Abstract
Organismal phenotypes often co-vary with environmental variables across broad geographic ranges. Less is known about the extent to which phenotypes match local conditions when multiple biotic and abiotic stressors vary at fine spatial scales. Bittercress (Brassicaceae: Cardamine cordifolia), a perennial forb, grows across a microgeographic mosaic of two contrasting herbivory regimes: high herbivory in meadows (sun habitats) and low herbivory in deeply shaded forest understories (shade habitats). We tested for local phenotypic differentiation in plant size, leaf morphology, and anti-herbivore defense (realized resistance and defensive chemicals, i.e., glucosinolates) across this habitat mosaic through reciprocal transplant–common garden experiments with clonally propagated rhizomes. We found habitat-specific divergence in morphological and defensive phenotypes that manifested as contrasting responses to growth in shade common gardens: weak petiole elongation and attenuated defenses in populations from shade habitats, and strong petiole elongation and elevated defenses in populations from sun habitats. These divergent phenotypes are generally consistent with reciprocal local adaptation: plants from shade habitats that naturally experience low herbivory show reduced investment in defense and an attenuated shade avoidance response, owing to its ineffectiveness within forest understories. By contrast, plants from sun habitats with high herbivory show shade-induced elongation, but no evidence of attenuated defenses canonically associated with elongation in shade-intolerant plant species. Finally, we observed differences in flowering phenology between habitat types that could potentially contribute to inter-habitat divergence by reducing gene flow. This study illuminates how clonally-heritable plant phenotypes track a fine-grained mosaic of herbivore pressure and light availability in a native plant.
Keywords: common garden, microgeographic divergence, phenotypic plasticity, shade avoidance syndrome, Brassicaceae
INTRODUCTION
In species distributed across environmental gradients, individuals often exhibit phenotypic clines (Linhart and Grant 1996; Richardson et al. 2014). These clines arise when natural selection acts divergently across habitats, but their emergence and persistence is also heavily influenced by gene flow (Kawecki and Ebert 2004). Specifically, gene flow among interspersed habitat patches acts as a homogenizing force that opposes divergence (Haldane 1930; Lenormand 2002). As environmental variation becomes increasingly fine-grained, more opportunities exist for dispersal and/or gene-flow across habitat types to prevent habitat-associated divergence and local adaptation. As result of this expectation, our knowledge of how populations diverge among habitats has been disproportionately shaped by studies at coarse spatial scales, where conditions for habitat-associated divergence are more favorable (Richardson et al. 2014).
While the concept is not new, much less attention has been paid to the problem of how local adaptation proceeds at fine-grained spatial scales (microgeographic divergence) in the presence of gene flow (Epling and Dobzhansky 1942; Ehrlich and Raven 1969; Linhart and Baker 1973; Selander and Kaufman 1975). This is trend is beginning to change, and there is growing evidence of microgeographic divergence in species spanning the diversity of life, from algae, fungi, insects, snails, frogs, fish, and flowering plants (Richardson et al. 2014). In many cases, the selective agents that have driven well-studied examples of adaptive microgeographic divergence are abiotic, such as seasonal water level variation in vernal pools (Richardson et al. 2014). But for Darwin, the strongest features of the tangled bank were biotic interactions—competition, predation, parasitism, and mutualism. He hypothesized that these biotic interactions, rather than purely abiotic factors, acted as the prevailing selective agents shaping adaptive phenotypes in the “struggle for existence” (Reznick and Ricklefs 2009, Darwin 1859). According to this perspective, strong biotic agents of selection may be key for generating adaptive divergence at microgeographic scales.
Much of the early pioneering research on variation in species distributions (e.g. Greig-Smith 1952) and phenotypic and genetic differentiation across fine-grained habitat mosaics focused on plants. While local adaptation to abiotic conditions clearly shapes plant distributions and phenotypes in many cases (reviewed in Linhart and Grant 1996), there is considerable evidence that plant consumers also play a central role (Fine et al. 2004, Maron and Crone 2006). Defoliation by insect herbivore communities exerts strong natural selection on plant phenotypes (Louda 1984; Prasad et al. 2012; Agrawal et al. 2012). In mustards (Brassicaceae), polymorphisms in genes encoding enzymes that modify defensive chemicals underlie adaptation to regional herbivore communities (Prasad et al. 2012; Züst et al. 2012), and the magnitude of geographic divergence at such loci is extreme compared to loci across the rest of the genome (Brachi et al. 2015). Much of the strongest evidence for local adaptation to herbivory is from populations separated by large distances, on the scale of hundreds to thousands of kilometers (km), even in studies on clines (Woods et al. 2012). But heritable divergence in herbivore resistance or defensive traits can correlate with variation in herbivore pressure at relatively fine spatial scales as well, ranging from a few km to only 500 m (Galen et al. 1991; Sork et al. 1993; Pellissier et al. 2014; Dostálek et al. 2016; Sato and Kudoh 2017). Thus, in line with Darwin’s view, herbivores exert selective pressures strong enough to impact the distribution and abundance of plants, as well as the genetic and phenotypic composition of plant populations, at both coarse- and fine-grained spatial scales.
Biotic and abiotic conditions are rarely uncorrelated, however, even at fine spatial scales. For plants, the composition of herbivore communities and the overall intensity of herbivory often vary across habitats that also differ along abiotic dimensions (Louda et al. 1987; Fine et al. 2004; Strauss and Cacho 2013). In fact, such co-variation between biotic and abiotic agents of selection, if strong enough, can amplify the total strength of divergent selection between habitats and promote microgeographic divergence (Nosil and Harmon 2009). Studies of traits involved in coping with both biotic and abiotic stresses can therefore offer a more complete view of microgeographic divergence.
In this study, we revisited a textbook study system (Ricklefs and Miller 2000) used to understand the ecological consequences of microgeographic variation in plant–herbivore interactions and light availability. Bittercress (Brassicaceae: Cardamine cordifolia Gray), a forb native to montane regions of western North America, grows across a mosaic of open meadow habitats (sun) and deeply shaded habitats (shade). Two notable ecological features reliably distinguish sun and shade habitats for bittercress (ESM §1 Fig. S1) and thus could promote habitat-specific phenotypic divergence. First, damage by the herbivore community of bittercress—and specifically by Scaptomyza nigrita (Drosophilidae), a specialist leaf-miner that can heavily defoliate stands of bittercress (Collinge and Louda 1989a; Gloss et al. 2014)—is primarily restricted to sun habitats, and this has been consistently observed throughout many growing season across decades of study (Louda and Rodman 1983a,b; Louda 1984; Collinge and Louda 1989a,b; Louda and Rodman 1996; Alexandre et al. 2018). Sun-biased herbivore pressure likely arises from a strong habitat preference of the herbivore (Alexandre et al. 2018), which is thought to promote colonization of shade habitats by bittercress. As a consequence, the enemy free space associated with shade habitats would be expected to favor lower investment in anti-herbivore defenses (Agrawal et al. 2012; Mooney et al. 2016). Indeed, Louda and Collinge (1992) hypothesized that the uneven distribution of herbivory between sun and shade habitats could lead to fine-grain spatial structuring of bittercress genotypes varying in herbivore resistance, but this intriguing hypothesis has not been addressed.
A second major feature distinguishing sun from shade habitats is that the growth form of neighboring plants that filter incoming light, either forbs or canopy trees, differs between habitats. Thus, traits involved in capturing and/or competing for light are likely to be selected differently in sun and shade habitats. Sunlight that passes through (or reflects off of) plant leaves has characteristic signatures that differ from unfiltered sunlight, including a reduced ratio of red to far-red light (R:FR) and diminished levels of blue light, UV-B light, and photosynthetically active radiation (PAR) (Ballaré and Pierik 2017). Plants perceive and respond to these reliable cues of canopy and neighbor shading through photoreceptors, which in turn regulate signaling pathways that control plant growth and physiology (Ballaré and Pierik 2017). Plants that are intolerant of shade express a suite of phenotypes upon perception of shade cues, termed the shade avoidance syndrome (SAS), that elevate photosynthesizing leaves to positions where light conditions are more favorable (Keuskamp et al. 2010). Although SAS manifests through many traits, one of the most conspicuous changes is the elongation of stems and petioles to overgrow competitors (Ballaré and Pierik 2017). Conspecific and heterospecific forbs grow at high densities around bittercress sun habitats (Electronic Supplemental Material [ESM] §1 Fig. S1), and SAS is frequently observed to be adaptive in this context (Keuskamp et al. 2010).
In contrast, large evergreen trees filter the majority of light from bittercress growing in shade habitats. Shading by the forest canopy cannot be overcome by elongating stems or petioles, and plants living under canopy shade benefit from reduced SAS expression (Dudley and Schmitt 1995, 1996; Schmitt et al. 2003; Donohue et al. 2000; Bell and Galloway 2008). Instead, understory-adapted plants typically cope with shade through a shade tolerance strategy, which involves morphological (e.g., expanded leaf area per unit mass) and physiological changes thought to optimize light capture and photosynthesis in low light conditions (Valladares & Niinemets 2008; Gommers et al. 2013). Differences in shade responses within species are an example of locally adaptive plasticity (Schmitt et al. 2003; Keuskamp et al. 2010). It is not known whether bittercress expresses SAS in response to canopy shading or if the strength of these responses has diverged between bittercress populations in sun and shade environments.
Together, the enemy-free space and consistently low light quality and availability for bittercress populations living in chronic shade might favor an attenuation of both anti-herbivore defenses and shade avoidance phenotypes, either constitutively or in response to shading. Thus, bittercress presents an opportunity to test for microgeographic divergence in both constitutive and plastic foliar morphology and herbivore resistance phenotypes. Through replicated reciprocal transplant–common garden experiments in the field and greenhouse, we tested if bittercress from shade and sun habitats differed in foliar morphological phenotypes, including investment of leaf mass into leaf area, and petiole elongation, two traits that are commonly responsive to shading (Valladares & Niinemets 2008; Ballaré and Pierik 2017). This design allowed us to measure the effect of phenotypic plasticity caused by light environment, and to test if plants from sun and shade populations differ in the average expression and/or the plasticity of these traits. In parallel, we tested for divergence between habitats in plant investment in anti-herbivore defenses. We focused on glucosinolates (GSL), the major defensive chemicals in plants of the family Brassicaceae (Halkier and Gershenzon 2006; Hopkins et al. 2009). GSL are hydrolyzed into a number of bioactive compounds upon tissue disruption, including isothiocyanates, which are generally toxic to insects (Hopkins et al. 2009). Importantly, patterns of herbivory in the field (e.g., Louda and Rodman 1983a,b; Collinge and Louda 1988a; Louda and Rodman 1996, Humphrey et al. 2014) and in controlled laboratory experiments (Humphrey et al. 2016) suggest that isothiocyanate-yielding GSL (IYG) in bittercress confer resistance to its herbivore community, including S. nigrita. To investigate whether divergence in IYG concentrations correlated with resistance to herbivory, we also assayed the performance of larval S. nigrita on plants in the common gardens. Given that S. nigrita is susceptible to GSL and to jasmonic acid (JA)-induced (i.e., wounding-induced) defenses more generally, we expected differences in resistance to track foliar IYG concentration across garden and source habitats. The biological effects of two other GSL classes, oxazolidine-2-thione-yielding GSL (OYG) and indole GSL, are less understood in both bittercress and other mustards (Agerbirk et al. 2009; Winde and Wittstock 2011). Finally, we tested if flowering phenology differs between habitats in a manner that could facilitate microgeographic divergence by reducing opportunities for gene flow across habitats.
MATERIALS AND METHODS
Experimental design and data collection
Common gardens in the field
This study was conducted at the Rocky Mountain Biological Laboratory (RMBL) in Gothic, Colorado, USA, between 2011–2014. In 2011, we planted a reciprocal transplant-common garden to test if habitat of origin (sun vs. shade) impacted plant size, leaf morphological traits, defensive chemistry, and realized herbivore resistance, either constitutively or in response to shading (Fig. S1). We chose nine sun and nine shade source sites from which to sample bittercress rhizomes for planting in common gardens that were either in meadows (sun) or under evergreen forest canopies (shade) (ESM §1 Fig. S2). At source and common garden sites, we recorded photosynthetically active radiation (PAR) using a light meter (Spectrum Technologies, Inc.), percent canopy cover using a densiometer, diameter at breast height (dbh) of the four largest trees within four meters, and latitude, longitude, and elevation (Garmin GPS) (ESM §1 Table S1). Source sites significantly differed in PAR, canopy cover, and dbh (ANOVA, all p<0.05) but did not differ in elevation (p>0.05; ESM §1 Table S2).
