Abstract
Brucellosis is a serious infectious disease that continues to be a significant cause of morbidity worldwide and across all ages. Despite early diagnosis and treatment, 10–30% of patients develop chronic brucellosis. Although there have been recent advances in our knowledge of Brucella virulence factors and hosts’ immune response to the infection, there is a lack of clear data regarding how the infection bypasses the immune system and becomes chronic. The present study investigated immunological factors and their roles in the transition of brucellosis from an acute to a chronic infection in CD4+ T cells. CD4+ T cells sorted from peripheral blood samples of patients with acute or chronic brucellosis and healthy controls using flow cytometry as well as more than 2000 miRNAs were screened using the GeneSpring GX (Agilent) 13.0 miRNA microarray software and were validated using reverse transcription polymerase chain reaction (RT-qPCR). Compared to acute cases, the expression levels of 28 miRNAs were significantly altered in chronic cases. Apart from one miRNA (miR-4649-3p), 27 miRNAs were not expressed in the acute cases (p <0.05, fold change> 2). According to KEGG pathway analysis, these miRNAs are involved in the regulation of target genes that were previously involved in the MAPK signalling pathway, regulation of the actin cytoskeleton, endocytosis, and protein processing in the endoplasmic reticulum. This indicates the potential role of these miRNAs in the development of chronic brucellosis. We suggest that these miRNAs can be used as markers to determine the transition of the disease into chronicity. This is the first study of miRNA expression that analyses human CD4+ T cells to clarify the mechanism of chronicity in brucellosis.
Introduction
Brucellosis is one of the most common diseases among humans and animals. The disease is caused by bacteria of the Brucella genus. These bacteria are facultative, intracellular, Gram-negative, non-spore-forming, non-motile coccobacilli that infect humans as well as domestic mammals (cattle, sheep, goat, swine, etc.) and wild mammals (deer, bison, etc.) [1]. Humans can become infected in different ways, including through direct contact with animals infected with Brucella and consumption of products derived from animals infected with Brucella. The majority of human cases are caused by Brucella melitensis, although Brucella abortus, Brucella suis, Brucella canis, and, more recently, Brucella pinnipedialis and Brucella ceti have also been associated with the acquisition of brucellosis in humans [2, 3]. In humans, the Brucella infection is considered either a generalized, febrile illness that does not impact organ systems or a focal disease in which one or more organs are involve, such as in the most common form of the disease-osteoarticular brucellosis [4, 5].
In acute brucellosis patients, the acute phase of the disease follows the eradication of the bacteria by the immune system [6]. However, despite early diagnosis and treatment, a failure to clear the brucellosis infection in its acute form leads the disease to transition to its chronic form, which is observed in almost 10–30% of patients [7, 8]. The diagnosis of chronicity is mainly based on clinical symptoms and high IgG titres in serological tests [9]. However, IgG titres may remain positive for years following the successful resolution of symptoms because of low specificity of the assay [7, 10]. Therefore, markers are needed for the clinical prediction, accurate diagnosis, and follow-up of brucellosis and for administering successful therapeutic approaches. Additionally, the mechanisms that lead to chronicity in brucellosis are not completely established. To understand the mechanism by which the infection evades the host’s immune response, we previously evaluated the molecular markers in the peripheral blood mononuclear cells (PBMCs) and CD8 (+) T lymphocytes of patients with acute and chronic brucellosis [11, 12]. However, the molecular markers expressed in CD4 (+) T lymphocytes remain unknown.
CD4+ T cells play a major role in regulating the immune system by helping to activate the cells of the innate immune system and B-cells and helping to generate cytotoxic CD8+ T cells and nonimmune cells. Moreover, CD4+ T cells play critical role in the suppression of immune reactions [13]. Several studies have identified new subsets of CD4+ cells besides the well-known T-helper 1 (TH1) and T-helper 2 (TH2) cells. TH1 cells are involved in host defence against intracellular pathogens and tumour cells. TH2 cells are responsible for coordinating humoral immunity and eosinophilic inflammation and are involved in host defence against extracellular parasites. New subsets of CD4+ cells include T-helper (TH9), T-helper 17 (TH 17), T-helper 22 (TH22), follicular helper T cell (THFH), Natural Killer T cell (NKT), induced T-regulatory cells (iTreg), regulatory type 1 cells (Tr1), and CD4+ Cytotoxic T cells (CD4+ CTL) [14–16]. Studies have shown that IFN-γ-mediated TH1 responses are essential for the clearance of intracellular bacteria such as Brucella spp. [17]. In a recent study, Martirosyan et al. show that, with high levels of IFN-γ and granzyme B expression, CD4+ CTL participated in the early phase clearance of the Brucella infection [16]. Thus, to provide valuable information regarding the pathogenesis of brucellosis and to develop new therapeutic approaches for treating brucellosis and preventing the transition to the chronic form of the disease, the molecular markers of all immune system components related to brucellosis must be identified.
MicroRNAs (miRNAs) are small, non-coding RNAs of 18–25 nucleotides in length that bind to complementary UTR regions of target mRNAs and regulate the transcriptional activity of the target gene [18, 19]. miRNAs have been implicated in several basic metabolic pathways and some biological processes, such as the regulation of haematopoiesis, cellular proliferation, and programmed cell death. Differentiation, organogenesis, and abnormalities in these miRNAs contribute to the development of diseases, such as cancerogenesis, and infections [20–26].
In our previous studies, we determined the changes in the miRNA expression of the PBMCs and CD4+ T cells that play important roles in the establishment and clearance of chronic Brucella infections [11, 12]. In this study, we aimed to investigate the potential miRNA biomarkers in CD4+ T cells that indicate whether a case of the Brucella infection turns into chronic form. To this end, the expression changes of the miRNAs involved in the immune responses of CD4+ T cells were comparatively examined in patients with acute and chronic brucellosis as well as healthy controls.
Materials and methods
Ethics statement
This study protocol was approved by the Committee on the Ethics of the University of Uludag, Faculty of Medicine (Permit number:2010-6/2). All participiants gave written informed consent for inclusion.
Patient cohort
A total of 15 patients with brucellosis (9 female and 6 male) with bone and joint involvement served as the experimental group, and a total of 8 healthy volunteers (4 female and 4 male) served as the control group. A diagnosis of brucellosis was made with the existence of one or more constitutional symptoms, such as fever, perspiration, and fatigue, as well as with the standard tube agglutination test titre in the Clinical Bacteriology and Infectious Diseases Department at Uludag University Faculty of Medicine [6]. Exclusion criteria were chronic granulomatous disease other than brucellosis, other infectious diseases, cardiovascular diseases, chronic renal insufficiency, chronic liver diseases, malignancy, collagen tissue disease, autoimmune disease. All cases were treated with the same therapy protocol as described previously [11]. Chronic brucellosis cases were followed up in outpatient clinic at least for 12 months. The duration of treatment for chronic brucellosis was three months and extended according to patient’s clinical status. The study was approved by the local ethics committee (approval number: 2010-6/2) and complied with the ethical standards of the Helsinki Declaration. All participants received information about the study, and all patients provided informed consent to participate in the study.
CD4+ T cell isolation
The CD4+ T cells of the cases were obtained from PBMCs, which were separated from 20 mL of peripheral blood using an intensity gradient centrifuge method with Ficoll (Biochrom, Germany). CD4+ T cells were sorted from the suspension of PBMCs using FACSAria (Beckton Dickinson [BD] Biosciences Cell-sorter, USA) after the cells were stained with CD4-FITC, CD8-PE, and CD3-PerCP antibodies (BD, Biosciences, USA). Briefly, live PBMCs were gated based on forward versus side-scatter profiles, followed by CD3+ based gating for T lymphocytes were identified. The CD3+ positive population was subdivided by CD8/CD4 expression and used to identify CD4+ (CD4+, CD8-) T cells. The purity of the resulting CD4+ T cells was 99.8%.
