Abstract
Transposable elements make important contributions to adaptation and evolution of their host genomes. The well-characterized transposase-derived transcription factor FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) and its homologue FAR-RED IMPAIRED RESPONSE1 (FAR1) have crucial functions in plant growth and development. In addition, FHY3 and FAR1 are the founding members of the FRS (FAR1-RELATED SEQUENCE) and FRF (FRS-RELATED FACTOR) families, which are conserved among land plants. Although the coding sequences of many putative FRS and FRF orthologs have been found in various clades of angiosperms, their physiological functions remain elusive. Here, we summarize recent progress toward characterizing the molecular mechanisms of FHY3 and FAR1, as well as other FRS-FRF family proteins, examining their roles in regulating plant growth and development. This review also suggests future directions for further functional characterization of other FRS-FRF family proteins in plants.
Keywords: light signal proteins, FHY3, FAR1, transcription factors, transcription, genetic
Introduction
Light is one of the most important environmental factors affecting plant growth and development. In the past few decades, ongoing research has identified multiple mutants that display defects in their response to various wavelengths of light. In 1993, the far-red elongated hypocotyls1 (fhy1), fhy2, and fhy3 mutants were shown to display an elongated hypocotyl in far-red light but not in white light (Whitelam et al., 1993). FHY1 and its homolog FHL (FHY1-LIKE) interact with phytochrome A (phyA) and are required for phyA translocation from the cytoplasm to the nucleus after exposure to far-red light (Desnos et al., 2001; Hiltbrunner et al., 2005; Zhou et al., 2005). FHY2 encodes a phyA that specifically responds to far-red light (Whitelam et al., 1993).
FHY3 and its homolog FAR1 encode transposase-derived transcription factors (Hudson et al., 1999; Wang and Deng, 2002; Lin et al., 2007). Although FHY3 and FAR1 were derived from transposases, they have evolved diverse and powerful physiological functions in adaptation and domestication. Recent studies have demonstrated that FHY3 and FAR1 play multiple roles in a wide range of cellular processes, including light signal transduction, photomorphogenesis (Wang and Deng, 2002; Lin et al., 2007), circadian clock and flowering time regulation (Li et al., 2011), shoot meristem and floral development (Li et al., 2016), chloroplast division (Ouyang et al., 2011), chlorophyll biosynthesis (Tang et al., 2012), starch synthesis (Ma et al., 2017), abscisic acid responses (Tang et al., 2013), oxidative stress responses (Ma et al., 2016), plant immunity (Wang et al., 2016), and the low-phosphate response (Liu Y. et al., 2017), indicating that FHY3 and FAR1 are crucial for plant growth and development (Table 1). In addition, 12 FAR1-RELATED SEQUENCE (FRS) and four FRS-RELATED FACTOR (FRF) family proteins were identified in Arabidopsis thaliana (Lin and Wang, 2004; Aguilar-Martinez et al., 2015). Besides FHY3 and FAR1, the physiological and molecular mechanisms of most FRS and FRF family proteins remain largely unknown in plants.
Table 1.
The physiological functions of FRS-FRF family proteins in Arabidopsis.
| Cellular process | Targets | Transcriptional regulation | Reference |
|---|---|---|---|
| Light signal transduction |
FHY1/FHL COP1 |
FHY3/FAR1 (+), HY5 (-) FHY3/FAR1 (+), HY5 (+) |
Lin et al., 2007; Li et al., 2010; Huang et al., 2012 |
| Chloroplast division | ARC5 | FHY3/FAR1(+), FRS4 (+) |
Ouyang et al., 2011; Gao et al., 2013 |
| Chlorophyll biosynthesis Immunity response |
HEMB1 | FHY3/FAR1 (+), PIF1(-) |
Tang et al., 2012; Wang et al., 2016 |
| Myo-inositol synthesis Oxidative response |
MIPS1/MIPS2 | FHY3/FAR1 (+) | Ma et al., 2016 |
| Starch synthesis | ISA2 | FHY3/FAR1 (+) | Ma et al., 2017 |
| Circadian clock Flowering time |
ELF4 | FHY3/FAR1/HY5 (+), CCA1/LHY (-) | Li et al., 2011 |
| Diurnal growth Flowering time regulation |
PIF4 Gl | FRS7 (-), FRS12 (-) | Ritter et al., 2017 |
| Floral development |
CLV3 SEP2 STM |
FHY3 (-) FHY3 (+) FRF1 (?) |
Li et al., 2016; Aguilar-Martinez et al., 2015 |
| Drought stress ABA response |
ABI5 | FHY3/FAR1 (+) | Tang et al., 2013 |
| Low phosphate response | PHR1 | FHY3/FAR1/EIN3 (+), HY5 (-) | Liu Y. et al., 2017 |
+, positive regulation; -, negative regulation.
Protein Structures
Multiple FRS family members, including FHY3 and FAR1, are transcription factors derived from Mutator-like element (MULE) transposases. Transposases are usually encode by transposable elements and are responsible for cutting and pasting the transposable elements from their original sites to new sites in the chromosome (Joly-Lopez and Bureau, 2014). An analysis of transposase protein structures revealed the presence of an N-terminal DNA-binding domain and a C-terminal catalytic domain. The N-terminal DNA-binding domains recognize specific DNA motifs in the terminal inverted repeats of the transposon and the C-terminal catalytic domains cleave the double-stranded DNA and insert the transferable element into a new genomic location (Feschotte et al., 2002; Makarova et al., 2002).
