Supplemental Digital Content is available in the text
Keywords: bioinformatics, diagnostic probe, lung metastasis, microRNA
Abstract
Presuming the stage of metastatic lung cancer is divided by its location, an intermediate state of ≤5 cumulative metastasis is defined as oligometastases (OM) and a widespread state of >5 cumulative metastasis as polymetastases (PM). According to the phenotypes, the different metastatic cancer patients can be treated with different methods: the OM patients can be treated by a metastasis-directed local therapy method, whereas the PM patients are not recommended to take such a treatment. It is also believed that the patients at the initial OM stage may progress to the PM stage. Currently, the OM- and PM-metastatic cancer patients can be identified by traditional imaging methods. However, the current methods are found to be insufficient for the discrimination. It hence is meaningful and important to develop new diagnostic methods for a better prediction to the patients following by selecting a correct metastasis-directed treatment.
MicroRNAs (miRNAs) can be used as the genetic probes for the new diagnostic methods. In this study, a bioinformatics strategy was employed to screen the microRNAs as potential diagnostic probes for distinguishing the OM and PM lung metastases patients. The expression profiles of microarray data of GSE38698 were downloaded from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) including the information from 63 patients: 24 PM and 39 OM patients. The microRNA expression patterns of tumor samples were identified for the OM and PM patients who were treated with the high-dose radiotherapy. Followed by analyzing the functional enrichment pathways, an early diagnosis model of OM and PM groups was identified with different expression genes (DEGs). The ratios of PM/OM were calculated by setting a high significance in the expressions of 377 mature miRNAs in the profile [log2 (PM/OM) >1 and P < .05]. Through a high combination power [area under the curve (AUC) ≥ 0.875] with the superior sensitivity and specificity, a panel of 10 miRNAs including 7 upregulation and 3 downregulation expressions were identified as potential probes for discriminating the PM and OM patients from the receiving operation characteristic (ROC). Considering the possible involvements of cancer progress, the interconnected axon guidance, cancer metastasis pathways, proteoglycans, and Mitogen-activated protein kinases signaling pathway and endocytosis were suggested for the subsequent miRNA target analysis. The results may reveal a biological significance that a profile of miRNAs can be used as the potential probes to identify the patients at the OM or PM stages and figure out the metastasis-directed treatment methods for the patients at the different metastasis stages.
1. Introduction
Known as the primary determinant of cancer-related death, the metastases of cancer is an importance factor for the prognosis of cancer patients when considering the tumor cell invasion and metastasis.[1,2] A fatal outcome with rare exceptions can occur for the metastases of solid tumors including soft tissue sarcoma and osteosarcoma.[3] However, as the cell proportion may limit a potential for the dissemination, the distant metastases of cancer are not always numerous and widespread.[4,5] As a clinically limited number ≤5, the metastases is defined as an intermediate state of oligometastases (OM), and as the number is over 5, the metastases is defined as a widespread state of polymetastases (PM).[6,7] The OM and PM patients are often treated in different therapeutic methods: the OM patients are treated by the surge method and the PM patients by the radiotherapy method.[8]
The lung metastasis is often caused by blood or lymph to the lung tissue, also known as a malignant tumor,[2] so it may occur at variable distances among the patients who died of malignancy. The lung metastases can progress very rapidly, and in some cases, may even occur earlier than the primary tumors,[9] such as breast tumor, bone tumor, digestive, and urogenital system tumor. Thus, the lung metastases are known with the multiple lung lesions, different sizes, uniform density, etc.[10,11] Currently, the single and multiple metastatic lesions of lung cancer can be diagnosed by traditional imaging methods, including lung enhancement computed tomography, magnetic resonance imaging, positron emission tomography, etc. However, these imaging methods are often less efficient for such identifications.[12] As a result, it becomes essential and necessary for developing new assays for distinguishing the lung metastasis patients at different prognostic stages. Furthermore, searching for the specific and efficient probes becomes basic and important.
We are interested in searching for novel biological markers particularly gene markers for predicting the multiple and single metastases of lung patients and furthermore use these gene markers to develop new diagnostic methods for the lung metastases prognosis.[13] As a class of small noncoding RNAs [14] and being able to control a wide range of biological functions including diagnosing the lung metastases at early stage, microRNAs (miRNAs) are well known to express typical and significant regulation among the hundreds of target genes in the cancer development and progression.[15,16] They hence can be emerged as the tissue-specific probes for the lung cancer identifications.[17] Digging the data of biochip in the database, it is very meaningful and important to explore the biological function of miRNAs, especially their effects from the single to multiple lesions.
In an early publication, the miRNAs were reported to be able to work as potential diagnostic probes for the cells, tissues, and serum samples from the lung metastatic patients,[18] including the miRNA expression profiles, screens of difference markers, and construction of diagnostic models. Interestingly, relying on the target downstream genes, dozens of miRNAs have been observed to express differentially in the lung cancer patients, either over- or under-regulated.[18] Hence, there is a high interest in collating the outcomes of miRNAs expression profiling and ascertain their differential expressions including normalization or significance thresholds. The results probably allow us to get insight into the expression profiling of lung cancer miRNAs[19,20] and furthermore find the potential probes for distinguishing the lung oligo- and polymetastases. However, even though the results display a kind of positive effects, the clinical diagnosis model using the miRNA probes is still not well established in the early publication, and particularly, the randomized controlled trials (RCTs) are not well documented yet.
