SUMMARY
The splicing factor SRSF1 promotes nonsense-mediated mRNA decay (NMD), a quality control mechanism that degrades mRNAs with premature termination codons (PTCs). Here we show that transcript-bound SRSF1 increases the binding of NMD factor UPF1 to mRNAs while in, or associated with, the nucleus, bypassing UPF2 recruitment and promoting NMD. SRSF1 promotes NMD when positioned downstream of a PTC, which resembles the mode of action of exon junction complex (EJC) and NMD factors. Moreover, splicing and/or EJC deposition increase the effect of SRSF1 on NMD. Lastly, SRSF1 enhances NMD of PTC-containing endogenous transcripts that result from various events. Our findings reveal an alternative mechanism for UPF1 recruitment, uncovering an additional connection between splicing and NMD. SRSF1’s role in the mRNA’s journey from splicing to decay has broad implications for gene expression regulation and genetic diseases.
Graphical abstract
In Brief: Aznarez et al. describe the mechanism behind the enhancement of nonsense-mediated mRNA decay (NMD) by the splicing factor SRSF1. Through its early association with the mRNA in the nucleus and its direct recruitment of UPF1, SRSF1 bypasses some of the steps necessary for the canonical NMD pathway.

INTRODUCTION
Nonsense-mediated mRNA decay (NMD) is a quality control pathway that blocks the generation of potentially deleterious truncated proteins. In the absence of NMD, transcripts with premature termination codons (PTCs)—arising from nonsense mutations, genomic rearrangements, errors in splicing, and regulated splicing—can give rise to toxic proteins with dominant-negative effects (Hwang and Maquat, 2011; Miller and Pearce, 2014). Transcripts are targeted for NMD through either exon junction complex (EJC)-dependent or -independent pathways (Kurosaki and Maquat, 2016). Generally, EJC-dependent NMD is highly efficient and is elicited by PTCs located >50–55 nt upstream of the last exon-exon junction in the spliced mRNA (Hug et al., 2016). In contrast, EJC-independent NMD is elicited by other messenger ribonucleoprotein (mRNP) features, such as a long 3′ UTR that may include UPF1-recruiting motifs, and it is increasingly recognized to play an integral role in global transcriptome regulation (Imamachi et al., 2017; Kurosaki and Maquat, 2016; Metze et al., 2013).
Pre-mRNA splicing is a prerequisite for EJC-dependent NMD, as EJCs are deposited onto the mRNA at the completion of splicing (Le Hir et al., 2000). Moreover, intronless gene mRNAs are not bound by EJCs, and those that carry PTCs fail to elicit NMD (Maquat and Li, 2001). The majority of EJCs (50%–80%) are deposited 20–24 nt upstream of an exon-exon junction (Hauer et al., 2016; Saulière et al., 2012; Singh et al., 2012). The EJC core consists of four proteins: the anchor eIF4A3, the Y14/MAGOH heterodimer, and Barentsz (BTZ or MLN51) (Le Hir et al., 2016); the first three components are recruited to the mRNA in the nucleus, whereas MLN51 is recruited to the EJC once the mRNP complex is exported to the cytoplasm (Gehring et al., 2009). The EJC serves as an anchor point for NMD factors, and it is thus the link between splicing and EJC-dependent NMD. UPF3B is the first NMD factor recruited in the nucleus to the spliced mRNA, through interaction with the EJC surface, comprising MAGOH-Y14 and eIF4A3 (Chamieh et al., 2008; Kim et al., 2001). The mRNP is then exported to the cytoplasm, where UPF2 is recruited through interactions between its MIF4G domain and the N-terminal region of UPF3B (Kadlec et al., 2004; Llorca, 2013).
Once in the cytoplasm, the mRNP bound by either the nuclear cap-binding complex (CBP80-CBP20) or eIF4E undergoes translation, a prerequisite for NMD (Hentze and Izaurralde, 2013); thus, blocking translation at any stage inhibits NMD (Maquat et al., 2010). During translation, EJCs are disassembled as the ribosome translocates along the mRNP (Maquat et al., 2010). When a ribosome encounters a stop codon, the transient SURF complex—consisting of SMG1, the key NMD factor UPF1, and the release factors eRF1 and eRF3—is recruited to the mRNP (Kashima et al., 2006). The recruitment of the SURF complex to the site of premature translation termination, marked by one or more downstream EJCs, distinguishes a PTC from the authentic termination signal, and it triggers the SMG1-mediated phosphorylation of the RNA helicase UPF1 (Kashima et al., 2006). This, in turn, signals the PTC-containing mRNA for degradation, carried out by endo- and exo-nucleases (Lykke-Andersen and Jensen, 2015).
In addition to the splicing-dependent deposition of EJCs on mRNA, splicing is further involved in NMD. Exon definition influences NMD efficiency, such that weak splice sites downstream (but not upstream) of a PTC decrease NMD efficiency (Gudikote et al., 2005). Moreover, overexpression of SRSF1—a member of the SR protein family of splicing factors (Long and Cáceres, 2009; Manley and Krainer, 2010)—increases the NMD efficiency of reporters with PTCs (Zhang and Krainer, 2004), and it can switch the pioneer round of translation of PTC-containing GPX1 mRNA from cytoplasmic to nuclear-associated, thereby enhancing NMD (Sato et al., 2008). In flies, EJC deposition on certain mRNAs depends on cis-acting sequences that promote NMD (Saulière et al., 2010). Non-canonical EJC deposition overlaps with SRSF1 motifs, suggesting its involvement in EJC recruitment/stabilization (Saulière et al., 2012; Singh et al., 2012). However, the mechanisms through which exon definition, SR proteins, and cis-acting sequences in certain transcripts influence the efficiency of NMD have remained unknown.
Here we investigated the molecular mechanisms by which SRSF1 acts as a positive regulator of NMD. Our results indicate that it does so by enhancing UPF1 binding to mRNAs in the nucleus and that this effect is most pronounced when SRSF1 binds downstream of a PTC in an intron-containing transcript.
RESULTS
SRSF1 Promotes NMD When Tethered Downstream of a PTC
Overexpression of SRSF1 promotes NMD of reporters carrying the HBB Ter39 mutation that causes a recessive form of β-thalassemia or the GPX1 Ter46 mutation associated with hemolytic anemia (Zhang and Krainer, 2004). Another SR protein, SRSF2, also promotes NMD of these reporters (Zhang and Krainer, 2004); here we tested whether additional members of the SR protein family have a similar stimulatory effect on NMD. To this end, we overexpressed T7-SRSF2, T7-SRSF3, untagged SRSF4, T7-SRSF6, and T7-SRSF9 in U2OS cells (Figure S1A), and we co-transfected the HBB wild-type (HBB-WT) or the Ter39 (HBB-Ter39) reporters (Figure 1A). Radioactive RT-PCR showed that all these proteins promote NMD, albeit to different extents (Figure S1B), suggesting a common function for SR proteins in NMD. We ensured that our RT-PCR conditions were in the linear range of amplification (Figure S2).
Figure 1. SRSF1 Promotes NMD When Tethered Downstream of a PTC and Requires Its RRM2.
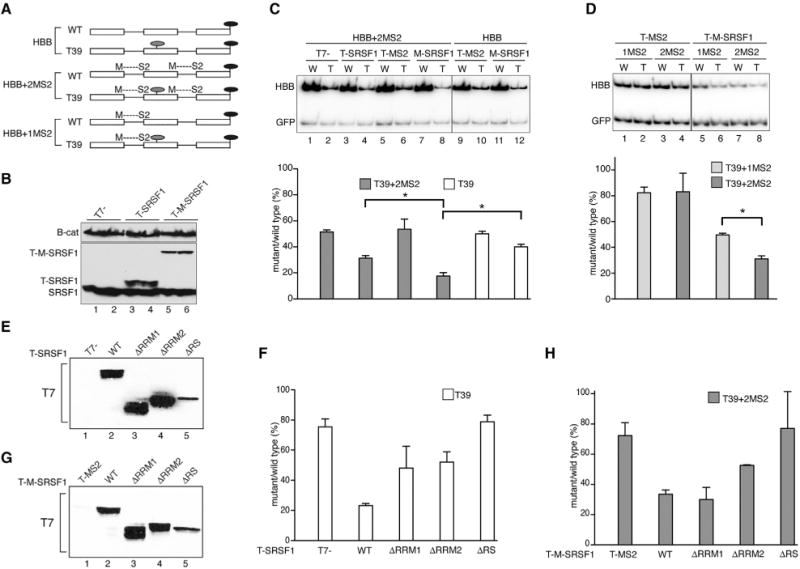
(A) β-globin (HBB) wild-type (WT) and mutant (T39) reporter constructs without or with MS2-binding sites formed after splicing at the first (1MS2) or first and second exon-exon junctions (2MS2). Light gray ovals, PTCs; black ovals, stop codons. Diagrams are not to scale.
