Abstract
This protocol describes a workflow for creating structural models of proteins or protein complexes using distance restraints derived from cross-linking mass spectrometry experiments. The distance restraints are used (i) to adjust preliminary models that are calculated based on a homologous template and primary sequence and (ii) to select the model that is in best agreement with the experimental data. In the case of protein complexes, the cross-linking data is further used to dock the subunits to one another to generate models of the interacting proteins. Predicting models in such a manner has the potential to indicate multiple conformations and dynamic changes that occur in solution. This modeling protocol is compatible with many cross-linking workflows and uses open-source programs or programs that are free for academic users and do not require expertise in computational modeling. This protocol is an excellent additional application with which to use cross-linking results for building structural models of proteins. The established protocol is expected to take 6-12 days to complete, depending on the size of the proteins and the complexity of the cross-linking data.
Keywords: cross-linking mass spectrometry, cross-link, mass spectrometry, XL-MS, protein subunit, protein structure, protein structure prediction, molecular modeling, structural proteomics, proteomics, protein model, protein complex, I-TASSER, Xwalk, CPORT, HADDOCK
Introduction
Proteins are large, complex molecules that have many critical roles in cells, thereby determining the structure, characteristics and functions of tissues and organs. Information regarding the structure and dynamic behavior of proteins often reveals mechanistic details of their function, which are important in understanding how they carry out their designated tasks. Proteins either act alone or as part of complexes, and uncovering their interaction partners and patterns is of high priority in biochemical research. To date, more than 120,000 structures of proteins and their complexes have been deposited in the global Protein Data Bank (PDB) archive1. Nevertheless, the number of solved protein structures is an order of magnitude lower than the number of known protein sequences. Hybrid methods that combine various types of low-resolution experimental data, partial high-resolution structures and computational structure prediction techniques are promising to accelerate the accrual of protein structural data.
Chemical cross-linking mass spectrometry (XL-MS) is an increasingly popular method with high analytical sensitivity that complements high-resolution structural approaches such as X-ray crystallography (XRC) and electron microscopy2. XL-MS provides information that two specific amino acids, present in either the same protein or in different proteins, can come within a certain distance of each other in 3D space in solution. Cross-linkers are specific toward certain types of residues, and can be of different lengths to bridge residues of different distances3. Distance restraints between specific residues that have been experimentally determined during XL-MS experiments serve as ideal parameters to guide molecular modeling of proteins and protein complexes. This approach has been demonstrated during the characterization of several important biological systems, including the protein phosphatase 2A network4,5, the yeast mediator complex6 and the nuclear pore complex7.
A current limitation of cross-link-guided molecular modeling has, to date, been the complexity of the computational workflow. We therefore developed a simple approach that can be followed by researchers without in-depth knowledge of computational modeling8. In this approach, cross-linking (XL) mass spectrometry data are integrated into a modeling workflow to refine protein structure predictions, and to define the position and orientation of proteins within a complex. It is of note that the term ‘refinement,’ as used in this paper, refers to the process of recalculating the structural model using spatial restraints from XL-MS results. XL-derived distance restraints are first applied to guide the structural prediction of the individual subunits. Second, they are used to evaluate how well the predicted model fits to the experimental cross-linking data. Third, the XL-derived distance restraints are applied during the docking of the subunits into the model of the intact complex. The programs I-TASSER (iterative threading assembly refinement)9,10 and HADDOCK (high-ambiguity-driven protein–protein docking)11–13 were selected for protein structural prediction and docking, respectively, because (i) they both allow distance restraints to be specified by the user, and (ii) they were identified as top-scoring modeling algorithms in molecular-modeling challenges14,15. Moreover, HADDOCK allows conformational change of the individual subunits during complex docking of the protein backbone as well as of the side chains, therefore accounting for some protein flexibility.
Overview of the Procedure
In this protocol, we describe the step-by-step procedures for the modeling of proteins and protein complexes, and provide guidelines for evaluating the resulting structures and for selecting the most appropriate models. All applied algorithms and programs for data analysis and for modeling are freely available for academic users. The described approach is a valuable guide for structural analysis and will facilitate the generation of protein models, especially for cases in which NMR, XRC or similar techniques cannot be applied.
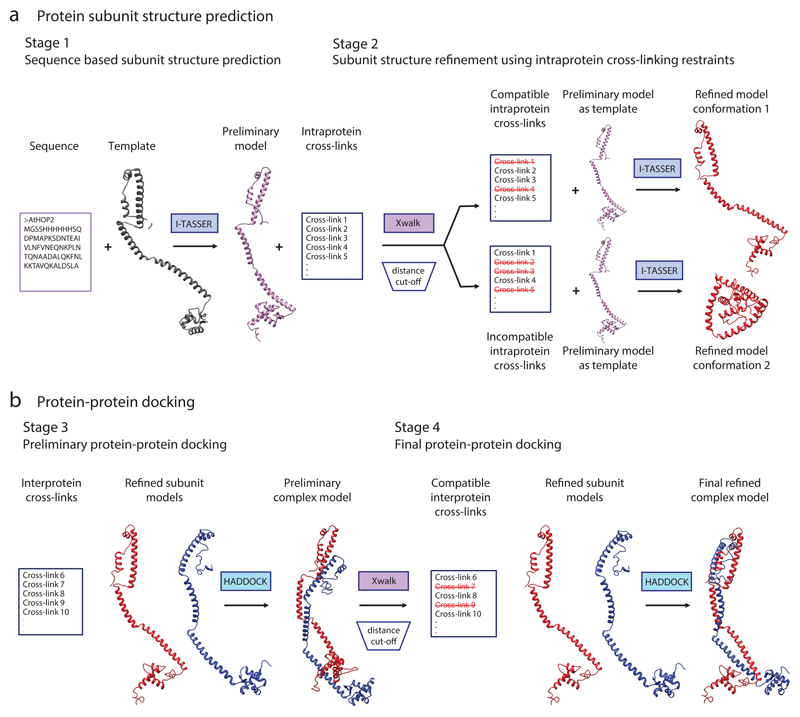
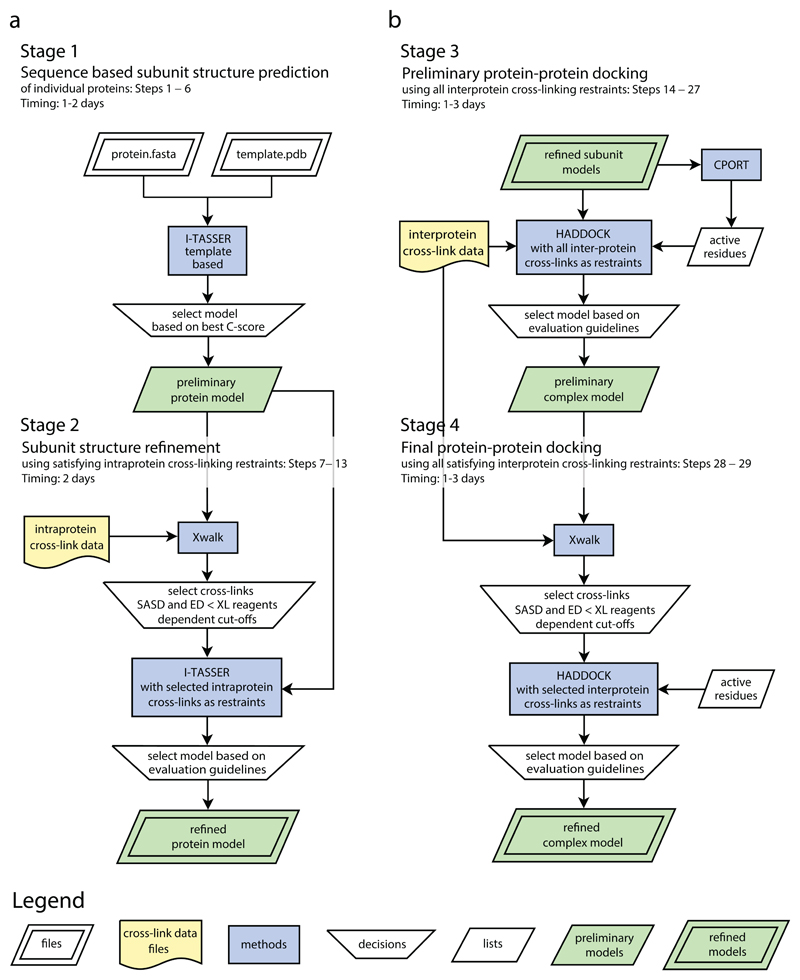
First, a preliminary model of the protein or protein subunit is predicted on the basis of a primary amino acid sequence and the solved structure of a homologous template (Steps 1–6, Figs. 1 and 2, Stage 1). This preliminary subunit model is then adjusted, using the experimental cross-links as distance restraints, to a conformation that is in better agreement with the cross-linking data derived from the protein in solution (Steps 7–13, Figs. 1 and 2, Stage 2). In the case of protein complexes, a structure of the complex is predicted from the individual subunit models, using cross-links between the subunits as distance restraints (Steps 14–29, Figs. 1 and 2, Stages 3 and 4). Evaluation guidelines for selecting the best model are provided in Box 1. Visualization of the final models displaying the experimental cross-links supports the evaluation, and is described in Steps 30–37.
Figure 1. Overview of the structural modeling workflow using XL-MS data.
