Abstract
Background
The dominant hypothesis about the pathogenesis of Alzheimer’s disease (AD) is the “amyloid cascade” concept and modulating the expression of proteins involved in the metabolism of amyloid-beta (Aβ) is proposed as an effective strategy for the prevention and therapy of AD. Recently, we found that an antibiotic ceftriaxone (CEF), which possesses neuroprotective activity, reduced cognitive deficits and neurodegenerative changes in OXYS rats, a model of sporadic AD. The molecular mechanisms of this effect are not completely clear, we suggested that the drug might serve as the regulator of the expression of the genes involved in the metabolism of Aβ and the pathogenesis of AD. The study was aimed to determine the effects of CEF on mRNA levels of Bace1 (encoding β-secretase BACE1 involved in Aβ production), Mme, Ide, Ece1, Ace2 (encoding enzymes involved in Aβ degradation), Epo (encoding erythropoietin related to endothelial function and clearance of Aβ across the blood brain barrier) in the frontal cortex, hippocampus, striatum, hypothalamus, and amygdala of OXYS and Wistar (control strain) male rats. Starting from the age of 14 weeks, animals received CEF (100 mg/kg/day, i.p., 36 days) or saline. mRNA levels were evaluated with RT-qPCR method. Biochemical parameters of plasma were measured for control of system effects of the treatment.
Results
To better understand strain variations studied here, we compared the gene expression between untreated OXYS and Wistar rats. This comparison showed a significant decrease in mRNA levels of Ace2 in the frontal cortex and hypothalamus, and of Actb in the amygdala of untreated OXYS rats. Analysis of potential effects of CEF revealed its novel targets. In the compound-treated OXYS cohort, CEF diminished mRNA levels of Bace1 and Ace2 in the hypothalamus, and Aktb in the frontal cortex. Furthermore, CEF augmented Mme, Ide, and Epo mRNA levels in the amygdala as well as the levels of Ece1 and Aktb in the striatum. Finally, CEF also attenuated the activity of ALT and AST in plasma of OXYS rats.
Conclusion
Those findings disclosed novel targets for CEF action that might be involved into neuroprotective mechanisms at early, pre-plaque stages of AD-like pathology development.
Keywords: Ceftriaxone, Neuroprotection, Alzheimer’s disease, Amyloid metabolism, Gene expression, mRNA, Rats
Background
Alzheimer’s disease (AD) is one of the most common neurodegenerative disorders of the central nervous system. Loss of neurons in multiple brain regions (mainly in the cortical and subcortical areas and hippocampus) leads to cognitive dysfunctions including memory impairment as well as to behavioral and physiological changes. The dominant hypothesis of the pathogenesis of AD is the “amyloid cascade” concept. The excessive accumulation and aggregation of amyloid-beta (Aβ) in AD is assigned as a key role in triggering the chain of pathological neurodegenerative processes [1, 2]. Major components of the Aβ deposits are 38–43 amino acids-long fragments derived from proteolytic processing of the amyloid precursor protein (APP) [3]. Especially Aβ42 elevation is related to AD progression due to its high tendency to aggregate. Oligomeric Aβ species represent the most toxic forms causing impaired synaptic and neuronal functions [4, 5].
The Aβ levels in the brain depend on its de novo formation by amyloidogenic APP processing with β-secretase BACE1 and also on its elimination via different mechanisms including its proteolytic degradation, transport processes, cell mediated clearance, and its deposition into insoluble aggregates [2, 6]. It is believed that the imbalance between the Aβ production and removal contributes to the abnormal Aβ accumulation, and those mechanisms become impaired with aging and disease [7]. Hence, modulating the expression of proteins involved in the metabolism of Aβ has attracted increasing attention and is regarded as quite a promising strategy for AD therapy by lowering Aβ deposition in the brain [6, 8]. It should be noted that there is no one specific way of Aβ catabolism. A range of the potential amyloid-degrading proteolytic enzymes have been identified that hydrolyse a range of other physiological substrates as well. In the present study we focused on those enzymes that showed a connection with AD and Aβ degrading activity in vivo, including neprilysin (NEP) peptidase that is considered as a major Aβ degrading enzyme in the brain [8, 9].
OXYS rat strain with hereditary defined accelerated senescence was produced in the Institute of Cytology and Genetics SB RAS (Novosibirsk, Russia) with selective breeding of Wistar rats that were highly sensitive to cataractogenic effect of d-galactose. The OXYS rats are known for shortened lifespan and an early development of age-related pathological phenotypes [10]. Recently found signs of early neurodegeneration, cognitive decline, Aβ deposits, and increased tau phosphorylation in rats of the OXYS strain [11–14] along with the absence of specific for the early familial forms of AD mutations in App, Psen1, and Psen2 genes in the OXYS genome [12] lend themselves to be a faithful model of sporadic AD.
Recently, we found that an antibiotic ceftriaxone (CEF), which possesses neuroprotective activity, reduced cognitive and neuronal deficits in OXYS rats [15]. The molecular mechanisms of this effect are not completely clear, therefore suggest that the drug might serve as the regulator of the gene expression involved in the metabolism of Aβ and the pathogenesis of AD. Hence, the present study was aimed to determine mRNA levels of Bace1 (gene of β-secretase BACE1 involved in Aβ production), Mme, Ece1, Ide, Ace2 (genes of enzymes involved in Aβ degradation), and Epo (gene for erythropoietin related to endothelial function and clearance of Aβ across the blood brain barrier) in the frontal cortex, hippocampus, striatum, hypothalamus, and amygdala of Wistar and OXYS rats as well as the potential effects of CEF on their modulation.
Results
Analysis of mRNA levels of CEF target genes in the amygdala
Abundance of mRNA species in the amygdala is summarized in Fig. 1.
Fig. 1.

Effects of CEF on mRNA levels of Ace2, Bace1, Ece1, Epo, Ide, Mme, and Actb in the amygdala of OXYS and Wistar rats. The data represent the relative mRNA levels of target genes to geometrical mean of B2m, Hprt1, and Mapk6 mRNA levels. The data are expressed as the Mean ± S.E.M. of the values obtained in an independent group of animals (n = 7–10 per group). Statistically significant differences: *P < 0.05 versus “Wistar + saline” group; #P < 0.05; ##P < 0.01 versus “Wistar + CEF” group; $P < 0.05 versus “OXYS + saline” group
Ace2
According to two-way ANOVA, there was a tendency in influence of the “Ceftriaxone” factor [F(1, 32) = 3.167, P = 0.085] and no significant effect of the “Genotype” factor [F(1, 32) = 1.851, P > 0.05] or of the interaction between the factors [F(1, 32) = 2.653, P > 0.05] on the mRNA levels of Ace2 in the amygdala of the rats.
Bace1
Two-way ANOVA did not show a significant effect of the “Genotype” [F(1, 32) < 1] or “Ceftriaxone” [F(1, 32) < 1] factor as well as of the interaction between the factors [F(1, 32) = 1.517, P > 0.05] on mRNA levels of Bace1.
Ece1
Two-way ANOVA did not show a significant effect of the “Genotype” [F(1, 34) < 1] or “Ceftriaxone” [F(1, 34) < 1] factor as well as of the interaction between the factors [F(1, 34) < 1] on the mRNA levels of Ece1.
Epo
According to two-way ANOVA, there was a significant influence of the “Genotype” factor [F(1, 22) = 4.314, P < 0.05] and of the interaction between the factors [F(1, 22) = 8.383, P < 0.01] but no significant effect of the “Ceftriaxone” factor [F(1, 22) = 1.452, P > 0.05] on the mRNA levels of Epo in the amygdala of the rats. LSD post hoc test revealed that the parameter was significantly higher in the rats of “OXYS + CEF” group versus that in Wistar rats of “Wistar + saline” (P < 0.05) and “Wistar + CEF” (P < 0.01) or “OXYS + saline” (P < 0.05) group.
Ide
According to two-way ANOVA, there was a significant influence of the “Genotype” factor [F(1, 32) = 6.163, P < 0.05], a tendency for the interaction between the factors [F(1, 32) = 2.935, P = 0.096] and no significant effect for the “Ceftriaxone” factor [F(1, 32) < 1] on the mRNA levels of Ide in the amygdala of the rats. LSD post hoc test revealed that the parameter was significantly higher in the rats of “OXYS + CEF” group versus that in Wistar rats of “Wistar + saline” (P < 0.05) or “Wistar + CEF” (P < 0.01) group.
