Abstract
Human milk is a unique and complex fluid that provides infant nutrition and delivers an array of bioactive molecules that serve various functions. Glycans, abundant in milk, can be found as free oligosaccharides or as glycoconjugates. Milk glycans are increasingly linked to beneficial outcomes in neonates through protection from pathogens and modulation of the immune system. Indeed, these glycans influence the development of the infant and the infant-gut microbiota. Bifidobacterium species commonly are enriched in breastfed infants and are among a limited group of bacteria that readily consume human milk oligosaccharides (HMOs) and milk glycoconjugates. Given the importance of bifidobacteria in infant health, numerous studies have examined the molecular mechanisms they employ to consume HMOs and milk glycans, thus providing insight into this unique enrichment and shedding light on a range of translational opportunities to benefit at-risk infants.
Keywords: milk oligosaccharides, glycans, infant microbiota, Bifidobacterium
INTRODUCTION
The development of a beneficial host-microbe symbiosis in the neonate gastrointestinal tract is a highly complex and important biological process (Scholtens et al. 2012). Numerous factors shape the developing gut microbiome, including diet, antimicrobial exposure, and mode of delivery as well as other influences linked to cultural practices and geographic location (Lewis & Mills 2017, Scholtens et al. 2012). During the first years of life, the infant microbiota is still changing, and the establishment of the intestinal microbiota is significantly impacted by diet (Koenig et al. 2011, Scholtens et al. 2012, Yatsunenko et al. 2012). Understanding this exquisite, yet complicated, relationship between neonate diet and the development of the microbiota in the early years of life is of significant interest.
Breastfeeding is associated with numerous beneficial effects on the neonate such as providing protection from infection and diarrhea (Duijts et al. 2010, Ruiz-Palacios et al. 1990). Through milk, the infant acquires all of the necessary elements for growth and nourishment. In human milk, lactose, lipids, and proteins are abundant components and serve as nutrient sources for the infant. In addition to providing nutrition, human milk delivers an array of unique bioactive components.
One class of bioactive molecules present in human milk are glycans. Glycans are highly abundant in human milk and are found as free human milk oligosaccharides (HMOs) or conjugated to proteins or lipids. Milk oligosaccharides have remarkable structural diversity and complexity, with hundreds of structures determined within human milk pools (Bode 2012, Petherick 2010). HMOs are thought to benefit the neonate via various direct and indirect roles. In addition to free oligosaccharides, approximately 70% of human milk proteins are glycosylated. This review examines the free glycans and glycoconjugates found in milk and their interactions with the neonate microbiota.
HUMAN MILK OLIGOSACCHARIDES: STRUCTURES AND COMPOSITION
HMOs are an abundant component of human milk (Bode et al. 2016, Petherick 2010), with approximately 5–20 g/L of unbound oligosaccharides present in mature milk (Bode 2006, Hong et al. 2014, McGuire et al. 2017). Surveys of different human milks have revealed tremendous structural diversity, with intricate molecular structures among and within HMO pools (Kunz et al. 2000; Wu et al. 2010, 2011). HMOs are composed of five monomers: glucose (Glc), galactose (Gal), N-acetylglucosamine (GlcNAc), fucose (Fuc), and N-acetylneuraminic acid (NeuAc, a sialic acid). Structurally, HMOs can be linear or branched, and nearly all structures contain a lactose core at the reducing end (Ruhaak & Lebrilla 2012; Wu et al. 2010, 2011). The lactose core can be extended by lacto-N-biose (LNB; Galβ1-3GlcNAc) or N-acetyllactosamine (Galβ1-4GlcNAc) (Ruhaak & Lebrilla 2012). HMOs extended with LNB are known as type I HMOs, and HMOs extended with N-acetyllactosamine are known as type II HMOs (Ruhaak & Lebrilla 2012). Fucose can be attached to HMO structures in α1-2, α1-3, and α1-4 linkages, while sialic acid can be attached by α2-3 and α2-6 linkages, thus providing additional opportunities for complexity and activity. Smaller HMO structures such as 2′-fucosyllactose (2′-FL), 3-fucosyllactose (3-FL), 3′-sialyllactose (3′-SL) and 6′-sialyllactose (6′-SL) are composed of one fucose or sialic acid residue attached to a lactose core with various linkages (Bode & Jantscher-Krenn 2012). HMOs can be categorized as acidic or neutral, with acidic HMOs referring to structures that contain sialic acid (Bode & Jantscher-Krenn 2012, Wu et al. 2011).
Variation in HMOs exists between different individuals (Ruhaak & Lebrilla 2012). For example, HMO fucosylation is determined by a mother’s secretor and Lewis blood group statuses (Bode & Jantscher-Krenn 2012). The FUT2 gene, which is also known as the secretor gene, catalyzes the transfer of fucose to terminal galactose via an α1-2 linkage (Bode & Jantscher-Krenn 2012). Another fucosyltransferase is encoded by the FUT3 gene, which transfers fucose with α1-3 and α1-4 linkages (Bode & Jantscher-Krenn 2012). Expression of FUT3 produces the Lewis b antigen in secretors and the Lewis a antigen in nonsecretors (Bode & Jantscher-Krenn 2012). In addition to variation from a woman’s secretor and Lewis blood group statuses, there is variation in HMOs over the course of lactation (Ruhaak & Lebrilla 2012). One example is that the number of HMOs is highest in colostrum compared to mature milk (Bode & Jantscher-Krenn 2012). In addition, the HMO synthetic capacity can be influenced by the gestational stage of the mother during pregnancy. In a recent profiling of milk from mothers who delivered prematurely, Underwood et al. (2015) revealed that the fractions of fucosylated and sialylated HMOs were highly variable in comparison to those from the milk of mothers who delivered at term.
BOVINE MILK OLIGOSACCHARIDES: STRUCTURES AND COMPOSITION
Milk from a number of animals, including cow (Albrecht et al. 2014; Barile et al. 2010; Tao et al. 2008, 2009), sheep (Albrecht et al. 2014, Martinez-Ferez et al. 2006), goat (Albrecht et al. 2014, Chaturvedi & Sharma 1988, Martin-Ortiz et al. 2016, Martinez-Ferez et al. 2006, Thum et al. 2015), pig (Albrecht et al. 2014, Mudd et al. 2016, Salcedo et al. 2016, Tao et al. 2010), horse (Albrecht et al. 2014, Nakamura et al. 2001, Urashima et al. 1991), and camel (Albrecht et al. 2014, Alhaj et al. 2013, Fukuda et al. 2010), contains oligosaccharides similar to those found in human milk. There is a growing interest in examining the structures and concentrations of these animal milk oligosaccharides to identify additional sources of compounds with similar bioactivity to HMOs. Comprehensive glycomic studies of bovine and porcine milks have shown a diverse collection of structures, with the relative proportion of acidic oligosaccharides being higher than that of human milk (Mudd et al. 2016, Tao et al. 2009). Although the type of animal used for industrial milk production can vary by country, bovine milk is generally produced in large quantities and is a potential source for industrial-scale oligosaccharide recovery.
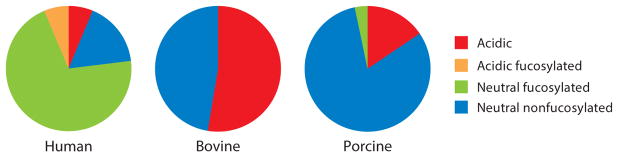
Bovine milk contains many oligosaccharides that are identical to those found in human milk, such as 3′-sialyllactose, 6′-sialyllactose, and lacto-N-neotetraose (LNnT), as well as numerous oligosaccharides that are unique in both monosaccharide composition and structure (Aldredge et al. 2013). Furthermore, several fucosylated structures, ranging in monosaccharide composition from 2Hex1Fuc to 3Hex6HexNAc1Fuc, have been observed in bovine milk; however, the total amount of fucosylation is small (Aldredge et al. 2013). In addition to differences in fucosylation, bovine milk contains a greater proportion of sialylated oligosaccharides than human milk (Ninonuevo et al. 2006, Tao et al. 2008, Wu et al. 2011). Relative abundances of acidic, acidic fucosylated, neutral fucosylated, and neutral nonfucosylated oligosaccharides in human, bovine, and porcine milk are shown in Figure 1. Although the predominant form of sialic acid is N-acetylneuraminic acid, bovine milk and many other mammal milks also contain trace levels of N-glycolylneuraminic acid (Bode 2006). Oligosaccharides with completely elucidated structures that are known to exist in both human and bovine milk are summarized in Table 1.
Figure 1.
Relative abundances of acidic, acidic fucosylated, neutral fucosylated, and neutral nonfucosylated oligosaccharides in human, bovine, and porcine milk, as demonstrated by selected glycomics studies (Mudd et al. 2016, Ninonuevo et al. 2006, Tao et al. 2009).
Table 1.
Oligosaccharides with known structures found in both human and bovine milk
| Name | Monosaccharide composition | Structure | Reference(s) | ||||
|---|---|---|---|---|---|---|---|
| Hex | HexNAc | Fuc | NeuAc | NeuGc | |||
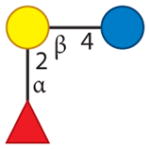
| 2′-Fucosyllactose | 2 | 0 | 1 | 0 | 0 |
|
Aldredge et al. 2013 |
| 3-Fucosyllactose | 2 | 0 | 1 | 0 | 0 |
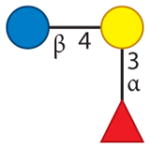
|
Aldredge et al. 2013 |
| Lacto-N-neohexaose | 4 | 2 | 0 | 0 | 0 |
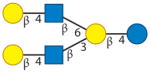
|
Aldredge et al. 2013, Kobata & Ginsburg 1972 |
| Lacto-N-tetraose | 3 | 1 | 0 | 0 | 0 |
|
Kunz et al. 2000 |
| Lacto-N-neotetraose | 3 | 1 | 0 | 0 | 0 |
|
Aldredge et al. 2013, Kunz et al. 2000 |
| 3′-Sialyllactosamine | 1 | 1 | 0 | 1 | 0 |
|
Aldredge et al. 2013, Wu et al. 2011 |
| 6′-Sialyllactosamine | 1 | 1 | 0 | 1 | 0 |
|
Aldredge et al. 2013, Wu et al. 2011 |
| 3′-Sialyllactose | 2 | 0 | 0 | 1 | 0 |
|
Aldredge et al. 2013, Martin-Sosa et al. 2003 |
| 6′-Sialyllactose | 2 | 0 | 0 | 1 | 0 |
|
Aldredge et al. 2013, Martin-Sosa et al. 2003 |
| Gal(β1-6)Gal(β1-4)Glc | 3 | 0 | 0 | 0 | 0 |
|
Aldredge et al. 2013 |
Abbreviations: Fuc, Fucose; Hex, Hexose; HexNAc, N-acetylhexosamine; NeuAc, N-acetylneuraminic acid; NeuGc, N-glycolylneuraminic acid.
