Summary
Human stem cell-derived models of development and neurodegenerative diseases are challenged by cellular immaturity in vitro. Microengineered organ-on-chip (or Organ-Chip) systems are designed to emulate microvolume cytoarchitecture and enable co-culture of distinct cell types. Brain microvascular endothelial cells (BMECs) share common signaling pathways with neurons early in development, but their contribution to human neuronal maturation is largely unknown. To study this interaction and influence of microculture, we derived both spinal motor neurons and BMECs from human induced pluripotent stem cells and observed increased calcium transient function and Chip-specific gene expression in Organ-Chips compared with 96-well plates. Seeding BMECs in the Organ-Chip led to vascular-neural interaction and specific gene activation that further enhanced neuronal function and in vivo-like signatures. The results show that the vascular system has specific maturation effects on spinal cord neural tissue, and the use of Organ-Chips can move stem cell models closer to an in vivo condition.
Keywords: organ-on-chip, spinal cord, iPSC, disease modeling, amyotrophic lateral sclerosis, microphysiological system, brain microvascular endothelial cells, spinal motor neurons, vasculature, microfluidic device
Highlights
-
•
iPSC-derived neural and vascular tissue interact in Organ-Chip
-
•
Chip culture enhances neuron function and signaling
-
•
iPSC-derived vasculature affects neuron development and neuron-vasculature pathways
-
•
BMECs co-cultured on Chip co-culture activate in vivo spinal cord developmental genes
Sances et al. combine Organ-Chip technology with human induced pluripotent stem cell-derived spinal motor neurons to study the maturation effects of Organ-Chip culture. By including microvascular cells also derived from the same patient line, the authors show enhancement of neuronal function, reproduction of vascular-neuron pathways, and specific gene activation that resembles in vivo spinal cord development.
Introduction
A wide variety of human tissues can be generated from human induced pluripotent stem cells (iPSCs) (Shi et al., 2017). However, iPSC-derived cells generated in vitro in most culture systems remain fetal-like in nature (Avior et al., 2016), potentially limiting their utility for research and disease modeling. Comparative analysis of developing spinal cord tissue and iPSC-derived motor neuron cultures have revealed that even with prolonged time in culture, spinal motor neurons generated from iPSCs do not possess functional and molecular maturation signatures beyond 4–6 gestational weeks (Ho et al., 2016). Novel culture systems that aim to enhance the function and maturation of iPSC-derived spinal motor neurons (spMNs) in vitro are of considerable interest, especially for the study of spMN dysfunction in adult-onset disorders such as amyotrophic lateral sclerosis (ALS).
Unlike conventional cell culture plates, organ-on-chip (or Organ-Chip) systems (Bhatia and Ingber, 2014) offer 3-dimensional microengineered platforms that aim to better recapitulate cellular microenvironment including cell-cell interactions of the lung (Huh et al., 2010) and gut (Kim et al., 2016). Due to reduced media volume and non-convection geometry, microengineered cultures may provide greater concentration of soluble signals compared with 96-well plates (Przybyla and Voldman, 2012). While groups have described these microvolume effects on various cell types (Domenech et al., 2009, Yu et al., 2007), the neurogenic effects of microvolume culture on iPSC-derived neurons remain unknown, prompting us to explore them here.
Developing spMNs rely on other cell types to provide signaling cues critical to maturation (Jessell, 2000). Neuromuscular junction formation and astrocyte emergence begin at 9 and 15 weeks post fertilization, respectively, and astrocytes continue to proliferate into post-natal development (Guo et al., 2015, Hesselmans et al., 1993). While the importance of the interaction of these cell types in improving spMN function has been demonstrated in vitro, this is not sufficient to mature them to adult counterparts (Demestre et al., 2015, Montoya et al., 2009, Rushton et al., 2013). Of note, brain microvascular endothelial cells (BMECs) invade the neural tube from the perineural vascular plexus beginning at 4 weeks post fertilization and form a primitive blood-brain barrier that directly interacts with developing neural tube progenitors and neurons prior to astrocyte emergence (Engelhardt and Liebner, 2014, Kurz, 2009) (Figure S1A). During development post fertalization, vascular angiogenesis and axon neurite outgrowth share common morphogenic mechanisms known as angioneurins (Wälchli et al., 2015, Zacchigna et al., 2008). In the adult context, BMECs have also been shown to influence neurogenesis (Louissaint et al., 2002, Palmer et al., 2000, Shen et al., 2004). The concordant developmental timing of the two cell types, as well as shared developmental signaling pathways, prompted our hypothesis that iPSC-derived BMECs could interact with and mature iPSC-derived spMN cultures in vitro.
In the current report, we differentiated human iPSCs into spinal neural progenitor cells (spNPCs), which were subsequently studied in a Spinal Cord-Chip system. Compared with a 96-well plate, cultures in the Chip had increased neuronal activity and expressed enhanced neuronal differentiation genes. When iPSC-derived BMECs were included, we observed further increases in neuronal activity, induction of vascular-neural interaction genes, and a developmental gene expression profile that indicated a more in vivo-like signature. This platform provides insight into endogenous signaling of spinal neural cultures and BMEC interaction during early human development and establishes a vascularized in vitro model of motor neuron tissue from human iPSCs to better understand and potentially treat motor neuron-related diseases.
Results
Neurons in Spinal Cord-Chip Have Spontaneous Activity
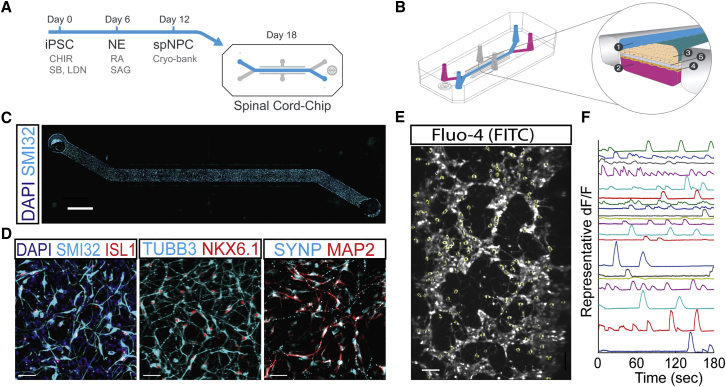
To study the consequences of an Organ-Chip microenvironment, we used a rapid protocol to differentiate healthy human iPSCs (83iCTR line) into spMNs, which was based on a combination of previously defined methods (Sances et al., 2016). First, iPSCs were differentiated to neural ectoderm (NE) then subsequently directed toward ventralized spNPCs over a 12-day period (Figure 1A). To test how spNPCs would develop in a microenvironment, we seeded them into the top channel of the dual-channel Spinal Cord-Chip constructed of polydimethylsiloxane (PDMS) (Emulate) (Figure 1B). Top and bottom channels of the Chip are separated by a 50-μm-thick membrane perforated by 7-μm diameter pores spaced 40 μm apart from center to center. Within 6 days of incubation, mixed neural cultures expressed spMN marker phosphorylated neurofilament heavy chain (SMI32) along the entire top channel (Figure 1C). Neuronal markers NKX6.1 and TUBB3, marked early spMN fate, and islet-1 (ISL1) indicated post-mitotic spMNs (Figures 1D). Neural cultures also stained positive for MAP2 and synaptophysin, indicating that synaptogenesis was initiated within the Spinal Cord-Chip.
Figure 1.
spNPCs Survive and Mature in the Chip Microenvironment
(A) Schematic of spinal neural progenitor cells (spNPC) differentiation from induced pluripotent stem cell (iPSC) cultures. Cells were fated to neural ectoderm (NE) using WNT agonist CHIR99021 and SMAD inhibitors LDN193189 and SB431542 for 6 days and then patterned to ventral spinal neurons using retinoic acid (RA) and sonic hedgehog agonist (SAG) in 6-well plates. At day 12, spNPCs were frozen, banked, and thawed for experiments (Cryo-bank). spNPCs were seeded into the top channel of the Spinal Cord-Chip and incubated for 6 days.
(B) Schematic of dual-channel Chip geometry (left) and magnified cross-section (right). Top (1) and bottom (2) channels can contain distinct cultures (3 and 4), and are separated by a porous membrane (5).
(C) Phosphorylated neurofilament heavy chain (SMI32) is enriched in spinal motor neurons (spMNs) and expressed in cells populating the entire top channel. Cells stained with nuclear dye DAPI. Scale bar, 200 μm.