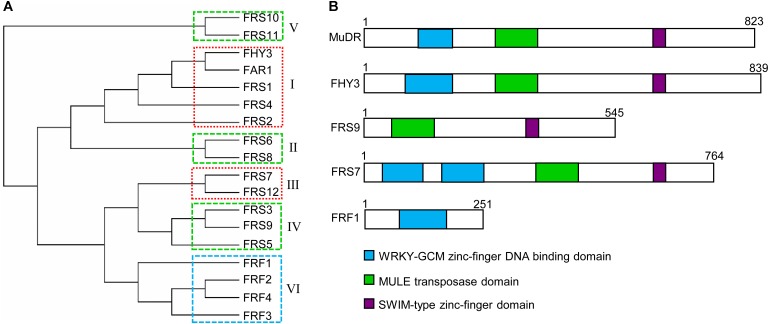
Most members of all FRS subgroups have an N-terminal C2H2 zinc-finger domain (also called a FAR1 DNA-binding domain), a central putative transposase domain similar to MULE transposases, and a C-terminal SWIM (SWI2/SNF2 and MuDR transposases) zinc-finger domain (Lin and Wang, 2004; Lin et al., 2007) (Figure 1B). Putative nuclear localization signal (NLS) motifs have been identified in most members of the FRS family, including FHY3, FAR1, and FRS2, but not FRS1, FRS8, and FRS9 (Lin and Wang, 2004). The N-terminal FAR1 DNA-binding domain is a type of C2H2 zinc-finger domain from the WRKY-Glial Cell Missing1 (WRKY-GCM1) superfamily, which bind to specific cis-elements in the promoter regions of diverse targets (Makarova et al., 2002; Lin et al., 2007).
FIGURE 1.
Phylogenetic and protein domain structures of the FRS-FRF family in Arabidopsis. (A) Multiple alignment of FRS and FRF family proteins were performed using MAFFT (V6.864, http://www.genome.jp/tools-bin/mafft). Phylogenetic analysis was generated using the neighbor-joining method (PHYLYP, V3.66) and displayed with MEGA (v7.0). FRS-FRF family proteins were classified into six subgroups (I–VI) based on their protein structures. The boxes with dashed lines indicate FRS-FRF members in the same clade that might form homodimers or heterodimers to coordinately regulate the transcription of their target genes. (B) The conserved protein domains in select FRS-FRF family proteins are shown.
The putative core transposase domains of FRS family proteins share significant sequence similarity with the transposase domain of MuDR family transposases from elements such as Jittery and MuDRA of maize, and LOM1 of rice (Lin et al., 2007). The transposase activity of FHY3, FAR1, and other FRS family proteins was most likely lost in Arabidopsis thaliana despite the high sequence similarity between their core transposase domains and MULE transposases (Hudson et al., 2003). By contrast, in maize, the terminal inverted repeats of a putative FRS5-LIKE gene were identified as an active transposon inserted in the coding region of ZmTOM1 in a mutant allele of yellow striped 3 (ys3). This suggests that the transposase activity of the core transposase domain of FRS family proteins might have been retained in maize (Chan-Rodriguez and Walker, 2018).
Mutations of the evolutionarily conserved amino acids in the core transposase domain or C-terminal SWIM domain failed to activate the expression of their target genes, indicating that these two domains are essential for the transcriptional activity, although the underlying molecular mechanism for this regulation remains unclear (Lin et al., 2007). Interestingly, ectopic overexpression of either the N-terminal or C-terminal domains of FHY3 in a wild-type background results in a long-hypocotyl phenotype, but only overexpression of the C-terminal of FHY3 results in a completely etiolated phenotype under far-red light, indistinguishable from the phenotype of phyA null mutants. These results indicated that overexpression of C-terminal fragments impairs the function of endogenous FHY3, and also produces a dosage-dependent dominant-negative effect on phyA signaling (Wang and Deng, 2002). It should be noted that FHY3 has been identified as associating with phyA under FR, which suggested that FHY3 might interact with phyA directly (Saijo et al., 2008). In addition, the FHY3 DNA-binding domain is located in the N-terminal, which directly binds to FHY1 and FHL promoters and promotes their gene expression, thus indirectly affecting the translocation of phyA into the nucleus (Lin et al., 2007).
Classification of FRS-FRF Family Proteins
Multiple sequence alignment and conserved protein motif analyses revealed that Arabidopsis FRS and FRF family proteins can be divided into 6 subgroups (Figure 1A), consistent with previous studies (Lin et al., 2007; Aguilar-Martinez et al., 2015; Joly-Lopez et al., 2016). These are: subgroup 1 (FHY3, FAR1, FRS1, FRS2, and FRS4), subgroup 2 (FRS6 and FRS8), subgroup 3 (FRS7 and FRS12), subgroup 4 (FRS3, FRS5 and FRS9), subgroup 5 (FRS10 and FRS11, which have unknown functions), and subgroup 6 (FRF1, FRF2, FRF3, and FRF4). Generally, most FRS-FRF family proteins have a DNA-binding domain in their N-terminal regions. As exceptions, in subgroup 3, FRS7 and FRS12 have two DNA-binding domains, and FRS9 of subgroup 4 has no DNA-binding domain in the N-terminal (Figure 1B).