(a) Schematic illustration of an example modeling procedure for protein subunit structure. Stage 1: a preliminary model of the protein is generated using the I-TASSER algorithm. Here, the structure is predicted on the basis of the primary amino acid sequence and the structure of a homologous template. Stage 2: the distances between the residues of the intraprotein cross-links are calculated within the preliminary model with Xwalk. The cross-links are subsequently divided according to their compatibility with the preliminary structure; those that bridge residues that are close together in 3D space are compatible with the model and are used to generate refined model 1. The cross-links that do not satisfy the model can be used to predict an alternative conformation (conformation 2, Stage 2). In this case, refined model conformation 1 corresponds to an open conformation, whereas refined model conformation 2 reflects a closed conformation. (b) Schematic illustration of the docking procedure for a protein dimer. Stage 3: a preliminary complex model is generated using the HADDOCK protein–protein docking tool, which combines the subunit structures obtained from Stage 2 with interprotein cross-link data. Stage 4: interprotein cross-links are divided according to their compatibility with the preliminary complex model. Compatible cross-links are used for a second round of protein complex docking with HADDOCK, generating the final refined protein complex model.
Figure 2. Cross-link data driven modeling workflow for predicting structures of proteins and protein complexes.
(a) Comparative modeling of individual protein structures using XL-MS data. Stage 1: preliminary structural models of the protein subunits are generated using the I-TASSER algorithm, the amino acid sequence and a structural template. Stage 2: after selection of the best model on the basis of the C-score, the distances of the cross-links are calculated with Xwalk. The cross-links are subsequently sorted according to their compatibility with the predicted model. Selected cross-links are used for a second I-TASSER structure prediction run. The best model is selected on the basis of the C-score and the compatibility of the cross-links with the model (Box 1). (b) Two-step protein–protein docking leading to the refined complex structure. Stage 3: the preliminary complex structure is built with the HADDOCK protein–protein docking tool using the refined individual protein models, the interprotein cross-link data and the active residue information, generated by CPORT. The best preliminary model is selected on the basis of the HADDOCK score and the compatibility of the cross-links with the model. Stage 4: Optionally, a refined complex model is generated by a second HADDOCK run using selected cross-links. The final refined protein complex model is chosen according to the evaluation guidelines. Adapted with permission from ref. 8, American Chemical Society. Flowcharts were generated using Dia v0.97.2.
Box 1. Evaluation guidelines for the selection of best structural models • TIMING 1 d.
For selection of an optimal model, different aspects are combined including the score from the prediction software and the compatibility of the model with the experimental cross-links.
1. Rank the models according to the HADDOCK or I-TASSER scores obtained in Steps 5,12 and 24 respectively.
2. Determine the number of compatible cross-links for all resulting models. The models that are in agreement with a higher number cross-links are considered to be the best. From this step, it may be possible to select the best model. If there are several models with similar scores and the same number of compatible cross-links, the best model can be predicted using the XL score as follows.
Δ CRITICAL STEP: You may find it useful to amalgamate all values in a table, such as in the evaluation.tsv file, which can be found in the Supplementary data 1-7.
3. Calculate XL-scores for a selection of the highest-scoring structural models. The XL-score can be calculated on the level of unique cross-links or PSMs by using either the SASDs or EDs calculated with Xwalk. Apply Xwalk to calculate the SASDs (or ED) of the experimental cross-links for each predicted structural model. See Steps 7-9 and Step 25. First sort unique cross-links by the calculated distance in ascending order. For each unique distance value, calculate the cumulative number of unique cross-links (or PSMs) up to this distance. Define an upper distance cutoff, depending on the spacer length (see Step 9), and discard cross-links exceeding the cutoff.
4. Create a graph with ‘distance between residues’ on the x axis and ‘cumulative counts’ on the y axis (Fig. 8).
5. Calculate the XL score as the area under the curve using, for example, the trapezoidal rule.
6. (Optional) Calculate a normalized XL-score by dividing the XL score by the product of the cutoff and the total number of counts. Alternatively, a normalized XL score can be calculated by dividing all XL cores by the highest obtained XL score. The normalized XL score will have a value between 0 and 1.
Δ CRITICAL STEP: A higher score indicates that a higher number of cross-links fit to a model with a shorter distance.
7. If a structure of a homologous complex is available, superimpose the resulting models in order to enable visual inspection of the differences and to reflect structural flexibility. This manual verification can serve as another possibility of evaluation. See Steps 36 and 37.
Generation of cross-linking data
The increasing popularity of XL-MS has resulted in several protocols describing experimental methodologies16,17. Leitner et al.16 published a protocol in connection with their xQuest/xProphet software pipeline, and a comparative XL-MS workflow was described by Schmidt and Robinson17 to delineate conformational changes of proteins, for example, upon ligand binding or post-translational modifications.
In a typical XL-MS experiment, a cross-linking reagent is added to a protein or protein complex of interest in order to form a covalent bond between two specific amino acid residues that are in close proximity to each other. The cross-linked protein is then enzymatically digested, and the resulting peptides are measured using liquid chromatography–MS (LC–MS)/MS. Careful analysis of the MS/MS data reveals which residues in the primary sequence have been cross-linked. This provides information that two specific amino acids, present in either the same protein or in different proteins, reside within a certain distance (depending on the chemical nature of the cross-linker) of each other in 3D space in solution. Besides the identification of interaction partners, protein interfaces and relative orientations in complexes, XL-MS has also been used to map entire protein interaction networks4,18–21 and to determine conformational changes induced by post-translational modifications and binding of small molecules17,22.
The digested sample of a cross-linked protein complex is of high complexity due to the mixture of cross-linked peptides, linear peptides and incompletely digested polypeptides. This presents a major challenge in the XL-MS workflow and currently limits its use as a high-throughput technique. Chromatographic methods that can be readily applied as XL-enrichment strategies currently include size-exclusion chromatography and strong cation exchange chromatography8,23,24. MS-cleavable XL reagents and stable isotope-labeled XL reagents facilitate the identification of XL peptides from the acquired MS/MS spectra17,25–29. Combining results from technical replicates and applying filters during the data interpretation will increase the confidence of the cross-linking data set. For example, one can filter MS2 spectra on the basis of the precursor mass accuracy and the probabilistic score provided by the algorithm of the software used. The number of peptide spectrum matches (PSMs) giving rise to a particular cross-link, as well as the number of technical replicates, can also be an indication of the confidence of each unique cross-link location.
Several search algorithms for XL identification have been developed in the past decade, including pLink30, xQuest16 and XlinkX26, as well as many more31–33. Interpretation of the obtained data is commonly facilitated by visualization tools such as customized circular, bar and network plots. These provide a clear schematic representation of the cross-links by integrating XL data, sequence data and domain annotations. Information on subunit orientation, major interaction sites (both inter- and intraprotein) and flexible regions can be easily communicated through such means. Several dedicated open-source software packages and web-based programs such as xVis34, xiNet35, ProXL36 and CX-circos (http://www.cx-circos.net) have been developed for this purpose.
To date, XL-MS-derived restraints have been integrated into molecular-modeling procedures in a limited number of publications4,7,37–39. XL results are often communicated by the graphical representations described above, and by mapping the cross-links onto experimentally solved or computationally predicted protein structures. As modeling performance strongly depends on the number of cross-links and their localization in the structures, a relatively large number of cross-links are required for molecular modeling. The number of identified cross-links for molecular modeling can be improved by performing replicate experiments and by the use of complementary XL reagents, as successfully demonstrated in our8 and other studies40.
Comparison with other approaches
A study concerning the organization of ribonucleoprotein particles implemented a similar modeling workflow, but used a targeted cross-linking approach in which one end of the XL reagent was anchored to a known site in the protein complex41. Kahraman et al. describe a cross-link-guided modeling method using ROSETTA, which is an alternative software package for protein structure prediction that requires more detailed knowledge of the command-line user interface and higher amounts of computational power5. A modeling method similar to ours is described in the detailed supplementary material of the publication from Herzog et al.4 concerning the protein Phosphatase 2A Network. In addition, Gaik et al. used I-TASSER to predict the structure of a missing domain of the Nup82 complex39 on the basis of sequence alone. They subsequently used HADDOCK to predict details of protein interactions within the overall protein complex. In their work, they used XL data as distance restraints at the docking steps, but not to refine the model of the isolated missing domain or to select the most relevant protein structure model. Although we selected I-TASSER and HADDOCK on the basis of usability and their high performance in published assessments of modeling programs14,15, other programs are also available and can be integrated into the protocol according to the needs of the user. Of note, Modeller42 can be used as an alternative to I-TASSER. The official version of Modeller requires a command line user interface in Anaconda Python, although graphical user interfaces have been developed by additional users43. An alternative tool to measure the solvent-accessible surface distance (SASD) between cross-linked residues is described by Matthew Allen Bullock et al.44. The tool, named Jwalk, is provided under public license, and although it performs well in regard to calculation time, its application is limited to Unix or Linux-based operating systems.
Limitations
In our published example of the methodology concerning the HOP2–MND1 heterodimer, the generation and identification of a high number of cross-links that cover the majority of the protein interaction interface were instrumental in the success of the structure calculation. This was in part due to the elongated protein structure with high proportions of solvent-accessible surface area, and also due to our ability to produce high amounts of purified protein. In the cases of larger proteins or a complex mixture of proteins, for example, a whole-cell lysate, it remains challenging to identify a comprehensive set of interprotein cross-links. In some proteins, the uneven distribution of reactive sites and differences in their accessibility result in irregular coverage of crosslinking data. The use of complementary XL reagents alleviates these limitations to some extent8,40. With the development of novel cross-linking technologies and detection capabilities29,45,46, such limitations are continuing to be addressed, which will increase the applicability of our modeling method.
