Abstract
Numerous genetic alterations of HSA 11q13 are found frequently in several cancer types, including breast cancer (BC). The 11q13 locus harbors FADS2 encoding Δ6 desaturation which is not functional in several cancer cell lines, including hormone positive MCF7 BC cells. In vitro, the non-functional FADS2 activity unmasks 18:2n-6 elongation to 20:2n-6 and 5-desaturation by FADS1 to yield 5Z,11Z,14Z-20:3 (sciadonic acid) rather than 5Z,8Z,11Z,14Z-20:4 (arachidonic acid). In this pilot study we aimed to determine whether 5,11,14-20:3 appears in vivo in hormone positive human BC tissue. Fatty acids were profiled in surgically removed human breast tumor and adjacent normal tissue (n=9). Sciadonic acid was detected in three of nine breast tumor samples and was below detect limits in normal breast tissue. The internal Δ8 double bond of arachidonic acid is required for normal eicosanoid synthesis but is missing in sciadonic acid. This pilot study demonstrates for the first time in vivo sciadonic acid in hormone positive BC tissue, warranting a larger survey study to further evaluate its appearance and the functional implications.
Keywords: Breast Cancer, Eicosanoid, Docosanoid, Estrogen, Fatty acid desaturase 2, Cell signaling
INTRODUCTION
Human breast cancer (BC) is the most common cancer among women in US and worldwide, with 2.4 million new cases diagnosed in 2015 [1, 2]. It is the second most common cause of death from cancer in women in US, with estimated deaths of 40,610 in 2017 [2]. The large percentage (~70%) of BC are endocrine-related and ovarian sex hormone estrogen is regarded as both initiator and promoter of BC [3–6]. Another ovarian sex hormone progesterone and its metabolites are also considered to promote BC [7, 8]. The BC expressing estrogen receptors (ER) and/or progesterone receptors (PR) responds to hormone therapy [9].
The human chromosome 11q13 (HSA 11q13) region is well known to be a major cancer hotspot, harboring potential oncogenic driver(s) [10–12]. Various genetic alterations of 11q13 (amplifications, deletions, insertions and translocations) are frequently found events in several cancer types, such as BC, ovarian cancer, cervical cancer, numerous types of squamous cell carcinoma, endocrine tumors, lymphomas and myelomas [13–20]. The biologically active eicosanoids and their metabolites are linked to tumor progression via several mechanisms including dysregulation of cell signaling [21–24]. In humans fatty acid desaturases, FADS1, FADS2, and FADS3 are three enzyme-coding genes localized to human chromosome 11q13 [25] and are required for the biosynthesis of 20 and 22 carbon polyunsaturated fatty acids (PUFA) that are direct cell signaling eicosanoid and docosanoid precursors [26–28]. In several cancer cell lines, including hormone positive MCF7 BC cells, FADS2 encoded Δ6 desaturation is not functional [29–31].
As FADS2 catalyzes the first critical step for the eicosanoid and docosanoid precursor biosynthesis, we hypothesized the depletion/dysregulation of normal eicosanoid and/or docosanoid cell signaling precursor milieu is a potential oncogenic driving event in certain cancer types, including BC. Wild type MCF7 cells have no bioactivity towards the polyunsaturated fatty acids 18:2n-6 and 18:3n-3, whereas transient transfection with FADS2 restores activity [29]. In normal cells, Δ6 desaturation catalyzing 18:2n-6 → 18:3n-6 masks the lower activity competing elongation pathway 18:2n-6 → 20:2n-6. In MCF7 cells, the absence of Δ6 desaturation activity unmasks elongation to 20:2n-6 (11,14-20:2), which then accumulates at modest levels. FADS1 coded Δ5 desaturation then operates on 11,14-20:2 → 5,11,14-20:3, sciadonic acid, which is otherwise below detection limits in normal tissue [29, 32, 33]. The main purpose of the present pilot study is to test for the appearance of the rare (all cis double bonds) 5Z,11Z,14Z-20:3 fatty acid in hormone positive BC tissue in vivo, thereby replicating our in vitro cell culture findings to provide insight to possible metabolic derangement in fatty acids of hormone positive BC.
Materials and Methods
Study Approvals
The study was approved by The Cornell University Institutional Review Board and The Cayuga Medical Center at Ithaca Institutional Review Board for human participants. A written informed consent was obtained from all nine women participants in this pilot study. All the nine participants were diagnosed with estrogen receptor (ER) and progesterone receptor (PR) positive breast tumors. From each participant surgically removed fresh 50 mg of the breast tumor and 50 mg of the adjacent noncancerous breast tissues were used for fatty acid analysis.
Fatty acid analysis
Breast tumor and adjacent normal breast tissues (Figure 1) were used for fatty acid extraction and analysis. Fatty acid methyl esters (FAME) were prepared using modified one-step method of Garces and Mancha [34] and were analyzed quantitatively using a Hewlett Packard 5890 series II gas chromatograph-flame ionization detector (GC-FID) equipped with a BPX 70 column (25 m, 0.22-mm inner diameter, 0.25 μm film; SGE, Austin, TX) using an equal weight mixture for response factor calibration. The peak structures were positively identified by GC-covalent adduct chemical ionization tandem mass spectrometry (GC-CACI-MS/MS) as previously described [35, 36].
Figure 1.