In this study, on the basis of a homogeneous group of patients with the resected lung metastases, we created an analysis model for predicting the clinical outcome with the rate of metastatic progression as well as the resection of patients presenting ≤5 metastases. In addition, the rates of metastatic progression and/or metastatic colonization in the lung cancer patients were also analyzed and the statistical results were discussed to associate with the miRNAs that were identified from our work. The current results were compared with those achieved from the miRNA datasets. The miRNA probes identified from the current research are suggested to service as the potential probes for predicting the lung cancer patients at the different metastasis stages and furthermore provide guidance for their metastasis-directed treatments.
2. Methods
2.1. Microarray data
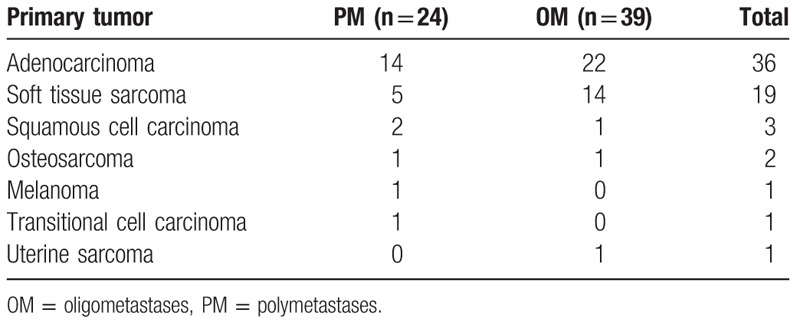
The miRNA expression profiles of microarray data GSE38698, which were obtained on the basis of platforms of GPL11239 TaqMan Array miRNA Card-A, were downloaded from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/).[21] The tumor samples were collected from 63 patients who had the metastasis between 1 and 5 sites. The collected samples included metastatic presentation and no clinical or radiologic evidence of metastases in the pleural, peritoneal, pericardial, or retroperitoneal cavities, every site of known metastases treated with definitive intent at the time of lung surgery, and after surgery, a minimum of 16 months of follow-up was required. All RNAs were derived from the FFPE metastatic tissue samples. The patients were divided into 2 groups: oligometastatic (OM) and polymetastatic (PM) groups on the basis of their profiles of prognosis mechanism. The information on the primary tumor of lung metastases is listed in Table 1.
Table 1.
The primary tumor of lung metastasis.
2.2. DEG screening
The microarray data from the GSE38698 dataset were normalized as described earlier.[22] Then, the DEGs of samples between the OM and PM groups were screened using LIMMA (linear models for microarray data) package (http://www.bioconductor.org). The cut-off criteria in the analysis was the adjusted P value of < .05 and |logfold change (FC)| >1. Including the inputting and preprocessing of the data when using the LIMMA package, a low-level analysis was performed in the R statistical computing environment. The empirical Bayes were used to moderate t statistics implemented using LIMMA to identify miRNAs that had different expressions. A comparison of interest was extracted as contrasts when a linear model was employed to fit to the data. The samples were clustered according to their miRNA expression levels. Then, the hierarchical cluster was supervised with the complete linkage followed by using both Euclidean and Pearson correlation as distance metrics.
A group comparison (PM vs OM) was made on the basis of a reliable subset of miRNAs that are specific for lung metastasis. Only those miRNAs that met the specific requirements [log2(PM/OM) > 1 and P < .05] were selected for the validation and discussion in this study. Using a function of “p.adjust” and a false discovery rate (FDR) method, all P values were adjusted for the multiple testing.[23] Here, the data visualization was also discussed including dynamic principle component analysis (PCA; based on variance filtering, significance level, and FDR).
2.3. Model construction and receiver operation of characteristic (ROC) curve analysis
Receiver operating characteristic (ROC) curves were identified and the areas under the curves (AUC) were calculated. According to the results, a panel of 10 miRNAs for the discrimination between the PM and OM groups was achieved. The sensitivity was obtained by plotting the binary classifier (both PM and OM) against the specificity. An AUC of 100% denotes a perfect discrimination by the miRNA, whereas an AUC of 50% denotes a completion with a lack of discrimination of miRNA. AUC of each miRNA was calculated with a 95% corresponding confidence interval.
A logistic regression model was employed to fit the data achieved from each miRNA sample with the probability of PM versus OM. Using this model, we could obtain the odds ratios and 95% confidence intervals as well as the ROC curves. The AUC values hence could be estimated for determining the overall diagnostic ability.
2.4. Biological function, pathway analysis, and network visualization
The miRNA tap package (available at http://bioconductor.org) was used to determine the targeted pathways of identified miRNAs. miRNAtap Targets gene are aggregated from 4 most commonly cited prediction algorithms: DIANA, Miranda, PicTar, and TargetScan.[24,25] Cytoscape software, Institute for Systems Biology, (version 3.4.0, The Cytoscape Consortium, New York, NY) was also used to perform a detail functional analysis and furthermore identify the different-expression miRNAs and target genes, miRNA mediated, cancer-related and statistically significant networks, biological functions, and canonical signaling pathways.