The described protocol is mainly applicable to guide structural predictions of proteins and complexes that have a similar structure to that of a related protein that is used as a template. In such a case, cross-linking data can be used to adjust the preliminary structure of the protein in question to an orientation that is in better agreement with the cross-linking data, and hence is more likely to exist in solution. In the case that the experimentally obtained cross-links are not in agreement with the structure of the template, the outlined protocol is not applicable. Nevertheless, establishing prevalent disagreement between XL distance restraints and the structure of the template still represents valuable information, as it indicates that the protein or protein complex in question is predominantly present in a markedly different configuration from that of the template structure. Of course, this hypothesis assumes that cross-linking results are reflective of the native protein conformation and are not derived from aggregated forms of the protein or other nonspecific interactions. Guidelines on optimization of the cross-linking protocol to avoid such artifactual cross-links can be found in appropriate Nature Protocols articles16,17.
Applications of the method
The described method was successfully applied in our previous study on the HOP2–MND1 complex from Arabidopsis thaliana8.
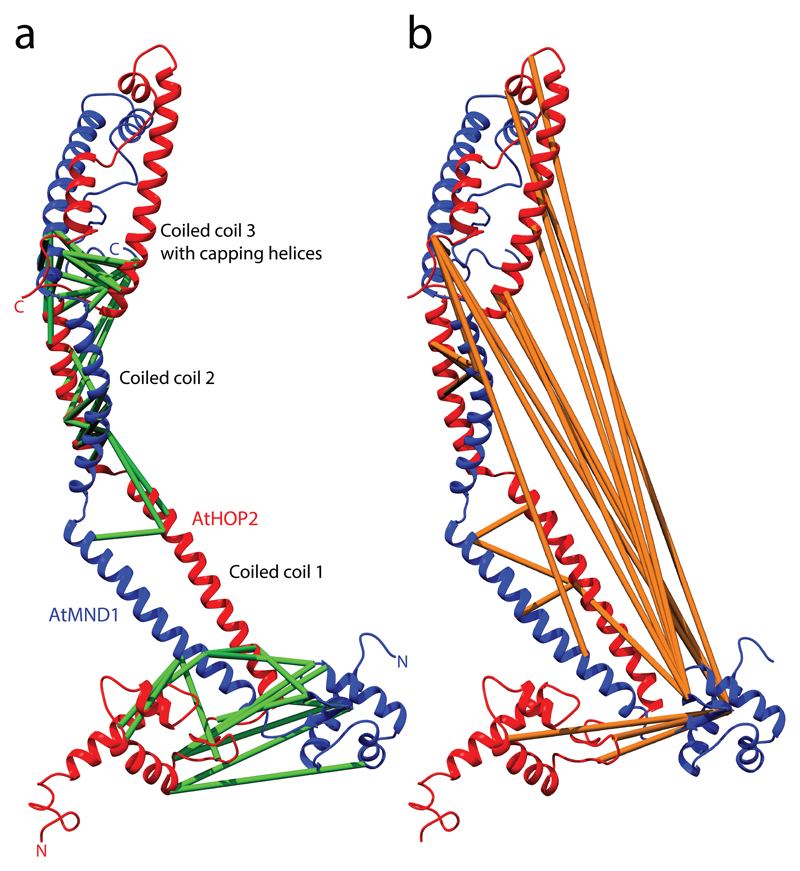
Results from the XL-MS workflow revealed a parallel orientation of the A. thaliana Hop2–Mnd1 heterodimer (Fig. 3). Furthermore, through implementation of the iterative comparative modeling approach described here, the structure of a dominant open conformation of the protein was predicted, which is similar to a crystal structure of the related HOP2–MND1 complex from Giardia lamblia8,47. Interestingly, our data also suggested a co-existing closed conformation that was not captured during XRC analysis of the related complex. This demonstrates the applicability of XL-MS to structural analysis, and also highlights its capacity to reveal dynamic changes in protein complexes and alternative conformations that are elusive to detection by other methods.
Figure 3. Chemical cross-links mapped onto the resulting comparative open protein-protein model of the HOP2-MND1 heterodimeric complex.
The distances between the cross-linked residues are depicted on the calculated structure. (a) A model of an ‘open’ conformation that is most similar to the template structure 4y66, with 45 matching interprotein cross-links shown in green. (b) The 22 incompatible cross-links shown in orange indicate the existence of a closed conformation. a adapted with permission from ref. 8, Copyright 2015 American Chemical Society.
We anticipate that our protocol will be highly applicable to the analysis of small protein complexes, in which it will allow the user to predict the relative orientation of the subunits and potential binding surfaces. It may also be applicable to multidomain proteins, in which orientation of different domains within the protein can be suggested.
As shown in the supplementary data sets and described in the ‘Anticipated Results’ section, we have applied our methodology to a selection of additional data sets to demonstrate further applications. We used cross-links obtained by Yilmaz et al.31 to predict a model of full-length calmodulin (see Anticipated Results and Fig. 4), and to dock the structure of N-terminal calmodulin to its binding partner, the plectin ABD-actin-binding domain (see Anticipated Results and Fig. 5).
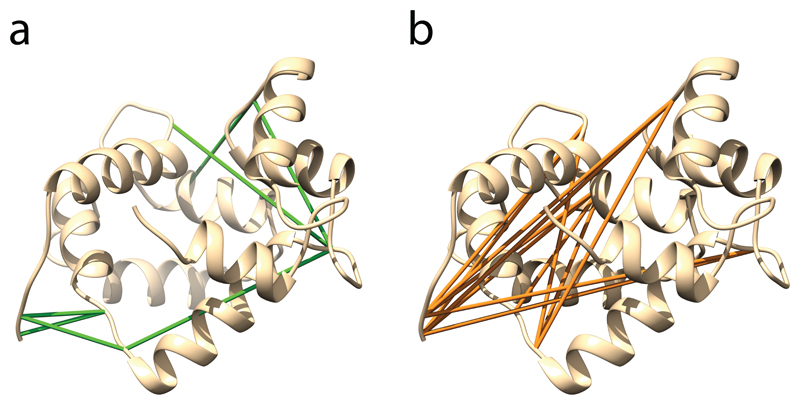
Figure 4. Predicted protein structure of calmodulin.
(a) The 8 cross-links compatible with the structure are shown in green. (b) 12 incompatible cross-links are depicted in orange.
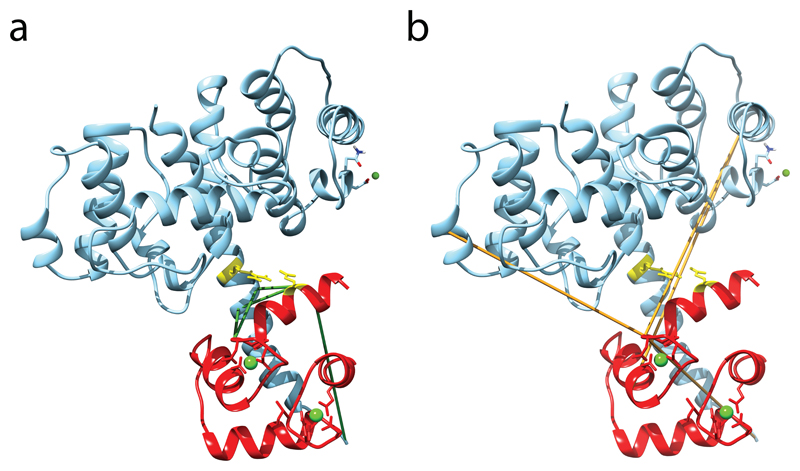
Figure 5. Predicted structure of the plectin ABD–calmodulin complex.
(a,b) Compatible (a) and incompatible (b) cross-links mapped onto the selected model of the complex between plectin ABD and the N-terminal lobe of calmodulin. The calmodulin chain is colored red. Compatible and incompatible cross-links are colored green and orange, respectively, and calcium and magnesium ions are shown as green balls. Residues E14 of calmodulin and R40 of plectin, the residues involved in the known salt bridge formation, are shown as sticks and are colored yellow.
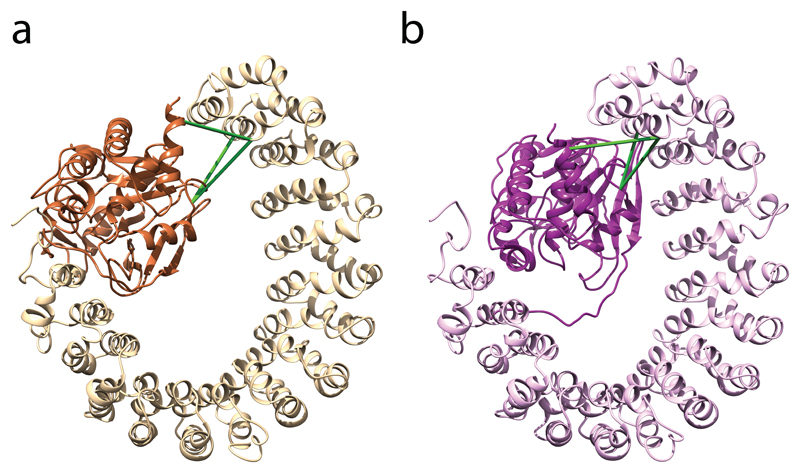
We also used our protocol to predict details of the interaction between the PPP2R1A and PPP2CA proteins within the protein phosphatase 2A complex (see Anticipated Results and Fig. 6). In this example, just three cross-links between the interaction domains were sufficient to correctly predict the orientation of the subunits. However, this limited data set was insufficient to position the interacting residues exactly as they are shown in the crystal structure.
Figure 6. Comparison of the predicted structure and the crystal structure of the PPP2R1A–PPP2CA complex.
(a) The predicted structure is the highest-scoring model that was calculated by HADDOCK using the cross-links shown in green as distance restraints. PPP2R1A and PPP2CA are colored light and dark, respectively. (b) The experimental cross-links are also shown on the crystal structure.