Surgically removed human breast tissue samples. A) Normal breast tissue B) Breast cancer tissue. Residual blue dye is present in some samples.
Chemicals
Solvents and reagents for fatty acid extractions were HPLC grade from Sigma-Aldrich (St. Louis, MO, USA) or Burdick & Jackson (Muskegon, MI, USA).
Results and Discussion
Previously, we have shown that ER and PR positive MCF7 breast cancer cells lack the initial FADS2-encoded desaturation activity step in the biosynthesis of long chain polyunsaturated fatty acids from the 18 carbon precursors that are abundant in the human diet. In vitro MCF7 cells biosynthesize 5,11,14-20:3 via elongation and FADS1 activity [29]. The 5,11,14-20:3 fatty acid is structurally identical to arachidonic acid (ARA; 20:4n-6; 5,8,11,14-20:4) except it lacks the internal Δ8 double bond required for prostaglandin and leukotriene synthesis, among other eicosanoids. A clear question is whether 5,11,14-20:3 is present in vivo, and specifically in ER and PR positive breast cancer tissue. We find 5,11,14-20:3 presence in 3 out of 9 breast tumor samples analyzed (Figure 2, Top). None of the adjacent noncancerous samples showed 5,11,14-20:3 above detection limits (Figure 2, Bottom).
Figure 2.
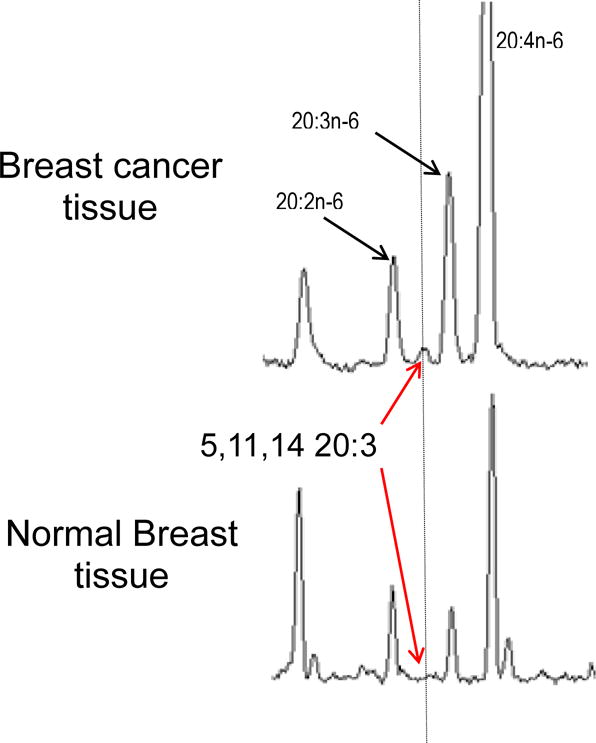
Gas chromatography results, normalized to the peak for 20:2n-6, from breast cancer and normal breast tissues. Top) Breast cancer tissue. Unusual 5,11,14-20:3 sciadonic acid detected in BC tissue at about 10% (area percent) of the precursor 20:2. 5,11,14-20:3 was found in three of nine samples. Bottom) Normal breast tissue. 5,11,14-20:3 is below detection limits.
Figure 3A presents the CACI-MS spectrum of 20:3, showing the familiar peaks at m/z 374, 321, 289, and 271, corresponding to the [M+54]+, [MH]+, [M+54-32]+, and [MH+54-32-18]+ ions, respectively, characteristic of a 20:3 FAME. Figure 3B is the collisionally activated dissociation spectrum (MS/MS) of [M+54]+ of 5,11,14-20:3, yielding ions at m/z 304 and 192 corresponding to the α and ω diagnostic ions, respectively, positively identifying the internal diene double bond structure of this fatty acid. The additional peak at m/z 272 locates the isolated 5-6 double bond.
Figure 3.
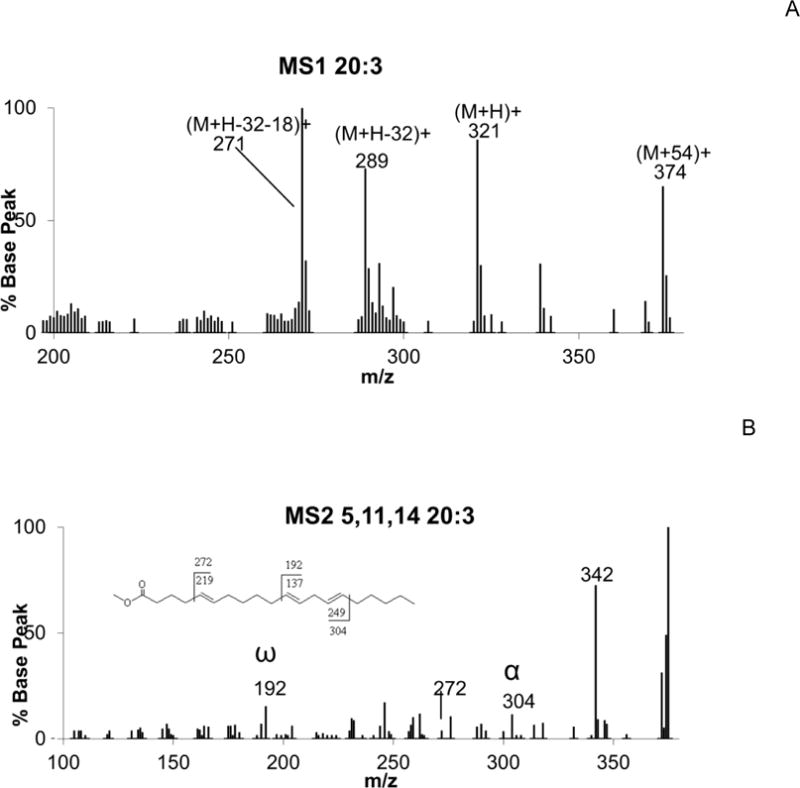
CACI-MS1 and CACI-MS2 spectra of 20:3 FAME. A) CACI-MS1 spectrum showing m/z 374, 321, 289, and 271 characteristic ions of 20:3 FAME. B) CACI-MS/MS spectrum of the m/z 374 ion positively identifying 5,11,14-20:3 based on the detection of diagnostic ions m/z 304 and 192 for the diene and m/z 272 vinylic to the isolated double bond, for the peak at the expected chromatographic retention time.