(D) Immunostaining of main channel of the Chip of markers for spMNs SMI32, nuclear marker islet1 (ISL1), Beta 3 tubulin (TUBB3), NKX6.1, neurofilament marker microtubule-associated protein 2 (MAP2), and synaptic marker synaptophysin (SYNP). Cells stained with nuclear dye DAPI. Scale bar, 40 μm.
(E) Representative image of Spinal Cord-Chip neurons treated with Fluo-4 calcium activated dye and acquired live in fluorescein isothiocyanate (FITC) channel. Scale bar, 100 μm.
(F) Florescence of individual active neurons normalized to baseline florescence and plotted over time (dF/F).
To determine whether the Spinal Cord-Chip culture showed spontaneous neuronal activity, we treated cultures with the calcium-activated dye Fluo-4 and acquired fluorescent activity at 20 Hz for 3 min (Figure 1E). By plotting the change in fluorescence with respect to time (dF/F), neuron-specific calcium transient events could be characterized by fast onset and slow decay, consistent with developing neurons (Warp et al., 2012). Calcium transient event detection showed extensive activity in the 18-day cultures, providing evidence of neuronal activity and connectivity in the Spinal Cord-Chip (Figure 1F). Together these data show that iPSC-derived spNPCs can survive and mature to an active neuronal phenotype within the Spinal Cord-Chip microenvironment.
iPSC-Derived BMECs Survive and Populate the Vascular Spinal Cord-Chip Channel
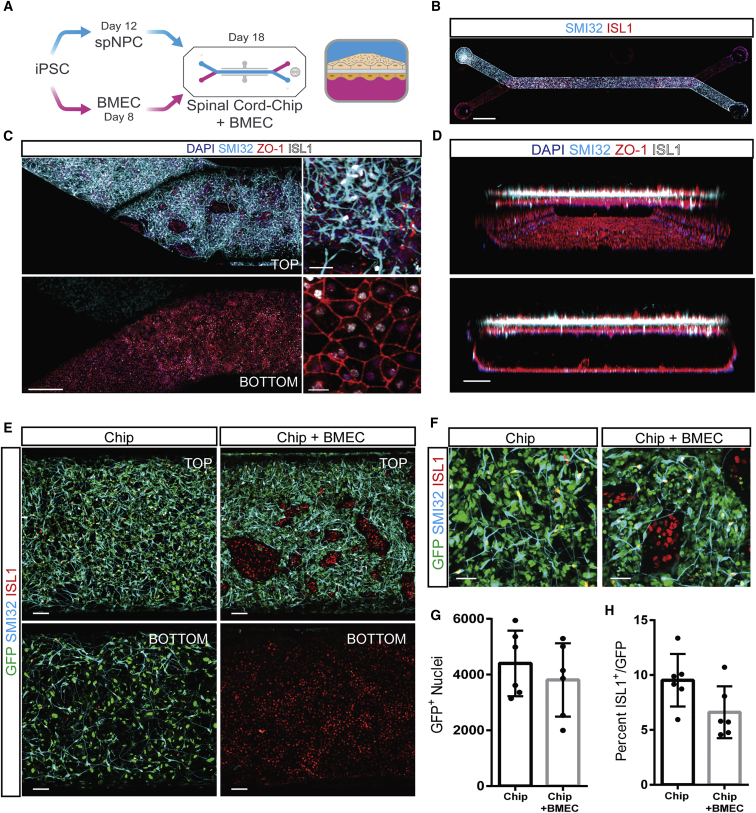
Given that BMECs have been shown to influence neurogenesis, we next wanted to recreate vascular-neural interactions in vitro by deriving BMECs from the same 83iCTR iPSC line using an established protocol (Lippmann et al., 2014). Initial attempts to co-culture both cell types in 96-well plates were unsuccessful as BMECs quickly overcrowded neural cultures (data not shown). We therefore seeded BMECs into the bottom channel of the Spinal Cord-Chip (Figure 2A), followed by seeding of spNPCs into the top channel. After 6 days in culture, immunostaining revealed clear separation of the two cultures in the top (neural) and bottom (endothelial) channels along the entire Spinal Cord-Chip (Figures 2B–2D). BMECs exhibited cobblestone morphology typical of endothelial culture in vitro and expressed endothelial tight junction proteins zona occludens 1 (ZO-1), glucose transporter 1 (GLUT-1), and Occludin (OCLDN) (Figures 2B–2D and S1B). BMEC cultures also expressed nuclear ISL1, previously unreported in human brain vasculature endothelium (Figure 2C). To confirm vascular ISL1 expression in vivo, we immunostained human fetal spinal cord sections at post-conception days 53 and 67 for ISL1 and GLUT-1 and found them to be co-expressed within infiltrating vasculature (Figure S1C). This expression was validated by transcript expression from a previously published dataset of iPSC-derived BMECs (Vatine et al., 2017) (Figure S1D). While SMI32-positive soma were not found in the bottom channel, ZO-1-positive SMI32-negative regions in the top channel revealed infiltration of BMECs (Figures 2C and 2D). Confocal 3-dimensional reconstruction of the main channel displayed clear distinction of the two cultures with confluent BMECs surrounding the entire bottom channel that formed a continuous vessel-like tube (Figure 2D).
Figure 2.
Co-culture of iPSC-Derived spMNs and BMECs in Spinal Cord-Chip
(A) Schematic of dual differentiation and seeding paradigm in Spinal Cord-Chip seeded with BMECs. Both spNPCs and BMECs are generated from human iPSCs and seeded into top and bottom channels, respectively. Transverse section of the Spinal Cord-Chip seeded with BMECs (right) shows two compartments separated by porous membrane.
(B) Immunostaining of whole Spinal Cord-Chip with spMNs expressing SMI32 in top channel and BMECs expressing ISL1 in the bottom channel. Scale bar, 1500 μm.
(C) Maximum projection images cropped at membrane z plane show top and bottom compartments of seeding end of Spinal Cord-Chip immunostained with spMN markers SMI32 and islet 1 (ISL1), and tight junction marker zona occludens 1 (ZO-1). Scale bars, 400 μm (left) and 40 μm (right).
(D) Confocal optical reconstruction along z-axis of Spinal Cord-Chip with computer-generated perspective view (top) exhibits confluent layer of BMECs surrounding entire bottom channel. Orthogonal view (bottom) exhibits distinct separation of cultures at 6 days separated by porous PDMS membrane. Scale bar, 100 μm.
(E) Representative images of cells attached to porous membrane in the main channel (top and bottom planes). Spinal Cord-Chip was seeded either with 83iGFP spNPCs alone in the top channel (Chip) or with the addition of non-GFP BMECs in the bottom compartment. Scale bar, 100 μm.
(F) High magnification of top compartment shows GFP-negative, ISL1-positive clusters indicating BMEC infiltration. Scale bar, 50 μm.
(G and H) Quantification of GFP and ISL1 populations in top compartment. Error bars represent standard deviation. Plots were determined from six individual Chips from three culture rounds (dots).
To further discriminate the infiltration of both cell types in the Spinal Cord-Chip co-culture, we engineered the 83iCTR iPSC line to constitutively express GFP (83iGFP) with a nuclear localization signal, by inserting the cassette into the AAVS1 locus using a zinc-finger nuclease. 83iGFP iPSCs were then differentiated into SMI32-expressing spNPCs and seeded into the top channel as before, while BMECs were generated from the 83iCTR iPSC line and seeded into the bottom channel (Figure 2E). This allowed for clear distinction of GFP-positive spMN cultures from GFP-negative, ISL1-positive expressing BMECs. 83iGFP spNPCs seeded into the top channel without BMECs in the bottom channel readily infiltrated through the membrane pores into the bottom channel (Chip). In contrast, when BMECs were included in the bottom channel, GFP-positive spNPC infiltration was effectively blocked (Figure 2E). Defined regions of GFP-negative/ISL1-positive nuclei were observed in the top channel, which is consistent with ZO-1 staining showing BMEC infiltration from the bottom channel (Figure 2C). To determine the influence of BMEC contact on SMI32-expressing spMN culture differentiation, we analyzed top-channel GFP-positive nuclei as well as ISL1-positive/GFP-positive expression (Figure 2F). Similar numbers of GFP-positive nuclei survived (Figure 2G) in the Chip over three experimental replicates (Figure 2G). The number of ISL1+ spMN nuclei differentiated alone in the Chip (9.5% ± 2%) did not significantly differ in the presence of BMECs (6.6% ± 2%) (Figure 2H). To confirm this result, we repeated the culture paradigm in an additional non-transgenic line (00iCTR) (Figure S2A). High magnification allowed quantification of SMI32-positive neurons that co-expressed ISL1 in both the 83iGFP and 00iCTR lines (Figure S2B). Indeed, similar numbers of ISL1+/SMI32+ spMNs in each condition indicated that co-culturing with BMECs did not affect overall spMN numbers.