Microarray analysis
miRNA was isolated from CD4+ T cells using an miRNeasy Kit (Qiagen, Germany) and was labelled with Cy3 during transformation into cDNA using an RNA Agilent miRNA Labeling Kit (Agilent, UK) and Spike Kit (Agilent, UK). cDNAs were hybridized to Human miRNA Microarray, Release 19.0, 8x60K(v19) microarray slides (Agilent, UK) according to Agilent microRNA Hybridization Kit protocol (Agilent, UK) in duplicate and scanned using an Nimblegen MS200 array scanner. Data were normalized using the Plier algorithm and were log transformed and analysed using GeneSpring GX 13.0 (Agilent, UK). Significant miRNAs were determined based on P-values < 0.05 or fold changes in the range of >2 and <-2.
Data availability: The microarraydata that supported the findings of this study were deposited in the Gene Expression Omnibus with an accession number of GSE107554.
qRT-PCR
Microarray analyses were validated using qRT-PCR for ten randomly selected miRNAs using the same miRNA stock samples isolated for microarray analysis. Using a Universal cDNA Synthesis Kit (Exiqon, Denmark), 5ng/μL of miRNA samples were reverse transcribed into cDNA. qRT-PCR was performed using LightCycler 480 II (Roche, Germany) microRNA LNA™ primer sets and an ExiLENT SYBR1 Green Master Mix Kit (Exiqon, Denmark), as described previously [11].
Identification of miRNAs, target proteins, and related pathways
Biological potency and functional classification of predicted miRNA targets were identified using pathway enrichment analysis. Selected miRNAs were entered into KEGG (Kyoto Encyclopedia of Genes and Genomes) (http://www.genome.jp/kegg/pathway.html) to elucidate the number of predicted miRNA target genes [27]. The signalling pathways associated with the predicted target genes of the miRNAs were determined using a web-based enrichment analysis tool known as WebGestalt (WEB-based GEneSeTAnaLysis Toolkit) (http://bioinfo.vanderbilt.edu/webgestalt/) and KEGG [27, 28].
Results
Clinical data
Based on the onset time of the symptoms, 7 of the cases (3 female and 4 male) whose onset was in a range of 0–2 months were diagnosed with acute brucellosis, and 8 of the cases (6 female and 2 male) whose onset was >12 months were diagnosed with chronic brucellosis [29, 30]. The median age of cases with acute brucellosis was 52.14 ±10.40, and the median age of cases with chronic brucellosis was 45.00 ±11.48. The median age of the control group was 39.62 ± 7.74.
miRNA expression pattern of CD4+ T cells in brucellosis
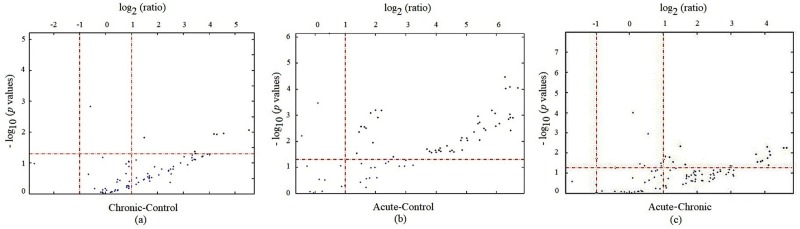
According to the microarray analyses, miRNAs with more than twofold differences between the samples and control were evaluated, and those that expressed with less than twofold differentiation were assumed as negligible (Fig 1). Based on the results of an independent samples t-test, 11 miRNAs were altered in all brucellosis cases (whether acute or chronic) compared to the control cases (Table 1). Besides these 11 miRNAs, the expression levels of the 41 miRNAs were different from those of the control cases, but these miRNAs demonstrated distinct expression patterns between the acute and chronic brucellosis cases (Table 2).
Fig 1. Differential expressions of miRNAs evaluated in (a) chronic versus control, (b) acute versus control, and (c) acute versus chronic cases depend on miRNA microarray analysis (p<0.05, cut off = 2).
Table 1. List of significantly altered miRNA expressions in both acute and chronic brucellosis cases compared to control cases.
| Chronic vs Control | Acute vs Control | Chronic vs Acute | |||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | Fold change | Fold regulation | P value* | Fold change | Fold regulation | p value* | Fold change | Fold regulation | P value* |
| hsa-miR-4530 | 46.11 | 46.11 | 0.0085 | 88.44 | 88.44 | 0.0037 | 0.52 | -1.92 | 0.1294 |
| hsa-miR-4739 | 18.12 | 18.12 | 0.0113 | 17.18 | 17.18 | 0.0225 | 1.05 | 1.05 | 0.9648 |
| hsa-miR-762 | 14.67 | 14.67 | 0.0114 | 27.37 | 27.37 | 0.0091 | 0.54 | -1.87 | 0.6060 |
| hsa-miR-4787-5p | 8.15 | 8.15 | 0.0629 | 15.74 | 15.74 | 0.0220 | 0.52 | -1.93 | 0.0459 |
| hsa-miR-940 | 8.02 | 8.02 | 0.0337 | 10.45 | 10.45 | 0.0309 | 0.77 | -1.30 | 0.8215 |
| hsa-miR-3676-5p | 4.19 | 4.19 | 0.1823 | 3.11 | 3.11 | 0.1725 | 1.35 | 1.35 | 0.8099 |
| hsa-miR-6090 | 3.70 | 3.70 | 0.1313 | 5.05 | 5.05 | 0.0695 | 0.73 | -1.37 | 0.2925 |
| hsa-miR-150-5p | 3.09 | 3.09 | 0.2224 | 3.24 | 3.24 | 0.2746 | 0.95 | -1.05 | 0.9478 |
| hsa-miR-4516 | 2.82 | 2.82 | 0.0151 | 4.22 | 4.22 | 0.0012 | 0.67 | -1.50 | 0.2302 |
| hsa-miR-4284 | 2.41 | 2.41 | 0.4221 | 4.13 | 4.13 | 0.2489 | 0.58 | -1.72 | 0.5832 |
| hsa-miR-3656 | 2.29 | 2.29 | 0.3029 | 3.94 | 3.94 | 0.0998 | 0.58 | -1.72 | 0.0486 |
*P values were calculated using an independent samples t-test.
Table 2. List of miRNAs that were differently regulated between acute and chronic brucellosis.