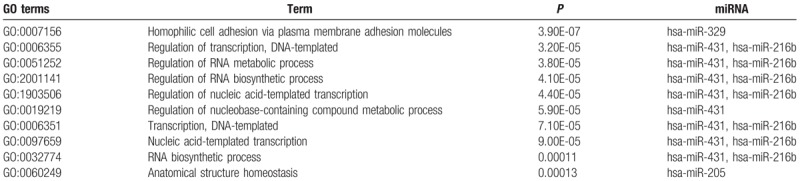
The biological mechanisms related to these miRNAs were interpreted in the functional enrichment pathways of known miRNA targets. The miRNet web tool was used to identify the biological pathways, processes, molecular functions, and cellular components for the most interesting miRNAs. The targets miRNAs that were identified from the research were used to identify the enrichment KEGG pathways and Gene Ontology (GO) terms. These target miRNAs were also validated in the operation, so the results were believed to have a higher reliability.
3. Results
3.1. Identification DEGs screening of OM and PM groups
According to the NCCN guidance, the polymetastatic lung cancer patients are not suggested to treat by the surgery method. As a result, the samples of polymetastatic lung cancer were obtained only when the patients were diagnosed as the oligometastases of lung cancer but finally confirmed as the polymetastases in the surgery. Such situations are very rare in the clinical treatments, and thus, it becomes very difficult to obtain a large number of polymetastatic lung cancer samples. In addition, it is hard to diagnose the polymetastatic lung cancers by the traditional imaging methods that are widely used in the current clinical diagnosis. In this study, we intend to screen the miRNA probes to find the potential probes and then use these genetic probes for distinguishing the oligo- and polymetastatic lung cancer to solve this unmet problem. A total of 63 patients is the largest sample number that we could find in this study.
Using the samples from the lung metastasis patients, a general expression of miRNAs was analyzed and due to their high significances [log2 (PM/OM) >1 and P < .05], a panel of 10 miRNAs were finally selected among 377 mature miRNAs for distinguishing the OM and PM groups. It was noticed that the analysis results displayed a miRNA signature in 2-way hierarchical cluster as shown in Fig. 1A. With 7 upregulated miRNAs and 3 downregulated miRNAs in a ratio of PM/OM, a total of 10 miRNAs were discovered to display different expressions (Tables 2 and 3). In addition, a greater variation in the miRNA expression was observed in the rectal cancer samples (around 50% higher SD among all PM samples than among OM samples). By the PCA plots, the variation was found to occur more frequently in the OM group, which is probably due to a more scattered distribution of tumor samples (red) in comparison with a distribution of normal samples (green) (Fig. 1B): the OM group is believed to have a more heterogeneous distribution than the PM group.
Figure 1.
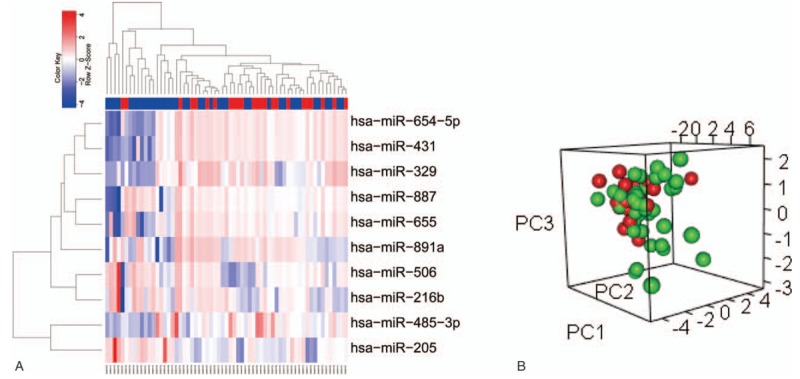
Identification consensus DEGs screening of PM and OM in lung metastasis patients. (A) The 10 DE miRNAs PM (red label) versus OM (green label) of GSE38698 set. Each column is a sample and each row is the expression level of a miRNA. The color scale represents the raw Z-score ranging from blue (low expression) to red (high expression). Dendrograms beside each heatmap correspond to the hierarchical clustering by expression of the 10 DE miRNAs. (B) PCA plot showing complete, unsupervised separation of 63 array samples into 24 PM (red) and 39 OM (green) samples.
Table 2.
miRNAs that are up- or downregulated for the expression of OM over PM tissues.
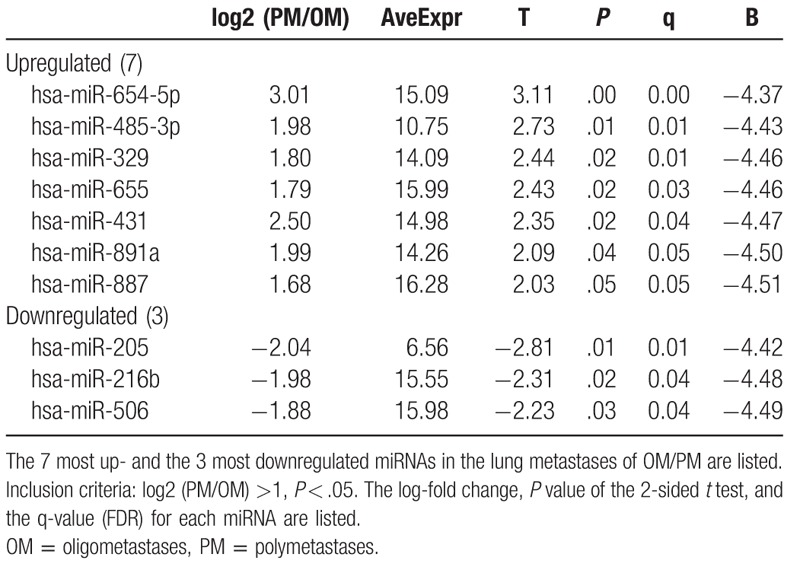
Table 3.
