | 0.4 | 14.4 |
| 32 | 22 | 1.3 | 15.7 |
| 33 | 7 | 0.4 | 16.2 |
| 34 | 10 | 0.6 | 16.8 |
| 35 | 74 | 4.4 | 21.2 |
| 36 | 16 | 1.0 | 22.1 |
| 37 | 15 | 0.9 | 23.0 |
| 38 | 27 | 1.6 | 24.7 |
| 39 | 13 | 0.8 | 25.4 |
| 40 | 94 | 5.6 | 31.1 |
| 41 | 13 | 0.8 | 31.8 |
| 42 | 18 | 1.1 | 32.9 |
| 43 | 15 | 0.9 | 33.8 |
| 44 | 11 | 0.7 | 34.5 |
| 45 | 74 | 4.4 | 38.9 |
| 46 | 18 | 1.1 | 40.0 |
| 47 | 13 | 0.8 | 40.8 |
| 48 | 32 | 1.9 | 42.7 |
| 49 | 11 | 0.7 | 43.3 |
| 50 | 134 | 8.0 | 51.3 |
| 51 | 12 | 0.7 | 52.1 |
| 52 | 23 | 1.4 | 53.4 |
| 53 | 19 | 1.1 | 54.6 |
| 54 | 23 | 1.4 | 56.0 |
| 55 | 94 | 5.6 | 61.6 |
| 56 | 26 | 1.6 | 63.1 |
| 57 | 18 | 1.1 | 64.2 |
| 58 | 19 | 1.1 | 65.4 |
| 59 | 7 | 0.4 | 65.8 |
| 60 | 161 | 9.6 | 75.4 |
| 61 | 9 | 0.5 | 75.9 |
| 62 | 13 | 0.8 | 76.7 |
| 63 | 9 | 0.5 | 77.3 |
| 64 | 8 | 0.5 | 77.7 |
| 65 | 82 | 4.9 | 82.6 |
| 66 | 6 | 0.4 | 83.0 |
| 67 | 10 | 0.6 | 83.6 |
| 68 | 16 | 1.0 | 84.6 |
| 69 | 2 | 0.1 | 84.7 |
| 70 | 128 | 7.7 | 92.3 |
| 71 | 5 | 0.3 | 92.6 |
| 72 | 8 | 0.5 | 93.1 |
| 73 | 4 | 0.2 | 93.4 |
| 74 | 3 | 0.2 | 93.5 |
| 75 | 26 | 1.6 | 95.1 |
| 76 | 5 | 0.3 | 95.4 |
| 77 | 5 | 0.3 | 95.7 |
| 78 | 6 | 0.4 | 96.1 |
| 79 | 2 | 0.1 | 96.2 |
| 80 | 36 | 2.2 | 98.3 |
| 81 | 1 | 0.1 | 98.4 |
| 82 | 1 | 0.1 | 98.4 |
| 83 | 1 | 0.1 | 98.5 |
| 84 | 2 | 0.1 | 98.6 |
| 85 | 13 | 0.8 | 99.4 |
| 86 | 2 | 0.1 | 99.5 |
| 90 | 6 | 0.4 | 99.9 |
| 93 | 1 | 0.1 | 99.9 |
| 95 | 1 | 0.1 | 100.0 |
| Total | 1671 | 100.0 |
The various parts of the body affected by cancer incidences and the number of people affected (frequencies) are indicated in Table 4.
Table 4.
Parts of the body affected by the various types of cancer.
| Topological (Top) location of cancer | Frequency | Percent | Cumulative Percent | |
|---|---|---|---|---|
| Valid | C77.9 Lymph node, NOS | 9 | 0.5 | 0.5 |
| C26.9 Gastrointestinal tract, NOS | 9 | 0.5 | 1.1 | |
| C20.9 Rectum, NOS | 54 | 3.2 | 4.3 | |
| C44.9 Skin, NOS | 47 | 2.8 | 7.1 | |
| C61.9 Prostate gland | 253 | 15.1 | 22.3 | |
| C63.9 Male genital organs, NOS | 1 | 0.1 | 22.3 | |
| C49.6 Soft tissues of trunk | 5 | 0.3 | 22.6 | |
| C50.9 Breast, NOS | 92 | 5.5 | 28.1 | |
| C77.3 Lymph nodes of axilla or arm | 2 | 0.1 | 28.2 | |
| C57.9 Female genital tract, NOS | 15 | 0.9 | 29.1 | |
| C53.9 Cervix uteri | 76 | 4.5 | 33.7 | |
| C22.0 Liver | 31 | 1.9 | 35.5 | |
| C77.0 Lymph nodes of head, face and | 6 | 0.4 | 35.9 | |
| C40.9 Bone of limb, NOS | 4 | 0.2 | 36.1 | |
| C53.8 Overl. lesion of cervix uteri | 1 | 0.1 | 36.2 | |
| C49.2 Soft tissues of lower limb an | 7 | 0.4 | 36.6 | |