| Chronic vs Control | Acute vs Control | Chronic vs Acute | |||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | Fold change | Fold regulation | P value* | Fold change | Fold regulation | P value* | Fold change | Fold regulation | P value* |
| hsa-miR-6068 | 23.45 | 23.45 | 0.0110 | 86.89 | 86.89 | 0.0009 | 0.27 | -3.70 | 0.0975 |
| hsa-miR-4497 | 19.62 | 19.62 | 0.0117 | 86.96 | 86.96 | 8.15E-05 | 0.23 | -4.43 | 0.1161 |
| hsa-miR-1207-5p | 16.26 | 16.26 | 0.0538 | 63.06 | 63.06 | 0.0026 | 0.26 | -3.88 | 0.2013 |
| hsa-miR-197-5p | 15.29 | 15.29 | 0.0517 | 92.97 | 92.97 | 0.0012 | 0.16 | -6.08 | 0.0920 |
| hsa-miR-642a-3p | 13.53 | 13.53 | 0.0591 | 87.52 | 87.52 | 0.0012 | 0.15 | -6.47 | 0.0817 |
| hsa-miR-3162-5p | 13.16 | 13.16 | 0.0627 | 84.64 | 84.64 | 0.0014 | 0.16 | -6.43 | 0.0818 |
| hsa-miR-4687-3p | 11.03 | 11.03 | 0.0489 | 61.77 | 61.77 | 0.0008 | 0.18 | -5.60 | 0.0724 |
| hsa-miR-3196 | 10.87 | 10.87 | 0.0421 | 41.87 | 41.87 | 0.0020 | 0.26 | -3.85 | 0.2665 |
| hsa-miR-371b-5p | 10.65 | 10.65 | 0.0768 | 68.32 | 68.32 | 0.0020 | 0.16 | -6.42 | 0.0650 |
| hsa-miR-3665 | 10.15 | 10.15 | 0.0807 | 32.77 | 32.77 | 0.0092 | 0.31 | -3.23 | 0.1531 |
| hsa-miR-4466 | 10.12 | 10.12 | 0.0551 | 38.25 | 38.25 | 0.0044 | 0.26 | -3.78 | 0.0781 |
| hsa-miR-574-5p | 10.05 | 10.05 | 0.0673 | 32.45 | 32.45 | 0.0075 | 0.31 | -3.23 | 0.1254 |
| hsa-miR-1225-5p | 8.96 | 8.96 | 0.1155 | 49.89 | 49.89 | 0.0037 | 0.18 | -5.57 | 0.1063 |
| hsa-miR-4485 | 8.60 | 8.60 | 0.0341 | 25.67 | 25.67 | 0.0087 | 0.33 | -2.99 | 0.3709 |
| hsa-miR-572 | 7.78 | 7.78 | 0.0332 | 40.92 | 40.92 | 0.0010 | 0.19 | -5.26 | 0.1215 |
| hsa-miR-5001-5p | 7.46 | 7.46 | 0.0984 | 28.89 | 28.89 | 0.0073 | 0.26 | -3.87 | 0.0857 |
| hsa-miR-4672 | 6.15 | 6.15 | 0.1297 | 104.31 | 104.31 | 8.96E-05 | 0.06 | -16.95 | 0.0182 |
| hsa-miR-1234-5p | 6.09 | 6.09 | 0.1573 | 20.53 | 20.53 | 0.0149 | 0.30 | -3.37 | 0.1528 |
| hsa-miR-4465 | 6.01 | 6.01 | 0.0798 | 50.85 | 50.85 | 0.0086 | 0.12 | -8.46 | 0.1389 |
| hsa-miR-4299 | 5.78 | 5.78 | 0.2277 | 43.53 | 43.53 | 0.0091 | 0.13 | -7.53 | 0.0606 |
| hsa-miR-6132 | 5.67 | 5.67 | 0.1578 | 29.19 | 29.19 | 0.0218 | 0.19 | -5.15 | 0.2663 |
| hsa-miR-4763-3p | 5.37 | 5.37 | 0.1745 | 43.50 | 43.50 | 0.0018 | 0.12 | -8.10 | 0.0465 |
| hsa-miR-937-5p | 5.36 | 5.36 | 0.1815 | 17.03 | 17.03 | 0.0219 | 0.31 | -3.17 | 0.3272 |
| hsa-miR-5585-3p | 4.98 | 4.98 | 0.0809 | 26.03 | 26.03 | 0.0087 | 0.19 | -5.23 | 0.1785 |
| hsa-miR-3679-5p | 4.48 | 4.48 | 0.1570 | 78.89 | 78.89 | 9.51E-05 | 0.06 | -17.61 | 0.0083 |
| hsa-miR-3940-5p | 4.26 | 4.26 | 0.0797 | 71.52 | 71.52 | 5.56E-08 | 0.06 | -16.79 | 0.0050 |
| hsa-miR-6088 | 4.16 | 4.16 | 0.2430 | 17.49 | 17.49 | 0.0181 | 0.24 | -4.20 | 0.0799 |
| hsa-miR-638 | 3.43 | 3.43 | 0.2978 | 13.00 | 13.00 | 0.0200 | 0.26 | -3.79 | 0.1218 |
| hsa-miR-6127 | 3.24 | 3.24 | 0.3862 | 47.74 | 47.74 | 0.0032 | 0.07 | -14.72 | 0.0268 |
| hsa-miR-4486 | 3.22 | 3.22 | 0.2627 | 6.86 | 6.86 | 0.0910 | 0.47 | -2.13 | 0.5447 |
| hsa-miR-1202 | 3.22 | 3.22 | 0.2973 | 17.77 | 17.77 | 0.0245 | 0.18 | -5.52 | 0.2000 |
| hsa-miR-4428 | 3.21 | 3.21 | 0.3204 | 57.54 | 57.54 | 0.0006 | 0.06 | -17.90 | 0.0124 |
| hsa-miR-5787 | 3.11 | 3.11 | 0.1781 | 13.96 | 13.96 | 0.0328 | 0.22 | -4.49 | 0.2420 |
| hsa-miR-4721 | 3.10 | 3.10 | 0.2778 | 77.63 | 77.63 | 3.39E-05 | 0.04 | -25.03 | 0.0055 |
| hsa-miR-1973 | 2.85 | 2.85 | 0.1706 | 24.12 | 24.12 | 0.0092 | 0.12 | -8.47 | 0.0725 |
| hsa-miR-5739 | 2.82 | 2.82 | 0.3329 | 15.99 | 15.99 | 0.0264 | 0.18 | -5.67 | 0.1735 |
| hsa-miR-4532 | 2.82 | 2.82 | 0.1706 | 10.25 | 10.25 | 0.0304 | 0.28 | -3.64 | 0.2504 |
| hsa-miR-3651 | 2.77 | 2.77 | 0.3462 | 23.08 | 23.08 | 0.0221 | 0.12 | -8.34 | 0.1226 |
| hsa-miR-1275 | 2.65 | 2.65 | 0.1709 | 35.41 | 35.41 | 0.0011 | 0.07 | -13.35 | 0.0117 |
| hsa-miR-4728-5p | 2.64 | 2.64 | 0.1705 | 20.48 | 20.48 | 0.0092 | 0.13 | -7.76 | 0.0682 |
| hsa-miR-1915-3p | 2.26 | 2.26 | 0.4917 | 7.97 | 7.97 | 0.0543 | 0.28 | -3.53 | 0.1282 |
*P values were calculated using an independent samples t-test.
Unique miRNA expression pattern of CD4+ T cells in acute or chronic forms of brucellosis
Twenty-eight miRNAs were significantly altered in the chronic cases. Twenty-seven of these miRNAs were not expressed in the acute cases, and one of them (miR-4649-3p) did not demonstrate significant differences between the acute cases and control cases (Table 3). However, 18 miRNAs were significantly altered in the acute cases. The expressions of these miRNAs were similar in the chronic cases and control cases (Table 4).
Table 3. List of significantly altered miRNA expressions in chronic brucellosis cases.
| Chronic vs Control | Acute vs Control | Chronic vs Acute | |||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | Fold change | Fold regulation | P value* | Fold change | Fold regulation | P value* | Fold change | Fold regulation | P value* |
| hsa-miR-4649-3p | 5.51 | 5.51 | 0.0859 | 1.70 | 1.70 | 0.3559 | 3.24 | 3.24 | 0.2660 |
| hsa-miR-885-5p | 2.16 | 2.16 | 0.3506 | - | - | - | 2.16 | 2.16 | 0.3506 |
| hsa-miR-766-3p | 2.02 | 2.02 | 0.3506 | - | - | - | 2.02 | 2.02 | 0.3506 |
| hsa-miR-6511b-3p | 2.04 | 2.04 | 0.3506 | - | - | - | 2.04 | 2.04 | 0.3506 |
| hsa-miR-6511a-3p | 2.13 | 2.13 | 0.3506 | - | - | - | 2.13 | 2.13 | 0.3506 |
| hsa-miR-5584-3p | 2.02 | 2.02 | 0.3506 | - | - | - | 2.02 | 2.02 | 0.3506 |
| hsa-miR-483-3p | 2.09 | 2.09 | 0.3506 | - | - | - | 2.09 | 2.09 | 0.3506 |
| hsa-miR-4763-5p | 2.06 | 2.06 | 0.3506 | - | - | - | 2.06 | 2.06 | 0.3506 |
| hsa-miR-4695-3p | 2.08 | 2.08 | 0.3506 | - | - | - | 2.08 | 2.08 | 0.3506 |
| hsa-miR-449b-3p | 2.07 | 2.07 | 0.3506 | - | - | - | 2.07 | 2.07 | 0.3506 |
| hsa-miR-4436b-5p | 2.03 | 2.03 | 0.3506 | - | - | - | 2.03 | 2.03 | 0.3506 |
| hsa-miR-3622b-3p | 2.01 | 2.01 | 0.3506 | - | - | - | 2.01 | 2.01 | 0.3506 |
| hsa-miR-3617-3p | 2.12 | 2.12 | 0.3506 | - | - | - | 2.12 | 2.12 | 0.3506 |
| hsa-miR-328 | 2.21 | 2.21 | 0.3506 | - | - | - | 2.21 | 2.21 | 0.3506 |
| hsa-miR-3195 | 2.99 | 2.99 | 0.1716 | - | - | - | 2.99 | 2.99 | 0.1716 |
| hsa-miR-2277-3p | 2.02 | 2.02 | 0.3506 | - | - | - | 2.02 | 2.02 | 0.3506 |
| hsa-miR-211-5p | 2.09 | 2.09 | 0.3506 | - | - | - | 2.09 | 2.09 | 0.3506 |
| hsa-miR-204-5p | 2.05 | 2.05 | 0.3506 | - | - | - | 2.05 | 2.05 | 0.3506 |
| hsa-miR-1976 | 2.02 | 2.02 | 0.3506 | - | - | - | 2.02 | 2.02 | 0.3506 |
| hsa-miR-197-3p | 2.11 | 2.11 | 0.3506 | - | - | - | 2.11 | 2.11 | 0.3506 |
| hsa-miR-1825 | 2.86 | 2.86 | 0.1727 | - | - | - | 2.86 | 2.86 | 0.1727 |
| hsa-miR-1296 | 2.01 | 2.01 | 0.3506 | - | - | - | 2.01 | 2.01 | 0.3506 |
| hsa-miR-129-1-3p | 3.07 | 3.07 | 0.1705 | - | - | - | 3.07 | 3.07 | 0.1705 |
| hsa-miR-1281 | 2.87 | 2.87 | 0.1729 | - | - | - | 2.87 | 2.87 | 0.1729 |
| hsa-miR-1260a | 2.09 | 2.09 | 0.3506 | - | - | - | 2.09 | 2.09 | 0.3506 |
| hsa-let-7e-3p | 2.06 | 2.06 | 0.3506 | - | - | - | 2.06 | 2.06 | 0.3506 |
| hsa-let-7d-3p | 2.11 | 2.11 | 0.3506 | - | - | - | 2.11 | 2.11 | 0.3506 |
| hsa-let-7b-3p | 3.22 | 3.22 | 0.1791 | - | - | - | 3.22 | 3.22 | 0.1791 |
*P values were calculated using independent sample T test.