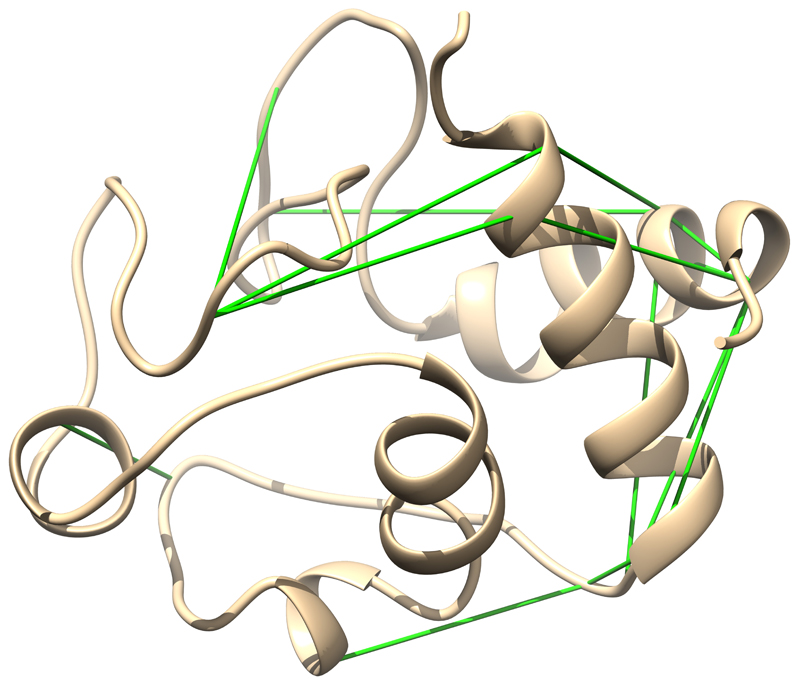
Another application of the protocol is demonstrated in Supplementary Data 1, in which it was used to show that all cross-links derived from bovine cytochrome c are in agreement with the preliminary model, indicating that the protein in solution has the same structure as the template crystal (Fig. 7).
Figure 7. Predicted protein structure of bovine cytochrome C.
All 16 cross-links are compatible with the structure and are shown in green.
Experimental design
This cross-link-guided molecular-modeling workflow, illustrated in Figure 1, was developed to enable the widespread use of XL restraint–guided structural modeling. It provides guidelines to allow researchers without extensive modeling experience to generate structural models of proteins and protein complexes; it also provides instructions for adapting the protocol depending on specific requirements in terms of protein characteristics and cross-linking data. The comparative modeling workflow is based on protein structure prediction with I-TASSER (Steps 1–13, Figs. 1a and 2a) and subsequent protein–protein docking with HADDOCK (Steps 14–29, Figs. 1b and 2b). In addition, evaluation and visualization guidelines are described in Box 1 and Steps 30–37 of the PROCEDURE, respectively.
Sequence-based structure prediction using I-TASSER (Steps 1-6)
In the first stage of the modeling workflow, I-TASSER is used to predict the possible structure of an individual protein or subunit of a protein complex, on the basis of its primary sequence and a homologous template (Fig. 2a, Stage 1). I-TASSER assigns a confidence score (C-score) to each model that reflects the confidence of the model with respect to the template. The best model is selected according to this C-score.
Subunit structure refinement using intraprotein cross-linking restraints with Xwalk and I-TASSER (Steps 7-13)
In the second stage, the distances of the experimental intraprotein cross-links are calculated with Xwalk48 to determine if they are in agreement with the model. As the linear Euclidean distance (the straight-line distance between two atoms) can be inappropriate because of penetration of the molecule’s surface, the distances between the cross-linked residues are calculated as the SASD, implemented in the Xwalk algorithm. A selection of cross-links, which are chosen according to provided guidelines, is subsequently used to refine the preliminary model (Fig. 2a, Stage 2). Data derived from multiple cross-linking reagents can be used in two ways. They can either be amalgamated in order to carry out a single refinement step (combined method), or they can be used separately in multiple refinement steps, whereby the ‘previously refined’ model is used for the next prediction step (iterative method). Optionally, any noncompatible cross-links that do not satisfy the model can be used to build an alternative conformation, in either the combined or the iterative method (Step 11C). The best model is selected according to the C-score and by how well the cross-links agree with the model (Box 1).
Preliminary protein-protein docking using HADDOCK and CPORT (Steps 14-27)
In the third stage, the structures of the individual subunits are used for a first round of protein–protein docking (Fig. 2b, Stage 3). Active residues (those that are involved in the protein–protein interface) are predicted by applying the CPORT program49. The active residues and the distances derived from the interprotein cross-links are used as an input for the preliminary docking at the expert interface of HADDOCK v2.2. The best models are chosen on the basis of their HADDOCK score, which is calculated on the basis of factors that include surface complementarities, electrostatic interactions, van der Waals repulsion and complementarity between the experimental crosslinks and the resulting model (Box 1).
Final protein-protein docking in HADDOCK (Steps 28 and 29)
The cross-links are sorted using distance restraints calculated with Xwalk, as described previously for the intraprotein cross-link data, (Steps 25 and 26) and used for the final docking step (Steps 28 and 29) (Fig. 2b, stage 4). The quality of the models is validated as in previous steps (Box 1) and the best model is selected for structure and cross-link visualization using UCSF Chimera50 (Fig. 3).
Evaluation guidelines for the selection of best structural models
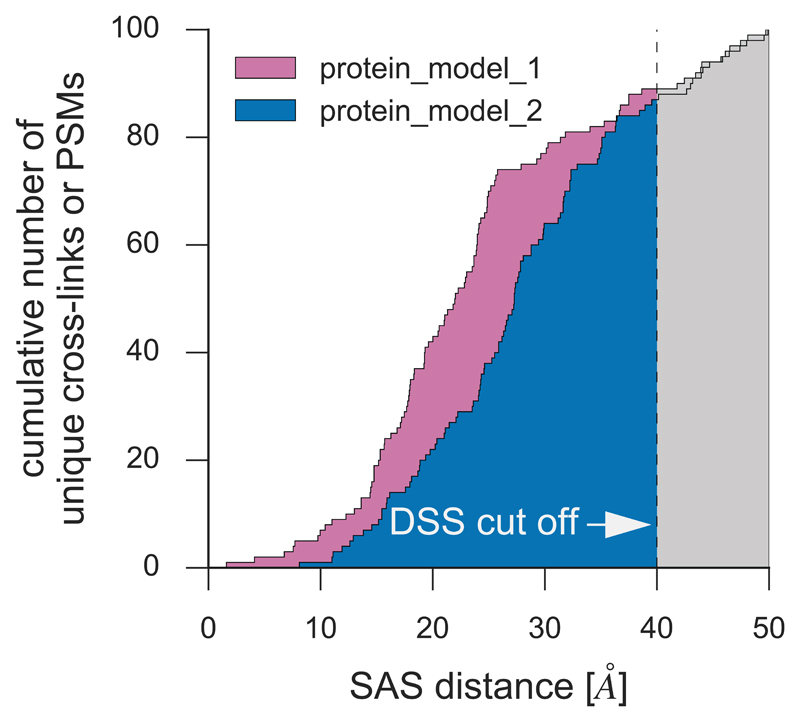
Comparative protein modeling and protein-protein docking approaches generate a large variety of predicted protein structures. Some of these structures are very similar, whereas others can show substantial differences. Software used to generate such protein models provide a score that helps the user to select the most appropriate model. However, these scores do not represent how well a model satisfies the cross-linking results, and thus, selecting an optimal protein model is not a trivial task. To assist with this, we introduce a simple method to calculate a cross-link score (XL-score) that describes how well a protein model satisfies the cross-links, taking into consideration the distance of matching cross-links (Fig. 8). To select an optimal protein model, we suggest consideration of the score provided by the prediction software, the number of cross-links that are satisfied by the model below a defined distance cut-off, and the XL-score as well as other experimental data, such as results from different biochemical experiments (Box1).
Figure 8. Simulated distribution of alpha-carbon distances between cross-linked residues in two resulting comparative models.
The plot depicts the cumulative number of DSS-derived cross-links that have an SASD less than or equal to a specified distance. Comparison of the plots for different structures (e.g. by calculating the normalized area under the resulting curve below a defined cut off value) facilitates best model selection at the evaluation step of the modeling workflow.
Visualization of protein and protein complex models and distances
We describe the use of UCSF Chimera50 for visualizing the predicted protein models along with the experimental cross-links. The structural model of the protein or protein complex of interest is first loaded,and the list of atom pairs of the cross-linked residues is fed to the program via its command line (Steps 30-32). From these steps the Euclidean distance of each cross-link is reported (Step 33), and an image of the resulting figure can be saved (Steps 34 and 35). Optionally, for the comparison of similar structures, superimposition of the protein molecules can be carried out (Steps 36 and 37).