(B) Representative western blot analysis of T7-tagged, T7-MS2-fused, and endogenous SRSF1, with β-catenin as a loading control.
(C) Representative radioactive RT-PCR analysis of HeLa cells co-transfected with WT (W) or T39 (T) reporters, with (2MS2, gray bars) or without MS2- (white bars) binding sites, and T-SRSF1 or T-M-SRSF1 constructs. HBB bands were quantified, normalized to GFP, and plotted as T/W × 100 (mean and SEM) using GraphPad Prism. n = 3.
(D) Representative radioactive RT-PCR analysis of U2OS cells co-transfected with HBB-WT or -T39 (T) reporters, with one MS2-binding site upstream of the T39 PTC (1MS2, light gray bars) or one MS2-binding site upstream and one downstream of PTC (2MS2, dark gray bars), and T-M-SRSF1 or empty vector control (T-MS2). Bands were quantified and plotted as above. n = 3. *p = 0.05 (Mann-Whitney).
(E) Representative western blot probed with anti-T7 antibody, showing the expression of T-SRSF1 WT and deletion mutants.
(F) Quantitation of radioactive RT-PCR of U2OS cells co-transfected with HBB-WT or Ter39 reporters and SRSF1 WT, deletion mutants, or T7-empty vector control. n = 3.
(G) Representative western blot showing the expression of T-M-SRSF1 WT and deletion mutants.
(H) Radioactive RT-PCR analysis as in (B) but with HBB-WT or Ter39 reporters with 2MS2-binding sites (2MS2) and T-MS2, T-M-SRSF1 WT, and deletion mutants. n = 3.
Error bars represent SEM.
To investigate the mode of action of SRSF1 and other SR proteins, we designed a set of reporter constructs to tether SR proteins to the transcript at different positions with respect to PTCs. We used HBB-WT and HBB-Ter39 genes with MS2 hairpins inserted upstream, or upstream and downstream, of the nonsense codons (Figure 1A). The MS2 hairpins were inserted such that a complete and functional hairpin was formed only after splicing at the first or second exon junction (Figure 1A; Lacadie et al., 2006). The experiments were carried out in HeLa (higher NMD efficiency; Linde et al., 2007) or U2OS cells (lower NMD efficiency).
First, we co-transfected into HeLa cells HBB-WT and HBB-Ter39 constructs with (+2MS2) or without MS2-hairpin sequences, with T7-tagged SRSF1 cDNA (T-SRSF1) or a T7-tagged MS2-SRSF1 fusion construct (T-M-SRSF1), representing free and tethered versions of SRSF1, respectively, as well as the respective T7-empty and T7-MS2 controls (Figure 1B). Radioactive RT-PCR showed that T-M-SRSF1 tethered to both exon junctions of HBB-Ter39 significantly increased the extent of NMD, compared to free T-SRSF1 or T-M-SRSF1 co-transfected with HBB-Ter39 without MS2-hairpin sequences (Figure 1C, lane 8 versus lane 4 or 12). The effect of free SRSF1 on NMD (Figure 1C, cf. lanes 2 and 4 or lanes 10 and 12) presumably reflects its binding to SRSF1 target sites present throughout the HBB mRNA (Schaal and Maniatis, 1999). However, the high-affinity binding between MS2 coat protein and the MS2 hairpins is expected to confer greater RNA-binding stability to T-M-SRSF1, leading to an enhanced effect on NMD.
To determine whether the location of SRSF1 or other SR proteins on the transcript with respect to a PTC is important for their effect on NMD, MS2-SR protein fusions were tethered upstream of the HBB Ter39 (+1MS2) mutation, or upstream and downstream of Ter39 (+2MS2) (Figure 1A). When SRSF1 was tethered downstream, its effect on NMD increased significantly, compared to tethering it only upstream (Figure 1D, cf. lanes 7 and 9, respectively). Tethering of the other SR proteins to the HBB reporters likewise promoted NMD when they were positioned downstream of a PTC (Figures S1C and S1D), with the exception of SRSF4 and SRSF6, which were not functional when fused to MS2 (data not shown). SRSF3, which enhanced NMD most potently, did not appear to enhance NMD any further with tethering, possibly due to its already maximal effect.
We conclude the following: (1) other members of the SR protein family also promote NMD; (2) to promote NMD of a PTC-containing transcript, SRSF1 needs to be bound to the transcript; and (3) SRSF1 and other SR proteins promote NMD when positioned downstream of a PTC.
SRSF1’s RRM2 Is Involved in NMD
To determine which domains are required for SRSF1’s function in NMD, we employed three domain mutants: ΔRRM1, ΔRRM2, and ΔRS (Cáceres and Krainer, 1993). The RRMs are involved in binding to RNA sequence motifs, whereas the RS domain is mainly involved in protein-protein interactions, subcellular localization, and recruitment of spliceosome components, though there is also evidence of RNA binding (Long and Cáceres, 2009; Daubner et al., 2013). We co-transfected plasmid vectors expressing each of these T7-tagged mutants and the T7-empty vector (Figure 1E) into U2OS cells, together with the HBB-WT or HBB-Ter39 reporters. Radioactive RT-PCR showed that all deletion mutants were defective to different extents in promoting NMD, compared to WT SRSF1 (Figure 1F). These results confirm our previous findings about the inability of the ΔRRM2 and ΔRS mutants to promote NMD (Zhang and Krainer, 2004), and they further show that the ΔRRM1 mutant (not previously tested for NMD) is also impaired in the NMD function.
To address the mechanisms underlying the NMD defects of the SRSF1 deletion mutants, we fused each mutant to MS2 protein (Figure 1G) and tethered it to HBB-WT and HBB-Ter39 reporters with MS2 hairpins at both exon junctions (+2MS2; Figure 1A). T7-MS2 was co-transfected as a tethering control. Radioactive RT-PCR of RNA from co-transfected U2OS cells indicated that tethering the T-M-ΔRRM1 fusion to the mRNA restored the NMD enhancement effect, suggesting that the ΔRRM1 mutant is defective in binding to HBB mRNA (Figure 1H, cf. bars corresponding to WT and ΔRRM1). Moreover, the MS2-fused ΔRRM1 mutant remained ineffective in promoting NMD in the absence of MS2 hairpins in the HBB mRNA (Figure S3). In contrast, tethering the T-M-ΔRRM2 or T-M-ΔRS fusions to the HBB mRNA did not restore the effect on NMD (Figure 1H, cf. bars corresponding to ΔRRM2 and ΔRS to that of WT and to the results in Figure 1F). However, both the free and fused versions of the ΔRS protein were expressed at lower levels than SRSF1 WT or other mutants, potentially accounting for the lack of an effect (Figures 1E and 1G). In light of the dose-dependent effect of SRSF1 on NMD (Zhang and Krainer, 2004), we tested whether increasing the amount of the ΔRS mutant protein might restore the NMD effect. To this end, we doubled the concentration of the ΔRS mutant plasmid, and the NMD effect was partially restored (Figure S4A). In addition, as the RS domain is involved in subcellular localization and the ΔRS mutant partially accumulates in the cytoplasm (Cáceres et al., 1998), the lower expression level, in combination with the inability to efficiently localize to the nucleus, may account for the poor performance of this mutant in NMD.
Considering that the NMD function of the ΔRRM1 mutant is rescued by restoring its binding to the transcript, we infer that RRM1 normally functions as an RNA-binding domain in the context of β-globin NMD. In contrast, tethering the RRM2 deletion mutant to the β-globin transcript does not fully rescue its function in NMD, suggesting additional functions of this domain.
SRSF1 Interacts with EJC Components
To further dissect the mechanism by which SRSF1 promotes NMD, we investigated the role of EJC components. We first performed co-immunoprecipitations (coIPs) to determine whether SRSF1 interacts with EJC core components. We transfected HeLa cells with WT T-SRSF1, T-ΔRRM2, T-ΔRS, or T7-empty vector control. We included ΔRRM2 to further dissect its inability to enhance NMD and ΔRS to exclude the possibility of any additional function of the RS domain. We performed IPs from nuclease-treated whole-cell lysate via the T7 tag, and we probed the immunoprecipitates for endogenous eIF4A3, Y14, and MAGOH. Western blotting showed interactions between both WT SRSF1 and the ΔRRM2 mutant and all three EJC proteins (Figure 2A, cf. lanes 5 and 6 to 4, top three panels). The ΔRS mutant also interacted with all three proteins (Figure S4B). These results suggest that neither RRM2 nor the RS domain is involved in these interactions. Moreover, as SRSF1 interacted with these proteins in an RNA-independent manner, this suggests direct protein-protein interaction(s) with the EJC.