Dysfunctional endogenous and de novo fatty-acid synthesis has long been recognized as a characteristic of human cancers, including BC [37, 38]. The detection of 5,11,14-20:3 in noncancerous tissues has not been reported, however, it has been detected in mammalian tissues and cells in animals fed 11,14-20:2 or 5,11,14-20:3 [29, 32, 33]. Feeding mice with small quantities of 5,11,14–20:3 significantly replaced the normal eicosanoid precursor ARA in the phosphatidylinositol (PI) fraction [39]. The acyl composition of PI is highly resistant to dietary modifications reflecting PI’s role as a major messaging lipid. Replacement of ARA in PI pools by 5,11,14-20:3 may lead to the production of novel secondary messengers [39]. In another mouse study, feeding 5,11,14-20:3 for a 2-week period resulted in 50% reduction in the levels of ARA in hepatic PI fractions. This reduction in the ARA content may have an influence on eicosanoid signaling since eicosanoid synthesis has long been known to be related to the abundance of ARA. In the same study 5,11,14-20:3 was extensively incorporated into hepatic phosphatidylinositol bisphosphate (PIP2), a precursor of second messengers. Similarly, 5,11,14-20:3 reduced the ARA content of the PI fraction in the HepG2 cells which may have influence on PI-originating bioactive lipids [40, 41]. The phosphoinositide signaling pathway is the most frequently altered in BC and alterations in this pathway are associated with resistance to hormone therapy [42, 43]. PhospholipaseA2 has poor affinity for 5,11,14-20:3, suggesting that 5,11,14-20:3 accumulate in membrane phospholipid (PL) pools even with moderate availability [44].
Swiss 3T3 cells, when cultured with 5,11,14-20:3 and stimulated using 100 nM of bombesin, produced 1-stearoyl-2-sciadonoyl-glycerol. 1-Stearoyl-2-sciadonoyl-glycerol was found to activate diacylglycerol-protein kinase C (PKC) signaling [45]. Similarly, in NG108-15 cells 1-stearoyl-2-sciadonoyl-glycerol (G) induced monoacylglycerol signaling, through a CBI receptor-dependent mechanism [46]. Topical application of 5,11,14-20:3 to mouse ear reduced ARA induced edema [47]. In colon cancer and a few other cell lines, exogenous ARA treatment caused apoptosis, suggestive of unesterified ARA signals induction of apoptosis [48–50].
Several studies have reported a positive correlation between amplification/overexpression of certain genes on 11q13 and estrogen receptor (ER) positive status [51–53]. Microdissection of tissue sections from same patients (15 of 15) showed loss of heterozygosity (LOH) of the identical allele at 11q13 in early stage and invasive BC, suggesting that important tumor suppressor gene(s) at 11q13 are associated with the development of BC [54] and micro-cell mediated transfer of HSA11 into MCF7 cells reduced tumorigenicity [55]. FADS2 is a multifunctional enzyme with known substrate specificities for 16, 18, 20, 22 and 24 carbon fatty acids [26]. Functional loss of FADS2 affects saturated, monounsaturated and polyunsaturated fatty acid levels, directly influencing eicosanoid and docosanoid pathways and signaling. FADS2 has not been carefully considered as an 11q13 candidate to be a tumor suppressor in neoplastic disorders. The dysregulation of ARA-based eicosanoid signaling has been implicated in the development and progression of BC [56, 57].
Conventional electrospray-MS based lipidomics does not discern fatty acid isomers (positional, geometric, or branched chain) and therefore 5,11,14-20:3 would only be detected as a mixture of 20:3 isomers, normally dominated by 20:3n-6 (dihomo-gamma-linolenic acid). GC-CACI-MS/MS method, developed in our lab, identifies the position and geometry of double bonds directly in FAME. Alternative online methods for double bond localization by electrospray MS are in progress but have not been comprehensively demonstrated for polyunsaturates of complex double bond structure[58].