Mme
According to two-way ANOVA, there was a significant influence of the “Genotype” factor [F(1, 31) = 7.488, P < 0.05], and no significant effect for the “Ceftriaxone” factor [F(1, 31) < 1] or for the interaction between the factors [F(1, 31) = 2.255, P > 0.05] on the mRNA levels of Mme in the amygdala of the rats. LSD post hoc test revealed that the parameter was significantly higher in the rats of “OXYS + CEF” group versus that in Wistar rats of “Wistar + saline” (P < 0.05) or “Wistar + CEF” (P < 0.01) group.
Actb
According to two-way ANOVA, there was a significant influence of the “Genotype” factor [F(1, 31) = 7.610, P < 0.01], and no significant effect for the “Ceftriaxone” factor [F(1, 31) < 1] or for the interaction between the factors [F(1, 31) < 1] on the mRNA levels of Actb in the amygdala of the rats. LSD post hoc test revealed that the parameter was significantly higher in the rats of “Wistar + CEF” group versus that in OXYS rats of “OXYS + saline” (P < 0.05) or “OXYS + CEF” (P < 0.05) group.
Analysis of mRNA levels of CEF target genes in the hypothalamus
The results on the mRNA abundance in the hypothalamus are summarized in Fig. 2.
Fig. 2.

Effects of CEF on mRNA levels of Ace2, Bace1, Ece1, Epo, Ide, Mme, and Actb in the hypothalamus of OXYS and Wistar rats. The data represent the relative mRNA levels of target genes to geometrical mean of B2m, Hprt1, and Mapk6 mRNA levels. The data are expressed as the Mean ± S.E.M. of the values obtained in an independent group of animals (n = 7–10 per group). Statistically significant differences: *P < 0.05; **P < 0.01; ***P < 0.001 versus “Wistar + saline” group; $P < 0.05 versus “OXYS + saline” group
Ace2
According to two-way ANOVA, there were a significant influence of the “Genotype” factor [F(1, 28) = 10.643, P < 0.01] and of the “Ceftriaxone” factor [F(1, 28) = 4.955, P < 0.05] but no significant effect of the interaction between the factors [F(1, 28) = 1.973, P > 0.05] on the mRNA levels of Ace2 in the hypothalamus of the rats. LSD post hoc test revealed that the parameter was significantly higher in the rats of “Wistar + saline” group versus that in rats of “Wistar + CEF” (P < 0.05) and “OXYS + saline” (P < 0.01) or “OXYS + CEF” (P < 0.001) group.
Bace1
According to two-way ANOVA, there was a significant influence of the interaction between the factors [F(1, 27) = 5.491, P < 0.05] and no significant effects of the “Genotype” [F(1, 27) < 1] or of the “Ceftriaxone” [F(1, 27) = 2.339, P > 0.05] factors on the mRNA levels of Bace1 in the hypothalamus of the rats. LSD post hoc test revealed that the parameter was significantly lower in the rats of “OXYS + CEF” group versus that in rats of “OXYS + saline” (P < 0.05) group.
Due to the lack of normal distribution of the data in the studied groups, Kruskal–Wallis ANOVA by Ranks was applied and revealed no significant differences between the groups for mRNA levels of Ece1 [H(3, 33) = 2.429, P > 0.05], Epo [H(3, 32) = 1.726, P > 0.05], and Ide [H(3, 34) = 1.461, P > 0.05]. For Mme, Kruskal–Wallis ANOVA by Ranks showed a significant difference between the groups: [H(3, 33) = 8.304, P = 0.040]. However, multiple comparisons of mean ranks for all groups demonstrated no significant intergroup differences.
Actb
Two-way ANOVA did not show a significant effect of the “Genotype” [F(1, 29) < 1] or “Ceftriaxone” [F(1, 29) = 2.070, P > 0.05] factor as well as of the interaction between the factors [F(1, 29) < 1] on the mRNA levels of Actb.
Analysis of mRNA levels of CEF target genes in the hippocampus
Abundance of mRNA species in the hippocampus is summarized in Fig. 3.
Fig. 3.

Effects of CEF on mRNA levels of Ace2, Bace1, Ece1, Epo, Ide, Mme, and Actb in the hippocampus of OXYS and Wistar rats. The data represent the relative mRNA levels of target genes to geometrical mean of B2m, Hprt1, and Mapk6 mRNA levels. The data are expressed as the Mean ± S.E.M. of the values obtained in an independent group of animals (n = 6–9 per group). Statistically significant differences: #P < 0.05 versus “Wistar + CEF” group
Due to the lack of normal distribution of the data in the studied groups, Kruskal–Wallis ANOVA by Ranks was applied and revealed no significant differences between the groups for mRNA levels of Ace2 [H(3, 30) = 1.657, P > 0.05], Bace1 [H(3, 34) = 2.317, P > 0.05], Epo [H(3, 22) = 5.239, P > 0.05], and Actb [H(3, 32) = 1.433, P > 0.05].
Ece1
According to two-way ANOVA, there was a significant influence of the “Genotype” factor [F(1, 27) = 5.462, P < 0.05], a tendency for the interaction between the factors [F(1, 27) = 3.511, P = 0.072] and no significant effect of the “Ceftriaxone” factor [F(1, 27) < 1] on the mRNA levels of Ece1 in the hippocampus of the rats. LSD post hoc test revealed that the parameter was significantly lower in the rats of “OXYS + CEF” group versus that in rats of “Wistar + CEF” group (P < 0.01).
Ide
Two-way ANOVA did not show a significant effect of the “Genotype” [F(1, 29) < 1] or “Ceftriaxone” [F(1, 29) = 2.858, P > 0.05] factor as well as of the interaction between the factors [F(1, 29) = 1.677, P > 0.05].
Mme
According to two-way ANOVA, there was a tendency for influence of the interaction between the factors [F(1, 27) = 3.261, P = 0.082] and no significant effect of the “Genotype” factor [F(1, 27) = 1.785, P > 0.05] or of the “Ceftriaxone” factor [F(1, 27) < 1] on the mRNA levels of Mme in the hippocampus of the rats.
Analysis of mRNA levels of CEF target genes in the striatum
Abundance of mRNA species in the striatum is summarized in Fig. 4.
Fig. 4.

Effects of CEF on mRNA levels of Ace2, Bace1, Ece1, Epo, Ide, Mme, and Actb in the striatum of OXYS and Wistar rats. The data represent the relative mRNA levels of target genes to geometrical mean of B2m, Hprt1, and Mapk6 mRNA levels. The data are expressed as the Mean ± S.E.M. of the values obtained in an independent group of animals (n = 6–9 per group). Statistically significant differences: *P < 0.05 versus “Wistar + saline” group; #P < 0.05 versus “Wistar + CEF” group
Due to the lack of normal distribution of the data in the studied groups, Kruskal–Wallis ANOVA by Ranks was applied and revealed no significant differences between the groups for mRNA levels of Ace2 [H(3, 30) = 2.069, P > 0.05], Bace1 [H(3, 30) = 0.623, P > 0.05], Epo [H(3, 27) = 2.162, P > 0.05], and Mme [H(3, 31) = 2.085, P > 0.05]. For Ece1, Kruskal–Wallis ANOVA by Ranks showed a significant difference between the groups [H(3, 31) = 8.654, P = 0.034]. Multiple comparisons of mean ranks for all groups revealed that the parameter was significantly higher in the rats of “OXYS + CEF” group versus that in rats of “Wistar + saline” group (P < 0.05).
Ide
Two-way ANOVA did not show a significant effect of the “Genotype” [F(1, 29) = 1.385, P > 0.05] or “Ceftriaxone” [F(1, 29) < 1] factor as well as of the interaction between the factors [F(1, 29) < 1].
Actb
According to two-way ANOVA, there was a significant influence of the “Genotype” factor [F(1, 25) = 7.084, P < 0.05], and no significant effects of the “Ceftriaxone” factor [F(1, 25) = 2.401, P > 0.05] and of the interaction between the factors [F(1, 25) < 1] on the mRNA levels of Actb in the striatum of the rats. LSD post hoc test revealed that the parameter was significantly higher in the rats of “OXYS + CEF” group versus that in Wistar rats of “Wistar + saline” (P < 0.05) or “Wistar + CEF” (P < 0.05) group.