| C49.9 Other soft tissues | 18 | 1.1 | 37.7 | |
| C67.9 Urinary bladder, NOS | 32 | 1.9 | 39.6 | |
| C56.9 Ovary | 60 | 3.6 | 43.2 | |
| C40.2 Long bones of lower limb | 1 | 0.1 | 43.3 | |
| C44.2 External ear | 1 | 0.1 | 43.3 | |
| C49.0 Soft tissues of head, face, & | 9 | 0.5 | 43.9 | |
| C44.7 Skin of lower limb and hip | 6 | 0.4 | 44.2 | |
| C39.9 Ill-defined sites within resp | 15 | 0.9 | 45.1 | |
| C49.1 Soft tissues of upper limb, s | 4 | 0.2 | 45.4 | |
| C44.6 Skin of upper limb and shoulder | 3 | 0.2 | 45.5 | |
| C19.9 Rectosigmoid junction | 4 | 0.2 | 45.8 | |
| C64.9 Kidney, NOS | 20 | 1.2 | 47.0 | |
| C40.8 Overl. lesion of bones of lim | 1 | 0.1 | 47.0 | |
| C41.0 Bones of skull and face | 2 | 0.1 | 47.2 | |
| C44.4 Skin of scalp and neck | 6 | 0.4 | 47.5 | |
| C16.3 Gastric antrum | 6 | 0.4 | 47.9 | |
| C18.0 Cecum | 20 | 1.2 | 49.1 | |
| C16.9 Stomach, NOS | 7 | 0.4 | 49.5 | |
| C49.5 Soft tissues of pelvis | 3 | 0.2 | 49.7 | |
| C04.9 Floor of mouth, NOS | 2 | 0.1 | 49.8 | |
| C73.9 Thyroid gland | 14 | 0.8 | 50.6 | |
| C77.1 Intrathoracic lymph nodes | 1 | 0.1 | 50.7 | |
| C52.9 Vagina, NOS | 8 | 0.5 | 51.2 | |
| C10.2 Lateral wall of oropharynx | 1 | 0.1 | 51.2 | |
| C44.5 Skin of trunk | 2 | 0.1 | 51.3 | |
| C69.0 Conjunctiva | 14 | 0.8 | 52.2 | |
| C21.8 Overl. lesion rectum, anal ca | 9 | 0.5 | 52.7 | |
| C49.4 Soft tissues of abdomen | 4 | 0.2 | 53.0 | |
| C18.4 Transverse colon | 1 | 0.1 | 53.0 | |
| C41.9 Bone, NOS | 1 | 0.1 | 53.1 | |
| C76.2 Abdomen, NOS | 1 | 0.1 | 53.1 | |
| C76.5 Lower limb, NOS | 1 | 0.1 | 53.2 | |
| C69.6 Orbit, NOS | 1 | 0.1 | 53.3 | |
| C49.3 Soft tissues of thorax | 3 | 0.2 | 53.4 | |
| C55.9 Uterus, NOS | 30 | 1.8 | 55.2 | |
| C44.8 Overl. lesion of skin | 1 | 0.1 | 55.3 | |
| C51.9 Vulva, NOS | 1 | 0.1 | 55.4 | |
| C10.9 Oropharynx, NOS | 2 | 0.1 | 55.5 | |
| C30.1 Middle ear | 1 | 0.1 | 55.5 | |
| C62.9 Testis, NOS | 2 | 0.1 | 55.7 | |
| C15.0 Cervical esophagus | 12 | 0.7 | 56.4 | |
| C18.7 Sigmoid colon | 1 | 0.1 | 56.4 | |
| C80.9 Unknown primary site | 200 | 12.0 | 68.4 | |
| C77.2 Intra-abdominal lymph nodes | 1 | 0.1 | 68.5 | |
| C11.9 Nasopharynx, NOS | 3 | 0.2 | 68.6 | |
| C50.0 Nipple | 168 | 10.1 | 78.7 | |
| C53.0 Endocervix | 105 | 6.3 | 85.0 | |
| C53.1 Exocervix | 1 | 0.1 | 85.0 | |
| C67.4 Posterior wall of urinary bla | 8 | 0.5 | 85.5 | |
| C16.0 Cardia, NOS | 33 | 2.0 | 87.5 | |
| C21.0 Anus, NOS | 17 | 1.0 | 88.5 | |
| C51.0 Labium majus | 3 | 0.2 | 88.7 | |
| C67.0 Trigone of urinary bladder | 57 | 3.4 | 92.1 | |