Table 4. List of significantly altered miRNA expressions in acute brucellosis cases.
| Chronic vs Control | Acute vs Control | Chronic vs Acute | |||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | Fold change | Fold regulation | P value* | Fold change | Fold regulation | P value* | Fold change | Fold regulation | P value* |
| hsa-miR-939-5p | - | - | - | 5.64 | 5.64 | 0.0782 | 0.18 | -5.64 | 0.0782 |
| hsa-miR-6724-5p | 1.85 | 1.85 | 0.6528 | 28.13 | 28.13 | 0.0092 | 0.07 | -15.21 | 0.0244 |
| hsa-miR-6125 | 1.38 | 1.38 | 0.3405 | 2.87 | 2.87 | 0.0027 | 0.48 | -2.08 | 0.0152 |
| hsa-miR-6075 | - | - | - | 9.40 | 9.40 | 0.0303 | 0.11 | -9.40 | 0.0303 |
| hsa-miR-5006-5p | - | - | - | 2.03 | 2.03 | 0.3559 | 0.49 | -2.03 | 0.3559 |
| hsa-miR-494 | 1.87 | 1.87 | 0.1288 | 4.57 | 4.57 | 0.0006 | 0.41 | -2.44 | 0.0283 |
| hsa-miR-4746-3p | 1.87 | 1.87 | 0.3506 | 10.54 | 10.54 | 0.0311 | 0.18 | -5.65 | 0.1253 |
| hsa-miR-4665-3p | - | - | - | 2.89 | 2.89 | 0.1723 | 0.35 | -2.89 | 0.1723 |
| hsa-miR-4507 | 1.38 | 1.38 | 0.7716 | 5.37 | 5.37 | 0.0608 | 0.26 | -3.89 | 0.1322 |
| hsa-miR-4459 | 1.74 | 1.74 | 0.2177 | 3.99 | 3.99 | 0.0006 | 0.44 | -2.30 | 0.0739 |
| hsa-miR-4430 | 1.74 | 1.74 | 0.3506 | 23.90 | 23.90 | 0.0106 | 0.07 | -13.77 | 0.0281 |
| hsa-miR-4281 | 1.87 | 1.87 | 0.5650 | 6.00 | 6.00 | 0.0400 | 0.31 | -3.21 | 0.1870 |
| hsa-miR-3653 | 1.40 | 1.40 | 0.7411 | 24.11 | 24.11 | 0.0257 | 0.06 | -17.28 | 0.0400 |
| hsa-miR-342-3p | 0.98 | -1.02 | 0.9800 | 3.48 | 3.48 | 0.2577 | 0.28 | -3.55 | 0.2463 |
| hsa-miR-3135b | 1.81 | 1.81 | 0.5380 | 13.81 | 13.81 | 0.0247 | 0.13 | -7.61 | 0.0958 |
| hsa-miR-2861 | 1.36 | 1.36 | 0.4150 | 3.10 | 3.10 | 0.0028 | 0.44 | -2.28 | 0.0161 |
| hsa-miR-1587 | 1.89 | 1.89 | 0.3963 | 3.60 | 3.60 | 0.1031 | 0.53 | -1.90 | 0.0331 |
| hsa-let-7b-5p | 1.76 | 1.76 | 0.5515 | 7.74 | 7.74 | 0.0893 | 0.23 | -4.41 | 0.2377 |
*P values were calculated using an independent samples t-test.
Predicted target pathways of miRNA
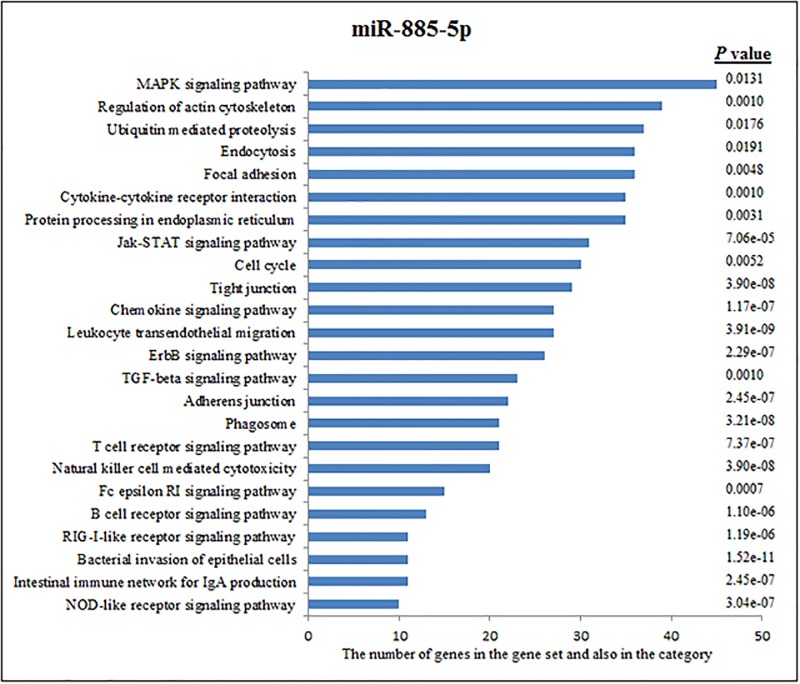
The predicted target genes of miRNAs that uniquely expressed in the chronic cases, including miR-885-5p, miR-483-3p, and miR-328, were analysed. The target genes of these miRNAs and the immunologically effective pathways that are potentially manipulated by these miRNAs are presented in Figs 2–4. According to these analyses, miR-885-5p is involved in the regulation of 3085 gene expressions. According to KEGG function annotations, these mutual genes play a role in several signalling pathways, such as the MAPK signalling pathway, regulation of the actin cytoskeleton, ubiquitin-mediated proteolysis, endocytosis, focal adhesion, cytokine-cytokine receptor interactions, protein processing in the endoplasmic reticulum, the JAK-STAT signalling pathway, cell the cycle, tight junction, the chemokine signalling pathway, and leukocyte transendothelial migration pathways (Fig 2).
Fig 2. Pathway analysis of miR-885-5p according to KEGG function annotations.