Our ability to collect samples limited this pilot study. Microdissection of tumor tissues to acquire maximum tumor load was not carried out because at least 50 mg of tissue was required for fatty acid analysis. As the microdissections were not carried out, piecing of tumor versus necrotic/apoptotic and normal tissue was not possible. In our study the ARA peak was found to be several fold larger in the tumor breast tissue compared to the normal breast tissue (Figure 2). We normalized the peaks to 20:2n-6 FA, the immediate precursor of 5,11,14-20:3. It is well known that the tumor microenvironment contributes to tumor heterogeneity. The cellular environment of a developing tumor consists of a scaffold of extracellular matrix composed of a dynamic variety of non-cancerous immune and stromal cells required for tumor growth, invasion and metastasis [59]. Several studies have indicated that unesterified arachidonic acid (AA) in cells can serve as a second messenger and yields a proapoptotic signal [48, 60, 61]. Tumor Necrosis Factor-induced ARA release in tumor cells has been reported [62] and a recent study has shown accumulation of ARA on the outer edge of the colorectal cancer [63]. Normal as well as necrotic/apoptotic tissue may have dominated the tumor sample in the six samples which we did not find sciadonic acid. Future survey studies with larger sample size and maximum tumor load samples are warranted to delineate the appearance of 5,11,14-20:3 in breast tumor tissues. Existing knowledge about the upstream synthesis pathway for 5,11,14-20:3 points to a genetic aberration at 11q13 and a specific switch to an alternative pathway for its synthesis. Implication for deranged signaling due to interference with ARA derived normal signals suggests that the presence of this unusual fatty acid in membranes may have functional implications for cell-to-cell signaling.
Highlights.
Sciadonic acid, 5Z,11Z,14Z-20:3, is an arachidonic acid analogue in cells missing 6-desaturation.
For the first time, sciadonic acid is detected in human breast cancer but not in adjacent healthy tissue.
Sciadonic acid is not a substrate for prostaglandins and its substitution for arachidonic acid may affect cell-cell signaling.
Acknowledgments
Sources of Support. This work was supported by NIH grant R01 AT007003 from the National Center for Complementary and Integrative Health (NCCIH) and the Office of Dietary Supplements (ODS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
JTB and KSDK conceived the research; KSDK, JTB, CF and SD designed the research protocol; CF, DS, DAS, JM and SD executed clinical aspects and sampling, HP and JYZ executed the laboratory research; JTB and KSDK contributed new reagents/analytic tools; HP, JYZ, PL, KSDK and JTB analyzed and interpreted the data; KSDK and JTB wrote the first draft; all authors approved the final draft.
Conflict of Interest
All authors declare no conflict of interest.
References
- 1.C. Global Burden of Disease Cancer. Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, Fleming T, Forouzanfar MH, Hancock J, Hay RJ, Hunter-Merrill R, Huynh C, Hosgood HD, Johnson CO, Jonas JB, Khubchandani J, Kumar GA, Kutz M, Lan Q, Larson HJ, Liang X, Lim SS, Lopez AD, MacIntyre MF, Marczak L, Marquez N, Mokdad AH, Pinho C, Pourmalek F, Salomon JA, Sanabria JR, Sandar L, Sartorius B, Schwartz SM, Shackelford KA, Shibuya K, Stanaway J, Steiner C, Sun J, Takahashi K, Vollset SE, Vos T, Wagner JA, Wang H, Westerman R, Zeeb H, Zoeckler L, Abd-Allah F, Ahmed MB, Alabed S, Alam NK, Aldhahri SF, Alem G, Alemayohu MA, Ali R, Al-Raddadi R, Amare A, Amoako Y, Artaman A, Asayesh H, Atnafu N, Awasthi A, Saleem HB, Barac A, Bedi N, Bensenor I, Berhane A, Bernabe E, Betsu B, Binagwaho A, Boneya D, Campos-Nonato I, Castaneda-Orjuela C, Catala-Lopez F, Chiang P, Chibueze C, Chitheer A, Choi JY, Cowie B, Damtew S, Neves J das, Dey S, Dharmaratne S, Dhillon P, Ding E, Driscoll T, Ekwueme D, Endries AY, Farvid M, Farzadfar F, Fernandes J, Fischer F, TT GH, Gebru A, Gopalani S, Hailu A, Horino M, Horita N, Husseini A, Huybrechts I, Inoue M, Islami F, Jakovljevic M, James S, Javanbakht M, Jee SH, Kasaeian A, Kedir MS, Khader YS, Khang YH, Kim D, Leigh J, Linn S, Lunevicius R, Razek HMA El, Malekzadeh R, Malta DC, Marcenes W, Markos D, Melaku YA, Meles KG, Mendoza W, Mengiste DT, Meretoja TJ, Miller TR, Mohammad KA, Mohammadi A, Mohammed S, Moradi-Lakeh M, Nagel G, Nand D, Nguyen Q Le, Nolte S, Ogbo FA, Oladimeji KE, Oren E, Pa M, Park EK, Pereira DM, Plass D, Qorbani M, Radfar A, Rafay A, Rahman M, Rana SM, Soreide K, Satpathy M, Sawhney M, Sepanlou SG, Shaikh MA, She J, Shiue I, Shore HR, Shrime MG, So S, Soneji S, Stathopoulou V, Stroumpoulis K, Sufiyan MB, Sykes BL, Tabares-Seisdedos R, Tadese F, Tedla BA, Tessema GA, Thakur JS, Tran BX, Ukwaja KN, Uzochukwu BSC, Vlassov VV, Weiderpass E, Terefe M Wubshet, Yebyo HG, Yimam HH, Yonemoto N, Younis MZ, Yu C, Zaidi Z, Zaki MES, Zenebe ZM, Murray CJL, Naghavi M. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Lumachi F, Brunello A, Maruzzo M, Basso U, Basso SM. Treatment of estrogen receptor-positive breast cancer. Curr Med Chem. 2013;20:596–604. doi: 10.2174/092986713804999303. [DOI] [PubMed] [Google Scholar]
- 4.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 5.Breast cancer and hormone replacement therapy: collaborative reanalysis of data from 51 epidemiological studies of 52,705 women with breast cancer and 108,411 women without breast cancer. Collaborative Group on Hormonal Factors in Breast Cancer. Lancet. 1997;350:1047–1059. [PubMed] [Google Scholar]
- 6.Winchester DJ. Hormone replacement therapy: a promoter and modulator of breast cancer. Ann Surg Oncol. 2004;11:9–10. doi: 10.1007/BF02524338. [DOI] [PubMed] [Google Scholar]
- 7.Li CI, Malone KE, Porter PL, Weiss NS, Tang MT, Cushing-Haugen KL, Daling JR. Relationship between long durations and different regimens of hormone therapy and risk of breast cancer. JAMA. 2003;289:3254–3263. doi: 10.1001/jama.289.24.3254. [DOI] [PubMed] [Google Scholar]
- 8.Wiebe JP. Progesterone metabolites in breast cancer. Endocr Relat Cancer. 2006;13:717–738. doi: 10.1677/erc.1.01010. [DOI] [PubMed] [Google Scholar]
- 9.Rastelli F, Crispino S. Factors predictive of response to hormone therapy in breast cancer. Tumori. 2008;94:370–383. doi: 10.1177/030089160809400314. [DOI] [PubMed] [Google Scholar]
- 10.Koreth J, Bakkenist CJ, McGee JO. Chromosomes, 11Q and cancer: a review. J Pathol. 1999;187:28–38. doi: 10.1002/(SICI)1096-9896(199901)187:1<28::AID-PATH166>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 11.Wilkerson PM, Reis-Filho JS. The 11q13-q14 amplicon: clinicopathological correlations and potential drivers. Genes Chromosomes Cancer. 2013;52:333–355. doi: 10.1002/gcc.22037. [DOI] [PubMed] [Google Scholar]
- 12.French JD, Ghoussaini M, Edwards SL, Meyer KB, Michailidou K, Ahmed S, Khan S, Maranian MJ, O’Reilly M, Hillman KM, Betts JA, Carroll T, Bailey PJ, Dicks E, Beesley J, Tyrer J, Maia AT, Beck A, Knoblauch NW, Chen C, Kraft P, Barnes D, Gonzalez-Neira A, Alonso MR, Herrero D, Tessier DC, Vincent D, Bacot F, Luccarini C, Baynes C, Conroy D, Dennis J, Bolla MK, Wang Q, Hopper JL, Southey MC, Schmidt MK, Broeks A, Verhoef S, Cornelissen S, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Fasching PA, Loehberg CR, Ekici AB, Beckmann MW, Peto J, dos Santos Silva I, Johnson N, Aitken Z, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Guenel P, Truong T, Laurent-Puig P, Menegaux F, Bojesen SE, Nordestgaard BG, Nielsen SF, Flyger H, Milne RL, Zamora MP, Perez JI Arias, Benitez J, Anton-Culver H, Brenner H, Muller H, Arndt V, Stegmaier C, Meindl A, Lichtner P, Schmutzler RK, Engel C, Brauch H, Hamann U, Justenhoven C, G. Network. Aaltonen K, Heikkila P, Aittomaki K, Blomqvist C, Matsuo K, Ito H, Iwata H, Sueta A, Bogdanova NV, Antonenkova NN, Dork T, Lindblom A, Margolin S, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, kConFab I, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Lambrechts D, Peeters S, Smeets A, Floris G, Chang-Claude J, Rudolph A, Nickels S, Flesch-Janys D, Radice P, Peterlongo P, Bonanni B, Sardella D, Couch FJ, Wang X, Pankratz VS, Lee A, Giles GG, Severi G, Baglietto L, Haiman CA, Henderson BE, Schumacher F, Le Marchand L, Simard J, Goldberg MS, Labreche F, Dumont M, Teo SH, Yip CH, Ng CH, Vithana EN, Kristensen V, Zheng W, Deming-Halverson S, Shrubsole M, Long J, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Devilee P, Seynaeve C, Garcia-Closas M, Figueroa J, Chanock SJ, Lissowska J, Czene K, Klevebring D, Schoof N, Hooning MJ, Martens JW, Collee JM, Tilanus-Linthorst M, Hall P, Li J, Liu J, Humphreys K, Shu XO, Lu W, Gao YT, Cai H, Cox A, Balasubramanian SP, Blot W, Signorello LB, Cai Q, Pharoah PD, Healey CS, Shah M, Pooley KA, Kang D, Yoo KY, Noh DY, Hartman M, Miao H, Sng JH, Sim X, Jakubowska A, Lubinski J, Jaworska-Bieniek K, Durda K, Sangrajrang S, Gaborieau V, McKay J, Toland AE, Ambrosone CB, Yannoukakos D, Godwin AK, Shen CY, Hsiung CN, Wu PE, Chen ST, Swerdlow A, Ashworth A, Orr N, Schoemaker MJ, Ponder BA, Nevanlinna H, Brown MA, Chenevix-Trench G, Easton DF, Dunning AM. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet. 2013;92:489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ormandy CJ, Musgrove EA, Hui R, Daly RJ, Sutherland RL. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res Treat. 2003;78:323–335. doi: 10.1023/a:1023033708204. [DOI] [PubMed] [Google Scholar]