| C44.0 Skin of lip, NOS | 15 | 0.9 | 93.0 | |
| C11.0 Superior wall of nasopharynx | 16 | 1.0 | 94.0 | |
| C08.0 Submandibular gland | 3 | 0.2 | 94.1 | |
| C14.0 Pharynx, NOS | 5 | 0.3 | 94.4 | |
| C26.0 Intestinal tract, NOS | 7 | 0.4 | 94.9 | |
| C65.9 Renal pelvis | 4 | 0.2 | 95.1 | |
| C10.0 Vallecula | 6 | 0.4 | 95.5 | |
| C25.0 Head of pancreas | 5 | 0.3 | 95.8 | |
| C60.0 Prepuce | 4 | 0.2 | 96.0 | |
| C21.2 Cloacogenic zone | 4 | 0.2 | 96.2 | |
| C18.6 Descending colon | 1 | 0.1 | 96.3 | |
| C66.9 Ureter | 1 | 0.1 | 96.3 | |
| C50.1 Central portion of breast | 1 | 0.1 | 96.4 | |
| C34.0 Main bronchus | 1 | 0.1 | 96.5 | |
| C21.1 Anal canal | 3 | 0.2 | 96.6 | |
| C18.9 Colon, NOS | 1 | 0.1 | 96.7 | |
| C01.9 Base of tongue, NOS | 3 | 0.2 | 96.9 | |
| C62.0 Undescended testis | 4 | 0.2 | 97.1 | |
| C11.2 Lateral wall of nasopharynx | 1 | 0.1 | 97.2 | |
| C50.6 Axillary tail of breast | 1 | 0.1 | 97.2 | |
| C54.1 Endometrium | 2 | 0.1 | 97.4 | |
| C25.9 Pancreas, NOS | 1 | 0.1 | 97.4 | |
| C30.0 Nasal cavity | 1 | 0.1 | 97.5 | |
| C00.9 Lip, NOS | 1 | 0.1 | 97.5 | |
| C54.2 Myometrium | 1 | 0.1 | 97.6 | |
| C48.8 Overl. lesion of retroperiton | 1 | 0.1 | 97.7 | |
| C76.7 Other ill-defined sites | 1 | 0.1 | 97.7 | |
| C03.0 Upper gum | 2 | 0.1 | 97.8 | |
| C15.9 Oesophagus, NOS | 1 | 0.1 | 97.9 | |
| C69.9 Eye, NOS | 1 | 0.1 | 98.0 | |
| C16.4 Pylorus | 1 | 0.1 | 98.0 | |
| C07.9 Parotid gland | 2 | 0.1 | 98.1 | |
| C67.5 Bladder neck | 1 | 0.1 | 98.2 | |
| C57.4 Uterine adnexa | 1 | 0.1 | 98.3 | |
| C16.2 Body of stomach | 1 | 0.1 | 98.3 | |
| C13.0 Postcricoid region | 7 | 0.4 | 98.7 | |
| C37.9 Thymus | 1 | 0.1 | 98.8 | |
| C17.0 Duodenum | 1 | 0.1 | 98.9 | |
| C06.0 Cheek mucosa | 1 | 0.1 | 98.9 | |
| C04.0 Anterior floor of mouth | 4 | 0.2 | 99.2 | |
| C47.0 Per. nerves & A.N.S. of head, | 3 | 0.2 | 99.3 | |
| C09.0 Tonsillar fossa | 2 | 0.1 | 99.5 | |
| C38.4 Pleura, NOS | 1 | 0.1 | 99.5 | |
| C38.0 Heart | 4 | 0.2 | 99.8 | |
| C67.1 Dome of urinary bladder | 1 | 0.1 | 99.8 | |
| C22.1 Intrahepatic bile duct | 1 | 0.1 | 99.9 | |
| C76.0 Head, face or neck, NOS | 1 | 0.1 | 99.9 | |
| C23.9 Gallbladder | 1 | 0.1 | 100.0 | |
| Total | 1671 | 100.0 | ||
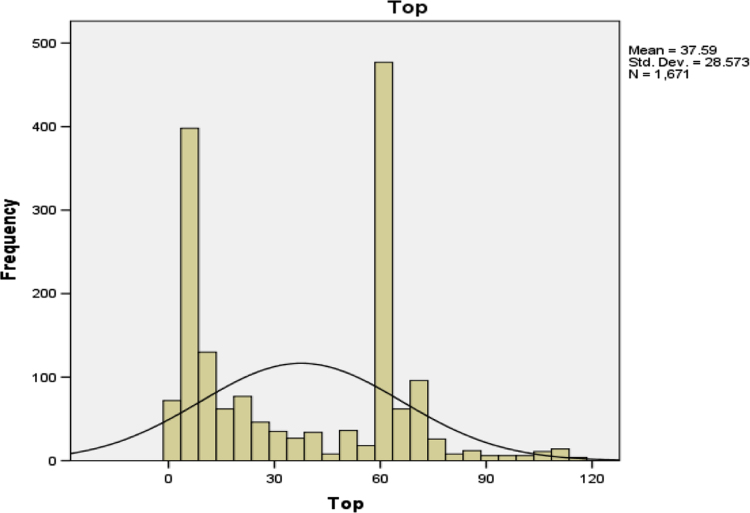
Table 4 shows that the part of the body affected mostly is the prostate gland. This is represented graphically in Fig. 3.
Fig. 1.
Gender of the patients.
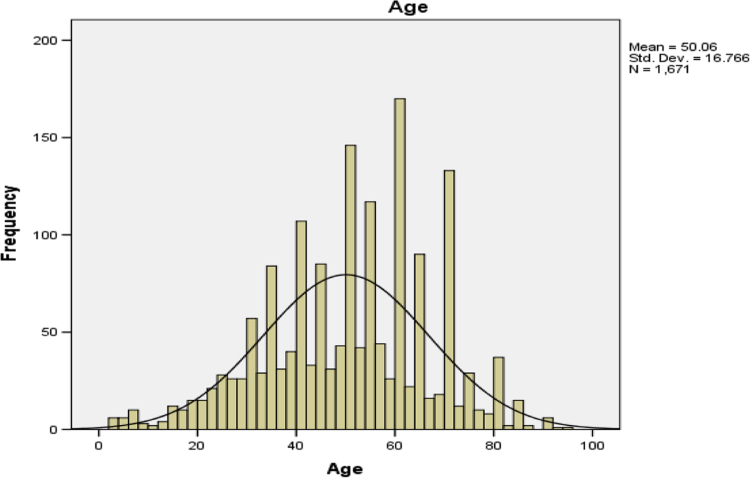
Fig. 2.
Age of the patients.
Fig. 3.
Diagrammatic presentation of the parts of the body affected by cancer.