Like HMOs, bovine milk oligosaccharide (BMO) abundances change during lactation (Tao et al. 2009). However, the total oligosaccharide concentration in bovine milk is significantly lower. Mature human milk contains a total oligosaccharide concentration of 5–20 g/L, whereas BMOs are present at 1–2 g/L in colostrum; this concentration decreases to approximately 50–100 mg/L in mature milk (Bode 2006, Fong et al. 2011, McGuire et al. 2017, McJarrow & van Amelsfort-Schoonbeek 2004, Nakamura et al. 2003).
Dairy streams resulting from cheese making are potential sources of milk oligosaccharides for large-scale extraction (Zivkovic & Barile 2011). Whey permeate, a low-value by-product obtained after whey proteins have been recovered, contains BMOs and has the potential to serve as a raw material for large-scale oligosaccharide isolation (Dallas et al. 2014, Zivkovic & Barile 2011). Membrane filtration can be used to recover oligosaccharides present in whey permeate (Cohen et al. 2017, de Moura Bell et al. 2016), making these compounds more readily available as a functional food ingredient. Such oligosaccharides recovered from dairy streams could serve as ingredients for infant formula, as their composition is similar to that of HMOs and such an ensemble cannot currently be produced synthetically (Meli et al. 2014, Simeoni et al. 2016).
HUMAN AND BOVINE MILK GLYCOCONJUGATES
Glycosylation is a common type of post-translational protein modification in which a glycan is attached to select amino acids in the protein (Moremen et al. 2012). In eukaryotes, there are two major types of glycosylation: asparagine-linked (N-linked) glycans and serine- or threonine-linked (O-linked) glycans. Milk-protein glycosylation is associated with different roles; e.g., it enables resistance to proteolytic digestion thereby facilitating the release of encrypted bioactive peptides, and it provides passive protection from pathogen binding to intestinal cells (van Berkel et al. 1995). Similar to that observed for free glycans, protein glycosylation varies during lactation (Barboza et al. 2012, Froehlich et al. 2010). Glycoproteins in human milk include lactoferrin, lysozyme, mucins, bile salt–stimulated lipase (BSSL), sIgA, casein, and α-lactalbumin (Newburg 2013).
Glycolipids are another type of glycoconjugate found in human milk. In particular, glycosphingolipids containing sialic acid, also known as gangliosides, are a primary form of glycolipids found in human milk (Newburg 2013). Human milk gangliosides are associated with the milk-fat-globule membrane (MFGM) (Lee et al. 2011). The glycolipids GD3 [Neu5Ac(α2-8)Neu5Ac(α2-3)Gal(β1-4)Glc(β1-1) ceramide] and GM3 [Neu5Ac(α2-3)Gal(β1-4)Glc(β1-1) ceramide] are gangliosides in human milk (Lee et al. 2011, Newburg 2013). Ganglioside composition changes over the course of lactation, with GM3 increasing and GD3 decreasing (Ma et al. 2015, Pan & Izumi 2000).
Bovine milk contains some of the same glycoconjugates as human milk, such as lactoferrin, immunoglobulins, glycoproteins from the MFGM, and gangliosides (O’Riordan et al. 2014). Additionally, dairy streams contain significant amounts of glycomacropeptide (GMP), which is a heavily sialylated glycopeptide derived from hydrolysis of κ-casein during cheesemaking (Neelima et al. 2013). Nwosu et al. (2012) recently compared the N-glycome of human and bovine milk and found substantial differences in N-glycan fucosylation. Seventy-five percent of human milk N-glycan abundances corresponded to fucosylated glycans, whereas in bovine milk, 31% of N-glycan abundances were from fucosylated glycans (Nwosu et al. 2012). The bovine N-glycome was more highly sialylated with 68% of N-glycans corresponded to sialylated compounds compared to 57% in human milk (Nwosu et al. 2012). Similar to free oligosaccharides, some N-glycans containing N-glycolylneuraminic acid (NeuGc) were found in bovine milk but not human milk (Nwosu et al. 2012). Lactoferrin is present in bovine milk in lower concentrations than in human milk and has five potential N-glycosylation sites, whereas human lactoferrin has only three potential N-glycosylation sites (Baker & Baker 2005, O’Riordan et al. 2014).
STRUCTURE-FUNCTION ASSOCIATIONS OF MILK GLYCANS
The functional roles of glycans in milk are varied and of significant interest. Glycan interactions can occur directly with host tissue or host-associated microbes. This latter interaction can also result in indirect effects whereby glycan-mediated modulation of microbial activity subsequently influences host functions. Some structure-function relationships of HMOs have been demonstrated, such as providing defense against pathogens and enriching the beneficial gut microbiota (Garrido et al. 2012a, Morrow et al. 2005, Smilowitz et al. 2014). Furthermore, HMOs are associated with modulation of intestinal epithelial cell responses in a direct manner (Bode 2012; Kuntz et al. 2008, 2009). Additionally, HMOs are linked to directly affecting the immune system (Eiwegger et al. 2004, Newburg 2009). Specific HMO structures, including lacto-N-fucopentaose (LNFP) III and LNnT, have been reported for their relationship to immune modulation (Atochina et al. 2001, Terrazas et al. 2001). HMOs are also associated with improved learning and memory (Vazquez et al. 2015).
BMOs appear to have biological functions that are similar to those associated with HMOs. BMOs and bovine milk glycoconjugates can inhibit adhesion of pathogens to intestinal cells and tissues (Douellou et al. 2017, Maldonado-Gomez et al. 2015). Associations between sialylated BMOs and microbiota-dependent growth promotion have been described (Charbonneau et al. 2016b). BMOs are associated with various health-enhancing functional roles such as decreasing gut permeability and reducing inflammation (Boudry et al. 2017). Milk-associated glycoconjugates are also linked to a range of functions. For example, lactoferrin and the κ-casein-derived peptide GMP have been linked to various activities such as immunomodulatory, antimicrobial, and prebiotic activities (Azuma et al. 1984, Debbabi et al. 1998, O’Riordan et al. 2014, Rahman et al. 2009, Wang et al. 2007). Additionally, the sialic acid content of GMP has been attributed various functional roles, including prevention of pathogen binding and improved memory and learning in piglets (O’Riordan et al. 2014).
Milk Glycan–Mediated Protection from Pathogens and Infection
Milk glycans provide a defense against pathogens and can block pathogen adhesion (Morrow et al. 2005). Many pathogens adhere to the intestinal epithelial surfaces, and milk glycans have epitopes identical to the intestinal surfaces, as they are generated from similar enzymes (Smilowitz et al. 2014). As a consequence, pathogens bind to milk glycans instead of host cell ligands. Another possible mechanism of glycan protection from pathogens is the competitive binding of glycans with host cell surface receptors (Morrow et al. 2005).
Campylobacter jejuni–mediated intestinal infection is inhibited by milk glycans (Ruiz-Palacios et al. 2003). C. jejuni binds to the intestinal H2 antigen, which contains a terminal α1-2-linked fucose residue and both in vitro and ex vivo studies have demonstrated that adherence of C. jejuni to intestinal epithelial cells is prevented by prior incubation of C. jejuni with 2′-fucosyllactose (Ruiz-Palacios et al. 2003, Weichert et al. 2013). This work provides a possible mechanism behind the reduced incidence of infection observed among the infants receiving milk from secretor mothers whereby the presence of α1-2-linked fucose on HMOs can act as a decoy for pathogen binding (Morrow et al. 2004). Additionally, Jiang et al. (2004) found that secretor and Lewis, but not A or B antigens, block noroviruses from binding to receptors.
Milk glycoconjugates lactoferrin, secretory IgA, mucins, κ-casein, and gangliosides have been examined for their ability to protect against pathogen infection (Liu & Newburg 2013). Lactoferrin is a major bioactive protein in human milk known to possess bacteriostatic activity via iron-deprivation activities as well as bactericidal activity (Lonnerdal 2013). Barboza et al. (2012) demonstrated that the changing glycosylation of lactoferrin throughout lactation modulated its interaction with pathogens. Secretory IgA (sIgA), a predominant antibody in human milk with a known mechanism for pathogen clearance (Liu & Newburg 2013), is also a glycosylated protein with both N-linkages and O-linkages (Pacheco et al. 2015). K-casein has been shown to prevent Helicobacter pylori from binding to the gastric mucosa (Stromqvist et al. 1995). Other glycoproteins present in human milk, such as bile salt–stimulated lipase and lactadherin are also known for protective properties (Liu & Newburg 2013). Specifically, bile salt–stimulated lipase hydrolyzes a variety of substrates, contributes to the efficient utilization of milk fat in newborn infants, and has been associated with inhibition of Norwalk virus capsids binding to their carbohydrate ligands (Li et al. 2007, Ruvoen-Clouet et al. 2006).
Milk oligosaccharides have also been shown to alter the intracellular fate of pathogens. For example, Lin et al. (2014) showed that pooled HMOs protect bladder epithelial cells against uropathogenic Escherichia coli (UPEC) invasion and cytotoxicity. Although the presence of HMOs did not significantly alter the adhesion to the host cells, a significant reduction was observed in bacterial internalization. Pretreatment with HMOs produced a reduction in UPEC-mediated activation of key signaling proteins such as MAP kinase and the master regulator NF-κB, thereby protecting the bladder epithelial cells from the proinflammatory effects of the bacterial infection (Lin et al. 2014). Additionally, a study that tested the influence of HMOs on E. coli–infection-mediated secretion of IL-8 (a cytokine) resulted in the identification of specific proinflammatory signaling molecules modulated by HMOs and determined the mechanism of action, i.e., pooled HMOs and 2′-FL are able to directly inhibit gram-negative, lipopolysaccharide-mediated inflammation during enterotoxigenic E. coli invasion (He et al. 2016). More recently, it has been shown that 2′-FL can function to attenuate the severity of experimental necrotizing enterocolitis (NEC). NEC is a common disease in premature infants characterized by abdominal distention and intestinal necrosis. 2′-FL has been shown to protect against NEC by enhancing mesenteric perfusion via increased endothelial nitric oxide synthase expression in the neonatal intestine, and these observations suggest that 2′-FL can mediate the protective benefits of milk in a clinical setting (Good et al. 2016).