Figure 2. SRSF1 Co-immunoprecipitates with EJC Components and Splicing Enhances Its Effect on NMD.
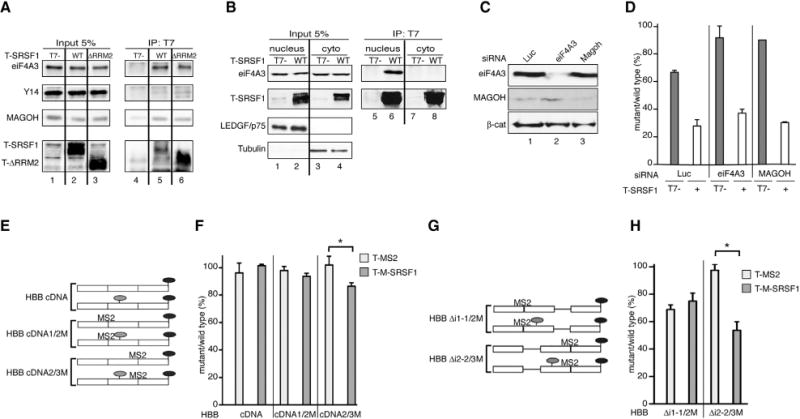
(A) Representative IP-western blot of nuclease-treated extracts from HeLa cells overexpressing T-SRSF1, T-ΔRRM2, or T7-empty vector control, immunoprecipitated with anti-T7 antibody and probed for endogenous EJC components with the indicated antibodies. SRSF1 appears as several distinct bands due to its extensive phosphorylation (Hanamura et al., 1998).
(B) Representative IP-western blot of nuclease-treated nuclear and cytoplasmic fractions from HeLa cells overexpressing T-SRSF1 or T7-empty vector control, immunoprecipitated with anti-T7 antibody and probed for endogenous eIF4A3. LEDGF/p75 and Tubulin were used as nuclear and cytoplasmic markers, respectively.
(C) Representative western blot showing siRNA-mediated knockdown of eiF4A3 (lane 2) and MAGOH (lane 3). β-catenin (β-cat) was used as a loading control.
(D) Radioactive RT-PCR analysis of HeLa cells co-transfected with HBB-WT or Ter39 reporters and T-SRSF1 (+) or T7-empty vector control (−), in the presence of the indicated siRNAs.
(E) HBB intronless cDNA WT and mutant (T39) reporter constructs without MS2-binding sites or with one MS2-binding site at the first (1/2M) or second (2/3M) exon-exon junction. Light gray ovals, PTCs; black ovals, stop codons.
(F) Radioactive RT-PCR analysis of U2OS cells co-transfected with WT or Ter39 versions of HBB-cDNA, cDNA1/2M or cDNA2/3M reporters and T-M-SRSF1 (dark gray bars) or T-MS2 control (light gray bars). *p = 0.05 (Mann-Whitney). n = 3.
(G) HBB with first or second intron deleted WT and mutant (T39) reporter constructs, with one MS2 binding site at the first (1/2M) or second (2/3M) exon-exon junction. Light-gray ovals: PTCs; black ovals: stop codons.
(H) Radioactive RT-PCR analysis of HeLa cells co-transfected with WT or Ter39 versions of HBB-Δi1-1/2M or Δi2-2/3M reporters and T-M-SRSF1 (dark gray bars) or T-MS2 control (light gray bars). *p < 0.05 (Mann-Whitney). n = 3.
Error bars represent SEM.
We verified that SRSF1 associates specifically with the EJC anchor protein eIF4A3 in cells, as opposed to a spurious post-lysis interaction (Riley et al., 2012). This was done by lysing HeLa cells that overexpress either T-SRSF1 or FLAG-eIF4A3 and pooling the lysates prior to IP. IP of T-SRSF1 led to coIP of endogenous eIF4A3 expressed in the same cells, but not F-eIF4A3, which was not expressed in the same cells and was only present in the combined lysate (Figure S5A). These results support a direct and specific interaction between SRSF1 and eIF4A3.
As EJC components are found in both the nucleus and cytoplasm, we performed IP-western blots of nuclear and cytoplasmic fractions from T-SRSF1-overexpressing HeLa cells to determine where the observed interactions occur. SRSF1 co-immunoprecipitated with the EJC anchor protein eIF4A3 only in the nuclear fraction (Figure 2B, cf. lanes 6 and 8, top panel).
To systematically search for EJC-related, putative interacting partners of SRSF1, we analyzed mass spectrometry (MS) data obtained by T7-tag IPs from lysates of T7-SRSF1-overexpressing HeLa cells, with and without nuclease treatment (Akerman et al., 2015). Table S1 lists all EJC and EJC-associated proteins found to co-immunoprecipitate with SRSF1. Most of them co-immunoprecipitated in an RNA-independent manner (Table S1), confirming our observation that SRSF1 appears to interact directly with the EJC. The nuclease treatment efficiently degraded RNA (Figure S5B), consistent with direct protein-protein interaction. The combined results from the IP-western and the IP-MS show that all EJC core components, except cytoplasmic MLN51, and most of the known associated proteins (Tange et al., 2005) co-immunoprecipitated with SRSF1, consistent with a functional relationship between SRSF1 and the EJC in the nucleus.
To assess whether the EJC-SRSF1 interactions are required for the effect of SRSF1 on NMD, we used short interfering RNA (siRNA) to silence the expression of eIF4A3 and MAGOH, including luciferase siRNA as a non-targeting control (Figure 2C), in the presence or absence of overexpressed T-SRSF1. Knocking down these EJC components in HeLa cells inhibited NMD of the HBB-Ter39 reporter (Figure 2D, gray bars). However, overexpression of SRSF1 still promoted NMD in the absence of each of the proteins (Figure 2D, white bars). These results suggest that the individual EJC components are not essential for SRSF1 to enhance NMD, even though they physically interact with it.
Splicing Enhances the Effect of SRSF1 on NMD
Next, we assessed whether splicing and EJC deposition are necessary for the effect of SRSF1 on NMD. We generated HBB intronless reporters corresponding to the WT or the T39 allele, lacking MS2-binding sites (cDNA), with one MS2-binding site spanning the first exon junction (cDNA1/2M), or with one MS2-binding site spanning the second exon junction (cDNA2/3M) (Figure 2E). Each intronless reporter was co-transfected into U2OS cells with T-MS2 or T-M-SRSF1 plasmid. SRSF1 could not promote NMD in the absence of an MS2-binding site or when tethered upstream of the T39 PTC (Figure 2F). However, SRSF1 appeared to destabilize the intronless T39 transcript when tethered downstream of the PTC (Figure 2F). These results are consistent with our finding (Figure 1D) that SRSF1 promoted NMD when positioned downstream of a PTC, but they also suggest that splicing/EJC deposition is not essential for the effect of SRSF1 on NMD. However, the effect of SRSF1 tethered downstream of the T39 PTC in the HBB intronless reporter, albeit significant, is not as pronounced as when the introns are present (cf. Figure 1D, lane 9 and the last bar in Figure 2F).
To further understand the contribution of splicing and EJC deposition to SRSF1’s function in NMD, we generated HBB reporters lacking either intron 1 (Δi1-1/2M) or 2 (Δi2-2/3M) and possessing an MS2-binding site at the exon-exon boundary resulting from deleting either intron (Figure 2G). First, transfection of the WT and T39 versions showed that deletion of intron 1 had no effect on NMD, whereas deletion of intron 2 abrogated NMD, as expected (Figure 2H, cf. light bars). Tethering T-MS2 or T-MS2-SRSF1 upstream of the T39 mutation, at the intron 1 deletion site, had no effect on NMD, even though intron 2 was still present. However, tethering T-MS2-SRSF1 downstream of the T39 mutation in the presence of intron 1 and absence of intron 2 significantly promoted NMD (Figure 2G, cf. dark bars), whereas T-MS2 had no effect. This result provides further evidence that SRSF1 needs to be positioned downstream of a PTC to enhance NMD. Moreover, splicing greatly increases the effect of SRSF1 on NMD, even if the intron is located upstream of the PTC, as tethering SRSF1 to an intron-containing transcript (Δi2-2/3M) led to a much more pronounced effect than tethering it to an intronless transcript (cDNA2/3M) (cf. Figure 2F and Figure 2H). Taken together, these results suggest that splicing and EJC component deposition enhance but are not absolutely required for the effect of SRSF1 on NMD, as knocking down EJC components or eliminating EJC deposition by deleting introns does not abrogate the effect of SRSF1 on NMD.