Top 10 miRNAs expressions (based on P value) that are mostly affected by the lung metastasis patients of OM and PM groups (miRNA tap package).
When screening the miRNA expressions by LIMMA and comparing the results with the adjacent noncancerous samples, it was found that a total of 10 DE miRNAs displayed significantly different expressions, including 7 upregulated and 3 downregulated, completely consistent with the result from the miRNA general analysis. It is interesting to notice that the number of downregulated genes is lower than that of upregulated genes in the current case. The miRNA signatures hence are considered to be able to distinguish the PM and OM samples in a 2-way hierarchical cluster (Fig. 1A) as well as in a PCA plot (Fig. 1B). The average of individual expression level (box plots) for the panel of 10 miRNAs is presented in Fig. 2. The detail information can be found in Supplementary Table S1.
Figure 2.
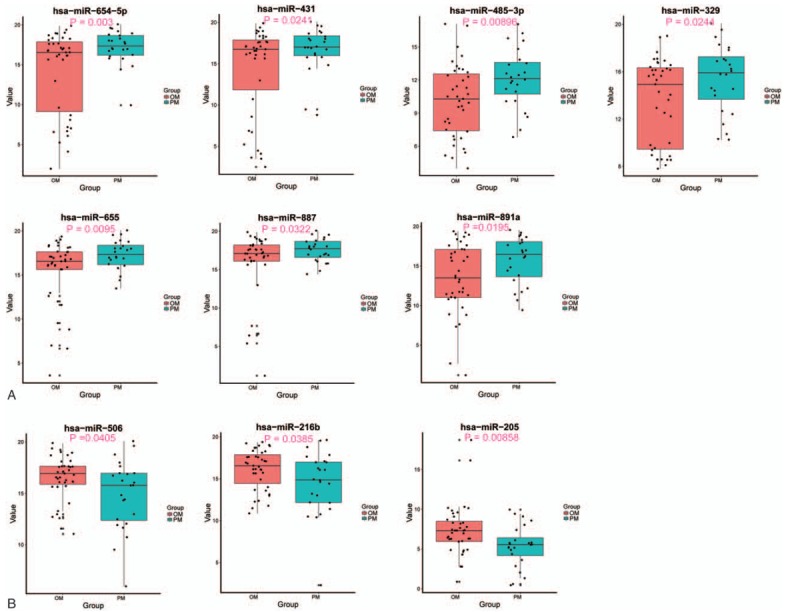
Expression levels of the 10 differentially expressed miRNAs observed between the PM and OM cases. In (A) and (B): miRNAs up- and downregulated, respectively, in the PM group of patients when compared with the OM group.
These top 10 up and downregulated miRNAs were also found to rely on a log2 fold change value, which reflected and compare the miRNA expression levels in the lung metastases for the both the OM and PM groups as summarized in Table 2 (t test; P < .05, FDR < 0.05).
3.2. Potential probe model construction and ROC curve analysis
In this work, some data from an early publication were employed for our bioinformation analysis.[21] However, different from the high rate of progression (HRP) and low rate of progression (LRP) by their rate of metastatic progression and survival, we worked on the analysis of miRNA panel as well as their biological functions on the signaling pathways, diagnosis methods at the early stage, gene regulations, etc. These results allow us to get insight into the functions among the hundreds of target genes in the cancer development and progression. In addition, the miRNAs were identified as the potential probes to distinguish the oligo- and polymetastases of lung cancer and furthermore develop a novel dynastical method that can be efficiently used in the clinical treatments.
Analyzing the individual ROC of 10 miRNAs that were identified as the panel, over 60% of the data were found to present a good power for the PM and OM groups of lung metastasis. With a high sensitivity and specificity (0.78 and 0.79, respectively), the panel of 10 miRNAs were also noticed to express a robust power in the discrimination of OP and MP groups using an AUC value of 0.875. With the AUC values for the 10 miRNAs, the ROC plots are presented in Fig. 3. The best performance on the accuracy could be obtained when using a linear regression model (AUC = 0.875): risk score = 0.243 × hsa-miR-485.3p + 0.138 × hsa-miR-329 -0.19564 × hsa-miR-654-5p -0.41265 × hsa-miR-205 + 0.39424 × hsa-miR-655 -0.03487 × hsa-miR-431 -0.34285 × hsa-miR-216b -0.17787 × hsa-miR-506 + 0.27648 × hsa-miR-891a + 0.06491 × hsa-miR-887 -2.75711.
Figure 3.