Milk Glycans and the Microbiota of Infants
The gastrointestinal tract of the infant is believed to be relatively sterile during gestation, and rapid colonization occurs with events associated with the process of giving birth. In the early stages of life, the formation of the microbiota is highly complex (Backhed et al. 2015, Ray 2016). Environmental factors, including mode of delivery, diet, and antibiotics, have been shown to affect bacterial diversity in the infant gut (Bokulich et al. 2016, Ray 2016). The initial colonization is dominated by facultatively anaerobic microbes, including staphylococci and members of the order Lactobacillales (e.g., enterococci and streptococci) (Backhed et al. 2015). Because these organisms also populate freshly expressed mother’s milk (Cabrera-Rubio et al. 2012), breastfeeding is considered the major and preferred vector for seeding the infant-gut microbiota, although the overall intimate interaction between mothers and infants may provide for other routes as well. Once this initial period has passed, Bifidobacterium, Bacteroides, and Clostridium become the signature early colonizers of the breastfed infant gut (Adlerberth & Wold 2009) with Bifidobacterium often dominating within the first three months (Yatsunenko et al. 2012).
Recent studies directly examined the relationship between milk glycans and the developing infant microbiota. De Leoz et al. (2014) first reported a significant decrease in fecal HMOs coinciding with emergent gut populations of HMO-consuming Bacteroidaceae and Bifidobacteriaceae, suggesting the active role of these clades in HMO consumption in situ. Lewis et al. (2015) similarly reported high levels of bifidobacteria positively correlating with lower levels of fecal HMOs. Davis et al. (2016) recently profiled oligosaccharides in the milk and feces of a mother/infant dyad and noted measurable decreases in specific HMO structures in the fecal samples compared to the mother’s milk. In this later work, a specific β-galactosidase from Bifidobacterium longum subsp. longum (B. longum) was used to digest HMOs in vitro, and the digested oligosaccharide fragments matched with compounds identified in infant fecal samples (Davis et al. 2016). In another study, Wang et al. (2015) correlated breast-milk HMO profiles with emergent fecal microbiota and noted specific correlations between select HMO species in the milk [such as LNFP I, monofucosyllacto-N-hexaose III (MFLNH III), sialyllacto-N-tetraose b (LSTb), and disialyllacto-N-tetraose (DSLNT)] and the level of bifidobacteria in the feces.
Various groups have profiled consumption of HMOs by bacterial groups in vitro. Ward et al. (2006) first demonstrated that Bifidobacterium longum subsp. infantis (B. infantis) ATCC 15697 can ferment HMOs as a sole carbon source. Locascio et al. (2009) subsequently showed differential consumption of HMOs by different species of bifidobacteria. More recently, Garrido et al. (2015) examined isolates of B. infantis and Bifidobacterium bifidum for their ability to use pooled HMOs and individual HMO sugars. All B. infantis isolates in this study grew well on pooled HMOs and grew well overall on individual HMO sugars. The growth of B. bifidum on individual HMOs was more variable, as some examined strains could not grow with 2′-FL and 6′-SL as a sole carbon source (Garrido et al. 2015).
Marcobal et al. (2010) first demonstrated that species of Bacteroides were able to consume HMOs; however, this phenotype was not observed with other gut-associated bacteria, including strains of Enterococcus, Streptococcus, Clostridium, and E. coli. These results were later extended by Yu et al. (2013) showing lack of growth of Clostridium spp., E. coli K12, Enterobacter spp., Enterococcus faecalis, and Staphylococcus spp. on HMOs. Hunt et al. (2012) reported that HMOs have growth-stimulating properties on Staphylococcus aureus and Staphylococcus epidermidis; however, analysis showed no HMO consumption from the spent media, suggesting factors other than HMOs caused the growth stimulation. Bacteroides thetaiotaomicron and Bacteroides fragilis utilize HMOs by inducing the same genes that are involved in mucin utilization (Marcobal et al. 2011). In this study, Marcobal et al. (2011) showed that in a gnotobiotic mouse model LNnT provided a significant growth advantage to B. infantis over B. thetaiotaomicron.
MOLECULAR MECHANISMS OF UTILIZATION OF HUMAN MILK OLIGOSACCHARIDES BY BIFIDOBACTERIA
Advances in DNA sequencing in the past decade have resulted in a flurry of genomic analysis of the bifidobacterial clade (Kwak et al. 2016, Milani et al. 2014) as well as pangenomic analysis of specific infant-oriented clades, with a particular focus on genes involved in HMO degradation (Bottacini et al. 2014, Duranti et al. 2015, Milani et al. 2015b, O’Callaghan et al. 2015). Milani et al. (2015b) inferred glycan consumption capacity in forty-seven representative Bifidobacterium species segregated into three large groups based on differences in their predicted ability to degrade plant polysaccharides. This work also revealed that certain bifidobacteria (Bifidobacterium scardovii, B. infantis, and B. bifidum) have an abundance of genes related to degradation of host-derived glycans, including sialidases, fucosidase, hexosaminidases, α-N-acetylgalactosaminidases, α-mannosidases, and lacto-N-biosidases. Given that the linkages in milk glycans and glycoconjugates require these enzymes for cleavage (Garrido et al. 2013), one would predict that functional analyses of breastfed infant microbiomes show an enrichment in these activities. Indeed, this appears to be the case, as metagenomics and metatranscriptomics of infant feces clearly reveal an abundance of expression of genes linked to host glycan degradation (Milani et al. 2015a, Yatsunenko et al. 2012).
The mechanism of host glycan consumption has been profiled extensively in two prototypical infant-borne bifidobacteria, B. infantis and B. bifidum. B. infantis possesses a 43-kb HMO gene cluster containing glycosyl hydrolases and ABC transporters (Sela et al. 2008). B. infantis consumes HMOs by using an array of ABC transporters to facilitate importation and then deconstructs HMOs via an array of intracellular glycosyl hydrolases (Garrido et al. 2011, 2012c; Sela et al. 2011, 2012; Yoshida et al. 2012). B. infantis ATCC 15697 expresses family 1 solute-binding proteins, a component of ABC transporters, when grown on HMOs, and these binding proteins have an affinity for mammalian oligosaccharides (Garrido et al. 2011). The genome of B. infantis ATCC 15697 has five intracellular α-L-fucosidases, with two of these enzymes located in the HMO cluster (Sela et al. 2012). Of the five α-L-fucosidases, there are four GH29 α-L-fucosidases and one GH95 α-L-fucosidase (Sela et al. 2012). Fucosidases located in the HMO cluster encoded by Blon_2335 and Blon_2336 were both found to have activity on fucosylated HMOs (Sela et al. 2012). Sela et al. (2011) examined two sialidases from B. infantis ATCC 15697. Sialidase NanH2 from B. infantis digested purified milk sialyloligosaccharides; however, sialidase NanH1 did not (Sela et al. 2011). It is possible this latter enzyme has activity on other types of sialylated glycans (Sela et al. 2011). B. infantis employs two different β-galactosidases to digest type-1 and type-2 HMOs (Yoshida et al. 2012). Additionally, B. infantis possesses three N-acetyl-β-D-hexosaminidases that cleave the GlcNAcβ1-3 linkage found in the HMO lacto-N-tetraose (LNT) (Garrido et al. 2012c).
The mechanism of fucose metabolism in B. infantis and other bifidobacteria has previously been unclear. Fucose is not detected in the supernatant during B. infantis growth on HMOs (Asakuma et al. 2011, Ward et al. 2007). The genome of B. infantis ATCC 15697 possesses two gene clusters containing genes that are similar to a fucose pathway involving nonphosphorylated intermediates that have been previously described in C. jejuni and Xanthomonas campestris (Sela et al. 2012, Yew et al. 2006). Bunesova et al. (2016) showed that fermentation end products lactate, acetate, and 1,2-propanediol were formed during growth of B. infantis and Bifidobacterium longum subsp. suis on L-fucose. This work proposed an L-fucose pathway similar to X. campestris and C. jejuni involving the formation of nonphosphorylated intermediates (Bunesova et al. 2016).
B. bifidum uses a process vastly different than B. infantis to catabolize HMOs by employing an array of extracellular glycosyl hydrolases (Ashida et al. 2009, Kitaoka 2012, Kiyohara et al. 2011, Turroni et al. 2010). A membrane-associated GH95 α-L-fucosidase, AfcA, with α-1,2-specificity has activity on oligosaccharides and glycoproteins (Katayama et al. 2004). Another membrane-associated fucosidase, AfcB, has α-1,3/4 specificity and releases α-1,3- and α-1,4-fucosyl residues from various substrates such as 3-FL, LNFP II, and LNFP III (Ashida et al. 2009). A membrane-associated exo-α-sialidase, SiaBb2, can free sialic acid from sialylated HMOs, glycoproteins, and gangliosides (Kiyohara et al. 2011). An extracellular lacto-N-biosidase has a substrate preference for unmodified β-linked LNB I (Wada et al. 2008). This latter pathway of type I HMO degradation contrasts with mechanisms employed by B. infantis.
Previous genome analysis of B. bifidum has described strategies for host glycan utilization, including methods of hydrolysis of fucosylated and sialylated moieties (Turroni et al. 2010). Transcriptomics of B. bifidum during growth on fucose revealed upregulation of genes associated with host glycan degradation, including genes encoding fucosidases, sialidases, and N-acetylhexosaminidases (Duranti et al. 2015). Garrido et al. (2015) showed that genes associated with HMO utilization are upregulated in both B. bifidum and B. infantis when these species are grown on HMOs. In this study, transcriptional analysis found that in the case of B. bifidum, the transcriptional response during growth on the individual HMO sugars 2′-FL, 3-FL, and 6′-SL was similar to that of lactose and was unlike the transcriptional response of B. infantis during growth on these substrates (Garrido et al. 2015). Interestingly, when B. infantis was grown on 2′-FL as the sole carbon source, the expression of the HMO cluster was at basal expression levels only, whereas other gene clusters associated with FL import, catabolism, and fucose metabolism appeared to be induced on these substrates (Garrido et al. 2015).
