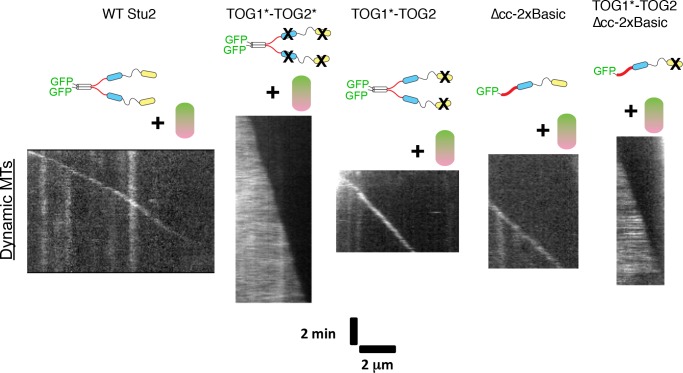
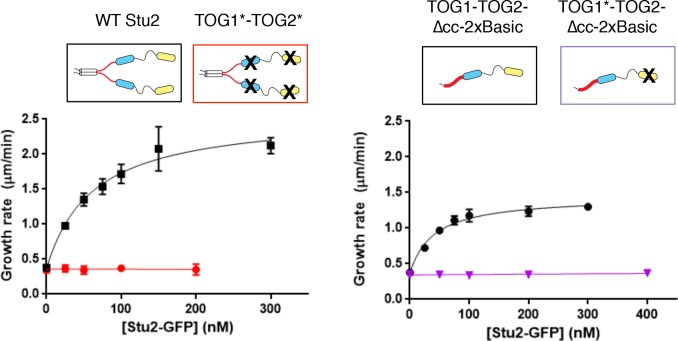
Figure 4. Polymerase elements required for preferential plus-end localization of Stu2.
Polymerase assays were carried out using wild-type tubulin (0.8 μM) and a panel of polymerase mutants (200 nM each). Representative kymographs are shown for Stu2 variants. Plus-end localization requires at least two tubulin-binding TOGs (WT, TOG1*-TOG2, ∆cc-2xBasic) and is not sensitive to how they are linked (compare TOG1*-TOG2 to ∆cc-2xBasic). Unexpectedly, polymerases with 0 (TOG1*-TOG2*) or one active TOGs (TOG1*-TOG2-∆cc-2xBasic) robustly coat the body of the microtubule. See also Figure 4—figure supplement 1.