2. Experimental design, materials and methods
The data set was obtained from the patients’ records at the data center of the University of Maiduguri teaching hospital. The hospital as stated earlier serves a large population from the six Northeastern states of Nigeria and beyond. The Northeastern region in particular and the entire northern region of the country is in variance with their natural endowments such as vast fertile lands, rivers and lakes for irrigation, mineral resources and abundant sunshine for renewable energy. The weak social structure of the region has resulted to excruciating poverty which often manifest as homelessness and destitution, insurgency, violence and crime [1]. The region has high poverty index, low human development index, lack of portable drinking water, electoral violence, dearth of medical personnel, high mortality, low life expectancy, decayed infrastructure and also an epicenter for joblessness, underage and teenage pregnancy, female genital mutilation, epidemics, illiteracy, malnutrition and now terrorism which comes in form of coordinated attacks on military, police formations and remote villages, guerrilla attacks, kidnappings, regicide, suicide bombings, mass killings, abduction of school girls, extra-judicial killings and summary execution, hypnotizing and forced conscriptions, indoctrination and forceful conversion to Islam and so on. The decadence is assumed to be as a result of corruption, tribalism, military intervention in governance, inequality, misappropriation, financial recklessness, bankrupt of ideas and dearth of developmental agendas, reduction of allocation of capital due to shortfalls of Nigeria revenue as a result of decline in crude oil price. Globally, efforts towards improving the healthcare and reducing the incidence of cancer have yielded desired results except in some developing countries. Hence, cancer related deaths remain stubbornly high in those countries. Cancer awareness, screening, prevention, management, treatment strategies are very low in the region/area studied in this article. Regrettably, capital allocations to the health sector are inadequate and the available funds are often allegedly diverted by corrupt government officials.
In addition, maternal death is one area that is currently affected by the Boko haram insurgency in that region as reported by [2]. Moreover, other areas have been seriously affected; for example; food security and dynamics, under five malnutrition, child mortality, escalation of cholera outbreaks, infections, sexually transmitted diseases, unsafe birth practices and abortion, child prostitution, sex for food at the displaced persons camps, increase in polio cases, See [3], [4], [5], [6], [7], [8] for details. Some related article can also be explored [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31].
Next, we analyze the dataset collected using Chi-square test of independence and curve estimation.
2.1. Chi-square test of independence
Chi-square test of independence was used to investigate the relationship between the location of the cancer (top), gender and age of patients.
2.1.1. Test of independency between “Top” and gender of the patients
Hypothesis Testing I:
H0: There is no significant association between the topological location of cancer and the gender of the patients.
Versus.
H1: There is a significant association between the topological location of cancer and the gender of the patients.
The result of the analysis is presented in Table 5.
Table 5.
Result of the chi-square test between gender and “Top”.
| Chi-Square Tests | Value | df | Asymp. Sig. (2-sided) |
|---|---|---|---|
| Pearson Chi-Square | 928.735 | 116 | 0.000 |
| Likelihood Ratio | 1214.083 | 116 | 0.000 |
| Linear-by-Linear Association | 64.659 | 1 | 0.000 |
| N of Valid Cases | 1670 |
Remarks: The null hypothesis (H0) is rejected since the p-value (0.000) is less than the level of significance (0.05). Therefore, it can be concluded that there is a significant association between the topological location of cancer and the gender of the patients.
The information about the correlation coefficient and its corresponding p-value is presented in Table 6.
Table 6.
Correlation coefficient.
| Symmetric Measures | Value | Asymp. Std. Error | Approx. T | Approx. Sig. | |
|---|---|---|---|---|---|
| Interval by Interval | Pearson's R | 0.197 | 0.024 | 8.199 | 0.000 |
| Ordinal by Ordinal | Spearman Correlation | 0.253 | 0.024 | 10.661 | 0.000 |
| N of Valid Cases | 1670 | ||||
2.1.2. Test of independency between “Top” and age of the patients
Hypothesis Testing II:
H0: There is no significant association between topological location of cancer is not dependent on the age of the patients.
Versus.
H1: There is a significant association between topological location of cancer is dependent on the age of the patients.
The result of the analysis is presented in Table 7.
Table 7.
Result of the chi-square test between age and “Top”.
| Chi-Square Tests | Value | Df | Asymp. Sig. (2-sided) |
|---|---|---|---|
| Pearson Chi-Square | 10762.735 | 9628 | 0.000 |
| Likelihood Ratio | 3148.516 | 9628 | 1.000 |
| Linear-by-Linear Association | 50.758 | 1 | 0.000 |
| N of Valid Cases | 1671 |
Remarks: Since the p-value is also less than 0.05, we conclude that there is a significant association between the topological location of cancer and the age of the patients.
Information about the correlation coefficient and its corresponding p-value is presented in Table 8.
Table 8.
Correlation coefficient result.
| Symmetric Measures | Value | Asymp. Std. Error | Approx. T | Approx. Sig. | |
|---|---|---|---|---|---|
| Interval by Interval | Pearson's R | −.174 | 0.024 | −7.233 | 0.000 |
| Ordinal by Ordinal | Spearman Correlation | −.189 | 0.025 | −7.881 | 0.000 |
| N of Valid Cases | 1671 | ||||
2.2. Curve estimation
Linear, logarithmic, inverse, quadratic, cubic, power, growth, exponential and logistic regression models were fitted to the dataset. “Top” is the dependent variable while Age is the independent variable. The summary of the variables used is presented in Table 9.
Table 9.
Summary of the variables.
| Variable Processing Summary | Variables |
||
|---|---|---|---|
| Dependent | Independent | ||
| Top | Age | ||
| Number of Positive Values | 1671 | 1671 | |
| Number of Zeros | 0 | 0 | |
| Number of Negative Values | 0 | 0 | |
| Number of Missing Values | User-Missing | 0 | 0 |
| System-Missing | 0 | 0 | |
2.2.1. Simple linear regression
The summary of the simple linear regression model is presented in Table 10.