Newer studies examined the ability of select Bifidobacterium breve strains to utilize HMOs (Ruiz-Moyano et al. 2013, James et al. 2016). Comparative genomics of B. breve shows that this species contains clusters related to HMO utilization, including those related to sialic acid, fucose, and LNB (Bottacini et al. 2014). B. breve exhibits strain-specific growth on HMOs; however, most strains grow well on LNT (Ruiz-Moyano et al. 2013). James et al. (2016) used transcriptome analysis and functional studies to examine pathways involved in LNT and LNnT utilization by B. breve UCC 2003. In this work, galactosidases and solute-binding proteins from B. breve UCC 2003 associated with type I and II HMO utilization were characterized, and this strain used separate pathways that have overlapping properties to metabolize LNT and LNnT (James et al. 2016). Growth on 2′-FL is strain dependent in B. breve (Ruiz-Moyano et al. 2013). Matsuki et al. (2016) examined bifidobacterial genomes and identified a putative fucosyllactose utilization pathway, which was associated with elevated acetate in the gut and altered microbiota composition. This work identified an ABC transporter as a component for fucosyllactose consumption in bifidobacteria (Matsuki et al. 2016). The fucosyllactose solute-binding protein gene associated with this ABC transporter was knocked out in a B. breve strain, and the knockout strain showed limited growth on HMO (Matsuki et al. 2016).
Other studies have examined B. longum mechanisms of HMO utilization. Unlike B. breve, most B. longum strains can grow moderately well on HMOs and well on LNT as a sole carbon source (Garrido et al. 2016). In B. longum SC596, the LNB/GNB cluster was shown to be induced on the HMOs LNT and LNnT, which suggests the involvement of the LNB/GNB cluster in HMO utilization in this strain (Garrido et al. 2016). From this strain, two galactosidases were found to be associated with HMO utilization. One of the galactosidases exhibited type I specificity, and the other had type II specificity (Garrido et al. 2016). Furthermore, B. longum SC596 family 1 solute-binding proteins were induced on HMO and possessed binding affinity to various HMOs, including LNT, type I–like HMOs, and 2′-FL (Garrido et al. 2016). Growth on 2′-FL and 3-FL as the sole carbon sources is strain dependent in B. longum (Garrido et al. 2016). B. longum SC596 grew vigorously on 2′-FL and 3-FL as a sole carbon source (Garrido et al. 2016). Glycoprofiling analysis revealed that this strain utilized fucosylated HMOs at higher levels compared with other B. longum strains examined in the study (Garrido et al. 2016). A gene cluster similar to that identified by Matsuki et al. (2016) was identified from the strain B. longum SC596 that allowed for the consumption of fucosylated HMOs (Garrido et al. 2016). This cluster, which was highly induced during growth on 2′-FL, contained a family 1 solute-binding protein with specificity for binding 2′-FL and two adjacent fucosidases with complementary α-1-2- and α-1-3/4-linkage specificities (Garrido et al. 2016). A comparison of modes of glycan utilization by B. infantis, B. breve, B. longum, and B. bifidum is presented in Figure 2.
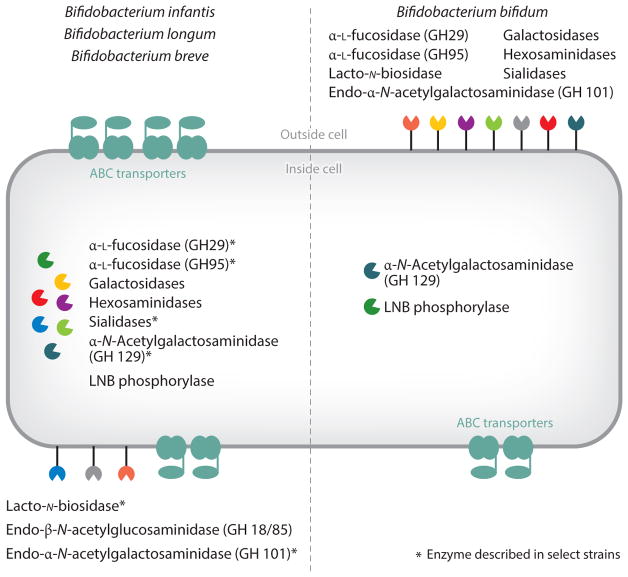
Figure 2.
Possible strategies of glycan utilization by Bifidobacterium infantis, Bifidobacterium breve, Bifidobacterium longum, and Bifidobacterium bifidum. (Left) B. infantis: galactosidases, hexosaminidases, α-L-fucosidases (GH29), α-L-fucosidase (GH95), sialidases, and lacto-N-biose (LNB) phosphorylase. Described in select strains of B. infantis: α-N-acetylgalactosaminidase (GH 129) and endo-β-N-acetylglucosaminidase (GH 18/85). B. breve: galactosidases, hexosaminidase, α-L-fucosidase (GH95), sialidase, and LNB phosphorylases. Described in select strains of B. breve: α-L-fucosidase (GH29), α-N-Acetylgalactosaminidase (GH 129), endo-β-N-acetylglucosaminidase (GH 18/85), and endo-α-N-acetylgalactosaminidase (GH101). B. longum: galactosidases, hexosaminidase, and LNB phosphorylases. Described in select strains of B. longum: α-L-fucosidase (GH95), α-L-fucosidase (GH29), lacto-N-biosidase, α-N-acetylgalactosaminidase (GH 129), endo-β-N-acetylglucosaminidase (GH 18/85), and endo-α-N-acetylgalactosaminidase (GH101). (Right) B. bifidum: galactosidases, hexosaminidases, α-L-fucosidase (GH29), α-L-fucosidase (GH95), sialidases, LNB phosphorylases, and lacto-N-biosidase. Described in select strains of B. bifidum: α-N-acetylgalactosaminidase (GH 129) and endo-α-N-acetylgalactosaminidase (GH101). Asterisks indicate that the enzyme has been described in select strains.
MECHANISMS OF UTILIZATION OF MILK GLYCOCONJUGATES BY BIFIDOBACTERIA
Mechanisms of deglycosylation and degradation of glycoproteins have previously been described in various bacteria such as Bacteroides, Streptococcus oralis, Streptococcus pneumoniae, E. faecalis, and Bifidobacterium (Byers et al. 1999, Collin & Fischetti 2004, Karav et al. 2016, Martens et al. 2009, Muramatsu et al. 2001, Renzi et al. 2011). Bacteroides uses a strategy involving membrane-bound extracellular glycosyl hydrolases to degrade mucin (Martens et al. 2009).
Molecular mechanisms of degradation and utilization of glycoconjugates by bifidobacteria have been investigated, and some bifidobacteria have the ability to degrade mucin (Crociani et al. 1994, Kiyohara et al. 2012, Ruas-Madiedo et al. 2008). MUC2 mucin can be hydrolyzed into Tn antigen by bifidobacterial glycosyl hydrolases (Kiyohara et al. 2012). Kiyohara et al. (2012) identified an α-N-acetylgalactosaminidase from B. bifidum that has activity on the Tn antigen. Garrido et al. (2012b) examined isolates of bifidobacteria for the ability to degrade glycoproteins. In that study, an endo-β-N-acetylglucosaminidase, EndoBI-1, from B. infantis was identified and shown to have activity on N-linked glycans (Garrido et al. 2012b). This enzyme has the ability to cleave N-linked glycans from human lactoferrin, IgA, and IgG, and, additionally, the activity of this enzyme is not affected by core fucosylation or sialylation of substrates (Garrido et al. 2012b).
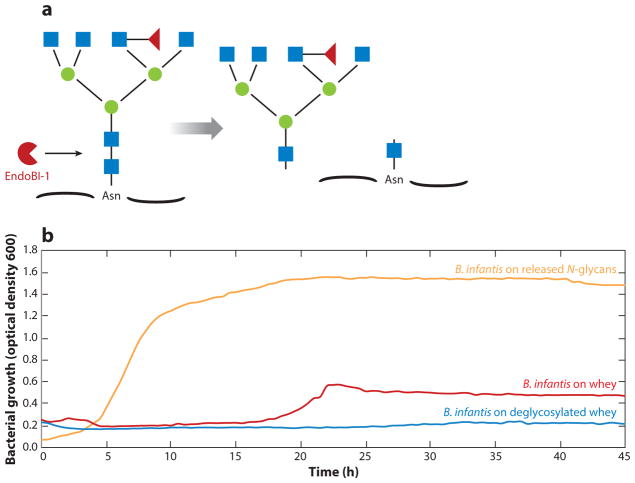
Bifidobacteria are able to use glycoproteins as a growth substrate (Garrido et al. 2012b, Kim et al. 2004). Bovine lactoferrin has been reported to stimulate growth of strains of B. infantis, B. breve, and B. bifidum (Kim et al. 2004). In another study, Garrido et al. (2012b) reported growth of bifidobacteria on yeast mannoproteins. Until recently, the degree of the glycan versus protein component contribution to milk glycoproteins’ prebiotic effect on bifidobacterial enrichment has been poorly understood (Karav et al. 2016, Oda et al. 2013). Karav et al. (2016) used EndoBI-1 to cleave conjugated N-glycans from glycoproteins and showed that B. infantis grew vigorously on these released glycans as a sole carbon source, as presented in Figure 3. Conversely, Bifidobacterium animalis subsp. lactis (B. lactis), a species that does not grow well on HMOs, did not grow on the released N-glycans (Karav et al. 2016). Mass spectrometry confirmed these phenotypes; B. infantis consumed 73% of neutral and 92% of sialylated N-glycans, and B. lactis consumed only 11% of neutral and less than 1% of the sialylated N-glycans. Lastly, the deglycosylated milk-protein fraction, in the absence of released glycans, did not support the growth of B. infantis (Karav et al. 2016). This suggests that released N-glycans from glycoproteins can serve as selective oligosaccharides that enrich only certain bifidobacterial species such as B. infantis (Karav et al. 2016) and that the role of the protein component in the gut remains to be elucidated.
Figure 3.
(a) Endo-β-N-acetylglucosaminidase (EndoBI-1) cleavage of a representative conjugated N-glycan structure. (b) Bifidobacterium infantis growth on whey, released N-glycans from whey, or deglycosylated whey as the sole carbon source. Released N-glycans did not support growth of Bifidobacterium animalis subsp. lactis (data not shown). Adapted from Karav et al. (2016).
Bifidobacteria also degrade milk gangliosides. Lee et al. (2014) reported that B. infantis and B. bifidum catabolized the two milk gangliosides GM3 and GD3, whereas select B. longum and B. lactis isolates poorly degraded these glycoconjugates (Lee et al. 2014). Additional work on gangliosides by Kiyohara et al. (2011) demonstrated that a specific exosialidase, SaBb2, from B. bifidum readily cleaved GD1a.
