Abstract
Duchenne muscular dystrophy (DMD) is an X-linked progressive muscle degenerative disease caused by mutations in the dystrophin gene. We generated induced pluripotent stem cells (iPSCs) from a 13-year-old male patient carrying a deletion mutation of exons 45–50; iPSCs were subsequently differentiated into cardiomyocytes. iPSCs exhibit expression of the pluripotent markers (SOX2, NANOG, OCT4), differentiation capacity into the three germ layers, normal karyotype, genetic identity to the skin biopsy dermal fibroblasts and the patient-specific dystrophin mutation.
Resource Table
| Unique stem cell line identifier | IITi001-A |
| Alternative name(s) of stem cell line | 29.2 |
| Institution | Technion – Israel Institute of Technology (IIT), Haifa, Israel. University of Michigan, Ann Arbor, Michigan, USA. |
| Contact information of distributor | Daniel Michele, PhD dmichele@umich.edu |
| Type of cell line | iPSC |
| Origin | Human |
| Additional origin info | Age: 13 Sex: male |
| Cell Source | Fibroblasts |
| Clonality | Clonal |
| Method of reprogramming | Sendai virus |
| Genetic Modification | NO |
| Type of Modification | N/A |
| Associated disease | Duchenne muscular dystrophy (DMD) |
| Gene/locus | Dystrophin del. Ex. 45–50 |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | 17/07/17 |
| Cell line repository/bank | N/A |
| Ethical approval | Consent was obtained according to IRB HUM00030934 approved by the University of Michigan Human IRB Committee |
Resource utility
This iPSC line was generated to study DMD disease pathophysiology and the electrophysiological abnormalities of iPSC-derived cardiomyocytes (iPSC-CMs) using methods such as: patch clamp, microelectrode array, calcium-contraction imaging.
Resource Details
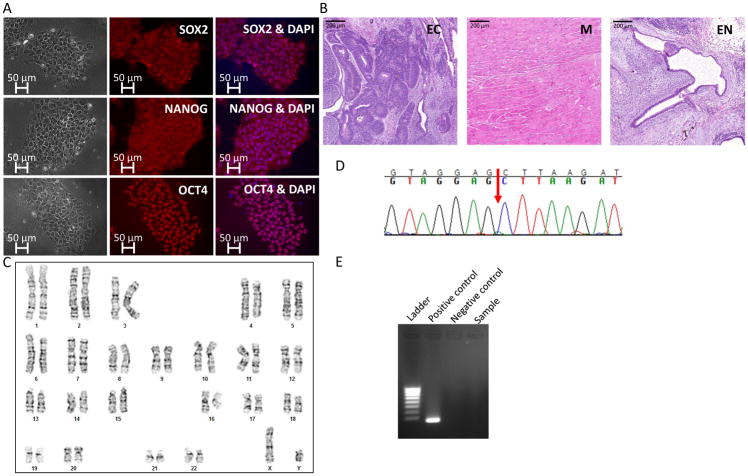
Duchenne muscular dystrophy (DMD) is an X-linked muscle degenerative disease resulting in progressive weakness, loss of ambulation in the early teen years, and death by the end of the 3rd decade of life due to respiratory and cardiac failure1. DMD is caused by mutations in the dystrophin gene, the longest gene in the human DNA2; loss of dystrophin expression leads to destabilization of the dystrophin glycoprotein complex (DGC), sarcolemma instability and fibrotic replacement of degenerated muscle3. To date, there are no curative treatments for DMD, and dilated cardiomyopathy (DCM) is a major cause of morbidity and mortality1. The most common murine model for DMD is the mdx mouse1; however, while DMD patients develop DCM, mdx mice develop extremely mild cardiomyopathic phenotypes3. Furthermore, mdx mice cardiomyocytes are unstable in primary culture preventing adaptation of this model for in vitro research. Therefore, the mdx model carries major limitations specifically regarding the cardiac pathology. To investigate the DMD cardiac manifestations using a cellular model, we generated human patient iPSCs that were subsequently differentiated into cardiomyocytes. After informed consent form was obtained according to the Helsinki Committee at the University of Michigan, a skin biopsy was obtained from a 13-year-old patient carrying a deletion of