From each source site, we collected rhizome tissue from each of 30 ramets and stored them at 4–10°C in low ambient light until planting. Rhizomes were collected from a single genet (i.e. clump) in most cases, or otherwise along short transects of no more than a few meters per site when large genets were not present. Using a randomized complete block design, 540 rhizomes—five from each of the 18 source sites (90 in total per garden; see ESM §1 Table S1–S2 for site locales and habitat features)—were planted in each of six common gardens (three in sun and three in shade) and spaced 16 cm apart to avoid shading from neighboring bittercress. Gardens exhibited light conditions consistent with source sites (ESM §1 Fig. S3) and were watered every 24–48 h with nearby water from a snowmelt stream. Plants were shrouded using fine mesh cloth to prevent herbivory throughout the experiment.
After five weeks, we counted the number of leaves >10 mm2 growing from each rhizome, and we removed and photographed the largest leaf. We measured petiole length and leaf area using ImageJ (Abràmoff et al. 2004) and leaf mass using a Sartorius CPA225D balance (Sartorius AG, Goettingen, Germany) after oven drying at 65°C for 2 d. Subsequently, we transplanted a single S. nigrita larva into the largest remaining leaf. Larvae were collected from mined leaves along Copper Creek near the RMBL (ESM §1 Fig. S2). Early instar larvae were excluded to approximately standardize larval developmental stage, but larvae were not weighed in order to minimize their manipulation. Thus, the amount of variance within treatments is likely inflated by variation in initial larval mass. After 48 h, we removed mined leaves, extracted larvae, and measured larval mass as a measure of herbivore resistance (Whiteman et al. 2011; Whiteman et al. 2012). At the same time, we removed the largest remaining unmined leaf from each plant for GSL analysis. These leaves were immediately frozen it in liquid nitrogen and stored at −80°C prior to GSL profiling (described below). Because leaves for GSL profiling were sampled 48 h after feeding by S. nigrita elsewhere on the same plant, GSL concentrations reflect the sum of constitutive and systemic inducible GSL.
Greenhouse experiment
To measure the elongation response in bittercress under more controlled conditions, we also performed greenhouse experiments with seed-grown plants in which we subjected seedlings to either natural light or filtered light with a lower ratio of red to far-red (R:FR) light, a reliable indicator of shading by overhanging plant leaves. In August 2012, we collected seeds from a single parental ramet of bittercress growing in each of two sun and three shade sites near the source sites for the common gardens by tying bags to developing racemes. From these five locales we recorded percent canopy cover using a densiometer and latitude, longitude, and elevation of each collection site as above (ESM §1 Table S3). Seeds were transported to the University of Arizona and surface-sterilized with a solution of 50% bleach and 0.5% Triton-X-100 and stratified on moist filter paper in petri dishes for five weeks at 4°C. Seeds were germinated in the greenhouse in moist filter paper in petri dishes, after which we planted them individually in soil in plastic pots. Seedlings from each source site (5 source sites, n=37 plants total) were split randomly between the two simulated light environments. Canopy shade was simulated by growing 19 plants under a blue plastic filter (Lee filter 115) that reduces the ratio of R:FR light available to plants (Runkle and Heins 2001). To simulate sun habitats, the remaining 18 plants were grown under a clear filter that does not alter the quality of light (Lee filter 130). Sample sizes from each source family are given in ESM §1 Table S3. The simulated shade treatment was applied as a single apparatus, held adjacent to the mock filter apparatus on the same greenhouse table, conducted simultaneously, and all plants were randomly assigned to position within and between each of the treatment setups. To minimize ontogenetic effects, we compared petiole length across seedlings at the eight-leaf stage, using measurements of fully-expanded leaf positions 1–4 only. We did not have access to S. nigrita larvae for the greenhouse experiment to measure realized herbivore resistance.
Field surveys of flowering phenology
In 2015, we counted floral abundance through time in sun and shade habitats in a sample of 240 bittercress plants spread across two sites (401 Trail and Copper Creek). At each site, we designated roughly equal numbers of plants in open sun, intermediate shade, and deep-shade and tracked plant-level flower abundance every 2–3 days for four weeks (between Jul 8 and Aug 5, 2015). From these data, we designated a first, peak, and last flowering Julian day for each stem. In many cases, focal stems flowered before the observation window, and these stems were assigned a Julian date of 1 prior to the first observation day. Plants with no flowers or fruits by the end of the observation window were assigned a no-flowering status and were excluded from the analysis. We also accounted for the presence of developing fruits, which allowed us to designate plants with no observed flowers as having completed flowering by the first observation date, thus distinguishing this class from those plants that had neither flowered nor fruited during the observation window. Many plants had just begun to flower by the end of the observation widow; the ‘peak’ and ‘last’ flowering events for these stems was assigned a Julian date of the last observation day +1.
Glucosinolate analysis
Frozen leaves (mean wet mass: 95 mg ± 50 mg standard deviation [sd]) were individually ground in liquid nitrogen in 1.7 mL microcentrifuge tubes. GSL were extracted from the frozen leaf powder in 1.5 mL 80% methanol, which inhibits endogenous myrosinase activity (Doheny-Adams et al. 2017), at 4°C for 4 hours. Samples were auto-injected through an Agilent Poroshell 120 SB-C18 3.0×150 mm, 2.7 μm column on an Agilent 6410BQQQ LC-MS with the following parameters: 350°C gas temperature, 7.0 L/min gas flow, 30 PSI nebulizer, 3000 V capillary, 20 eV fixed collision energy, 140 V fragmentor. Samples were eluted with 0.1% formic acid in water (A) and 100% Acetonitrile (B) using the following separation gradient: 2 min of 98% A followed by a gradient from 98% to 70% A (2% to 30% B) over 10 min, and a wash with 99% B for 3 minutes with a 4 minute post-run re-equilibration to 98% A. Flow rate was maintained at 0.3 mL/min throughout. The mass spectrometer was run in precursor negative-ion electrospray mode, monitoring all parent ions from m/z 350 to 500 with daughter ions of m/z 97, which correspond to the sulfate moiety of the GSL analytes. An external standard of 1 mM sinigrin in water was run every fifth sample to ensure detection intensities did not vary among samples.
GSL compounds previously characterized in C. cordifolia (Louda and Rodman 1983a) were identified using parent ion mass and relative retention times of these molecules in other studies (Agerbirk et al. 2010; Liu et al. 2016a). Compound abundance was quantified as base peak area (i.e., total counts) of the parent ion with Agilent Mass Hunter Software (Qualitative Analysis B06.00 Build 6.0.633.02012). Response factors that relate intact GSL detection intensities to molar equivalents are unknown, and pure standards were unavailable for comparison, for most GSL compounds in bittercress. To enable estimation of total isothiocyanate-yielding GSL (IYG) content per leaf, we weighted peak areas for each IYG compound so that its mean percent contribution to total IYG abundance across our experimental plants was equal to the mean relative abundance of the same compound in basal leaves in natural bittercress populations in mid-August, which Rodman and Louda (1985) quantified using using purified preparations of each compound as standards. Weighting factors (ESM §4 Table S6) did not vary considerably among compounds, and the conclusions of downstream analyses were qualitatively unaffected by the use of raw parent ion counts vs. counts weighted to approximately reflect relative molar concentrations.
Statistical analyses
We conducted all statistical analyses using R v.3.3 (R Core Team 2017). All data and R scripts can be found in the Dryad data repository (doi pending).
Common gardens in the field
Using data collected for all plants surviving after five weeks, we constructed hierarchical mixed effects regression models to estimate the effects of source and garden types (and their interaction) on foliar morphology, defensive chemistry, and herbivore resistance phenotypes in the common gardens. A detailed description of our regression model structure, including model formulae and coefficient definitions, can be found in ESM §2.1.
Overall approach
Our main goal was to estimate if plants from sun and shade source habitats expressed different morphological and defensive phenotypes when grown in experimental sun and shade common gardens which mimicked the abiotic differences between the two source habitat types. Thus, statistical interactions between source and garden type were of primary interest. For each response variable, we estimated coefficients for fixed effects of source habitat (two levels), common garden habitat (two levels), and their interaction, using linear mixed effects models implemented in R package lme4 (Bates et al. 2015). We also included group-level (i.e. random) effects that captured among-garden plot and among-genet differences in average expression of plant phenotypes (ESM §2.1 Eq. 1). These random effects also reflected the nested experimental design, with rhizomes (clones, n=9) nested within genets (n=1 genet per source site), and the blocking structure of the experiment, with clones from each genet measured across six separate gardens. We also estimated an effect of among-genet variation in plasticity across garden types (i.e. a random slope term) and evaluated statistical support for its inclusion in the model via likelihood ratio tests. In no case was this term retained, either because its effects were not significant (likelihood ratio test, p≥0.1) or because likelihood profiling for confidence intervals for this term (details below) produced negative values.
Many traits—including foliar morphology, defenses, and resistance to herbivory—vary with ontogeny and plant size (Poorter 1999; Valladares & Niinemets 2008; Boege and Marquis 2005; Barton and Boege 2017). Pairwise trait correlation plots revealed moderate to high correlations among potential covariates (ESM §3.1 Fig. S5) which would manifest as multicollinearity if included in a multiple regression. While multicollinearity among several predictor variables does not affect overall goodness of fit of multiple regression models, it inflates error variance in the coefficient estimates for correlated yet biologically meaningful covariates, leading to downwardly-biased parameter estimates and increased Type II error (Graham 2003). We assessed multicollinearity among our four measured foliar traits by calculating variance inflation factors (VIF) using R package mctest v.1.1.1 (Ullah et al. 2017). Several traits showed VIF ≥1.5, indicating moderate multicollinearity. Avoiding inflated error variance in covariate coefficient estimation was a priority because we included covariates to test how they interacted with source and garden type, rather than to increase overall model goodness-of-fit.
We took two approaches to analyze inter-correlated trait data. First, we implemented a series of focal trait regression models where we orthogonalized the response variable and any relevant covariate trait against a common variable (Mosteller and Tukey 1977, Freckleton 2002). This approach avoids the loss-of-power induced by multicollinearity to estimate regression coefficients for secondary predictors (and their interaction with additional factors) while preserving the biological interpretability of coefficient estimates [approach (I), below]. We also modeled each untransformed trait vector according to Eq. 1 in ESM §2.1 to document systematic differences in plant trait distributions between source and/or garden types in a manner naive to covariation (ESM §3). Second, as a complementary and standard multivariate approach, we conducted principal components analysis on four plant traits (number of leaves per plant, as well as the dry mass, leaf area, and petiole length of the largest leaf). This allowed us to transform inter-correlated variables into statistically orthogonal vectors which could be analyzed as response variables in our regression models described above [approach (II), below].
(I) Focal traits regression analysis
We orthogonalized the response and the covariate with respect to plant size, as described for each trait below, by taking the residuals from ordinary least squares regression. This subtracts the mean effect of plant size from each trait vector, such that each transformed trait vector is orthogonal to the vector of plant size (Mosteller and Tukey 1977). The subsequent focal regression models estimate the extent of correlations between trait deviations from each of their expected relationship with plant size. Importantly, if underlying differences in plant size were driving all variation in other focal traits, we would not detect differences between source and/or garden types using this approach.