- 14.Brown LA, Kalloger SE, Miller MA, Ie M Shih, McKinney SE, Santos JL, Swenerton K, Spellman PT, Gray J, Gilks CB, Huntsman DG. Amplification of 11q13 in ovarian carcinoma. Genes Chromosomes Cancer. 2008;47:481–489. doi: 10.1002/gcc.20549. [DOI] [PubMed] [Google Scholar]
- 15.Srivatsan ES, Chakrabarti R, Zainabadi K, Pack SD, Benyamini P, Mendonca MS, Yang PK, Kang K, Motamedi D, Sawicki MP, Zhuang Z, Jesudasan RA, Bengtsson U, Sun C, Roe BA, Stanbridge EJ, Wilczynski SP, Redpath JL. Localization of deletion to a 300 Kb interval of chromosome 11q13 in cervical cancer. Oncogene. 2002;21:5631–5642. doi: 10.1038/sj.onc.1205698. [DOI] [PubMed] [Google Scholar]
- 16.Rodrigo JP, Garcia-Carracedo D, Garcia LA, Menendez S, Allonca E, Gonzalez MV, Fresno MF, Suarez C, Garcia-Pedrero JM. Distinctive clinicopathological associations of amplification of the cortactin gene at 11q13 in head and neck squamous cell carcinomas. J Pathol. 2009;217:516–523. doi: 10.1002/path.2462. [DOI] [PubMed] [Google Scholar]
- 17.Debelenko LV, Zhuang Z, Emmert-Buck MR, Chandrasekharappa SC, Manickam P, Guru SC, Marx SJ, Skarulis MC, Spiegel AM, Collins FS, Jensen RT, Liotta LA, Lubensky IA. Allelic deletions on chromosome 11q13 in multiple endocrine neoplasia type 1-associated and sporadic gastrinomas and pancreatic endocrine tumors. Cancer Res. 1997;57:2238–2243. [PubMed] [Google Scholar]
- 18.Rosa-Rosa JM, Pita G, Gonzalez-Neira A, Milne RL, Fernandez V, Ruivenkamp C, van Asperen CJ, Devilee P, Benitez J. A 7 Mb region within 11q13 may contain a high penetrance gene for breast cancer. Breast Cancer Res Treat. 2009;118:151–159. doi: 10.1007/s10549-009-0317-1. [DOI] [PubMed] [Google Scholar]
- 19.Ronchetti D, Finelli P, Richelda R, Baldini L, Rocchi M, Viggiano L, Cuneo A, Bogni S, Fabris S, Lombardi L, Maiolo AT, Neri A. Molecular analysis of 11q13 breakpoints in multiple myeloma. Blood. 1999;93:1330–1337. [PubMed] [Google Scholar]
- 20.Ramos-Garcia P, Ruiz-Avila I, Gil-Montoya JA, Ayen A, Gonzalez-Ruiz L, Navarro-Trivino FJ, Gonzalez-Moles MA. Relevance of chromosomal band 11q13 in oral carcinogenesis: An update of current knowledge. Oral Oncol. 2017;72:7–16. doi: 10.1016/j.oraloncology.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panigrahy D, Greene ER, Pozzi A, Wang DW, Zeldin DC. EET signaling in cancer. Cancer Metastasis Rev. 2011;30:525–540. doi: 10.1007/s10555-011-9315-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Greene ER, Huang S, Serhan CN, Panigrahy D. Regulation of inflammation in cancer by eicosanoids. Prostaglandins Other Lipid Mediat. 2011;96:27–36. doi: 10.1016/j.prostaglandins.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cathcart MC, Lysaght J, Pidgeon GP. Eicosanoid signalling pathways in the development and progression of colorectal cancer: novel approaches for prevention/intervention. Cancer Metastasis Rev. 2011;30:363–385. doi: 10.1007/s10555-011-9324-x. [DOI] [PubMed] [Google Scholar]
- 25.Marquardt A, Stohr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 26.Park HG, Park WJ, Kothapalli KS, Brenna JT. The fatty acid desaturase 2 (FADS2) gene product catalyzes Delta4 desaturation to yield n-3 docosahexaenoic acid and n-6 docosapentaenoic acid in human cells. FASEB J. 2015;29:3911–3919. doi: 10.1096/fj.15-271783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang JY, Kothapalli KS, Brenna JT. Desaturase and elongase-limiting endogenous long-chain polyunsaturated fatty acid biosynthesis. Curr Opin Clin Nutr Metab Care. 2016;19:103–110. doi: 10.1097/MCO.0000000000000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kothapalli KS, Ye K, Gadgil MS, Carlson SE, O’Brien KO, Zhang JY, Park HG, Ojukwu K, Zou J, Hyon SS, Joshi KS, Gu Z, Keinan A, Brenna JT. Positive Selection on a Regulatory Insertion-Deletion Polymorphism in FADS2 Influences Apparent Endogenous Synthesis of Arachidonic Acid. Mol Biol Evol. 2016;33:1726–1739. doi: 10.1093/molbev/msw049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park