Spinal Cord-Chip and BMECs Increase Spontaneous Neuronal Activity of spMN Cultures
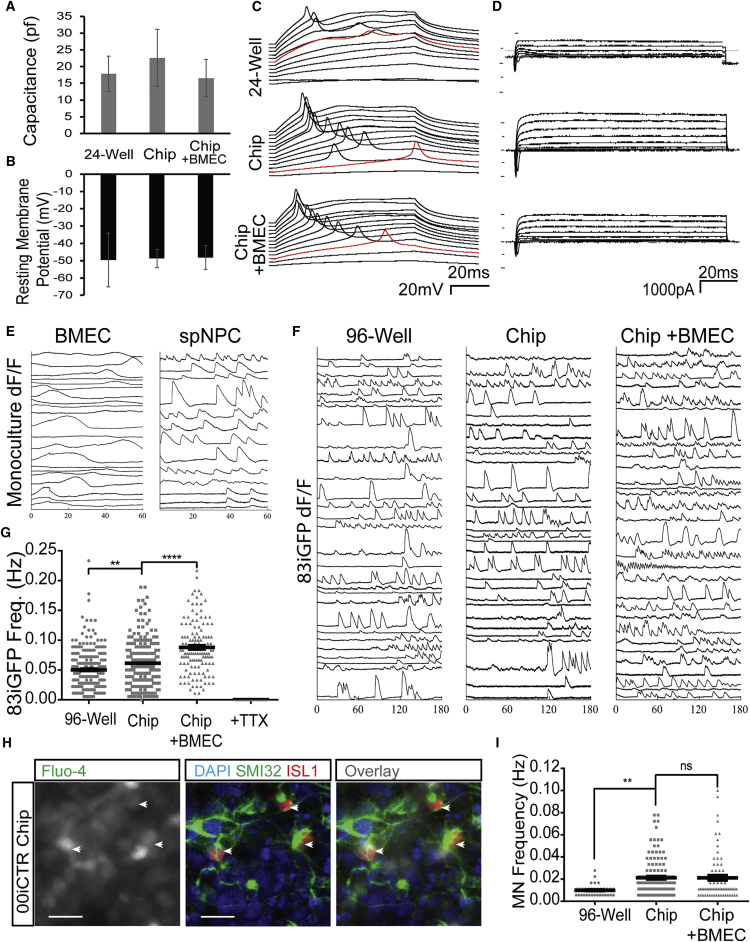
The intrinsic electrophysiological properties of developing spMNs can provide direct measures of function and maturation. As such, we asked whether exposure of spNPCs to either the Spinal Cord-Chip microenvironment and/or BMECs led to enhanced spMN physiological function, compared with a culture plate. Attempts to conduct whole cell patch-clamp using an openable Chip containing the PDMS membrane were unsuccessful; however, polyester membrane-containing Chips allowed sufficient stability for electrophysiological recording of neurons in the top compartment. Of note, the 1-μm pore size in the polyester chips prohibited direct contact between BMECs and neurons (data not shown). Cells from these Spinal Cord-Chips had resting membrane potentials and capacitance values (Figures 3A and 3B) consistent with developing iPSC-derived spMNs neurons in vitro (Takazawa et al., 2012). Current injection steps elicited action potentials and voltage-clamp recordings indicative of neuronal voltage-gated sodium and potassium channel currents (Figures 3C and 3D). Quantification of current densities associated with voltage-gated sodium and voltage-gated potassium channels, maximum action potential amplitude and minimum current necessary to fire an action potential, were similar across culture conditions (Figures S2C–S2F). These data demonstrate that physiologically active spMNs can be cultured within the Spinal Cord-Chip and that basic electrophysiological properties are similar for culture in the Spinal Cord-Chip, with or without BMECs, compared with a culture plate.
Figure 3.
Spinal Cord-Chip Environment Increases Spontaneous Neuronal Activity
(A and B) Average capacitance and resting membrane potential calculated at time of access to neuron during whole-cell current-clamp recording across three culture conditions: 24-well plate, Chip, and Chip containing BMECs (n = 5, 4, and 6 neurons, respectively).
(C) Membrane voltage recordings plotted over time during current-clamp. Traces are sequentially staggered for each 10-pA sweep. Red trace denotes minimum current sweep that reached action potential membrane threshold of 0 mV.
(D) Voltage-clamp recordings of membrane current over time. Picofarads (pf), milliseconds (ms), millivolts (mV), picoamperes (pA).
(E) Individual representative calcium transient activity of spNPC (top) and BMEC (bottom) cultures acquired at the seeding compartments where cultures do not interact. Change in florescence intensity over background (dF/F) plotted with respect to time totaling 60 s.
(F) Calcium transient activity plots of 30 representative neurons plotted over time in seconds and derived from 83iGFP spNPC cultures in each culture condition.
(G) Transient frequency plot of 128, 226, and 232 83iGFP-derived neurons cultured in 96-well plates, Chips, and Chips containing BMECs, respectively. Neuron activity was ablated with the administration of tetrodotoxin (TTX).
(H) Immunocytochemistry staining of islet1 (ISL1) and SMI32 (right) of site previously acquired for live calcium transients using Fluo-4 dye (left). ISL1-positive neurons (arrowheads) are superimposed to determine spMN firing specificity. Scale bars, 200 μm.
(I) Activity of ISL1 SMI32 double-positive neurons in three culture conditions plotted as frequency in hertz.
In graphs, means are represented by black bars and error bars represent standard error of the mean. Significance was calculated by one-way ANOVA: ∗∗p < 0.01, ∗∗∗∗p < 0.001; ns, not significant.
To further assess neuronal physiological function, we next performed live calcium transient imaging on density-matched 83iCTR spNPCs exposed for 6 days to the Spinal Cord-Chip microenvironment, either with or without BMECs in the bottom channel, compared with a 96-well plate. Immunostaining of a 96-well plate showed cell density and neuronal marker expression comparable with those of Spinal Cord-Chip (Figure S2G). Calcium transient signatures of isolated BMECs analyzed at the seeding end were characterized by gradual influx and efflux that was distinct from the neural compartment (Figure 3E). To ensure that only neuron activity was analyzed in all conditions, we filtered out putative BMEC activity by requiring events to return to baseline within 20 s. Transient events for 148–300 neurons from each condition were plotted (Figure 3F) and quantification determined a significant increase in the average number of spontaneous neuronal transients in the Spinal Cord-Chip cultures compared with 96-well cultures (Figure 3G). Of note, neurons cultured with BMECs in the Spinal Cord-Chip had a significantly increased frequency of calcium transients compared with culture in the Spinal Cord-Chip alone. Transient fluorescence was effectively blocked in the presence of the voltage-gated sodium channel antagonist tetrodotoxin (TTX), indicating fluorescence specificity to channel activity. These findings were reproduced using an additional control iPSC line, 00iCTR (Figure S2H–S2I).
As spNPC cultures differentiate into not only spMNs but also contain a mixture of both inhibitory and excitatory interneurons that can modulate network activity (Du et al., 2015), we next wanted to determine whether spMNs themselves were specifically activated in the Spinal Cord-Chip. To achieve population identification of calcium imaging data, 00iCTR cultures following the above activity study were immunostained for spMN markers SMI32 and ISL1 and merged with the same site of live calcium acquisition (Figure 3H). Comparing 116 SMI32+/Isl1+ spMNs with 232 representative active neurons from the same acquisition site showed that ISL1+/SMI32+ spMNs had significantly fewer detected calcium events when compared with overall neuronal activity (Figure S2J). Of the identified SMI32+/ISL1+ spMNs in the three culture conditions, the Spinal Cord-Chip showed increased activity when compared with 96-well plates (Figure 3I). However, the addition of BMECs to the Spinal Cord-Chip did not increase spMN activity, suggesting that specific spMN activity did not significantly contribute to the overall increased activity in neurons cultured with BMECs. Taken together, these data show that culture in the Spinal Cord-Chip maintains intrinsic electrophysiological properties that are consistent with developing neurons but enhances neuronal spontaneous activity.