Materials
Equipment
Datasets
• List of identified cross-links found within a protein or protein complex
• Length and residue specificity of the XL reagent
• Amino acid sequences of the proteins of interest
Software
• Xwalk (free command line version at http://www.xwalk.org)
• Structure-viewing program (for example UCSF Chimera50, version 1.11.2 available at https://www.cgl.ucsf.edu/chimera/, free software for academic users)
• Internet access and a web browser
• Java Runtime Environment (minimum version 1.4 at https://java.com/)
Example files
• Supplementary Data 1: I-TASSER prediction of bovine cytochrome c (uses XL data originally published by Kao et al.25)
• Supplementary Data 2: I-TASSER prediction of HOP2 (uses XL data originally published by Rampler et al.8)
• Supplementary Data 3: I-TASSER prediction of full-length calmodulin (uses XL data originally published by Yilmaz et al.31)
• Supplementary Data 4: I-TASSER prediction of MND1 (uses XL data originally published by Rampler et al.8)
• Supplementary Data 5: HADDOCK docking of calmodulin to plectin (uses XL data originally published by Yilmaz et al.31 and Song et al.51)
• Supplementary Data 6: HADDOCK docking of PPP2R1A to PPP2CA (uses XL data originally published by Herzog et al. 4)
• Supplementary Data 7: HADDOCK docking of HOP2 to MND1 (uses XL data originally published by Rampler et al. 8)
• Supplementary Data 8: Windows batch script that can be used to automatically run Xwalk on a series of PDB files.
• Supplementary Data 9: Description of structure and content of supplementary data sets.
Procedure
Sequence-based structure prediction using I-TASSER • TIMING 1-2 d per protein
Δ CRITICAL Each step of the subunit structure prediction (Steps 1–12) can be tested using the example files provided in Supplementary Data 1–4. For data sets 1 and 4, use option A of Step 11. For data sets 2 and 3, use option B of Step 11. Description of the structure and content of the supplemental data sets is provided in Supplementary Data 9.
1| Go to the I-TASSER homepage (http://zhanglab.ccmb.med.umich.edu/I-TASSER/), register as academic user and log in.
2| Paste the amino acid sequence of the protein of interest into the provided form, or upload a file containing the sequence in FASTA format. To date, the server only accepts protein sequences 10-1,500 amino aa in length.
3| Provide an e-mail address, a password and an ID for the structure prediction.
4| (Optional) Specify a solved protein structure as the ‘template without alignment’, by uploading the PDB file or by providing the PDB ID and chain ID under ‘Option I’. If no template is specified, I-TASSER selects the best templates from the PDB. It is possible to exclude homologous or specific template structures under ‘Option II’. In general, excluding homologous templates will decrease the quality of I-TASSER modeling.
5| To submit the sequence, click the ‘Run I-TASSER’ button. After successful submission, a notification message that includes a job identification number will appear. Once the prediction has been completed, an e-mail is sent containing the ‘job information’, figures of the predicted models and a link to a web page with more detailed results. These remain accessible on the server for three months. The results include protein secondary structure predictions for the top five predicted models (PDB files) and the respective C-scores. The I-TASSER ‘Forum’, ‘Annotation’ and ‘FAQ’ pages at http://zhanglab.ccmb.med.umich.edu/ I-TASSER are helpful additional resources for this step.
Δ CRITICAL STEP: Only one job per user and IP address of your computer can be submitted at a time. As an alternative, it is possible to download the I-TASSER suite software to run it on a local server. Depending on the computing capacity, several days can be saved. For local installations, knowledge of the command line is advantageous. As I-TASSER performs Monte Carlo simulations, several CPU cores and free disk space of more than 60 G are recommended.
? TROUBLESHOOTING
6| Choose models with the highest I-TASSER C-score, which ranges between -5 and 2. A higher score reflects a model with higher quality.
Δ CRITICAL STEP: We observed that model 1 (of the five provided) usually has the highest C-score but is often biased further towards the template structure than towards the experimental distance restraints, of which it satisfies only a few. The root-mean-square deviation (RMSD) of model 1 from the given template structure, which can be measured by superimposition of the two structures (Steps 36 and 37), is often the smallest. In such cases, one may consider selecting an alternative model, guidelines for which are provided in the section ‘Evaluation guidelines for the selection of best structural models’ section (Box 1).
Subunit structure refinement using intraprotein cross-linking restraints using Xwalk and I-TASSER • TIMING 2 d per protein
7| Measurement of the experimental cross-links on the preliminary model using Xwalk. Prepare an input file for Xwalk containing the cross-linked residues and the chain as defined in the PDB file of the protein of interest. Xwalk will then calculate the distances between the β-carbon atoms of the cross-linked residues. It is recommended to name this file:
xwalk_crosslink_input.tsv
Cross-link data must be presented as shown in the table below and saved as a tab-separated text file
| 1 | LYS-202-B-CB | LYS-206-B-CB |
This example shows that cross-link number 1 is between Lysine 202 of chain B and Lysine 206 of chain B. The term ‘CB’ specifies that the distance between the β-carbon atom of the respective residues will be measured in Xwalk. Note that the cross-linked residues must be specified in column three and four. The second column must remain empty; it will contain the name of the model in the output.
8| Calculate the Euclidean distance (ED) and SASD between the cross-linked residues on the chosen model from Step 6 in Xwalk, either using command lines option A) or, for those without knowledge of using command lines, using the online graphical user interface (GUI) (option B).
A) Calculation of the ED and SASD in Xwalk using command lines
i) Open the web page of Xwalk (http://www.xwalk.org/) and download the free Xwalk software.
ii) Enter the following command line:
java -Xmx1024m -cp Xwalk\Xwalk_v0.6\bin\ Xwalk -infile yourprotein.pdb -out yourprotein.xwalk.tsv -dist xwalk_crosslink_input.tsv -max 50 -bb -f
Refer to the README file in the downloaded folder of Xwalk folder for a complete overview of useful explanations or use explanations from the following short list adapted from the Xwalk README file:
| Explanation | Description |
|---|---|
| cp <path> | Location of the Xwalk "bin" folder from the Xwalk installation directory. |
| infile <path> | Your input PDB file. |
| out <path> | Xwalk writes the output to this file. |
| dist <path> | File which will be used to extract the indices and the residue pairs for the distance calculation (from Step 7). |
| max <double> | Calculates distances in Angstrom, only up-to this value |
| bb [switch] | Reads in only backbone and beta carbon atom coordinates from the input file and increases the solvent radius for calculating the solvent accessible surface area to 2. |
| f [switch] | Forces output to be written into a file, overwriting existing files. |
Δ CRITICAL STEP: In cases in which you are measuring XL distances on multiple models, it may be easier to use a batch file to automatically run Xwalk for a series of PDB files. The batch script can be found in Supplementary Data 8. Make sure to adapt directories as described in the batch script before running.
Δ CRITICAL STEP: Calculating the SASD between the ß-carbon atoms of long-distance residue pairs can be time consuming. We therefore implement a cutoff value of 50 Å for the maximum distance that should be measured.
B) Calculation of the ED and SASD in Xwalk using the online GUI
i) Go to http://www.xwalk.org/ and choose the desired running mode: ‘Production Mode’.
ii) Choose the input file by uploading your selected model from Step 6.
iii) Set the desired cross-link parameters: choose the reactive amino acid of the first and second residues in cross-links, according to the used reagent (e.g., Lys and Lys when using BS2G cross-linkers) and choose ‘Intra XL’.
iv) Set the maximum distance to the defined cutoff value mentioned in Step 8 A ii.
v) Click ’Run Xwalk” to create a list of the number of residues between the cross-linked residues in the primary sequence, the Euclidean distance of the XL (Å) and the SASD of the XL (Å).
| 1 | model.pdb | LYS-202-B-CB | LYS-206-B-CB | 4 | 4.2 | 4.6 |
? TROUBLESHOOTING
9| Decide the cutoff values for your XL reagents. This is defined as the maximum distance between cross-linked residues. Suggested values are provided in the table below:
| XL-reagent | ED (Å) | SASD (Å) |
|---|---|---|
| DSS | < 35 | < 40 |
| BS2G | < 30 | < 35 |
| EDC | < 23 | < 28 |
EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride.
The SASD cutoff values will subsequently be used to determine if the cross-links fit a predicted model, while the ED cutoff values will be used for structure prediction, protein subunit docking, evaluation of models and visualization of cross-links the resulting protein models.
Δ CRITICAL STEP The recommended Euclidean cutoff for disuccinimidyl suberate (DSS) is 30 Å (Herzog et al.4) to 35 Å (Hall et al.52), which is calculated as the sum of the length of the two extended lysine side chains (2 x 5.5 Å), the spacer length (11.4 Å) and the remaining 7.6 - 12.6 Å allowing for conformational dynamics5. We therefore recommend the cutoff distances for other cross-linkers to be calculated as the length of the two extended cross-link-reactive side-chains, plus the spacer length of the cross-linker and ~13 Å for conformational dynamics.
10| Categorize the cross-links in the Xwalk output list according to their compatibility with the preliminary model. Those that have an ED and SASD that are below the cutoff values defined in Step 9 are in agreement with the preliminary model (compatible cross-links), whereas those that have distances above the cutoff values are incompatible. In the unlikely case that most or all of the cross-links are incompatible with the preliminary model, it is not recommended to continue with the protocol.
? TROUBLESHOOTING
11| In the case that all cross-links are compatible with the preliminary model, continue with the refinement as described in option A. This indicates that the protein structure in solution is similar to that of the preliminary model. In the case that there are both compatible and noncompatible cross-links, perform separate refinement steps using the full list of cross-links (option A), only the compatible (option B) and only the noncompatible cross-links (option C).
A) Refinement of the model using all cross-links as distance restraints
i) Repeat Steps 1-3 of the PROCEDURE.