Analysis of mRNA levels of CEF target genes in the frontal cortex
The results on the mRNA abundance in the frontal cortex are summarized in Fig. 5.
Fig. 5.
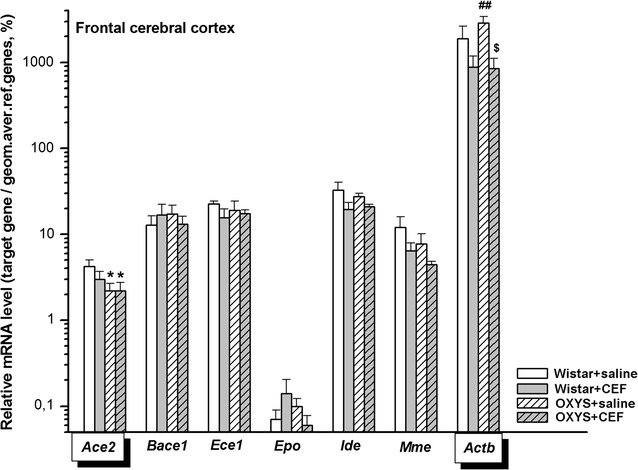
Effects of CEF on mRNA levels of Ace2, Bace1, Ece1, Epo, Ide, Mme, and Actb in the frontal cerebral cortex of OXYS and Wistar rats. The data represent the relative mRNA levels of target genes to geometrical mean of B2m, Hprt1, and Mapk6 mRNA levels. The data are expressed as the Mean ± S.E.M. of the values obtained in an independent group of animals (n = 6–9 per group). Statistically significant differences: *P < 0.05 versus “Wistar + saline” group; ##P < 0.01 versus “Wistar + CEF” group; $P < 0.05 versus “OXYS + saline” group
Ace2
According to two-way ANOVA, there was a significant influence of the “Genotype” factor [F(1, 24) = 4.416, P < 0.05], and no significant effect of the “Ceftriaxone” factor [F(1, 25) < 1] or of the interaction between the factors [F(1, 25) < 1] on the mRNA levels of Ace2 in the frontal cortex of the rats. LSD post hoc test revealed that the parameter was significantly higher in the rats of “Wistar + saline” group versus that in rats of “OXYS + saline” (P < 0.05) or “OXYS + CEF” (P < 0.05) group.
Bace1
Due to the lack of normal distribution of the data in the studied groups, Kruskal–Wallis ANOVA by Ranks was applied and revealed no significant differences between the groups [H(3, 30) = 0.780, P > 0.05].
Ece1
Two-way ANOVA did not show a significant effect of the “Genotype” [F(1, 26) < 1] or “Ceftriaxone” [F(1, 26) = 1.587, P > 0.05] factor as well as of the interaction between the factors [F(1, 26) < 1].
Epo
According to two-way ANOVA, there was a tendency for influence of the interaction between the factors [F(1, 22) = 3.288, P = 0.083] and no significant effects of the “Genotype” [F(1, 22) < 1] and “Ceftriaxone” [F(1, 22) < 1] factors on the mRNA levels of Epo in the frontal cortex of the rats.
Ide
Two-way ANOVA did not show a significant effect of the “Genotype” [F(1, 22) < 1] or “Ceftriaxone” [F(1, 22) = 2.740, P > 0.05] factor as well as of the interaction between the factors [F(1, 22) < 1] on the mRNA levels of Ide in the frontal cortex of the rats.
Mme
Two-way ANOVA did not show a significant effect of the “Genotype” [F(1, 25) = 1.404, P > 0.05] or “Ceftriaxone” [F(1, 25) = 2.898, P > 0.05] factor as well as of the interaction between the factors [F(1, 25) < 1] on the mRNA levels of Mme in the frontal cortex of the rats.
Actb
According to two-way ANOVA, there was a significant influence of the “Ceftriaxone” factor [F(1, 21) = 9.455, P < 0.01], and no significant effect of the “Genotype” factor [F(1, 21) < 1] or of the interaction between the factors [F(1, 21) = 1.102, P > 0.05] on the mRNA levels of Actb in the frontal cortex of the rats. LSD post hoc test revealed that the parameter was significantly higher in the rats of “OXYS + saline” group versus that in Wistar rats of “Wistar + CEF” (P < 0.01) or “OXYS + CEF” (P < 0.05) group.
Effects on body weight gain and biochemical parameters of plasma
We found no adverse side effects of CEF treatment on general condition and body weight gain of the animals. Body weight at sacrifice was 485.00 ± 9.81, 436.57 ± 18.18, 368.20 ± 18.49, or 364.60 ± 7.08 g in rats of “Wistar + saline”, “Wistar + CEF”, “OXYS + saline”, or “OXYS + CEF” group, respectively. Biochemical features of the groups are summarized in Table 1. While the levels of uric acid, creatinine, phosphate, and total bilirubin in plasma were not significantly affected by the factors, significant differences were revealed in the activity of ALT and AST, levels of Ca2+ and parameters of plasma lipid profile.
Table 1.
Biochemical parameters of plasma in Wistar and OXYS rats treated chronically with CEF
| Biochemical parameters | Wistar | OXYS | ||
|---|---|---|---|---|
| Control (saline) | CEF (100 mg/kg/day, 36 days) | Control (saline) | CEF (100 mg/kg/day, 36 days) | |
| Uric acid (mmol/L) | 7.48 ± 0.41 | 7.56 ± 0.26 | 7.70 ± 0.18 | 8.19 ± 0.42 |
| Creatinine (µmol/L) | 59.21 ± 0.44 | 57.70 ± 1.04 | 60.55 ± 0.71 | 58.74 ± 0.93 |
| ALT, U/L | 72.33 ± 4.03 | 70.56 ± 4.72 | 79.14 ± 2.58 | 64.00 ± 1.77$ |
| AST, U/L | 132.66 ± 3.34 | 137.64 ± 6.01 | 187.13 ± 15.04***,### | 148.72 ± 4.51$$ |
| Total bilirubin (µmol/L) | 2.38 ± 0.21 | 1.99 ± 0.31 | 2.10 ± 0.21 | 1.79 ± 0.24 |
| Ca2+ (mmol/L) | 2.78 ± 0.03 | 2.70 ± 0.04 | 2.68 ± 0.03 | 2.59 ± 0.06** |
| Phosphate (mmol/L) | 2.49 ± 0.06 | 2.46 ± 0.10 | 2.58 ± 0.17 | 2.30 ± 0.06 |
| Total cholesterol (mmol/L) | 1.82 ± 0.06 | 1.74 ± 0.14 | 1.42 ± 0.14*,# | 1.56 ± 0.05 |
| Triglycerides, g/L | 1.07 ± 0.17 | 1.14 ± 0.12 | 1.30 ± 0.10 | 1.59 ± 0.15*,# |
| LDL-C (µmol/L) | 0.463 ± 0.075 | 0.495 ± 0.089 | 0.213 ± 0.079*,# | 0.200 ± 0.031*,## |
| HDL (mmol/L) | 0.869 ± 0.042 | 0.843 ± 0.066 | 0.704 ± 0.060* | 0.774 ± 0.045 |
Data are presented as the Mean ± S.E.M. of the values obtained in an independent group of animals (n = 7–8 per group)
Statistically significant differences: *P < 0.05, ***P < 0.001 versus “Wistar + saline” group; #P < 0.05, ##P < 0.01, ###P < 0.001 versus “Wistar + CEF” group; $P < 0.05, $$P < 0.01 versus “OXYS + saline” group
Two-way ANOVA showed significant influence of the “Genotype” factor [F(1, 27) = 13.857, P < 0.001] and the interaction of the factors [F(1, 27) = 6.069, P < 0.05] on AST activity, and there was a tendency for the “Ceftriaxone” factor [F(1, 27) = 3.603, P = 0.068]. CEF treatment significantly reduced the augmented AST activity in OXYS rats. Similarly, chronic treatment with CEF attenuated the activity of ALT in OXYS rats. Two-way ANOVA revealed significant effect of the “Ceftriaxone” factor [F(1, 27) = 5.603, P < 0.05] and a tendency for the interaction of the factors [F(1, 27) = 3.510, P = 0.072] but not of “Genotype” factor [F(1, 27) < 1] on ALT activity. For the levels of Ca2+, two-way ANOVA showed significant influence of the “Genotype” [F(1, 27) = 7.14, P < 0.05] and “Ceftriaxone” [F(1, 27) = 4.36, P < 0.05] factors while the interaction of the factors was insignificant [F(1, 27) < 1]. LSD post hoc test revealed that the parameter was significantly lower in the rats of “OXYS + CEF” group versus that in Wistar rats of “Wistar + saline” (P < 0.01) group.