Table 10.
Model summary.
| R | R Square | Adjusted R Square | Std. Error of the Estimate |
|---|---|---|---|
| 0.174 | 0.030 | 0.030 | 28.144 |
The independent variable is Age.
The corresponding analysis of variance (ANOVA) table testing for the fitness of the model is presented in Table 11.
Table 11.
ANOVA table for the linear model.
|
ANOVA | |||||
|---|---|---|---|---|---|
| Sum of Squares | df | Mean Square | F | Sig. | |
| Regression | 41440.679 | 1 | 41440.679 | 52.318 | 0.000 |
| Residual | 1321998.748 | 1669 | 792.090 | ||
| Total | 1363439.427 | 1670 | |||
The independent variable is Age.
The linear regression model is significant at 0.05 level of significance and with R-square value of 3%.
2.2.2. Logarithmic model
The summary of the logarithmic model is presented in Table 12.
Table 12.
Model summary for the logarithmic model.
| R | R Square | Adjusted R Square | Std. Error of the Estimate |
|---|---|---|---|
| 0.130 | 0.017 | 0.016 | 28.340 |
The independent variable is Age.
Estimating the model parameter gives the result in Table 13.
Table 13.
Parameter estimation for the logarithmic model.
| Coefficients | Unstandardized Coefficients |
Standardized Coefficients | T | Sig. | |
|---|---|---|---|---|---|
| B | Std. Error | Beta | |||
| ln(Age) | −8.130 | 1.520 | −0.130 | −5.349 | 0.000 |
| (Constant) | 68.755 | 5.869 | 11.716 | 0.000 | |
The ANOVA table for the logarithmic model is presented in Table 14.
Table 14.
ANOVA table for the logarithmic model.
|
ANOVA | |||||
|---|---|---|---|---|---|
| Sum of Squares | df | Mean Square | F | Sig. | |
| Regression | 22977.216 | 1 | 22977.216 | 28.609 | 0.000 |
| Residual | 1340462.210 | 1669 | 803.153 | ||
| Total | 1363439.427 | 1670 | |||
The independent variable is Age.
The logarithmic model is significant at 0.05 level of significance and with R-square value of 1.7%.
2.2.3. Inverse model
The summary of the inverse model is presented in Table 15.
Table 15.
Summary of the inverse model.
| R | R Square | Adjusted R Square | Std. Error of the Estimate |
|---|---|---|---|
| 0.047 | 0.002 | 0.002 | 28.550 |
The independent variable is age.
The result for the estimation of parameters using the inverse model is presented in Table 16.
Table 16.
Parameter estimation using inverse model.
| Coefficients | Unstandardized Coefficients |
Standardized Coefficients | t | Sig. | |
|---|---|---|---|---|---|
| B | Std. Error | Beta | |||
| 1/Age | 49.544 | 25.664 | 0.047 | 1.930 | 0.054 |
| (Constant) | 36.327 | 0.956 | 38.018 | 0.000 | |
The corresponding ANOVA table is presented in Table 17.
Table 17.
The ANOVA table for the inverse model.
|
ANOVA | |||||
|---|---|---|---|---|---|
| Sum of Squares | df | Mean Square | F | Sig. | |
| Regression | 3037.719 | 1 | 3037.719 | 3.727 | 0.054 |
| Residual | 1360401.707 | 1669 | 815.100 | ||
| Total | 1363439.427 | 1670 | |||
The independent variable is age.
The inverse model is not significant as its p-value is greater than the level of significance (0.05).
2.2.4. Quadratic model
The summary for the quadratic model is presented in Table 18.
Table 18.
Summary for the quadratic model.
| R | R Square | Adjusted R Square | Std. Error of the Estimate |
|---|---|---|---|
| 0.195 | 0.038 | 0.037 | 28.043 |
The independent variable is age.
The result for the estimation of parameter using the quadratic model is presented in Table 19.
Table 19.
Parameter estimation for the quadratic model.
| Coefficients | Unstandardized Coefficients |
Standardized Coefficients | t | Sig. | |
|---|---|---|---|---|---|
| B | Std. Error | Beta | |||
| Age | 0.348 | 0.183 | 0.204 | 1.897 | 0.058 |
| Age ** 2 | −0.007 | 0.002 | −0.388 | −3.607 | 0.000 |
| (Constant) | 38.929 | 4.329 | 8.992 | 0.000 | |
The corresponding ANOVA table is presented in Table 20.
Table 20.
ANOVA table for the quadratic model.
|
ANOVA | |||||
|---|---|---|---|---|---|
| Sum of Squares | df | Mean Square | F | Sig. | |
| Regression | 51674.289 | 2 | 25837.144 | 32.854 | 0.000 |
| Residual | 1311765.138 | 1668 | 786.430 | ||
| Total | 1363439.427 | 1670 | |||
The independent variable is age.
The quadratic model is significant at 0.05 level of significance and with R-square value of 3.8%.
2.2.5. Cubic model
The summary for the cubic model is presented in Table 21.
Table 21.
Summary for the cubic model.
| R | R Square | Adjusted R Square | Std. Error of the Estimate |
|---|---|---|---|
| 0.197 | 0.039 | 0.037 | 28.036 |
The independent variable is age.
The result for the estimation of parameter for the cubic model is presented in Table 22.
Table 22.