CROSS-FEEDING OF MILK GLYCANS AND GLYCOCONJUGATES
Various models for the degradation of complex polysaccharides by gut commensals reveal a constellation of primary and secondary consumers with the latter clade feeding on the previously unavailable degradation products of polysaccharides (White et al. 2014). Given the complexity of milk glycans and glycoconjugates, it is expected that a similar cross-feeding of glycan components occurs in situ (Sela & Mills 2010). Indeed, cross-feeding of milk glycans between Bacteroides (primary degrader) and Enterobacteriaceae (secondary consumer) has been demonstrated in mice fed sialyllactose (Huang et al. 2015) and gnotobiotic mice and piglets containing transplanted human microbiota fed BMOs (Charbonneau et al. 2016b); cross-feeding activity was predicted from metagenomic analysis of nursing piglets (Frese et al. 2015).
Recent work has also demonstrated cross-feeding of various glycans among individual bifidobacteria in vitro (Milani et al. 2015a, Turroni et al. 2015). For example, spent medium from B. bifidum grown on mucin (Egan et al. 2014a) or sialyllactose (Egan et al. 2014b) cross-fed B. breve. In a different study, B. bifidum grown on resistant starch and/or xylan released sugars that enabled growth of B. breve, B. adolescentis, and Bifidobacterium thermophilum (Turroni et al. 2015). Such cross-feeding might explain why consortia of bifidobacterial species are found in some infants. Although this cross-feeding among bifidobacteria has been described as “mutualistic” behavior, it remains to be determined whether the primary consumer, in this case B. bifidum, is benefiting from this interaction. More alarmingly, several groups have clearly demonstrated that such glycan-driven cross-feeding can just as readily enrich enteric pathogens (Huang et al. 2015, Ng et al. 2013). Overall, the context by which the primary degrader consumes the milk glycans—i.e., complete internalization of the glycans versus extracellular degradation prior to internalization—may be a critical competitive strategy that shapes the trajectory and composition of the infant-gut microbiome (Charbonneau et al. 2016a, Sela & Mills 2010).
DIETS SUPPLEMENTED WITH EXOGENOUS MILK OLIGOSACCHARIDES
Infant formula manufacturers aim to better mimic human milk complexity by increasing the number of components that are normally found only in human breast milk (Hernell 2011). Infant formula has in the past contained mostly bovine milk components. However, critical bioactive components found in human breast milk may be lacking in infant formula, and to address this, the composition of infant formula has changed over time (Lonnerdal 2014). Present-day infant formula is now supplemented with components such as oils, starches, and vitamins to better align with human milk (Ackerman et al. 2017). However, human milk contains an array of diverse bioactive structures, and the production of structurally and compositionally similar molecules remains a challenging task.
Because HMOs are not readily available as an ingredient, infant formulas are now commonly supplemented with inulin and galactooligosaccharides (GOSs) or fructooligosaccharides (FOSs) (Ackerman et al. 2017), and these commercial prebiotics have been shown to increase bifidobacterial abundance in vivo (Haarman & Knol 2005, Knol et al. 2005, Ramirez-Farias et al. 2009). Although these carbohydrates are prebiotic, emerging research is showing that they do not possess other important bioactivities of milk oligosaccharides. Recent studies have examined the effects of infant formula supplemented with a specific HMO species (Donovan & Comstock 2016). Goehring et al. (2016) reported that infants fed a formula containing 2′-FL had lower levels of inflammatory cytokines compared to those fed, which is similar to infants that are breastfed. Steenhout et al. (2016) demonstrated that infants fed formula supplemented with 2′-FL and LNnT possessed a stool microbiota that more closely resembled that of breastfed infants.
As mentioned earlier, bovine dairy streams are another promising source for larger-scale isolation of milk oligosaccharides (Barile et al. 2009, Cohen et al. 2017). A recent clinical study showed that consumption of BMOs by adult volunteers was remarkably well-tolerated, even at dosages of up to 35% of daily fiber intake (Smilowitz et al. 2017). Additional studies have provided some mechanistic insight into possible benefits of BMO. Hamilton et al. (2017) showed that provision of BMOs attenuated weight gain, decreased adiposity, and decreased caloric intake within a mouse model for high-fat-diet-induced obesity. In this study, BMOs also completely abolished the high-fat-diet-induced increase in paracellular and transcellular permeability in both the small and large intestine, coinciding with increases in bifidobacteria and lactobacilli in the ileum. Boudry et al. (2017) witnessed a similar BMO-driven reduction in intestinal permeability in diet-induced obese mice. Charbonneau et al. (2016b) reported that provision of sialylated BMOs dramatically repaired growth phenotypes within gnotobiotic mice and pigs transplanted with microbiota from malnourished Malawian children. Amazingly, this study showed dramatically improved lean body mass, altered bone morphology, and modified liver, muscle, and brain metabolism, suggesting a greater ability to garner nutrients for anabolism linked to the interaction between the supplemented BMOs and the Malawian infant-gut microbiota. Because these improvements were not observed with inulin supplementation, the specific chemical nature of the milk oligosaccharides provides a key metabolic factor endowing this benefit, albeit the mechanism for this remains to be determined.
Although opportunities to supplement formula with milk oligosaccharides represent a significant advance to match the glycan content of breast milk, much study is still needed to understand their impact on the developing infant-gut microbiota. Given that cross-feeding of milk glycans might include enrichment of problematic clades such as Enterobacteriaceae, caution is warranted (Charbonneau et al. 2016a, Shin et al. 2015). In particular, studies using formula containing milk oligosaccharides need to present individual neonate microbiota changes rather than solely showing summarized changes among all study subjects. Given the individualized nature of the gut microbiota, it is critical to follow each test subject’s response to the milk oligosaccharide interventions to assess whether any subpopulations of infants exhibit signs of cross-feeding and to probe associations with health outcomes.
CONCLUSIONS AND FUTURE STUDIES
In addition to providing nutrition for the infant, human breast milk provides an array of intricate bioactive components that modulate the developing infant-gut microbiota. The relationship between the formation of the intestinal environment and milk glycans is of significant interest. Glycans in milk are associated with various functional roles, including pathogen deflection and modulation of immunity. Human milk glycans also shape the enrichment of a protective intestinal microbiota. Recent work has investigated molecular mechanisms involved in milk oligosaccharides and milk glycoconjugate utilization by infant-associated bifidobacteria. The identification of milk glycans coupled with research investigating these mechanisms of bifidobacterial glycan utilization has provided insight into the interactions between bifidobacteria and milk glycans. However, the vastly complex relationship between glycans and the human microbiota remains poorly described. Future research on these glycans will aid our understanding of infant development, the design of synbiotic partners, modulation of the intestinal microbiota, and treatment of intestinal dysbiosis.
Acknowledgments
We acknowledge all the researchers in the UC Davis Foods for Health Institute and the Milk Bioactives Program for their enthusiasm, imagination, and collective contribution to this subject matter. Work by the Milk Bioactives Program has been supported by the UC Davis Research Investments in the Sciences and Engineering Program, the Bill & Melinda Gates Foundation, China Mengiu Dairy Company Limited, and National Institutes of Health awards HD059127, HD065122, HD061923, AT006180, AT007079, and AT008759. D.A.M. acknowledges support as the Peter J. Shields Endowed Chair in Dairy Food Science.
Footnotes
DISCLOSURE STATEMENT
D.A.M. and D.B. are cofounders of Evolve Biosystems, a company focused on diet-based manipulation of the gut microbiota. Evolve Biosystems played no role in the design, execution, interpretation, or publication of this work.