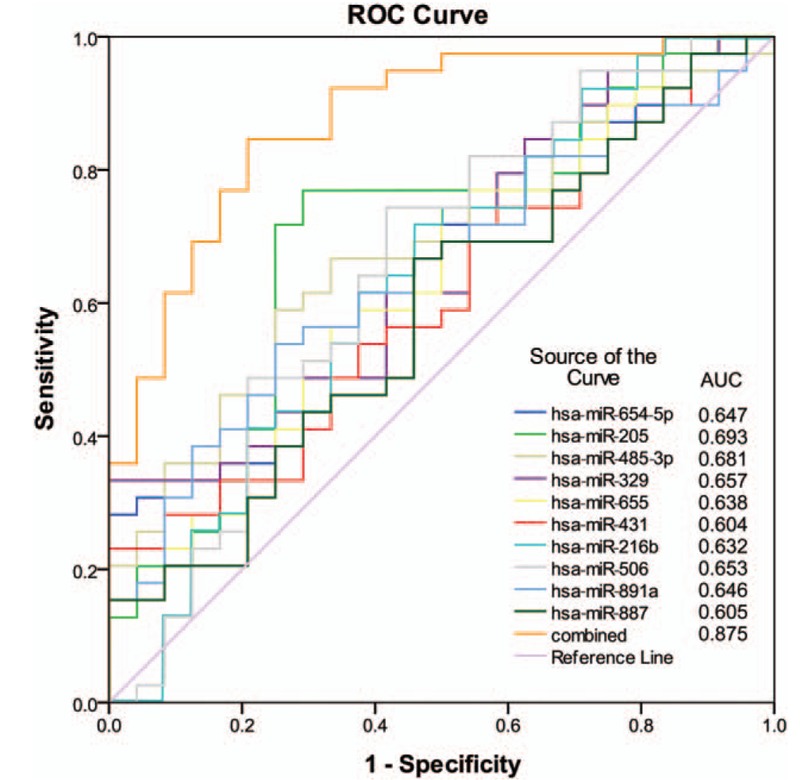
ROC curves to assess the accuracy of the model composed of the 10 miRNAs signature. Analysis of data implied that most of the dysregulated miRNAs have potential as diagnostic probes for PM detection, with high sensitivity and specificity.
3.3. Functional enriched pathways
In this case, only the patients with the limited number of lung metastases were selected with a clinical criterion.[26,27] The survival rate of these patients was only approx. 25%, revealing that a majority of them were not curable due to their tumor aggressive local metastases. The patients were furthermore accurately classified into the OM and PM groups. The OM and PM groups of patients were also suggested to treat with the different local and systemic therapies. With a potentially curative resection, the patients who had significantly worse prognosis were found to have increased expressions of 7 miRNAs and reduced expression of 3 miRNAs. The expressions of these regulated miRNAs were believed to inhibit the growth of lung cancer cells. The areas under the ROC curves (AUC) were estimated and the value was 0.875. The biological process and KEGG pathways of these 10 miRNAs are presented in Fig. 4 as well as in Supplementary Table S3. The regulation of their network structures is shown in Fig. 5.
Figure 4.
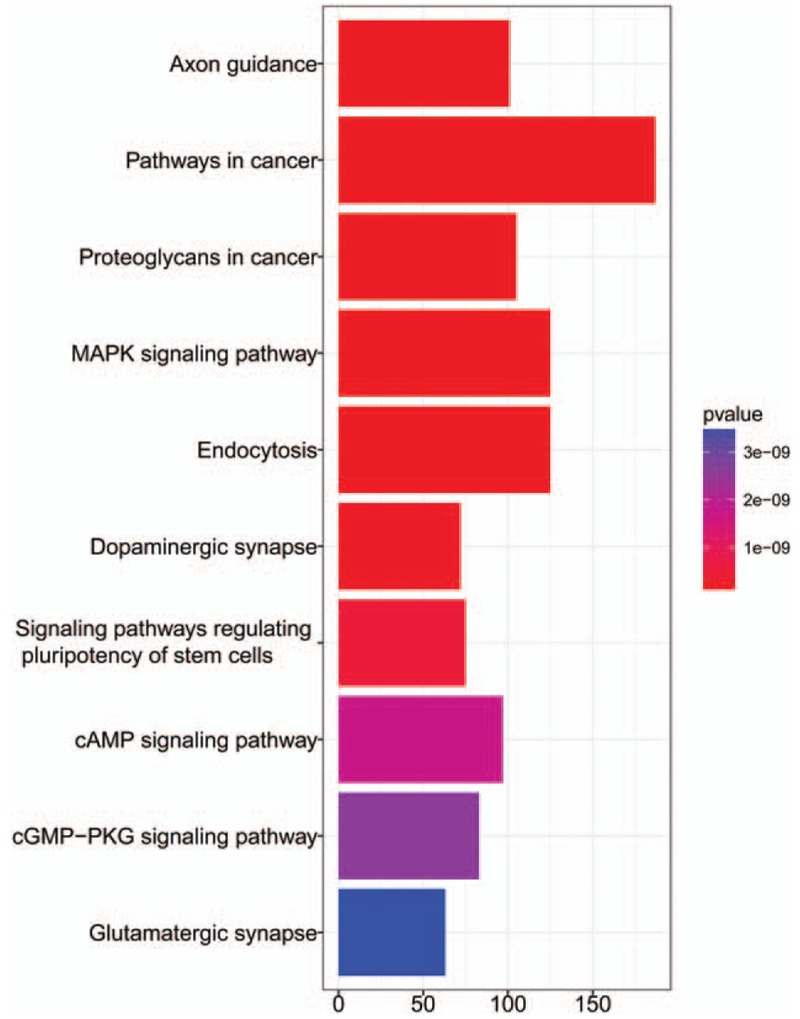
KEGG pathway of 10 miRNA targeted genes.