We modeled leaf area expansion by including the plant size-adjusted residual vector of dry leaf mass (log g) as a covariate, along with all covariate interactions with source and garden type (according to Eq. 4 in ESM §2.1). The response variable was the residual vector of (log) cm2 leaf area after linear regression against log leaf number. We next modeled petiole elongation by including the plant size-adjusted vector of leaf mass (petiole length model ‘A’) as a covariate. In this case, the petiole length (mm) response variable was also converted to a residual vector from a linear regression with log leaf number. Finally, we tested the strength of the correlation between residual leaf area and residual petiole length, to determine if petiole elongation correlated with leaf area expansion (petiole length model ‘B’). In this model, we used the plant size- and leaf mass-adjusted residual vector of (log) cm2 leaf area as a covariate. The petiole length response variable was the residual vector from a linear regression with plant size-corrected leaf mass. Thus, this model focused on identifying deviations in the covariance between leaf area and petiole length across source and/or garden types, independent of how leaf mass, leaf area, and petiole length scale on average with plant size.
For each orthogonalized response variable, we first estimated all coefficients in a fixed effects interaction model without covariate terms, as specified in Eq. 1 (ESM §2.1), and then elaborated this model by adding the focal orthogonalized covariate and and its interaction with the fixed effects, recovering the full model specified by Eq. 4 (ESM §2.1). We then constructed nested simplified models that dropped higher order covariate terms and compared goodness-of-fit of each model using corrected Akaike’s Information Criterion (AICc). We report parameter estimates of models with the lowest AICc for which simplification yielded no additional reduction in AICc. From such models, we estimated 95% confidence limits on fixed and random effect coefficients via likelihood profiling, as implemented in lme4. P-values for coefficient estimates and for overall model effects in ANOVA-style linear hypothesis testing were calculated using F tests with the Kenward–Roger approximation for denominator degrees of freedom (Kenward and Roger 1997; Luke 2017), as implemented in package lmerTest (Kuznetsova et al. 2017). Finally, we used post-hoc pairwise least squares mean contrasts for source × garden interactions using lmerTest. Coefficient of determination for fixed effects ( ) and fixed + random effects ( ) was calculated according to Nakagawa and Schielzeth (2013) using package MuMIn (Bartón 2017).
(II) Principal components analysis (PCA)
We used as PCA inputs the scaled and centered trait vectors of leaf number, leaf mass (g), leaf area (cm2), and petiole length (mm). We conducted PCA with log-transformed input variables, which increased homogeneity of variance. For regression analysis, we modeled PC1 and PC2 (separately) as response variables according to Eq. 1 (ESM §2.1). For PC2, we then modeled covariate interactions with PC1 according to Eq. 4 (ESM §2.1), as described above.
Plant defenses and herbivore resistance
Glucosinolates (GSL)
We approached our analysis of foliar GSL content from the perspective of plant investment in defense. This required us to account for differences in how average GSL concentration changed with plant size and ontogeny (Boege and Marquis 2005; Barton and Boege 2017). To account for the effects of plant size (total number of leaves) and focal leaf size (wet mass), GSL concentrations and leaf wet mass were first regressed against (log transformed) leaf number. We then estimated effects of source and garden habitat as described earlier (following Eq. 4) using GSL concentration as the response variable and leaf mass as a covariate, with both vectors being orthogonal to plant size.
Our analysis was conducted separately for three GSL classes found in bittercress. The presence of one GSL class (OYG) was largely consistent within genets but polymorphic within the population (see Results). We therefore excluded data points with total OYG of zero to avoid conflating concentrations of GSL class with the production of that GSL class altogether. Models were constructed, compared, and analyzed as described above.
Next, as a complementary multivariate approach, we conducted PCA on centered and scaled log-transformed GSL concentrations for each of the eight measured compounds (ESM§4.1 Table S6). The PCs reflect the relative contribution of these eight compounds to a plant’s full GSL profile (rather than the total abundance of each compound). Each PC was modeled as a function of source and/or garden types, again including all model terms (without covariates) represented in Eq. 4 (ESM §2.1). The goal of this analysis was to investigate the effects of plant genet, source habitat, and garden habitat on variation in GSL profiles between plants in our common gardens.
Realized herbivore resistance
We analyzed S. nigrita larval mass (mg) as our response variable according to Eq. 1 (i.e. without any plant covariates) as an indirect measure of overall plant resistance to S. nigrita herbivory (“realized” resistance).
Greenhouse experiment
We analyzed leaf area (cm2) and petiole length (mm) of the four fully-expanded and earliest-developing leaves of seed-grown plants (n=37) at a single developmental point: immediately following the emergence of the seventh and eight leaves in each plant. Because seeds were only collected from each of five source sites, we restricted our analysis to examine the main effect of light environment (simulated high FR:R shade or a mock filter) and not source habitat. We capitalize on our nested design to estimate the variance contributed by genetic family (n=5 families, with between 4 and 11 separate plants reared from seed collected from siliques on a single parental ramet; ESM §1 Table S3). Because we measured foliar traits on multiple leaves per plant, we also included plant ID and leaf position as random intercept terms in the model. Confidence limits on fixed and random effects were constructed via likelihood profiling, as above.
Field survey of flowering phenology
To compare flowering distributions between sun and shade habitats at each of two sites, we compared the accumulation of each flowering event (first, peak, last) through time using nonparametric two-sample Kolmogorov–Smirnov (KS) tests. KS test p-values were obtained from 1000 replicate permutations of the identity of the source habitat across plants.
RESULTS
Effects of source habitat and light environment on foliar traits
Of the 540 rhizomes transplanted into the common gardens, 274 (~50%) survived transplantation and had ≥ 2 leaves after five weeks of growth. All of these plants were included in the foliar trait analyses below.
(I) Focal trait regression analysis
When grown in the shade, sun-derived plants had larger leaf area and longer petioles than shade-derived plants (ESM §3.1 Fig. S4C–D), while plants from both habitats were largely indistinguishable when grown in sun gardens, indicating a strong source × garden interaction (ESM §3.1 Tables S4–S5). Sun- and shade-derived plants also differed in average plant size and average mass of the largest leaf (Fig. S4A–B, Tables S4–S5); these traits varied among gardens as well. Without accounting for differences in plant and leaf size, the effects of source and garden habitat type on leaf traits cannot be disentangled from effects of plant and leaf ontogeny (Poorter 1999; Valladares & Niinemets 2008). Thus, below we present the results from an analysis that sought to control for the ways that plant size, and its relationship leaf mass, impact the distribution of leaf area and petiole length among plants.
Leaf area
We first determined how leaf area differed between source and garden types independent of differences in plant size and mass of leaves. For plots of this response variable against the leaf mass covariate see ESM §3.2 Fig. S6B. The leaf mass covariate strongly and positively predicted our leaf area response variable (γ0= 0.77 [0.7, 0.84], p<0.001, Table 1): heavier than expected leaves also have larger than expected leaf area. Additionally, the effect of garden habitat on leaf area differed between plants from sun and shade source habitats (Fig. 1A, source × garden overall effect, p<0.001, Table 3). Specifically, in shade gardens, leaves from sun-derived plants were 20% larger than expected based on their size and leaf mass than those from shade-derived plants (α1 = 0.08 [0.03, 0.13], p<0.001, Table 1). This difference between source types dropped to ~0 in sun gardens (β1 = −0.09 [−0.14, −0.05], p<0.001, Table 1), resulting in plants from each source habitat showing low mass-corrected leaf area in sun gardens (β0 = −0.08 [−0.16,0], 0.05≤p<0.1, Table 1; overall garden effect p=0.02, Table 3; ESM §3.3 Fig. S7A–B). Notably, the rate at which sun-derived plants gained leaf area per unit leaf mass was 30% greater than for shade-derived plants on average (γ1 = 0.12 [0.03, 0.21], p<0.05, Table 1). This indicates that source types differed in how leaf mass was translated into leaf area (covariate × source interaction, p=0.012, Table 3). Fixed effects explained ~80% of the variation in the data for the leaf area model reported in Table 1.
Table 1.
Coefficient estimates for models of plant foliar phenotypes from field common garden experiment.
| Term type | Coefficient estimate | sym1 | (log) leaf area (cm2)4 | petiole length (mm) A5 | petiole length (mm) B6 | PC1 | PC2 | |
|---|---|---|---|---|---|---|---|---|
| Main effects | Intercept [shade] | α0 | 0.02 (−0.04, 0.08) | 1.2 (−7.7, 10.0) | −1.0 (−7.3, 5.3) | −1.94 (−2.6, –1.3)*** | 0.44 (−0.08, 0.96) | |
| source [sun] | αk | 0.08 (0.03, 0.13)*** | 11.8 (6.4, 17.2)*** | 4.0 (−0.4, 8.4)• | 1.29 (0.6, 2.0)*** | 0.39 (0.15, 0.64)*** | ||
| garden [sun] | β0 | −0.08 (−0.16, 0)• | −11.6 (−23.8, 0.60) | −4.8 (−14.0, 4.0) | 2.3 (1.4, 3.1)*** | −0.92 (−1.65, −0.2)• | ||
| source * garden [sun, sun] | βk | −0.09 (−0.14, –0.05)*** | −6.5 (−12.4, −0.8)* | 0.9 (−4.4, 6.2) | −0.9 (−1.5, −0.3)*** | −0.44 (−0.77, −0.12)* | ||
| Covariate terms | x | γ0 | 0.77 (0.7, 0.84)*** | 23.6 (10.3, 37.1)*** | 50 (24, 76)*** | |||
| x × source [sun] | γ1 | 0.12 (0.03, 0.21)* | 15.6 (−2.8, 33.8)• | 39 (5.4, 73)* | ||||
| x × garden [sun] | γ2 | – | 2.4 (−16.09, 20.45) | 7.8 (−29.0, −44.0) | ||||
| x × source [sun] × garden [sun] | γ12 | – | −16.3 (−40, 7.15) | −36.5 (−83.61-9.48) | ||||
| Random effects2 | source ID | τα(j) | 0.04 (0.02, 0.06) | 3.1 (1.1, 5.3) | 1.6 (0, 3.16) | 0.5 (0.2, 0.8) | 0 (0, 0.16) | |
| garden ID | τβ(l) | 0.04 (0.02, 0.08) | 7.3 (3.5, 12.3) | 5.2 (2.3, 8.7) | 0.45 (0.2, 0.8) | 0.44 (0.2, 0.7) | ||
| residual error | σe | 0.08 (0.08, 0.09) | 10.5 (9.5, 11.3) | 9.2 (8.4, 10) | 1.2 (1.1, 1.3) | 0.65 (0.6, 0.7) | ||
| Model statistics |
|
0.79 | 0.33 | 0.47 | 0.31 | 0.38 | ||
|
|
0.86 | 0.58 | 0.61 | 0.47 | 0.57 |
p-values indicated by asterisks:
0.05<p<0.1,
p<0.05,
p<0.01,
p<0.001 from t tests with Kenward–Roger approximated ddf; confidence intervals calculated via profile likelihood.
see ESM section 2.1 for symbolic representation of model coefficients in context of regression equations.
group-level random effect estimates are presented on the standard deviation scale.
response = residual(log leaf area ~ log leaf number); covariate = residual(log leaf mass ~ log leaf number).
response = residual(petiole length ~ log leaf number); covariate = residual(log leaf mass ~ log leaf number)
response = residual(petiole length ~ log leaf number + log leaf mass); covariate = residual(log leaf area ~ log leaf number + log leaf mass)
Fig. 1. Foliar phenotypes are differentiated between bittercress from sun and shade source habitats when grown in shade gardens.