Parameter estimation for the cubic model.
| Coefficients | Unstandardized Coefficients |
Standardized Coefficients | t | Sig. | |
|---|---|---|---|---|---|
| B | Std. Error | Beta | |||
| Age | 0.951 | 0.477 | 0.558 | 1.993 | 0.046 |
| Age ** 2 | −0.021 | 0.011 | −1.230 | −1.970 | 0.049 |
| Age ** 3 | 0.000 | 0.000 | 0.504 | 1.369 | 0.171 |
| (Constant) | 32.108 | 6.601 | 4.864 | 0.000 | |
The corresponding ANOVA table is presented in Table 23.
Table 23.
ANOVA table for the cubic model.
|
ANOVA | |||||
|---|---|---|---|---|---|
| Sum of Squares | df | Mean Square | F | Sig. | |
| Regression | 53146.668 | 3 | 17715.556 | 22.538 | 0.000 |
| Residual | 1310292.759 | 1667 | 786.018 | ||
| Total | 1363439.427 | 1670 | |||
The independent variable is age.
The cubic model is significant and with R-square value of 3.9%.
2.2.6. Power model
The summary for the power model is presented in Table 24.
Table 24.
Summary for the power model.
| R | R Square | Adjusted R Square | Std. Error of the Estimate |
|---|---|---|---|
| 0.159 | 0.025 | 0.025 | 1.125 |
The independent variable is age.
The result for the estimation of parameter for the power model is presented in Table 25.
Table 25.
Parameter estimation for the power model.
| Coefficients | Unstandardized Coefficients |
Standardized Coefficients | t | Sig. | |
|---|---|---|---|---|---|
| B | Std. Error | Beta | |||
| ln(Age) | −0.397 | 0.060 | −0.159 | −6.583 | 0.000 |
| (Constant) | 105.955 | 24.692 | 4.291 | 0.000 | |
The dependent variable is ln(Top).
The corresponding ANOVA table is presented in Table 26.
Table 26.
ANOVA table for the power model.
|
ANOVA | |||||
|---|---|---|---|---|---|
| Sum of Squares | df | Mean Square | F | Sig. | |
| Regression | 54.875 | 1 | 54.875 | 43.330 | 0.000 |
| Residual | 2113.710 | 1669 | 1.266 | ||
| Total | 2168.585 | 1670 | |||
The independent variable is age.
The power model is significant at 0.05 level of significance and with R-square value of 2.5%.
2.2.7. Growth model
The model summary for the growth model is presented in Table 27.
Table 27.
Summary for the growth model.
| R | R Square | Adjusted R Square | Std. Error of the Estimate |
|---|---|---|---|
| 0.216 | 0.047 | 0.046 | 1.113 |
The independent variable is age.
The result for the estimation of parameter of the growth model is presented in Table 28.
Table 28.
Parameter estimation for the growth model.
| Coefficients | Unstandardized Coefficients |
Standardized Coefficients | t | Sig. | |
|---|---|---|---|---|---|
| B | Std. Error | Beta | |||
| Age | −0.015 | 0.002 | −0.216 | −9.038 | 0.000 |
| (Constant) | 3.875 | 0.086 | 45.180 | 0.000 | |
The dependent variable is ln(Top).
The corresponding ANOVA table is presented in Table 29.
Table 29.
ANOVA table for the growth model.
|
ANOVA | |||||
|---|---|---|---|---|---|
| Sum of Squares | df | Mean Square | F | Sig. | |
| Regression | 101.181 | 1 | 101.181 | 81.683 | 0.000 |
| Residual | 2067.404 | 1669 | 1.239 | ||
| Total | 2168.585 | 1670 | |||
The independent variable is age.
The growth model is significant at 0.05 level of significance and with R-square value of 4.7%.
2.2.8. Exponential model
The model summary for the exponential model is presented in Table 30.
Table 30.
Summary for the exponential model.
| R | R Square | Adjusted R Square | Std. Error of the Estimate |
|---|---|---|---|
| 0.216 | 0.047 | 0.046 | 1.113 |
The independent variable is age.
The result for the estimation of parameter for the exponential model is presented in Table 31.
Table 31.
Parameter estimation for the exponential model.
| Coefficients | Unstandardized Coefficients |
Standardized Coefficients | t | Sig. | |
|---|---|---|---|---|---|
| B | Std. Error | Beta | |||
| Age | −0.015 | 0.002 | −0.216 | −9.038 | 0.000 |
| (Constant) | 48.173 | 4.132 | 11.660 | 0.000 | |
The dependent variable is ln(Top).
The corresponding ANOVA table is presented in Table 32.
Table 32.
ANOVA table for the exponential model.
|
ANOVA | |||||
|---|---|---|---|---|---|
| Sum of Squares | df | Mean Square | F | Sig. | |
| Regression | 101.181 | 1 | 101.181 | 81.683 | 0.000 |
| Residual | 2067.404 | 1669 | 1.239 | ||
| Total | 2168.585 | 1670 | |||
The independent variable is age.
The exponential model is significant at 0.05 level of significance and with R-square value of 4.7%.
2.2.9. Logistic model
The model summary for the logistic model is presented in Table 33.
Table 33.
Summary for the logistic model.
| R | R Square | Adjusted R Square | Std. Error of the Estimate |
|---|---|---|---|
| 0.216 | 0.047 | 0.046 | 1.113 |
The independent variable is age.
The estimation of parameters for the logistic model is presented in Table 34.
Table 34.
