Summary
Multilocular silique is a desirable agricultural trait with great potential for the development of high‐yield varieties of Brassica. To date, no spontaneous or induced multilocular mutants have been reported in Brassica napus, which likely reflects its allotetraploid nature and the extremely low probability of the simultaneous random mutagenesis of multiple gene copies with functional redundancy. Here, we present evidence for the efficient knockout of rapeseed homologues of CLAVATA3 (CLV3) for a secreted peptide and its related receptors CLV1 and CLV2 in the CLV signalling pathway using the CRISPR/Cas9 system and achieved stable transmission of the mutations across three generations. Each BnCLV gene has two copies located in two subgenomes. The multilocular phenotype can be recovered only in knockout mutations of both copies of each BnCLV gene, illustrating that the simultaneous alteration of multiple gene copies by CRISPR/Cas9 mutagenesis has great potential in generating agronomically important mutations in rapeseed. The mutagenesis efficiency varied widely from 0% to 48.65% in T0 with different single‐guide RNAs (sgRNAs), indicating that the appropriate selection of the sgRNA is important for effectively generating indels in rapeseed. The double mutation of BnCLV3 produced more leaves and multilocular siliques with a significantly higher number of seeds per silique and a higher seed weight than the wild‐type and single mutant plants, potentially contributing to increased seed production. We also assessed the efficiency of the horizontal transfer of Cas9/gRNA cassettes by pollination. Our findings reveal the potential for plant breeding strategies to improve yield traits in currently cultivated rapeseed varieties.
Keywords: Brassica napus, genome editing, CRISPR/Cas9, CLAVATA genes, multilocular silique
Introduction
Rapeseed (Brassica napus L., AACC, 2n = 38), one of the most important oil crops worldwide, provides edible oil for human diets, protein‐rich feed for animals and raw materials for industrial processes, such as biodiesel production. Achieving high yields and genetic improvements has always been the major goal in rapeseed production. The siliques of oilseed rape contain seeds that serve not only as the productive organs for the life cycle but also as storage compartments for oils and proteins, which are the predominant products of this crop. Therefore, the silique and its related traits, that is, the number of siliques per plant, the number of seeds per silique (NSS) and the seed weight (SW), are important factors for improving yield (Liu, 2000).
Similar to Arabidopsis, the silique of B. napus develops from the gynoecium, which typically comprises two carpels that are separated by a false septum, and thus has two locules (bilocular). A few multilocular (more than two carpels) lines of Brassica have been identified in nature, such as the multilocular yellow sarson in B. rapa and Santong, Silun, Duoshi in B. juncea (Liu, 2000). A multilocular silique is a desirable agricultural trait that has great potential in developing high‐yield varieties of Brassica due to the potentially greater NSS and better shatter resistance to avoid seed loss during mechanical harvest (Katiyar et al., 1998; Lv et al., 2012; Varshney, 1987; Xiao et al., 2013; Xu et al., 2013). However, only a few studies have investigated the multilocular trait in B. napus due to the lack of mutants with stable multilocular traits. Thus far, no multilocular trait has been applied to rapeseed breeding.
One and two recessive nuclear genes are responsible for the multilocular trait in B. rapa and B. juncea, respectively (He et al., 2003; Lv et al., 2012; Xiao et al., 2013). A single‐nucleotide mutation in CLAVATA3 (CLV3) gene homologue and insertion of a copia‐LTR retrotransposable element in CLAVATA1 (CLV1) gene homologue interrupt the function of the target genes and control the multilocular trait in B. rapa and B. juncea, respectively (Fan et al., 2014; Xu et al., 2017). In Arabidopsis, CLV3 acts as a small secreted peptide that interacts with a CLV1‐CLV2‐CORYNE (CRN)‐RECEPTOR‐LIKE PROTEIN KINASE2 (RPK2) receptor kinase‐mediated pathway to repress the expression of the stem cell‐promoting homeodomain transcription factor WUSCHEL in shoot apical meristems (SAMs) (Clark et al., 1993, 1997; Jeong et al., 1999; Kayes and Clark, 1998; Kinoshita et al., 2010; Müller et al., 2008). CLV1 encodes a member of the leucine‐rich repeat (LRR) receptor kinase family (Clark et al., 1997); CLV2 encodes a LRR receptor‐like protein lacking a cytoplasmic domain and acts together with a membrane‐associated protein kinase, CRN/SUPPRESSOR OF LLP1 2 (SOL2), to transmit the CLV3 signal (Jeong et al., 1999). RPK2 is another key receptor‐like kinase in the CLV pathway (Kinoshita et al., 2010). Mutations in CLV pathway genes result in expanded SAMs, an increased number of floral organs and multilocular siliques (Clark et al., 1995; Fletcher et al., 1999). The CLV pathway is functionally conserved in plants. In tomato, mutations in the homologues of CLV1, CLV2 and CLV3 increase the locule number and thus increase the fruit size (Xu et al., 2015). Mutations in the homologous genes in the CLV signalling pathway in maize and rice, such as FASCIATED EAR2 (FAE2), THICK TASSEL DWARF1 (TD1), FLORAL ORGAN NUMBER1 (FON1) and FLORAL ORGAN NUMBER4 (FON4), also increase the kernel row number in maize and the seed number per inflorescence in rice (Bommert et al., 2005, 2013; Chu et al., 2006; Suzaki et al., 2004; Taguchi‐Shiobara et al., 2001). Thus, CLV pathway genes are attractive targets for the genomic engineering of Brassica species to improve yield‐related traits.
Brassica napus resulted from a recent allopolyploidy between ancestors of B. rapa (2n = 20, AA) and B. oleracea (2n = 18, CC). In addition to more ancient polyploidization events, this recent allopolyploidy conferred an aggregate 72× genome multiplication since the origin of angiosperms (Chalhoub et al., 2014). Therefore, obtaining rapeseed mutants is challenging due to the high genetic redundancy, and genetic analyses are critical for determining gene function in both basic and applied research studies. Therefore, technologies that can target one specific copy or several homologous gene copies are needed to characterize and improve the agronomic traits of rapeseed.
Recently, various genome‐editing methods, particularly methods involving sequence‐specific nucleases (SSNs) for creating targeted double‐strand breaks (DSBs), have emerged as major breakthroughs in site‐specific genome editing (Cong et al., 2013; Li et al., 2011; Wood et al., 2011). In particular, CRISPR/CRISPR‐associated 9 (CRISPR/Cas9) is considered the most simple and effective SSN developed thus far and has been used for genome editing in major crops, including rice (Jiang et al., 2013; Miao et al., 2013; Shan et al., 2013), sorghum (Jiang et al., 2013), tobacco (Gao et al., 2015), wheat (Shan et al., 2013; Wang et al., 2014), maize (Liang et al., 2014), barley (Lawrenson et al., 2015), cotton (Wang et al., 2017), tomato (Brooks et al., 2014), soya bean (Li et al., 2015) and camelina (Jiang et al., 2017). To date, few studies have reported successful site‐directed mutagenesis using the Cas9/single‐guide RNA (sgRNA) system in rapeseed (Braatz et al., 2017). Therefore, genome‐editing methods in rapeseed have not been fully established, and the further relevant studies are required for the utilization of these methods on a practical level.
Here, we used the CRISPR/Cas9 system to generate efficient knockouts of three key genes in the CLV signalling pathway with stable transformation in rapeseed. The CRISPR/Cas9‐induced mutations of both copies of each BnCLV gene could result in multilocular siliques. In particular, the double mutation of BnCLV3 produced heritable multilocular siliques that could increase seed production. Thus, the CRISPR/Cas9 system can advance rapeseed functional genomic research studies and has the potential to improve plant breeding strategies to yield beneficial traits in currently cultivated varieties.
Results
Design of sgRNAs to knock out homologues of the CLV genes in B. napus
We have shown that B. rapa harbouring loss‐of‐function clv3 alleles produces more locules and higher seed yield (Fan et al., 2014). Thus, modifications of CLV3 in B. napus may provide an opportunity to breed high‐yield varieties. However, attempts to introduce clv3 null alleles from B. rapa into B. napus do not result in the desired traits due to the multiple dominant CLV3 alleles in the subgenomes. According to the released rapeseed genome information, B. napus cultivar Darmor‐bzh contains three CLV3 copies, that is, BnA04.CLV3 (BnaA04g15710D), BnC04.CLV3 (BnaC04g38990D) and BnC02.CLV3 (BnaC02g15230D) (http://www.genoscope.cns.fr/brassicanapus/). We confirmed the sequences of BnA04.CLV3 and BnC04.CLV3 in the B. napus pure line J9707, which is amenable to Agrobacterium‐mediated transformation; however, we were unable to amplify BnC02.CLV3. The result of genomic Southern blotting analysis verified only two copies of BnCLV3 gene in J9707 and the Darmor‐bzh reference genome (Figure S1). Further analysis of the recently released genome of B. napus cultivar ZS11 also showed that it contains only the copies of BnA04.CLV3 and BnC04.CLV3 (Sun et al., 2017). Collectively, these results indicated the lack of BnC02.CLV3 in the B. napus genome. BnA04.CLV3 and BnC04.CLV3 are 94.8% and 97.8% identical at the nucleotide and protein levels, respectively, suggesting that these genes may share similar functions. After careful analysis on the sequences of these two BnCLV3 copies, we were able to distinguish the origins of these copies by means of several single‐nucleotide polymorphisms (SNPs) (Figure S2).