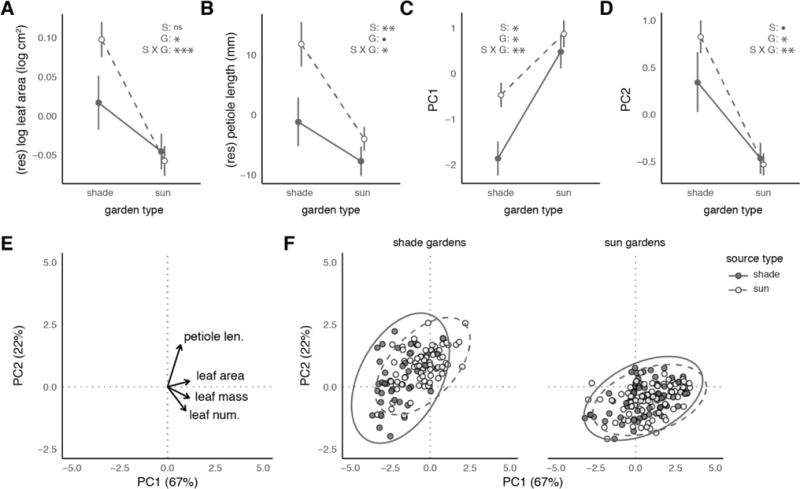
Sun source plants exhibit increased leaf area (A) and petiole length (B) compared to shade source plants when grown in shade gardens, independent of overall plant size and the mass of focal leaves (see Methods). Principal components analysis (PCA; E–F) reveals average differences in PC1 (C) and PC2 (D) shade gardens but not sun gardens. E. Vector loadings of four log-transformed input trait vectors, with leaf area and petiole length largely oriented towards upper right quadrant. P-values correspond to source (‘S’), garden (‘G’), or source × garden (‘S × G’) effects in Table 3. F. 95% ellipsoids are drawn around data points from each source type.
Table 3.
ANOVA-style hypothesis tests for significance of model terms.
| Response | Model term | MSE | F | P-value |
|---|---|---|---|---|
| (res) leaf area (log cm2) | Source | 0.02 | F(1,14) = 2.6 | 0.131 |
| Garden | 0.08 | F(1,4) = 11.1 | 0.027 | |
| source × garden | 0.10 | F(1,257) = 15.0 | <0.001 | |
| (res) leaf mass (log g) | 7.98 | F(1,261) = 1169.2 | <0.001 | |
| (res) leaf mass (log g) × source | 0.04 | F(1,260) = 6.4 | 0.012 | |
| (res) petiole length (mm) A2 | source | 1702 | F(1,18) = 15.5 | 0.001 |
| garden | 637 | F(1,4.1) = 5.8 | 0.072 | |
| source × garden | 520 | F(1,259) = 4.8 | 0.030 | |
| (res) leaf mass (log g) | 9098 | F(1,261) = 83.1 | <0.001 | |
| (res) leaf mass (log g) × source | 162 | F(1,260) = 1.5 | 0.225 | |
| (res) leaf mass (log g) × garden | 94 | F(1,260) = 0.9 | 0.355 | |
| (res) leaf mass (log g) × source × garden | 194 | F(1,259) = 1.8 | 0.184 | |
| (res) petiole length (mm) B3 | source | 8121 | F(1,256.0) = 95.2 | <0.001 |
| garden | 674 | F(1,21.6) = 7.9 | 0.010 | |
| source × garden | 79 | F(1,4.2) = 0.9 | 0.388 | |
| (res) leaf area (log cm2) | 10 | F(1,260.4) = 0.1 | 0.738 | |
| (res) leaf area (log cm2) × source | 241 | F(1,257.8) = 2.8 | 0.094 | |
| (res) leaf area (log cm2) × garden | 57 | F(1,263.8) = 0.7 | 0.415 | |
| (res) leaf area (log cm2) × source × garden | 197 | F(1,259.4) = 2.3 | 0.130 | |
| PC1 | source | 12.52 | F(1,15) = 8.4 | 0.011 |
| garden | 31.09 | F(1,4) = 20 | 0.01 | |
| source × garden | 11.98 | F(1,256) = 8.1 | 0.005 | |
| PC2 | source | 1.75 | F(1,15) = 4.1 | 0.059 |
| garden | 3.97 | F(1,4) = 9.4 | 0.037 | |
| source × garden | 3.01 | F(1,262) = 7.1 | 0.008 | |
| (res) IYG (log AUC1) | source | 0.035 | F(1,18) = 0.8 | 0.398 |
| garden | 0.028 | F(1,7) = 0.6 | 0.467 | |
| source × garden | 0.509 | F(1,85) = 10.1 | 0.001 | |
| (res) leaf mass (log g) | 0.618 | F(1,84) = 13.1 | 0.001 | |
| (res) I3M (log AUC) | source | 0.151 | F(1,17) = 1.4 | 0.258 |
| (res) leaf mass (log g) | 0.003 | F(1,88) = 0.1 | 0.873 | |
| (res) leaf mass (log g) × source | 0.879 | F(1,95) = 8.0 | 0.006 | |
| Larval mass (mg) | source | 0.495 | F(1,15) = 1.6 | 0.228 |
| garden | 0.634 | F(1,3.7) = 2.0 | 0.234 | |
| source × garden | 0.911 | F(1,143) = 2.9 | 0.091 | |
| larval batch | 0.571 | F(1,3.3) = 1.8 | 0.263 |
AUC = area under curve
Petiole length (A)
Next, we examined whether petioles elongated differently between source types across garden types, using plant size-corrected leaf mass as a covariate. For plots of this response variable against the leaf mass covariate see ESM §3.2 Fig. S6C. In shade gardens, independent of the effects of plant and leaf size, the petioles of sun-derived plants were >11 mm longer on average (77% of one population standard deviation, σ = 15 mm) than those from shade-derived plants (α1 = 11.8 [6.4,17.2], p<0.001, Table 1). While petioles on shade-source plants showed a 11 mm marginally significant reduction in sun gardens (β0 = −11.6 [−24,0.6], 0.05≤p<0.01, Table 1), sun-derived plants showed a 40% stronger (6.5 mm) reduction in their petioles in sun gardens, (β1 = −6.5 [−12.4, −0.8], p<0.05, Table 1), indicating an overall source × garden interaction (p=0.03, Table 3). Finally, the scaling between leaf mass and petiole length was marginally significantly different between source types when growing in the shade: for every unit increase in (log) mass invested in leaves, petioles elongated >60% more on average for sun-compared to shade-derived plants (ESM §3.3 Fig. S7C; γ1 = 16 [−3, 34], 0.05≤p<0.1, Table 1). These patterns indicate that plasticity in petiole length across garden types was higher in sun-source than shade-source plants (Fig. S7C–D). Together, fixed effects explained 33% of the variation in the data for this model (Table 1).
Petiole length (B)
We next addressed whether differential petiole elongation was correlated with leaf area differences noted above. For plots of this response variable against the leaf area covariate see ESM §3.2 Fig. S6D. On average, residual differences in leaf area largely explained residual differences in petiole length for shade-derived plants (γ0 = 49 [24, 76], p<0.001, Table 1). However, for sun-derived plants growing in shade gardens, the rate at which residual petiole length increased with (log) residual leaf area was ~75% steeper than for shade-derived plants (ESM §3.3 Fig. S7E; γ1 = 39 [5, 73], p<0.05, Table 1), and this difference was reduced in sun gardens (γ12 = −36 [−83, 10], p>0.1, Table 1). While such thee-way covariate interaction terms were not statistically significant by ANOVA (Table 3), their inclusion in the final regression model was well-supported by ΔAICc = −6.4 compared to the next simplest model, and ΔAICc = −15 over a model lacking all covariate interactions. These patterns indicate that differences in petiole elongation between sun- and shade-source plants cannot be explained solely as a by-product of differences in leaf area.
For the preceding three models, the magnitude of the variance un-absorbed by the fixed effects (τα(j) + τβ() + σe) was apportioned similarly or more to plot-to-plot variation among gardens (τα(j)) compared to among-genet (τβ()) variation (Table 1).
(II) Principal components analysis
Together, PC1 (67%) and PC2 (22%) explained ~90% of variation in the set of four input traits (number of leaves, and mass, area, and petiole length of the largest leaf). Petiole length loaded almost directly along PC2, while PC1 represented loadings from leaf area, leaf mass, and leaf number (Fig. 1E). In shade gardens, sun-derived plants had higher average values of PC1 compared to shade-derived plants (Fig. 1C; α1 = 1.3 [0.6, 2.0], p<0.001, Table 1). Growth in sun gardens increased values of PC1 for shade-derived plants (β0 = 2.3 [1.4, 3.1I], p<0.001, Table 1) but did so less for sun-derived plants (β1 = −0.9 [−1.5, −0.3], p<0.001, Table 1). This difference in phenotype between plants from the sun and shade sources in sun gardens is reflected in an overall source × garden interaction (p=0.005, Table 2), beyond the average garden type effect (p=0.01, Table 2). Fixed effects explained 31% of variation in PC1 (Table 1).
Table 2.
Coefficient estimates for models of plant defense chemistry and realized herbivore resistance.
| Term type | Coefficient estimate | sym1 | (log) IYG3,4 | (log) OYG3,4 | (log) I3M3,4 | larval mass (mg)5 | |
|---|---|---|---|---|---|---|---|
| Main effects | Intercept [shade] | α0 | −0.2 (−0.44, 0.03) | −0.06 (−0.55, 0.43) | 0.05 (−0.1, 60.19) | 1.58 (1.34, 1.8)*** | |
| Source [sun] | αk | 0.3 (0.02, 0.58)• | – | −0.11 (−0.3, 0.07) | −0.29 (−0.53, −0.01)* | ||
| Garden [sun] | β0 | 0.24 (0.04, 0.45)* | – | – | −0.36 (−0.68, −0.03)• | ||
| Source [sun] * garden [sun] | βk | −0.39 (−0.62, −0.16)*** | – | – | 0.33 (−0.05, 0.7)• | ||
| Covariate terms | x | γ0 | −0.54 (−0.82, −0.27)*** | – | 0.6 (−0.11, 1.37) | 0.18 (−0.01, 0.38) | |
| x × source [sun] | γ1 | – | – | −1.28 (−2.23, −0.44)* | – | ||
| x × garden [sun] | γ2 | – | – | – | – | ||
| x × source [sun] × garden [sun] | γ12 | – | – | – | – | ||
| Random effects2 | genet | τα(j) | 0.21 (0.13, 0.31) | 0.91 (0.62, 1.34) | 0.12 (0, 0.21) | 0 (0, 0.14) | |
| garden plot | τβ() | 0 (0, 0.08) | 0.05 (0, 0.35) | 0.03 (0, 0.16) | 0.1 (0, 0.18) | ||
| residual error | σe | 0.22 (0.18, 0.25) | 0.49 (0.41, 0.59) | 0.33 (0.28, 0.38) | 0.56 (0.5, 0.62) | ||
| Model statistics |
|
0.14 | 0 | 0.12 | 0.05 | ||
|
|
0.56 | 0.78 | 0.23 | 0.08 |
p-values indicated by asterisks:
0.05<p<0.1,
p<0.05,
p<0.01,
p<0.001 based on t tests using Kenward–Roger approximation for ddf
confidence intervals calculated via profile likelihood.
see ESM section 2.1 for symbolic representation of model coefficients in context of regression equations.
group-level effect estimates are presented on the standard deviation scale.
covariate = residual(log leaf mass ~ log leaf number)
response = residual(log [GSL] ~ log leaf number)
covariate = larval batch (A or B).
Sun-derived plants had higher values of PC2 than shade-derived plants when growing in the shade (Fig. 1D; α1 = 0.39 [0.15, 0.64], p<0.001, Table 1). This difference was not present in sun gardens (β1 = −0.44 [−0.77, −0.12], p=0.01, Table 1), reflected in an overall source × garden interaction (p=0.008, Table 3), beyond the average garden type effect (p=0.04, Table 3). Fixed effects explained 38% of variation in PC2 (Table 1).
Simulated neighbor shade response in bittercress grown from seed
Under simulated neighbor shade, bittercress seedlings showed a 2.3-fold average reduction in leaf area and grew ~1.5-fold longer petioles (β = −26 [−31, −21], p=<0.001, ESM §5 Fig. S12, Tables S7–S8). This indicates that bittercress does indeed exhibit an elongation response consistent with shade avoidance, and that this effect was evident across five separate maternal plants (e.g., families). Among-family differences explained ~15% of the variation un-explained by fixed effects in models of petiole length and leaf area (Table S7), indicating the potential for genetically-based variation in foliar traits. Because any given pair of seedling within a family are expected to be half or full sibs (and thus share 50% of the maternal and possibly paternal haplotypes), the phenotypic variance apportioned among families can be interpreted as a conservative lower bound on the broad-sense heritability of shade-responsive foliar traits. However, this experimental design conflates genetic and maternal effects and thus might overestimate heritability if petiole length is strongly influenced by maternal effects.