Parameter estimation for the logistic model.
| Coefficients | Unstandardized Coefficients |
Standardized Coefficients | t | Sig. | |
|---|---|---|---|---|---|
| B | Std. Error | Beta | |||
| Age | 1.015 | 0.002 | 1.241 | 615.592 | 0.000 |
| (Constant) | 0.021 | 0.002 | 11.660 | 0.000 | |
The dependent variable is ln(1 / Top).
The corresponding ANOVA table is presented in Table 35.
Table 35.
ANOVA table for the logistic model.
|
ANOVA | |||||
|---|---|---|---|---|---|
| Sum of Squares | df | Mean Square | F | Sig. | |
| Regression | 101.181 | 1 | 101.181 | 81.683 | 0.000 |
| Residual | 2067.404 | 1669 | 1.239 | ||
| Total | 2168.585 | 1670 | |||
The independent variable is age.
The logistic model is also significant at 0.05 level of significance and with R-square value of 4.7%.
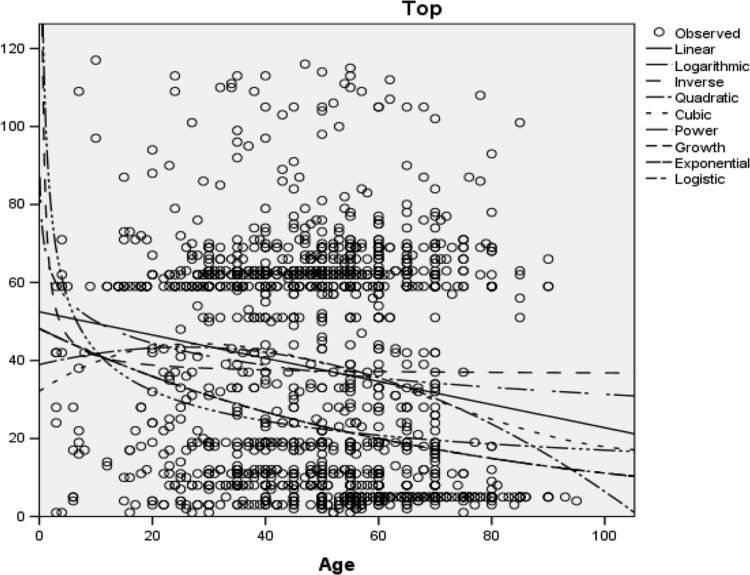
Lastly, all the fitted models are illustrated in Fig. 4.
Fig. 4.
The fitted model with respect to the data set.
Important points
-
•
More females are infected with cancer than men.
-
•
The age with the highest record (or incidence) of cancer is 60 years old.
-
•
The part of the body that is mostly affected by cancer is the prostate gland (based on the data set collected).
-
•
There is a significant association between the topological location of cancer and the gender of the patients.
-
•
There is a significant association between the topological location of cancer and the age of the patients.
-
•
All the models fitted to the data produced low R-square values; nevertheless, the models that best fit the data based on their R-square values are growth model, exponential model and logistic model.
Acknowledgement
The authors are grateful to the reviewers and to Covenant University for providing an enabling environment for this research.
Footnotes
Transparency data associated with this article can be found in the online version at doi:10.1016/j.dib.2018.04.135.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2018.04.135.
Contributor Information
Patience I. Adamu, Email: peluemman@yahoo.com.
Pelumi E. Oguntunde, Email: pelumi.oguntunde@covenantuniversity.edu.ng.
Transparency document. Supplementary material
Supplementary material
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Khan A., Cheri L. An examination of poverty as the foundation of crisis in Northern Nigeria. Insight Afr. 2016;8(1):59–71. [Google Scholar]
- 2.Adamu P.I., Adamu M.O., Okagbue H.I. Data in support of high rate of pregnancy related deaths in Maiduguri, Borno State, Northeast Nigeria. Data Brief. 2018;18:409–414. doi: 10.1016/j.dib.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adebayo O., Olagunju K., Kabir S.K., Adeyemi O. Social crisis, terrorism and food poverty dynamics: evidence from Northern Nigeria. Int. J. Econ. Finan. Issues. 2016;6(4):1865–1872. [Google Scholar]
- 4.Olajide F., Adeshakin K. Towards the investigation of using social network analysis for counter terrorism in West Africa: case study of Boko Haram in Nigeria. J. Eng. Sci. Technol. 2016;11(11):1629–1638. [Google Scholar]
- 5.Cumber S.N., Jaila S., Nancy B., Tsoka-Gwegweni J.M. Under five malnutrition crises in the Boko Haram area of Cameroon. S. Afr. J. Clin. Nutr. 2017;30(2):41–42. [Google Scholar]
- 6.Bigna J.J.R. Polio eradication efforts in regions of geopolitical strife: the Boko Haram threat to efforts in sub-Saharan Africa. Afr. Health Sci. 2016;16(2):584–587. doi: 10.4314/ahs.v16i2.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamisu A.W., Johnson T.M., Craig K., Mkanda P., Banda R., Tegegne S.G., Oyetunji A., Ningi N., Mohammed S.M., Adamu M.I., Abdulrahim K., Nsubuga P., Vaz R.G., Muhammed A.J.G. Strategies for improving polio surveillance performance in the security-challenged Nigerian States of Adamawa, Borno, and Yobe During 2009–2014. J. Infect. Dis. 2016;213:5136–5139. doi: 10.1093/infdis/jiv530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Omole O., Welye H., Abimbola S. Boko Haram insurgency: implications for public health. Lancet. 2015;385(9972):941. doi: 10.1016/S0140-6736(15)60207-0. [DOI] [PubMed] [Google Scholar]
- 9.Oguntunde P.E., Adejumo A.O., Okagbue H.I. Breast cancer patients in Nigeria: data exploration approach. Data Brief. 2017;15:47–57. doi: 10.1016/j.dib.2017.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adejumo A.O., Ikoba N.A., Suleiman E.A., Okagbue H.I., Oguntunde P.E., Odetunmibi O.A., Job O. Quantitative exploration of factors influencing psychotic disorder ailments in Nigeria. Data Brief. 2017;14:175–185. doi: 10.1016/j.dib.2017.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adejumo A.O., Suleiman E.A., Okagbue H.I., Oguntunde P.E., Odetunmibi O.A. Quantitative evaluation of pregnant women delivery status' records in Akure, Nigeria. Data Brief. 2018;16:127–134. doi: 10.1016/j.dib.2017.