LITERATURE CITED
- Ackerman DL, Craft KM, Townsend SD. Infant food applications of complex carbohydrates: structure, synthesis, and function. Carbohydr Res. 2017;437:16–27. doi: 10.1016/j.carres.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98:229–38. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- Albrecht S, Lane JA, Marino K, Al Busadah KA, Carrington SD, et al. A comparative study of free oligosaccharides in the milk of domestic animals. Br J Nutr. 2014;111:1313–28. doi: 10.1017/S0007114513003772. [DOI] [PubMed] [Google Scholar]
- Aldredge DL, Geronimo MR, Hua S, Nwosu CC, Lebrilla CB, Barile D. Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures. Glycobiology. 2013;23:664–76. doi: 10.1093/glycob/cwt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhaj OA, Taufik E, Handa Y, Fukuda K, Saito T, Urashima T. Chemical characterisation of oligosaccharides in commercially pasteurised dromedary camel (Camelus dromedarius) milk. Int Dairy J. 2013;28:70–75. [Google Scholar]
- Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, et al. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. 2011;286:34583–92. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, et al. Two distinct α-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19:1010–17. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- Atochina O, Daly-Engel T, Piskorska D, McGuire E, Harn DA. A schistosome-expressed immunomodulatory glycoconjugate expands peritoneal Gr1+ macrophages that suppress naive CD4+ T cell proliferation via an IFN-γ and nitric oxide-dependent mechanism. J Immunol. 2001;167:4293–302. doi: 10.4049/jimmunol.167.8.4293. [DOI] [PubMed] [Google Scholar]
- Azuma N, Yamauchi K, Mitsuoka T. Bifidus growth-promoting activity of a glycomacropeptide derived from human K-casein. Agric Biol Chem. 1984;48:2159–62. [Google Scholar]
- Backhed F, Roswall J, Peng Y, Feng Q, Jia H, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Baker EN, Baker HM. Molecular structure, binding properties and dynamics of lactoferrin. Cell Mol Life Sci. 2005;62:2531–39. doi: 10.1007/s00018-005-5368-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza M, Pinzon J, Wickramasinghe S, Froehlich JW, Moeller I, et al. Glycosylation of human milk lactoferrin exhibits dynamic changes during early lactation enhancing its role in pathogenic bacteria-host interactions. Mol Cell Proteom. 2012;11:M111015248. doi: 10.1074/mcp.M111.015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile D, Marotta M, Chu C, Mehra R, Grimm R, et al. Neutral and acidic oligosaccharides in Holstein-Friesian colostrum during the first 3 days of lactation measured by high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. J Dairy Sci. 2010;93:3940–99. doi: 10.3168/jds.2010-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile D, Tao N, Lebrilla CB, Coisson JD, Arlorio M, German JB. Permeate from cheese whey ultra-filtration is a source of milk oligosaccharides. Int Dairy J. 2009;19:524–30. doi: 10.1016/j.idairyj.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode L. Recent advances on structure, metabolism, and function of human milk oligosaccharides. J Nutr. 2006;136:2127–30. doi: 10.1093/jn/136.8.2127. [DOI] [PubMed] [Google Scholar]
- Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22:1147–62. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode L, Contractor N, Barile D, Pohl N, Prudden AR, et al. Overcoming the limited availability of human milk oligosaccharides: challenges and opportunities for research and application. Nutr Rev. 2016;74:635–44. doi: 10.1093/nutrit/nuw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode L, Jantscher-Krenn E. Structure-function relationships of human milk oligosaccharides. Adv Nutr. 2012;3:383S–91. doi: 10.3945/an.111.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottacini F, O’Connell Motherway M, Kuczynski J, O’Connell KJ, Serafini F, et al. Comparative genomics of the Bifidobacterium breve taxon. BMC Genom. 2014;15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudry G, Hamilton MK, Chichlowski M, Wickramasinghe S, Barile D, et al. Bovine milk oligosaccharides decrease gut permeability and improve inflammation and microbial dysbiosis in diet-induced obese mice. J Dairy Sci. 2017;100:2471–81. doi: 10.3168/jds.2016-11890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunesova V, Lacroix C, Schwab C. Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol. 2016;16:248. doi: 10.1186/s12866-016-0867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers HL, Tarelli E, Homer KA, Beighton D. Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human α1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology. 1999;9:469–79. doi: 10.1093/glycob/9.5.469. [DOI] [PubMed] [Google Scholar]
- Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96:544–51. doi: 10.3945/ajcn.112.037382. [DOI] [PubMed] [Google Scholar]
- Charbonneau MR, Blanton LV, DiGiulio DB, Relman DA, Lebrilla CB, et al. A microbial perspective of human developmental biology. Nature. 2016a;535:48–55. doi: 10.1038/nature18845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau MR, O’Donnell D, Blanton LV, Totten SM, Davis JC, et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell. 2016b;164:859–71. doi: 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P, Sharma CB. Goat milk oligosaccharides: purification and characterization by HPLC and high-field 1H-NMR spectroscopy. Biochim Biophys Acta. 1988;967:115–21. doi: 10.1016/0304-4165(88)90196-1. [DOI] [PubMed] [Google Scholar]
- Cohen JL, Barile D, Liu Y, de Moura Bell JMLN. Role of pH in the recovery of bovine milk oligosaccharides from colostrum whey permeate by nanofiltration. Int Dairy J. 2017;66:68–75. doi: 10.1016/j.idairyj.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin M, Fischetti VA. A novel secreted endoglycosidase from Enterococcus faecalis with activity on human immunoglobulin G and ribonuclease B. J Biol Chem. 2004;279:22558–70. doi: 10.1074/jbc.M402156200. [DOI] [PubMed] [Google Scholar]
- Crociani F, Alessandrini A, Mucci MM, Biavati B. Degradation of complex carbohydrates by Bifidobacterium spp. Int J Food Microbiol. 1994;24:199–210. doi: 10.1016/0168-1605(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Dallas DC, Weinborn V, de Moura Bell JM, Wang M, Parker EA, et al. Comprehensive peptidomic and glycomic evaluation reveals that sweet whey permeate from colostrum is a source of milk protein–derived peptides and oligosaccharides. Food Res Int. 2014;63:203–9. doi: 10.1016/j.foodres.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JC, Totten SM, Huang JO, Nagshbandi S, Kirmiz N, et al. Identification of oligosaccharides in feces of breast-fed infants and their correlation with the gut microbial community. Mol Cell Proteom. 2016;15(9):2987–3002. doi: 10.1074/mcp.M116.060665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbabi H, Dubarry M, Rautureau M, Tome D. Bovine lactoferrin induces both mucosal and systemic immune response in mice. J Dairy Res. 1998;65:283–93. doi: 10.1017/s0022029997002732. [DOI] [PubMed] [Google Scholar]
- De Leoz ML, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, et al. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J Proteome Res. 2014;14:491–502. doi: 10.1021/pr500759e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moura Bell JM, Aquino LF, Liu Y, Cohen JL, Lee H, et al. Modeling lactose hydrolysis for efficiency and selectivity: toward the preservation of sialyloligosaccharides in bovine colostrum whey permeate. J Dairy Sci. 2016;99:6157–63. doi: 10.3168/jds.2016-11065. [DOI] [PubMed] [Google Scholar]
- Donovan SM, Comstock SS. Human milk oligosaccharides influence neonatal mucosal and systemic immunity. Ann Nutr Metabol. 2016;69(Suppl 2):42–51. doi: 10.1159/000452818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douellou T, Montel MC, Thevenot Sergentet D. Invited review: anti-adhesive properties of bovine oligosaccharides and bovine milk fat globule membrane-associated glycoconjugates against bacterial food enteropathogens. J Dairy Sci. 2017;100:3348–59. doi: 10.3168/jds.2016-11611. [DOI] [PubMed] [Google Scholar]
- Duijts L, Jaddoe VW, Hofman A, Moll HA. Prolonged and exclusive breastfeeding reduces the risk of infectious diseases in infancy. Pediatrics. 2010;126:e18–25. doi: 10.1542/peds.2008-3256. [DOI] [PubMed] [Google Scholar]
- Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, et al. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ Microbiol. 2015;17:2515–31. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- Egan M, Motherway MO, Kilcoyne M, Kane M, Joshi L, et al. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 2014a;14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M, Motherway MO, Ventura M, van Sinderen D. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2014b;80:4414–26. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiwegger T, Stahl B, Schmitt J, Boehm G, Gerstmayr M, et al. Human milk–derived oligosaccharides and plant-derived oligosaccharides stimulate cytokine production of cord blood T-cells in vitro. Pediatr Res. 2004;56:536–40. doi: 10.1203/01.PDR.0000139411.35619.B4. [DOI] [PubMed] [Google Scholar]
- Fong B, Ma K, McJarrow P. Quantification of bovine milk oligosaccharides using liquid chromatography-selected reaction monitoring-mass spectrometry. J Agric Food Chem. 2011;59:9788–95. doi: 10.1021/jf202035m. [DOI] [PubMed] [Google Scholar]
- Frese SA, Parker K, Calvert CC, Mills DA. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome. 2015;3:28. doi: 10.1186/s40168-015-0091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich JW, Dodds ED, Barboza M, McJimpsey EL, Seipert RR, et al. Glycoprotein expression in human milk during lactation. J Agric Food Chem. 2010;58:6440–48. doi: 10.1021/jf100112x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Yamamoto A, Ganzorig K, Khuukhenbaatar J, Senda A, et al. Chemical characterization of the oligosaccharides in Bactrian camel (Camelus bactrianus) milk and colostrum. J Dairy Sci. 2010;93:5572–87. doi: 10.3168/jds.2010-3151. [DOI] [PubMed] [Google Scholar]
- Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv Nutr. 2012a;3:415S–21. doi: 10.3945/an.111.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Dallas DC, Mills DA. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology. 2013;159:649–64. doi: 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Kim JH, German JB, Raybould HE, Mills DA. Oligosaccharide binding proteins from Bifidobacterium longum subsp infantis reveal a preference for host glycans. PLOS ONE. 2011;6:e17315. doi: 10.1371/journal.pone.0017315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Nwosu C, Ruiz-Moyano S, Aldredge D, German JB, et al. Endo-β-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins. Mol Cell Proteom. 2012b;11:775–85. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Ruiz-Moyano S, Kirmiz N, Davis JC, Totten SM, et al. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp longum SC596. Sci Rep. 2016;6:35045. doi: 10.1038/srep35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Ruiz-Moyano S, Lemay DG, Sela DA, German JB, Mills DA. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci Rep. 2015;5:13517. doi: 10.1038/srep13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Ruiz-Moyano S, Mills DA. Release and utilization of N-acetyl-D-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp infantis. Anaerobe. 2012c;18:430–35. doi: 10.1016/j.anaerobe.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring KC, Marriage BJ, Oliver JS, Wilder JA, Barrett EG, Buck RH. Similar to those who are breastfed, infants fed a formula containing 2′-fucosyllactose have lower inflammatory cytokines in a randomized controlled trial. J Nutr. 2016;146:2559–66. doi: 10.3945/jn.116.236919. [DOI] [PubMed] [Google Scholar]