Effects source habitat and light environment on plant defense and herbivore resistance
Herbivore-inducible plant secondary compounds
For IYG (the dominant class of GSL in bittercress), sun-derived plants exhibited a marginally significant 2-fold increase in size- and mass-corrected IYG concentration compared to shade-derived plants when grown in shade gardens (Fig. 2A; α1 = 0.3 [0.02, 0.6], 0.05≤p<0.1, Table 2). This effect was reversed in sun gardens (β1 = −0.39 [−0.62, 0.16], p<0.001, Table 2), which accounts for the overall significant source × garden effect in this model (p<0.001, Table 3; post-hoc contrasts in ESM §4.1 Fig. S9A–B). Across all plants collectively, the relationship between the leaf mass covariate and IYG concentration was negative after accounting for leaf number (γ0 = −0.54 [−0.82, 0.27], p<0.001, Table 2). Thus, smaller than average leaves had lower IYG concentrations. Statistical interactions between the covariate and source or garden terms were not supported by model comparisons (gains via ΔAICc < 2 upon their addition). Total variation explained by fixed effects was modest ( ), but among-genet variance captured 50% of the remaining unexplained variance, increasing to 0.56. This indicated a large degree of among-genet variation unexplained by measured factors (Table 2).
Fig. 2. Plant chemical defenses and realized herbivore resistance across source and garden types.
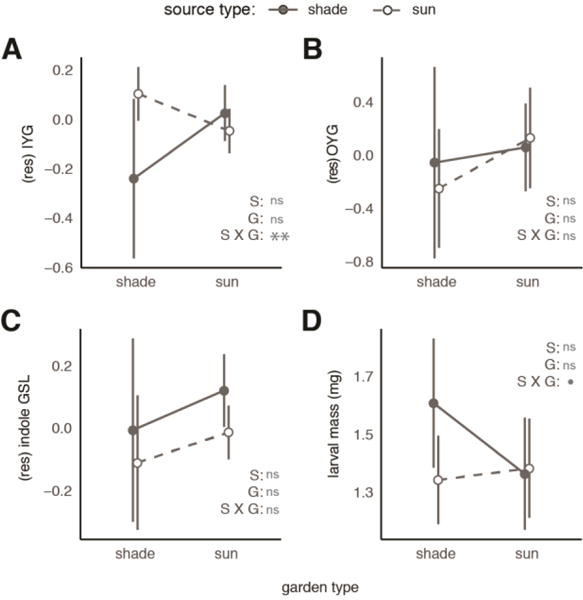
Isothiocyanate-yielding glucosinolate (IYG) concentrations are differentiated between source habitats in shade gardens (A), while oxazolidine-2-thione-yielding glucosinolates (OYG) (B) and indole glucosinolates (C) show no differentiation between source or garden types. P-values correspond to source (‘S’), garden (‘G’), or source × garden (‘S × G’) effects in Table 3.
In contrast to IYG, concentrations of the two other GSL classes—which do not predominantly yield isothiocyanates and have not yet been linked to herbivore resistance in bittercress—did not vary by plant source habitat or garden habitat (Fig. 2B). Instead, we noted idiosyncratic differences among plant genets in OYG concentrations (τβ() = 0.91, or 62% of the total variation; total model
; Tables 2–3). Indole GSL concentrations were similarly idiosyncratic among genets (Fig. 2C; ), and among-genet variance explained 25% of the remaining variance (Tables 2–3). Indole GSL concentrations decreased more with leaf mass in sun-derived than shade-derived plants (γ1 = −1.3 [−2.2, −0.4], p=0.01, Table 2), yielding a source × covariate effect (p=0.006, Table 3; ESM §4.1 Fig. S9C).
When we considered relative concentrations of each of the eight GSL compounds together through PCA (see Methods), we found that among-genet variation explained 65, 56, and 70% of all variation in PCs 1–3, respectively (ESM §4.1.2 Fig. S10). This indicates a high degree of heritability (in the broad sense) in GSL composition among clonal plant groups separated at small spatial scales.
Realized herbivore resistance
Residual variation in larval mass unexplained by model terms was extensive (R2 of full model = 0.08). Nonetheless, in shade gardens, larvae that fed on sun-derived plants were on average 18% lower in mass compared to those that fed on shade-derived plants (1.29 vs.1.58 mg; Fig. 2D; α1 = −0.29 [−0.53, −0.01], p = 0.04; Table 1). This difference in larval mass between source plant types was only observed in shade gardens (Fig. 2D; β0 = −0.36 [−0.68, −0.03]; β1 = 0.33 [−0.05,0.7], both 0.05≤p<0.1; Table 1), reflected in a marginally significant source × garden interaction (p=0.09, Table 2). The only detectable source × garden contrast (p<0.05) was between source types in shade gardens (ESM §4.2 Fig. S11).
Effects of source habitat on flowering phenology in the field
At the 401 Trail site, plants in all habitats exhibited very similar flowering curves through time (K–S tests, all comparisons p>0.05; Fig. 3A–B). In contrast, at the Copper Creek site, timing of first, peak, and last flowering was delayed for bittercress growing in intermediate shade and deep shade compared to open sun habitats (sun vs. intermediate or deep shade all p<0.05; Fig. 3C–D). Examined another way, the abundance of flowering stems in each habitat through time strongly overlapped among habitats at the 401 Trail site, while plants in deep shade habitats reached peak flower abundance later than those in open sun and intermediate shade at Copper Creek (Fig. 3B).
Fig. 3. Differences in flowering phenology between sun and shade habitats.
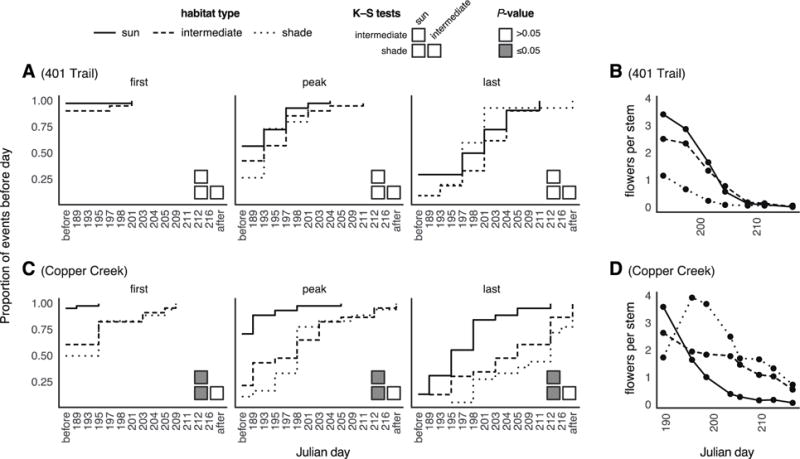
(A) Phenological progression through first, peak, and last flowering, depicted as empirical cumulative distributions, indicate faster flowering for plants in the sun at Copper Creek but not 401 Trail site. Plots show the proportion of flowering events event on or before a given Julian day. P-values are from pairwise two-sample, two-tailed Kolmogorov–Smirnov (K–S) tests. (B) Pooled average flower density through time shows delayed flowering peak in shade habitats at Copper Creek but not 401 Trail site.
DISCUSSION
Bittercress is distributed across a fine-grained habitat mosaic, ranging from high light intensity, high herbivory, and dense neighbor competition in meadows (sun habitat), to low light availability and low herbivory in the deep shade created by evergreen forest canopies (shade habitat). Through reciprocal-transplant common gardens, we found evidence for microgeographic phenotypic variation that tracks these distinct light and herbivory environments. Divergence was detected across traits indicative of plant size, leaf morphology, defense, and realized resistance to a dominant herbivore. The expression of these differences was dependent on garden habitat, typically manifesting only in response to shade. Because we sampled interspersed source sites separated by a few km at most (and often only a few hundred m), the significant effects of source habitat in our common gardens could only arise through phenotypic divergence among sun- and shade-inhabiting populations that occurs relatively consistently, and at fine-spatial scales, across the landscape. Although we did not measure plant fitness, and thus did not explicitly test for local adaptation, patterns of divergence generally mirrored well-studied cases of local adaptation to light and herbivory in other plant species (as described below). Overall, our study illuminates how variation in biotic and abiotic stressors across a fine-grained habitat mosaic may influence divergence in light- and herbivore-responsive traits. Additionally, partially non-overlapping flowering phenology observed between habitat types may be at least one factor that helps to reinforce habitat-specific divergence in bittercress.
Heritable divergence in plant size, leaf morphology, and herbivore resistance traits across a fine-grained habitat mosaic
Divergence in plant size and leaf traits
Overall, our common garden and greenhouse experiments revealed that (1) leaf morphological traits (leaf area and petiole length) exhibit plastic responses to light habitat in bittercress, and (2) the magnitudes of these plastic responses differ between bittercress from sun and shade source sites. Specifically, after accounting for the effect of plant and leaf ontogeny on petiole length, we found that bittercress derived from both sun and shade populations grew larger leaves (i.e., larger leaf area relative to leaf mass) and longer petioles in shade vs. sun common gardens. Notably, however, the shade-induced leaf area expansion and petiole elongation responses were exaggerated in sun source plants (Fig. 1A–B). These patterns were further reflected in an analysis of plant variation in the context of PCA, where growth in shade gardens led to differentiation by garden and source habitat type along both PC1 and PC2.
In general, our findings mirror heritable differences in the response to shading that have been observed between plant populations and between closely-related species that occupy different light habitats. This is typified by an attenuated (or absent) petiole elongation response in populations from consistently shaded or sparsely vegetated habitats where SAS is unlikely to offer a competitive advantage (Dudley and Schmitt 1995; van Hinsberg et al. 1997; Donohue et al. 2000; Schmitt et al. 2003; Bell and Galloway 2008; Sasidharan et al. 2008; Gommers et al. 2017). The stronger elongation response observed in bittercress from sun habitats may aid in outcompeting herbaceous competitors in meadows. In contrast, the weaker response in bittercress from shade habitats may help avoid the cost of allocating resources to a growth strategy that offers no competitive advantage in forest understories. Evaluating such adaptive hypotheses for the observed patterns of phenotypic differentiation awaits further study.
The biological significance of differences in leaf area expansion between habitat types is less clear. An increased ratio of leaf area relative to leaf mass is frequently observed in response to shading across plants, and this is hypothesized to be one strategy for optimizing photosynthesis in low light conditions (Valladares & Niinemets 2008; Gommers et al. 2013). However, an alternative hypothesis is that an increase in leaf area per leaf mass in response to shading may reflect an inability to maintain biomass under low resource conditions and be a poor indicator of shade tolerance (Liu et al. 2016b). Further, physiological rather than morphological changes may be more important for shade tolerance (Valladares & Niinemets 2008; Gommers et al. 2013). More comprehensive metrics, such as the relative allocation of biomass across tissue types, may offer a clearer picture of how bittercress tolerates (or suffers from) shading.
Bittercress from shade populations were also smaller in both garden types (Fig. S8, Tables S4–S5). This pattern is consistent with predictions that slower growth may facilitate shade tolerance (Gommers et al. 2013). Alternatively, plants from shade populations might be smaller because they simply use light resources less efficiently, and this may not necessarily be adaptive (Liu et al. 2016b). Dry mass of the largest leaf (independent of plant size) was also higher for sun-derived plants, but only in shade gardens (Fig. S8, Tables S4–S5). This suggests that the relationship between plant size and resources invested in leaves systematically varies between plants from sun and shade habitats when grown in shade gardens, which adds to the evidence of multi-modal phenotypic divergence in bittercress across habitats.
Divergence in defensive chemistry and anti-herbivore resistance
Our results indicate two important features of how chemical defenses vary at fine spatial scales in bittercress. First, investment in foliar IYGs varies as a function of light environment, and bittercress from sun and shade populations exhibit average differences in the extent to which this occurs. Second, such changes in average IYG concentrations occur against a backdrop of high GSL compositional variation (e.g., clonally-heritable variation in the relative abundance of different GSL compounds) among bittercress genotypes separated by as little as < 1km. In parallel, we found that performance of a dominant herbivore, S. nigrita, differed between bittercress from sun shade habitats. These differences only emerged in shade common gardens, where plants from shade sources had relatively lower levels of foliar IYGs and lower resistance to S. nigrita than plants from sun sources. These patterns detected within shade gardens had only modest statistical support due to low replication of IYG profiles among shade source plants and because our resistance bio-assay was inherently noisy (see Methods). Nonetheless, the result is consistent with the expected positive effect of IYGs on resistance to S. nigrita.