SRSF1 Interacts with UPF3B but Still Promotes NMD in the Absence of UPF2 or UPF3B
To explore other interaction partners that might mediate SRSF1’s enhancement of NMD, we tested whether SRSF1 interacts with the NMD factors UPF3B and UPF2. Nuclease-treated extracts from T-SRSF1-overexpressing or T7-empty vector-transfected HeLa cells were immunoprecipitated with T7-specific antibody. In addition, we used extracts derived from HeLa cells overexpressing the T-ΔRRM2 and T-ΔRS mutants. We probed the IP-western blots for endogenous UPF2 and UPF3B. WT SRSF1 and the ΔRRM2 mutant co-immunoprecipitated with UPF3B in an RNA-independent manner (Figure 3A, middle panel, cf. lanes 5 and 6 to lane 4). Moreover, the ΔRS mutant gave a similar result (Figure S4C). In contrast, we did not detect interactions between SRSF1 or the mutants and UPF2 (Figure 3A, top panel). Thus, RRM2 and the RS domain are not required for the interaction between SRSF1 and UPF3B, and the lack of NMD-promoting activity of the ΔRRM2 mutant must be due to a different property of the deleted domain. Moreover, similar to the SRSF1-EJC interaction, IP-western analysis of nuclease-treated nuclear and cytoplasmic fractions from T-SRSF1-overexpressing HeLa cells indicated that the interaction between SRSF1 and UPF3B occurs only in the nuclear fraction (Figure 3B, middle panel, cf. lanes 6 and 8). We observed no interaction between SRSF1 and UPF2 in either nuclear or cytoplasmic fractions (Figure 3B, top panel), confirming the above findings.
Figure 3. SRSF1/UPF3B Interaction Occurs in the Nucleus but Is Not Required to Stimulate NMD.
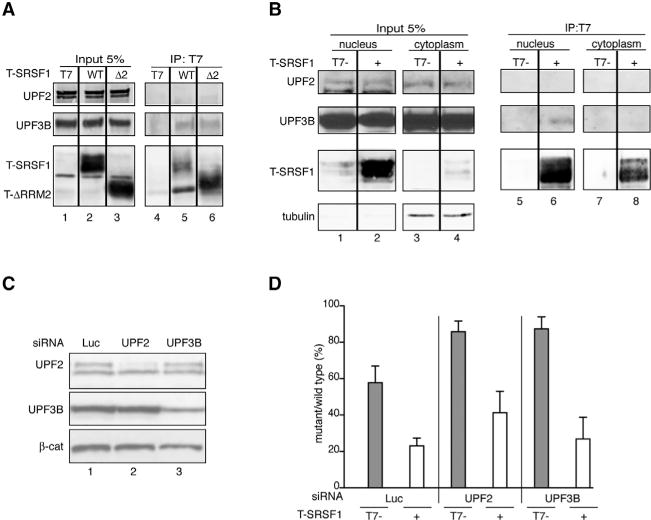
(A) Representative IP-western blot of nuclease-treated HeLa cell extracts overexpressing T-SRSF1 WT, T-ΔRRM2 (Δ2), or T7-empty vector control, immunoprecipitated with anti-T7 antibody and probed for endogenous UPF2 and UPF3B.
(B) Representative IP-western blot of nuclease-treated nuclear (left panels) and cytoplasmic (right panels) fractions from HeLa cells overexpressing T-SRSF1 or T7-empty vector control, immunoprecipitated with anti-T7 antibody and probed for endogenous UPF2 and UPF3B. Tubulin was used as a cytoplasmic marker.
(C) Representative western blot showing siRNA-mediated knockdown of UPF2 (lane 2) and UPF3B (lane 3). β-catenin (β-cat) was used as a loading control.
(D) Radioactive RT-PCR analysis of HeLa cells co-transfected with HBB-WT or Ter39 reporters and T-SRSF1 (+) or T7-empty vector control (−) in the presence of the indicated siRNAs. n = 3.
Error bars represent SEM.
To determine whether UPF3B is required for the effect of SRSF1 on NMD, we silenced its expression in HeLa cells using siRNA (Figure 3C) in the presence or absence of SRSF1 overexpression. Even though an interaction between UPF2 and SRSF1 was not evident, as UPF2 is involved in interacting with UPF1 to form the DECID complex (Kashima et al., 2006), we also knocked down UPF2 (Figure 3C), and we measured NMD of the HBB reporters in the context of overexpressed SRSF1. Radioactive RT-PCR confirmed that partially silencing either UPF3B or UPF2 inhibited NMD of the HBB-Ter39 transcript in the absence of SRSF1 overexpression, compared to control non-targeting siRNA (Figure 3D, gray bars). However, under the same conditions, overexpression of SRSF1 restored NMD of the HBB-Ter39 transcript to levels comparable to the non-targeting siRNA control (Figure 3D). It is possible that the reduced level of UPF3B, albeit insufficient to trigger NMD of the HBBTer39 transcript, is sufficient to be recruited by SRSF1 and contribute to its effect on NMD. Alternatively, UPF3A could be acting in place of UPF3B.
Taken together, these results indicate that UPF2 is not required for the effect of SRSF1 on NMD, as it not only does not interact with SRSF1 but also reducing its expression does not completely abrogate the effect of SRSF1 on NMD. Moreover, we reason that, as UPF3B is recruited to the EJC in the nucleus and SRSF1 co-immunoprecipitates with both UPF3B and EJC components, SRSF1 might promote their recruitment to the transcript in the nucleus. However, the recruitment of UPF3B to the transcript, though likely contributing, does not seem to be an absolute requirement for SRSF1’s effect on NMD.
SRSF1 Interacts with UPF1 and Enhances Its Binding to mRNA
The effect of SRSF1 on NMD is UPF1 dependent, as either silencing UPF1 expression or overexpressing a dominant-negative form of UPF1 inhibits SRSF1’s enhancement of NMD (Zhang and Krainer, 2004). This observation, together with the above findings, led us to hypothesize the existence of a direct connection between SRSF1 and UPF1, bypassing the requirement for other NMD factors and possibly the EJC. To test our hypothesis, we generated nuclease-treated nuclear and cytoplasmic fractions of T-SRSF1-overexpressing or T7-empty vector-transfected HeLa cells, and we immunoprecipitated protein extracts from each fraction. Western blotting of endogenous UPF1 demonstrated that SRSF1 co-immunoprecipitated with UPF1, both in the nuclear and cytoplasmic fractions, in an RNA-independent manner (Figure 4A, top panel, lanes 6 and 8). The results also indicate that the SRSF1-UPF1 interaction was more prevalent in the nucleus than in the cytoplasm, as UPF1 was less abundant in the input nuclear fraction but similar levels were co-immunoprecipitated by SRSF1 from both fractions. Moreover, as shown by the tubulin blots, the nuclear fraction appeared to have little to no contamination from the cytoplasmic fraction, suggesting that the SRSF1/UPF1 interaction was truly occurring in the nucleus or in association with the nuclear membrane. Nevertheless, we confirmed the nuclear interaction between SRSF1 and UPF1 by IP-western analysis of T-SRSF1 fused to a nuclear retention signal (NRS; Cazalla et al., 2002), and we verified that nuclear-retained SRSF1 also co-immunoprecipitated with endogenous UPF1 (Figure 4B). As SRSF1-NRS can still promote NMD (Zhang and Krainer, 2004), we conclude that the first communication between SRSF1 and UPF1 occurs in the nucleus or at the nuclear membrane and is essential for the effect of SRSF1 on NMD.
Figure 4. SRSF1 Interacts with UPF1 in the Nucleus and Cytoplasm and Enhances UPF1 Binding to mRNA.
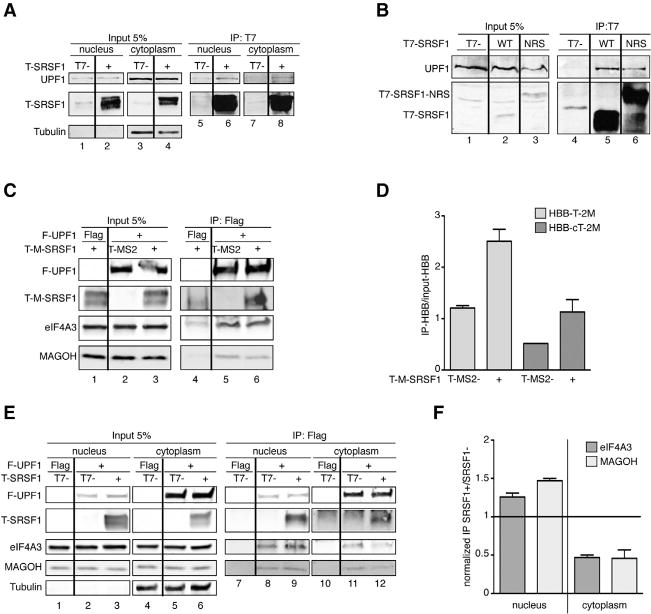
(A) Representative IP-western blot of nuclease-treated nuclear and cytoplasmic fractions from HeLa cells overexpressing T-SRSF1 or T7-empty vector control, immunoprecipitated with anti-T7 antibody and probed for endogenous UPF1. Tubulin was used as a cytoplasmic marker. The interaction occurs in both the nucleus and cytoplasm (same samples were also used in Figure 2B).