- Good M, Sodhi CP, Yamaguchi Y, Jia H, Lu P, et al. The human milk oligosaccharide 2′-fucosyllactose attenuates the severity of experimental necrotising enterocolitis by enhancing mesenteric perfusion in the neonatal intestine. Br J Nutr. 2016;116:1175–87. doi: 10.1017/S0007114516002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarman M, Knol J. Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl Environ Microbiol. 2005;71:2318–24. doi: 10.1128/AEM.71.5.2318-2324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MK, Ronveaux CC, Rust BM, Newman JW, Hawley M, et al. Prebiotic milk oligosaccharides prevent development of obese phenotype, impairment of gut permeability, and microbial dysbiosis in high fat-fed mice. Am J Physiol Gastrointest Liver Physiol. 2017;312:G474–87. doi: 10.1152/ajpgi.00427.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Liu S, Kling DE, Leone S, Lawlor NT, et al. The human milk oligosaccharide 2′-fucosyllactose modulates CD14 expression in human enterocytes, thereby attenuating LPS-induced inflammation. Gut. 2016;65:33–46. doi: 10.1136/gutjnl-2014-307544. [DOI] [PubMed] [Google Scholar]
- Hernell O. Human milk vs. cow’s milk and the evolution of infant formulas. Nestle Nutr Workshop Ser Pediatr Progr. 2011;67:17–28. doi: 10.1159/000325572. [DOI] [PubMed] [Google Scholar]
- Hong Q, Ruhaak LR, Totten SM, Smilowitz JT, German JB, Lebrilla CB. Label-free absolute quantitation of oligosaccharides using multiple reaction monitoring. Anal Chem. 2014;86:2640–47. doi: 10.1021/ac404006z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YL, Chassard C, Hausmann M, von Itzstein M, Hennet T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat Commun. 2015;6:8141. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt KM, Preuss J, Nissan C, Davlin CA, Williams JE, et al. Human milk oligosaccharides promote the growth of staphylococci. Appl Environ Microbiol. 2012;78:4763–70. doi: 10.1128/AEM.00477-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James K, Motherway MO, Bottacini F, van Sinderen D. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci Rep. 2016;6:38560. doi: 10.1038/srep38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Huang P, Zhong W, Tan M, Farkas T, et al. Human milk contains elements that block binding of noroviruses to human histo-blood group antigens in saliva. J Infect Dis. 2004;190:1850–59. doi: 10.1086/425159. [DOI] [PubMed] [Google Scholar]
- Karav S, Le Parc A, de Moura Bell JMLN, Frese SA, Kirmiz N, et al. Oligosaccharides released from milk glycoproteins are selective growth substrates for infant-associated bifidobacteria. Appl Environ Microbiol. 2016;82:3622–30. doi: 10.1128/AEM.00547-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, et al. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-β-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95) J Bacteriol. 2004;186:4885–93. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WS, Ohashi M, Tanaka T, Kumura H, Kim GY, et al. Growth-promoting effects of lactoferrin on L. acidophilus and Bifidobacterium spp. BioMetals. 2004;17:279–83. doi: 10.1023/b:biom.0000027705.57430.f1. [DOI] [PubMed] [Google Scholar]
- Kitaoka M. Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Adv Nutr. 2012;3:422S–29. doi: 10.3945/an.111.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara M, Nakatomi T, Kurihara S, Fushinobu S, Suzuki H, et al. α-N-Acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J Biol Chem. 2012;287:693–700. doi: 10.1074/jbc.M111.277384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara M, Tanigawa K, Chaiwangsri T, Katayama T, Ashida H, Yamamoto K. An exo-α-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology. 2011;21:437–47. doi: 10.1093/glycob/cwq175. [DOI] [PubMed] [Google Scholar]
- Knol J, Scholtens P, Kafka C, Steenbakkers J, Gro S, et al. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J Pediatr Gastroenterol Nutr. 2005;40:36–42. doi: 10.1097/00005176-200501000-00007. [DOI] [PubMed] [Google Scholar]
- Kobata A, Ginsburg V. Oligosaccharides of human milk. IV Isolation and characterization of a new hexasaccharide, lacto-N-neohexaose. Arch Biochem Biophys. 1972;150:273–81. doi: 10.1016/0003-9861(72)90036-7. [DOI] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, et al. Succession of microbial consortia in the developing infant gut microbiome. PNAS. 2011;108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz S, Kunz C, Rudloff S. Oligosaccharides from human milk induce growth arrest via G2/M by influencing growth-related cell cycle genes in intestinal epithelial cells. Br J Nutr. 2009;101:1306–15. doi: 10.1017/S0007114508079622. [DOI] [PubMed] [Google Scholar]
- Kuntz S, Rudloff S, Kunz C. Oligosaccharides from human milk influence growth-related characteristics of intestinally transformed and non-transformed intestinal cells. Br J Nutr. 2008;99:462–71. doi: 10.1017/S0007114507824068. [DOI] [PubMed] [Google Scholar]
- Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- Kwak MJ, Kwon SK, Yoon JK, Song JY, Seo JG, et al. Evolutionary architecture of the infant-adapted group of Bifidobacterium species associated with the probiotic function. Syst Appl Microbiol. 2016;39:429–39. doi: 10.1016/j.syapm.2016.07.004. [DOI] [PubMed] [Google Scholar]
- Lee H, An HJ, Lerno LA, Jr, German JB, Lebrilla CB. Rapid profiling of bovine and human milk gangliosides by matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry. Int J Mass Spectrom. 2011;305:138–50. doi: 10.1016/j.ijms.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Garrido D, Mills DA, Barile D. Hydrolysis of milk gangliosides by infant-gut associated bifidobacteria determined by microfluidic chips and high-resolution mass spectrometry. Electrophoresis. 2014;35:1742–50. doi: 10.1002/elps.201300653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZT, Mills DA. Differential establishment of bifidobacteria in the breastfed infant gut. Nestle Nutr Inst Workshop Ser. 2017;88:149–59. doi: 10.1159/000455399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:13. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lindquist S, Lowe M, Noppa L, Hernell O. Bile salt–stimulated lipase and pancreatic lipase–related protein 2 are the dominating lipases in neonatal fat digestion in mice and rats. Pediatr Res. 2007;62:537–41. doi: 10.1203/PDR.0b013e3181559e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AE, Autran CA, Espanola SD, Bode L, Nizet V. Human milk oligosaccharides protect bladder epithelial cells against uropathogenic Escherichia coli invasion and cytotoxicity. J Infect Dis. 2014;209:389–98. doi: 10.1093/infdis/jit464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Newburg DS. Human milk glycoproteins protect infants against human pathogens. Breastfeed Med. 2013;8:354–62. doi: 10.1089/bfm.2013.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio R, Ninonuevo M, Kronewitter S, Freeman S, German J, et al. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb Biotechnol. 2009;2:333–42. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonnerdal B. Bioactive proteins in breast milk. J Paediatr Child Health. 2013;49(Suppl 1):1–7. doi: 10.1111/jpc.12104. [DOI] [PubMed] [Google Scholar]
- Lonnerdal B. Infant formula and infant nutrition: bioactive proteins of human milk and implications for composition of infant formulas. Am J Clin Nutr. 2014;99:712S–17. doi: 10.3945/ajcn.113.071993. [DOI] [PubMed] [Google Scholar]
- Ma L, Liu X, MacGibbon AK, Rowan A, McJarrow P, Fong BY. Lactational changes in concentration and distribution of ganglioside molecular species in human breast milk from Chinese mothers. Lipids. 2015;50:1145–54. doi: 10.1007/s11745-015-4073-1. [DOI] [PubMed] [Google Scholar]
- Maldonado-Gomez MX, Lee H, Barile D, Lu M, Hutkins RW. Adherence inhibition of enteric pathogens to epithelial cells by bovine colostrum fractions. Int Dairy J. 2015;40:24–32. [Google Scholar]
- Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–40. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–14. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem. 2009;284:18445–57. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Ortiz A, Salcedo J, Barile D, Bunyatratchata A, Moreno FJ, et al. Characterization of goat colostrum oligosaccharides by nano-liquid chromatography on chip quadrupole time-of-flight mass spectrometry and hydrophilic interaction liquid chromatography-quadrupole mass spectrometry. J Chromatogr A. 2016;1428:143–53. doi: 10.1016/j.chroma.2015.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Sosa S, Martin MJ, Garcia-Pardo LA, Hueso P. Sialyloligosaccharides in human and bovine milk and in infant formulas: variations with the progression of lactation. J Dairy Sci. 2003;86:52–59. doi: 10.3168/jds.S0022-0302(03)73583-8. [DOI] [PubMed] [Google Scholar]
- Martinez-Ferez A, Rudloff S, Guadix A, Henkel CA, Pohlentz G, et al. Goats’ milk as a natural source of lactose-derived oligosaccharides: isolation by membrane technology. Int Dairy J. 2006;16:173–81. [Google Scholar]
- Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun. 2016;7:11939. doi: 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire MK, Meehan CL, McGuire MA, Williams JE. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr. 2017;105:1086–100. doi: 10.3945/ajcn.116.139980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJarrow P, van Amelsfort-Schoonbeek J. Bovine sialyl oligosaccharides: seasonal variations in their concentrations in milk, and a comparison of the colostrums of Jersey and Friesian cows. Int Dairy J. 2004;14:571–79. [Google Scholar]
- Meli F, Puccio G, Cajozzo C, Ricottone GL, Pecquet S, et al. Growth and safety evaluation of infant formulae containing oligosaccharides derived from bovine milk: a randomized, double-blind, noninferiority trial. BMC Pediatr. 2014;14:306. doi: 10.1186/s12887-014-0306-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, et al. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol. 2014;80:6290–302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. 2015a;5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, et al. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol. 2015b;82:980–91. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–62. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, et al. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J Pediatr. 2004;145:297–303. doi: 10.1016/j.jpeds.2004.04.054. [DOI] [PubMed] [Google Scholar]
- Morrow AL, Ruiz-Palacios GM, Jiang X, Newburg DS. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J Nutr. 2005;135:1304–7. doi: 10.1093/jn/135.5.1304. [DOI] [PubMed] [Google Scholar]
- Mudd AT, Salcedo J, Alexander LS, Johnson SK, Getty CM, et al. Porcine milk oligosaccharides and sialic acid concentrations vary throughout lactation. Front Nutr. 2016;3:39. doi: 10.3389/fnut.2016.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu H, Tachikui H, Ushida H, Song X, Qiu Y, et al. Molecular cloning and expression of endo-β-N-acetylglucosaminidase D, which acts on the core structure of complex type asparagine-linked oligosaccharides. J Biochem. 2001;129:923–28. doi: 10.1093/oxfordjournals.jbchem.a002938. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Amikawa S, Harada T, Saito T, Arai I, Urashima T. Occurrence of an unusual phospho-rylated N-acetyllactosamine in horse colostrum. Biochim Biophys Acta. 2001;1525:13–18. doi: 10.1016/s0304-4165(00)00148-3. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Kawase H, Kimura K, Watanabe Y, Ohtani M, et al. Concentrations of sialyloligosaccharides in bovine colostrum and milk during the prepartum and early lactation. J Dairy Sci. 2003;86:1315–20. doi: 10.3168/jds.S0022-0302(03)73715-1. [DOI] [PubMed] [Google Scholar]
- Neelima, Sharma R, Rajput YS, Mann B. Chemical and functional properties of glycomacropeptide (GMP) and its role in the detection of cheese whey adulteration in milk: a review. Dairy Sci Technol. 2013;93:21–43. doi: 10.1007/s13594-012-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburg DS. Neonatal protection by an innate immune system of human milk consisting of oligosaccharides and glycans. J Anim Sci. 2009;87:26–34. doi: 10.2527/jas.2008-1347. [DOI] [PubMed] [Google Scholar]