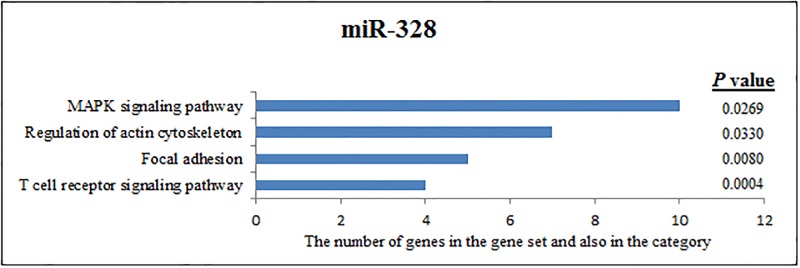
Fig 4. Pathway analysis of miR-328 according to KEGG function annotations.
The results showed that 165 predicted genes were annotated by miR-483-3p. These genes have functions in the regulation of the actin cytoskeleton, the ErbB signalling pathway, focal adhesion, endocytosis, protein processing in the endoplasmic reticulum, tight junction, and the TGF-beta signalling pathway (Fig 3).
Fig 3. Pathway analysis of miR-483-3p according to KEGG function annotations.
According to KEGG function annotations, 332 predicted genes have a binding site for miR-328. These genes are involved in the MAPK signalling pathway, regulation of the actin cytoskeleton, focal adhesion, and T cell receptor signalling pathway (Fig 4).
Moreover, not only were 41 miRNA expressions different from the control group, but they also displayed a difference between the acute and chronic brucellosis groups (Table 2).
Compared to the acute cases, the expression levels of 28 miRNAs were significantly altered in the chronic cases. All of these miRNAs were up-regulated in the chronic cases compared to in the control cases. Apart from 1 miRNA (miR-4649-3p), 27 miRNAs were not expressed in the acute cases (Table 3). In addition, the expression levels of 18 miRNAs were significantly altered in the acute cases. The expressions of these miRNAs were similar in the chronic cases and control cases (Table 4).
Predicted target pathways of miRNA
To understand the role of immunological and genetic factors involved in the transition of brucellosis from an acute to a chronic infection, target pathway prediction of some miRNAs was performed according to KEGG function annotations. One increased miRNA (miR-885-5p) that targets genes of immunologically effective pathways is shown in Fig 2. Other increased miRNAs (miR-483-3p and miR-328) that target genes of immunologically effective pathways are shown in Figs 3 and 4, respectively.
Approximately 3,085 genes were regulated by miR-885-5p. According to KEGG function annotations, these mutual genes play a role in multiple signalling pathways, such as the MAPK signalling pathway, regulation of actin cytoskeleton, ubiquitin-mediated proteolysis, endocytosis, focal adhesion, cytokine-cytokine receptor interactions, protein processing in the endoplasmic reticulum, the JAK-STAT signalling pathway, the cell cycle, tight junction, the chemokine signalling pathway, and leukocyte transendothelial migration pathways (Fig 2).
The results showed that about 312 predicted genes were annotated by miR-483-3p. These genes are active in the regulation of the actin cytoskeleton, the ErbB signalling pathway, focal adhesion, endocytosis, protein processing in the endoplasmic reticulum, tight junction, and the TGF-beta signalling pathway (Fig 3).
The miR-328 that increased in the chronic cases was not expressed in the acute cases. The results showed that about 332 predicted genes were annotated with the genes MAPK signalling pathway, regulation of the actin cytoskeleton, focal adhesion and the T cell receptor signalling pathway (Fig 4).
Discussion and conclusion
Brucellosis is a disease that is prevalent throughout the world. Although we know that brucellosis can convert from an acute to a chronic form, our understanding of the mechanisms of that transition remains incomplete. Multiple bacterial and host factors may be involved in this complex process [31]. Some studies have proposed that CD4+ T cells can play an important role in the regulation of the Brucella infection [32, 33]. Moreover, some studies have provided evidence that the gamma interferon (IFN-γ)-mediated T helper 1 (Th1) immune response is crucial for the regulation of the Brucella infection [34].
In the current study, we focused on miRNAs that are involved in the pathogenesis of the Brucella infection. The expression levels of these miRNAs demonstrate distinct patterns within individual cases across age, gender or clinical phenotype independent manner.
Analysing miRNA expressions in CD4+ T cells sorted from chronic, acute, and control cases, we investigated several miRNAs that are involved in the post-transcriptional regulation of gene expressions, which is associated with the immunological basis of chronicity.
So far, only a few studies have investigated the role of miRNAs in Brucella infections. One of these previous studies demonstrated the role of eight miRNAs in the Brucella–host interactions in-vitro conditions using mock- and Brucella-infected RAW264.7 cells and high-throughput sequencing. It was determined that the target genes of these miRNAs were involved in regulating apoptosis and autophagy [35].
Liu and his colleagues defined the role of miR-125b-5p-mediated A20 (TNFAIP3) regulation in macrophages activation of B. abortus-infection. According to their findings, reduced miR-125b-5p expression leads to increase in the A20 protein level, suppress the activation of NF-kB and leads to intracellular bacterial survival [36]. In another study, to identify miRNAs affected by the Brucella spp. infection in the absence CD14, Rong et al. analysed the miRNA expression profiles of CD14 knockdown RAW264.7 cells affected by B. melitensis M5-90 using an miRNA array. The authors observed up-regulation of mmu-miR-199a-3p and mmu-miR-183-5p. Interestingly, among the predicted target genes of mmu-miR-199a-3p and mmu-miR-183-5p, the significantly enriched gene ontology terms were the apoptosis process, immune response, inflammatory response, innate immune response, anti-apoptosis, cytokine production, and cytokine-mediated signalling pathway [37]. In one of our previous studies, more than 2,000 miRNAs were screened in peripheral blood mononuclear cells of patients with acute or chronic brucellosis, and we determined that while the expression level of miR-1238-3p increased, miR-494, miR-6069, and miR-139-3p decreased in the chronic group compared to the acute group. The involvement of differentially expressed miRNAs and their predicated target genes involved in endocytosis, regulation of actin cytoskeleton, the MAPK signaling pathway, and the cytokine-cytokine receptor interaction as well as its chemokine signaling pathway indicates the miRNAs’ potential roles in chronic brucellosis and its progression [11]. In another one of our previous studies, the regulatory roles of 2000 miRNA were evaluated in human CD8+ T cells. We determined that there are 42 miRNAs involved in the Brucella infection. Two of these miRNAs uniquely expressed in chronic brucellosis and five miRNAs uniquely expressed in acute brucellosis. The differentially expressed miRNAs in chronic brucellosis and their predicted target genes are involved in the MAPK signalling pathway, cytokine-cytokine receptor interactions, endocytosis, regulation of actin cytoskeleton, and focal adhesion [12].
The patients enrolled in the present study were treated with same antibiotic regimen of doxycycline and rifampicin. In two previous studies, the potential effect of rifampicin on the expression levels of miRNA was demonstrated [38, 39]. However, besides those two studies, the effects of antibiotics on miRNA expressions in other types of cells have not been examined. Because all the patients received the same antibiotic regimen, this issue was avoided in this study.
In the current study, the regulatory role of 2,000 miRNAs was evaluated in human CD4+ T cells. We determined that 11 miRNAs are involved in the Brucella infection. We also determined that 41 miRNAs were differently regulated between the acute and chronic brucellosis cases, 28 miRNAs were uniquely expressed in chronic brucellosis cases, and 18 miRNAs were uniquely expressed in acute brucellosis cases.
Similarly expressed miRNAs in acute and chronic brucelosis
Eleven miRNAs (miRNA-4530, miRNA-4739, miRNA-762, miRNA-4787-5p, miRNA-940, miRNA-3676-5p, miRNA-6090, miRNA-150-5p, miRNA-4516, miRNA-4284, miRNA-3656) demonstrated a similar expression trend in both the acute and chronic groups. This indicated that both acute and chronic brucellosis may share, at least partly, similar regulatory mechanisms.
Differently expressed miRNAs in acute brucellosis compared to in chronic brucellosis
In order to identify the miRNAs in PBMCs that may be correlated with chronicity, we focused on the differentially expressed miRNAs between the acute group and the chronic group. According to our findings, there were 41 dysregulated miRNAs between the two groups. According to our findings, all of the miRNAs were decreased in the chronic group compared with in the acute group.