exons 45–50 in the dystrophin gene. The mutation was first diagnosed by multiplex PCR amplification and a deletion of exons 45–50 was confirmed independently by MLPA and Southern blotting of exons 44 and 512. Human dermal fibroblasts were transfected with four Yamanaka’s factors resulting in iPSCs exhibiting pluripotent stem cell-like morphology. To characterize the iPSCs, we performed immunofluorescence staining for the pluripotent markers SOX2, NANOG, OCT4 (Figure 1A). Furthermore, teratoma assay analysis demonstrated differentiation capacity to the three germ layers: ectoderm, represented by neuronal rosettes; mesoderm, represented by striated muscle; and endoderm, represented by epithelial lining (Figure 1B). Karyotype analysis revealed normal 46 XY chromosomes (Figure 1C). To verify that the iPSCs are genetically identical to the human dermal fibroblasts from which they were generated, short tandem repeat (STR) profiling of 24 loci was performed, and complete match was found (to protect the patient’s identity, data is available with the authors). To investigate the phenotypes of dystrophin-mutated cardiomyocytes, iPSCs were differentiated into contracting cardiomyocytes using a directed differentiation protocol4, 5. To verify that the deletion mutation of exons 45–50 was preserved through the process of reprograming, Sanger sequencing was performed on a dystrophin RT-PCR product (spanning exons 44–51) generated from iPSC cardiomyocyte RNA (Figure 1D, red arrow indicates the deletion point). Furthermore, mycoplasma test was performed and the result was negative (Figure 1E).
Figure 1.
Materials and Methods
Generation of induced Pluripotent Stem Cells (iPSCs) from the patient
iPSCs were generated from the patient’s dermal fibroblasts using Sendai virus CytoTune-iPS 2.0 Sendai Reprogramming Kit, #A16517 (Thermo Fisher, Waltham, MA, USA) for the transfection of Yamanaka’s 4 factors: OCT4, KLF4, c-Myc, SOX2. Subsequently, iPSCs were cultured on Matrigel (GFR, BD Biosciences, Franklin Lakes, NJ, USA) coated 6-well plates with mTeSR1 medium (Stemcell Technologies, Vancouver, Canada) and cultured in 37°C with 5% CO2. Every 5 days, iPSCs were passaged in a ratio of 1:6 by dissociation using 1 ml/well Versene solution (Invitrogen, Life Technologies, Woburn, MA, USA) at 37°C for 7 minutes.
Karyotype analysis
Karyotype analysis was performed using standard G-banding chromosome analysis for 20 metaphase spreads by the Rambam Health Center cytogenetic laboratory according to standard procedures.
Genotyping
Extracted RNA was achieved using PureLink RNA mini kit (Thermo Fisher, Waltham, MA, USA). Subsequently, cDNA was produced using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher, Waltham, MA, USA). PCR was performed (35 cycles; denaturation: 94° C for 30 sec, annealing: 61° C for 60 sec, elongation: 72° C for 45 sec; Biometra 2005; Analytik Jena, Jena, Germany) to the dystrophin gene using the primers listed in Table 2 resulting in a product of 340 base pairs in length. Next, Sanger sequencing for the PCR product containing the mutation area within the DMD gene confirmed the deletion of exons 45–50.
Table 2.