In subgroup 6, FRF1–FRF4 have been considered as truncated FRS family proteins since they only contain the FAR1 DNA-binding domain but not the putative core transposase and C-terminal SWIM domains (Aguilar-Martinez et al., 2015). It has been suggested that FRFs might compete with FRSs for the DNA-binding sites of their targets, or regulate the transcription of targets by interacting with other transcription factors; however, determining the underlying molecular mechanism requires further functional studies (Aguilar-Martinez et al., 2015).
DNA-Binding Activity
The representative FRS family proteins FHY3 and FAR1 bind the FHY3/FAR1-binding site (FBS, CACGCGC) cis-elements that reside in the promoter regions of various target genes, including FHY1, FHL, EARLY-FLOWERING4 (ELF4), ACCUMULATION AND REPLICATION OF CHLOROPLASTS5 (ARC5), HEMB1, CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), ABA-INSENSITIVE5 (ABI5), CLAVATA3 (CLV3), SEPALLATA2 (SEP2), myo-INOSITOL-1-PHOSPHATE SYNTHASE (MIPS1), ISOAMYLASE2 (ISA2), and PHOSPHATE STARVATION RESPONSE1(PHR1) (Lin et al., 2007; Li et al., 2011, 2016; Ouyang et al., 2011; Huang et al., 2012; Stirnberg et al., 2012; Tang et al., 2012, 2013; Gao et al., 2013; Wang and Wang, 2015a; Ma et al., 2016, 2017; Liu Y. et al., 2017). Genome-wide chromatin immunoprecipitation-sequencing (ChIP-seq) or DNA affinity purification-sequencing (DAP-seq) analyses have confirmed that FHY3 and FAR1 specifically bind to FBS cis-elements in the promoter regions of over 1000 genes in Arabidopsis (Ouyang et al., 2011; Li et al., 2016; O’Malley et al., 2016). A recent study revealed that FRS4/CPD25, another member of subgroup 1, could also bind to FBS or FBL (FBS-Like) cis-elements in the ARC5 promoter (Gao et al., 2013). When the essential amino acid in the DNA-binding domain was mutated in FHY3, the mutant protein failed to bind to the FBS cis-elements in the FHY1 and ARC5 promoters, which indicates that the N-terminal FAR1 DNA-binding domain is responsible for binding to DNA (Lin et al., 2007; Gao et al., 2013).
Although both FRS6 and FRS8 of subgroup 2 have been shown to play a role in flowering time regulation, the cis-elements they recognize and the genes they regulate are unknown (Lin and Wang, 2004). FRS6 and FRS8 may recognize the same cis-elements in the promoter region of their various targets, since they contain the same essential amino acid in their N-terminal domains. Tandem chromatin affinity purification followed by next-generation sequencing with the subgroup 3 member FRS12 demonstrated that it specifically recognizes FRS12-BOX cis-elements (FRB1, TGTGTG; FRB2, TATATATATATATATATAT; FRB3, TATACATA) in the PIF4, GI, and PIL1 promoters (Ritter et al., 2017). However, it is still unclear whether subgroup 4 member FRS9 can associate with DNA since it does not have a N-terminal FAR1 DNA-binding domain, even though it contains the putative core transposase and C-terminal SWIM zinc-finger domains. FRS9 might interact with other FRS members to form heterodimers in the nucleus to regulate the transcription of target genes in various cellular processes.
FRF1 from subgroup 6 can bind to the large RB-box in the SHOOT MERISTEMLESS promoter (Aguilar-Martinez et al., 2015), however, the specific cis-elements and nucleotide sequences required for FRF1 binding to this region have not been identified. There is still no information available on the cis-elements recognized by subgroup 5 FRS-FRF members, since FRS10 and FRS11 have not been functionally characterized.
Physiological Functions of FRS-FRF Family Proteins
Light Signal Transduction and Photomorphogenesis
Several types of photoreceptors have evolved in plants to perceive environmental light signals. In Arabidopsis, phytochromes (phyA and phyB) primarily absorb red or far-red light; cryptochromes (cry1 and cry2) and phototropins (phot1 and phot2) perceive blue and ultraviolet A (UV-A) light; and UVR8 specific perceive UV-B light (Wang and Wang, 2015b). A forward genetic screen identified FHY3 and FAR1 as key components in far-red light signal transduction (Whitelam et al., 1993; Hudson et al., 1999; Wang and Deng, 2002). FHY3 and FAR1 form homodimers or heterodimers to directly bind to promoters and activate transcription of FHY1 and FHL, which encode two key regulators of phyA translocation into the nucleus under far-red light. Therefore, FHY3 and FAR1 indirectly affect phyA nuclear translocation (Lin et al., 2007). It should be noted that FHY3 and FAR1 affect phyA nuclear translocation indirectly and in association with phyA, thus playing a role in protecting insufficiently phosphorylated phyA from proteosomal degradation mediated by the COP1 E3 ligase (Saijo et al., 2008). Interestingly, the transcript levels of both FHY3 and FAR1 are repressed by far-red light and in phyA-dependent manner by an unknown mechanism (Wang and Deng, 2002; Lin et al., 2007). In addition, ELONGATED HYPOCOTYL (HY5), a well-characterized bZIP transcription factor, physically interacts with both FHY3 and FAR1 directly, and antagonizes their transcriptional activation on FHY1 and FHL, thus affecting far-red light signal transduction (Li et al., 2010).