Nineteen of the downregulated miRNAs in chronic patients were found to be associated with carcinogenesis in various organs and tissues, such as miR-1207 [40], miR-3162-5p [41, 42], miR-3196 [43, 44], miR-371b-5p [45], miR-574-5p [46, 47], miR-1225-5p [48], miR-4485 [49], miR-572 [50], miR-4299 [51], miR-3679-5p [52], miR-3940-5p [53], miR-638 [54, 55], miR-1202 [56], miR-5787 [57], miR-1973 [58], miR-4532 [59], miR-1275 [60], miR-4728-5p [61], and miR-1915-3p [62].
Specifically altered miRNAs in chronic brucellosis
Determining the prognostic factors associated with the chronicity of brucellosis is necessary for developing follow-up treatments and improving alternative therapeutic protocols for treating the disease. In the current study, 28 miRNAs which displayed a statistically significant change in chronic cases compared to both the acute cases and the control cases were identified. All of the miRNAs were up-regulated in chronic cases compared to the control cases. Only miR-4649-3p was expressed in the acute cases, while the remaining 27 miRNAs were not.
One of the 28 miRNAs in which the expression was upregulated in chronic brucellosis is miR-885-5p. Afanasyeva et al. show that miR-885-5p inhibits cell proliferation and survival and promotes cellular senescence and apoptosis. They demonstrated that miR-885-5p downregulates cyclin-dependent kinase (CDK2) and that the mini-chromosome maintenance protein (MCM5) activates p53. They provided evidence that CDK2, which participates in cell cycle regulation and is especially critical during the G1 to S phase transition, and MCM5, which is involved in the initiation of DNA replication, are direct miR-885-5p targets [63]. In our study, this miRNA, which was only upregulated in the chronic group, may have played a significant role in the immunological response to Brucella. Yan et al. showed that miR-885-5p inhibited the expression of MMP9 indirectly [64]. MMM9 may play an essential role in the local proteolysis of the extracellular matrix, leukocyte migration, and bone osteoclastic resorption.
According to KEGG pathways analysis, miR-885-5p was linked to the MAPK signalling pathway, regulation of the actin cytoskeleton, ubiquitin-mediated proteolysis, endocytosis, focal adhesion, cytokine-cytokine receptor interactions, protein processing in the endoplasmic reticulum, the JAK-STAT signalling pathway, the cell cycle, tight junction, the chemokine signalling pathway, and leukocyte transendothelial migration pathways in chronic brucellosis.
Among the 28 miRNAs, the miR-483-3p upregulated in the chronic group in comparison to the control group. There was no expression in the acute infection group. Ni et al. showed that miR-483-3p is identified as a critical regulator of the expression of Insulin-like growth factor 1 (IGF-1) in natural killer cells [65]. According to present knowledge, IGF-1 is involved in the regulation of immunity and inflammation in several ways, such as regulating haematopoiesis, direct effector functions of cells of the innate and acquired immune systems, and functions of lymphocytes and monocytes via interactions with IGF-1R [65–68]. The majority of immune system cells carry the IGF-1R protein; thus, T lymphocytes are one of the targets of IGF-1 [66].
The function of IGF-1 in human natural killer cell development and cytotoxicity was demonstrated previously [65]. High miR-483-3p levels blockade IGF-1 and reduce natural killer cell cytotoxicity. These findings indicate that miR-483-3p is a regulator of natural killer cell functions [65]. According to our results that miR-483-3p upregulated in the chronic group compared to in the control group and that there was no expression in the acute group is remarkable. Dornand et al. showed that NK cells are activated by B. suis-infected macrophages and that they inhibit the intracellular multiplication of the bacteria by lysing the infected cells. Therefore, NK cells could be one actor for controlling the development of Brucella in humans [69].
One of the 28 miRNAs, miR-328, is involved in cancer [70, 71], autoimmune [72], and neuronal diseases [73]. Tay et al. demonstrated another role of miR-328 in the regulation of innate immune cell function by showing that suppression of miR-328 improved bacterial clearance in the lungs. According to their findings, miR-328 is involved in the regulation of phagocytosis and survival of bacteria through modifying the uptake of live, heat-killed, non-typeable Haemophilus influenzae [74]. They also suggested that bacterial killing enhanced by an oxygen-independent pathway depends on their increased cathepsin D observation in macrophages treated with anti-328 [74]. Thus, based on previous findings, miR-328 may have a strong potential to play a critical role in the microbial host defence of innate immune cells by augmenting phagocytosis as well as the production of ROS and microbicidal activity. In our study, the expression of miR-328 was 2.21-fold in the chronic group compared to in the control group. There was no expression in the acute group. Increased miR-328 levels in the chronic cases may be associated with fail of Brucella clearance and establishment of chronicity of Brucella infections. According to KEGG pathways analysis, miR-328 was linked to the MAPK signalling pathway, regulation of the actin cytoskeleton, focal adhesion, and the T cell receptor signalling pathway.
Identifying the prognostic factors of brucellosis is essential for ensuring more accurate prognoses as well as developing satisfactory follow-up and alternative therapeutic approaches to the disease. In the present study, we uniquely determined enhanced expressions of 28 miRNAs in CD4 (+) T cells which were significantly associated with chronicity in brucellosis. Due to these miRNAs identified in a small subset of individuals, the obtained data required to be validated in a larger group. In additions, further studies are needed to clarify the potential target genes and signalling pathways of these miRNAs and their relation to chronicity, but these preliminary findings suggest that the altered expression of these miRNAs in CD4 (+) T cells may serve as biomarkers that indicate progression to chronicity.
Data Availability
The microarray data that support the findings of this study are deposited in the Gene Expression Omnibus with an accession number of GSE107554.
Funding Statement
Ferah Budak, Haluk Barbaros Oral, Emin Halis Akalın were the authors who were supported by a funder. They were supported by the the same grand. The name of the funder is: Türkiye Bilimsel ve Teknolojik Araştırma Kurumu. The link of the funder: (https://www.tubitak.gov.tr). The grand number: SBAG-111S183. This funder (Türkiye Bilimsel ve Teknolojik Araştırma Kurumu) had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The rest of the authors (Salih Haldun Bal, Gulcin Tezcan, Abdullah Yılmaz, Pınar Hız) were not supported by any funder.