Two complementary interpretations suggest divergence we observed in foliar IYG concentration and herbivore resistance may be locally adaptive, given the context in which shade and herbivory are experienced in meadow and shade habitats by bittercress. First, our results are consistent with a reduction of investment in IYGs upon perception of shade by bittercress populations from shade habitats. Shade generated by the dense forest canopy is a reliable indicator of low levels of herbivory for bittercress. The production of chemical defenses (including GSL) is metabolically costly (Züst et al. 2011; Bekaert et al. 2012; Züst and Agrawal 2017), so for bittercress populations in deeply shaded habitats, attenuated defenses upon shade perception may enable reallocation of resources to other physiological processes without substantially increasing the risk of defoliation. Second, our results are consistent with a lack of shade-induced plasticity in IYG production (or even a modest shade-induced increase in IYGs) by bittercress from sun habitats. In sun habitats, shade is caused by herbaceous plants and is not a reliable indicator of herbivore abundance, since herbivory is not reduced on bittercress in shaded microhabitats in meadows (Collinge and Louda 1988b). Attenuated investment in defenses following shade perception may therefore risk heavy defoliation in sun habitats, and this may explain why bittercress from sun populations did not exhibit this pattern.
More extensive assays, coupled with measurements of plant fitness, are needed to test if the phenotypic divergence we observed in IYG levels and realized anti-herbivore resistance are locally adaptive. Inter-habitat variation in factors such as resource availability (Coley 1985) or abiotic stress (e.g., higher water stress for bittercress in sun; Louda and Collinge 1992) may interact with herbivore abundance to determine the costs and benefits of investment in defense. Interestingly, Mooney et al. (2016) found that plants (Ligusticum porteri, Apiaceae) in meadows near our field sites had higher aphid abundances than conspecifics in the forest understory due to the effect of higher ant activity and ant tending of aphids in meadows, which provided protection against aphid predators. Whether habitat-dependent effects on indirect defenses and tri-trophic interactions also shape herbivory and plant fitness across habitats is not known in bittercress.
No evidence that a SAS–defense tradeoff constrains habitat-associated phenotypic divergence
Negative phenotypic correlations between SAS and defensive traits have been observed in shade-intolerant plants, such as Arabidopsis thaliana (Arabidopsis), that mount a morphological elongation response upon perception of shade (Wit et al. 2013; Ballaré 2014). In fact, the trait (petiole length) we measured as a metric for a shade avoidance response is itself modified in the opposite direction by wounding-induced signaling in Arabidopsis (Cipollini 2005). Observations in our common gardens suggest that while bittercress does mount a shade-induced petiole elongation response indicative of SAS, any SAS–defense tradeoff that might exist in bittercress did not prevent sun-derived plants in our common gardens from mounting a robust defense response upon perception of shade. Specifically, in shaded common gardens, bittercress derived from sun habitats exhibited stronger SAS, yet accumulated higher foliar IYG concentrations and were more resistant to S. nigrita, than bittercress from shade habitats.
Our results are consistent with extensive evidence that a tradeoff between SAS and defense can be decoupled through mutation and natural selection (reviewed in, e.g., Ballaré 2014; Ballaré and Pierik 2017). Prolonged exposure to different regimes of herbivore pressure alters correlations between traits involved in competitive growth and resistance to herbivory in experimentally evolved plant populations (Uesugi et al. 2017). Functional genetic studies in Arabidopsis have further elucidated how SAS and defense can be uncoupled at the molecular level. Upon perception of light with reduced R:FR, phyB (a light-sensing phytochrome protein) both triggers SAS growth responses and desensitizes plants to JA, a potent regulator of plant defenses against herbivores and necrotrophs (Moreno et al. 2009). phyB-mediated destabilization of JAZ proteins, which repress JA-dependent defenses, plays a key role in the shade-induced attenuation of defenses (Leone et al. 2014; Chico et al. 2014). Accordingly, plants bearing specific mutations affecting the phyB and/or JAZ proteins can simultaneously mount SAS and an unimpaired or even elevated defense response (Campos et al. 2016; Cerrudo et al. 2017). Future studies could test if habitat-associated divergence in bittercress elongation and defense phenotypes have evolved through genetic or epigenetic mutations to these and other well-characterized genes that mediate SAS–defense tradeoffs in Arabidopsis.
Our results do not provide evidence for a decoupling of SAS and defense in bittercress. Tradeoffs that arise through pleiotropy or resource allocation constraints can be masked when genotypes vary in their efficiency of resource acquisition or use (Agrawal et al. 2010; Züst and Agrawal 2017). Although rhizome size was standardized in our common garden experiment, the quality or quantity of stored resources may differ among rhizomes collected from different habitat sites. Such differences, and their effects on unmeasured traits that impact plant defense and herbivore performance, might explain why plants from sun sources exhibited SAS without attenuated GSL concentrations or resistance to S. nigrita.
Potential mechanisms underlying microgeographic variation in bittercress
Maternal inheritance of habitat-induced phenotypes is a potential driver of habitat differentiation in bittercress revealed by our study. Transgenerational effects of parental environment on the phenotypes of clonally propagated offspring (Schwaegerle et al. 2000; Latzel and Klimešová 2010) and seed propagated offspring (Galloway 2005; Galloway and Etterson 2007) have been well documented in plants. These effects can shape offspring resistance against herbivory (Agrawal 2001; Rasmann et al. 2012; Colicchio 2017), and epigenetic variants induced by (and affecting) resistance phenotypes can persist for multiple generations (Rasmann et al. 2012). Habitat matching of heritable phenotypes in bittercress could therefore be driven simply by parental experience, without requiring natural selection to filter locally unfit genotypes from sun and shade habitats.
Alternatively, if microgeographic divergence in the expression of growth and defense has a genetic basis in bittercress, matching of these genotypes to local habitats would require strong, spatially varying selection to overcome homogenizing effects of gene flow across fine-grained habitat mosaics (Lenormand 2002). There is evidence that this filtering process can occur in plants, sometimes at the scale of only a few meters (Waser and Price 1989; Schmitt and Gamble 1990; Antonovics 2006; Schemske and Bierzychudek 2007; Hendrick et al. 2016). If such differentiation does in fact have an additive genetic basis in bittercress, differences in flowering time among habitat patches, coupled with natural filtering unfit phenotypes migrating across habitats, could facilitate the buildup of locally adaptive genetic variants (Levin 2009).
We monitored flowering time between adjacent sun and shade habitats at two sites and found a marked difference in one of these sites, with shade plants flowering later on average than those in sun. Flowering distributions still overlapped even at the site where these average differences were observed; thus, microgeographic divergence in bittercress is unlikely to be the simple product of phenological asynchrony preventing gene flow between habitats. Nevertheless, the earlier initiation, peak, and conclusion of flowering time in sun habitats, where it occurs, would be expected to reduce inter-habitat gene flow and thus could facilitate microgeographic divergence. Furthermore, mechanisms other than plant phenology, such as spatial variation in pollinator distributions, might also reduce inter-habitat gene flow in bittercress. In fact, the composition of the pollinator community associated with bittercress varies between sun and shade habitats (Louda and Rodman 1996), driven by higher pierid visitation in the sun.
Conclusion
We found that bittercress exhibits growth, morphological, and defense phenotypes that track parental habitat across a fine-grained (i.e., microgeographic) habitat mosaic. Plants from sun and shade populations primarily differed in their phenotypically plastic responses to shade: a weak elongation response and attenuated defenses in populations from shade habitats, and a strong elongation response and elevated defenses in populations from sun habitats. Phenotypic divergence was studied in reciprocal transplant—common gardens using clonally-propagated rhizomes, and studies in seed-propagated offspring are needed to disentangle effects of genetic variation from transgenerational effects of parental habitat on offspring phenotypes. Nonetheless, if this phenotypic divergence causes offspring to have higher viability or fecundity in their natal habitats, it could be an important mechanism that reduces rates of inter-habitat recruitment and gene flow. This in turn could facilitate the maintenance of genetic and epigenetic variation within bittercress populations (Hedrick 2006). The ecological, physiological, and evolutionary mechanisms underlying microgeographic phenotype-habitat matching in bittercress—and their generality across plant systems—await future study.
Supplementary Material
Acknowledgments
We acknowledge Ian Billick (RMBL), Jennifer Reithel (RMBL), Kailen Mooney (UC-Irvine), and Carol Boggs (University of South Carolina) for advice during the design and data collection phases of this project. We also thank Timothy Morton (University of Chicago) for assistance with glucosinolate profiling. Financial support was provided through the Research Experience for Undergraduate Site Program at the National Science Foundation Division of Environmental Biology (0753774) at the RMBL, to N.K.W. by the RMBL (Research Fellowships 2010–2013), NSF DEB (1256758), the John Templeton Foundation (41855) and the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM119816; to P.T.H. by RMBL Graduate Fellowships (2011–2013), the University of Arizona Center for Insect Science, and NSF DEB (1309493); to A.D.G. by NSF DEB (1405966), an NSF Graduate Research Fellowship, and an RMBL Graduate Fellowship (2011); and to J.F. and S.F. by RMBL undergraduate research awards.
Footnotes
Author contributions: ADG, ACND, SF and NKW designed the experiments. ADG, JF, ACND, SF and NKW carried out the work. PTH and ADG conducted the analyses. PTH, ADG and NKW wrote the paper.