Significant influence of the “Genotype” factor was found on the levels of total cholesterol [F(1, 27) = 6.985, P < 0.05], triglycerides [F(1, 27) = 6.055, P < 0.05], LDL-C [F(1, 27) = 13.622, P < 0.001], and HDL [F(1, 27) = 4.483, P < 0.05] while the effects of “Ceftriaxone” factor or the interaction of the factors were insignificant.
Discussion
In our laboratory, comparing Wistar to OXYS rats, we found that at the age of 5 months, OXYS rats demonstrated impaired novel object recognition and retarded learning in a T-maze test [14]. Furthermore, parallel studies illustrated that compared to Wistar, OXYS rats presented with age-related somatic alterations, early signs of mental decline similar to AD pathology, such as learning and memory deficits, gradual deterioration of cognitive function along with the neurodegenerative changes. At the age of 3 months, OXYS rats displayed significant associative learning deficits in a single-trial passive avoidance test [16]. Moreover, OXYS rats were characterized by a progressive decline in spatial learning and memory at the age of 3–16 m.o. in a Morris water maze [10, 13, 17]. Those cognitive deficits in OXYS rats occurred along with changes in the hippocampal synaptic plasticity including a deficit in long-term potentiation (LTP) [18] and demyelinating lesions detected using MRI starting at the age of 3 m.o. Furthermore, a 4% reduction in the brain volume was observed by the age of 12 m.o. [11]. Histological and immunohistochemical studies of young OXYS rats (3–5 m.o.) revealed neurodegenerative changes in the CA1, CA3, and dentate gyrus regions of the hippocampus including the increased percentage of dead or damaged neurons [15, 19, 20]. A trend of a decrease in the density of neuronal somas and fibers in the nigrostriatal DAergic system was also noted in OXYS rats [15]. At ultrastructural and molecular levels, young OXYS rats possessed mitochondrial and synaptic degeneration [12].
Additional studies illustrated marked extracellular and vascular Aβ deposits in the brain of OXYS rats that were detected at the age of 15–18 or 24 m.o., in rare cases—at the age of 3–7 m.o. Most brain structures (cerebral cortex, hippocampus, thalamus, hypothalamus, brain stem) except for the cerebellum were affected by Aβ plaque pathology, with the highest Aβ load in the cerebral cortex. Noteworthy that the levels of soluble oligomeric Aβ1–42 fractions, the most neurotoxic Aβ forms, were significantly augmented in OXYS rats at the age of 7, 12, and 24 m.o. [12]. Thus, one may conclude that Aβ metabolism is altered in OXYS rats and we may expect to find changes in the enzymes involved in Aβ production, degradation, or clearance in the rats even at young age.
Earlier, comparative SNP analyses disclosed the absence of specific for the early familial forms of AD mutations in App, Psen1, and Psen2 genes in the OXYS genome [12]. At the same time, comparison of OXYS and Wistar rats by next-generation RNA sequencing revealed significant differences in gene expression of more than 900 genes at the age of 5 m.o. (and more than 2000 genes at the age of 18 m.o.) in the prefrontal cortex. Most of those genes were related to neuronal plasticity, protein phosphorylation, Ca2+ homeostasis, hypoxia, immune processes, and apoptosis. Among genes related to Aβ metabolism, mRNA levels of Mme encoding neprilysin (NEP) were decreased in the prefrontal cortex of OXYS with age and did not differ from Wistar rats at the age of 5 m.o. [21]. Here we also did not find significant differences in mRNA levels of Mme in the frontal cortex as well as other brain structures studied in 5-month-old OXYS rats as compared to Wistar controls. Noteworthy, mRNA levels of Ece1 and Ide encoding other main amyloid-degrading enzymes, endothelin-converting enzyme (ECE)-1 and insulin-degrading enzyme (IDE), respectively, as well as of Bace1 encoding β-secretase BACE1 did not vary significantly between control OXYS and Wistar rats. These results indicate that the expression of genes encoding the main enzymes of Aβ synthesis and degradation is not substantially affected (or compensated) in OXYS rats at young age and other mechanisms are likely to define the progression of neurodegenerative pathology at this stage.
Although some studies showed that angiotensin-converting enzyme (ACE) degrades Aβ42 to Aβ40 [22–24], overall data on the contribution of ACE to Aβ clearance are rather contradictory. Current data provide conflicting information on whether activation or inhibition of ACE could be beneficial in AD. Moreover, the efficiency of hydrolysis of Aβ by ACE is much inferior to NEP, IDE, and ECE and hence correlations between ACE levels and AD may relate more to effects on the renin-angiotensin system and brain vascular changes than to a major contribution to Aβ clearance [9]. Hence, we did not include Ace into the present study. However, we found a significant decrease in mRNA levels of Ace2 encoding angiotensin-converting enzyme 2 (ACE2) in the frontal cortex and hypothalamus of control OXYS rats. These results are in a good agreement with previous findings demonstrating that ACE2 activity was significantly reduced in the brain of AD patients compared with age-matched controls and correlated inversely with levels of Aβ and phosphorylated tau [25], ACE2 activity also showed a tendency to decrease in the serum of AD patients compared with normal controls [24]. ACE2 is considered to influence Aβ metabolism as converting the longer and neurotoxic forms of Aβ (Aβ43) to the shorter, less toxic forms of Aβ [24] as well as ameliorating Aβ-induced inflammatory response [26]. Thus, we suggest that the disturbance in Ace2 expression in the brain of OXYS may partially contribute to Aβ metabolism alterations in rats of this strain.
We also revealed that the expression levels of Actb encoding β-actin varied substantially between the experimental groups and brain structures in our experiment, hence we further analyzed Actb as a target gene. β-actin specifically controls cell growth and migration [27] as well as axon growth and collateral branch formation in neurons [28]. A significant decrease of Actb mRNA levels in the amygdala of OXYS rats may indicate the marked neurodegenerative changes at neurite level.
Here we also studied the potential effects of the antibiotic drug CEF, which has neuroprotective activity, on the modulation of mRNA levels of genes related to the system of Aβ metabolism in the brain. Recently, we found that CEF revised cognitive and neuronal deficits in OXYS rats [15]. In the present study we applied the dose of 100 mg/kg/day and treatment course of 36 daily drug injections that appeared to be effective for correction of both behavioral and neuronal deficits in OXYS rats. As in the previous study, we found no adverse side effects on general condition and body weight gain of the animals. Moreover, here we checked the effects of CEF on biochemical parameters of plasma. Similar to our previous results [14], interstrain differences were revealed in the activity of AST and ALT, levels of Ca2+, and parameters of plasma lipid profile. CEF did not have any significant effects on the indices of Wistar rats and normalized levels of AST and ALT in OXYS rats. The latter finding may indicate hepatoprotective properties of CEF.
The effects of CEF on AD-related pathology were also studied earlier using transgenic mouse models, namely, 3xTg-AD [29] and APPPS1 [30] strains. In middle aged 12-month-old 3xTg-AD mice, CEF rescued cognitive decline but had no pronounced effect on APP processing, overall Aβ species levels (except for the increase in Aβ40 levels in the CEF-treated mice), or plaque pathology. CEF neuroprotective effects were attributed to the restoration of synaptic proteins and inhibition of tau accumulation via attenuation of glutamatergic excitotoxicity induced by Aβ deposits [29]. The study by Hefendehl et al. [30] was focused on the pathological changes in glutamate dynamics in the immediate vicinity of Aβ deposits in APPPS1 transgenic mice and altered neuronal activity in such microenvironment that were corrected by CEF treatment. Noteworthy, both studies implied the models of advanced stages of AD-like pathology with highly expressed Aβ plaque pathology.