Spinal Cord-Chip and BMECs Induce Enhanced Developmental Transcriptomic Signatures
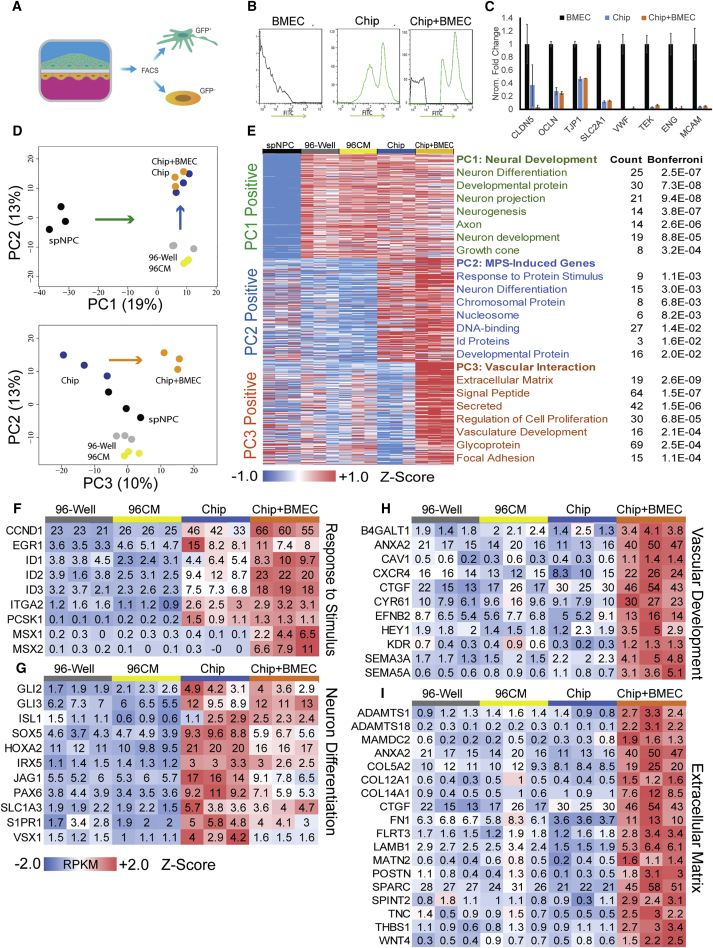
In addition to electrophysiology as an important marker of neuronal activity, cellular transcriptomics can elucidate activated signaling pathways relating to neuronal maturation and cell-cell interactions. To address this, we performed RNA sequencing (RNA-seq) to compare spNPCs cultured in 96-well plates, in the Spinal Cord-Chip alone, or in the Spinal Cord-Chip with BMECs. Day-12 GFP-positive spNPC cultures were thawed and then cultured for 6 days in four conditions: 96-well plate, 96-well plate with BMEC conditioned medium (96CM), Spinal Cord-Chip seeded with spNPCs alone (Chip), or with the addition of BMECs (Chip + BMEC). After 6 days, cells were dissociated and the GFP-positive (neural) population was separated from the GFP-negative population (BMEC) by fluorescence-activated cell sorting (FACS) (Figures 4A and 4B). Thawed spNPCs were also included to determine differences in gene expression resulting from the 6 days in culture. A total of 10,002 genes expressing at least 0.5 reads per kilobase per million (RPKM) in at least one condition were included in the dataset, and RPKM data were quantile normalized across all samples (Figure S3A). Pearson correlation analysis showed high reproducibility among sample replicates (Figure S3B). Effective removal of BMECs from the co-cultured neural samples was indicated by a comparison with the previously published BMEC RNA-seq dataset (Vatine et al., 2017) (black), which showed no increased expression in Spinal Cord-Chip samples that had been co-cultured with BMECs (orange) (Figure 4C).
Figure 4.
Spinal Cord-Chip Induces Neuronal Differentiation and Vascular Interaction Gene Expression
(A) GFP-spMN isolation schematic. Nuclear GFP expressing spNPCs seeded on top channel with isogenic non-GFP expressing BMECs on bottom. FACS conducted on FITC to purify neural cells after BMEC co-culture.
(B) Single population histogram of Chips seeded only with non-GFP BMECs (black) or GFP-spMNs (green) that defined negative and positive fractions, respectively. Number of events is displayed on y axis and FITC intensity on x axis.
(C) Mean RPKM data of previously published BMECs (black), purified neural cells incubated in the Spinal Cord-Chip alone (blue) or in the presence of BMECs (orange). Canonical markers are claudin 5 (CLDN5), occluding (OCLN), Tight junction protein 1 (TJP1), Glut-1 (SLC2A1), von Willebrand factor (VWF), Tie2 receptor (TEK), endoglin (ENG), and melanoma cell adhesion molecule (MCAM).
(D) Principal component (PC) analysis plots of PCs 1 and 2 (top) and PCs 2 and 3 (bottom). Arrows indicate weighting along the axis of each respective PC.
(E) Top 200 ranked genes from each PC displayed as Z score calculated across all conditions for each row and indicated by colorimetric scale. Each PC was entered into DAVID ontology pathway analysis and the top seven categories listed for each PC. The number of genes (count) in each category is displayed with corrected significance values from DAVID analysis (Bonferroni).
(F and G) Differentially expressed genes contributing to the “Response to Protein Stimulus” and “Neural Differentiation” DAVID terms enriched in PC2. RPKM data are averaged across sample replicates and normalized to 96-well plate control.
(H and I) Differentially expressed genes contributing to “Vascular Development” and “Extracellular Matrix” DAVID terms enriched in PC3.
To discover pathways affected in each condition, we performed principal component analysis (PCA) on all samples, and entered the top 200 ranked genes from the top three components into the network-based pathway analysis database DAVID (Huang da et al., 2009) (Figures 4D and 4E). The greatest variance (PC1) separated the day-12 spNPCs from day 18 in all other conditions (Figure 4D). Day-18 neural cells had higher expression of gene pathways involved in neural differentiation, neurogenesis, and axonal growth (Figure 4E). The next greatest variance (PC2) separated Chip culture conditions (Chip and Chip + BMEC) from the two 96-well cultures (96-well and 96CM) and was driven by genes associated with neuronal differentiation pathways that were specifically upregulated in the Chip microenvironment. However, PC2 did not show a notable difference between the 96-well plate and 96CM conditions, suggesting only a small effect of conditioned medium on neural cultures grown in 96-well plates. Finally, PC3 was defined by genes that were differentially expressed when spNPCs were directly co-cultured with BMECs within the Chips (Chip + BMEC). DAVID analysis highlighted genes associated with extracellular matrix, secreted signaling peptides, and endothelial cell interaction pathways.
Spinal Cord-Chip Microenvironment Enhances Neurogenic Programs in spNPC Cultures
Because PC2 elicited “Response to Protein Stimulus” and “Neuron Differentiation” DAVID gene ontology categories, we further investigated Spinal Cord-Chip induced genes relating to neuronal maturation. Relative expression of genes contributing to both categories was compared by calculating a Z score for each gene across all samples (Figures 4F–4I). Genes associated with “Response to Protein Stimulus” were upregulated in both Spinal Cord-Chip conditions, suggesting increased endogenous signaling activity resulting from the Spinal Cord-Chip microenvironment. Among the induced genes, early growth response gene 1 (EGR1) is known to be rapidly transcribed in response to environmental stimulation and neuronal activation (Lacar et al., 2016). Cyclin D1 (CCND1) has been shown to promote spMN neurogenesis in the developing spinal cord in a cell cycle-independent manner, acting through a Notch-signaling effector pathway (Lukaszewicz and Anderson, 2011). Within the “Neuron Differentiation” category, transcription factors involved in spinal cord fate specification (Ericson et al., 1997) were also induced (Figure 4G). PAX6 is an intermediate neural progenitor differentiation marker that regulates neuronal cell cycle exit in the spinal cord. GLI2 and GLI3 were also induced and act to regulate dorsal-ventral patterning in the spinal cord (Persson et al., 2002). Upregulation of ISL1 expression confirmed the staining in Spinal Cord-Chips and further supported increased differentiation resulting from the Spinal Cord-Chip microenvironment. SLC1A3, also known as glutamate aspartate transporter or GLAST, was increased in Spinal Cord-Chip conditions, indicating a potential increase in glutamate transport. A panel of neuronal subtype markers determined in the mouse embryo (Lu et al., 2015) was then probed to identify potential contributors to this increased activity. The Spinal Cord-Chip significantly increased expression of V2a interneuron subdomain markers (Figure S3D). When BMECs were included, the signature shifted to V2b interneurons indicated by NOTCH1 and GATA3 expression.