FHY3 and FAR1 are also involved in UV-B light signal transduction, where they act by directly binding to the promoter of COP1 and activating its transcription in response to UV-B (Huang et al., 2012). Although FRS4 recognizes the same cis-elements as FHY3, it has not been found to be involved in far-red light signal transduction (Gao et al., 2013). Suppression of FRS9 transcription resulted in a short hypocotyl under red light, but not far-red, or blue light conditions; this observation suggested that FRS9 is also involved in light signal transduction (Lin and Wang, 2004). However, the underlying molecular mechanism is still unclear.
Chloroplast Division
In higher plants, chloroplasts are the major site for photosynthesis and for the biosynthesis of numerous important components. FRS family proteins affect chloroplast function by regulating chloroplast biogenesis, chlorophyll biosynthesis, and starch synthesis (Ouyang et al., 2011; Tang et al., 2012; Gao et al., 2013; Ma et al., 2017). Chloroplast division determines the number of chloroplasts in each cell and plays an important role in cell expansion, division, and retrograde signal transduction from the chloroplast to the cytoplasm and the nucleus (Gao et al., 2003; Maple and Moller, 2007; Osteryoung and Pyke, 2014). Although many critical components involved in chloroplast division such as ARC5, ARC6, PLASTID DIVISION1 (PDV1), and PDV2, have been identified, the regulation of chloroplast division is still mysterious (Miyagishima et al., 2006; Glynn et al., 2008).
ARC5 encodes a dynamin-related protein that is essential for chloroplast division, since disruption of ARC5 results in enlarged, dumbbell-shaped chloroplasts (Gao et al., 2003). Recent studies unexpectedly revealed that FRS family proteins have important roles in the transcriptional regulation of ARC5 (Ouyang et al., 2011; Gao et al., 2013; Chang et al., 2015). Disruption of either FHY3, FAR1, or FRS4 results in enlarged, dumbbell-shaped chloroplasts, while constitutive overexpression of ARC5 in these mutants restores normal chloroplast morphology, demonstrating that these three FBSs are all required for the transcriptional activation of ARC5, albeit by distinct molecular mechanisms (Ouyang et al., 2011; Gao et al., 2013). FHY3 and FAR1 form homodimers or heterodimers, which bind directly to the FBS and FBS-Like (FBL) cis-elements in the ARC5 promoter to activate its expression (Ouyang et al., 2011). Another possibility is that FRS4 might regulate the transcription of ARC5 by forming heterodimers with FHY3 in the nucleus, since FRS4 alone does not have the ability to regulate transcription (Gao et al., 2013).
Chlorophyll Biosynthesis
Chlorophyll biosynthesis is strictly regulated by environmental light-dark cycles through a series of enzymatic reactions (Kobayashi and Masuda, 2016; Larkin, 2016). In the dark, chlorophyll biosynthesis is terminated by the accumulation of the intermediate metabolite protochlorophyllide (Pchlide). In the light, Pchlide is rapidly converted to chlorophyllide and ultimately chlorophyll (Larkin, 2016; Liu X. et al., 2017). The excessive accumulation of Pchlide in plants triggers the production of reactive oxygen species (ROS), which cause cell death and photobleaching (Larkin, 2016; Liu X. et al., 2017).
The etiolated seedlings of fhy3 and fhy3 far1 mutants greening rapidly after transferred to light, due to with less Pchlide accumulation by the end of the night, which reduces the production of ROS and increases plant survival and fitness after light exposure (Tang et al., 2012). Furthermore, FHY3 and FAR1 directly bind to the HEMB1 promoter to activate its transcription. HEMB1 encodes a 5-aminolevulinic acid dehydratase (ALAD) that catalyzes the synthesis of porphobilinogen (PBG) from 5-aminolevulinic acid (ALA). Thus, FHY3 and FAR1 both act as positive regulators of chlorophyll biosynthesis (Tang et al., 2012). Besides HEMB1, the expression level of other chlorophyll biosynthesis-related genes including HEMA1, HEMA3, FERROCHELATASE 2 (FC2), and HEME OXYGENASE 1 (HO1) are also significantly affected in the fhy3 and fhy3 far1 mutants, which indicates that FHY3 and FAR1 might regulate chlorophyll biosynthesis through transcriptional regulation of multiple diverse genes (McCormac and Terry, 2002; Tang et al., 2012).
PHYTOCHROME-INTERACTING FACTOR1 (PIF1) is a negative regulator of photomorphogenesis and chlorophyll biosynthesis that physically interacts with FHY3 and partially inhibits transcription of FHY3, which in turn regulates the expression of HEMB1 and chlorophyll biosynthesis (Tang et al., 2012). In addition, ETHYLENE-INSENSITIVE 3 (EIN3) and EIN3-LIKE 1 (EIL1), two transcription factors of the ethylene signaling pathway, directly bind to the promoters and activate the transcription of PROTOCHLOROPHYLLIDE OXIDOREDUCTASE A (PORA) and PORB, which are also essential in chlorophyll biosynthesis (Zhong et al., 2009). It is worthwhile to note that EIN3/EIL1 physically interacts with FHY3/FAR1 and PIF1 (Tang et al., 2012; Liu Y. et al., 2017); however, how these transcription factors work together to coordinate transcriptional regulation of chlorophyll biosynthesis-related genes in early seedling development requires further investigation.