References
- 1.Ferooz J, Letesson JJ. Morphological analysis of the sheathed flagellum of Brucella melitensis. BMC Research Notes. 2010;3(1): 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norman FF, Monge-Maillo B, Chamorro-Tojeiro S, Pérez-Molina J, López-Vélez R. Imported brucellosis: a case series and literature review, travel medicine and infectious disease. 2016;14(3): 182–199. doi: 10.1016/j.tmaid.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 3.Yüce A, Alp-Çavus S. Brucellosis in Turkey, A review. Klimik Dergisi. 2006;19: 87–97. [Google Scholar]
- 4.de Figueiredo P, Ficht TA, Rice-Ficht A, Rossetti CA, Adams LG. Pathogenesis and immunobiology of brucellosis review of Brucella-host interactions. Am J Path. 2015; 185: 1505–1517. doi: 10.1016/j.ajpath.2015.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turan H, Serefhanoglu K, Karadeli E, Togan T, Arslan H. Osteoarticular involvement among 202 brucellosis cases identified in Central Anatolia region of Turkey. Intern. Med. 2011;50: 421–428. [DOI] [PubMed] [Google Scholar]
- 6.Franco MP, Mulder M, Gilman RH, Smits HL. Human brucellosis. Lancet Infect Dis. 2007;7(12): 775–786. doi: 10.1016/S1473-3099(07)70286-4 [DOI] [PubMed] [Google Scholar]
- 7.Galińska EM, Zagórski J. Brucellosis in humans-etiology, diagnostics, clinical forms. Ann Agric Environ Med. 2013;20(2): 233–238. [PubMed] [Google Scholar]
- 8.Ulu-Kılıç A, Metan G, Alp E. Clinical presentations and diagnosis of brucellosis. Recent Patents on Anti-Infective Drug Discovery. 2013;8(1): 34–41. [PubMed] [Google Scholar]
- 9.Ariza J, Pellicer T, Pallarés R, Foz A, Gudiol F. Specific antibody profile in human brucellosis. Clin Infect Dis. 1992;14: 131–140. [DOI] [PubMed] [Google Scholar]
- 10.Araj GF. Update on laboratory diagnosis of human brucellosis. Int J Antimicrob Agents. 2010;36(1): 12–17. [DOI] [PubMed] [Google Scholar]
- 11.Budak F, Bal SH, Tezcan G, Akalın H, Göral G, Oral HB. Altered expressions of miR-1238-3p, miR-494, miR-6069, and miR-139-3p in the formation of chronic brucellosis. J. of Immunol. Research. 2016. http://dx.doi.org/10.1155/2016/4591468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budak F, Bal SH, Tezcan G, Guvenc F, Akalın H, Göral G, et al. MicroRNA expression patterns of CD8+ T cells in acute and chronic brucellosis. PLoS One. 2016;11(11): e0165138 doi: 10.1371/journal.pone.0165138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luckheeram RV, Zhou R, Verma AD, Xia B. CD4+T cells: differentiation and functions. Clinical and Developmental Immunology. 2012;2012: 925135 doi: 10.1155/2012/925135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6: 1123–1132. doi: 10.1038/ni1254 [DOI] [PubMed] [Google Scholar]
- 15.Hori S, Takahashi T, Sakaguchi S. Control of autoimmunity by naturally arising regulatory CD4+ T cells. Adv Immunol. 2003;81: 331–371. [DOI] [PubMed] [Google Scholar]
- 16.Martirosyan A, Bargen KV, Gorvel VA, Zhao W, Hanniffy S, Bonnardel J. In vivo identification and characterization of CD4+ cytotoxic T cells induced by virulent Brucella abortus infection. PLoS One. 2013;8(12): e82508 doi: 10.1371/journal.pone.0082508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldwin CL, Goenka R. Host immune responses to the intracellular bacteria Brucella: does the bacteria instruct the host to facilitate chronic infection? Crit Rev Immunol. 2006;26: 407–442. [DOI] [PubMed] [Google Scholar]
- 18.Ambros V. The functions of animal microRNAs. Nature. 2004;431: 350–355. doi: 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116: 281–297. [DOI] [PubMed] [Google Scholar]
- 20.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113: 25–36. [DOI] [PubMed] [Google Scholar]
- 21.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33: 1290–1297. doi: 10.1093/nar/gki200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9: 1274–1281. doi: 10.1261/rna.5980303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309: 310–311. doi: 10.1126/science.1114519 [DOI] [PubMed] [Google Scholar]
- 24.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6: 590–610. doi: 10.1016/j.molonc.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wahlquist C, Jeong D, Rojas-Munoz A, Kho C, Lee A, Mitsuyama S. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature. 2014;508: 531–535. doi: 10.1038/nature13073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skalsky RL, Cullen BR, Viruses, microRNAs, and host interactions. Ann Rev Microbiol. 2010;64: 123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1): 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Duncan D, Shi Z, Zhang B. Web-based Gene Set Analysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;Web server issue: W77–83. doi: 10.1093/nar/gkt439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young EJ. Brucella species, In: Mandell GL, Bennett JE, Dolin R, editors. Princible and practice of infectious diseases, 7th ed Philadelphia: Churchill Livingstone Elsevier; 2010. pp. 2921–2925. [Google Scholar]
- 30.Doğanay AM, Aygen B. Human brucellosis: an overview. Int J Infect Dis. 2003;7: 173–182. [Google Scholar]
- 31.Waqas A, Zheng K, Liu ZF. Establishment of chronic infection: Brucella’s stealth strategy. Frontiers in Cellular and Infection Microbiology. 2016; 6(30). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cassataro J, Estein SM, Pasquevich KA, Velikovsky CA, de la Barrera R, Bowden R, et al. Vaccination with the recombinant Brucella outer membrane protein 31 or a derived 27-amino-acid synthetic peptide elicits a CD4+ T helper 1 response that protects against Brucella melitensis infection. Infect. Immun. 2005;73(12): 8079–8088. doi: 10.1128/IAI.73.12.8079-8088.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira SC, Harms JS, Banai M, Splitter GA. Recombinant Brucella abortus proteins that induce proliferation and gamma-interferon secretion by CD4+ T cells from Brucella-vaccinated mice and delayed-type hypersensitivity in sensitized guinea pigs. Cell. Immunol. 1996;172: 262–268. doi: 10.1006/cimm.1996.0241 [DOI] [PubMed] [Google Scholar]
- 34.Vitry MA, De Trez C, Goriely S, Dumoutier L, Akira S, Ryffel B, et al. Crucial role of gamma interferon-producing CD4+ Th1 cells but dispensable function of CD8+ T Cell, B Cell, Th2, and Th17 responses in the control of Brucella melitensis infection in mice. Infect Immun. 2012. December;80(12): 4271–4280. doi: 10.1128/IAI.00761-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng K, Chen DS, Wu YQ, Xu XJ, Zhang H, Chen HC, et al. MicroRNA expression profile in RAW264.7 cells in response to Brucella melitensis infection. Int J of Bio Sci. 2012;8: 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu N, Wang L, Sun C. microRNA-125b-5p suppresses Brucella abortus intracellular survival via control of A20 expression. BMC Microbiol. 2016;16: 171–180. doi: 10.1186/s12866-016-0788-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rong H, Jiao H, Hao Y, Pang F, Li G, Peng D, et al. CD14 gene silencing alters the microRNA expression profile of RAW264.7 cells stimulated by Brucella melitensis infection. Innate Immun. 2017. July;23(5): 424–431. doi: 10.1177/1753425917707025 [DOI] [PubMed] [Google Scholar]
- 38.Takahashi K, Tatsumi N, Fukami T, Yokoi T, Nakajima M. Integrated analysis of rifampicin-induced microRNA and gene expression changes in human hepatocytes. Drug Metab Pharmokinet. 2014;29(4): 333–340. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Wang Y, Wang L, Liang H, Feng W, Meng X. Integrative network analysis of rifampin-regulated miRNAs and their functions in human hepatocytes. Bio-Medical Materials and Engineering. 2015;26(1): S1985–S1991. [DOI] [PubMed] [Google Scholar]
- 40.Wu G, Liu A, Zhu J, Lei F, Wu S, Zhang X, et al. MiR-1207 overexpression promotes cancer stem cell-like traits in ovarian cancer by activating the Wnt/β-catenin signaling pathway. Oncotarget. 2015;6(30): 28882–94. doi: 10.18632/oncotarget.4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stegeman S, Amankwah E, Klein K, O’Mara TA, Kim D, Lin HY, et al. A large-scale analysis of genetic variants within putative miRNA binding sites in prostate cancer. Cancer Discov. 2015;5(4): 368–79. doi: 10.1158/2159-8290.CD-14-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Yao D, Li Y, Chen H, He C, Ding N, et al. Serum microRNA expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. Int J Mol Med. 2013;32(3): 557–67. doi: 10.3892/ijmm.2013.1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, Li J, Sun M, Sun L, Zhang X. miRNA expression in breast cancer varies with lymph node metastasis and other clinicopathologic features. IUBMB Life. 2014;66(5): 371–7. doi: 10.1002/iub.1273 [DOI] [PubMed] [Google Scholar]