BMECs Induce Angioneurin and Extracellular Matrix Gene Expression in Developing Neural Cultures in Spinal Cord-Chip
Angioneurin signaling pathways related to vascular endothelial growth factor (VEGF), Notch, and axon guidance mechanisms are known to impact heavily on both neuronal and vascular maturation in vivo. We therefore next determined whether BMECs activated these pathways when co-cultured with spNPCs in the Spinal Cord-Chip. Results showed that within the “Vascular Development” catagory the VEGFA receptor, KDR, was induced in response to BMEC contact (Figure 4H). Finally, DLK1, a potent inhibitor of Notch signaling, was increased, and the Notch-activating ligands JAG1, DLL1, DLL3, and DLL4 were all downregulated in response to BMEC co-culture in the Spinal Cord-Chip (Figure S3C).
Within the “Vascular Development” category, cell surface proteins important for both vascular infiltration and neuronal outgrowth were expressed specifically in response to BMEC co-culture (Figure 4H). Semaphorins can have opposing effects on angiogenesis in vivo (Gu and Giraudo, 2013), and in neurons are required for axon guidance, synaptogenesis, and neuronal maturation (Pasterkamp, 2012). Thrombospondin domain-containing semaphorins 3A and 5A transcripts increased 2- and 5-fold with the addition of BMECs. Chemokine receptor (CXCR4) and ephrin B2 (EFNB2) are cell surface guidance molecules that are required for motor neuron axon outgrowth, and were also increased in response to BMEC co-culture.
Extracellular matrix proteins that are implicated in neuronal maturation showed some of the greatest induction in response to BMEC interaction. Thrombospondin 1 (THBS1) of astrocyte origin has been shown to facilitate synaptogenesis and maturation of iPSC-derived neurons in vitro (Christopherson et al., 2005, Rushton et al., 2013). Both THBS1 and its cleavage partners ADAMTS1/18 were significantly upregulated in the BMEC containing Spinal Cord-Chip (Figure 4I). Finally, extracellular matrix proteins in the collagen family are important constituents of vascular basement membrane and neurovascular niche. Expression of collagen family members COL5A2, COL12A1, and most significantly COL14A1 (over 10-fold expression) were significantly upregulated by neural cultures in response to BMEC inclusion in the Spinal Cord-Chip (Figure 4I).
Spinal Cord-Chip Induces Genes that Are Associated with Fetal Development
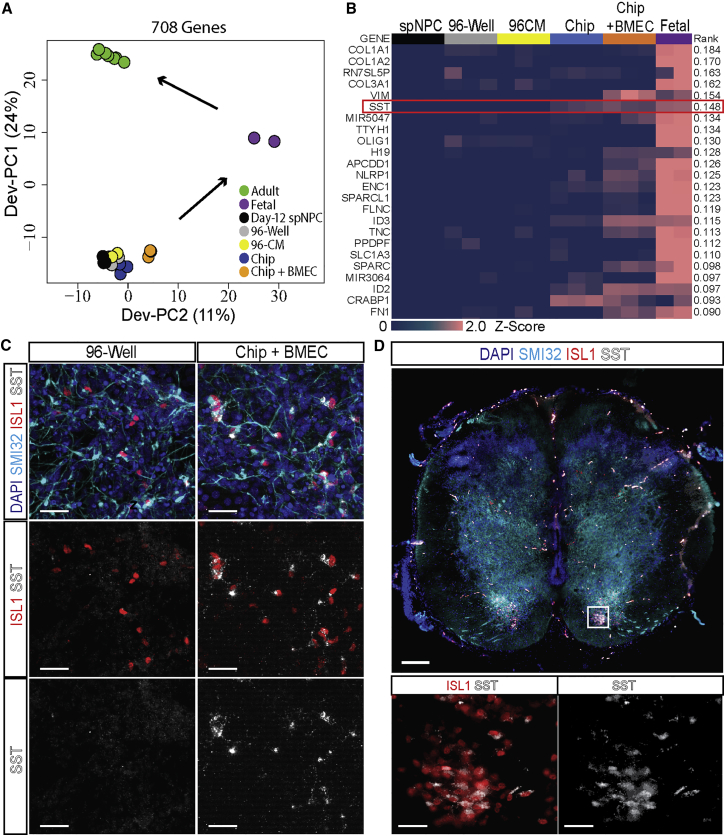
The pathways induced in response to BMEC co-culture in the Spinal Cord-Chip are known to be present in the developing central nervous system. However, most studies were primarily generated using animal models. To ensure that induced pathways reflected those in the human developing spinal cord and to determine the extent to which these activated genes contributed to in vivo maturation, we compared whole transcript data from the culture conditions with datasets generated from primary human spinal cord tissue. Published transcriptomic profiles of fetal spinal cord at days 96 and 105 (purple) (Chadwick, 2012) and adult laser-captured spinal motor neurons of a non-diseased cohort (green) (Batra et al., 2016) were included and filtered to genes that were expressed in both datasets, leaving 9,835 genes in the analysis (Figure S4A). PCA including in vivo and in vitro datasets illustrated no differential effects among culture conditions (Figure S4B). However, a focused PCA based on the top and bottom 200 ranked gene loadings from PC2 and PC3 distinguished the culture conditions with respect to in vivo fetal and adult spinal tissues (Figure 5A). In this reduced dataset, more discrete separation of experimental replicates could be resolved. Along PC1, thawed day-12 spNPCs (black) were furthest from the fetal samples, followed by cultured 96-well (gray) and 96CM (yellow) in the positive direction. The Spinal Cord-Chip (blue) and Spinal Cord-Chip containing BMECs (orange) conditions registered more positively along the Dev-PC2 axis and closer to the fetal samples, indicating that this condition induces gene expression changes relevant to and promoting in vivo spinal tissue development.
Figure 5.
BMECs Induce Vascular Interaction Gene Expression in Spinal Cord-Chip
(A) Principle component (PC) analysis comparing expression data of differentially expressed genes from PC2 and PC3 and including in vivo adult laser-captured spMNs (green), in vivo fetal spinal cord (purple), and in vitro experimental data (black, gray, yellow, blue, and orange).
(B) Heatmap of top ranking genes from Dev-PC2 that describe variance in fetal gene expression. Z score calculated from log2 RPKM data per gene row and displayed by colorimetric scale.
(C) Representative images of 96-well plate (96-well) and Spinal Cord-Chips co-cultured with BMECs (EC/Spinal Cord-Chip) immunostained with SST and spMN markers SMI32 and ISL1. Scale bars, 40 μm.
(D) Whole-mount image of day-67 human fetal spinal cord (top) immunostained with SST, SMI32, and ISL1; (bottom) ventral horn ISL1-positive spMN pool (box) co-expressing SST. Scale bars, 200 μm (top) and 40 μm (bottom).
Spinal Cord-Chip Microenvironment Induces Somatostatin Expression in iPSC-Derived spMNs
Dev-PC2 provided a ranked list of genes that contributed to the in vivo fetal signature along the most positively weighted genes. We therefore sought to determine specific genes induced by Spinal Cord-Chip culture that promoted an in vivo-like spinal cord signature. The 50 highest-weighted Dev-PC2 genes were compared across all iPSC-derived cultures as before (Figure 5B). Angioneurin gene SEMA5A approached levels similar to those of in vivo samples; however, expression could not be confirmed by antibody staining (data not shown). The sixth-ranking gene in PC2 somatostatin (SST) was highly expressed in the fetal spinal cord samples. In brain samples both in vitro and in vivo, SST has been shown to have trophic factor activity, influence synaptogenesis and axonal pathfinding, and regulate cortical circuit maturation (Ferriero et al., 1994, Tuncdemir et al., 2016). Compared with 96-well samples, Spinal Cord-Chip containing spNPCs alone and in the presence of BMECs induced 9- and 15-fold increased SST expression, respectively. To confirm protein expression in vitro, we cultured 96-well plate and EC/Spinal Cord-Chips as before and stained them with SST antibody along with MN markers ISL1 and SMI32. Pronounced SST staining was observed in the Spinal Cord-Chip compared to 96-well both in the absence of BMECs, and to a greater extent with BMEC co-culture (Figure 5C). Expression was co-localized with SMI32-positive staining and proximal to ISL1 nuclear staining in the mixed culture, providing evidence that SST expression was specific to iPSC-derived spMNs. To determine whether MN localization of SST was consistent with developing spMN pools in vivo, we immunostained human fetal spinal cord at day 67 for SST and MN markers ISL1 and SMI32 (Figure 5D). ISL1-positive spMN pools could be visualized in the ventral horn that again contained positive staining for SST, confirming MN specificity in vivo. Thus the Spinal Cord-Chip had captured the expression kinetic of a key developmental molecular marker in human fetal spinal motor neurons.