Starch Synthesis and Starch Granule Formation
In the daytime, sugar produced by photosynthesis in the leaves provides the energy to maintain all kinds of metabolic activities, and any unused sugar is stored for the short term as starch (Streb and Zeeman, 2012). In the night, the transient starch in plant leaves is degraded into sugar to maintain plant growth and metabolism (Streb and Zeeman, 2012). Starch metabolism is regulated by the environmental light-dark cycle and endogenous sugar content, yet little is known about the underlying molecular mechanism (Smith et al., 2005; Graf and Smith, 2011).
A recent study revealed that FHY3 and FAR1 are crucial for starch synthesis but not turnover (Ma et al., 2017). Disruption of FHY3 and FAR1 resulted in a decrease in starch content and an increase in water-soluble polysaccharide content in the leaf at end of a light period. In addition, the highly ordered starch granule structure was dramatically disrupted in fhy3 and fhy3 far1 mutants, a phenotype that is very similar to that of isoamylase 2 (isa2) mutant plants (Ma et al., 2017). Arabidopsis ISA1 and ISA2 encode isoamylase-type debranching enzymes that are essential for starch granule biosynthesis (Delatte et al., 2005; Wattebled et al., 2005). Further investigation showed that FHY3 and FAR1 regulate starch synthesis through transcriptional activation of ISA2 thus mediate light-induced regulation of starch synthesis during the day (Ma et al., 2017).
The Circadian Clock and Flowering Time Regulation
Many biological processes have an endogenous oscillation of about 24 h, aligning with the day-night cycle; this oscillation is driven by the endogenous circadian clock (McClung et al., 2002; Doherty and Kay, 2010; Oakenfull and Davis, 2017). In plants, environmental light signals are among the most important factors affecting the entrainment of the circadian clock (Devlin and Kay, 2001; McClung et al., 2002). Multiple components governing light signal transduction, including phyA, phyB, cry1, and cry2 are involved in light entrainment of the circadian clock in Arabidopsis (Devlin and Kay, 2001; Harmer, 2009). FHY3 was originally identified for its role in gating the red light signal for clock resetting (Allen et al., 2006). Disruption of FHY3 causes arrhythmicity of CAB2 expression under continuous red light but not under continuous blue light (Allen et al., 2006). Further studies revealed that the expression levels of multiple circadian clock-related genes, including CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and ELF4, are significantly altered in fhy3, far1, and fhy3 far1 mutants. Therefore, FHY3 and FAR1 were proposed to be part of the light input pathway for the circadian clock (Li et al., 2011).
FHY3 and FAR1 directly bind to FBS cis-elements in the promoter of the central clock gene ELF4 to activate its transcription and promote flowering. HY5 also directly binds to the ELF4 promoter to induce its expression. CCA1 and LHY, two MYB type transcription factors, directly bind to the evening elements (EE) in the ELF4 promoter to repress its expression at dawn. Furthermore, CCA1 and LHY physically interact with the transcriptional activators FHY3, FAR1, and HY5, to suppress their activation of ELF4 transcription during the day (Li et al., 2011). FHY3 and FAR1 buffer the transcript level of ELF4 in the evening in a red light-dependent manner, and act downstream of the light-stable phytochromes phyB, phyD, and phyE (Siddiqui et al., 2016). FHY3 was found to associate with phyA in vivo (Saijo et al., 2008). These observations encourage further investigations that aim to identify other phytochromes or photoreceptors that FHY3 associates with in vivo.
Multiple FRS family members, including FHY3, FAR1, FRS6, FRS7, FRS8, and FRS12, negatively regulate flowering time by regulating the transcription of a diverse set of genes (Lin and Wang, 2004; Li et al., 2011; Ritter et al., 2017). FHY3 and FAR1 negatively regulate flowering time by activating the transcription of ELF4, with FHY3 playing a primary role (Li et al., 2011). FRS7 and its paralog FRS12 negatively regulate flowering time by repressing the transcription of GIGANTEA, and FRS7 plays a primary role (Ritter et al., 2017). Since mutating FRS7 or FRS12 has no effect on the expression of the central circadian clock genes such as LHY and TOC1, the FRS7–FRS12 complex might regulate plant growth and development by affecting the output of the circadian clock (Ritter et al., 2017). FRS6 and FRS8 also negatively regulate flowering time, as the disruption of FRS6 and FRS8 results in early flowering. However, the downstream targets and underlying molecular mechanism remain unclear (Lin and Wang, 2004).
Shoot Apical Meristem and Floral Development
In addition to playing multiple roles in early seedling development and flowering time regulation, FHY3 also participates in reproduction in Arabidopsis. Genetic evidence has revealed that FHY3 can promote shoot branching and is necessary for shoot meristem determinacy and maintenance (Stirnberg et al., 2012; Li et al., 2016). A genetic screen for suppressors of the highly branched mutant max2-1 (more axillary branching2-1) identified three recessive alleles at the FHY3 locus, indicating that FHY3 promotes shoot branching (Stirnberg et al., 2012). Mutating FAR1 also slightly suppressed the highly branched max2-1 phenotype, but mutating FHY1 or PHYA did not. In addition, a fhy3-12 mutant displayed reduced axillary bud activity in the strigolactone-deficient mutant backgrounds, max2-1 and max4-1, and cytokinin-overproducing mutant amp1 (altered meristem programme1), but not in the auxin-related axr1-3 (auxin resistant1-3) mutant background. This suggests that FHY3 might be involved in attenuating the auxin-regulated inhibition of bud outgrowth (Stirnberg et al., 2012). However, the molecular mechanism for FHY3-dependent activation of shoot branching requires further investigation.