To generate Cas9‐induced mutations in both copies of BnCLV3, two sgRNAs, that is, sgRNA1 (S1) and sgRNA2 (S2), that target the first exon and the C‐terminal conserved CLV3/ESR‐related (CLE) domain, respectively, were designed using the CRISPR‐P program (Lei et al., 2014; Figure 1a,b). The sgRNAs precisely matched all CLV3 copies, except for BnC04.CLV3, which has a SNP located 15‐bp upstream of the corresponding protospacer adjacent motif (PAM), whereas sgRNA2 matched well with both copies of BnCLV3 (Figure 1a). These two sgRNAs were expressed with PU3b and PU6‐1. To determine which promoter was better for Cas9 protein expression in rapeseed, two binary constructs carrying Cas9p driven by different promoters, that is, Pubi:Cas9‐BnCLV3 (referred to as UCLV3) and P35s:Cas9‐BnCLV3 (referred to as SCLV3), were generated based on the CRISPR/Cas9 multiplex genome‐editing vector as previously described by Ma et al. (2015b).
Figure 1.
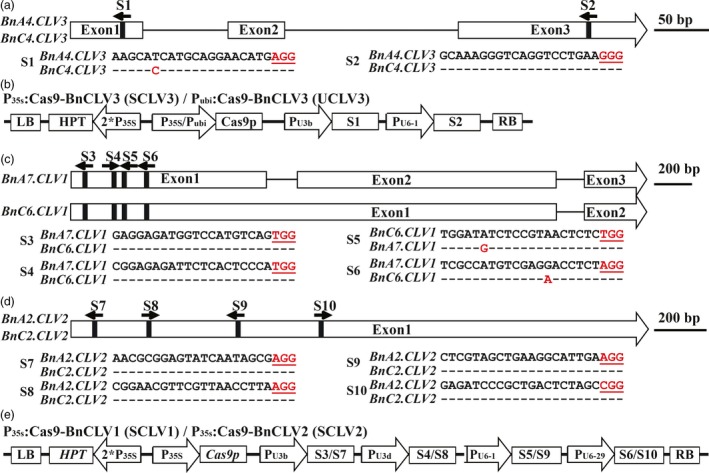
BnCLV gene models with target sequences and schematics of binary plasmid vectors. (a) The BnCLV3 gene model includes three exons (white box) separated by two introns (represented by the solid line). The vertical line in the gene model indicates the target site, and the arrow indicates the sgRNA direction. The target sequences are shown with the PAM highlighted in red. (b) The constructs of SCLV3 and UCLV3 house the following: a hygromycin resistance cassette consisting of the hygromycin phosphotransferase coding sequence driven by the cauliflower mosaic virus 35S promoter; a Cas9 expression cassette comprising the sequence encoding Cas9 driven by P35S or a ubiquitin promoter from maize; and two sgRNAs S1 and S2 driven by the U3b and U6‐1 promoters from Arabidopsis, respectively. (c, b) The BnCLV1 gene model with target sites S3 to S6 and the BnCLV2 gene model with target sites S7 to S10. (e) The binary constructs SCLV1 and SCLV2 with four sgRNAs driven by the U3b, U3d, U6‐1 and U6‐29 promoters from Arabidopsis. [Colour figure can be viewed at http://wileyonlinelibrary.com]
The rapeseed homologues of CLV1 and CLV2 were also targeted for knockout in J9707. According to the rapeseed genome information, there are two copies for each gene, that is BnA07.CLV1 (BnaA07g32120D) and BnC06.CLV1 (BnaC06g36500D) for BnCLV1 with 96.1% identity at the nucleotide level and BnA02.CLV2 (BnaA02g12070D) and BnC02.CLV2 (BnaC02g45200D) for BnCLV2 with 95.3% identity at the nucleotide level. According to the sequence alignment of these two copies of the BnCLV1 and BnCLV2 genes, polymorphisms distinguished the origins of these gene copies (Figures S3 and S4).
Two binary constructs carrying four sgRNAs within each target gene with Cas9p driven by P35S, that is P35s:Cas9‐BnCLV1 (containing sgRNA3‐sgRNA6, referred to as SCLV1) and P35s:Cas9‐BnCLV2 (containing sgRNA7‐sgRNA10, referred to as SCLV2), were generated as previously described by Ma et al. (2015b). These sgRNAs matched well with both copies of BnCLV1 and BnCLV2, except for S5 and S6, which contain an SNP located 15 bp upstream of the corresponding PAM motif in BnA07.CLV1 and 7 bp upstream of the corresponding PAM motif in BnC06.CLV1, respectively (Figure 1c‐e). Each site was located in the 5′ portion of each gene and was deliberately selected to ensure that gene disruptions altering the reading frame would produce a translation product lacking enzymatic activity.
Rapid identification of the edited lines using native PAGE screening
These four constructs were independently transformed into J9707 using Agrobacterium‐mediated transformation and generated 335, 366, 119 and 40 independent lines for SCLV3, UCLV3, SCLV1 and SCLV2, respectively. According to a PCR examination using NPTII gene‐specific primers, 74.6% (250/335), 66.7% (244/366), 84.9% (101/119) and 92.5% (37/40) of these T0 lines carried T‐DNA insertions (Figure 2a).
Figure 2.
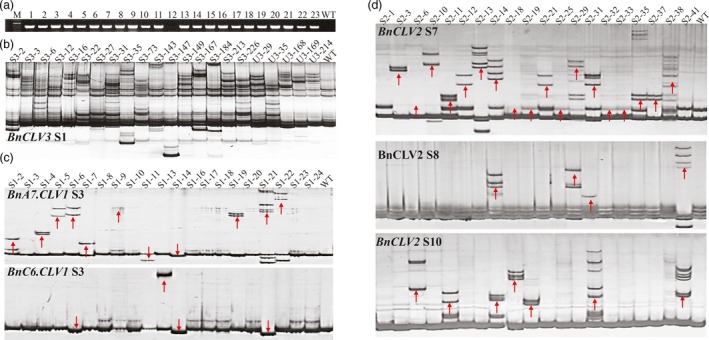
Detection of mutations in BnCLVs using PAGE method. (a) The transgenic positive detection in T0 regenerated plants via a PCR assay using NPT II gene‐specific primers and J9707 (WT) as a negative control. (b) Detection of the targeted mutations in T0 plants with the WT as a negative control. (c) Detection of mutations in different targets in the two BnCLV1 copies in T0 plants with the WT as a negative control. (d) Detection of mutations in different targets in the two BnCLV2 copies in T0 plants with the WT as a negative control. The numbers ‘S#‐#’ and ‘U#‐#’ above the PAGE gels represents the corresponding individual IDs ‘SCLV#‐#’ and ‘UCLV#‐#’, respectively. ‘S#’ represents the specific targets of BnCLVs. A red arrow indicates that the tested target has been edited. [Colour figure can be viewed at http://wileyonlinelibrary.com]
We performed polyacrylamide gel electrophoresis (PAGE) to identify the edited lines. Thus, the PCR products of each target site were denatured, renatured and subsequently separated using native PAGE. This method is based on the slower migration of heteroduplex DNA (with mutation) than homoduplex DNA (without mutation) on native PAGE (Zhu et al., 2014). Many plants displayed band profiles that differed from the profiles displayed by the wild‐type (WT) plants on the nondenaturing PAGE gels, indicating the presence of mutations in the target sites (Figure 2b‐d). According to the PAGE screening, 36/250 (14.4%), 15/244 (6.1%), 47/101 (46.5%) and 21/37 (56.8%) mutant plants were detected for SCLV3, UCLV3, SCLV1 and SCLV2, respectively (Tables S2‐S4). The editing efficiency of SCLV3 was higher than for UCLV3, suggesting that P35s has higher activity than Pubi in rapeseed. Interestingly, the contributions of the ten selected sgRNAs in directing Cas9 and mutating the target genes were not equal; the highest efficiency of mutagenesis was observed at S7 (48.7%) and S3 (46.5%), while an intermediate efficiency of mutagenesis was observed at S10 (24.3%) and S8 (13.5%), a lower efficiency of mutagenesis was observed at S1 (14.4% in SCLV3 and 6.2% in UCLV3) and S4&S5 (5.0%), and no mutation was detected at S2, S6 and S9 (Tables 1; S5 and S6), indicating that the appropriate selection of sgRNA pairs is important for effectively generating indels. The average mutagenesis efficiency of the sgRNAs driven by different promoters ranged widely from 0% to 47.6% (Table 1), indicating that not all promoters were effective in driving genome editing in rapeseed.
Table 1.