WJ, Kothapalli KS, Lawrence P, Brenna JT. FADS2 function loss at the cancer hotspot 11q13 locus diverts lipid signaling precursor synthesis to unusual eicosanoid fatty acids. PLoS One. 2011;6:e28186. doi: 10.1371/journal.pone.0028186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaudszus A, Degen C, Barth SW, Klempt M, Schlormann W, Roth A, Rohrer C, Sauerwein H, Sachse K, Jahreis G. Loss of FADS2 function severely impairs the use of HeLa cells as an in vitro model for host response studies involving fatty acid effects. PLoS One. 2014;9:e115610. doi: 10.1371/journal.pone.0115610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grammatikos SI, Subbaiah PV, Victor TA, Miller WM. Diversity in the ability of cultured cells to elongate and desaturate essential (n-6 and n-3) fatty acids. Ann N Y Acad Sci. 1994;745:92–105. doi: 10.1111/j.1749-6632.1994.tb44366.x. [DOI] [PubMed] [Google Scholar]
- 32.Endo Y, Osada Y, Kimura F, Fujimoto K. Effects of Japanese torreya (Torreya nucifera) seed oil on lipid metabolism in rats. Nutrition. 2006;22:553–558. doi: 10.1016/j.nut.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Ullman D, Sprecher H. An in vitro and in vivo study of the conversion of eicosa-11,14-dienoic acid to eicosa-5,11,14-trienoic acid and of the conversion of eicosa-11-enoic acid to eicosa-5,11-dienoic acid in the rat. Biochim Biophys Acta. 1971;248:186–197. doi: 10.1016/0005-2760(71)90006-3. [DOI] [PubMed] [Google Scholar]
- 34.Garces R, Mancha M. One-step lipid extraction and fatty acid methyl esters preparation from fresh plant tissues. Anal Biochem. 1993;211:139–143. doi: 10.1006/abio.1993.1244. [DOI] [PubMed] [Google Scholar]
- 35.Lawrence P, Brenna JT. Acetonitrile covalent adduct chemical ionization mass spectrometry for double bond localization in non-methylene-interrupted polyene fatty acid methyl esters. Anal Chem. 2006;78:1312–1317. doi: 10.1021/ac0516584. [DOI] [PubMed] [Google Scholar]
- 36.Van Pelt CK, Brenna JT. Acetonitrile chemical ionization tandem mass spectrometry to locate double bonds in polyunsaturated fatty acid methyl esters. Anal Chem. 1999;71:1981–1989. doi: 10.1021/ac981387f. [DOI] [PubMed] [Google Scholar]
- 37.Mashima T, Seimiya H, Tsuruo T. De novo fatty-acid synthesis and related pathways as molecular targets for cancer therapy. Br J Cancer. 2009;100:1369–1372. doi: 10.1038/sj.bjc.6605007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaidi N, Lupien L, Kuemmerle NB, Kinlaw WB, Swinnen JV, Smans K. Lipogenesis and lipolysis: the pathways exploited by the cancer cells to acquire fatty acids. Prog Lipid Res. 2013;52:585–589. doi: 10.1016/j.plipres.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger A, German JB. Extensive incorporation of dietary delta-5,11,14 eicosatrienoate into the phosphatidylinositol pool. Biochim Biophys Acta. 1991;1085:371–376. doi: 10.1016/0005-2760(91)90142-5. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka T, Takimoto T, Morishige J, Kikuta Y, Sugiura T, Satouchi K. Non-methylene-interrupted polyunsaturated fatty acids: effective substitute for arachidonate of phosphatidylinositol. Biochem Biophys Res Commun. 1999;264:683–688. doi: 10.1006/bbrc.1999.1559. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka T, Morishige J, Takimoto T, Takai Y, Satouchi K. Metabolic characterization of sciadonic acid (5c,11c,14c-eicosatrienoic acid) as an effective substitute for arachidonate of phosphatidylinositol. Eur J Biochem. 2001;268:4928–4939. doi: 10.1046/j.0014-2956.2001.02423.x. [DOI] [PubMed] [Google Scholar]
- 42.Bunney TD, Katan M. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat Rev Cancer. 2010;10:342–352. doi: 10.1038/nrc2842. [DOI] [PubMed] [Google Scholar]
- 43.Miller TW, Balko JM, Arteaga CL. Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. J Clin Oncol. 2011;29:4452–4461. doi: 10.1200/JCO.2010.34.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenthal MD, Garcia MC, Sprecher H. Substrate specificity of the agonist-stimulated release of polyunsaturated fatty acids from vascular endothelial cells. Arch Biochem Biophys. 1989;274:590–600. doi: 10.1016/0003-9861(89)90474-8. [DOI] [PubMed] [Google Scholar]
- 45.Morishige J, Takai Y, Hirano K, Tanaka T, Satouchi K. Production and protein kinase C activation of diacylglycerols containing polymethylene-interrupted PUFA. Lipids. 2005;40:155–162. doi: 10.1007/s11745-005-1370-8. [DOI] [PubMed] [Google Scholar]