In a genetic screen for second-site suppressors of ag-10 (agamous-10), FHY3 was also identified as an enhancer of floral meristem determinacy (Li et al., 2016). The mutants alleles of fhy3-27, fhy3-39, fhy3-46, and fhy3-68 display small petals, sterile anthers, and very short, bulged siliques, which suggests that FHY3 is necessary for seed reproduction (Li et al., 2016). A combination of ChIP-seq and RNA-seq identified hundreds of direct FHY3 targets that are involved in floral development. Interestingly, FHY3 directly represses the transcription of CLV3, but activates the transcription of SEP2 to ultimately promote floral meristem formation in the shoot apical meristem (Li et al., 2016). In addition, FHY3 might act as a transcriptional repressor of shoot apical meristem and floral meristem development, which is distinct from its roles as a transcriptional activator in light signaling, or light entrainment of the circadian clock during seedling development (Wang and Wang, 2015a; Li et al., 2016).
Oxidative Stress, Plant Immunity, and Cell Death
FHY3 and FAR1 also negatively regulate ROS accumulation and oxidative stress-induced cell death (Stirnberg et al., 2012; Ma et al., 2016; Wang et al., 2016). The adult fhy3 far1 mutant plants had slow, stunted growth, accumulated ROS, and displayed severe cell death under short-day or extended darkness conditions (Stirnberg et al., 2012; Ma et al., 2016; Wang et al., 2016). This cell death phenotype can be rescued by overexpressing SA-3-hydroxylase (S3H) to reduce accumulation of salicylic acid (SA), or by crossing fhy3 far1 plants with either SA metabolism mutants or signal transduction-related mutants (including pad4, eds1, sid2 and NahG), which suggests that the cell death phenotype in fhy3 far1 mutants is largely dependent on the accumulation of SA (Ma et al., 2016; Wang et al., 2016).
Interestingly, loss of FHY3 and FAR1 function enhances the expression of defense-responsive genes, thus increasing resistance to Pseudomonas syringae bacteria, which indicates that FHY3 and FAR1 are also involved in modulating plant immunity (Wang et al., 2016). Overexpression of HEMB1, one of the chlorophyll biosynthesis genes regulated by FHY3 and FAR1, also rescues the cell death phenotype in fhy3 far1 mutants. However, reducing the expression of HEMB1 increases the expression of defense-response genes and results in a lesion-mimic phenotype. This genetic and molecular evidence demonstrates that FHY3 and FAR1 negatively modulate plant immunity and cell death, possibly by interfering with biosynthesis of chlorophyll and SA signaling (Wang et al., 2016).
In addition, FHY3 and FAR1 suppress the accumulation of ROS and oxidative-stress-induced cell death partially through positively regulating myo-inositol biosynthesis (Ma et al., 2016). Myo-inositol is the precursor for the biosynthesis of many inositol derivatives including ascorbate acid, and is essential for plant growth and development (Gillaspy, 2011; Munnik and Nielsen, 2011). In plants, myo-inositol 1-phosphate synthase (MIPS1) catalyzes the rate-limiting step in myo-inositol synthesis (Donahue et al., 2010; Gillaspy, 2011; Munnik and Nielsen, 2011; Valluru and Van den Ende, 2011). Disruption of MIPS1 results in reduction of myo-inositol biosynthesis, enhanced expression of plant defense genes, and a severe cell death phenotype (Meng et al., 2009; Donahue et al., 2010; Luo et al., 2011). The transcript abundance of MIPS1 and MIPS2, and myo-inositol contents, are dramatically reduced in fhy3 and fhy3 far1 seedlings, which indicates that FHY3 and FAR1 positively regulate the transcription of MIPS1/2 and the biosynthesis of myo-inositol (Ma et al., 2016). Further evidence indicates that FHY3 and FAR1 directly bind to the MIPS1 and MIPS2 promoters to activate their transcription and thus promote the biosynthesis of myo-inositol under light conditions. In addition, constitutive overexpression of MIPS1 partially rescued inositol contents and the oxidative stress-induced cell death phenotype in the fhy3 far1 mutant background. Therefore, FHY3 and FAR1 improve resistance to oxidative stress and suppress plant cell death also by positively regulating the biosynthesis of myo-inositol (Ma et al., 2016).
One surprising observation is that disruption of both FRS7 and FRS12 resulted in larger rosette leaves and plant size, and overexpression of FRS7 or FRS12 resulted in stunted growth under both long-day and short-day conditions, which indicate that FRS7 and FRS12 negatively regulate rosette leaf growth (Ritter et al., 2017). The molecular mechanism of how FRS7 and FRS12 negatively regulate rosette leaf growth is still unclear. It is worth noting that FRS7–FRS12 and FHY3–FAR1 have antagonistic roles in the growth of rosette leaves, since fhy3 far1 and frs7 frs12 adult plants display opposite growth phenotypes (Ma et al., 2016; Wang et al., 2016; Ritter et al., 2017).