Here we found that CEF influenced mRNA levels of Bace1, Mme, Ece1, Ide, Ace2, Epo, and Actb. The following effects may contribute to neuroprotective properties of CEF as decreasing Aβ burden: CEF diminished Bace1 mRNA levels in the hypothalamus, augmented Ide and Mme mRNA levels in the amygdala as well as the levels of Ece1 in the striatum of OXYS rats. Moreover, CEF treatment increased Epo mRNA levels in the amygdala. Erythropoietin (EPO) encoded by Epo gene is a growth hormone and cytokine with wide range of biological activity. EPO and its receptors are found in the CNS of mammals, and are essential for neurodevelopment, adult neurogenesis, and neuroprotection. In animal models of neurodegenerative diseases including AD, neuroprotective and neuroregenerative mechanisms of EPO are related to inhibiting apoptosis, reducing oxidative stress and inflammation, promoting the angiogenesis and neurogenesis, and maintaining blood brain barrier (BBB) integrity [31]. The latter effect might be especially important mechanism in OXYS model due to neurovascular disturbances registered in OXYS rats [10, 32, 33]. Moreover, recent evidence suggests that cerebrovascular degeneration contributes to the pathogenesis of AD by causing faulty clearance of Aβ across the BBB [34]. It should be also noted that recently we revealed synergistic effect of combined treatment with CEF and exogenous EPO on neuroprotection and improvement in cognition in a MPTP-induced PD rat model [35].
Interestingly, these results showed that CEF had the most pronounced effect in the amygdala augmenting Epo, Ide, and Mme mRNA levels. Many studies have determined the main cognitive impairment in the preclinical phase of AD is episodic memory, in which a network of the hippocampus, entorhinal cortex, and amygdala are involved. In a recent study by Rasero et al. [36], the authors found progressive brain dysfunction using Diffusion-Tensor brain networks across severity stages in AD, namely, variations in connectivity patterns that start in a module network clearly associated with memory function (including part of the hippocampus, amygdala, entorhinal cortex, fusiform gyrus, inferior and middle temporal gyrus, parahippocampal gyrus and temporal pole) and later on, alterations were found widespread along the entire brain. One may suggest that CEF effects in the amygdala found in our study may be related to restoration of episodic memory in the novel object recognition test in OXYS rats [15].
At present, the CNS target responsible for CEF neuroprotective properties is unknown. Some studies have suggested possible interactors such as GFAP [37], alpha-synuclein [38], or just recently revealed high-affinity binding partner of CEF, metallo-β-lactamase domain-containing protein 1 (MBLAC1) [39], although further pathways and their contribution to CEF neuroprotection via modulation of gene and/or protein expression should be further investigated. On the other hand, it is generally assumed that the up-regulation of glutamate transporter EAAT2 in glial cells is responsible for CEF-mediated neuroprotection via its ability to reduce extracellular glutamate levels and subsequent excitotoxicity [40]. This effect was found to involve NF-κB-mediated EAAT2 promoter activation [41]. Further studies revealed another mechanism of CEF neuroprotection against oxidative glutamate toxicity. It was related to robust induction of the expression of the glutamate/cystine exchanger, system xc− , through Nrf2/ARE transcriptional program in different neuronal or glial cell culture models [42]. The present study disclosed the novel targets for CEF action that are expressed in neuronal or/and glial (Bace1, Mme, Ece1, Ide, Ace2, and Epo) or endothelial (Ece1, Ace2) cells [2, 9, 43, 44]. In this study, most changes in mRNA levels after CEF treatment were registered in OXYS but not in Wistar rats thus emphasizing the relation of those effects to AD-like pathogenetic processes. The exact mechanisms through which CEF modulates the expression of those genes and their cross-talk with the known mechanisms of CEF neuroprotection as well as brain structure specificity of the effects need to be elucidated in further studies. We cannot exclude the interaction of CEF with epigenetic elements directly involved in the regulation of gene expression.
Conclusion
The results of the study demonstrated a significant decrease in mRNA levels of Ace2 in the frontal cortex and hypothalamus in control OXYS rats compared to control Wistar rats while mRNA levels of Mme, Ece1, and Ide encoding main amyloid-degrading enzymes, as well as of Bace1 encoding β-secretase BACE1 in the brain structures studied did not vary significantly between control OXYS and Wistar rats at young age of 5 m.o. A profound reduction of Actb mRNA levels found in the amygdala of OXYS rats may indicate the marked neurodegenerative changes at neurite level.
The study on the potential effects of CEF in the modulation of mRNA levels of genes related to the system of Aβ metabolism in the brain revealed that CEF diminished Bace1 mRNA levels in the hypothalamus, augmented Ide, Mme, and Epo mRNA levels in the amygdala as well as the levels of Ece1 in the striatum. Thus, those findings disclosed novel targets for CEF action that might be involved into neuroprotective mechanisms at early, pre-plaque stages of AD-like pathology development.
Methods
Experimental animals
14-week-old male Wistar rats and OXYS male rats of the same age from The Federal Research Center “Institute of Cytology and Genetics”, Siberian Branch of the Russian Academy of Sciences (Novosibirsk, Russia) were used. Rats were housed in groups of five in acrylic cages (40 × 60 × 20 cm) in an animal room under standard conditions (a natural light–dark cycle (16 h light and 8 h dark), temperature: 18–22 °C, relative humidity: 50–60%, standard food and water ad libitum). Each animal was handled for 5 min/day on three consecutive days, before taking into experiment. Rats were divided into four experimental groups: Control (Saline-treated) Wistar males (“Wistar + saline” group, n = 10), Ceftriaxone-treated at a dose of 100 mg/kg Wistar males (“Wistar + CEF” group, n = 10), Control (Saline-treated) OXYS males (“OXYS + saline” group, n = 10), and Ceftriaxone-treated at a dose of 100 mg/kg OXYS males (“OXYS + CEF” group, n = 10).
General procedures and drug administration
Ceftriaxone was purchased from Roche (Switzerland). Starting from the age of 14-week old (day 1), rats received 36 daily intraperitoneal (i.p.) injections of saline (1 mL/kg; “Wistar + saline” and “OXYS + saline” groups) or 36 daily i.p. injections of CEF (100 mg/kg/day). Rats were weighed weekly during the experiment to correct drug dosages. The rationale behind the CEF dosage (100 mg/kg/day) adopted in the current study was based on our recent study showing neuroprotective effects of CEF in correcting behavioral and neuronal deficits in OXYS rats [15].
On the day 37 the rats were sacrificed by exposure to CO2 and decapitation. Although certain influence of hypoxia on mRNA levels of some of the target genes were found earlier [45, 46], those effects were registered after prolonged hypoxic treatment (for at least 24 h). In our experiment, rats were sacrificed within 5 min after exposure to CO2. To our knowledge, no significant effects of hypoxia on the expression of the genes involved into the present study were observed after such a short period of exposure. Immediately after sacrifice, blood samples were taken for biochemical assays and brain structures (frontal cortex, hippocampus, amygdala, hypothalamus, and striatum) were rapidly dissected on ice, frozen in liquid nitrogen, and stored at − 70 °C until the total RNA extraction.
Quantitative real-time PCR (qPCR)
The relative amount of target mRNA was measured by qPCR.
Sequences of specific primers for the reference genes (Actb, B2m, Hprt1, Mapk6) and target genes (Ace2, Bace1, Ece1, Epo, Mme, and Ide) were designed using the Primer 3 (NCBI) (Table 2). Specificity of the primers was checked by search of homologous sequences in rat Refseq mRNA database by means of the software PrimerBlast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Each pair of primers was tested using RT-PCR with fluorescent SYBR Green dye on the cDNA matrix of known structure in the thermocycler CFX96 (Bio-Rad, USA) followed by determination of the melting temperature of the reaction product. The PCR product of each primer pair was analyzed in polyacrylamide gel electrophoresis as well to verify the quality and length of produced amplicons.
Table 2.