Δ CRITICAL STEP: When data derived from multiple cross-linking reagents are available, they can be combined to be used in one single refinement step. Alternatively, they may be used separately to perform iterative refinement steps. Both may be tried during optimization of the protocol for different proteins and cross-linking data.
ii) Click on the ‘Option I’ menu and specify a template without alignment: upload the best-scoring model from the prediction in Step 6 in standard PDB format.
iii) Prepare a .txt file containing the list of all cross-links, along with the cutoff ED for the specific XL-reagent (e.g. 35 Å for DSS, see Step 9). Use the following syntax to define the atoms of experimentally observed cross-links:
DIST /t <1st residue position> /t CA /t <2nd residue position> /t CA /t <Euclidean distance>
Example for DSS:
| DIST | 41 | CA | 65 | CA | 35 |
Example for EDC and DSS:
| DIST | 32 | CA | 78 | CA | 23 |
| DIST | 41 | CA | 65 | CA | 35 |
Δ CRITICAL STEP: If combining XL data obtained from the use of different reagents it is preferred to prepare a combined list at this step.
iv) Upload the restraints file prepared in Step 11Aiii to I-TASSER. This is done by expanding the ‘Option I (assign additional restraints & templates to guide I-TASSER modeling)’ submenu, and uploading the file in the ‘Assign contact/distance restraints’ section.
v) To submit the sequence, click the ‘Run I-TASSER’ button. After successful submission, a notification message including a job identification number will appear. The output from I-TASSER includes protein secondary structure predictions for the top five predicted models (PDB files) and the respective C-scores, as described in Step 5. Refer back to Step 5 for extra information.
B) Refinement of the model with compatible cross-links
i) To refine the preliminary model (from Step 6) using compatible cross-links as distance restraints (from Step 10), carry out Step 11A (i-v) of the PROCEDURE with one modification; at Step 11Aiii prepare a .txt file containing only compatible cross-links from Step 10, along with the cutoff ED for the specific XL reagent (e.g., 35 Å for DSS, see Step 9).
C) Refinement of the model with noncompatible cross-links
i) If you have a group of noncompatible cross-links from Step 10, use them to calculate a possible alternative structure of the subunit. Such cross-links may belong to an alternative conformation. Apply them to the preliminary model from Step 6 as described in Step 11B.
Δ CRITICAL STEP: The alternative model should be critically evaluated according to Box 1 (Evaluation guidelines for the selection of best structural models).
12| The output of Step 11 is a refined structural model of a subunit that can be used for data-driven biomolecular docking (Steps 14–29). Select the best model using a combination of aspects that are described in Box 1.
13 | Repeat Steps 1-12 for each subunit in the protein complex of interest.
Preliminary protein-protein docking using HADDOCK and CPORT • TIMING 1-3 d
Δ CRITICAL The inputs for HADDOCK are the structures of the subunits. These may be obtained from the I-TASSER modeling section (Step 12) or from other sources. For a detailed description of HADDOCK, see deVries et al.11.
Δ CRITICAL Each step of the protein–protein docking (Steps 14–29) can be tested using the example files provided in Supplementary Data 5, 6, 7. For data sets 5 and 7, use option A of Step 28. For data set 6, use option D of Step 28. Description of the structure and content of the supplemental data sets is provided in Supplementary Data 9.
14| Calculate the active and passive residues of the refined subunit structure from Step 12 using CPORT at http://milou.science.uu.nl/services/CPORT/. Unfold the ‘Protein structure to predict’ menu. In the ‘Where is the structure provided’ menu, select ‘I’m submitting it’, specify the chain and choose the PDB file of the best model selected at Step 12. Unfold the ‘Sequence alignment’ menu and set the ‘Threshold’ under ‘Prediction threshold to use’ to ‘Very sensitive’ (recommended for HADDOCK) and submit. A web page will then open that provides a link to the results. Click on the link. When the calculation is finished, the predicted active residues and the surrounding passive residues will be listed.
Δ CRITICAL STEP: For a more detailed description on how to use CPORT, see de Vries and Bonvin49.
Δ CRITICAL STEP: Residues that are known to participate in a protein-protein interaction should additionally be defined as active. Such knowledge can come from previous experimental evidence.
15| Go to the HADDOCK registration site at http://haddock.science.uu.nl/services/HADDOCK2.2/signup.html, fill out the Account Signup Form, agree to the user conditions and submit. A confirmation mail will be sent with a pending administrator approval and shortly afterwards an email containing a login link and a username will be sent. The account can then be unlocked by the administrator to allow use of the HADDOCK 2.2 webserver ‘s ‘Easy Interface’. Request to upgrade your account to expert level to implement experimentally obtained distance restraint data.
16| Once your expert access level is active, go to the ‘Expert Interface’ at http://haddock.science.uu.nl/services/HADDOCK2.2/haddockserver-expert.html and give the docking run a name.
17| Unfold the ‘First molecule’ menu and choose ‘I’m submitting it’, specify the chain and select the appropriate PDB file.
18| Copy the active and passive residues from the CPORT prediction (Step 14) and paste them into the ‘Restraint definition’ menu. In addition, choose ‘Protein’ as the ‘kind of molecule’.
19| Unfold the ‘Histidine protonation states’ and ‘Semi-flexible segments’ menus and set them as automatically guessed (default settings).
20| Repeat Steps 16-19 using the individual protein structure of other subunits of the complex.
CRITICAL: The following steps (Step 21 and 22) are compatible only with the’ Expert/Guru’ interface.
21| Unfold the ‘Distance restraints’ menu. Here, supply your HADDOCK restraints TBL file (as described below), defining your experimental interprotein cross-linking restraints as unambiguous. If data from multiple reagents are available, combine the cross-links into one list. The format of a distance restraint for the TBL file is as follows.
assign (resid <residue number> and segid <1st molecule chain>) (resid <residue number> and segid <2nd molecule chain>) <target distance> <lower distance margin> <higher distance margin>
An example for three restraints in the TBL format is provided below (first two rows for DSS cross-links and the third for an EDC cross-link):
assign (resid 152 and segid B) (resid134 and segid A) 35 35 0 assign (resid 152 and segid B) (resid137 and segid A) 35 35 0 assign (resid 235 and segid B) (resid147 and segid A) 23 23 0
22| Specify username and password and submit. It is possible to use the grid-enabled submission page for faster docking.
23| After submission, a link to the parameter file will be available. Save this file, because it can be used as a template for subsequent submissions.
24| After the docking calculations are completed (which may take several days), an email will be sent containing the link to the ‘results page’. Follow the link to the results page. The clustered docking solutions will be displayed, sorted by their HADDOCK scores. A lower HADDOCK score corresponds to a better solution for the complex structure. The results of the docking run with the predicted structures can be downloaded. It is recommended to download all the results, as the file will be deleted from the server after 1 week.
? TROUBLESHOOTING
25| Use Xwalk to measure the ED and SASD between the β-carbon atoms of the interprotein cross-linked residues as described in Steps 7-9.
Δ CRITICAL STEP: In the input file for Xwalk that lists the cross-linked residues, ensure that the chain label is the same as that used in the PDB file.
? TROUBLESHOOTING
26| Categorize the interprotein cross-links in the Xwalk output list according to their compatibility with the preliminary complex models from Step 24. Those that have an ED and SASD that are below the cutoff value defined in Step 9 are in agreement with the preliminary model.
27| Choose the best preliminary complex model based on the HADDOCK score and a combination of other aspects. See section Box 1. In the unlikely case that most or all the cross-links are incompatible with the preliminary model, it is not recommended to continue with the protocol.
? TROUBLESHOOTING
Structural refinement of the protein complex using Xwalk, HADDOCK and XL data: • TIMING 1-3 d
28| In the case that there are both compatible and noncompatible interprotein cross-links from Step 26, try separate refinement steps using only the compatible (option A) and only the noncompatible cross-links (option B) or no XL-derived distance restraints (option C). The resulting models from all refinement steps should be evaluated according to the evaluation guidelines (Box 1).
In the case that all cross-links are compatible with the preliminary complex model, the result indicates that the structure of the protein complex in solution is in agreement with the preliminary model (option D). Therefore, such a result represents experimental validation of the model, and there is no need to continue further with the protocol.
A) Refinement of the model (from step 27) using the compatible cross-links as distance restraints
i) Repeat the docking according to the first HADDOCK docking (Steps 14-24) with one modification: unfold the ‘Distance restraints’ menu at Step 21 and supply the HADDOCK restraints TBL file defining your compatible experimental interprotein cross-linking restraints (from Step 26) as unambiguous.
B) Refinement the model (from step 27) using the noncompatible cross-links as distance restraints
i) Repeat the docking according to the first HADDOCK docking (Steps 14–24), with one modification: unfold the ‘Distance restraints’ menu at Step 21 and supply the HADDOCK restraints TBL file defining your noncompatible experimental interprotein cross-linking restraints (from Step 26) as unambiguous.
C) Docking of the model with no cross-links
i) Repeat docking according to the first HADDOCK docking (Steps 14–24), with one modification: do not unfold the ‘Distance restraints’ menu at Step 21.
D) Preliminary model is the final model
i) In the case that all cross-links are compatible with the preliminary complex model, the result indicates that the structure of the protein complex in solution is in agreement with the preliminary model. There is no need to repeat the docking. Continue with the evaluation (Box 1) and visualization steps (Steps 30–37).
29| Choose the best complex model using a combination of aspects (see Box 1).
Visualization of protein and protein complex models and distances • TIMING 1 h per model
Δ CRITICAL STEP: There are several visualization programs for protein structures. We used USCF Chimera50, because it provides a means for interactive visualization and analysis of molecular structures. Distances between residues can be measured, and high-quality images can be generated.
30| Download and install Chimera at https://www.cgl.ucsf.edu/chimera/download.html.