Abscisic Acid Signal Transduction and Stress Responses
Abscisic acid (ABA) plays multiple essential roles in plant growth and development including seed maturation, germination, gene expression, and stress responses (Wasilewska et al., 2008). Seed germination and seedling establishment are also regulated by light signals, yet how light and ABA synergistically regulate plant growth and development is still unclear (Lau and Deng, 2010). Recently, studies revealed that ABI5, a basic leucine zipper transcription factor, responds to both ABA and light signals (Finkelstein and Lynch, 2000; Tang et al., 2013).
Disruption of FHY3 and FAR1 reduced the ABA-dependent inhibition of seed germination, seedling greening, and root elongation. Compared with wild-type control plants, the mutant plants of fhy3 are less sensitive to salinity and osmotic stresses. In addition, mutants with disruption function of FHY3 and FAR1 were less sensitive to ABA-induced stomatal closure, thus FHY3 and FAR1 are required for stomatal movement and drought response (Tang et al., 2013). Further investigation showed the transcript abundance of ABI5 decreases in fhy3, far1, and fhy3 far1 mutant seedlings, and overexpression of ABI5 restores the fhy3 mutant phenotype to wild-type levels, indicating that ABI5 is regulated by FHY3 and FAR1. Therefore, FHY3 and FAR1 bind to the ABI5 promoter and activate its transcription thus mediating ABA signal transduction and abiotic stress responses (Tang et al., 2013).
Nutrient Absorption
Phosphorus (Pi) is an essential macronutrient and the limiting factor for plant growth, development, and metabolism. The MYB-type transcription factor PHOSPHATE STARVATION RESPONSE1 (PHR1) is crucial for the plant response to Pi deficiency (Rubio et al., 2001; Nilsson et al., 2007). Recent studies revealed that environmental light signals and ethylene in the soil work together to regulate PHR1 transcription and ultimately the Pi starvation response (Liu Y. et al., 2017).
The light-signaling proteins FHY3 and FAR1 directly bind to the PHR1 promoter to mediate the light-induced expression of PHR1 and the Pi starvation response. Meanwhile, EIN3 and its closest homolog EIL1, two master transcription factors in the ethylene signal transduction pathway, also bind directly to the PHR1 promoter to activate its expression and control the Pi starvation response. Disrupting both FHY3 and EIN3 significantly reduces Pi uptake in fhy3 ein3 seedlings compared to wild-type control plants (Liu Y. et al., 2017). However, the bZIP transcription factor HY5 negatively regulates PHR1 transcription. Thus, PHR1 is positively regulated by FHY3 and FAR1 and negatively regulated by HY5 in response to light above-ground and ethylene stimuli in the soil. FHY3, FAR1, HY5, and EIN3 work together to regulate PHR1 transcription and ultimately mediate the plant Pi starvation response (Liu Y. et al., 2017).
The Protein Interaction Network of FRS-FRF Family Proteins
FHY3 has been found to physically interact with FAR1, FRS4, HY5, CCA1, LHY, PIF1, and EIN3 to regulate the transcription of various targets (Table 1; Wang and Deng, 2002; Li et al., 2010, 2011, 2016; Tang et al., 2012; Gao et al., 2013). Generally, FHY3 and FAR1 interact with each other to form homodimers or heterodimers and regulate the transcription of various target genes, but FHY3 plays the primary role in this regulation (Wang and Deng, 2002). CCA1, LHY, and PIF1 interact with FHY3 and suppress its transcriptional activation of ELF4 and HEMB1 (Li et al., 2011; Tang et al., 2012). HY5 interacts with FHY3 and coordinately promotes the expression of ELF4 and COP1 (Li et al., 2011; Huang et al., 2012), but suppresses the transcriptional activation of FHY3 on FHY1 and PHR1 (Li et al., 2010; Liu Y. et al., 2017). EIN3 interacts with FHY3 and coordinately promotes the expression of PHR1 (Liu Y. et al., 2017). FRS4 interacts with FHY3 to promote the expression of ARC5 (Gao et al., 2013), and FRS3 interacts with JAZ3 and ZML2, as shown by a proteome-wide protein–protein interaction network analysis (Arabidopsis Interactome Mapping Consortium, 2011). In addition, tandem affinity purification-mass spectrometry showed that FRS7, HON4 (Histone-like protein 4), and AHL14 (AT-Hook motif nuclear localized protein 14) co-purified with FRS12, and that FRS7–FRS12 acts as part of a transcriptional repressor complex (Ritter et al., 2017). Many proteins have been found to interact with FHY3 or other FRS family proteins, and most of them are transcription factors (Table 1). It remains to be determined how different FRS family members interact with other transcriptional regulators, such as regulators of histone modification or other signal transduction proteins, to coordinate the regulation of the transcription of various genes.
FRS-FKF Family Proteins in Other Plant Species
Over 1000 putative FRS homologs have been predicted in diverse higher plant species, which indicates that FRS-FRF family proteins are evolutionarily conserved in angiosperms (Lin et al., 2007; Joly-Lopez et al., 2016). Beside Arabidopsis thaliana, the physiological functions of FRS family members have not been investigated in other higher plant groups. However, recent studies have suggested that putative FRS members are the candidate genes for Panicle and Spikelet Degeneration (PSD) gene in rice (Zhang et al., 2015), and a photoperiod-dependent flowering time regulator in wheat (Kiseleva et al., 2017). In addition, genomic sequencing and population genomic analysis of silver birch (Betula pendula) also demonstrated that a putative FRS10 member might be the candidate gene correlated with the adaptation to environment (Salojarvi et al., 2017). Thus, it would be interesting to investigate the physiological function of FRS-FRF family protein in other species. Indeed, in Aspergillus nidulans, VipA contains a FAR1-like DNA-binding domain and modulates light-regulated heme biosynthesis through direct association with the hemB promoter, indicating that FRS family proteins also function in filamentous fungi (Rohrig et al., 2017).