(B) IP-western blot analysis of HeLa cell extracts overexpressing T-SRSF1, T-SRSF1-NRS, or empty vector control, immunoprecipitated with anti-T7 antibody and probed for endogenous UPF1 (first panel). The results show that a nuclear-retained form of SRSF1 interacts with UPF1.
(C) Representative IP-western blot of HeLa cell extracts overexpressing FLAG-empty vector control or FLAG-UPF1, T-MS2 or T-MS2-SRSF1, and HBB-T39 or HBB-cDNAT39-2MS2, immunoprecipitated with anti-FLAG antibody and probed with the indicated antibodies.
(D) Radioactive RT-PCR analysis of RNA extracted from the IPs shown in (B) of HBB-Ter39-2MS2 (light gray bars) and HBB-cDNATer39-2MS2 (dark gray bars). The ratio of IP/input was calculated and plotted. n = 3.
(E) Representative IP-western blot of nuclear and cytoplasmic fractions from HeLa cells overexpressing FLAG-empty vector control or FLAG-SRSF1 and T7-empty vector control or T-SRSF1, immunoprecipitated with anti-FLAG antibody and probed for endogenous eIF4A3 and MAGOH. Tubulin was used as a cytoplasmic marker.
(F) The intensities of bands corresponding to eIF4A3 (dark gray bars) and MAGOH (light gray bars) from both fractions were quantified and normalized first to input and then to IP efficiency, and the ratio of SRSF1+/SRSF1 was calculated. A ratio of 1 (line) indicates no change. n = 2.
Error bars represent SEM.
To rigorously test whether the observed RNA-independent interaction between SRSF1 and UPF1 is direct, we performed an in vitro pull-down assay with recombinant proteins. Purified GST-UPF1 or GST alone was immobilized and incubated with purified MBP-SRSF1 or MBP alone in the presence of nucleases. SRSF1 was pulled down by GST-UPF1, but not GST alone (Figure S6A, cf. lanes 3 and 6), indicating that the interaction between SRSF1 and UPF1 is direct.
In addition to being recruited by EJC and UPF proteins, UPF1 can bind to mRNAs to sense 3′ UTR length, and this binding is cycloheximide resistant, suggesting that UPF1 binds to mRNAs prior to translation elongation (Hogg and Goff, 2010). As we found that SRSF1 and UPF1 interact directly and co-immunoprecipitate in both the nucleus and cytoplasm, we hypothesized that SRSF1 could be involved in either recruiting UPF1 or stabilizing its binding to mRNAs. To test this hypothesis, we immunoprecipitated FLAG-tagged UPF1 in the presence of T-MS2-SRSF1, or T-MS2 as a control, from HeLa cells expressing HBB-T39-2MS2 (Figure 1A) or HBB-cDNAT39-2MS2 (an intronless version of HBB-T39-2MS2). First, the IP-western results showed that SRSF1 co-immunoprecipitated with UPF1 in the presence of RNA (Figure 4C, second panel from top, lane 6). We then performed radioactive RT-PCR from cell extract input and IPs to assess whether the HBB transcript was co-immunoprecipitated with UPF1 and whether the presence of tethered SRSF1 influenced the UPF1 interaction with HBB RNA. The RT-PCR results were quantified and plotted, and they showed that both versions of the HBB reporters co-immunoprecipitated with UPF1, but a significantly higher proportion of the intron-containing reporter was detected (Figure 4D, cf. first and third bars). This result agrees with the notion that splicing/EJC deposition is important for UPF1 binding to mRNA (Le Hir et al., 2000). Remarkably, upon tethering T-MS2-SRSF1 to the HBB reporters, the amount of HBB mRNA associated with UPF1 doubled (Figure 4D, cf. first to second and third to fourth bars), indicating that the presence of SRSF1 on the mRNA increased or stabilized the binding of UPF1.
As splicing/EJC deposition appears to enhance UPF1 binding to the HBB transcript, we examined whether SRSF1 overexpression affected the interaction of UPF1 with eIF4A3 and MAGOH. FLAG-UPF1 immunoprecipitates were probed by western blotting, which showed that both EJC factors co-immunoprecipitate with UPF1 (Figure 4C, third and fourth panels, lane 5); however, there was a slight reduction in eIF4A3 and MAGOH co-immunoprecipitated with UPF1 in the presence of RNA upon SRSF1 overexpression (Figure 4C, cf. lanes 5 and 6, third and fourth panel). To determine in which compartment this reduction occurred, we immunoprecipitated FLAG-UPF1 from HeLa cells overexpressing T-SRSF1, or T7-empty vector control, from nuclear and cytoplasmic fractions, and we probed for eIF4A3 and MAGOH. SRSF1 (second panel), eIF4A3 (third panel), and MAGOH (fourth panel) co-immunoprecipitated with UPF1 both in the nuclear and cytoplasmic fractions in the presence of RNA (Figure 4E, lanes 9 and 12, 8 and 9, and 11 and 12, respectively). We then quantified the bands and plotted the ratio between immunoprecipitated eIF4A3 (dark gray) or MAGOH (light gray) in the presence of T-SRSF1, over immunoprecipitated eIF4A3 or MAGOH in the absence of T-SRSF1, in the nucleus and cytoplasm. Interestingly, the coIP of UPF1 and EJC factors in the presence of mRNAs in the nucleus increased upon overexpression of SRSF1 (Figure 4F, first two bars, ratio > 1). The converse occurred in the cytoplasm, as less eIF4A3 and MAGOH co-immunoprecipitated with UPF1 in the presence of SRSF1 (Figure 4F, last two bars, ratio < 1), suggesting that, in the nucleus, SRSF1 promotes the recruitment of EJC factors and UPF1, whereas, in the cytoplasm, the interaction between UPF1 and the EJC is disrupted, further suggesting that NMD is enhanced in the presence of SRSF1.
SRSF1 RRM2 Involvement in NMD
To determine whether RRM2 or the RS domain of SRSF1 mediates its interaction with UPF1, we immunoprecipitated nuclease-treated protein extracts from HeLa cells overexpressing T-SRSF1, the T-ΔRRM2 and T-ΔRS mutants, or the T7-empty vector control. IP-western analysis showed that deletion of RRM2 (Figure 5A, lane 6, top panel) or the RS domain (Figure S4D) did not abrogate the interaction with UPF1. We noticed, however, that a slower-migrating band (Figure 5A, top panel, lane 6, marked by an asterisk), possibly corresponding to a hyperphosphorylated form of UPF1, was increased in the T-ΔRRM2 mutant IP compared to WT SRSF1 (lane 5). To examine this further, we tested whether phosphorylated UPF1 (p-UPF1) co-immunoprecipitates with SRSF1 and whether this interaction requires RRM2. To this end, we used two different antibodies that detect p-UPF1. The first antibody recognizes phosphorylated targets of the ATR/ATM kinases and specifically detects p-UPF1 (Stalder and Mühlemann, 2009). Western blotting showed that p-UPF1 co-immunoprecipitated with both SRSF1 and the ΔRRM2 mutant; however, hyperphosphorylated UPF1 was further enriched in the T-ΔRRM2 mutant IP (Figure 5A, second panel, cf. lanes 5 and 6). Likewise, western blotting with a second antibody that specifically recognizes UPF1’s p-Thr28 (Ohnishi et al., 2003) indicated that the interaction between T-ΔRRM2 and p-T28-UPF1 was enhanced, compared to WT SRSF1 (Figure 5A, third panel, cf. lanes 5 and 6).
Figure 5. SRSF1’s RRM2 Is Involved in UPF1 Dephosphorylation.