31| Open USCF Chimera and open a PDB file by using the menu ‘File’ > ‘Open’. The selected model will be visible. You can interactively turn, move or enlarge the model. For detailed instructions see the User’s Guide at https://www.cgl.ucsf.edu/chimera/docs/UsersGuide/. The simplest way to map and measure distances simultaneously is to copy a list of atom pairs of the cross-linked residues to the chimera command line. First prepare the list of the atom pairs corresponding to the cross-linked residues. An example for measuring the distance between the C α atom of residue 235 of chain b in model 0 and the C α atom of residue 154 of chain a in model 0 is shown below:
distance #0: 235 .b@CA #0: 154 .a@CA
32| Go to ‘Favorites’ and then ‘Command Line’ and insert the list. Press ‘enter’, and a line connecting the specified atoms will be displayed
33| In the ‘Structure Measurement’ window (‘Tools’ > ‘Structure Analysis’ > ‘Distances’), the Euclidean distance values are listed. For example,
| ID | Atom1 | Atom2 | Distance |
|---|---|---|---|
| 1 | LYS 235.B CA | LYS 154.A CA | 15.380 Å |
34| Go to the ‘Structure Measurement’ window to change the style (e.g., change the line thickness for better recognition of distances) if this is desired.
35| To save an image, go to the path ‘File’ > ‘Save Image’. In the ‘Image Size’ window, check the box ‘Use print units’, and specify the resolution. In the ‘Image Options’ window, set ‘Rendering’ to ‘POV-Ray’ and select ‘Transparent background’ (optionally).
36| If a homologous structure is available, superimpose this onto the predicted complex model by opening the PDB file of the template structure and the PDB file of your predicted protein model in the same window.
37| Define ‘Reference structure’ and ‘Structure(s)’ to match by opening ‘Tools’ > ‘Structure Comparison’ > ‘MatchMaker’. In the ‘Matchmaker’ window, select ‘Best aligning pair of chains between reference and match structure’. Use, for example, the default settings: ‘Needleman-Wunsch’ as ‘Alignment algorithm’ and ‘Blosum-62 matrix’. RMSD values and additional information on the superimposition can be found at ‘Favorites’ > ‘Reply log’.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 5 | No structure of the individual protein is predicted by the I-TASSER protein structure prediction | No homologues templates are available | Provide restraint information immediately at first prediction step. Try to repeat the first prediction step with I-TASSER using all cross-linking restraints of the corresponding intraprotein cross-links |
| The number of predicted models is < 5 | The identified templates are very similar | Choose the model with the highest C-score | |
| 8, 25 | Distance calculations can fail under certain conditions. In such a case, negative distance values will be reported |
|
The values -2, -3, -4 and -5 should be replaced with the Euclidian distance between the two atoms. The value -1 should be replaced with a value higher than the maximum cutoff (e.g., 100) for future categorization (Step 10 and Step 26) |
| 10 and 27 | Most or all of the cross-links are incompatible with the preliminary model | Such a case is likely to be the result of a markedly different protein conformation in solution compared with the preliminary model. Alternatively, it could also indicate an unsuccessful cross-linking experiment | It is not recommended to continue with the protocol |
| 24 | Docking results at HADDOCK server have disappeared | Results are deleted at HADDOCK server after 1 week | Save all results immediately after getting the notification from the server about the completed job. Save the parameter file for each HADDOCK submission. It is possible to use the ‘File Upload’ interface for repeating the docking analysis at a later time point |
Timing
Steps 1-6, sequence based subunit prediction using I-TASSER: ~1-2 d per protein, depending on the sizes of the proteins and the number of jobs in the queue
Steps 7-13, subunit structure refinement using intraprotein cross-linking restraints using Xwalk and I-TASSER: 2 d per protein
Steps 14-27, preliminary protein-protein docking using HADDOCK and CPORT: 1-3 d
Steps 28-29, structural refinement of the protein complex using Xwalk, HADDOCK and XL data. 1-3 d; depending on the size of the complex and the number of jobs in the queue
Steps 30-37, visualization of protein and protein complex models and distances: 1h per model
Box 1, evaluation guidelines for the selection of best structural models at different steps of the protocol: 1 d
Anticipated Results
The heterologously expressed and purified Arabidopsis HOP2-MND1 heterodimer was used to exemplify an optimized workflow of comprehensive XL-MS, followed by comparative modeling8. The use of cross-linking reagents that vary in spacer length and reaction chemistry (EDC, DSS, BS2G) resulted in high cross-linking coverage across the complex. This cross-linking methodology, along with the availability of the existing homologous template structure, allowed modeling of the refined individual subunit structures on the basis of protein structural prediction via I-TASSER. We then modeled the structure of the whole complex using protein-protein docking with the data-driven HADDOCK approach.
A similar approach to ours was implemented by Shi et al.5, who used data from EDC and DSS cross-linking experiments to model the NUP84 protein complex. However, although we performed the modeling on individual subunits and on the protein complex, in their study, the entire protein complex was predicted by integrative modeling. The workflow described within the current protocol can be further extended with quantitative approaches, which can be achieved by using isotopically-labeled XL reagent17. This could allow investigation of conformational change induced by changes of the experimental conditions, for example the addition of a ligand.
The experimental modeling workflow that we present allowed us to determine the previously unknown orientation of the HOP2-MND1 complex partners. The results strongly suggested a parallel conformation for the plant HOP2-MND1 heterodimer, in agreement with the structure of the G. lamblia complex. In addition, our cross-linking results provide evidence for multiple conformations of the protein in solution (see Figure S5, Rampler et al.8). These two results also highlight the strength of the outlined protocol: first, it robustly decodes low-resolution structural information from the XL-MS data regarding the arrangement of subunits within protein complexes; second, it informs about alternative protein complex conformations in solution. The results for these calculations can be found in Supplementary Data 2, 4 and 7, along with resources corresponding to intermediate steps in the workflow for the modeling of each individual subunit and docking of the complex.
To test the applicability of the protocol, we used cross-links obtained by Yilmaz et al. 31 to 1) predict a model of full-length calmodulin and 2) dock the calmodulin structure to its binding partner, the plectin actin binding domain (ABD). I-TASSER was first used to create a preliminary structure based on the amino acid sequence, and the length of each experimentally observed cross-link was measured on this model (Steps 1-8). Distance restraints derived from all experimental cross-links were used to recalculate the structure (Step 11A), and this led to five different models, some of which are compact, and some extended. The cross-links were then categorized according to their fit with the preliminary structure, as described in Step 10, and subsequently only those in agreement were used in a further calculation to generate a refined model (Step 11B). The latter approach led to one single model that was in agreement with the compact structure (Supplementary Data 3, Fig. 4). This demonstrates different results that can be obtained upon use of all cross-links versus only cross-links that are compatible with the preliminary structure. Furthermore, docking steps were carried out between calmodulin and the plectin ABD using the solved crystal structure of the N-terminal lobe of calmodulin (PDB ID 4Q57). Here we utilized data from two different published data sets, obtained with different cross-linking reagents31,51. The results for the calculations using compatible cross-links can be found in Supplementary Data 5, along with resources corresponding to intermediate steps in the workflow. The best model of the interacting domains of plectin and calmodulin is depicted in Figure 5, along with the experimental cross-links.
We also used the modeling protocol to predict details of the interaction between the PPP2R1A and PPP2CA proteins within the protein phosphatase 2A complex. Three DSS cross-links between lysine residues that were obtained from the work of Herzog et al.4 were used as distance restraints during protein-protein docking by HADDOCK (Supplementary Data 6 and Fig. 6). The generated models show the correct orientation of the complex partners; however, the cross-links were insufficient to position the interacting residues as they are shown in the crystal structure (PDB ID 2IAE).
We also used the modeling workflow to predict a structure of bovine cytochrome c using Steps 1-12 of the protocol. During the calculation of the preliminary model, all templates of > 40% homology were excluded by I-TASSER. In this example, all cross-links are in agreement with the preliminary model, indicating that the protein in solution has a structure similar to that of template crystal (Supplementary Data 1, Fig. 7). In addition, the model predicted with I-TASSER is in strong agreement with the crystal structure of bovine cytochrome c.
Supplementary Material
Example input and output files along with instructions for comparative modeling of bovine cytochrome C using cross-linking data.
Example input and output files along with instructions for comparative modeling of HOP2 from A. thaliana using cross-linking data.
Example input and output files along with instructions for comparative modeling of human calmodulin using cross-linking data.
Example input and output files along with instructions for comparative modeling of MND1 from A. thaliana using cross-linking data.
Example input and output files along with instructions for protein-protein docking of human N-lobe of calmodulin and the actin binding domain of mouse plectin using cross-linking data.
Example input and output files along with instructions for protein-protein docking of mouse Ppp2r1a and human PPP2CA using cross-linking data.
Example input and output files along with instructions for protein-protein docking of HOP2 and MND1 from A. thaliana using cross-linking data.
A windows batch script that can be used to automatically run Xwalk on a series of PDB files. The batch file contains comments on how to use it. By changing the file type from .txt to .bat, the code is executable.
Description of structure and content of supplementary data sets. It is recommended to read this file before using the supplementary data sets.
Acknowledgements
This research was funded by the Austrian Science Fund (SFB F3402; P24685-B24; and TRP 308-N15) and the EU FP7 project MEIOsys (222883-2). We thank the Research Institute of Molecular Pathology (IMP) and the Institute of Molecular Biotechnology of the Austrian Academy of Sciences (IMBA) for general funding. We acknowledge M. Hartl and T. Gossenreiter for useful and inspiring scientific discussions. Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). Images in Chimera were created using Persistence of Vision Raytracer (v3.6), Persistence of Vision Pty. Ltd. (2004), and were retrieved from http://www.povray.org/download/.