Reference and target genes PCR primer sequences (5′–3′), amplicon size
| Symbol | Sequence (5′–3′) | Amplicon size (bp) | |
|---|---|---|---|
| Reference genes | |||
| Actb | GTTCGCCATGGATGACGATATCG | CATCACACCCTGGTGCCTAG | 139 |
| B2m | TGCCATTCAGAAAACTCCCCA | GAGGTGGGTGGAACTGAGACA | 104 |
| Hprt1 | ACAGGCCAGACTTTGTTGGA | TGGCTTTTCCACTTTCGCTG | 123 |
| Mapk6 | CTCGATGAGTCGGAGAAGTCCC | TGTTGGCTGACAGGTGTCTCT | 115 |
| Target genes | |||
| Bace1 | GGACAACCTGAGGGGAAAGTC | GTACTGGACAGCTGCCTTTGGTA | 168 |
| Ace2 | TCATTCGATATTACACAAGGACCA | AGCAACTTCTGCCCAGCTT | 130 |
| Mme | GCTGTGGGGAGGCTTTATGT | AATTGCCAGGGCCTTCTCTT | 165 |
| Ece1 | TTAGCGGGAGGTGCATCCAAA | CCTCGGAGAGTGAGTCCACC | 111 |
| Ide | GGCCTGAGCTATGATCTCCA | GATCGCATGTACGCCTCTTTG | 164 |
| Epo | ATGGGGGTGCCCGAACG | TACCTCTCCAGAACGCGACT | 122 |
Because of high variability, using just one reference gene should be avoided [47]. Four reference genes which are recommended by Qiagen (http://www.sabiosciences.com/rt_pcr_product/HTML/PARN-000Z.html#function) and other researchers [48, 49] were chosen as the most stable for the assessment of relative mRNA levels in rat brain structures. However, the values for the Actb in our experiment appeared to vary substantially between the experimental groups and brain structures, hence it was further analyzed as target gene. All the primers were purchased from Medigen Ltd. (Novosibirsk, Russia); exon spanning primer sets including large introns were used to eliminate the detection of residual genomic DNA.
Extraction of total RNA from fresh-frozen tissue was performed using RNeasy Mini kit in accordance with the manufacturer’s instructions (Qiagen, Germany). The amount of brain tissue of each sample was about 20 mg. The concentration of total RNA was determined using a NanoDrop 1000 microspectrophotometer (Nano-Drop Technologies, Wilmington, DE, USA). RNA sample quality was evaluated using the A260/A280 absorbance. The minimal RNA concentration was 24.6 ng/μl, the maximal—88.6 ng/μl. The amount of about 250 ng of total RNA was used in the further RT-PCR reactions.
The reverse transcriptase reaction volume was 30 µL. Composition of the reaction mixture for reverse transcription reaction included: 5 µL of RNA sample, 0.3 μM forward and reverse primers, 64 units of RNase inhibitor (RNasin®, Promega, USA), 160 units of reverse transcriptase (Biosan, Russia), 0.8 mM dNTP (Medigen, Russia), 1× RT Buffer (Biosan, Russia), 11.6 µL H2O. The protocol of reverse transcription consisted of 3 stages: 25 °C for 5 min, 42 °C for 30 min, and 95 °C for 5 min.
Polymerase chain reaction was carried out in 25 µL of reaction mixture. Reaction mixture contained: 0.2 mM dNTP (Medigen, Russia), 2.5 mM MgCl2 (Medigen, Russia), 0.04 μM forward and reverse primers, 1/500 V (V-reaction volume) SYBR Green I dye (Sigma-Aldrich, USA), 1x buffer for Taq DNA polymerase (Medigen, Russia), 1 unit of Smart Taq DNA polymerase (Medigen, Russia), 5 µL of cDNA. Amplification was started by an initial activation of the enzyme at 95 °C for 180 s. Each amplification cycle included denaturation at 95 °C for 10 s, annealing at 58 °C for 30 s, and elongation at 72 °C for 20 s with fluorescence signal acquisition. The maximum cycle threshold (Ct) value was set at 40.
Raw data were exported from the thermocycler CFX96 and the quantitative analysis of the qRT-PCR data was performed according to Ruijter et al. [50] to calculate the starting concentration (N0) of each cDNA template. Due to high variability of Actb mRNA values, levels of the target genes were normalized to the geometric average of three reference gene mRNA levels (B2m, Hprt1, Mapk6).
Biochemical assays
Trunk blood of a rat was collected into heparinized Green Vac-Tubes (Green Cross MS Corp., Korea) right after sacrifice, centrifuged for 20 min at 3000 rpm at + 4 °C, plasma was stored at − 24 °C until assay. Plasma levels of the uric acid, creatinine, calcium, phosphate, total cholesterol, low-density lipoprotein cholesterol (LDL-C), triglycerides and high-density lipoproteins (HDL) as well as the plasma activity of aminotransferases [aspartate aminotransferase (AST), alanine aminotransferase (ALT)] and the levels of total bilirubin were measured using clinical chemistry analyzer Konelab 30i and Konelab kits (Thermo Fisher Scientific Inc., USA) according to the manufacturer’s instructions [14].
Data analysis
All results were presented as mean ± SEM and compared using two-way ANOVA followed by post hoc Fisher LSD test or nonparametric Kruskal–Wallis ANOVA followed by multiple comparisons of mean ranks for all groups (in case of the lack of normal distribution of the data in the studied groups). The independent variables for two-way ANOVA were Genotype (Wistar or OXYS strain) and Treatment (Saline or CEF). The level of significance was defined as P < 0.05. STATISTICA 10.0 software was used to perform all the statistical analyses.
Declarations
About this supplement
This article has been published as part of BMC Neuroscience Volume 19 Supplement 1, 2018: Selected articles from Belyaev Conference 2017: neuroscience. The full contents of the supplement are available online at https://bmcneurosci.biomedcentral.com/articles/supplements/volume-19-supplement-1.
Authors’ contributions
MT made substantial contributions to design of the study and interpretation of data and was a major contributor in writing the manuscript; TA made significant contributions to design of the study and data interpretation, coordinated experimental activities and work on the manuscript; VB processed raw data of quantitative real time PCR, performed statistical analysis and helped to draft the manuscript; LF made substantial contributions to data analysis and interpretation; EP and AA performed quantitative real time PCR; NB made substantial contributions to conception and design of the study and proofreading of the text; SK, NK, and LA contributed significantly to conception of the study and participated in interpretation of data. All authors read and approved the final manuscript.
Acknowledgements
This work was partially supported by budget from the Project No. 0538-2014-0009 of the SRIPhBM, by NSU: Academic Strategic Unit “Neuroscience in Translational Medicine”, and Grant No. 15-04-05593-a from the Russian Foundation for Basic Research (Russia). We thank Dr. Svetlana Ya. Zhanaeva (SRIPhBM) for i.p. injections of ceftriaxone or saline to rats and Ms. Inna Yarantseva (NSU) for technical support with quantitative real time PCR.
Competing interests
The authors declare that they have no competing interests. The authors alone are responsible for the content and writing of the paper.
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All experimental procedures were performed according to the NIH Guide for the Care and Use of Laboratory Animals and approved by the Animal Care Committee of the Federal State Budgetary Scientific Institution “Scientific Research Institute of Physiology and Basic Medicine”. All efforts were made to minimize the number of animals used and their suffering.
Funding
Publication of this article has been funded by the Federal State Budgetary Scientific Institution “Scientific Research Institute of Physiology and Basic Medicine”.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Aβ
amyloid-beta
- ACE
angiotensin-converting enzyme
- Ace
gene of ACE
- ACE2
angiotensin-converting enzyme 2
- Ace2
gene of ACE2
- AD
Alzheimer’s disease
- Aktb
gene of β-actin
- ALT
alanine aminotransferase
- APP
amyloid precursor protein
- App
gene of APP
- ARE
antioxidant response element
- AST
aspartate aminotransferase
- B2m
gene of beta-2 microglobulin
- BACE1
β-secretase
- Bace1
gene of BACE1
- BBB
blood brain barrier
- CEF
ceftriaxone
- CNS
central nervous system
- EAAT2
excitatory amino acid transporter 2
- ECE1
endothelin-converting enzyme 1
- Ece1
gene of ECE1
- EPO
erythropoietin
- Epo
gene of EPO
- GFAP
glial fibrillary acidic protein
- HDL
high-density lipoproteins
- Hprt1
gene of hypoxanthine phosphoribosyltransferase 1
- IDE
insulin-degrading enzyme
- Ide
gene of IDE
- i.p.