Figure 5.
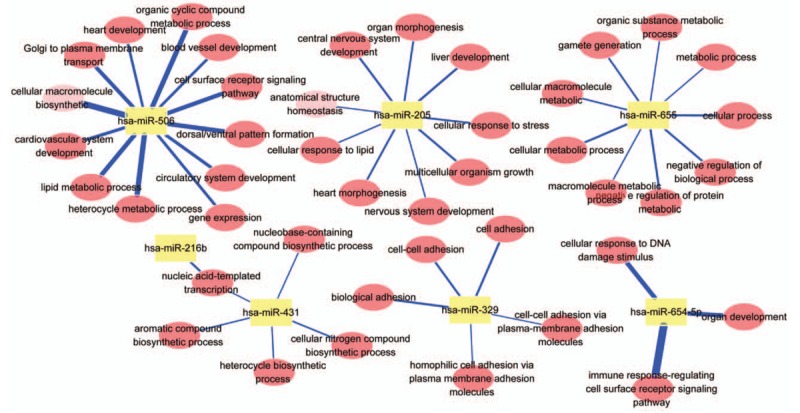
GO-Tree: red circles represent the BP involved in GO terms, and yellow squares represent miRNA in GO terms. Blue lines represent P values.
4. Discussion
In this study, only 63 lung tumor samples were collected and subsequently classified into the OM and PM groups. Thus, there were only limited data available for the miRNA expression profiles on the lung metastasis analysis. More samples are probably needed to deploy for reaching continuous phenotype analysis in our future work. According to the analysis to the data of all patients, it was found that the miRNAs were significantly expressed. The change on the average fold was a vote-counting strategy and a conservative analysis method was employed for a classification of discrete progression groups.
The different expressions of miRNAs from the PM and OM groups were investigated using both the rate of progression phenotype and binary OM/PM phenotypes.[21] A further investigation is suggested for a distinct separation of metastatic progression via a comparison of results of miRNAs between the OM and PM groups (Table 2 and Fig. 3). It was also noticed that over 60% of data on the expressions of a panel of 10 miRNAs have a good power in the discrimination of lung metastasis for the OM and PM groups. These 10 miRNAs were found to express with significant differences: 7 upregulated and 3 downregulated. For the diagnosis of lung metastases at the early stage, these miRNAs are believed to have a great potential to serve as the diagnostic probes for the novel bioassay development.
Supposing that a widespread reduction in the miRNA expression may promote a tumorigenesis, miRNA-329 is expected to display an underexpression in both the OM and PM groups.[28] In addition, it has been proven that miRNA-328 can work as a RNA decoy to interfere with the cell regulatory proteins in the chronic myelogenous leukemia (CML) even though miRNA-502-5p has not been well characterized in terms of cancer.[14] It is also interesting to notice that miRNA-328 is absence in the blast crisis of CML, and the expression restoration of miR-328 may decrease the survival of leukemic blasts.[29] In addition, a majority of downregulated miRNAs are found to have close associations with the tumor-suppression functions (Table 2, Table S4). Nevertheless, the results from this study may provide a support for analyzing the molecular pathways for discriminating the OM and PM phenotypes, which currently is not well-known.
From the above discussion, it is believed that the enrichment of KEGG pathway can be employed to analyze the lung metastases of PM and OM groups as well as insight their corresponding targets for understanding the functions of miRNAs listed in the current panel. Refereeing the pathways with the lowest P values, 3 pathways were suggested herein, including axon guidance pathway, molecular pathway in cancer, and proteoglycan formation pathway in cancer (Fig. 4 and Supplementary Table S2). As the interconnected target genes to involve in the regulation of network structure and associate with their special functions (Fig. 5), the biology processes and function pathways of miRNAs in the panel are discussed in 3 possible pathways at the cancer metastasis.
The first is an axon guidance pathway. The tumor metastasis is known to occur via removal of a single tumor cell from the primary site followed by a migration of tumor cell to other parts of body and then producing an uncontrollable proliferation of tumor cell.[2,9] Thus, in the molecular mechanism, the tumor cells are required to have a capability of penetrating the wall of lymphatic and blood vessel, circulating through the bloodstream (circulating tumor cells) to other sites in the body, repenetrating the vessel or wall for multiplication, and eventually forming other clinically detectable tumors at the second site. Hence, the tumor cell at the first site is believed to have a high mobility to remove from the primary tumor site and attach to the second site in the body for completing the metastasis.