Analysis of putative factors affecting editing efficiency at different sgRNA targets
| Promoter | sgRNA | Target gene | Target sequencea | GC% content (without PAM) | Continuous matching between target and sgRNA sequenceb | Editing efficiencyc | Average efficiency |
|---|---|---|---|---|---|---|---|
| AtU3b | S1 | BnCLV3 | AAGCATCATGCAGGAACATGAGG | 45.0% | 3 | 14.4% | 11.0% |
| S4 | BnCLV1 | CGGAGAGATTCTCACTCCCATGG | 55.0% | 0 | 5.0% | ||
| S8 | BnCLV2 | CGGAACGTTCGTTAACCTTAAGG | 45.0% | 5 | 13.5% | ||
| AtU3d | S3 | BnCLV1 | GAGGAGATGGTCCATGTCAGTGG | 55.0% | 8 | 46.5% | 47.6% |
| S7 | BnCLV2 | AACGCGGAGTATCAATAGCGAGG | 50.0% | 5 | 48.7% | ||
| AtU6_1 | S2 | BnCLV3 | GCAAAGGGTCAGGTCCTGAAGGG | 55.0% | 3 | 0.0% | 0.0% |
| S5 | BnCLV1 | TGGATATCTCCGTAACTCTCTGG | 45.0% | 7 | 0.0% | ||
| S9 | BnCLV2 | CTCGTAGCTGAAGGCATTGAAGG | 50.0% | 5 | 0.0% | ||
| AtU6_29 | S6 | BnCLV1 | TCGCCATGTCGAGGACCTCTAGG | 60.0% | 8 | 0.0% | 12.2% |
| S10 | BnCLV2 | GAGATCCCGCTGACTCTAGCCGG | 60.0% | 5 | 24.3% |
The PAM is underlined.
Counting the maximum number of continuous matching bases between target and sgRNA sequence.
The percentage of edited plants over the total number of tested plants for the corresponding targets.
To determine whether PAGE‐based assays can readily be used to distinguish indels of any length, we generated a set of indel plasmids with deletions ranging from 1 to 8 bp. These indels were PCR‐amplified and evaluated using PAGE analysis. All the PCR amplicons with different deletion sizes, except for 1‐bp indels, could be distinguished from WT (Figure S5).
Variety and frequency of mutations in BnCLV3
To confirm the PAGE screening results, the PCR products from 22 edited lines of BnCLV3 were sequenced. Various mutations, including the insertion and deletion of different nucleotides, were produced at the S1 target in all lines (Figure 3a), indicating that PAGE‐based screening is an efficient and simple method for identifying edited lines. Interestingly, S1 simultaneously targeted both copies of BnCLV3, even though the copy paired with the BnC04.CLV3 target had one mismatch. This finding is consistent with the reported Cas9 specificity in targeting DNA sites with 1–5 mismatches proximal to the PAM. Of the 22 T0 lines, 17 lines were double mutants of BnCLV3, while the mutations in four lines and one line were restricted to BnA04.CLV3 and BnC04.CLV3, respectively (Table S7). Of all the mutations examined, nearly half (48.7%, 19/39) of the BnCLV3 loci were putatively heterozygous mutations, 25.6% (10/39) of the loci were chimeric mutations, 20.5% (8/39) of the loci were putatively bi‐allelic mutations, and 5.1% (2/39) of the loci were putatively homozygous mutations (Table S7). According to the allele mutation types, 47.6% of the mutations were nucleotide insertions, 47.6% of the mutations were nucleotide deletions, and 4.8% of the mutations were simultaneous nucleotide deletions and insertions (Figure 3b). Of the insertion mutations, 100% were 1‐bp insertions, with a marked preference for C (70.0%, 21/30) nucleotide inserts over A (20.0%, 6/30), T (10.0%, 3/30) and G (0, 0/30) nucleotide inserts (Figure 3b), which was inconsistent with previous reports in rice, camelina and citrus, in which most 1‐bp insertions were A or T (Jia et al., 2017; Jiang et al., 2017; Ma et al., 2015b; Zhang et al., 2014). Most deletion mutations (63.3%, 19/30) were short (<10 bp) deletions, and the remaining 36.7% of the mutations (11/30) were longer deletions ranging from 10 bp to 91 bp (Figure 3b). Overall, approximately just over half (57.1%) of all mutations changed by only 1 bp. Expectedly, all 1‐bp indels occurred immediately upstream of the DSB position at the fourth base from the PAM site.
Figure 3.
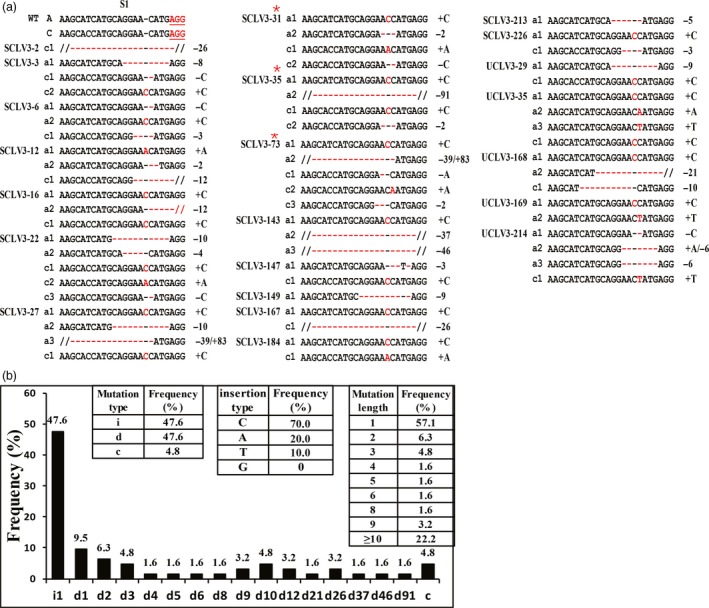
Detailed genotype analysis of mutations in BnCLV3 on S1 in T0 generation. (a) Sequencing results of 22 editing T0 plants at the S1 site. Insertions and deletions are indicated in red font and with red hyphens, respectively. Edited plants with red stars have multilocular phenotypes. On the left, A and C and the WT allele of the BnA04.CLV3 and Bn0C4.CLV3 copies, respectively; a# and c# show the mutant allele numbers. ‘−’ and ‘+’ indicate the deletion and insertion of the indicated number of nucleotides, respectively; ‘−/+’ indicates the simultaneous deletion and insertion of the indicated number of nucleotides. (b) Mutation types and frequency at the S1 target site in 22 T0 plants. In the left insert table, the occurrence of deletions (d), insertions (i) and combined (c) mutation types is shown. In the right insert table, the frequency of different mutation lengths is shown. In the middle insert table, frequency of different insertion types is shown. X‐axis: d#, # of base pair (bp) deleted from the target site; i#, # of bp inserted at the target site; c#, combined mutation. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Isolation of stable edited BnCLV3 lines without T‐DNA at generation T2
To obtain stable mutant lines, 11 T0 lines carrying mutations in BnCVL3 were self‐pollinated, and individual T1 progeny were genotyped via a direct sequence analysis of the PCR products of the target sites. The allelic mutations in the T0 mutant plants were transmitted to the T1 generation at an average transmission rate of 82.3% (Table 2). For instance, the mutations detected in the T1 progenies matched the mutations observed in the corresponding SCLV3‐31 and SCVL3‐35 T0 lines (Figure 4). Most lines produced T1 plants carrying homozygous mutations at one or two BnCLV3 loci, indicating that the mutations were stably inherited by the next generation and fixed as homozygous genotypes (Table 2). Interestingly, two new mutations that failed in 29 T0 lines were identified in 519 T1 progeny lines at the S1 site of BnC04.CLV3 (Figure S6), indicating that the WT site was further edited at low efficiency during the growth of the transgenic plants.
Table 2.