- 46.Nakane S, Tanaka T, Satouchi K, Kobayashi Y, Waku K, Sugiura T. Occurrence of a novel cannabimimetic molecule 2-sciadonoylglycerol (2-eicosa-5′,11′,14′-trienoylglycerol) in the umbrella pine Sciadopitys verticillata seeds. Biol Pharm Bull. 2000;23:758–761. doi: 10.1248/bpb.23.758. [DOI] [PubMed] [Google Scholar]
- 47.Berger A, Monnard I, Baur M, Charbonnet C, Safonova I, Jomard A. Epidermal anti-Inflammatory properties of 5,11,14 20:3: effects on mouse ear edema, PGE2 levels in cultured keratinocytes, and PPAR activation. Lipids Health Dis. 2002;1:5. doi: 10.1186/1476-511X-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao Y, Pearman AT, Zimmerman GA, McIntyre TM, Prescott SM. Intracellular unesterified arachidonic acid signals apoptosis. Proc Natl Acad Sci U S A. 2000;97:11280–11285. doi: 10.1073/pnas.200367597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hori T, Kashiyama S, Hayakawa M, Shibamoto S, Tsujimoto M, Oku N, Ito F. Tumor necrosis factor is cytotoxic to human fibroblasts in the presence of exogenous arachidonic acid. Exp Cell Res. 1989;185:41–49. doi: 10.1016/0014-4827(89)90035-9. [DOI] [PubMed] [Google Scholar]
- 50.Rizzo MT, Regazzi E, Garau D, Akard L, Dugan M, Boswell HS, Rizzoli V, Carlo-Stella C. Induction of apoptosis by arachidonic acid in chronic myeloid leukemia cells. Cancer Res. 1999;59:5047–5053. [PubMed] [Google Scholar]
- 51.Parl FF. Estrogens, Estrogen Receptor, and Breast Cancer. Amsterdam: IOS Press; 2000. [Google Scholar]
- 52.Fantl V, Richards MA, Smith R, Lammie GA, Johnstone G, Allen D, Gregory W, Peters G, Dickson C, Barnes DM. Gene amplification on chromosome band 11q13 and oestrogen receptor status in breast cancer. Eur J Cancer. 1990;26:423–429. doi: 10.1016/0277-5379(90)90009-i. [DOI] [PubMed] [Google Scholar]
- 53.Bieche I, Olivi M, Nogues C, Vidaud M, Lidereau R. Prognostic value of CCND1 gene status in sporadic breast tumours, as determined by real-time quantitative PCR assays. Br J Cancer. 2002;86:580–586. doi: 10.1038/sj.bjc.6600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhuang Z, Merino MJ, Chuaqui R, Liotta LA, Emmert-Buck MR. Identical allelic loss on chromosome 11q13 in microdissected in situ and invasive human breast cancer. Cancer Res. 1995;55:467–471. [PubMed] [Google Scholar]
- 55.Negrini M, Sabbioni S, Possati L, Rattan S, Corallini A, Barbanti-Brodano G, Croce CM. Suppression of tumorigenicity of breast cancer cells by microcell-mediated chromosome transfer: studies on chromosomes 6 and 11. Cancer Res. 1994;54:1331–1336. [PubMed] [Google Scholar]
- 56.Nakanishi M, Rosenberg DW. Roles of cPLA2alpha and arachidonic acid in cancer. Biochim Biophys Acta. 2006;1761:1335–1343. doi: 10.1016/j.bbalip.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas W, Caiazza F, Harvey BJ. Estrogen, phospholipase A and breast cancer. Front Biosci. 2008;13:2604–2613. doi: 10.2741/2869. [DOI] [PubMed] [Google Scholar]
- 58.Poad BL, Green MR, Kirk JM, Tomczyk N, Mitchell TW, Blanksby SJ. High-Pressure Ozone-Induced Dissociation for Lipid Structure Elucidation on Fast Chromatographic Timescales. Anal Chem. 2017;89:4223–4229. doi: 10.1021/acs.analchem.7b00268. [DOI] [PubMed] [Google Scholar]
- 59.Moore GY, Pidgeon GP. Cross-Talk between Cancer Cells and the Tumour Microenvironment: The Role of the 5-Lipoxygenase Pathway. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18020236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abramson SB, Leszczynska-Piziak J, Weissmann G. Arachidonic acid as a second messenger. Interactions with a GTP-binding protein of human neutrophils. J Immunol. 1991;147:231–236. [PubMed] [Google Scholar]
- 61.Khan WA, Blobe GC, Hannun YA. Arachidonic acid and free fatty acids as second messengers and the role of protein kinase C. Cell Signal. 1995;7:171–184. doi: 10.1016/0898-6568(94)00089-t. [DOI] [PubMed] [Google Scholar]
- 62.Foghsgaard L, Lademann U, Wissing D, Poulsen B, Jaattela M. Cathepsin B mediates tumor necrosis factor-induced arachidonic acid release in tumor cells. J Biol Chem. 2002;277:39499–39506. doi: 10.1074/jbc.M206669200. [DOI] [PubMed] [Google Scholar]
- 63.Hiraide T, Ikegami K, Sakaguchi T, Morita Y, Hayasaka T, Masaki N, Waki M, Sugiyama E, Shinriki S, Takeda M, Shibasaki Y, Miyazaki S, Kikuchi H, Okuyama H, Inoue M, Setou M, Konno H. Accumulation of arachidonic acid-containing phosphatidylinositol at the outer edge of colorectal cancer. Sci Rep. 2016;6:29935. doi: 10.1038/srep29935. [DOI] [PMC free article] [PubMed] [Google Scholar]