- Newburg DS. Glycobiology of human milk. Biochemistry. 2013;78:771–85. doi: 10.1134/S0006297913070092. [DOI] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninonuevo MR, Park Y, Yin H, Zhang J, Ward RE, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–80. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- Nwosu CC, Aldredge DL, Lee H, Lerno LA, Zivkovic AM, et al. Comparison of the human and bovine milk N-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J Proteome Res. 2012;11:2912–24. doi: 10.1021/pr300008u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan A, Bottacini F, Motherway MO, van Sinderen D. Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genom. 2015;16:832. doi: 10.1186/s12864-015-1968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Wakabayashi H, Yamauchi K, Sato T, Xiao JZ, et al. Isolation of a bifidogenic peptide from the pepsin hydrolysate of bovine lactoferrin. Appl Environ Microbiol. 2013;79:1843–49. doi: 10.1128/AEM.03343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan N, Kane M, Joshi L, Hickey RM. Structural and functional characteristics of bovine milk protein glycosylation. Glycobiology. 2014;24:220–36. doi: 10.1093/glycob/cwt162. [DOI] [PubMed] [Google Scholar]
- Pacheco AR, Barile D, Underwood MA, Mills DA. The impact of the milk glycobiome on the neonate gut microbiota. Annu Rev Anim Biosci. 2015;3:419–45. doi: 10.1146/annurev-animal-022114-111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan XL, Izumi T. Variation of the ganglioside compositions of human milk, cow’s milk and infant formulas. Early Hum Dev. 2000;57:25–31. doi: 10.1016/s0378-3782(99)00051-1. [DOI] [PubMed] [Google Scholar]
- Petherick A. Development: mother’s milk: a rich opportunity. Nature. 2010;468:S5–7. doi: 10.1038/468S5a. [DOI] [PubMed] [Google Scholar]
- Rahman MM, Kim WS, Ito T, Kumura H, Shimazaki K. Growth promotion and cell binding ability of bovine lactoferrin to Bifidobacterium longum. Anaerobe. 2009;15:133–37. doi: 10.1016/j.anaerobe.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. 2009;101:541–50. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
- Ray K. Gut microbiota: first steps in the infant gut microbiota. Nat Rev Gastroenterol Hepatol. 2016;13:437. doi: 10.1038/nrgastro.2016.108. [DOI] [PubMed] [Google Scholar]
- Renzi F, Manfredi P, Mally M, Moes S, Jeno P, Cornelis GR. The N-glycan glycoprotein deglycosylation complex (Gpd) from Capnocytophaga canimorsus deglycosylates human IgG. PLOS Pathog. 2011;7:e1002118. doi: 10.1371/journal.ppat.1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas-Madiedo P, Gueimonde M, Fernandez-Garcia M, de los Reyes-Gavilan CG, Margolles A. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl Environ Microbiol. 2008;74:1936–40. doi: 10.1128/AEM.02509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak LR, Lebrilla CB. Analysis and role of oligosaccharides in milk. BMB Rep. 2012;45:442–51. doi: 10.5483/BMBRep.2012.45.8.161. [DOI] [PubMed] [Google Scholar]
- Ruiz-Moyano S, Totten SM, Garrido DA, Smilowitz JT, German JB, et al. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl Environ Microbiol. 2013;79:6040–49. doi: 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Palacios GM, Calva JJ, Pickering LK, Lopez-Vidal Y, Volkow P, et al. Protection of breast-fed infants against Campylobacter diarrhea by antibodies in human milk. J Pediatr. 1990;116:707–13. doi: 10.1016/s0022-3476(05)82652-6. [DOI] [PubMed] [Google Scholar]
- Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fucα1, 2Galβ1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278:14112–20. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- Ruvoen-Clouet N, Mas E, Marionneau S, Guillon P, Lombardo D, Le Pendu J. Bile-salt-stimulated lipase and mucins from milk of “secretor” mothers inhibit the binding of Norwalk virus capsids to their carbohydrate ligands. Biochem J. 2006;393:627–34. doi: 10.1042/BJ20050898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo J, Frese SA, Mills DA, Barile D. Characterization of porcine milk oligosaccharides during early lactation and their relation to the fecal microbiome. J Dairy Sci. 2016;99:7733–43. doi: 10.3168/jds.2016-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtens PA, Oozeer R, Martin R, Amor KB, Knol J. The early settlers: intestinal microbiology in early life. Annu Rev Food Sci Technol. 2012;3:425–47. doi: 10.1146/annurev-food-022811-101120. [DOI] [PubMed] [Google Scholar]
- Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, et al. The genome sequence of Bifidobacterium longum subsp infantis reveals adaptations for milk utilization within the infant microbiome. PNAS. 2008;105:18964–69. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Garrido D, Lerno L, Wu S, Tan K, et al. Bifidobacterium longum subsp infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol. 2012;78:795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, et al. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem. 2011;286:11909–18. doi: 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Simeoni U, Berger B, Junick J, Blaut M, Pecquet S, et al. Gut microbiota analysis reveals a marked shift to bifidobacteria by a starter infant formula containing a synbiotic of bovine milk-derived oligosaccharides and Bifidobacterium animalis subsp lactis CNCM I-3446. Environ Microbiol. 2016;18:2185–95. doi: 10.1111/1462-2920.13144. [DOI] [PubMed] [Google Scholar]
- Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. Breast milk oligosaccharides: structure-function relationships in the neonate. Annu Rev Nutr. 2014;34:143–69. doi: 10.1146/annurev-nutr-071813-105721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilowitz JT, Lemay DG, Kalanetra KM, Chin EL, Zivkovic AM, et al. Tolerability and safety of the intake of bovine milk oligosaccharides extracted from cheese whey in healthy human adults. J Nutr Sci. 2017;6:e6. doi: 10.1017/jns.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenhout P, Sperisen P, Martin F-P, Sprenger N, Wernimont S, et al. Term infant formula supplemented with human milk oligosaccharides (2′fucosyllactose and lacto-N-neotetraose) shifts stool microbiota and metabolic signatures closer to that of breastfed infants. FASEB J. 2016;30:275–7. [Google Scholar]
- Stromqvist M, Falk P, Bergstrom S, Hansson L, Lonnerdal B, et al. Human milk kappa-casein and inhibition of Helicobacter pylori adhesion to human gastric mucosa. J Pediatr Gastroenterol Nutr. 1995;21:288–96. doi: 10.1097/00005176-199510000-00006. [DOI] [PubMed] [Google Scholar]
- Tao N, DePeters EJ, Freeman S, German JB, Grimm R, Lebrilla CB. Bovine milk glycome. J Dairy Sci. 2008;91:3768–78. doi: 10.3168/jds.2008-1305. [DOI] [PubMed] [Google Scholar]
- Tao N, DePeters EJ, German JB, Grimm R, Lebrilla CB. Variations in bovine milk oligosaccharides during early and middle lactation stages analyzed by high-performance liquid chromatography-chip/mass spectrometry. J Dairy Sci. 2009;92:2991–3001. doi: 10.3168/jds.2008-1642. [DOI] [PubMed] [Google Scholar]
- Tao N, Ochonicky KL, German JB, Donovan SM, Lebrilla CB. Structural determination and daily variations of porcine milk oligosaccharides. J Agric Food Chem. 2010;58:4653–59. doi: 10.1021/jf100398u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrazas LI, Walsh KL, Piskorska D, McGuire E, Harn DA., Jr The schistosome oligosaccharide lacto-N-neotetraose expands Gr1+ cells that secrete anti-inflammatory cytokines and inhibit proliferation of naive CD4+ cells: a potential mechanism for immune polarization in helminth infections. J Immunol. 2001;167:5294–303. doi: 10.4049/jimmunol.167.9.5294. [DOI] [PubMed] [Google Scholar]
- Thum C, Cookson A, McNabb WC, Roy NC, Otter D. Composition and enrichment of caprine milk oligosaccharides from New Zealand Saanen goat cheese whey. J Food Compos Anal. 2015;42:30–37. [Google Scholar]
- Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. PNAS. 2010;107:19514–19. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Ozcan E, Milani C, Mancabelli L, Viappiani A, et al. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front Microbiol. 2015;6:1030. doi: 10.3389/fmicb.2015.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood MA, Gaerlan S, De Leoz ML, Dimapasoc L, Kalanetra KM, et al. Human milk oligosaccharides in premature infants: absorption, excretion, and influence on the intestinal microbiota. Pediatr Res. 2015;78:670–77. doi: 10.1038/pr.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urashima T, Saito T, Kimura T. Chemical structures of three neutral oligosaccharides obtained from horse (thoroughbred) colostrum. Comp Biochem Physiol B. 1991;100:177–83. doi: 10.1016/0305-0491(91)90103-k. [DOI] [PubMed] [Google Scholar]
- van Berkel PH, Geerts ME, van Veen HA, Kooiman PM, Pieper FR, et al. Glycosylated and ungly-cosylated human lactoferrins both bind iron and show identical affinities towards human lysozyme and bacterial lipopolysaccharide, but differ in their susceptibilities towards tryptic proteolysis. Biochem J. 1995;312(Pt. 1):107–14. doi: 10.1042/bj3120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez E, Barranco A, Ramirez M, Gruart A, Delgado-Garcia JM, et al. Effects of a human milk oligosaccharide, 2′-fucosyllactose, on hippocampal long-term potentiation and learning capabilities in rodents. J Nutr Biochem. 2015;26:455–65. doi: 10.1016/j.jnutbio.2014.11.016. [DOI] [PubMed] [Google Scholar]
- Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, et al. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol. 2008;74:3996–4004. doi: 10.1128/AEM.00149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yu B, Karim M, Hu H, Sun Y, et al. Dietary sialic acid supplementation improves learning and memory in piglets. Am J Clin Nutr. 2007;85:561–69. doi: 10.1093/ajcn/85.2.561. [DOI] [PubMed] [Google Scholar]
- Wang M, Li M, Wu S, Lebrilla CB, Chapkin RS, et al. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J Pediatr Gastroenterol Nutr. 2015;60:825–33. doi: 10.1097/MPG.0000000000000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72:4497–99. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res. 2007;51:1398–405. doi: 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- Weichert S, Jennewein S, Hufner E, Weiss C, Borkowski J, et al. Bioengineered 2′-fucosyllactose and 3-fucosyllactose inhibit the adhesion of Pseudomonas aeruginosa and enteric pathogens to human intestinal and respiratory cell lines. Nutr Res. 2013;33:831–38. doi: 10.1016/j.nutres.2013.07.009. [DOI] [PubMed] [Google Scholar]
- White BA, Lamed R, Bayer EA, Flint HJ. Biomass utilization by gut microbiomes. Annu Rev Microbiol. 2014;68:279–96. doi: 10.1146/annurev-micro-092412-155618. [DOI] [PubMed] [Google Scholar]
- Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res. 2011;10:856–68. doi: 10.1021/pr101006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9:4138–51. doi: 10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–27. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew WS, Fedorov AA, Fedorov EV, Rakus JF, Pierce RW, et al. Evolution of enzymatic activities in the enolase superfamily: L-fuconate dehydratase from Xanthomonas campestris. Biochemistry. 2006;45:14582–97. doi: 10.1021/bi061687o. [DOI] [PubMed] [Google Scholar]
- Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, et al. Bifidobacterium longum subsp infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology. 2012;22:361–68. doi: 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- Yu ZT, Chen C, Newburg DS. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology. 2013;23:1281–92. doi: 10.1093/glycob/cwt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic AM, Barile D. Bovine milk as a source of functional oligosaccharides for improving human health. Adv Nutr. 2011;2:284–89. doi: 10.3945/an.111.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]