Molecular and genetic analysis of CRISPR/Cas9‐induced mutations in BnCLV3 and their transmission to the T1 and T2 generations
| Analysis of T0 plants | Mutation segregation in T1 plants | Mutation segregation in T2 plants | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plant ID | Genotype of BnCLV3 | Mutation detected (bp)a | No. of tested plants with editing | WT | Hetero | Homo | Mutation transmissionb | T‐DNA free | No. of tested plants with editing | WT | Hetero | Homo | Mutation transmission | T‐DNA free |
| SCLV3‐3 | Aa | −8 | 5 | 0 (AA) | 0 (Aa) | 5 (aa) | 100.0% | 1/35 (2.9%) | 19 | 0 (AA) | 0 (Aa) | 19 (aa) | 100.0% | 42/137 (0.7%) |
| Cc | +1, −1 | 2 (CC) | 3 (Cc) | 0 (cc) | 60.0% | 17 (CC) | 2 (Cc) | 0 (cc) | 10.5% | |||||
| SCLV3‐6 | Aa | −1, +1 | 4 | 1 (AA) | 2 (Aa) | 1 (aa) | 75.0% | 5/18 (11.1%) | 6 | 2 (AA) | 3 (Aa) | 1 (aa) | 66.7% | 9/9 (100.0%) |
| Cc | −3 | 1 (CC) | 2 (Cc) | 1 (cc) | 75.0% | 0 (CC) | 0 (Cc) | 6 (cc) | 100.0% | |||||
| SCLV3‐16 | Aa | +1, −11 | 3 | 0 (AA) | 0 (Aa) | 3 (aa) | 100.0% | 0/34 (0%) | 20 | 0 (AA) | 0 (Aa) | 20 (aa) | 100.0% | 2/65 (3.1%) |
| Cc | +1 | 0 (CC) | 3 (Cc) | 0 (cc) | 100.0% | 7 (CC) | 13 (Cc) | 0 (cc) | 65.0% | |||||
| SCLV3‐27 | Aa | +1, −10, −39/+83 | 4 | 0 (AA) | 2 (Aa) | 2 (aa) | 100.0% | 12/30 (40.0%) | 10 | 0 (AA) | 0 (Aa) | 10 (aa) | 100.0% | 23/31 (74.2%) |
| Cc | +1 | 4 (CC) | 0 (Cc) | 0 (cc) | 0.0% | 10 (CC) | 0 (Cc) | 0 (cc) | 0.0% | |||||
| SCLV3‐31 | aa | +1, −2 | 4 | 0 (AA) | 0 (Aa) | 4 (aa) | 100.0% | 9/35 (20.0%) | 5 | 0 (AA) | 0 (Aa) | 5 (aa) | 100.0% | 7/20 (35.0%) |
| cc | +1, −1 | 0 (CC) | 0 (Cc) | 4 (cc) | 100.0% | 0 (CC) | 0 (Cc) | 5 (cc) | 100.0% | |||||
| SCLV3‐35 | aa | +1, −91 | 8 | 0 (AA) | 0 (Aa) | 8 (aa) | 100.0% | 2/36 (5.5%) | 5 | 0 (AA) | 0 (Aa) | 5 (aa) | 100.0% | 3/15 (20.0%) |
| cc | +1, −2 | 0 (CC) | 0 (Cc) | 8 (cc) | 100.0% | 0 (CC) | 0 (Cc) | 5 (cc) | 100.0% | |||||
| SCLV3‐167 | Aa | +1 | 2 | 2 (AA) | 0 (Aa) | 0 (aa) | 0.0% | 2/9 (22.2%) | 9 | 9 (AA) | 0 (Aa) | 0 (aa) | 0.0% | 8/41 (19.5%) |
| Cc | −26 | 0 (CC) | 0 (Cc) | 2 (cc) | 100.0% | 0 (CC) | 0 (Cc) | 9 (cc) | 100.0% | |||||
| SCLV3‐184 | aa | +1 | 5 | 0 (AA) | 0 (Aa) | 5 (aa) | 100.0% | 2/35 (5.7%) | 14 | 0 (AA) | 0 (Aa) | 14 (aa) | 100.0% | 5/84 (6.0%) |
| Cc | +1 | 0 (CC) | 2 (Cc) | 3 (cc) | 100.0% | 4 (CC) | 3 (Cc) | 7(cc) | 71.4% | |||||
| UCLV3‐169 | aa | +1, +1 | 2 | 0 (AA) | 0 (Aa) | 2 (aa) | 100.0% | 3/11 (18.2%) | 6 | 0 (AA) | 0 (Aa) | 6 (aa) | 100.0% | 0/15 (0%) |
| Cc | −6 | 0 (CC) | 1 (Cc) | 1 (cc) | 100.0% | (CC) | 3 (Cc) | 3 (cc) | 100.0% | |||||
| UCLV3‐29 | Aa | −9 | 2 | 0 (AA) | 0 (Aa) | 2 (aa) | 100.0% | 9/18 (50.0%) | 2 | 0 (AA) | 0 (Aa) | 2 (aa) | 100.0% | 11/11 100.0%) |
| Cc | +1 | 1 (CC) | 1 (Cc) | 0 (cc) | 50.0% | 2 (CC) | 0 (Cc) | 0 (cc) | 0.0% | |||||
| UCLV3‐214 | aa | +1/−6, −1, −6 | 2 | 0 (AA) | 0 (Aa) | 2 (aa) | 100.0% | 4/18 (22.2%) | 2 | 0 (AA) | 0 (Aa) | 2 (aa) | 100.0% | 7/7 (100.0%) |
| Cc | +1 | 1 (CC) | 1 (Cc) | 0 (cc) | 50.0% | 2 (CC) | 0 (Cc) | 0 (cc) | 0.0% | |||||
Hetero, heterozygous; Homo, homozygous.
‘−’ and ‘+’ indicate the deletion and insertion of the indicated number of nucleotides, respectively; ‘−/+’ indicates the simultaneous deletion and insertion of the indicated number of nucleotides; ‘+,…’ indicates multiple types of insertions or deletions occurring in different mutation events at the same target site.
Based on the number of plants carrying the observed mutation over the total number of plants tested.
Figure 4.

Germ‐line transmission of CRISPR/Cas9‐induced mutations at the S1 target site of SCLV3 from the T0 generation to the T2 generation. CRISPR/Cas9‐induced insertions and deletions are indicated by red font and red hyphens, respectively. On the left, A and C and the WT allele of the BnA4.CLV3 and BnC4.CLV3 copies, respectively; a# and c# show the mutant allele numbers. ‘−’ and ‘+’ indicate the deletion and insertion of the indicated number of nucleotides, respectively. [Colour figure can be viewed at http://wileyonlinelibrary.com]
We further analysed the transmission of the mutations from these T1 plants to their T2 offspring. The allelic mutations in the T1 mutant plants were transmitted to the T2 generation at an average transmission rate of 73.4% (Table 2). Consistently, the mutations detected in the T2 progeny lines matched the mutations observed in the corresponding SCLV3‐31 and SCVL3‐35 T0 and T1 lines (Figure 4). Of the 98 T2 plants that were sequenced, 84 (85.7%) and 35 (35.7%) plants had homozygous mutations at BnA04.CLV3 and BnC04.CLV3, respectively (Table 2). Altogether, the T1 and T2 sequence data from 11 T0 lines provided strong evidence for stable germ‐line transmission of Cas9‐induced mutations in rapeseed.
To investigate the potential for achieving targeted modifications without incorporating foreign DNA into the rapeseed genome, we performed PCR assays of the T1 and T2 plants. The NPT II gene was not detected in 49 of 279 (17.6%) T1 plants and 117 of 435 (26.9%) T2 plants originating from 11 independent T0 lines (Table 2). Altogether, a variety of BnCLV3 single and double homozygous T‐DNA‐free mutants were obtained in the T2 generation (Table S8). Therefore, T‐DNA‐free plants carrying the desired gene modifications could be acquired through genetic segregation in rapeseed.
Multilocular phenotype can be recovered by knockout mutations of both copies of each BnCLV gene
Because all 1‐bp indels and most other indels change the reading frame of the gene, most mutations generated in the BnCLV3 genes are predicted to lead to gene knockouts. Expectedly, eight of the 494 T0‐positive transgenic plants showed a visible knockout phenotype with multilocular silique. Three of these plants (i.e. SCLV3‐31, SCLV3‐35 and SCLV3‐73) were sequenced, and all the BnCLV3 loci in these plants were devoid of any WT alleles at the S1 target site, showing either putative bi‐allelic mutations or chimeric mutations (Figure 3a; Table S7). Thus, the CRISPR/Cas9 system can efficiently generate targeted mutations in the rapeseed genome.
To obtain a stable knockout phenotype of BnCLV1, three T0 mutants were self‐pollinated and produced T1 progeny carrying various mutations at both BnCLV1 loci (Table 3). Expectedly, six of these plants could produce multilocular siliques with two to four locules, and the percentage of multilocular siliques ranged from 2.1% to 50.0% (Table 3). Sequencing analysis revealed that all these T1 plants have homozygous mutations in both copies of the BnCLV1 genes, which are predicted to lead to frameshift mutations that most likely result in nonfunctional proteins. As a control, the mutant SCLV1‐13‐4, with a 1‐bp homozygous insertion in BnA07.CLV1, showed a phenotype similar to WT (Table 3). Both SCLV1‐21‐12 and SCLV1‐22‐1, with frameshift mutations in BnC06.CLV1 and deletions in multiples of three nucleotides resulting the deletion of several amino acids in BnA07.CLV1, also showed a phenotype similar to WT (Table 3). Thus, both copies of the BnCLV1 gene function redundantly in multilocular trait development.
Table 3.