Basically, the cancer cell can switch among 3 kinds of motions, including collective motility, mesenchymal-type movement, and amoeboid movement. Thus, the tumor cell is generally removed from the adjoining tissue by degrading the proteins and making up the surrounding extracellular matrix (ECM), which is often mediated by the chemokines and transforming growth factor beta to escape from the first tumor site.[30] The axon is also known to have a highly motile structure at the growing tip so-called growth cone in the human cell.[31] The growth cone on the axon can bring up an excellent extracellular activity in the environment. The growth axon is also believed to rely on a variety of guidance cues. With an appropriate guidance, the growth cone of an extending axon can process the cue into an intricate system of signal interpretation and integration. Such cell mobility via axon guidance is suggested to severely associate with the initial OM at the early stage, and furthermore, progress to the PM stage. Although there is still lack of a report on the effect of axon guidance for the OM/PM lung metastases, the axon function in the tumor cell is regarded as an important pathway in the lung cancer metastases.
The pathway of cancer metastasis is another important factor that may influence the lung cancer progress.[32] As a metastatic tumor is very common in the late stage of cancer, the cancer metastases may frequently occur via 4 typical routes. For the first transcoelomic spread, the malignancy is allowed to get into the body cavities via a penetrating the surface of peritoneal, pleural, pericardial, or subarachnoid spaces. For the second lymphatic spread, the tumor cells can transport into the lymph nodes followed by other sites of the body. The third hematogenous spread is a typical route of metastasis for the sarcomas, which is also a favorite route for certain types of carcinoma such as the tumors original from the kidney (so-called renal cell carcinoma). It is because the veins have more opportunities for being invaded by the metastatic tumor cells than the arteries, and as a result, the tumor metastasis tends to the pattern of venous flow through their thinner walls. The fourth is the transplantation or implantation. The cancer cells can spread to the lymph nodes nearby the primary tumor, which is also named as the nodal involvement, positive node, or regional disease. Regarding to the OM/PM phenotypes and prognosis, the cancer cells can be spread within the bodies of lung metastatic patients.
The proteoglycans are suggested as the third pathway to associate with the tumor metastases. The proteoglycans are glycosylated proteins that own the glycosaminoglycan (GAG) chains as well as covalently bound protein cores.[33] Presence on the cell surfaces or in the ECM, the proteoglycans can maintain the tissue turgor in the interstitial matrixes and serve as the molecular sieves in the basement membranes. The proteoglycans can also associate with hyaluronan or fibrous matrix proteins to form the complexes. Thus, the proteoglycans have the capability to influence the activity and stability of proteins and signaling molecules within the matrix. As a result, the mobility of proteoglycans can be regulated through the interactions with the cations and water in the ECM. The proteoglycans can also interact with the growth factors, cytokines, growth factor receptors, lipoproteins, and viruses. Both the sugar chains and protein cores of the proteoglycans thus can possess the distinct and well-determined functions. Thus, the proteoglycans can mediate the blood-vascular progression not only in the lymphatic vasculature that may promote the lymph node metastasis such as tumor lymphangiogenesis but also the lymphatic vessel tumor invasion in the cancer metastasis. The proteoglycans are believed as an important pathway to associate with the tumor metastases.
In this work, we focus on the miRNA probe screening for the identification of oligo- and polymetastatic lung cancer patients. Three possible pathways for the lung cancer metastases were discussed. We also know that there is still lack of experimental results for supporting the discussion. Nevertheless, we intend to offer some scientific ideas for interpreting our observations in this work.
5. Conclusion
In this study, the miRNA-200 family from the patients who have developed to the lung cancer was analyzed and the unique prioritized features of these miRNAs were classified into the OM/PM groups via feature screening. A total of 10 DE miRNAs were selected using the LIMMA program from the OM and PM samples, including 7 upregulated and 3 downregulated, as compared with the adjacent noncancerous samples. The observation supports a biological basis using the miRNAs as the potential probes to identify the cancer metastatic patients with a different OM or PM phenotype. The results can also provide guidance for the metastasis-directed treatments of OM or PM patients. Through the miRNA analysis, 3 pathways of axon guidance, cancer metastasis, and proteoglycan are believed to contribute to the lung cancer metastases of OM and PM phenotypes. On the contrary, the mechanisms of these miRNAs functions still remain uncertain. A larger group of lung metastases patients are needed for their function analysis in future work.
Acknowledgment
We would like to thank NCBI GEO data project organizers as well as all study participants.
Author contributions
Formal analysis: Shaolin Gao.
Funding acquisition: Zhichao Wang.
Investigation: Zhichao Wang.
Methodology: Shaolin Gao, Xiaoyu Ma, Haiyong Zhu.
Project administration: Zhichao Wang.
Software: Shaolin Gao.
Supervision: Zhichao Wang.
Validation: Tiezhi Li.
Writing – original draft: Tiezhi Li, Xiaoyu Ma.
Writing – review & editing: Zhichao Wang, Tiezhi Li, Xiaoyu Ma, Hongjiang Yan.
Supplementary Material
Footnotes
Abbreviations: AUC = area under the curve, CML = chronic myelogenous leukemia, ECM = extracellular matrix, FDR = false discovery rate, GO = Gene Ontology, OM = oligometastases, PM = polymetastases, RCT = randomized controlled trials, ROC = receiver operating characteristic.