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarc E., Eichmann T.O., Zimmermann R., Petan T. Lipidomic data on lipid droplet triglyceride remodelling associated with protection of breast cancer cells from lipotoxic stress. Data Brief. 2018;18:234–240. doi: 10.1016/j.dib.2018.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong J.H., Ko Y.H., Kang K. RNA-seq data of invasive ductal carcinoma and adjacent normal tissues from a Korean patient with breast cancer. Data Brief. 2018;18:736–739. doi: 10.1016/j.dib.2018.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aka J.A., Calvo E.L., Lin S.X. Genomic data on breast cancer transcript profile modulation by 17beta-hydroxysteroid dehydrogenase type 1 and 17-beta-estradiol. Data Brief. 2016;9:1000–1012. doi: 10.1016/j.dib.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarvaiya H.A., Lazar I.M. Insulin stimulated MCF7 breast cancer cells: proteome dataset. Data Brief. 2016;9:579–584. doi: 10.1016/j.dib.2016.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoof E.M., Lechman E.R., Dick J.E. Global proteomics dataset of miR-126 overexpression in acute myeloid leukemia. Data Brief. 2016;9:57–61. doi: 10.1016/j.dib.2016.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aumsuwan P., Khan S.I., Khan I.A., Walker L.A., Dasmahapatra A.K. Gene expression profiling and pathway analysis data in MCF-7 and MDA-MB-231 human breast cancer cell lines treated with dioscin. Data Brief. 2016;8:272–279. doi: 10.1016/j.dib.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park E. Data on cell cycle in breast cancer cell line, MDA-MB-231 with ferulic acid treatment. Data Brief. 2016;7:107–110. doi: 10.1016/j.dib.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.To K.K., Poon D.C., Wei Y., Wang F., Lin G., Fu L.W. Data showing the circumvention of oxaliplatin resistance by vatalanib in colon cancer. Data Brief. 2016;7:437–444. doi: 10.1016/j.dib.2016.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angelopoulos N., Stebbing J., Xu Y., Giamas G., Zhang H. Proteome-wide dataset supporting functional study of tyrosine kinases in breast cancer. Data Brief. 2016;7:740–746. doi: 10.1016/j.dib.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuishi Y., Kimura Y., Arakawa N., Hirano H. Data for identification of GPI-anchored peptides and ω-sites in cancer cell lines. Data Brief. 2016;7:1302–1305. doi: 10.1016/j.dib.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Oliveira F.M., Carmona A.M., Ladeira C. Genotoxicity assessment data for exfoliated buccal cells exposed to mobile phone radiation. Data Brief. 2017;15:344–347. doi: 10.1016/j.dib.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park E. Data on the effects of anti-cancer drug of resveratrol in breast cancer cells, MDA-MB-231 cells. Data Brief. 2017;12:68–71. doi: 10.1016/j.dib.2017.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das D.K., Ali T., Krampis K., Ogunwobi O.O., O. O Fibronectin and androgen receptor expression data in prostate cancer obtained from a RNA-sequencing bioinformatics analysis. Data Brief. 2017;11:131–135. doi: 10.1016/j.dib.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konathala G., Mandarapu R., Godi S. Data on polymorphism of XRCC1 and cervical cancer risk from South India. Data Brief. 2017;10:11–13. doi: 10.1016/j.dib.2016.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikitina A.S., Sharova E.I., Danilenko S.A., Selezneva O.V., Butusova T.B., Vasiliev A.O., Kostryukova E.S. Datasets for next-generation sequencing of DNA and RNA from urine and plasma of patients with prostate cancer. Data Brief. 2017;10:369. doi: 10.1016/j.dib.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kel A.E. Data on master regulators and transcription factor binding sites found by upstream analysis of multi-omics data on methotrexate resistance of colon cancer. Data Brief. 2017;10:499–504. doi: 10.1016/j.dib.2016.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itou J., Tanaka S., Li W., Matsumoto Y., Sato F., Toi M. Data of a fluorescent imaging-based analysis of anti-cancer drug effects on three-dimensional cultures of breast cancer cells. Data Brief. 2015;5:429–433. doi: 10.1016/j.dib.2015.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Santos C., Taylor C., Carroll J.S., Mohammed H. RIME proteomics of estrogen and progesterone receptors in breast cancer. Data Brief. 2015;5:276–280. doi: 10.1016/j.dib.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moawad E.Y. Data to establish the optimal standard regimen and predicting the response to docetaxel therapy. Data Brief. 2015;5:439–446. doi: 10.1016/j.dib.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uziel O., Lahav M. Proteomic and microRNA data clarifying the effects of telomere shortening on cancer cells. Data Brief. 2015;2:48–51. doi: 10.1016/j.dib.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material