- 44.Sand M, Skrygan M, Sand D, Georgas D, Hahn SA, Gambichler T, et al. Expression of microRNAs in basal cell carcinoma. Br J Dermatol. 2012;167(4): 847–55. doi: 10.1111/j.1365-2133.2012.11022.x [DOI] [PubMed] [Google Scholar]
- 45.Mutlu S, Mutlu H, Kirkbes S, Eroglu S, Kabukcuoglu YS, Kabukcuoglu F. The expression of miR-181a-5p and miR-371b-5p in chondrosarcoma. Eur Rev Med Pharmacol Sci. 2015;19(13): 2384–8. [PubMed] [Google Scholar]
- 46.Zhou R, Zhou X, Yin Z, Guo J, Hu T, Jiang S. MicroRNA-574-5p promotes metastasis of non-small cell lung cancer by targeting PTPRU. Sci Rep. 2016;20(6): 35714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui Z, Tang J, Chen J, Wang Z. Hsa-miR-574-5p negatively regulates MACC-1 expression to suppress colorectal cancer liver metastasis. Cancer Cell Int. 2014;7(14): 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng H, Zhang F, Lin X, Huang C, Zhang Y, Li Y. MicroRNA-1225-5p inhibits proliferation and metastasis of gastric carcinoma through repressing insulin receptor substrate-1 and activation of β-catenin signaling, Oncotarget. 2016;7(4): 4647–63. doi: 10.18632/oncotarget.6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sripada L, Singh K, Lipatova AV, Singh A, Prajapati P, Tomar D, et al. hsa-miR-4485 regulates mitochondrial functions and inhibits the tumorigenicity of breast cancer cells. J Mol Med (Berl). 2017;95(6): 641–651. [DOI] [PubMed] [Google Scholar]
- 50.Wu AH, Huang YL, Zhang LZ, Tian G, Liao QZ, Chen SL. MiR-572 prompted cell proliferation of human ovarian cancer cells by suppressing PPP2R2C expression. Biomed Pharmacother. 2016;77: 92–7. doi: 10.1016/j.biopha.2015.12.005 [DOI] [PubMed] [Google Scholar]
- 51.Miao X, Jia L, Zhou H, Song X, Zhou M, Xu J, et al. miR-4299 mediates the invasive properties and tumorigenicity of human follicular thyroid carcinoma by targeting ST6GALNAC4. UBMB Life. 2016;68(2): 136–44. [DOI] [PubMed] [Google Scholar]
- 52.Xie Z, Yin X, Gong B, Nie W, Wu B, Zhang X, et al. Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev Res (Phila). 2015. February;8(2): 165–73. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Wang K, Dang N, Wang L, Zhang M. Downregulation of miR-3940-5p promotes T-cell activity by targeting the cytokine receptor IL-2R gamma on human cutaneous T-cell lines. Immunobiology. 2016. December;221(12): 1378–1381. doi: 10.1016/j.imbio.2016.07.008 [DOI] [PubMed] [Google Scholar]
- 54.Cheng J, Chen Y, Zhao P, Li N, Lu J, Li J, et al. Dysregulation of miR-638 in hepatocellular carcinoma and its clinical significance. Oncol Lett. 2017. May;13(5): 3859–3865. doi: 10.3892/ol.2017.5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao LY, Yao Y, Han J, Yang J, Wang XF, Tong DD, et al. miR-638 suppresses cell proliferation in gastric cancer by targeting Sp2. Dig Dis Sci. 2014;59(8): 1743–53. doi: 10.1007/s10620-014-3087-5 [DOI] [PubMed] [Google Scholar]
- 56.Quan Y, Song Q, Wang J, Zhao L, Lv J, Gong S. MiR-1202 functions as a tumor suppressor in glioma cells by targeting Rab1A. Tumour Biol. 2017;39(4): 1010428317697565. [DOI] [PubMed] [Google Scholar]
- 57.Yan S, Han B, Gao S, Wang X, Wang Z, Wang F, et al. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget. 2017; 8(36): 60149–60158. doi: 10.18632/oncotarget.18557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.El-Awady RA, Hersi F, Al-Tunaiji H, Saleh EM, Abdel-Wahab AH, Al Homssi A, et al. Epigenetics and miRNA as predictive markers and targets for lung cancer chemotherapy. Cancer Biol Ther. 2015;16(7): 1056–70. doi: 10.1080/15384047.2015.1046023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boo L, Ho WY, Ali NM, Yeap SK, Ky H, Chan KG, et al. MiRNA transcriptome profiling of spheroid-enriched cells with cancer stem cell properties in human breast MCF-7 cell line. Int J Biol Sci. 2016;12(4): 427–45. doi: 10.7150/ijbs.12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fawzy IO, Hamza MT, Hosny KA, Esmat G, El Tayebi HM, Abdelaziz AI. miR-1275: a single microRNA that targets the three IGF2-mRNA-binding proteins hindering tumor growth in hepatocellular carcinoma. FEBS Lett. 2015;589(17): 2257–65. doi: 10.1016/j.febslet.2015.06.038 [DOI] [PubMed] [Google Scholar]
- 61.Li H, Zhou X, Zhu J, Cheng W, Zhu W, Shu Y, et al. MiR-4728-3p could act as a marker of HER2 status. Cancer Biomark. 2015;15(6): 807–14. doi: 10.3233/CBM-150524 [DOI] [PubMed] [Google Scholar]
- 62.Borrelli N, Denaro M, Ugolini C, Poma AM, Miccoli M, Vitti P, et al. miRNA expression profiling of ‘noninvasive follicular thyroid neoplasms with papillary-like nuclear features’ compared with adenomas and infiltrative follicular variants of papillary thyroid carcinomas. Mod Pathol. 2017;30(1): 39–51. doi: 10.1038/modpathol.2016.157 [DOI] [PubMed] [Google Scholar]
- 63.Afanasyeva EA, Mestdagh P, Kumps C, Vandesompele J, Ehemann V, Theissen J, et al. MicroRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Differ. 2011;18(6): 974–984. doi: 10.1038/cdd.2010.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan W, Zhang W, Sun L, Liu Y, You G, Wang Y, et al. Identification of MMP-9 specific microRNA expression profile as potential targets of anti-invasion therapy in glioblastoma multiforme. Brain Res. 2011;1411: 108–115. doi: 10.1016/j.brainres.2011.07.002 [DOI] [PubMed] [Google Scholar]
- 65.Ni F, Sun R, Fu B, Wang F, Guo C, Tian Z, et al. IGF-1 promotes the development and cytotoxic activity of human NK cells. Nat Commun. 2013;4: 1479 doi: 10.1038/ncomms2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stuart CA, Meehan RT, Neale LS, Cintron NM, Furlanetto RW. Insulin-like growth factor-I binds selectively to human peripheral blood monocytes and B-lymphocytes. J. Clin. Endocrinol. Metab. 1991;72: 1117–1122. doi: 10.1210/jcem-72-5-1117 [DOI] [PubMed] [Google Scholar]
- 67.Kooijman R, Willems M, De Haas CJ, Rijkers GT, Schuurmans AL, Van Buul-ogffers SC, et al. Expression of type I insulin-like growth factor receptors on human peripheral blood mononuclear cells. Endocrinol. 1992;131, 2244–2250. [DOI] [PubMed] [Google Scholar]
- 68.Badolato R, Bond HM, Valerio G, Petrella A, Morrone G, Waters MJ, et al. Differential expression of surface membrane growth hormone receptor on human peripheral blood lymphocytes detected by dual fluorochrome flow cytometry. J. Clin. Endocrinol. Metab. 1994;79: 984–990. doi: 10.1210/jcem.79.4.7962309 [DOI] [PubMed] [Google Scholar]
- 69.Dornand J, Lafont V, Oliaro J, Terraza A, Castaneda-Roldan E, Liautard JP. Impairment of intramacrophagic Brucella suis multiplication by human natural killer cells through a contact-dependent mechanism, Infect Immun. 2004;72(4): 2303–2311. doi: 10.1128/IAI.72.4.2303-2311.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saberi A, Danyaei A, Neisi N, Dastoorpoor M, Tahmasbi Birgani MJ. MiR-328 may be considered as an oncogene in human invasive breast carcinoma. Iran Red Crescent Med J. 2016;7,18(11): e42360 eCollection 2016 Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu C, Zhang L, Huang Y, Lu K, Tao T, Chen S, et al. MicroRNA-328 directly targets p21-activated protein kinase 6 inhibiting prostate cancer proliferation and enhancing docetaxel sensitivity. Mol Med Rep. 2015;12(5): 7389–95. doi: 10.3892/mmr.2015.4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Padgett KA, Lan RY, Leung PC, Lleo A, Dawson K, Pfeiff J, et al. Primary biliary cirrhosis is associated with altered hepatic micro RNA expression. Journal of autoimmunity. 2009;32: 246–253. doi: 10.1016/j.jaut.2009.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and micro RNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol. Chem. 2009;284: 1971–1981. doi: 10.1074/jbc.M807530200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tay HL, Kaiko GE, Plank M, Li J, Maltby S, Essilfie AT, et al. Antagonism of miR- 328 increases the antimicrobial function of macrophages and neutrophils and rapid clearance of non-typeable Haemophilus influenzae (NTHi) from infected lung. PLoS Pathog. 2015;11(4): e1004549 doi: 10.1371/journal.ppat.1004549 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The microarray data that support the findings of this study are deposited in the Gene Expression Omnibus with an accession number of GSE107554.