Phenotypic and genotypic analysis of BnCLV1 and BnCLV2 mutants and their transmission to T1 generation
| Line | Generation | Multilocular siliques%a | BnA07.CLV1 | BnC06.CLV1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S3b | S4 | S5 | S6 | S3 | S4 | S5 | S6 | |||
| SCLV1‐13 | T0 | / | +A, Homo | WT | WT | WT | WT | WT | WT | WT |
| SCLV1‐13‐4 | T1 | 0.0 | +A, Homo | WT | WT | WT | WT | WT | WT | WT |
| SCLV1‐21 | T0 | / | Chimeric | / | / | / | −11 bp, Homo | WT | WT | WT |
| SCLV1‐21‐3 | T1 | 3.1 | −37 bp, Homo | WT | WT | WT | −11 bp, Homo | WT | WT | WT |
| SCLV1‐21‐24 | T1 | 40.7 | −37 bp, Homo | WT | WT | WT | −11 bp, Homo | WT | WT | WT |
| SCLV1‐21‐6 | T1 | 2.1 | −37 bp, +1 bp | WT | WT | WT | −11 bp, Homo | WT | WT | WT |
| SCLV1‐21‐2 | T1 | 18.2 | −37 bp, −15 bp, +1 bp | WT | WT | WT | −11 bp, Homo | WT | WT | WT |
| SCLV1‐21‐5 | T1 | 50.0 | −37 bp, −15 bp, +1 bp | WT | WT | WT | −11 bp, Homo | WT | WT | WT |
| SCLV1‐21‐25 | T1 | 4.0 | −37 bp, −15 bp, +1 bp | WT | WT | WT | −11 bp, Homo | WT | WT | WT |
| SCLV1‐21‐12 | T1 | 0.0 | −15 bp, Homo | WT | WT | WT | −11 bp, Homo | WT | WT | WT |
| SCLV1‐22 | T0 | / | Hetero | / | / | / | +G, Homo | WT | WT | WT |
| SCLV1‐22‐1 | T1 | 0.0 | −27 bp, Homo | WT | WT | WT | +G, Homo | WT | WT | WT |
| BnA02.CLV2 | BnC02.CLV2 | |||||||||
| S7 | S8 | S9 | S10 | S7 | S8 | S9 | S10 | |||
| SCLV2‐11 | T0 | / | −A, Homo | WT | WT | WT | Hetero | / | WT | WT |
| SCLV2‐11‐2 | T1 | 0.0 | −A, Homo | WT | WT | WT | −A, −3 bp | WT | WT | WT |
| SCLV2‐12 | T0 | / | Chimeric | / | WT | WT | Hetero | / | / | Hetero |
| SCLV2‐12‐9 | T1 | 10.0 | +A, Homo | +C, Homo | WT | WT | −A, Homo | WT | WT | WT |
| SCLV2‐12‐10 | T1 | 44.0 | +G, −3 bp | +C, Homo | WT | WT | −A, Homo | WT | WT | +T, Hetero |
| SCLV2‐12‐11 | T1 | 10.5 | +G, −3 bp | +C, Homo | WT | WT | −A, Homo | WT | WT | WT |
| SCLV2‐24 | T0 | / | WT | WT | WT | WT | Hetero | / | WT | WT |
| SCLV2‐24‐3 | T1 | 0.0 | WT | WT | WT | WT | +A, Homo | WT | WT | WT |
| SCLV2‐24‐4 | T1 | 0.0 | WT | WT | WT | WT | +A, Homo | WT | WT | WT |
See footnotes for Table 2.
Similarly, three T0 editing lines were self‐pollinated and produced T1 progeny carrying various mutations at different target sites at the BnCLV2 loci (Table 3). Consistently, three T1 plants originating from SCLV2‐12 could produce multilocular siliques with two to four locules, reflecting frameshift mutations in both BnCLV2 copies (Table 3). As a control, the mutants SCLV2‐24‐3 and SCLV2‐24‐4, each with a 1‐bp homozygous insertion in BnC02.CLV2, showed a phenotype similar to WT (Table 3). A T1 line (i.e. SCLV2‐11‐2) with a 1‐bp homozygous deletion in BnA02.CLV2 and heterozygous mutations in BnC02.CLV2 showed a phenotype similar to WT (Table 3). Thus, the multilocular trait is also controlled by both BnCLV2 copies with redundant functions in rapeseed.
The number of leaves, NSS and the SW were significantly increased in the BnCLV3 mutants
To characterize the phenotype of the BnCLV3 mutants, all homozygous mutant T2 lines with different frameshift mutations (Table S8) were grown in the field. The leaf number in the 30‐d‐old seedlings in the double mutants was significantly higher (P < 0.01) than in the WT control plants, whereas this trait in the two single mutants of BnCLV3 was comparable to WT control (Figure 5a,f). The leaf numbers and the size of the SAMs of inflorescences were also dramatically higher in the double mutants than in the WT control (Figure 5b and c). Consistent with this finding, the number of the four floral organs was significantly higher (P < 0.01) only in the double mutants (Figures 5d,f; S7). All the siliques in the double mutants were multilocular, ranging from 5.0 to 7.9 with a mean ± SD value of 6.6 ± 0.7 locules per silique, although the WT and both single mutants were bilocular siliques (Figure 5e,f). The multilocular siliques were shorter, rounder and thicker as reported in B. rapa (Fan et al., 2014; Figure 5e,f). The NSS and TSW were simultaneously increased to more than 10.8 and 0.92 g, compared with the WT average of 24.4 and 3.38 g, respectively; consequently, the SW per silique was increased by more than 74.4% on average, in contrast to 0.08 g in the WT (Figure 5f). Thus, BnA04.CLV3 and BnC04.CLV3 contribute to the increased leaf number and locule number of siliques, and the simultaneous mutation of the two homo‐alleles confers a multilocular trait with high‐yield potential. Therefore, the double mutants of BnCLV3 generated in the present study might provide excellent starting materials for high‐yield breeding in rapeseed.
Figure 5.
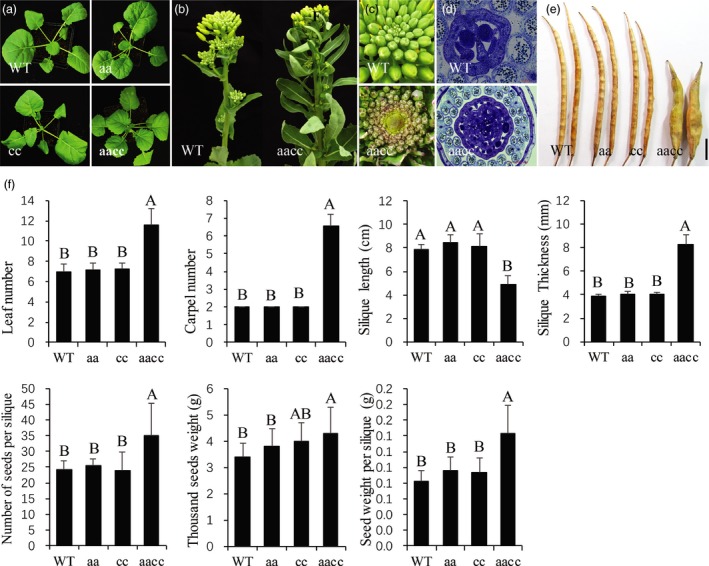
Phenotypes of the BnCLV3 mutants. (a) Leaf numbers of 30‐day‐old seedlings in WT, single and double homozygous mutants of BnCLV3. (b, c) The inflorescences (b) and SAM (c) in the WT and a double homozygous mutant of BnCLV3. (d) Cross sections of gynoecia in the WT and a double homozygous mutant of BnCLV3 at stages 9–10. (e) Siliques in the WT, single and double homozygous mutants of BnCLV3. Bar = 1 cm. (f) Statistical analysis of the leaf number, carpel number, silique length, silique thickness, NSS, thousand seeds weight and SW per silique in the WT and single and double homozygous mutants of BnCLV3. The data and error bars represent the mean ± SD (n ≥ 15 plants for each genotype). Upper‐case letters indicate a significant difference at the 0.01 probability level. aa, homozygous mutation of BnA04.CLV3; cc, homozygous mutation of BnC04.CLV3; aacc, double homozygous mutation of BnA04.CLV3 and BnC04.CLV3. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Off‐target activity of CRISPR/Cas9 in T0 transgenic B. napus plants
To determine whether off‐targeting occurred in the present study, we searched the B. napus genome for putative off‐target sites with a high homology to the five sgRNAs that detected on‐target mutations according to the CRISPR‐P program (Lei et al., 2014). These potential off‐target sites and their related genome positions are listed in Table S9. There were 17, 18, 11, 4 and 7 putative off‐target sites for S1, S3, S4, S7 and S8, respectively (Table 4).
Table 4.
Detection of potential off‐target effects for each sgRNA target site in T0 mutated plants
| On‐target site | Binary vector | No. of plants for sequencing | Putative off‐target sites | Off‐target editing |
|---|---|---|---|---|
| S1 | SCLV3 + UCLV3 | 47 | 17 | No |
| S3 | SCLV1 | 22 | 18 | No |
| S4 | SCLV1 | 22 | 11 | No |
| S7 | SCLV2 | 45 | 4 | No |
| S8 | SCLV2 | 45 | 7 | No |
High‐throughput sequencing of the PCR products of these 57 potential sites from many T0 mutated plants showed no mutations (Tables 4; S9), suggesting that the off‐target effect is negligible when the sgRNA specificity is considered well according to the genome sequence. Thus, the CRISPR/Cas9 system has a high specificity for targeted mutagenesis in B. napus.