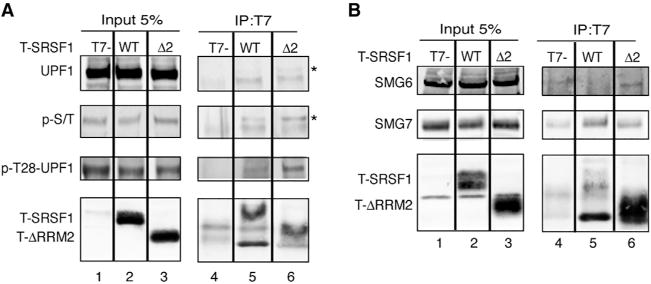
(A) Representative IP-western blot analysis of nuclease-treated HeLa cell extracts overexpressing T-SRSF1 WT, T-ΔRRM2, or T7-empty vector control, immunoprecipitated with anti-T7 antibody and probed with the indicated antibodies. SRSF1 WT and ΔRRM2 co-immunoprecipitate with UPF1; however, a slower-migrating band increases in the ΔRRM2 IP (asterisk) and corresponds to p-T28-UPF1.
(B) Representative IP-western blot of nuclease-treated extracts from HeLa cells overexpressing T-SRSF1 WT, T-ΔRRM2, or T7-empty vector control, immunoprecipitated with anti-T7 antibody and probed for endogenous SMG proteins with the indicated antibodies.
Phosphorylation of UPF1 by SMG1 on Thr28 and Ser1096 generates a scaffold for SMG6 and SMG5/SMG7 binding, which in turn promotes PP2A-mediated dephosphorylation of UPF1 (Okada-Katsuhata et al., 2012), and its subsequent release to form a new SURF complex. It was also independently shown that SRSF1 and PP2A co-immunoprecipitate (Michlewski et al., 2008). Thus, we wondered whether RRM2 of SRSF1 is involved in promoting UPF1 dephosphorylation and, perhaps, in additional interactions with SMG5/SMG7 or SMG6. To test this idea, we immunoprecipitated T-SRSF1 and T-ΔRRM2, and we probed for SMG6 and SMG7. Despite several attempts to reliably detect SMG6, we could not find a better antibody than the one employed in these experiments, and the results were inconclusive. Western blotting showed that both SRSF1 and ΔRRM2 interacted with SMG7 (Figure 5B, middle panel, lanes 5 and 6), but the interaction between ΔRRM2 and SMG7 appeared to be weaker.
Taken together, these results suggest that SRSF1 can form a complex with p-UPF1 and SMG7, potentially facilitating UPF1 dephosphorylation via PP2A through SRSF1’s RRM2. Based on these findings, we reasoned that T-SRSF1-NRS might promote NMD to a lesser extent than T-SRSF1, as dephosphorylation of p-UPF1 occurs in the cytoplasm and the former is retained in the nucleus, such that the cytoplasmic effect would be carried out by endogenous SRSF1. Radioactive RT-PCR analysis indicated that, indeed, T-SRSF1-NRS promoted NMD to a lesser extent than T-SRSF1 (Figure S6B), consistent with the involvement of SRSF1 in the dephosphorylation of UPF1.
SRSF1 Promotes NMD of PTC-Containing Endogenous Transcripts
This study and others showed that SRSF1 promotes NMD of HBB and GPX1 reporters (Sato et al., 2008; Zhang and Krainer, 2004). However, it was not known whether SRSF1 can promote NMD of endogenous targets. To answer this question, we selected a random subset of PTC-containing endogenous transcripts that are known NMD targets (Pan et al., 2006). These PTC-containing transcripts result from skipping of an exon, causing a frameshift, or from the inclusion of a PTC-containing exon (Figure 6A). We also analyzed an SRSF2 transcript that becomes an NMD target by splicing of intronic sequences in the 3′ UTR; NMD of this transcript is UPF2 independent (Gehring et al., 2005). In addition, we analyzed the transcript from the IDUA gene from skin fibroblasts of a patient with mucopolysaccharidosis type I, homozygous for the W402X nonsense mutation (Scott et al., 1993).
Figure 6. SRSF1 Promotes NMD of PTC-Containing Endogenous Transcripts.
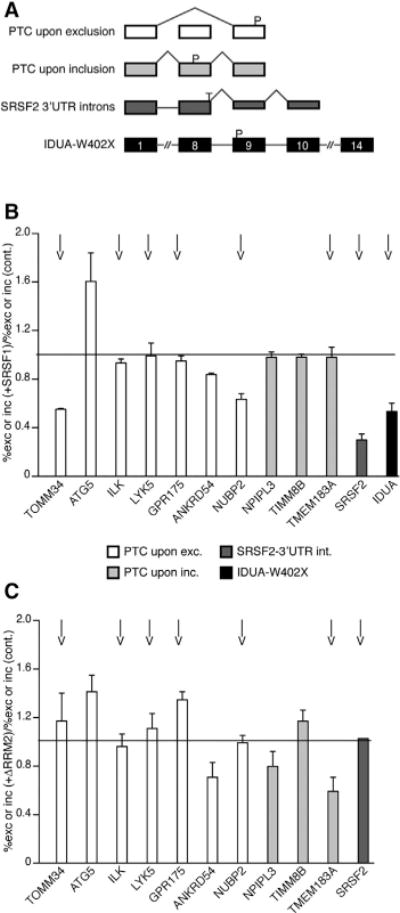
(A) Diagrams of four events that can introduce PTCs. White, PTC upon skipping of a cassette exon (Pan et al., 2006); light gray, PTC upon inclusion of a cassette exon (Pan et al., 2006); dark gray, splicing within the 3′ UTR; black, PTC introduced by a nonsense mutation (IDUA W402X; Scott et al., 1993).
(B) Radioactive RT-PCR analysis of GFP-sorted HeLa cells (white and gray bars) or patient fibroblasts (black) co-transfected with T-SRSF1 or empty vector control and GFP. The data were plotted as the ratio of percent skipped or included mRNA from SRSF1-transfected cells over percent skipped or included mRNA from control transfections. The IDUA mRNA level was normalized to endogenous actin, and the mRNA ratio of SRSF1-transfected over control-transfected cells was plotted. n ≥ 2.
(C) Same experimental design in HeLa cells as in (B) but expressing T-ΔRRM2. n = 2.
A ratio of 1 (line) indicates no change in mRNA level between control and SRSF1 or ΔRRM2 overexpression. Arrows indicate true NMD targets, sensitive to cycloheximide treatment. See also Figure S7.
Error bars represent SEM.
We co-transfected T-SRSF1 or an empty vector control and GFP cDNA into HeLa or patient fibroblast cells. GFP was used to sort the cells by flow cytometry to enrich for transfected cells. Radioactive RT-PCR (Figure S7) showed that overexpression of SRSF1 promoted NMD of the PTC-containing isoforms of TOMM34 and NUBP2, and to a lesser extent of ANKRD54, among those in the PTC-upon-skipping category (Figure 6B, white bars). We note that the decrease in the ratio of the percent skipping of “SRSF1+” over the percent skipping of “SRSF1−” resulted from a reduction of the PTC-containing product (exon skipping) upon SRSF1 overexpression, rather than from an increase in exon inclusion. Moreover, SRSF1 promoted NMD of the SRSF2 transcript in which 3′ UTR sequences were recognized as introns (Figure 6B, dark gray bar), as well as the IDUA W402X mutant mRNA (Figure 6B, black bar). In contrast, SRSF1 did not promote NMD of PTC-containing transcripts ATG5, ILK, LYK5, and GPR175 (Figure 6B, white bars) or those in the PTC-upon-inclusion category (Figure 6B, light gray bars).
To determine whether the PTC-containing endogenous isoforms that were reduced by overexpressing SRSF1 are true NMD targets, we treated cells with cycloheximide to block NMD and SRSF1’s effect. In addition, we tested the role of RRM2. First, the cycloheximide treatment abrogated NMD of eight of the PTC-containing isoforms (Figure 6B, arrows), four of which are SRSF1’s targets; in all these four cases, SRSF1’s effect was also inhibited. Only one putative SRSF1 target (ANKRD54) was not affected by cycloheximide, suggesting that it is not a bona fide NMD target. Second, deletion of RRM2 abrogated the effect of SRSF1 on three true NMD targets we tested (Figure 6C; TOMM34, NUBP2, and SRSF2).
To investigate whether the transcript-specific response to overexpression of SRSF1 could be due to the presence or absence of SRSF1-binding sites downstream of their PTCs, we analyzed enhanced cross-linking immunoprecipitation (eCLIP) data from K562 cells (Van Nostrand et al., 2016). Of the 12 genes listed, 9 were found to have SRSF1 occupancy downstream of a PTC, based on the eCLIP data; these include TOMM34, NUBP2, and SRSF2, which are bona fide NMD targets that markedly responded to SRSF1 overexpression (Table S2).