Discussion
Models of human neuronal physiology that direct differentiation of stem cells to specific neuronal fates in monoculture have yielded a great deal of insight into basic intracellular mechanisms governing potency, lineage commitment, and function. However, more complex organ-level physiology, and often disease pathogenesis, involve a variety of specialized cell types, and this intricate combination may be vital to accurately model disease in vitro. For instance, in ALS a specific neuronal subtype, the spMN, is highly vulnerable; however, additional cell types such as astrocytes (Haidet-Phillips et al., 2011, Zhong et al., 2008), microglia (O'Rourke et al., 2016), vasculature (Zhong et al., 2008), and other neuronal subtypes such as the cortical motor neuron (Thomsen et al., 2014) all contribute to pathophysiology. The Spinal Cord-Chip system permits the study of these multiple cell types as demonstrated here by the specific contribution of BMECs on neuronal maturation.
To address these inherent limitations in monolayer culture, self-organizing 3-dimensional neural clusters known as brain organoids can develop into highly diverse, semi-organized tissues, which contain many neural subtypes that emerge in patterns reminiscent of the developing brain (Birey et al., 2017, Lancaster et al., 2013). Even after long periods in culture, however, organoids stall developmentally (Akhtar et al., 2017). This has been attributed to a lack of vascular-mediated perfusion that has not been successfully implemented into 3-dimensional neural tissues. In addition, inherent variation between individual organoids, resulting from a lack of control in spatial organization and emergence of neuronal subtypes, presents a challenging setting in which to determine disease-specific mechanisms in patient-derived lines.
Previous in vitro studies that focus on the interplay of vascular endothelium through media conditioning have shown enhanced neurogenic potential of primary mouse cortical neural stem cells (Shen et al., 2004). However, soluble factors secreted by human iPSC-derived BMECs were not a major driver of transcript expression and spontaneous neuronal function in this study. Whether this difference is a result of iPSC origin and differentiation, or whether BMECs in the adult neurovascular stem cell niche are functionally distinct from microvasculature in a developing context, or whether the difference is due to specific aspects of the microenvironment offered by this Organ-Chip system, remains unclear. Previous Organ-Chip models involving human neural and endothelial interaction have used human umbilical vein endothelial cells (Griep et al., 2013, Xu et al., 2016, Ye et al., 2014) or, more recently, iPSC sources of endothelial cells (Schwartz et al., 2015, Wang et al., 2017). While these studies provided an important focus on the barrier properties of endothelial cells and toxicology, they lacked detailed assessment of neuronal function and maturation resulting from Chip co-culture.
The expression of transcription factor ISL1 in BMECs was a surprising finding and could mediate common signaling between the two cell types, such as angioneurins, that were also upregulated in co-culture. Although BMEC ISL1 activity on vascular development and interaction with the developing neural tube is unknown, ISL1 knockout mice with ablated spMN pools also displayed compromised vasculature (Pfaff et al., 1996), implicating ISL1 as required in both vascular and spMN development. Transcriptomic profiling of BMECs from human brain tissue showed a lack of ISL1 expression the adult, indicating potential developmental specificity (Zhang et al., 2014).
While several neuronal genes were upregulated in response to BMECs, SST was particularly intriguing as its presence and function in developing spMNs remains largely unknown. In the brain, SST expression is mainly present in GABAergic inhibitory interneurons (Urban-Ciecko and Barth, 2016) and is a key regulator of motor cortex activity during motor learning (Chen et al., 2015b). Electrophysiological signatures of SST-expressing neurons in the neocortex are characterized by a low threshold for action potential firing and high levels of spontaneous activity that can be activated by acetylcholine (Chen et al., 2015a). In a mouse model of ALS, SST interneuron hyperactivity led to sustained disinhibition of cortical motor neurons and cell death (Zhang et al., 2016). Within a developmental context, in a meta-analysis of published human embryonic and adult spinal cord microarray data, SST was determined to be a major contributor to spinal cord maturity (Ho et al., 2016). This, coupled with data here showing SST protein co-localization with ISL1 spMN pools in human fetal spinal cord, suggests that SST could be a critical neuropeptide in human spMN development and disease. The specific expression of SST resulting from Spinal Cord-Chip culture indicates that the Chip platform and co-culture may help increase the physiological relevance of iPSC-derived neurological models, thereby increasing the capacity to analyze functional and pathological mechanisms of the human central nervous system.
Pr is a very powerful model of a neural system wherein neural and endothelial cells co-exist to better mimic in vivo development. In particular, the 7-μm size of the membrane pores was ideal for interaction of BMECs with neural cells and allowed migration of the endothelial cells through the pores to the neural side of the device. That said, reducing the diameter size of the pores may be necessary to inhibit BMEC infiltration into the top channel for a more separated neural/endothelial system. However, this could also lead to lack of maturation as seen in the current study. The relatively short incubation time was chosen to provide reliable data for the initiation and influence of BMEC contact and microenvironment culture on iPSC-derived neural differentiation as is observed in vivo. However, longer-term cultures may provide additional enhancements in maturation. Critically, sustained Spinal Cord-Chip culture can provide a platform for controlled administration of prospective therapeutics through the vascularized compartment to the patient-derived neural tissue. This application would allow an unprecedented tool to study human blood-spinal cord barrier penetrance and neural activity modulation, and critically, to elucidate disease mechanisms for therapeutic discoveries.
Experimental Procedures
Induced Pluripotent Stem Cell Culture and Directed Differentiation into spMNs and BMECs
Two human iPSC lines (83iCTR and 00iCTR) used in this study were previously derived from non-diseased control patient fibroblasts under the direction and approval of Cedars-Sinai Internal Review Board using a non-integrating system (Mattis et al., 2015). Spinal motor neuron derivation was based on the previously published protocol (Du et al., 2015) with modifications. BMECs were differentiated as described previously (Lippmann et al., 2014).
Organ-Chip Culture
Chips were seeded with BMECs at a concentration of 25,000 cells/μL and allowed to attach to the membrane. Later spNPCs were thawed and seeded at a density of 6,000 cells/μL and incubated overnight before flushing with fresh medium. Differentiation medium was then replaced every other day with approximately 25 μL in the main channel and an extra 25 μL in the reservoir tips on either side for 6 days.
Imaging and Population Analysis
Confocal images were acquired using an A1 confocal microscope (Nikon) using a Plan Apo 10× objective at 1-μm increments. Figure 2D was generated from auto-stitching three confocal stacks and using maximum projection for each channel at 10× (Nikon Elements). For population and ISL1 nuclei analysis, three representative sites per Chip were acquired in 3 chips per condition over three experiments. Images were then cropped to only top channel using IMARIS software. GFP-positive nuclei were quantified for each site using a spots algorithm and averaged per Chip. ISL1-positive nuclei were quantified by filtering GFP-positive data by ISL1 co-localization. Population statistics were determined by one-way ANOVA using PRISM (GraphPad).
Transcriptomic Analysis
Three Chips or three wells of a 96-well plate were dissociated and pooled for each experimental replicate for a total of three replicates. Pooled samples were sorted using an Influx FACS sorter (Becton Dickinson). GFP-positive gating was established using Spinal Cord-Chip seeded exclusively with 83iGFP spMNs for a positive control and 83iCTR BMECs for a negative control. All samples were sorted using this established gating. Positive fraction cell pellets were frozen at −80°C until processing. RNA-seq libraries were generated using the Illumina TruSeq Kit, sequenced on a Nextseq500 by 75-bp single reads, and aligned to the hg19 human genome using TopHat2. Rseqtools was used to quantify UCSC annotated transcripts to RPKM, and these transcripts were annotated with unique HGNC gene symbols by taking the maximum RPKM value. Genes not detected in each replicate at above 0.5 RPKM for at least one condition were removed from the analysis to reduce false positives due to sequencing noise. To allow log transformation of zero values for plotting, we added 1 RPKM to all data. RPKM values were normalized by quantile normalization across all samples for cross-comparison and relevant statistics were calculated using R statistical software. PCA was generated from normalized log-transformed data by Cluster 3.0 software. Heatmaps were generated by calculating Z score and superimposing normalized RPKM data on each cell using Excel (Microsoft).