Genome editing of cultivars by crossing genome‐edited lines with other cultivars
Because few cultivars are amenable to transformation and introducing recessive alleles by crossing is tedious, we explored the efficiency of the horizontal transfer of the Cas9/gRNA cassettes using open pollination. The B. napus cultivar HY, which has a specific lobe‐leaf trait, was grown near the T2 SCLV3‐35 lines (homozygous mutation in BnA04.CLV3 and bi‐allelic mutation in BnC04.CLV3 at the S1 site) in an isolated area of the field to control pollination. Open‐pollination seeds from HY were germinated, and seedlings with different leaf shapes from HY, which is an indicator of hybridization with SCLV3‐35 for the incompletely dominant lobed‐leaf trait, were selected for further genotyping. In total, 90 of the 2980 seedlings were selected using this method for an approximately 3.02% natural outcrossing rate (Figure 6a). Of these 90 seedlings, 68 seedlings carried the transgene insertion transmitted from the SCLV3‐35 progeny. Mixed genomic DNA from these plants was used as the template for the amplification of the S1 site using BnCLV3‐specific primers. The products were purified to generate a DNA library for high‐throughput sequencing. WT plants (HY) were also included in the genome resequencing and subsequent analysis as controls. The reads from the S1 site of BnCLV3 were aligned to the WT DNA sequences to detect mutations, and approximately 1 WT:1 mutant reads were observed at the S1 site of each copy of BnCLV3 (Figure 6b,c). Almost all mutated reads matched the reads observed in the corresponding parental SCLV3‐35 line (original mutants, Figure 6b,c). Interestingly, various novel mutations were detected at the S1 site of BnA04.CLV3 in these progeny with 10.57% efficiency, which clearly showed that all novel mutations occurred in the HY allele based on the polymorphism between the HY and SCLV3‐35 lines in the amplicon fragment (Figure 6b,c; Table S10). This result indicated that the WT allele (HY) was further edited with a low efficiency in the presence of CRISPR‐Cas9 (Figure 6b,c; Table S10). Various novel mutations were also detected at the S1 site of BnC04.CLV3 in these progeny with 2.62% efficiency. However, it could not be determined in which allele the novel mutation had occurred because there were no polymorphisms between the HY and SCLV3‐35 lines in the amplicon fragment of BnC4.CLV3 (Table S10). Additionally, a few unexpected reads with novel mutations in the WT DNA were also observed, which might reflect primer mismatches during PCR because of the much lower frequency than that of the HY progeny (Figure 6b,c).
Figure 6.
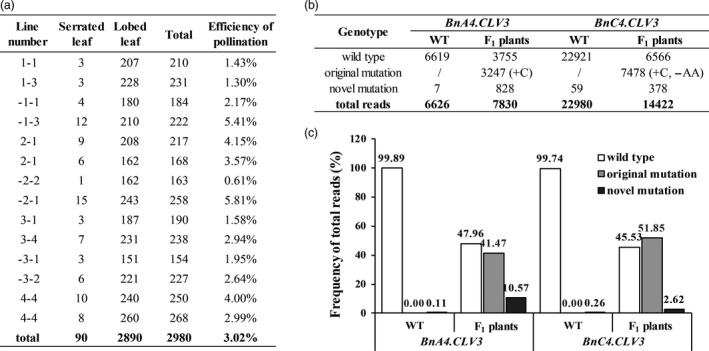
Genome editing of cultivars by crossing Cas9‐sgRNA lines. (a) Selection of the natural outcrossing plants from open‐pollination progeny of HY. Plants with serrated leaves are hybrids of HY and the double homozygous mutant SCLV3‐35 for the incompletely dominant lobed‐leaf trait. (b) The read numbers at the S1 site of BnCLV3 in mixed genomic DNA from F1 hybrid plants with T‐DNA are shown, and WT (HY) was included as a control. (c) Frequency of different genotype reads. Original mutant reads, reads with the same mutations detected in SCLV3‐35; novel mutant, reads with different mutations detected in SCLV3‐35.
Discussion
In this study, we used RNA‐guided Cas9 to induce targeted mutations in rapeseed and report the stable transmission of mutations across generations. P35s resulted in better Cas9 protein expression and a higher efficiency of mutagenesis in rapeseed than Pubi, which was inconsistent with a previous report by Ma et al. (2015b), who recommended Pubi rather than P35s for Cas9 protein expression in dicot plants. A visible multilocular silique can be recovered after knocking out all copies of each BnCLV gene in rapeseed, which is similar to observations in Arabidopsis and exemplifies the use of RNA‐guided Cas9 to target important traits in Brassica crops based on knowledge regarding gene function from model plants. We assayed 57 potential off‐target loci, and none of these genes showed evidence of a CRISPR/Cas9 system‐induced mutation, indicating that well‐designed specific sgRNAs do not target undesired sites. We obtained a variety of transgene‐free B. napus plants with homozygous mutations in the target gene, supporting the potential for further biotechnological applications.
Several crucial factors, including the expression levels of Cas9 and sgRNA, GC% content, targeting context and secondary structure of the target sgRNAs, may influence sgRNA efficacy in plants (Ma et al., 2015b; Makarova et al., 2011; Wang et al., 2014). In the present study, the mutagenesis efficiency had a wide range – from 0% to 48.7% in T0 for the ten sgRNAs (Table 1). To determine the parameters affecting the Cas9 targeting efficiency in rapeseed, we analysed the GC% content and secondary structures of all target sgRNAs, but we did not detect any association with their respective editing efficiency (Table 1). We further detected the expression of Cas9p and sgRNAs in T0‐positive transgenic lines with targeting of the S3‐S6 sites of the BnCLV1 genes. These gRNAs targeting the same gene exhibited dramatically variable genome‐editing efficiencies, such as 46.5% in S3, 5.0% in S4 and 0% in S5 and S6. The overall levels of Cas9 and sgRNAs in these plants were similar (Figure S8), although the S3 and S4 targets were edited in six plants, and S5 and S6 failed to show any editing. The expression levels of Cas9 and sgRNAs might not be the limiting factors of genome editing in B. napus T0 plants. In summary, the variations in the mutagenesis efficiency in different sgRNAs could most likely reflect differences in the nucleotide composition of the sgRNAs. Although prediction algorithms have been developed to evaluate the guide activity (Haeussler et al., 2016; Lei et al., 2014), the accuracy of these algorithms in different crops, such as B. napus, requires additional studies.
Plant genome‐editing techniques largely depend on plant genetic transformation. Compared with other crops, the transformation efficiency in most B. napus cultivars remains low. For instance, Braatz et al. (2017) reported site‐directed mutagenesis using the Cas9/sgRNA system in rapeseed using the spring cultivar Haydn as the transformation recipient and showed that only one transgenic plant regenerated at the first generation for a transformation rate of 0.9%. Thus, the application of CRISPR/Cas9‐induced mutations in rapeseed is likely limited in certain currently cultivated plants. In the present study, the pure B. napus line J9707 was a good transformation recipient at a transformation rate ranging from 66.7% to 92.5%. Thus, we hypothesize that crossing Cas9‐gRNA lines generated from a suitable transformation recipient with other cultivars, together with marker‐assisted selection, is an alternative for achieving desired gene modifications.
The allotetraploid B. napus contains two distinct but closely related homologous subgenomes. Consequently, certain important traits are controlled by several gene copies with redundant functions in both subgenomes in rapeseed. For instance, the multilocular trait in the present study was controlled by both copies of each BnCLV gene, and only double homozygous mutants showed this knockout phenotype. To date, no spontaneous or induced multilocular mutants have been reported in B. napus, likely reflecting its allotetraploid nature and the extremely low probability of double mutations in the same plant. Thus, the simultaneous alteration of multiple gene copies by CRISPR/Cas9 mutagenesis has great potential in revealing gene function and generating agronomically important mutations in crops. In a previous study, a single‐nucleotide mutation in a BrCLV3 gene homologue resulted in a weak mutation phenotype that produced siliques with 3‐4 locules and an increased NSS without a reduction in TSW (Fan et al., 2014). In the present study, the double homozygous mutants of the BnCLV3 genes likely exhibited a full clv3 loss‐of‐function phenotype, with siliques harbouring 5.0–7.9 locules, and significantly increased NSS and TSW (Figure 5d–f). Consequently, the SW per silique in the double mutants of BnCLV3 increased by more than 74.4% on average compared with that of WT (Figure 5f). In addition, an increase in the leaf number was also not observed in the clv3 mutants in B. rapa and Arabidopsis. Thus, the double mutants of BnCLV3 generated in the present study might provide excellent starting materials for further high‐yield breeding in rapeseed. In B. juncea, a mutation in the CLV1 gene homologue on the B genome exhibited four valves stably in trilocular siliques (Xu et al., 2017). Compared with the mutation phenotype of the same gene homologue in B. juncea, the phenotype of the double mutants of BnCLV1 generated in the present study was instable; that is, they produced a variable valve numbers in the siliques of the same plant and a lower percentage of multilocular siliques per plant (Table 3). The multilocular silique phenotype of the double mutants of BnCLV2 was also instable as that of BnCLV1 (Table 3). Thus, the genetic control of multilocular siliques was more complicated in B. napus. Based on the prevailing model for CLV signalling in Arabidopsis, CLV3 is recognized by at least three functionally redundant receptors, including CLV1, the CLV2/CRN complex and RPK2 (Clark et al., 1997; Jeong et al., 1999; Kinoshita et al., 2010). Thus, it is understandable that the mutation phenotypes of BnCLV1 and BnCLV2 are less stable than BnCLV3. The large and specific mutant diversity of BnCLV1 and BnCLV2 generated in the present study provided a valuable resource for further genetic studies in rapeseed.
In conclusion, our work presents a successful example to utilize CRISPR/Cas9‐induced mutations for revealing gene functions in polyploid species and also provides agronomically important crop mutations.