intraperitoneal
- LDL-C
low-density lipoprotein cholesterol
- LTP
long-term potentiation
- Mapk6
gene of mitogen-activated protein kinase 6
- MBLAC1
metallo-β-lactamase domain-containing protein 1
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- Mme
gene of membrane metalloendopeptidase
- NEP
neprilysin
- NF-κB
nuclear factor kappa B
- Nrf2
nuclear factor erythroid 2-related factor 2
- Psen1
gene of presenilin 1
- Psen2
gene of presenilin 2
Contributor Information
Maria A. Tikhonova, Email: tikhonovama@physiol.ru, Email: mar-a-tikh@mail.ru
Tamara G. Amstislavskaya, Email: amstislavskayatg@physiol.ru
Victor M. Belichenko, Email: belichenko@physiol.ru
Larisa A. Fedoseeva, Email: fedoseeva@bionet.nsc.ru
Sergey P. Kovalenko, Email: sp_kovalenko@yahoo.com
Ekaterina E. Pisareva, Email: katerina.pisareva@mail.ru
Alla S. Avdeeva, Email: leshchenkoalla@gmail.com
Nataliya G. Kolosova, Email: kolosova@bionet.nsc.ru
Nikolai D. Belyaev, Email: N.D.Belyaev@leeds.ac.uk
Lyubomir I. Aftanas, Email: liaftanas@gmail.com
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Singh SK, Srivastav S, Yadav AK, Srikrishna S, Perry G. Overview of Alzheimer’s disease and some therapeutic approaches targeting Abeta by using several synthetic and herbal compounds. Oxid Med Cell Longev. 2016;2016:7361613. doi: 10.1155/2016/7361613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguzzi A, Haass C. Games played by rogue proteins in prion disorders and Alzheimer’s disease. Science. 2003;302(5646):814–818. doi: 10.1126/science.1087348. [DOI] [PubMed] [Google Scholar]
- 4.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8(2):101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 5.Walsh DM, Selkoe DJ. A beta oligomers—a decade of discovery. J Neurochem. 2007;101(5):1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 6.Grimm MO, Mett J, Stahlmann CP, Haupenthal VJ, Zimmer VC, Hartmann T. Neprilysin and Abeta clearance: impact of the APP intracellular domain in NEP regulation and implications in Alzheimer’s disease. Front Aging Neurosci. 2013;5:98. doi: 10.3389/fnagi.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang DS, Dickson DW, Malter JS. beta-Amyloid degradation and Alzheimer’s disease. J Biomed Biotechnol. 2006;2006(3):58406. doi: 10.1155/JBB/2006/58406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nalivaeva NN, Belyaev ND, Turner AJ. New insights into epigenetic and pharmacological regulation of amyloid-degrading enzymes. Neurochem Res. 2016;41(3):620–630. doi: 10.1007/s11064-015-1703-1. [DOI] [PubMed] [Google Scholar]
- 9.Nalivaeva NN, Beckett C, Belyaev ND, Turner AJ. Are amyloid-degrading enzymes viable therapeutic targets in Alzheimer’s disease? J Neurochem. 2012;120(Suppl 1):167–185. doi: 10.1111/j.1471-4159.2011.07510.x. [DOI] [PubMed] [Google Scholar]
- 10.Kolosova NG, Stefanova NA, Sergeeva SV. OXYS rats: a prospective model for evaluation of antioxidant availability in prevention and therapy of accelerated aging and age-related cognitive decline. In: Gariepy Q, Menard R, editors. Handbook of cognitive aging: causes, proceses. NY: Nova Science; 2009. pp. 47–82. [Google Scholar]
- 11.Stefanova NA, Kozhevnikova OS, Vitovtov AO, Maksimova KY, Logvinov SV, Rudnitskaya EA, Korbolina EE, Muraleva NA, Kolosova NG. Senescence-accelerated OXYS rats: a model of age-related cognitive decline with relevance to abnormalities in Alzheimer disease. Cell Cycle. 2014;13(6):898–909. doi: 10.4161/cc.28255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefanova NA, Muraleva NA, Korbolina EE, Kiseleva E, Maksimova KY, Kolosova NG. Amyloid accumulation is a late event in sporadic Alzheimer’s disease-like pathology in nontransgenic rats. Oncotarget. 2015;6(3):1396–1413. doi: 10.18632/oncotarget.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefanova NA, Muraleva NA, Skulachev VP, Kolosova NG. Alzheimer’s disease-like pathology in senescence-accelerated OXYS rats can be partially retarded with mitochondria-targeted antioxidant SkQ1. J Alzheimers Dis. 2014;38(3):681–694. doi: 10.3233/JAD-131034. [DOI] [PubMed] [Google Scholar]
- 14.Tikhonova MA, Yu CH, Kolosova NG, Gerlinskaya LA, Maslennikova SO, Yudina AV, Amstislavskaya TG, Ho YJ. Comparison of behavioral and biochemical deficits in rats with hereditary defined or d-galactose-induced accelerated senescence: evaluating the protective effects of diosgenin. Pharmacol Biochem Behav. 2014;120:7–16. doi: 10.1016/j.pbb.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Tikhonova MA, Ho SC, Akopyan AA, Kolosova NG, Weng JC, Meng WY, Lin CL, Amstislavskaya TG, Ho YJ. Neuroprotective effects of ceftriaxone treatment on cognitive and neuronal deficits in a rat model of accelerated senescence. Behav Brain Res. 2017;330:8–16. doi: 10.1016/j.bbr.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Loskutova LV, Kolosova NG. Emotional state and one-trial learning in OXYS rats with hereditarily elevated production of oxygen radicals. Bull Exp Biol Med. 2000;130(8):746–748. doi: 10.1007/BF02766083. [DOI] [PubMed] [Google Scholar]
- 17.Stefanova NA, Fursova A, Kolosova NG. Behavioral effects induced by mitochondria-targeted antioxidant SkQ1 in Wistar and senescence-accelerated OXYS rats. J Alzheimers Dis. 2010;21(2):479–491. doi: 10.3233/JAD-2010-091675. [DOI] [PubMed] [Google Scholar]
- 18.Beregovoy NA, Sorokina NS, Starostina MV, Kolosova NG. Age-specific peculiarities of formation of long-term posttetanic potentiation in OXYS rats. Bull Exp Biol Med. 2011;151(1):71–73. doi: 10.1007/s10517-011-1262-7. [DOI] [PubMed] [Google Scholar]
- 19.Maksimova KY, Logvinov SV, Stefanova NA. Morphological characterization of Oxys and Wistar rat hippocampus in the aging process. Morfologiia. 2015;147(3):11–16. [PubMed] [Google Scholar]
- 20.Rudnitskaya EA, Maksimova KY, Muraleva NA, Logvinov SV, Yanshole LV, Kolosova NG, Stefanova NA. Beneficial effects of melatonin in a rat model of sporadic Alzheimer’s disease. Biogerontology. 2015;16(3):303–316. doi: 10.1007/s10522-014-9547-7. [DOI] [PubMed] [Google Scholar]
- 21.Stefanova NA, Korbolina EE, Ershov NI, Rogaev EI, Kolosova NG. Changes in the transcriptome of the prefrontal cortex of OXYS rats as the signs of Alzheimer’s disease development. Vavilov J Genet Breed. 2015;19(4):445–454. doi: 10.18699/VJ15.059. [DOI] [Google Scholar]
- 22.Zou K, Maeda T, Watanabe A, Liu J, Liu S, Oba R, Satoh Y, Komano H, Michikawa M. Abeta42-to-Abeta40- and angiotensin-converting activities in different domains of angiotensin-converting enzyme. J Biol Chem. 2009;284(46):31914–31920. doi: 10.1074/jbc.M109.011437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou K, Yamaguchi H, Akatsu H, Sakamoto T, Ko M, Mizoguchi K, Gong JS, Yu W, Yamamoto T, Kosaka K, et al. Angiotensin-converting enzyme converts amyloid beta-protein 1-42 (Abeta(1-42)) to Abeta(1-40), and its inhibition enhances brain Abeta deposition. J Neurosci. 2007;27(32):8628–8635. doi: 10.1523/JNEUROSCI.1549-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, Liu J, Miura Y, Tanabe C, Maeda T, Terayama Y, Turner AJ, Zou K, Komano H. Conversion of Abeta43 to Abeta40 by the successive action of angiotensin-converting enzyme 2 and angiotensin-converting enzyme. J Neurosci Res. 2014;92(9):1178–1186. doi: 10.1002/jnr.23404. [DOI] [PubMed] [Google Scholar]