References
- Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophoton Int. 2004;11:36–42. doi: 10.1002/9780470089941.eta03cs9. [DOI] [Google Scholar]
- Agerbirk N, De Vos M, Kim JH, Jander G. Indole glucosinolate breakdown and its biological effects. Phytochemistry Reviews. 2009;8:101–120. doi: 10.1007/s11101-008-9098-0. [DOI] [Google Scholar]
- Agerbirk N, Olsen CE, Chew FS, Ørgaard M. Variable glucosinolate profiles of Cardamine pratensis (Brassicaceae) with equal chromosome numbers. Journal of Agricultural and Food Chemistry. 2010;58(8):4693–4700. doi: 10.1021/jf904362m. [DOI] [PubMed] [Google Scholar]
- Agrawal AA, Hastings AP, Johnson MT, Maron JL, Salminen J. Insect herbivores drive real-time ecological and evolutionary change in plant populations. Science. 2012;338:113–116. doi: 10.1126/science.1225977. [DOI] [PubMed] [Google Scholar]
- Agrawal AA, Conner JK, Rasmann S. Tradeoffs and negative correlations in evolutionary ecology. In: Bell MA, Eanes WF, Futuyma DJ, Levinton JS, editors. Evolution After Darwin: The First 150 Years. Sinauer Associates; Sunderland, MA: 2010. [Google Scholar]
- Agrawal AA. Transgenerational consequences of plant responses to herbivory: An adaptive maternal effect? Am Nat. 2001;157:555–569. doi: 10.1086/319932. [DOI] [PubMed] [Google Scholar]
- Alexandre NM, Humphrey PT, Frazier J, Gloss AD, Lee J, Affeldt HA, Whiteman NK. Habitat preference of an herbivore shapes the habitat distribution of its host plant. bioRxiv. 2018:156240. doi: 10.1101/156240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovics J. Evolution in closely adjacent plant populations X: Long-term persistence of prereproductive isolation at a mine boundary. Heredity. 2006;97:33. doi: 10.1038/sj.hdy.6800835. [DOI] [PubMed] [Google Scholar]
- Ballaré CL, Pierik R. The shadeavoidance syndrome: multiple signals and ecological consequences. Plant, Cell & Environment. 2017;40:2530–2543. doi: 10.1111/pce.12914. [DOI] [PubMed] [Google Scholar]
- Ballaré CL. Light regulation of plant defense. Annual Review of Plant Biology. 2014;65:335–363. doi: 10.1146/annurev-arplant-050213-040145. [DOI] [PubMed] [Google Scholar]
- Bartoń K. MuMIn: Multi-Model Inference. 2017 R package version 1.40.0. [Google Scholar]
- Barton KE, Boege K. Future directions in the ontogeny of plant defence: understanding the evolutionary causes and consequences. Ecology Letters. 2017;20:403–11. doi: 10.1111/ele.12744. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. lme4: Linear mixed-effects models using Eigen and S4. 2014:1–23. R package v1.7. [Google Scholar]
- Bekaert M, Edger PP, Hudson CM, Pires JC, Conant GC. Metabolic and evolutionary costs of herbivory defense: systems biology of glucosinolate synthesis. New Phytologist. 2012;196:596–605. doi: 10.1111/j.1469-8137.2012.04302.x. [DOI] [PubMed] [Google Scholar]
- Bell DL, Galloway LF. Population differentiation for plasticity to light in an annual herb: Adaptation and cost. Am J Bot. 2008;95:59–65. doi: 10.3732/ajb.95.1.59. [DOI] [PubMed] [Google Scholar]
- Boege K, Marquis RJ. Facing herbivory as you grow up: the ontogeny of resistance in plants. Trends in Ecology & Evolution. 2005;20:441–8. doi: 10.1016/j.tree.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Brachi B, Meyer CG, Villoutreix R, Platt A, Morton TC, Roux F, Bergelson J. Co-selected genes determine adaptive variation in herbivore resistance throughout the native range of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2015;112:4032–4037. doi: 10.1073/pnas.1421416112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD, Howe GA. Rewiring of jasmonate and phytochrome B signaling uncouples plant growth-defense tradeoffs. Nature communications. 2016;7:12570. doi: 10.1038/ncomms12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrudo I, Caliri-Ortiz ME, Keller MM, Degano ME, Demkura PV, Ballaré CL. Exploring growth-defence tradeoffs in Arabidopsis: phytochrome B inactivation requires JAZ10 to suppress plant immunity but not to trigger shadeavoidance responses. Plant, Cell & Environment. 2017;40:635–44. doi: 10.1111/pce.12877. [DOI] [PubMed] [Google Scholar]
- Chico JM, Fernández-Barbero G, Chini A, Fernández-Calvo P, Díez-Díaz M, Solano R. Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. The Plant Cell. 2014;26:1967–1980. doi: 10.1105/tpc.114.125047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollini D. Interactive effects of lateral shading and jasmonic acid on morphology, phenology, seed production, and defense traits in Arabidopsis thaliana. International Journal of Plant Sciences. 2005;166:955–959. doi: 10.1086/432896. [DOI] [Google Scholar]
- Coley PD, Bryant JP, Chapin FS. Resource availability and plant anti-herbivore defense. Science. 1985;230:895–899. doi: 10.1126/science.230.4728.895. [DOI] [PubMed] [Google Scholar]
- Colicchio J. Transgenerational effects alter plant defence and resistance in nature. J Evol Biol. 2017;30:664–680. doi: 10.1111/jeb.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge SK, Louda SM. Patterns of resource use by a drosophilid (Diptera) leaf miner on a native crucifer. Annals of the Entomological Society of America. 1988a;81:733–741. doi: 10.1093/aesa/81.5.733. [DOI] [Google Scholar]
- Collinge SK, Louda SM. Herbivory by leaf miners in response to experimental shading of a native crucifer. Oecologia. 1988b;75:559–566. doi: 10.1007/BF00776420. [DOI] [PubMed] [Google Scholar]
- Collinge SK, Louda SM. Scaptomyza nigrita Wheeler (Diptera: Drosophilidae), a leaf miner of the native crucifer, Cardamine cordifolia A. gray (bittercress) J Kans Entomol Soc. 1989a;62:1–10. [Google Scholar]
- Collinge SK, Louda SM. Influence of plant phenology on the insect herbivore/bittercress interaction. Oecologia. 1989b;79:111–116. doi: 10.1007/BF00378247. [DOI] [PubMed] [Google Scholar]
- Darwin C. On the origin of species by means of natural selection, or, the preservation of favoured races in the struggle for life. London: J. Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- Doheny-Adams T, Redeker K, Kittipol V, Bancroft I, Hartley SE. Development of an efficient glucosinolate extraction method. Plant Methods. 2017;13:17. doi: 10.1186/s13007-017-0164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K, Messiqua D, Pyle EH, Heschel MS, Schmitt J. Evidence of adaptive divergence in plasticity: Density- and site-dependent selection on shade-avoidance responses in Impatiens capensis. Evolution. 2000;54:1956–1968. doi: 10.1111/j.0014-3820.2000.tb01240.x. [DOI] [PubMed] [Google Scholar]
- Dostálek T, Rokaya MB, Maršík P, Rezek J, Skuhrovec J, Pavela R, Münzbergová Z. Trade-off among different anti-herbivore defence strategies along an altitudinal gradient. AoB Plants. 2016;8:plw026. doi: 10.1093/aobpla/plw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley S, Schmitt J. Genetic differentiation in morphological responses to simulated foliage shade between populations of Impatiens capensis from open and woodland sites. Funct Ecol. 1995;9:655–666. doi: 10.2307/2390158. [DOI] [Google Scholar]
- Dudley SA, Schmitt J. Testing the adaptive plasticity hypothesis: Density-dependent selection on manipulated stem length in Impatiens capensis. Am Nat. 1996;147:445–465. doi: 10.1086/285860. [DOI] [Google Scholar]
- Ehrlich PR, Raven PH. Differentiation of populations. Science. 1969;165:1228–1232. doi: 10.1126/science.165.3899.1228. [DOI] [PubMed] [Google Scholar]
- Epling C, Dobzhansky T. Genetics of natural populations. VI. Microgeographic races in Linanthus parryae. Genetics. 1942;27:317–332. doi: 10.1093/genetics/27.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine PV, Mesones I, Coley PD. Herbivores promote habitat specialization by trees in Amazonian forests. Science. 2004;305:663–665. doi: 10.1126/science.1098982. [DOI] [PubMed] [Google Scholar]
- Freckleton RP. On the misuse of residuals in ecology: regression of residuals vs. multiple regression. Journal of Animal Ecology. 2002;71:542–545. doi: 10.1046/j.1365-2656.2002.00618.x. [DOI] [Google Scholar]
- Galen C, Shore JS, Deyoe H. Ecotypic divergence in alpine Polemonium viscosum: Genetic structure, quantitative variation, and local adaptation. Evolution. 1991;45:1218–1228. doi: 10.1111/j.1558-5646.1991.tb04388.x. [DOI] [PubMed] [Google Scholar]
- Galloway LF. Maternal effects provide phenotypic adaptation to local environmental conditions. New Phytol. 2005;166:93–100. doi: 10.1111/j.1469-8137.2004.01314.x. [DOI] [PubMed] [Google Scholar]
- Galloway LF, Etterson JR. Transgenerational plasticity is adaptive in the wild. Science. 2007;318:1134–1136. doi: 10.1126/science.1148766. [DOI] [PubMed] [Google Scholar]
- Gloss AD, Vassao DG, Hailey AL, Nelson Dittrich AC, Schramm K, Reichelt M, Rast TJ, Weichsel A, Cravens MG, Gershenzon J, Montfort WR, Whiteman NK. Evolution in an ancient detoxification pathway is coupled with a transition to herbivory in the Drosophilidae. Mol Biol Evol. 2014;31:2441–2456. doi: 10.1093/molbev/msu201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers CM, Visser EJ, St Onge KR, Voesenek LA, Pierik R. Shade tolerance: when growing tall is not an option. Trends in Plant Science. 2013;18:65–71. doi: 10.1016/j.tplants.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Gommers CM, Keuskamp DH, Buti S, Van Veen H, Koevoets IT, Reinen E, Voesenek LA, Pierik R. Molecular profiles of contrasting shade response strategies in wild plants: differential control of immunity and shoot elongation. The Plant Cell. 2017;29:331–44. doi: 10.1105/tpc.16.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham HM. Confronting multicollinearity in ecological multiple regression. Ecology. 2003;84:2809–15. doi: 10.1890/02-3114. [DOI] [Google Scholar]
- Greig-Smith P. The use of random and contiguous quadrats in the study of the structure of plant communities. Annals of Botany. 1952;16:293–316. doi: 10.1093/oxfordjournals.aob.a083317. [DOI] [Google Scholar]
- Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annual Review of Plant Biology. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. A mathematical theory of natural and artificial selection (Part VI, isolation) Proceedings of the Cambridge Philosophical Society. 1930;26:220–230. doi: 10.1017/S0305004100015450. [DOI] [Google Scholar]
- Hedrick PW. Genetic polymorphism in heterogeneous environments: The age of genomics. Annu Rev Ecol Evol Syst. 2006;37:67–93. doi: 10.1146/annurev.ecolsys.37.091305.110132. [DOI] [Google Scholar]
- Hendrick MF, Finseth FR, Mathiasson ME, Palmer KA, Broder EM, Breigenzer P, Fishman L. The genetics of extreme microgeographic adaptation: An integrated approach identifies a major gene underlying leaf trichome divergence in Yellowstone Mimulus guttatus. Mol Ecol. 2016;25:5647–5662. doi: 10.1111/mec.13753. [DOI] [PubMed] [Google Scholar]
- Hopkins RJ, van Dam NM, van Loon JJ. Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annual Review of Entomology. 2009;54:57–83. doi: 10.1146/annurev.ento.54.110807.090623. [DOI] [PubMed] [Google Scholar]
- Humphrey PT, Gloss AD, Alexandre NM, Villalobos MM, Fremgen MR, Groen SC, Meihls LN, Jander G, Whiteman NK. Aversion and attraction to harmful plant secondary compounds jointly shape the foraging ecology of a specialist herbivore. Ecology and evolution. 2016;6:3256–3268. doi: 10.1002/ece3.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey PT, Nguyen TT, Villalobos MM, Whiteman NK. Diversity and abundance of phyllosphere bacteria are linked to insect herbivory. Mol Ecol. 2014;23:1497–1515. doi: 10.1111/mec.12657. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. doi: 10.1111/j.1461-0248.2004.00684.x. [DOI] [Google Scholar]
- Kenward MG, Roger JH. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics. 1997;53:983–997. doi: 0.2307/2533558. [PubMed] [Google Scholar]
- Keuskamp DH, Sasidharan R, Pierik R. Physiological regulation and functional significance of shade avoidance responses to neighbors. Plant Signaling & Behavior. 2010;5:655–662. doi: 10.4161/psb.5.6.11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen RHB. lmerTest Package: Tests in Linear Mixed Effects Models. Journal of Statistical Software. 2017;82(13) doi: 10.18637/jss.v082.i13. [DOI] [Google Scholar]