Experimental procedures
Plant material
The semi‐winter B. napus pure line J9707 was used as the transformation receptor in this study. Another semi‐winter B. napus cv. HY, which has a lobed‐leaf phenotype, was used for the pollination testing along with the genome‐edited line SCLV3‐35. All seeds were obtained from the National Engineering Research Centre of Rapeseed, Wuhan, China.
CRISPR/Cas9 target locus selection and construct assembly
Sequence‐specific sgRNAs were designed using the web‐based tool CRISPR‐P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). Two or four output target sites were selected for each target gene based on their location in the gene, GC% content and putative off‐targets (Figure 1a,c,d). These targets were assessed using PCR and Sanger sequencing in J9707 to ensure that no polymorphisms existed between the sgRNAs and the corresponding target sequences.
The binary pYLCRIPSR/Cas9 multiplex genome targeting vector system, which was provided by Prof. Yaoguang Liu (South China Agriculture University), included pYLCRISPR/Cas9Pubi‐H and pYLCRISPR/Cas9P35S‐H, in which Cas9p is driven by the maize ubiquitin promoter (Pubi) and the cauliflower mosaic virus 35S promoter (P35S), and four plasmids with sgRNA cassettes driven by the promoters of AtU3b, AtU3d, AtU6‐1 and AtU6‐29; this system was used for construct assembly according to a method previously described by Ma et al. (2015b). The oligos used to construct the sgRNA vectors are listed in Table S1. The resulting constructs contained a Cas9p expression cassette, sgRNA expression cassettes with target sequences and a hygromycin resistance cassette (Figure 1b,e).
Agrobacterium‐mediated transformation of rapeseed
The Cas9/sgRNA‐expressing binary vectors were transformed into J9707 via the Agrobacterium tumefaciens‐mediated hypocotyl method (Zhou et al., 2002). Regenerated seedlings were selected according to their hygromycin resistance, further cultivated in a growth chamber under a 14‐h light/10‐h dark cycle at 25 °C and transferred to a field during the rapeseed growing season.
Identification of mutant transgenic plants
The presence of a T‐DNA construct was assessed by PCR using the NPT II gene‐specific primers 35S‐3/HPT F (Table S1).
PCR was performed to amplify the genomic region surrounding the CRISPR target sites using specific primers (Table S1), and the mutations were screened using the PAGE method previously described by Zhu et al. (2014). Briefly, the PCR products were denatured at 90°C for 5 min, followed by cooling to room temperature for renaturing. The renatured PCR products were separated using native PAGE. To confirm the results of the PAGE‐based genotyping, the PCR fragments were directly sequenced or cloned into the pEASY‐T vector (TransGen Biotech, Beijing, China) and subsequently sequenced using a Sanger method to identify the mutations. The sequences were compared to WT sequences to detect the presence of indels. The sequencing chromatograms were also examined to identify overlapping traces in the region surrounding the PAM, which are indicative of the presence of mutations. The bi‐allelic and heterozygous mutations were decoded using the degenerate sequence decoding (DSD) method (Ma et al., 2015a).
Phenotyping of B. napus transgenic lines
The WT and homozygous T2 mutant lines were grown during the winter‐type oilseed rape growing season on an experimental farm at Huazhong Agriculture University, Wuhan, China. The leaf number of 30‐day‐old seedlings was measured based on at least 30 plants per genotype. At the flowering stage, at least 30 flowers were randomly selected from each plant to count the floral organ number, including the number of sepals, petals, stamens and carpels. All mature siliques were collected from each plant to determine the percentage of multilocular siliques; thirty siliques were randomly sampled from each plant and used to measure the silique length, silique thickness and seed number per silique. The cleaned seeds were air‐dried for at least 4 weeks. The SW of each plant was measured based on 100 fully developed seeds with three replicates.
Analysis of potential off‐targets
The potential off‐target sites were identified using CRISPR‐P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). An approximately 300‐bp DNA sequence covering each off‐target site was amplified by PCR. The primers are listed in Table S1. For each target gene, mixed genomic DNA from T0 editing plants was used as the template, and WT DNA was included as a control. All PCR products were purified and mixed in equal amounts (50 ng for each) as one sample. The DNA library construction, sequencing using the Illumina HiSeq 3000 system and data analysis were conducted according to the methods previously described by Wang et al. (2017). The independent sequence reads of each off‐target site were aligned to the genomic WT sequence, which covered each off‐target site as a reference.
Southern blotting analysis
Genomic DNA was isolated from the young leaves by the cetyltrimethylammonium bromide method. A total of 30 μg of genomic DNA was digested with HindIII and then separated on a 0.8% agarose gel. After electrophoresis, the digested DNA was transferred onto a nylon membrane. For hybridization, a 293‐bp 3′‐terminus conserved BnCLV3 sequence was used as a probe.
Supporting information
Figure S1 Determination of BnCLV3 gene copy number by southern blotting analysis in J9707 and Darmor‐bzh.
Figure S2 Sequence alignment of two BnCLV3 gene copies.
Figure S3 Sequence alignment of the two BnCLV1 gene copies.
Figure S4 Sequence alignment of the two BnCLV2 gene copies.
Figure S5 Detection of a series of mutations with different indel sizes using a PAGE‐based method.
Figure S6 Novel mutations were detected in the T1 progeny with T‐DNA transmission.
Figure S7 Variations in the floral organs in the double homozygous mutants of BnCLV3.
Figure S8 Expression of Cas9p and sgRNAs in SCLV1. (a) The genotypes of eight T0 plants used for the gene expression analysis.
Table S1 Primers used in the present study.
Table S2 Mutations identified at the target sites in BnCLV3 T0 plants using PAGE‐based screening.
Table S3 Mutations identified at the target sites in BnCLV1 T0 plants using PAGE‐based screening.
Table S4 Mutations identified at the target sites in BnCLV2 T0 plants using PAGE‐based screening.
Table S5 Ratios of mutant genotype at the target sites in T0 BnCLV1 plants.
Table S6 Ratios of mutant genotype at the target sites in T0 BnCLV2 plants.
Table S7 Genotypes at the S1 target site in 22 T0 plants of BnCLV3 by sequencing after TA cloning.
Table S8 Variety of T‐DNA‐free BnCLV3 T2 generation homozygous mutants.
Table S9 Detection of potential off‐target effects at each sgRNA target site.
Table S10 Alignment of novel mutation sequences in the transmission analysis of CRISPR/Cas9‐induced mutations.
Acknowledgements
We thank Prof. Yaoguang Liu (South China Agriculture University) for kindly providing the sgRNA‐Cas9 plant expression vectors. This work was supported by the National Key Basic Research Program of China (2015CB150202), the National Natural Science Foundation of China (31371240, 31671279) and the Fundamental Research Funds for the Central Universities (Program No. 2662015PY172).
The authors declare no conflict of interest.