Reagents details
| Antibodies used for immunocytochemistry/flow-citometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Rabbit anti-SOX2 | 1:100 | Santa Cruz Biotechnology Cat# sc20088, RRID: AB_2255358 |
| Pluripotency Markers | Rabbit anti-NANOG | 1:100 | PeproTech Cat# 500-P236, RRID: AB_1268274 |
| Pluripotency Markers | Rabbit anti-OCT4 | 1:200 | Santa Cruz Biotechnology Cat# sc9081, RRID: AB_2167703 |
| Secondary antibodies | Cy2 Donkey Anti-Rabbit IgG | 1:100 | Jackson ImmunoResearch Cat# 711-225-152, RRID: AB_2340612 |
| Secondary antibodies | Alexa Fluor 555 Donkey Anti-Rabbit IgG | 1:100 | Life Technologies Cat# A31572, RRID: AB_162543 |
| Primers | |||
| Target | Forward/Reverse primer (5′-3′) | ||
| Genotyping | Dystrophin exons 44–51 | ATCTGTTGAGAAATGGCGGC/ GATGGTGGGTGACCTTGAGG | |
Immunofluorescence staining
Immunofluorescence staining was performed according to standard procedures. Briefly, 3 days after iPSCs passaging, growth medium was discarded and iPSCs were washed with PBS (Biological Industries, Kibbutz Beit-Haemek, Israel). Subsequently, fixation was performed using 4% paraformaldehyde for 30 minutes and washed 3 times with PBS followed by permeabilization with 0.5% Triton X-100 in PBS for 15 minutes. Goat serum (#50-062Z, Invitrogen, Life Technologies, Woburn, MA, USA) was used as a blocking solution for 15 minutes, followed by incubation with primary antibody (as listed in Table 2) in antibody dilutent (#ZUC025-100, Zytomed Systems, Berlin, Germany) overnight at 4° C. After 3 washes with PBS, secondary antibody was incubated for 1 hour at room temperature, followed by 3 washes and incubation with DAPI (1:500; Sigma Aldrich, St. Louis, MO, USA) for 5 minutes. Images were obtained using Zeiss Axio observer microscope (Zeiss, Oberkochen, Germany). SOX2-positive cells (500) were evaluated by cell-counting in immunofluorescence staining images.
Teratoma Formation
To evaluate the differentiation capacity of iPSCs in vivo, colonies from two 6-well plates were detached using 1 mg/ml type IV collagenase, washed 3 times in PBS and injected into the thigh muscle of severe combined immunodeficient (SCID) mice. Eight-to-12 weeks after injection, teratomas were observed, and images were obtained from formalin-fixed (4%) and paraffin-embedded teratoma sections stained with hematoxylin and eosin (H&E).
STR profiling
The analysis was performed using the Promega GenePrint 24 System in order to determine STR profile of 23 loci plus Amelogenin for gender determination (X or XY). In addition, the male-specific DYS391 locus is included to identify null Y allele results for Amelogenin. DNA sample from the kit (2800M Control DNA) was included in the analysis and served as positive control for the PCR step. The results were analyzed using the 3500xl Genetic Analyzer (Life Technologies, Carlsbad, CA, USA) and GeneMapper IDX software. Tested loci: AMEL, D3S1358, D1S1656, D2S441, D10S1248, D13S317, Penta E, D16S539, D18S51, D2S1338, CSF1PO, Penta D, TH01, vWA, D21S11, D7S820, D5S818, TPOX, DYS391, D8S1179, D12S391, D19S433, FGA, D22S1045.
Differentiation into cardiomyocytes
iPSC-CMs were generated according to the directed differentiation by modulating Wnt/β-catenin signaling as previously described5. Briefly, iPSCs were cultured on Matrigel (GFR, BD Biosciences, Franklin Lakes, NJ, USA) coated 6-well plates with mTeSR1 medium (Stemcell Technologies, Vancouver, Canada) for 5–6 days. To initiate differentiation, the cells were dissociated using 1 ml/well Versene solution (Invitrogen, Life Technologies, Woburn, MA, USA) at 37°C for 7 minutes and reseeded on Matrigel coated plate at 8.5x106/12 well plate density in mTeSR1 medium supplemented with 5 μmol/l ROCK inhibitor (Cayman Chemical, Ann Arbor, MI, USA). The medium was replaced daily, and after 2 days when the monolayer of cells reached 100% confluence, the medium was changed to RPMI supplemented with B27 minus insulin (Invitrogen, Life Technologies, Woburn, MA, USA) containing 10 μmol/l CHIR99021; this day was labelled as day 1 of differentiation. On the next day, the medium was changed to RPMI supplemented with B27 minus insulin. On day 4, the medium was changed to RPMI supplemented with B27 minus insulin, containing 10 μmol/l of IWP-4 or 2. On day 6, the medium was changed to RPMI supplemented with B27 minus insulin. Finally, from day 8 onwards, the medium was changed to RPMI supplemented with B27 complete supplement (Invitrogen, Life Technologies, Woburn, MA, USA).