The authors declare no conflict of interest.
Supplemental Digital Content is available for this article.
References
- [1].Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell 2006;127:679–95. [DOI] [PubMed] [Google Scholar]
- [2].Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 2011;331:1559–64. [DOI] [PubMed] [Google Scholar]
- [3].Spano D, Heck C, De Antonellis P, et al. Molecular networks that regulate cancer metastasis. Semin Cancer Biol 2012;22:234–49. [DOI] [PubMed] [Google Scholar]
- [4].Redig AJ, McAllister SS. Breast cancer as a systemic disease: a view of metastasis. J Intern Med 2013;274:113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hasebe T, Sasaki S, Imoto S, et al. Primary tumour-vessel tumour-nodal tumour classification for patients with invasive ductal carcinoma of the breast. Br J Cancer 2005;92:847–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Reyes DK, Pienta KJ. The biology and treatment of oligometastatic cancer. Oncotarget 2015;6:8491–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Milano MT, Katz AW, Muhs AG, et al. A prospective pilot study of curative-intent stereotactic body radiation therapy in patients with 5 or fewer oligometastatic lesions. Cancer 2008;112:650–8. [DOI] [PubMed] [Google Scholar]
- [8].Niibe Y, Hayakawa K. Oligometastases and oligo-recurrence: the new era of cancer therapy. Jpn J Clin Oncol 2010;40:107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Brooks SA, Lomax-Browne HJ, Carter TM, et al. Molecular interactions in cancer cell metastasis. Acta Histochem 2010;112:3–25. [DOI] [PubMed] [Google Scholar]
- [10].Weickhardt AJ, Scheier B, Burke JM, et al. Local ablative therapy of oligoprogressive disease prolongs disease control by tyrosine kinase inhibitors in oncogene-addicted non-small-cell lung cancer. J Thorac Oncol 2012;7:1807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pfannschmidt J, Dienemann H. Surgical treatment of oligometastatic non-small cell lung cancer. Lung Cancer 2010;69:251–8. [DOI] [PubMed] [Google Scholar]
- [12].Dwamena BA, Sonnad SS, Angobaldo JO, et al. Metastases from non-small cell lung cancer: mediastinal staging in the 1990s-meta-analytic comparison of PET and CT. Radiology 1999;213:530–6. [DOI] [PubMed] [Google Scholar]
- [13].D’Amico TA, Massey M, Herndon JE, 2nd, et al. A biologic risk model for stage I lung cancer: immunohistochemical analysis of 408 patients with the use of ten molecular markers. J Thorac Cardiovasc Surg 1999;117:736–43. [DOI] [PubMed] [Google Scholar]
- [14].Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834–8. [DOI] [PubMed] [Google Scholar]
- [15].Uppal A, Ferguson MK, Posner MC, et al. Towards a molecular basis of oligometastatic disease: potential role of micro-RNAs. Clin Exp Metastasis 2014;31:735–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang L, Wang J. MicroRNA-mediated breast cancer metastasis: from primary site to distant organs. Oncogene 2012;31:2499–511. [DOI] [PubMed] [Google Scholar]
- [17].Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res 2010;70:5923–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vosa U, Vooder T, Kolde R, et al. Meta-analysis of microRNA expression in lung cancer. Int J Cancer 2013;132:2884–93. [DOI] [PubMed] [Google Scholar]
- [20].Guan P, Yin Z, Li X, et al. Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J Exp Clin Cancer Res 2012;31:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lussier YA, Khodarev NN, Regan K, et al. Oligo-and polymetastatic progression in lung metastasis (es) patients is associated with specific microRNAs. PLoS One 2012;7:e50141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Moher D, Liberati A, Tetzlaff J. Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg 2010;8:336–41. [DOI] [PubMed] [Google Scholar]
- [23].Tarca AL, Draghici S, Khatri P, et al. A novel signaling pathway impact analysis. Bioinformatics 2009;25:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Maragkakis M, Reczko M, Simossis VA, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 2009. Jul; 37 (Web Server issue):W273–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vlachos IS, Paraskevopoulou MD, Karagkouni D, et al. DIANA-TarBase v7.0: indexing more than half amillion experimentally supported miRNA:mRNA interactions. Nucleic Acids Res 2015;43:D153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Crinò L, Weder W, van Meerbeeck J, et al. ESMO Guidelines Working Group. Early stage and locally advanced (non-metastatic) non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2010;21(suppl 5):v103–15. [DOI] [PubMed] [Google Scholar]
- [27].Peters S, Adjei AA, Gridelli C, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2012;23(suppl 7):vii56–64. [DOI] [PubMed] [Google Scholar]
- [28].Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 2007;39:673–7. [DOI] [PubMed] [Google Scholar]
- [29].Eiring AM, Harb JG, Neviani P, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 2010;140:652–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Balkwill F. Cancer and the chemokine network. Nat Rev Cancer 2004;4:540–50. [DOI] [PubMed] [Google Scholar]
- [31].Chang C, Werb Z. The many faces of metalloproteases: cell growth, invasion, angiogenesis and metastasis. Trends Cell Biol 2001;11:S37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2002;2:563–72. [DOI] [PubMed] [Google Scholar]
- [33].Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest 2001;108:341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