These results indicate that the effect of SRSF1 on NMD is not limited to HBB and GPX1 reporters but rather can be extended to a subset of PTC-containing endogenous transcripts. Much like the effect of SRSF1 on the HBB-T39 reporter, SRSF1’s effect on endogenous NMD targets was also dependent on RRM2.
DISCUSSION
We previously found that SRSF1 stimulates NMD (Zhang and Krainer, 2004). Here we investigated the underlying mechanism, using a combination of cell-based functional assays and biochemical assays. We obtained evidence that SRSF1 enhances UPF1 binding to the mRNA in the nucleus and is involved in UPF1 dephosphorylation after an initial round of translation in the cytoplasm.
In addition, SRSF1’s function in NMD is shared by other SR proteins, indicating an additional function for this class of multi-tasking splicing factors. While investigating SRSF1’s mode of action, we found that it resembles that of EJC components and NMD factors, whose binding downstream of a PTC promotes NMD. We showed this by tethering SRSF1 to the transcript at different positions, which indicated that SRSF1 promotes NMD more efficiently when positioned downstream of a PTC. Moreover, we also learned that SRSF1 needs to be bound to a transcript to promote its NMD. We showed this both by tethering SRSF1 to the transcript and by the lack of NMD-stimulatory function of the ΔRRM1 domain deletion mutant, which could be restored by forcing its binding to the transcript through tethering. SRSF1’s mostly nuclear localization is also key to its NMD function, as deleting its RS domain—which is necessary for efficient nuclear accumulation (Cáceres et al., 1998)—rendered SRSF1 partially inactive in NMD. This observation, together with the fact that a nuclear-retained form of SRSF1 maintains the NMD-stimulatory activity (Zhang and Krainer, 2004), led us to surmise that a nuclear function of SRSF1 is key to promoting NMD.
Searching for NMD-related factors that could mediate SRSF1’s activity in NMD, we found that SRSF1 has an intimate connection in the nucleus with EJC core components and associated factors, including UPF3B. Moreover, SRSF1’s function in NMD is enhanced by the presence of introns, suggesting an important role of splicing and/or EJC deposition. This tight relationship between SRSF1 and the EJC suggests that SRSF1 can promote the recruitment of the EJC to the transcript. The findings that point to cis-acting sequences in Drosophila transcripts as being essential for the recruitment of EJCs and for NMD (Saulière et al., 2010) argue in favor of this conclusion, as these cis-acting elements could represent splicing factor-binding sites. Moreover, non-canonical EJC deposition on transcripts notably coincides with locations that are rich in SRSF1 ESE motifs (Saulière et al., 2012; Singh et al., 2012). Together, these observations implicate SRSF1 in EJC deposition and/or stabilization on transcripts, and they indicate that the presence of the EJC enhances SRSF1’s effect on NMD. In turn, these findings could explain the variability in NMD efficiency observed for disease-associated nonsense mutations; for example, transcripts with more SRSF1-binding sites downstream of a PTC could have additional or more tightly bound EJCs, which could drive more efficient NMD.
Despite the interactions we observed between SRSF1 and the EJC/UPF3B, our findings suggest that neither individual EJC components nor UPF3B is essential for the effect of SRSF1 on NMD, although they might contribute to the overall effect. We also established that UPF2 is not involved in mediating the effect of SRSF1 on NMD. As the recruitment of UPF1 to transcripts during translation is thought to be mediated in part by the EJC-UPF3B-UPF2 complex (Chamieh et al., 2008), the lack of contribution of either of these factors to the effect of SRSF1 on NMD was puzzling; however, alternative NMD routes can be followed in the absence of UPF3B or UPF2 (Chan et al., 2007, Gehring et al., 2005, respectively), and UPF1 can bind mRNA even when translation is inhibited (Hogg and Goff, 2010). These findings led us to believe that SRSF1 can bypass the recruitment to the transcript of at least UPF2, by directly interacting with UPF1. Indeed, our findings suggest that this interaction occurs in both the nucleus and the cytoplasm and is independent of RNA. We believe that the nuclear interaction is key to SRSF1’s effect on NMD, as proper nuclear localization of SRSF1 is essential to promote NMD. It is important to note that, unlike UPF1, which interacts with SRSF1 both in the nucleus and the cytoplasm, EJC components and UPF3B appear to interact with SRSF1 only in the nucleus. Moreover, we found that the association of UPF1 with the EJC is increased in the nucleus but further reduced in the cytoplasm when SRSF1 is overexpressed. This finding suggests that higher occupancy of and/or more stably pre-bound UPF1 accelerates NMD, by releasing EJCs from the mRNA.
Evidence of cytoplasmic interaction between SRSF1 and UPF1 suggests that SRSF1’s involvement in NMD goes beyond recruiting/stabilizing UPF1 in the nucleus. Once translation begins, triggering UPF1 phosphorylation, SRSF1 appears to be involved in its dephosphorylation. This potential function of SRSF1 was revealed in our experiments by the accumulation of hyperphosphorylated UPF1 associated with expression of the SRSF1 ΔRRM2 mutant. It is possible that the lack of interaction between an RRM2 mutant and PP2A (Michlewski et al., 2008) could lock UPF1 in a hyperphosphorylated state, making it NMD inactive. We also found that SRSF1 interacts with SMG7 and this interaction is reduced by deleting RRM2, suggesting that SRSF1, PP2A, and SMG5/SMG7 can form a complex that could potentially facilitate UPF1 dephosphorylation.
The proposed mechanism underlying SRSF1’s involvement in NMD is summarized in Figure 7. We propose that SRSF1’s journey through the NMD pathway starts with enhancing the binding of UPF1 to the spliced mRNA in the nucleus. As the mRNP is exported to the cytoplasm, the increased presence of UPF1 may accelerate the NMD process (Figure 7A). Our findings are consistent with the suggestion that SRSF1 promotes NMD by switching the pioneer round of translation from cytoplasmic to nuclear associated (Sato et al., 2008), such that NMD occurs sooner, as the mRNP comes out of the nuclear pore in the presence of SRSF1, as opposed to later on elsewhere in the cytoplasm. Once a PTC is encountered, SMG1 phosphorylates UPF1 at the N and C termini, and these phosphorylation sites function as docks for SMG6 and SMG5/SMG7. At this point, SRSF1 assists with UPF1 dephosphorylation through interactions with SMG7 and PP2A, possibly mediated by RRM2 (Figure 7B), which allows for UPF1 release and its recycling into the SURF complex. Progressive hyperphosphorylation of UPF1, while it remains bound to a transcript, was proposed to act as a molecular clock that enhances the NMD activity of UPF1 with time, ensuring the decay of the transcript even when the downstream NMD effectors are relatively depleted (Durand et al., 2016). The hyperphosphorylated UPF1 seen in our experiments could, therefore, represent a loss of dephosphorylation activity and/or stalling of the NMD pathway through another mechanism that depends on SRSF1 activity.
Figure 7. SRSF1’s Functions in NMD.
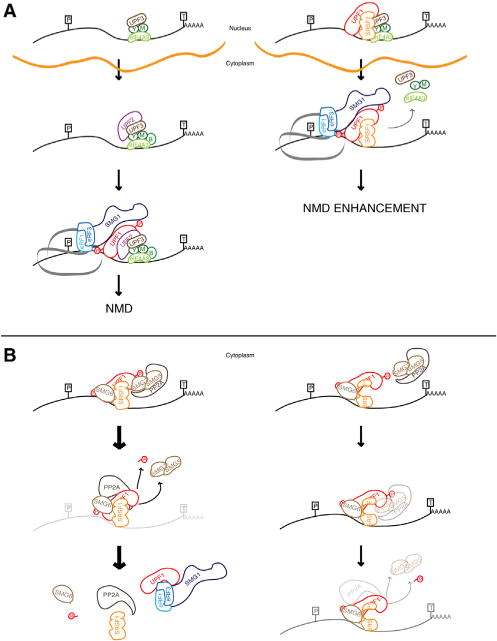
(A) A simplified diagram of the NMD pathway is shown on the left. On the right, SRSF1 recruits UPF1 and interacts with the EJC and UPF3B in the nucleus. As the mRNP complex is exported to the cytoplasm, translation occurs in the presence of UPF1, bypassing UPF2 recruitment and initiating NMD.
(B) On the left, SRSF1 assists in p-UPF1 dephosphorylation and release from the transcript via interactions with p-UPF1, SMG7, and PP2A (Michlewski et al., 2008). On the right, SRSF1 lacking RRM2 reduces the interaction between the RRM2 mutant and SMG7, decreasing PP2A-mediated dephos-phorylation of UPF1 and leading to a hyperphosphorylated form of UPF1. P, PTC; T, termination codon; green Y, Y14; green M, MAGOH; red Ps, phosphorylated residues in UPF1; orange line, nuclear membrane.