Live Calcium Transient Imaging Analysis
Calcium activity was counted by tracing region of interest (ROI) in blinded fashion and creating masks (ImageJ) for use in extracting florescence intensity data. Active neurons were selected by having at least one transient that returned to baseline within 20 s. Neurons were quantified across three separate experiments, two included spMNs derived from 83iGFP line with four biological replicates totaling 128–232 neurons per condition, and one included spMNs derived from 00iCTR line with two replicates totaling 144–278 neurons per condition. dF/F traces and transient events were generated from ROI using the modified MATLAB open-source package FluoroSNNAP and automated calcium event detection was accomplished through template libraries at a threshold of dF/F > 0.05 (Patel et al., 2015). Events were then curated manually by blinded counter. Statistics of calcium transient frequency were determined by multiple comparison-corrected one-way ANOVA using PRISM (GraphPad).
Author Contributions
S.S. and C.N.S. designed the experiments and wrote and edited the manuscript. S.S. performed in vitro experiments with assistance from G.V., D.W., A.M., P.S.K., and M.G. S.S. performed electrophysiology experiments. S.S. and R.H. performed transcriptomic analysis. S.S., B.M., and D.S. developed the motor neuron generation protocol. Chip engineering assistance was provided by A.L., N.W., C.H., and G.A.H.
Acknowledgments
We would like to thank Dr. Soshana Svendsen for critical editing. We would also like to acknowledge Steven Huang and Kathleen Kurowski for help with Spinal Cord-Chip culture and Brett Clair for providing Chip diagrams. RNA sequencing datasets were downloaded from the ROADMAPS Epigenomics Consortium website: http://nihroadmap.nih.gov/epigenomics/. This work was supported by the ALS Association grant ID18-SI-389, the NIH Tissue Consortium 2.0 1UG3NS105703-01, and the California Institute for Regenerative Medicine grant ID DISC1-08800. Cedars-Sinai owns a minority stock interest in Emulate, Inc., the company that produces the study's microfluidic devices (chips). An officer of Cedars-Sinai also serves on Emulate's Board of Directors. Emulate provides no financial support for this research.
Published: March 22, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.02.012.
Contributor Information
Samuel Sances, Email: samuel.sances@cshs.org.
Clive N. Svendsen, Email: clive.svendsen@cshs.org.
Accession Numbers
Transcriptomic datasets generated from iPSC-derived spMNs were uploaded to GEO under the series number GEO: GSE110826. Additional downloaded datasets were as follows: Fetal spinal cords from day 96, GEO: GSM1101679 and day 105, GEO: GSM1101711. Adult non-diseased control laser-capture microdissected tissue from series GEO: GSE76220 were GEO: GSM1977027, GSM1977028, GSM1977029, GSM1977030, GSM1977031, GSM1977032, GSM1977033, and GSM1977034.
Supplemental Information
References
- Akhtar A.A., Sances S., Barrett R., Breunig J.J. Organoid and organ-on-a-chip systems: new paradigms for modeling neurological and gastrointestinal disease. Curr. Stem Cell Rep. 2017;3:98–111. doi: 10.1007/s40778-017-0080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avior Y., Sagi I., Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- Batra R., Hutt K., Vu A., Rabin S.J., Baughn M.W., Libby R.T., Hoon S., Ravits J., Yeo G.W. Gene expression signatures of sporadic ALS motor neuron populations. bioRxiv. 2016 [Google Scholar]
- Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Birey F., Andersen J., Makinson C.D., Islam S., Wei W., Huber N., Fan H.C., Metzler K.R.C., Panagiotakos G., Thom N. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick L.H. The NIH roadmap epigenomics program data resource. Epigenomics. 2012;4:317–324. doi: 10.2217/epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Sugihara H., Sur M. An acetylcholine-activated microcircuit drives temporal dynamics of cortical activity. Nat. Neurosci. 2015;18:892–902. doi: 10.1038/nn.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.X., Kim A.N., Peters A.J., Komiyama T. Subtype-specific plasticity of inhibitory circuits in motor cortex during motor learning. Nat. Neurosci. 2015;18:1109–1115. doi: 10.1038/nn.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson K.S., Ullian E.M., Stokes C.C., Mullowney C.E., Hell J.W., Agah A., Lawler J., Mosher D.F., Bornstein P., Barres B.A. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Demestre M., Orth M., Fohr K.J., Achberger K., Ludolph A.C., Liebau S., Boeckers T.M. Formation and characterisation of neuromuscular junctions between hiPSC derived motoneurons and myotubes. Stem Cell Res. 2015;15:328–336. doi: 10.1016/j.scr.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Domenech M., Yu H., Warrick J., Badders N.M., Meyvantsson I., Alexander C.M., Beebe D.J. Cellular observations enabled by microculture: paracrine signaling and population demographics. Integr. Biol. (Camb.) 2009;1:267–274. doi: 10.1039/b823059e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z.W., Chen H., Liu H., Lu J., Qian K., Huang C.L., Zhong X., Fan F., Zhang S.C. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat. Commun. 2015;6:6626. doi: 10.1038/ncomms7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt B., Liebner S. Novel insights into the development and maintenance of the blood-brain barrier. Cell Tissue Res. 2014;355:687–699. doi: 10.1007/s00441-014-1811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J., Rashbass P., Schedl A., Brenner-Morton S., Kawakami A., van Heyningen V., Jessell T.M., Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Ferriero D.M., Sheldon R.A., Messing R.O. Somatostatin enhances nerve growth factor-induced neurite outgrowth in PC12 cells. Brain Res. Dev. Brain Res. 1994;80:13–18. doi: 10.1016/0165-3806(94)90084-1. [DOI] [PubMed] [Google Scholar]
- Griep L.M., Wolbers F., de Wagenaar B., ter Braak P.M., Weksler B.B., Romero I.A., Couraud P.O., Vermes I., van der Meer A.D., van den Berg A. BBB on chip: microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed. Microdevices. 2013;15:145–150. doi: 10.1007/s10544-012-9699-7. [DOI] [PubMed] [Google Scholar]
- Gu C., Giraudo E. The role of semaphorins and their receptors in vascular development and cancer. Exp. Cell Res. 2013;319:1306–1316. doi: 10.1016/j.yexcr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.H., Ma W., Yang J.W., Gao Y., Liang Z., Liu J., Wang D.Y., Luo T., Cheng J.R., Li L.Y. Expression pattern of NeuN and GFAP during human fetal spinal cord development. Childs Nerv. Syst. 2015;31:863–872. doi: 10.1007/s00381-015-2713-7. [DOI] [PubMed] [Google Scholar]
- Haidet-Phillips A.M., Hester M.E., Miranda C.J., Meyer K., Braun L., Frakes A., Song S., Likhite S., Murtha M.J., Foust K.D. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat. Biotechnol. 2011;29:824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselmans L.F., Jennekens F.G., Van den Oord C.J., Veldman H., Vincent A. Development of innervation of skeletal muscle fibers in man: relation to acetylcholine receptors. Anat. Rec. 1993;236:553–562. doi: 10.1002/ar.1092360315. [DOI] [PubMed] [Google Scholar]