- Latzel V, Klimešová J. Transgenerational plasticity in clonal plants. Evolutionary Ecology. 2010;24:1537–1543. doi: 10.1007/s10682-010-9385-2. [DOI] [Google Scholar]
- Lenormand T. Gene flow and the limits to natural selection. Trends in Ecology & Evolution. 2002;17:183–189. doi: 10.1016/S0169-5347(02)02497-7. [DOI] [Google Scholar]
- Leone M, Keller MM, Cerrudo I, Ballaré CL. To grow or defend? Low red:far-red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytologist. 2014;204:355–367. doi: 10.1111/nph.12971. [DOI] [PubMed] [Google Scholar]
- Levin DA. Flowering-time plasticity facilitates niche shifts in adjacent populations. New Phytol. 2009;183:661–666. doi: 10.1111/j.1469-8137.2009.02889.x. [DOI] [PubMed] [Google Scholar]
- Linhart YB, Baker I. Intra-population differentiation of physiological response to flooding in a population of Veronica peregrina L. Nature. 1973;242:275–276. doi: 10.1038/242275a0. [DOI] [Google Scholar]
- Linhart YB, Grant MC. Evolutionary significance of local genetic differentiation in plants. Annu Rev Ecol Syst. 1996;27:237–277. doi: 10.1146/annurev.ecolsys.27.1.237. [DOI] [Google Scholar]
- Liu T, Zhang X, Yang H, Agerbirk N, Qiu Y, Wang H, Shen D, Song J, Li X. Aromatic glucosinolate biosynthesis pathway in Barbarea vulgaris and its response to Plutella xylostella infestation. Frontiers in Plant Science. 2016a;7:83. doi: 10.3389/fpls.2016.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Dawson W, Prati D, Haeuser E, Feng Y, van Kleunen M. Does greater specific leaf area plasticity help plants to maintain a high performance when shaded? Annals of Botany. 2016b;118:1329–36. doi: 10.1093/aob/mcw180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louda SM, Rodman JE. Ecological patterns in the glucosinolate content of a native mustard, Cardamine cordifolia, in the Rocky Mountains. Journal of Chemical Ecology. 1983a;9:397–422. doi: 10.1007/BF00988458. [DOI] [PubMed] [Google Scholar]
- Louda SM, Rodman JE. Concentration of glucosinolates in relation to habitat and insect herbivory for the native crucifer Cardamine cordifolia. Biochemical Systematics and Ecology. 1983b;11:199–207. doi: 10.1016/0305-1978(83)90054-6. [DOI] [Google Scholar]
- Louda SM. Herbivore effect on stature, fruiting, and leaf dynamics of a native crucifer. Ecology. 1984;65:1379–1386. doi: 10.2307/1939118. [DOI] [Google Scholar]
- Louda SM, Dixon PM, Huntly NJ. Herbivory in sun versus shade at a natural meadow-woodland ecotone in the Rocky Mountains. Plant Ecol. 1987;72:141–149. [Google Scholar]
- Louda SM, Collinge SK. Plant resistance to insect herbivores: a field test of the environmental stress hypothesis. Ecology. 1992;73:153–169. doi: 10.2307/1938728. [DOI] [Google Scholar]
- Louda SM, Rodman JE. Insect herbivory as a major factor in the shade distribution of a native crucifer (Cardamine cordifolia A. Gray, bittercress) J Ecol. 1996;84:229–237. doi: 10.2307/2261358. [DOI] [Google Scholar]
- Luke SG. Evaluating significance in linear mixed-effects models in R. Behavior Research Methods. 2017;49:1494–1502. doi: 10.3758/s13428-016-0809-y. [DOI] [PubMed] [Google Scholar]
- Maron JL, Crone E. Herbivory: effects on plant abundance, distribution and population growth. Proceedings of the Royal Society of London B: Biological Sciences. 2006;273:2575–2584. doi: 10.1098/rspb.2006.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney EH, Phillips JS, Tillberg CV, Sandrow C, Nelson AS, Mooney KA. Abiotic mediation of a mutualism drives herbivore abundance. Ecology Letters. 2016;19:37–44. doi: 10.1111/ele.12540. [DOI] [PubMed] [Google Scholar]
- Moreno JE, Tao Y, Chory J, Ballare CL. Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc Natl Acad Sci U S A. 2009;106:4935–4940. doi: 10.1073/pnas.0900701106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosteller F, Tukey JW. Data analysis and regression: A second course in statistics. Reading, Mass: Addison-Wesley Pub. Co; 1977. [Google Scholar]
- Nakagawa S, Schielzeth H. A general and simple method for obtaining R2 from Generalized Linear Mixed-effects Models. Methods in Ecology and Evolution. 2013;4:133–142. doi: 10.1111/j.2041-210x.2012.00261.x. [DOI] [Google Scholar]
- Nosil P, Harmon L. Niche dimensionality and ecological speciation. In: Butlin R, Bridle J, Schluter D, editors. Speciation and Patterns of Diversity (Ecological Reviews) Cambridge: Cambridge University Press; 2009. pp. 127–154. [DOI] [Google Scholar]
- Pellissier L, Roger A, Bilat J, Rasmann S. High elevation Plantago lanceolata plants are less resistant to herbivory than their low elevation conspecifics: Is it just temperature? Ecography. 2014;37:950–959. doi: 10.1111/ecog.00833. [DOI] [Google Scholar]
- Poorter L. Growth responses of 15 rain-forest tree species to a light gradient: the relative importance of morphological and physiological traits. Functional Ecology. 1999;13:396–410. doi: 10.1046/j.1365-2435.1999.00332.x. [DOI] [Google Scholar]
- Prasad KV, Song BH, Olson-Manning C, Anderson JT, Lee CR, Schranz ME, Windsor AJ, Clauss MJ, Manzaneda AJ, Naqvi I, Reichelt M, Gershenzon J, Rupasinghe SG, Schuler MA, Mitchell-Olds T. A gain-of-function polymorphism controlling complex traits and fitness in nature. Science. 2012;337:1081–1084. doi: 10.1126/science.1221636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. v3.3. URL http://www.R-project.org/ [Google Scholar]
- Rasmann S, De Vos M, Casteel CL, Tian D, Halitschke R, Sun JY, Agrawal AA, Felton GW, Jander G. Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 2012;158:854–863. doi: 10.1104/pp.111.187831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick DN, Ricklefs RE. Darwin’s bridge between microevolution and macroevolution. Nature. 2009;457:837–842. doi: 10.1038/nature07894. [DOI] [PubMed] [Google Scholar]
- Richardson JL, Urban MC, Bolnick DI, Skelly DK. Microgeographic adaptation and the spatial scale of evolution. Trends in Ecology & Evolution. 2014;29:165–176. doi: 10.1016/j.tree.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Ricklefs R, Miller G. Ecology. 4th. W.H. Freeman; New York: 2000. [Google Scholar]
- Rodman JE, Louda SM. Seasonal flux of isothiocyanate-yielding glucosinolates in roots, stems and leaves of Cardamine cordifolia. Biochemical Systematics and Ecology. 1985;13:405–412. doi: 10.1016/0305-1978(85)90085-7. [DOI] [Google Scholar]
- Runkle ES, Heins RD. Specific functions of red, far red, and blue light in flowering and stem extension of long-day plants. J Am Soc Hort Sci. 2001;126:275–282. [Google Scholar]
- Sasidharan R, Chinnappa CC, Voesenek LA, Pierik R. The regulation of cell wall extensibility during shade avoidance: a study using two contrasting ecotypes of Stellaria longipes. Plant Physiology. 2008;148:1557–1569. doi: 10.1104/pp.108.125518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Kudoh H. Fine-scale frequency differentiation along a herbivory gradient in the trichome dimorphism of a wild Arabidopsis. Ecology and Evolution. 2017;7:2133–2141. doi: 10.1002/ece3.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schemske DW, Bierzychudek P. Spatial differentiation for flower color in the desert annual Linanthus parryae: Was Wright right? Evolution. 2007;61:2528–2543. doi: 10.1111/j.1558-5646.2007.00219.x. [DOI] [PubMed] [Google Scholar]
- Schmitt J, Stinchcombe JR, Heschel MS, Huber H. The adaptive evolution of plasticity: phytochrome-mediated shade avoidance responses. Integrative and Comparative Biology. 2003;43:459–469. doi: 10.1093/icb/43.3.459. [DOI] [PubMed] [Google Scholar]
- Schmitt J, Gamble SE. The effect of distance from the parental site on offspring performance and inbreeding depression in Impatiens capensis: A test of the local adaptation hypothesis. Evolution. 1990;44:2022–2030. doi: 10.1111/j.1558-5646.1990.tb04308.x. [DOI] [PubMed] [Google Scholar]
- Schwaegerle KE, McIntyre H, Swingley C. Quantitative genetics and the persistence of environmental effects in clonally propagated organisms. Evolution. 2000;54:452–461. doi: 10.1111/j.0014-3820.2000.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Selander RK, Kaufman DW. Genetic structure of populations of the brown snail (Helix aspersa). I. Microgeographic variation. Evolution. 1975;29:385–401. doi: 10.1111/j.1558-5646.1975.tb00829.x. [DOI] [PubMed] [Google Scholar]
- Sork VL, Stowe KA, Hochwender C. Evidence for local adaptation in closely adjacent subpopulations of northern red oak (Quercus rubra L.) expressed as resistance to leaf herbivores. Am Nat. 1993;142:928–936. doi: 10.1086/285581. [DOI] [PubMed] [Google Scholar]
- Strauss SY, Cacho NI. Nowhere to run, nowhere to hide: the importance of enemies and apparency in adaptation to harsh soil environments. Am Nat. 2013;182:1, E1–E14. doi: 10.1086/670754. [DOI] [PubMed] [Google Scholar]
- Uesugi A, Connallon T, Kessler A, Monro K. Relaxation of herbivore-mediated selection drives the evolution of genetic covariances between plant competitive and defense traits. Evolution. 2017;71:1700–1709. doi: 10.1111/evo.13247. [DOI] [PubMed] [Google Scholar]
- Ullah MI, Aslam DM. mctest: Multicollinearity Diagnostic Measures, 2017. 2017 R package version 1.1.1. [Google Scholar]
- Valladares F, Niinemets Ü. Shade tolerance, a key plant feature of complex nature and consequences. Annual Review of Ecology, Evolution, and Systematics. 2008;39:237–257. doi: 10.1146/annurev.ecolsys.39.110707.173506. [DOI] [Google Scholar]
- van Hinsberg A, van Tienderen P. Variation in growth form in relation to spectral light quality (red/far-red ratio) in Plantago lanceolata L. in sun and shade populations. Oecologia. 1997;111:452–459. doi: 10.1007/s004420050258. [DOI] [PubMed] [Google Scholar]
- Waser NM, Price MV. Optimal outcrossing in Ipomopsis aggregata: seed set and offspring fitness. Evolution. 1989;43:1097–1109. doi: 10.2307/2409589. [DOI] [PubMed] [Google Scholar]
- Winde I, Wittstock U. Insect herbivore counter adaptations to the plant glucosinolate-myrosinase system. Phytochemistry. 2011;72:1566–1575. doi: 10.1016/j.phytochem.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Whiteman NK, Gloss AD, Sackton TB, Groen SC, Humphrey PT, Lapoint RT, Sonderby IE, Halkier BA, Kocks C, Ausubel FM, Pierce NE. Genes involved in the evolution of herbivory by a leaf-mining, drosophilid fly. Genome Biol Evol. 2012;4:900–916. doi: 10.1093/gbe/evs063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman NK, Groen SC, Chevasco D, Bear A, Beckwith N, Gregory TR, Denoux C, Mammarella N, Ausubel FM, Pierce NE. Mining the plant-herbivore interface with a leafmining Drosophila of Arabidopsis. Mol Ecol. 2011;20:995–1014. doi: 10.1111/j.1365-294X.2010.04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wit M, Spoel SH, Sanchez-Perez GF, Gommers CM, Pieterse CM, Voesenek LA, Pierik R. Perception of low red: far-red ratio compromises both salicylic acid-and jasmonic acid-dependent pathogen defences in Arabidopsis. The Plant Journal. 2013;75:90–103. doi: 10.1111/tpj.12203. [DOI] [PubMed] [Google Scholar]
- Woods EC, Hastings AP, Turley NE, Heard SB, Agrawal AA. Adaptive geographical clines in the growth and defense of a native plant. Ecological Monographs. 2012;82:149–168. doi: 10.1890/11-1446.1. [DOI] [Google Scholar]
- Züst T, Joseph B, Shimizu KK, Kliebenstein DJ, Turnbull LA. Using knockout mutants to reveal the growth costs of defensive traits. Proceedings of the Royal Society of London B: Biological Sciences. 2011;278:2598–2603. doi: 10.1098/rspb.2010.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst T, Agrawal AA. Trade-offs between plant growth and defense against insect herbivory: An emerging mechanistic synthesis. Annual Review of Plant Biology. 2017;68:513–534. doi: 10.1146/annurev-arplant-042916-040856. [DOI] [PubMed] [Google Scholar]
- Züst T, Heichinger C, Grossniklaus U, Harrington R, Kliebenstein DJ, Turnbull LA. Natural enemies drive geographic variation in plant defenses. Science. 2012;338:116–119. doi: 10.1126/science.1226397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