Even though we dissected the functions of SRSF1 in NMD using an HBB reporter, we also found that SRSF1 promotes NMD of ~50% of the endogenous transcripts we tested that are true NMD targets. Moreover, this effect was abrogated by the deletion of RRM2. These endogenous transcripts, however, did not respond universally to the presence of SRSF1, which could potentially be attributed to the presence or absence of SRSF1-binding sites downstream of the PTC. These PTC-containing transcripts arise from different mechanisms, such as alternative splicing events, splicing coupled to NMD, and disease-causing mutations. These findings imply that SRSF1 has a broad function in NMD.
Overall, our work has uncovered the mechanism by which SRSF1 increases NMD efficiency, shedding light onto the connections between pre-mRNA splicing and NMD.
EXPERIMENTAL PROCEDURES
Further details and lists of reagents are in the Supplemental Experimental Procedures.
Plasmids
Plasmids HBB-WT and Ter39 were previously reported (Zhang and Krainer, 2004). The MS2-binding sites and removal of each intron were generated by PCR-mutagenesis. pCIneo-T7-MS2-SRSF1, -SRSF2, -SRSF3, -SRSF4, -SRSF6, and -SRSF9 were constructed by cloning the open reading frame (ORF) of each SR protein into BamHI/EcoRI sites downstream of MS2.
Transfection and RT-PCR
HeLa or U2OS cells were maintained in DMEM with 10% fetal bovine serum at 37°C in 5% CO2. Plasmids were transfected using FuGENE 6 (Promega), and RNA was extracted and analyzed by radioactive RT-PCR with 32P-α-dCTP for 26 cycles. Products were analyzed by 5% PAGE, and bands were quantified using a Typhoon FLA 7000 (GE Healthcare).
RNAi
siRNAs (100 nM) were transfected into HeLa cells using Oligofectamine (Invitrogen) for 72 hr.
Cell Fractionation, IP, and Western Blotting
Whole-cell extracts or nuclear and cytoplasmic fractions were obtained from two 15-cm plates of HeLa cells per condition, using IP-lysis buffer or centrifugation and high salt concentration, respectively. IP was performed with 200 mM NaCl, using anti-T7, anti-FLAG, or anti-V5 antibodies coupled to Dy-nabeads Protein G (Invitrogen), for 1 hr at 4°C. Western blotting was performed using IRDye-680LT- or 800CW-labeled secondary antibodies for detection in a LI-COR Odyssey instrument.
Protein Purification and In Vitro Pull-down
MBP, MBP-SRSF1, GST, and GST-UPF1 were purified from E. coli. Purification and pull-down protocols are described in the Supplemental Experimental Procedures.
Statistical Analysis
Results are presented as mean ± SEM. The means were compared using the Mann-Whitney U test, with p ≤ 0.05 as the cutoff for statistical significance.
Supplementary Material
Highlights.
SRSF1 promotes NMD when positioned downstream of a premature termination codon
Splicing and/or EJC deposition enhance the effect of SRSF1 on NMD
SRSF1 enhances NMD by promoting UPF1 binding to nuclear-associated mRNAs
Acknowledgments
We thank L. Maquat, E. Izaurralde, S. Ohno, and J. Lykke-Andersen for expression vectors; p-UPF1 antibodies; and UPF1, UPF2, and UPF3B antibodies. We also thank L. Maquat, M. Moore, J. Cáceres, and former and current lab members, especially O. Anczuków, M. Akerman, S. Das, and K.-T. Lin, for helpful advice. This work was supported by NIH grant R37GM42699 to A.R.K. and by Harvey Karp Foundation and Canadian Institutes of Health Research fellowships to I.A. We acknowledge assistance from the CSHL Shared Resources, funded in part by NCI Cancer Center Support Grant 5P30CA045508.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.04.039.
AUTHOR CONTRIBUTIONS
I.A. and A.R.K. designed the study. I.A., T.T.N., and M.A.R. performed cell culture-based experiments. J.T.-N. and H.R. performed in vitro pull-down experiments. O.F. performed IP-MS analysis. I.A., T.T.N., and A.R.K. wrote the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Akerman M, Fregoso OI, Das S, Ruse C, Jensen MA, Pappin DJ, Zhang MQ, Krainer AR. Differential connectivity of splicing activators and repressors to the human spliceosome. Genome Biol. 2015;16:119. doi: 10.1186/s13059-015-0682-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Cáceres JF. Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, 2nd, Wilkinson MF. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner GM, Cléry A, Allain FH. RRM-RNA recognition: NMR or crystallography and new findings. Curr Opin Struct Biol. 2013;23:100–108. doi: 10.1016/j.sbi.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Durand S, Franks TM, Lykke-Andersen J. Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat Commun. 2016;7:12434. doi: 10.1038/ncomms12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009;7:e1000120. doi: 10.1371/journal.pbio.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudikote JP, Imam JS, Garcia RF, Wilkinson MF. RNA splicing promotes translation and RNA surveillance. Nat Struct Mol Biol. 2005;12:801–809. doi: 10.1038/nsmb980. [DOI] [PubMed] [Google Scholar]
- Hanamura A, Cáceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- Hauer C, Sieber J, Schwarzl T, Hollerer I, Curk T, Alleaume AM, Hentze MW, Kulozik AE. Exon Junction Complexes Show a Distributional Bias toward Alternatively Spliced mRNAs and against mRNAs Coding for Ribosomal Proteins. Cell Rep. 2016;16:1588–1603. doi: 10.1016/j.celrep.2016.06.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Izaurralde E. Making sense of nonsense. Nat Struct Mol Biol. 2013;20:651–653. doi: 10.1038/nsmb.2601. [DOI] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell. 2010;143:379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug N, Longman D, Cáceres JF. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr Opin Genet Dev. 2011;21:422–430. doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamachi N, Salam KA, Suzuki Y, Akimitsu N. A GC-rich sequence feature in the 3′ UTR directs UPF1-dependent mRNA decay in mammalian cells. Genome Res. 2017;27:407–418. doi: 10.1101/gr.206060.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol. 2004;11:330–337. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 2016;129:461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Tardiff DF, Kadener S, Rosbash M. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 2006;20:2055–2066. doi: 10.1101/gad.1434706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Saulière J, Wang Z. The exon junction complex as a node of post-transcriptional networks. Nat Rev Mol Cell Biol. 2016;17:41–54. doi: 10.1038/nrm.2015.7. [DOI] [PubMed] [Google Scholar]
- Linde L, Boelz S, Neu-Yilik G, Kulozik AE, Kerem B. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur J Hum Genet. 2007;15:1156–1162. doi: 10.1038/sj.ejhg.5201889. [DOI] [PubMed] [Google Scholar]
- Llorca O. Structural insights into nonsense-mediated mRNA decay (NMD) by electron microscopy. Curr Opin Struct Biol. 2013;23:161–167. doi: 10.1016/j.sbi.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Long JC, Cáceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24:1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Li X. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA. 2001;7:445–456. doi: 10.1017/s1355838201002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metze S, Herzog VA, Ruepp MD, Mühlemann O. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA. 2013;19:1432–1448. doi: 10.1261/rna.038893.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Miller JN, Pearce DA. Nonsense-mediated decay in genetic disease: friend or foe? Mutat Res Rev Mutat Res. 2014;762:52–64. doi: 10.1016/j.mrrev.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40:1251–1266. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley KJ, Yario TA, Steitz JA. Association of Argonaute proteins and microRNAs can occur after cell lysis. RNA. 2012;18:1581–1585. doi: 10.1261/rna.034934.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Hosoda N, Maquat LE. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol Cell. 2008;29:255–262. doi: 10.1016/j.molcel.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Saulière J, Haque N, Harms S, Barbosa I, Blanchette M, Le Hir H. The exon junction complex differentially marks spliced junctions. Nat Struct Mol Biol. 2010;17:1269–1271. doi: 10.1038/nsmb.1890. [DOI] [PubMed] [Google Scholar]
- Saulière J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonquèze O, Audic Y, Paillard L, Roest Crollius H, Le Hir H. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19:1124–1131. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott HS, Litjens T, Nelson PV, Thompson PR, Brooks DA, Hopwood JJ, Morris CP. Identification of mutations in the alpha-L-iduronidase gene (IDUA) that cause Hurler and Scheie syndromes. Am J Hum Genet. 1993;53:973–986. [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Mühlemann O. Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA. 2009;15:1265–1273. doi: 10.1261/rna.1672509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange TØ, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