- 25.Kehoe PG, Wong S, Al Mulhim N, Palmer LE, Miners JS. Angiotensin-converting enzyme 2 is reduced in Alzheimer’s disease in association with increasing amyloid-beta and tau pathology. Alzheimers Res Ther. 2016;8(1):50. doi: 10.1186/s13195-016-0217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu X, Lin R, Qiu Y, Yu P, Lei B. Overexpression of angiotensin-converting enzyme 2 ameliorates amyloid beta-induced inflammatory response in human primary retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2017;58(7):3018–3028. doi: 10.1167/iovs.17-21546. [DOI] [PubMed] [Google Scholar]
- 27.Bunnell TM, Burbach BJ, Shimizu Y, Ervasti JM. beta-Actin specifically controls cell growth, migration, and the G-actin pool. Mol Biol Cell. 2011;22(21):4047–4058. doi: 10.1091/mbc.E11-06-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moradi M, Sivadasan R, Saal L, Luningschror P, Dombert B, Rathod RJ, Dieterich DC, Blum R, Sendtner M. Differential roles of alpha-, beta-, and gamma-actin in axon growth and collateral branch formation in motoneurons. J Cell Biol. 2017;216(3):793–814. doi: 10.1083/jcb.201604117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zumkehr J, Rodriguez-Ortiz CJ, Cheng D, Kieu Z, Wai T, Hawkins C, Kilian J, Lim SL, Medeiros R, Kitazawa M. Ceftriaxone ameliorates tau pathology and cognitive decline via restoration of glial glutamate transporter in a mouse model of Alzheimer’s disease. Neurobiol Aging. 2015;36(7):2260–2271. doi: 10.1016/j.neurobiolaging.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Hefendehl JK, LeDue J, Ko RW, Mahler J, Murphy TH, MacVicar BA. Mapping synaptic glutamate transporter dysfunction in vivo to regions surrounding Abeta plaques by iGluSnFR two-photon imaging. Nat Commun. 2016;7:13441. doi: 10.1038/ncomms13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cevik B, Solmaz V, Yigitturk G, Cavusoglu T, Peker G, Erbas O. Neuroprotective effects of erythropoietin on Alzheimer’s dementia model in rats. Adv Clin Exp Med. 2017;26(1):23–29. doi: 10.17219/acem/61044. [DOI] [PubMed] [Google Scholar]
- 32.Agafonova IG, Kotel’nikov VN, Mischenko NP, Kolosova NG. Evaluation of effects of histochrome and mexidol on structural and functional characteristics of the brain in senescence-accelerated OXYS rats by magnetic resonance imaging. Bull Exp Biol Med. 2011;150(6):739–743. doi: 10.1007/s10517-011-1238-7. [DOI] [PubMed] [Google Scholar]
- 33.Agafonova IG, Kolosova NG, Mishchenko NP, Chaikina EL, Stonik VA. Effect of histochrome on brain vessels and research and exploratory activity of senescence-accelerated OXYS rats. Bull Exp Biol Med. 2007;143(4):467–471. doi: 10.1007/s10517-007-0158-z. [DOI] [PubMed] [Google Scholar]
- 34.Lee ST, Chu K, Park JE, Jung KH, Jeon D, Lim JY, Lee SK, Kim M, Roh JK. Erythropoietin improves memory function with reducing endothelial dysfunction and amyloid-beta burden in Alzheimer’s disease models. J Neurochem. 2012;120(1):115–124. doi: 10.1111/j.1471-4159.2011.07534.x. [DOI] [PubMed] [Google Scholar]
- 35.Huang CK, Chang YT, Amstislavskaya TG, Tikhonova MA, Lin CL, Hung CS, Lai TJ, Ho YJ. Synergistic effects of ceftriaxone and erythropoietin on neuronal and behavioral deficits in an MPTP-induced animal model of Parkinson’s disease dementia. Behav Brain Res. 2015;294:198–207. doi: 10.1016/j.bbr.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 36.Rasero J, Alonso-Montes C, Diez I, Olabarrieta-Landa L, Remaki L, Escudero I, Mateos B, Bonifazi P, Fernandez M, Arango-Lasprilla JC, et al. Group-level progressive alterations in brain connectivity patterns revealed by diffusion-tensor brain networks across severity stages in Alzheimer’s disease. Front Aging Neurosci. 2017;9:215. doi: 10.3389/fnagi.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruzza P, Vitale RM, Hussain R, Biondi B, Amodeo P, Sechi G, Siligardi G. Interactions of GFAP with ceftriaxone and phenytoin: SRCD and molecular docking and dynamic simulation. Biochim Biophys Acta. 2016;1860(10):2239–2248. doi: 10.1016/j.bbagen.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 38.Ruzza P, Siligardi G, Hussain R, Marchiani A, Islami M, Bubacco L, Delogu G, Fabbri D, Dettori MA, Sechi M, et al. Ceftriaxone blocks the polymerization of alpha-synuclein and exerts neuroprotective effects in vitro. ACS Chem Neurosci. 2014;5(1):30–38. doi: 10.1021/cn400149k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Retzlaff CL, Kussrow A, Schorkopf T, Saetear P, Bornhop DJ, Hardaway JA, Sturgeon SM, Wright J, Blakely RD. Metallo-beta-lactamase domain-containing protein 1 (MBLAC1) is a specific, high-affinity target for the glutamate transporter inducer ceftriaxone. ACS Chem Neurosci. 2017 doi: 10.1021/acschemneuro.7b00232. [DOI] [PubMed] [Google Scholar]
- 40.Sheldon AL, Robinson MB. The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neurochem Int. 2007;51(6–7):333–355. doi: 10.1016/j.neuint.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SG, Su ZZ, Emdad L, Gupta P, Sarkar D, Borjabad A, Volsky DJ, Fisher PB. Mechanism of ceftriaxone induction of excitatory amino acid transporter-2 expression and glutamate uptake in primary human astrocytes. J Biol Chem. 2008;283(19):13116–13123. doi: 10.1074/jbc.M707697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewerenz J, Albrecht P, Tien ML, Henke N, Karumbayaram S, Kornblum HI, Wiedau-Pazos M, Schubert D, Maher P, Methner A. Induction of Nrf2 and xCT are involved in the action of the neuroprotective antibiotic ceftriaxone in vitro. J Neurochem. 2009;111(2):332–343. doi: 10.1111/j.1471-4159.2009.06347.x. [DOI] [PubMed] [Google Scholar]
- 43.Xia H, Lazartigues E. Angiotensin-converting enzyme 2 in the brain: properties and future directions. J Neurochem. 2008;107(6):1482–1494. doi: 10.1111/j.1471-4159.2008.05723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noguchi CT, Wang L, Rogers HM, Teng R, Jia Y. Survival and proliferative roles of erythropoietin beyond the erythroid lineage. Expert Rev Mol Med. 2008;10:e36. doi: 10.1017/S1462399408000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Yang D, Zhang X, Li T, Li J, Tang Y, Le W. Hypoxia-induced down-regulation of neprilysin by histone modification in mouse primary cortical and hippocampal neurons. PLoS ONE. 2011;6(4):e19229. doi: 10.1371/journal.pone.0019229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerridge C, Kozlova DI, Nalivaeva NN, Turner AJ. Hypoxia affects neprilysin expression through caspase activation and an app intracellular domain-dependent mechanism. Front Neurosci. 2015;9:426. doi: 10.3389/fnins.2015.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai J, Li T, Huang B, Cheng H, Ding H, Dong W, Xiao M, Liu L, Wang Z. The use of laser microdissection in the identification of suitable reference genes for normalization of quantitative real-time PCR in human FFPE epithelial ovarian tissue samples. PLoS ONE. 2014;9(4):e95974. doi: 10.1371/journal.pone.0095974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walder RY, Wattiez AS, White SR, Marquez de Prado B, Hamity MV, Hammond DL. Validation of four reference genes for quantitative mRNA expression studies in a rat model of inflammatory injury. Mol Pain. 2014;10:55. doi: 10.1186/1744-8069-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Julian GS, Oliveira RW, Perry JC, Tufik S, Chagas R. Validation of housekeeping genes in the brains of rats submitted to chronic intermittent hypoxia, a sleep apnea model. PLoS ONE. 2014;9(10):e109902. doi: 10.1371/journal.pone.0109902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37(6):e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.