Mycoplasma test
Supernatant was collected for mycoplasma test after 48 hours of iPSCs culture; the test was performed using the PCR-based EZ-PCR Mycoplasma Test Kit (Biological Industries, Kibbutz Beit-Haemek, Israel).
Table 1.
Characterization and validation
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | iPSCs exhibit pluripotent stem cell-like morphology | Figure 1A, left panel |
| Phenotype | Qualitative analysis (Immunofluorescent stainings) | Positive for pluripotency markers: OCT4, NANOG, SOX2 | Figure 1A |
| Quantitative analysis (Immunofluorescent counting) | 97% SOX2-positive iPSCs | N/A | |
| Genotype | Karyotype | 46XY, G-banding Resolution 400 | Figure 1C |
| Identity | STR analysis | Profile of 24 loci identifies complete match of the iPSCs and the human dermal fibroblasts (HDF) from which they were generated | Available with the authors |
| Mutation analysis (IF APPLICABLE) | Sequencing | Homozygous, deletion of exons 45–50 | Figure 1D |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by PCR, Negative | Figure 1E |
| Differentiation potential | Teratoma formation and Directed differentiation into cardiomyocytes | Teratoma assay analysis demonstrates differentiation capacity to the 3 germ layers (ectoderm, mesoderm, and endoderm). Directed differentiation into cardiomyocytes resulted in contracting iPSC-CMs | Figure 1B |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
Footnotes
Additional files:
STR analysis
Data is available with the authors.
References
- 1.McNally EM, Kaltman JR, Benson DW, Canter CE, Cripe LH, Duan D, Finder JD, Groh WJ, Hoffman EP, Judge DP, Kertesz N, Kinnett K, Kirsch R, Metzger JM, Pearson GD, Rafael-Fortney JA, Raman SV, Spurney CF, Targum SL, Wagner KR, Markham LW Working Group of the National Heart, Lung and BI, Parent Project Muscular Dystrophy. Contemporary cardiac issues in Duchenne muscular dystrophy. Working Group of the National Heart, Lung, and Blood Institute in collaboration with Parent Project Muscular Dystrophy. Circulation. 2015;131:1590–8. doi: 10.1161/CIRCULATIONAHA.114.015151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lalic T1, Vossen RH, Coffa J, Schouten JP, Guc-Scekic M, Radivojevic D, Djurisic M, Breuning MH, White SJ, den Dunnen JT. Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet. 2005;13:1231–4. doi: 10.1038/sj.ejhg.5201465. [DOI] [PubMed] [Google Scholar]
- 3.Khouzami L, Bourin M-C, Christov C, Damy T, Escoubet B, Caramelle P, Perier M, Wahbi K, Meune Christophe, Pavoine C, Pecker F Hospitalier Henri Mondor-Albert Chenevier G. Delayed cardiomyopathy in dystrophin deficient mdx mice relies on intrinsic glutathione resource. Am J Pathol. 2010;177:1356–1364. doi: 10.2353/ajpath.2010.090479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben Jehuda R, Eisen B, Shemer Y, Mekies LN, Szantai A, Reiter I, Cui H, Guan K, Haron-Khun S, Freimark D, Sperling SR, Gherghiceanu M, Arad M, Binah O. CRISPR correction of the PRKAG2 gene mutation in the patient’s induced pluripotent stem cell-derived cardiomyocytes eliminates electrophysiological and structural abnormalities. Hear Rhythm. 2018;15:267–276. doi: 10.1016/j.hrthm.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 5.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]