Footnotes
Author Contributions
Z.O.-N., E.R., D.M.H, T.S, and K.M. designed and developed the modeling workflow. J.D. and O.H performed data analysis and tested the workflow. Z.O.-N., R.B and D.M.H. wrote and edited the manuscript. P.S provided significant input into the method development.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leitner A, Faini M, Stengel F, Aebersold R. Crosslinking and Mass Spectrometry: An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends Biochem Sci. 2016;41:20–32. doi: 10.1016/j.tibs.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Holding AN. XL-MS: Protein cross-linking coupled with mass spectrometry. Methods. 2015;89:54–63. doi: 10.1016/j.ymeth.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Herzog F, et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 5.Kahraman A, et al. Cross-link guided molecular modeling with ROSETTA. PLoS One. 2013;8:e73411. doi: 10.1371/journal.pone.0073411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson PJ, et al. Molecular architecture of the yeast Mediator complex. Elife. 2015;4:e08719. doi: 10.7554/eLife.08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, et al. Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Mol Cell Proteomics. 2014;13:2927–2943. doi: 10.1074/mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rampler E, et al. Comprehensive Cross-Linking Mass Spectrometry Reveals Parallel Orientation and Flexible Conformations of Plant HOP2–MND1. J Proteome Res. 2015;14:5048–5062. doi: 10.1021/acs.jproteome.5b00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, et al. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Vries SJ, Van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 12.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 13.Rodrigues JP, Bonvin AM. Integrative computational modeling of protein interactions. FEBS J. 2014;281:1988–2003. doi: 10.1111/febs.12771. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins. 2009;77:100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Vries SJ, et al. HADDOCK versus HADDOCK: new features and performance of HADDOCK2. 0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 16.Leitner A, Walzthoeni T, Aebersold R. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat Protoc. 2014;9:120. doi: 10.1038/nprot.2013.168. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt C, Robinson CV. A comparative cross-linking strategy to probe conformational changes in protein complexes. Nat Protoc. 2014;9:2224–2236. doi: 10.1038/nprot.2014.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zorn M, Ihling CH, Golbik R, Sawers RG, Sinz A. Mapping cell envelope and periplasm protein interactions of Escherichia coli respiratory formate dehydrogenases by chemical cross-linking and mass spectrometry. J Proteome Res. 2014;13:5524–5535. doi: 10.1021/pr5004906. [DOI] [PubMed] [Google Scholar]
- 19.Zheng C, et al. Cross-linking measurements of in vivo protein complex topologies. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006841. M110. 006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lasker K, et al. Integrative structure modeling of macromolecular assemblies from proteomics data. Mol Cell Proteomics. 2010;9:1689–1702. doi: 10.1074/mcp.R110.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russel D, et al. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller MQ, et al. An innovative method to study target protein–drug interactions by mass spectrometry. J Med Chem. 2009;52:2875–2879. doi: 10.1021/jm9000665. [DOI] [PubMed] [Google Scholar]
- 23.Leitner A, et al. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014126. M111. 014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fritzsche R, Ihling CH, Götze M, Sinz A. Optimizing the enrichment of cross-linked products for mass spectrometric protein analysis. Rapid Commun Mass Spectrom. 2012;26:653–658. doi: 10.1002/rcm.6150. [DOI] [PubMed] [Google Scholar]
- 25.Kao A, et al. Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol Cell Proteomics, mcp. 2010 doi: 10.1074/mcp.M110.002212. M110. 002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Rijkers DT, Post H, Heck AJ. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat Methods. 2015 doi: 10.1038/nmeth.3603. [DOI] [PubMed] [Google Scholar]
- 27.Müller MQ, Dreiocker F, Ihling CH, Schäfer M, Sinz A. Cleavable cross-linker for protein structure analysis: reliable identification of cross-linking products by tandem MS. Anal Chem. 2010;82:6958–6968. doi: 10.1021/ac101241t. [DOI] [PubMed] [Google Scholar]
- 28.Chakrabarty JK, Naik AG, Fessler MB, Munske GR, Chowdhury SM. Differential tandem mass spectrometry-based cross-linker: a new approach for high confidence in identifying protein cross-linking. Anal Chem. 2016;88:10215–10222. doi: 10.1021/acs.analchem.6b02886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu C, et al. Developing a Multiplexed Quantitative Cross-Linking Mass Spectrometry Platform for Comparative Structural Analysis of Protein Complexes. Anal Chem. 2016;88:10301–10308. doi: 10.1021/acs.analchem.6b03148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang B, et al. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 31.Yılmaz Su, et al. Xilmass: A New Approach toward the Identification of Cross-Linked Peptides. Anal Chem. 2016;88:9949–9957. doi: 10.1021/acs.analchem.6b01585. [DOI] [PubMed] [Google Scholar]
- 32.Götze M, et al. StavroX—a software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom. 2012;23:76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
- 33.Lima DB, et al. SIM-XL: A powerful and user-friendly tool for peptide cross-linking analysis. J Proteomics. 2015;129:51–55. doi: 10.1016/j.jprot.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Grimm M, Zimniak T, Kahraman A, Herzog F. xVis: a web server for the schematic visualization and interpretation of crosslink-derived spatial restraints. Nucleic Acids Res. 2015;43:W362–W369. doi: 10.1093/nar/gkv463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Combe CW, Fischer L, Rappsilber J. xiNET: cross-link network maps with residue resolution. Mol Cell Proteomics. 2015;14:1137–1147. doi: 10.1074/mcp.O114.042259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riffle M, Jaschob D, Zelter A, Davis TN. ProXL (Protein Cross-Linking database): A platform for analysis, visualization, and sharing of protein Cross-Linking mass spectrometry data. J Proteome Res. 2016;15:2863–2870. doi: 10.1021/acs.jproteome.6b00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erzberger JP, et al. Molecular architecture of the 40SeIF1eIF3 translation initiation complex. Cell. 2014;158:1123–1135. doi: 10.1016/j.cell.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Politis A, et al. A mass spectrometry-based hybrid method for structural modeling of protein complexes. Nat Methods. 2014;11:403–406. doi: 10.1038/nmeth.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaik M, et al. Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold. J Cell Biol. 2015;208:283–297. doi: 10.1083/jcb.201411003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding Y-H, et al. Increasing the Depth of Mass-Spectrometry-Based Structural Analysis of Protein Complexes through the Use of Multiple Cross-Linkers. Anal Chem. 2016;88:4461–4469. doi: 10.1021/acs.analchem.6b00281. [DOI] [PubMed] [Google Scholar]
- 41.Trahan C, Oeffinger M. Targeted cross-linking-mass spectrometry determines vicinal interactomes within heterogeneous RNP complexes. Nucleic Acids Res. 2016;44:1354–1369. doi: 10.1093/nar/gkv1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webb B, Sali A. Protein Structure Prediction. Vol. 1137. Humana Press; New York, NY: 2014. Protein structure modeling with MODELLER; pp. 1–15. [DOI] [PubMed] [Google Scholar]
- 43.Kuntal BK, Aparoy P, Reddanna P. EasyModeller: A graphical interface to MODELLER. BMC Res Notes. 2010;3:226–226. doi: 10.1186/1756-0500-3-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthew Allen Bullock J, Schwab J, Thalassinos K, Topf M. The Importance of Non-accessible Crosslinks and Solvent Accessible Surface Distance in Modeling Proteins with Restraints From Crosslinking Mass Spectrometry. Mol Cell Proteomics. 2016;15:2491–2500. doi: 10.1074/mcp.M116.058560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinz A. Divide and conquer: cleavable cross-linkers to study protein conformation and protein-protein interactions. Anal Bioanal Chem. 2017;409:33–44. doi: 10.1007/s00216-016-9941-x. [DOI] [PubMed] [Google Scholar]
- 46.Tan D, et al. Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states. Elife. 2016;5 doi: 10.7554/eLife.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang H-A, et al. Crystal structure of Hop2–Mnd1 and mechanistic insights into its role in meiotic recombination. Nucleic Acids Res. 2015;43:3841–3856. doi: 10.1093/nar/gkv172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kahraman A, Malmström L, Aebersold R. Xwalk: computing and visualizing distances in cross-linking experiments. Bioinformatics. 2011;27:2163–2164. doi: 10.1093/bioinformatics/btr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Vries SJ, Bonvin AM. CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One. 2011;6:e17695. doi: 10.1371/journal.pone.0017695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Song J-G, et al. Structural Insights into Ca2+-Calmodulin Regulation of Plectin 1a-Integrin β4 Interaction in Hemidesmosomes. Structure. 2015;23:558–570. doi: 10.1016/j.str.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hall Z, Schmidt C, Politis A. Uncovering the Early Assembly Mechanism for Amyloidogenic beta2-Microglobulin Using Cross-linking and Native Mass Spectrometry. J Biol Chem. 2016;291:4626–4637. doi: 10.1074/jbc.M115.691063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example input and output files along with instructions for comparative modeling of bovine cytochrome C using cross-linking data.
Example input and output files along with instructions for comparative modeling of HOP2 from A. thaliana using cross-linking data.
Example input and output files along with instructions for comparative modeling of human calmodulin using cross-linking data.
Example input and output files along with instructions for comparative modeling of MND1 from A. thaliana using cross-linking data.
Example input and output files along with instructions for protein-protein docking of human N-lobe of calmodulin and the actin binding domain of mouse plectin using cross-linking data.
Example input and output files along with instructions for protein-protein docking of mouse Ppp2r1a and human PPP2CA using cross-linking data.
Example input and output files along with instructions for protein-protein docking of HOP2 and MND1 from A. thaliana using cross-linking data.
A windows batch script that can be used to automatically run Xwalk on a series of PDB files. The batch file contains comments on how to use it. By changing the file type from .txt to .bat, the code is executable.
Description of structure and content of supplementary data sets. It is recommended to read this file before using the supplementary data sets.