References
- Bommert, P. , Lunde, C. , Nardmann, J. , Vollbrecht, E. , Running, M. , Jackson, D. , Hake, S. et al (2005) Thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine‐rich repeat receptor‐like kinase. Development, 132, 1235–1245. [DOI] [PubMed] [Google Scholar]
- Bommert, P. , Nagasawa, N.S. and Jackson, D. (2013) Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 45, 334–337. [DOI] [PubMed] [Google Scholar]
- Braatz, J. , Harloff, H.J. , Mascher, M. , Stein, N. , Himmelbach, A. and Jung, C. (2017) CRISPR‐Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174, 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, C. , Nekrasov, V. , Lippman, Z.B. and Van Eck, J. (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR‐associated9 system. Plant Physiol. 166, 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S. , Parkin, I.A.P. , Tang, H. , Wang, X. , Chiquet, J. et al (2014) Early allopolyploid evolution in the post‐neolithic Brassica napus oilseed genome. Science, 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Chu, H. , Qian, Q. , Liang, W. , Yin, C. , Tan, H. , Yao, X. , Yuan, Z. et al (2006) The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 142, 1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E. , Running, M.P. and Meyerowitz, E.M. (1993) CLAVATA1, a regulator of meristem and flower development in Arabidopsis . Development, 119, 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S.E. , Running, M.P. and Meyerowitz, E.M. (1995) CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1 . Development, 121, 2057–2067. [Google Scholar]
- Clark, S.E. , Williams, R.W. and Meyerowitz, E.M. (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis . Cell, 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. , Hsu, P.D. et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. , Wu, Y. , Yang, Q. , Yang, Y. , Meng, Q. , Zhang, K. , Li, J. et al (2014) A novel single‐nucleotide mutation in a CLAVATA3 gene homolog controls a multilocular silique trait in Brassica rapa L. Mol. Plant, 7, 1788–1792. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C. , Brand, U. , Running, M.P. , Simon, R. and Meyerowitz, E.M. (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science, 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Gao, J. , Wang, G. , Ma, S. , Xie, X. , Wu, X. , Zhang, X. , Wu, Y. et al (2015) CRISPR/Cas9‐mediated targeted mutagenesis in Nicotiana tabacum . Plant Mol. Biol. 87, 99–110. [DOI] [PubMed] [Google Scholar]
- Haeussler, M. , Schonig, K. , Eckert, H. , Eschstruth, A. , Mianne, J. , Renaud, J.B. , Schneider‐Maunoury, S. et al (2016) Evaluation of off‐target and on‐target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y. , Long, W. , Hu, J. , Fu, T. , Li, D. , Chen, B. and Tu, J. (2003) Anatomic and genetic studies on multicapsular character in Brassica campestris L . Chin. J. Oil Crop Sci. 25, 1–4. [Google Scholar]
- Jeong, S. , Trotochaud, A.E. and Clark, S.E. (1999) The Arabidopsis CLAVATA2 gene encodes a receptor‐like protein required for the stability of the CLAVATA1 receptor‐like kinase. Plant Cell, 11, 1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, H. , Zhang, Y. , Orbovic, V. , Xu, J. , White, F.F. , Jones, J.B. and Wang, N. (2017) Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Zhou, H. , Bi, H. , Fromm, M. , Yang, B. and Weeks, D.P. (2013) Demonstration of CRISPR/Cas9/sgRNA‐mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41, e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Henry, I.M. , Lynagh, P.G. , Comai, L. , Cahoon, E.B. and Weeks, D.P. (2017) Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15, 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar, R.K. , Chamola, R. and Chopra, V.L. (1998) Tetralocular mustard, Brassica juncea: New promising variability through interspecific hybridization. Plant Breed. 117, 398–399. [Google Scholar]
- Kayes, J.M. and Clark, S.E. (1998) CLAVATA2, a regulator of meristem and organ development in Arabidopsis . Development, 125, 3843–3851. [DOI] [PubMed] [Google Scholar]
- Kinoshita, A. , Betsuyaku, S. , Osakabe, Y. , Mizuno, S. , Nagawa, S. , Stahl, Y. , Simon, R. et al (2010) RPK2 is an essential receptor‐like kinase that transmits the CLV3 signal in Arabidopsis . Development, 137, 3911–3920. [DOI] [PubMed] [Google Scholar]
- Lawrenson, T. , Shorinola, O. , Stacey, N. , Li, C. , Ostergaard, L. , Patron, N. , Uauy, C. et al (2015) Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA‐guided Cas9 nuclease. Genome Biol. 16, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Y. , Lu, L. , Liu, H.Y. , Li, S. , Xing, F. and Chen, L.L. (2014) CRISPR‐P: a web tool for synthetic single‐guide RNA design of CRISPR‐system in plants. Mol. Plant, 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li, T. , Huang, S. , Zhao, X. , Wright, D.A. , Carpenter, S. , Spalding, M.H. , Weeks, D.P. et al (2011) Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 39, 6315–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Liu, Z. , Xing, A. , Moon, B.P. , Koellhoffer, J.P. , Huang, L. , Ward, R.T. et al (2015) Cas9‐guide RNA directed genome editing in soybean. Plant Physiol. 169, 960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z. , Zhang, K. , Chen, K. and Gao, C. (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics, 41, 63–68. [DOI] [PubMed] [Google Scholar]
- Liu, H. (2000) Brassica napus Genetics and Breeding. Beijing: China Agricultural University Press. [Google Scholar]
- Lv, Z. , Xu, P. , Zhang, X. , Wen, J. , Yi, B. , Ma, C. , Tu, J. et al (2012) Primary study on anatomic and genetic characteristics of muti‐loculus in Brassica juncea . Chin. J. Oil Crop Sci. 34, 462–466. [Google Scholar]
- Ma, X. , Chen, L. , Zhu, Q. , Chen, Y. and Liu, Y.G. (2015a) Rapid decoding of sequence‐specific nuclease‐induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol. Plant, 8, 1285–1287. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. et al (2015b) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant, 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Makarova, K.S. , Haft, D.H. , Barrangou, R. , Brouns, S.J. , Charpentier, E. , Horvath, P. , Moineau, S. et al (2011) Evolution and classification of the CRISPR‐Cas systems. Nat. Rev. Microbiol. 9, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, J. , Guo, D. , Zhang, J. , Huang, Q. , Qin, G. , Zhang, X. , Wan, J. et al (2013) Targeted mutagenesis in rice using CRISPR‐Cas system. Cell Res. 23, 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, R. , Bleckmann, A. and Simon, R. (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell‐limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell, 20, 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q. , Wang, Y. , Li, J. , Zhang, Y. , Chen, K. , Liang, Z. , Zhang, K. et al (2013) Targeted genome modification of crop plants using a CRISPR‐Cas system. Nat. Biotechnol. 31, 686–688. [DOI] [PubMed] [Google Scholar]
- Sun, F. , Fan, G. , Hu, Q. , Zhou, Y. , Guan, M. , Tong, C. , Li, J. et al (2017) The high‐quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi‐winter morphotype. Plant J. 92, 452–468. [DOI] [PubMed] [Google Scholar]
- Suzaki, T. , Sato, M. , Ashikari, M. , Miyoshi, M. , Nagato, Y. and Hirano, H.Y. (2004) The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine‐rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1 . Development, 131, 5649–5657. [DOI] [PubMed] [Google Scholar]
- Taguchi‐Shiobara, F. , Yuan, Z. , Hake, S. and Jackson, D. (2001) The fasciated ear2 gene encodes a leucine‐rich repeat receptor‐like protein that regulates shoot meristem proliferation in maize. Genes Dev. 15, 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney, S.K. (1987) Inheritance of siliqua characters in Indian colza I. Locule number and siliqua position. Euphytica, 36, 541–544. [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.L. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Wang, P. , Zhang, J. , Sun, L. , Ma, Y. , Xu, J. , Liang, S. , Deng, J. et al (2017) High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. https://doi.org/10.1111/pbi.12755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, A.J. , Lo, T.W. , Zeitler, B. , Pickle, C.S. , Ralston, E.J. , Lee, A.H. , Amora, R. et al (2011) Targeted genome editing across species using ZFNs and TALENs. Science, 333, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, L. , Zhao, H. , Zhao, Z. , Du, D. , Xu, L. , Yao, Y. , Zhao, Z. et al (2013) Genetic and physical fine mapping of a multilocular gene Bjln1 in Brassica juncea to a 208‐kb region. Mol. Breeding, 32, 373–383. [Google Scholar]
- Xu, P. , Lv, Z. , Zhang, X. , Wang, X. , Pu, Y. , Wang, H. , Yi, B. et al (2013) Identification of molecular markers linked to trilocular gene (mc1) in Brassica juncea L . Mol. Breeding, 33, 425–434. [Google Scholar]
- Xu, C. , Liberatore, K.L. , MacAlister, C.A. , Huang, Z. , Chu, Y.H. , Jiang, K. , Brooks, C. et al (2015) A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47, 784–792. [DOI] [PubMed] [Google Scholar]
- Xu, P. , Cao, S. , Hu, K. , Wang, X. , Huang, W. , Wang, G. , Lv, Z. et al (2017) Trilocular phenotype in Brassica juncea L. resulted from interruption of CLAVATA1 gene homologue (BjMc1) transcription. Sci. Rep. 7, 3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, J. , Wei, P. , Zhang, B. , Gou, F. , Feng, Z. , Mao, Y. et al (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Wang, H. , Gilmer, S. , Whitwill, S. , Keller, W. and Fowke, L.C. (2002) Control of petal and pollen development by the plant cyclin‐dependent kinase inhibitor ICK1 in transgenic Brassica plants. Planta, 215, 248–257. [DOI] [PubMed] [Google Scholar]
- Zhu, X. , Xu, Y. , Yu, S. , Lu, L. , Ding, M. , Cheng, J. , Song, G. et al (2014) An efficient genotyping method for genome‐modified animals and human cells generated with CRISPR/Cas9 system. Sci. Rep. 4, 6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Determination of BnCLV3 gene copy number by southern blotting analysis in J9707 and Darmor‐bzh.
Figure S2 Sequence alignment of two BnCLV3 gene copies.
Figure S3 Sequence alignment of the two BnCLV1 gene copies.
Figure S4 Sequence alignment of the two BnCLV2 gene copies.
Figure S5 Detection of a series of mutations with different indel sizes using a PAGE‐based method.
Figure S6 Novel mutations were detected in the T1 progeny with T‐DNA transmission.
Figure S7 Variations in the floral organs in the double homozygous mutants of BnCLV3.
Figure S8 Expression of Cas9p and sgRNAs in SCLV1. (a) The genotypes of eight T0 plants used for the gene expression analysis.
Table S1 Primers used in the present study.
Table S2 Mutations identified at the target sites in BnCLV3 T0 plants using PAGE‐based screening.
Table S3 Mutations identified at the target sites in BnCLV1 T0 plants using PAGE‐based screening.
Table S4 Mutations identified at the target sites in BnCLV2 T0 plants using PAGE‐based screening.
Table S5 Ratios of mutant genotype at the target sites in T0 BnCLV1 plants.
Table S6 Ratios of mutant genotype at the target sites in T0 BnCLV2 plants.
Table S7 Genotypes at the S1 target site in 22 T0 plants of BnCLV3 by sequencing after TA cloning.
Table S8 Variety of T‐DNA‐free BnCLV3 T2 generation homozygous mutants.
Table S9 Detection of potential off‐target effects at each sgRNA target site.
Table S10 Alignment of novel mutation sequences in the transmission analysis of CRISPR/Cas9‐induced mutations.