- Ho R., Sances S., Gowing G., Amoroso M.W., O'Rourke J.G., Sahabian A., Wichterle H., Baloh R.H., Sareen D., Svendsen C.N. ALS disrupts spinal motor neuron maturation and aging pathways within gene co-expression networks. Nat. Neurosci. 2016;19:1256–1267. doi: 10.1038/nn.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huh D., Matthews B.D., Mammoto A., Montoya-Zavala M., Hsin H.Y., Ingber D.E. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell T.M. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Li H., Collins J.J., Ingber D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz H. Cell lineages and early patterns of embryonic CNS vascularization. Cell Adhes. Migration. 2009;3:205–210. doi: 10.4161/cam.3.2.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacar B., Linker S.B., Jaeger B.N., Krishnaswami S., Barron J., Kelder M., Parylak S., Paquola A., Venepally P., Novotny M. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.-A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann E.S., Al-Ahmad A., Azarin S.M., Palecek S.P., Shusta E.V. A retinoic acid-enhanced, multicellular human blood-brain barrier model derived from stem cell sources. Sci. Rep. 2014;4:4160. doi: 10.1038/srep04160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louissaint A., Jr., Rao S., Leventhal C., Goldman S.A. Coordinated interaction of neurogenesis and angiogenesis in the adult songbird brain. Neuron. 2002;34:945–960. doi: 10.1016/s0896-6273(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Lu D.C., Niu T., Alaynick W.A. Molecular and cellular development of spinal cord locomotor circuitry. Front. Mol. Neurosci. 2015;8:25. doi: 10.3389/fnmol.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A.I., Anderson D.J. Cyclin D1 promotes neurogenesis in the developing spinal cord in a cell cycle-independent manner. Proc. Natl. Acad. Sci. USA. 2011;108:11632–11637. doi: 10.1073/pnas.1106230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattis V.B., Tom C., Akimov S., Saeedian J., Østergaard M.E., Southwell A.L., Doty C.N., Ornelas L., Sahabian A., Lenaeus L. HD iPSC-derived neural progenitors accumulate in culture and are susceptible to BDNF withdrawal due to glutamate toxicity. Hum. Mol. Genet. 2015;24:3257–3271. doi: 10.1093/hmg/ddv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya G.J., Sutachan J.J., Chan W.S., Sideris A., Blanck T.J., Recio-Pinto E. Muscle-conditioned media and cAMP promote survival and neurite outgrowth of adult spinal cord motor neurons. Exp. Neurol. 2009;220:303–315. doi: 10.1016/j.expneurol.2009.09.003. [DOI] [PubMed] [Google Scholar]
- O'Rourke J.G., Bogdanik L., Yanez A., Lall D., Wolf A.J., Muhammad A.K., Ho R., Carmona S., Vit J.P., Zarrow J. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351:1324–1329. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer T.D., Willhoite A.R., Gage F.H. Vascular niche for adult hippocampal neurogenesis. J. Comp. Neurol. 2000;425:479–494. doi: 10.1002/1096-9861(20001002)425:4<479::aid-cne2>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Pasterkamp R.J. Getting neural circuits into shape with semaphorins. Nat. Rev. Neurosci. 2012;13:605–618. doi: 10.1038/nrn3302. [DOI] [PubMed] [Google Scholar]
- Patel T.P., Man K., Firestein B.L., Meaney D.F. Automated quantification of neuronal networks and single-cell calcium dynamics using calcium imaging. J. Neurosci. Methods. 2015;243:26–38. doi: 10.1016/j.jneumeth.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson M., Stamataki D., te Welscher P., Andersson E., Böse J., Rüther U., Ericson J., Briscoe J. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 2002;16:2865–2878. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff S.L., Mendelsohn M., Stewart C.L., Edlund T., Jessell T.M. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron– dependent step in interneuron differentiation. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Przybyla L., Voldman J. Probing embryonic stem cell autocrine and paracrine signaling using microfluidics. Annu. Rev. Anal. Chem. 2012;5:293–315. doi: 10.1146/annurev-anchem-062011-143122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton D.J., Mattis V.B., Svendsen C.N., Allen N.D., Kemp P.J. Stimulation of GABA-induced Ca2+ influx enhances maturation of human induced pluripotent stem cell-derived neurons. PLoS One. 2013;8:e81031. doi: 10.1371/journal.pone.0081031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sances S., Bruijn L.I., Chandran S., Eggan K., Ho R., Klim J.R., Livesey M.R., Lowry E., Macklis J.D., Rushton D. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat. Neurosci. 2016;19:542–553. doi: 10.1038/nn.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M.P., Hou Z., Propson N.E., Zhang J., Engstrom C.J., Costa V.S., Jiang P., Nguyen B.K., Bolin J.M., Daly W. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc. Natl. Acad. Sci. USA. 2015;112:12516–12521. doi: 10.1073/pnas.1516645112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Goderie S.K., Jin L., Karanth N., Sun Y., Abramova N., Vincent P., Pumiglia K., Temple S. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304:1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- Shi Y., Inoue H., Wu J.C., Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takazawa T., Croft G.F., Amoroso M.W., Studer L., Wichterle H., MacDermott A.B. Maturation of spinal motor neurons derived from human embryonic stem cells. PLoS One. 2012;7:e40154. doi: 10.1371/journal.pone.0040154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen G.M., Gowing G., Latter J., Chen M., Vit J.P., Staggenborg K., Avalos P., Alkaslasi M., Ferraiuolo L., Likhite S. Delayed disease onset and extended survival in the SOD1G93A rat model of amyotrophic lateral sclerosis after suppression of mutant SOD1 in the motor cortex. J. Neurosci. 2014;34:15587–15600. doi: 10.1523/JNEUROSCI.2037-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncdemir S.N., Wamsley B., Stam F.J., Osakada F., Goulding M., Callaway E.M., Rudy B., Fishell G. Early somatostatin interneuron connectivity mediates the maturation of deep layer cortical circuits. Neuron. 2016;89:521–535. doi: 10.1016/j.neuron.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban-Ciecko J., Barth A.L. Somatostatin-expressing neurons in cortical networks. Nat. Rev. Neurosci. 2016;17:401–409. doi: 10.1038/nrn.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatine G.D., Al-Ahmad A., Barriga B.K., Svendsen S., Salim A., Garcia L., Garcia V.J., Ho R., Yucer N., Qian T. Modeling psychomotor retardation using iPSCs from MCT8-deficient patients indicates a prominent role for the blood-brain barrier. Cell Stem Cell. 2017;20:831–843.e5. doi: 10.1016/j.stem.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wälchli T., Wacker A., Frei K., Regli L., Schwab M.E., Hoerstrup S.P., Gerhardt H., Engelhardt B. Wiring the vascular network with neural cues: a CNS perspective. Neuron. 2015;87:271–296. doi: 10.1016/j.neuron.2015.06.038. [DOI] [PubMed] [Google Scholar]
- Wang Y.I., Abaci H.E., Shuler M.L. Microfluidic blood-brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol. Bioeng. 2017;114:184–194. doi: 10.1002/bit.26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warp E., Agarwal G., Wyart C., Friedmann D., Oldfield C.S., Conner A., Del Bene F., Arrenberg A.B., Baier H., Isacoff E.Y. Emergence of patterned activity in the developing zebrafish spinal cord. Curr. Biol. 2012;22:93–102. doi: 10.1016/j.cub.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Li Z., Yu Y., Sizdahkhani S., Ho W.S., Yin F., Wang L., Zhu G., Zhang M., Jiang L. A dynamic in vivo-like organotypic blood-brain barrier model to probe metastatic brain tumors. Sci. Rep. 2016;6:36670. doi: 10.1038/srep36670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M., Sanchez H.M., Hultz M., Yang Z., Bogorad M., Wong A.D., Searson P.C. Brain microvascular endothelial cells resist elongation due to curvature and shear stress. Sci. Rep. 2014;4:4681. doi: 10.1038/srep04681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Alexander C.M., Beebe D.J. Understanding microchannel culture: parameters involved in soluble factor signaling. Lab Chip. 2007;7:726–730. doi: 10.1039/b618793e. [DOI] [PubMed] [Google Scholar]
- Zacchigna S., Lambrechts D., Carmeliet P. Neurovascular signalling defects in neurodegeneration. Nat. Rev. Neurosci. 2008;9:169–181. doi: 10.1038/nrn2336. [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhang L., Liang B., Schroeder D., Zhang Z.W., Cox G.A., Li Y., Lin D.T. Hyperactive somatostatin interneurons contribute to excitotoxicity in neurodegenerative disorders. Nat. Neurosci. 2016;19:557–559. doi: 10.1038/nn.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O'Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Deane R., Ali Z., Parisi M., Shapovalov Y., O’Banion K., Stojanovic K., Sagare A., Boillee S., Cleveland D.W. ALS-causing SOD1 mutants generate vascular changes prior to motor neuron degeneration. Nat. Neurosci. 2008;11:420–422. doi: 10.1038/nn2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.