Concluding Remarks and Future Perspectives
FHY3, FAR1, and other FRS family members regulate various cellular processes by transcriptionally activating or repressing the expression of diverse target genes. ChIP-seq, DAP-seq, and tandem chromatin affinity purification-sequencing have identified thousands of different target genes that are subject to transcriptional regulation by FHY3, FAR1, or FRS7; however, only a few of them have been functionally characterized so far (Ouyang et al., 2011; Li et al., 2016; O’Malley et al., 2016; Ritter et al., 2017). Therefore, the FRS family proteins might have roles that are more important than was originally thought. In addition, FHY3–FAR1 and FRS7–FRS12 recognize distinct cis-elements in the promoters of their target genes (Lin et al., 2007; Ouyang et al., 2011; Li et al., 2016; O’Malley et al., 2016; Ritter et al., 2017), suggesting that different FRS family members might have different DNA-binding activities and gene activation features. Further exploring the physiological roles of the FRS family proteins and their gene targets will expand our understanding of how different FRS family proteins regulate plant growth and development. Therefore, to characterize the new physiological functions of various FRS-FRF family proteins is one of the important future directions in plant biology research.
FHY3 and FAR1 primarily activate the transcription of light-induced target genes, while FHY3 also represses another set of genes through an unknown mechanism (Ouyang et al., 2011). The precise molecular mechanism of how FHY3 or other FRS members activate or repress the transcription of various targets has remained elusive, but other transcriptional regulators such as Polycomb Repressive Complex 2 or the Mediator complex might be involved in these processes. However, when and why FHY3 (or other FRS family proteins) act as either activators or repressors in different organs or development stages will need to be determined in the future.
FRS family proteins are derived from transposases. Numerous studies have demonstrated that transposable elements and selfish elements are powerful contributors to genome evolution and diversity in angiosperms (Oliver et al., 2013). Although FHY3 and FAR1 probably did not retain transposase activity in Arabidopsis, it remains to be explored whether the FRS family proteins in other plants have retained transposase activity or have developed novel activity as transcription factors. FHY3 and FAR1 are the best characterized plant transcription factors that were derived from transposons. How other transcription factors were derived from transposons and diversified into the key transcriptional regulators they are today is worthy of further investigation.
Author Contributions
LM and GL drafted and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by grants from the National Natural Science Foundation of China (31670249) and Funds of Shandong “Double Tops” Program (to GL).
References
- Aguilar-Martinez J. A., Uchida N., Townsley B., West D. A., Yanez A., Lynn N., et al. (2015). Transcriptional, posttranscriptional, and posttranslational regulation of SHOOT MERISTEMLESS gene expression in Arabidopsis determines gene function in the shoot apex. Plant Physiol. 167 424–442. 10.1104/pp.114.248625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T., Koustenis A., Theodorou G., Somers D. E., Kay S. A., Whitelam G. C., et al. (2006). Arabidopsis FHY3 specifically gates phytochrome signaling to the circadian clock. Plant Cell 18 2506–2516. 10.1105/tpc.105.037358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333 601–607. 10.1126/science.1203877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N., Gao Y., Zhao L., Liu X., Gao H. (2015). Arabidopsis FHY3/CPD45 regulates far-red light signaling and chloroplast division in parallel. Sci. Rep. 5:9612. 10.1038/srep09612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Rodriguez D., Walker E. L. (2018). Analysis of yellow striped mutants of zea mays reveals novel loci contributing to iron deficiency chlorosis. Front. Plant Sci. 9:157. 10.3389/fpls.2018.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte T., Trevisan M., Parker M. L., Zeeman S. C. (2005). Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J. 41 815–830. 10.1111/j.1365-313X.2005.02348.x [DOI] [PubMed] [Google Scholar]
- Desnos T., Puente P., Whitelam G. C., Harberd N. P. (2001). FHY1: a phytochrome A-specific signal transducer. Genes Dev. 15 2980–2990. 10.1101/gad.205401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin P. F., Kay S. A. (2001). Circadian photoperception. Annu. Rev. Physiol. 63 677–694. 10.1146/annurev.physiol.63.1.677 [DOI] [PubMed] [Google Scholar]
- Doherty C. J., Kay S. A. (2010). Circadian control of global gene expression patterns. Annu. Rev. Genet. 44 419–444. 10.1146/annurev-genet-102209-163432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue J. L., Alford S. R., Torabinejad J., Kerwin R. E., Nourbakhsh A., Ray W. K., et al. (2010). The Arabidopsis thaliana Myo-inositol 1-phosphate synthase1 gene is required for Myo-inositol synthesis and suppression of cell death. Plant Cell 22 888–903. 10.1105/tpc.109.071779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Jiang N., Wessler S. R. (2002). Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3 329–341. 10.1038/nrg793 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. R., Lynch T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. 10.1105/tpc.12.4.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Kadirjan-Kalbach D., Froehlich J. E., Osteryoung K. W. (2003). ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. U.S.A. 100 4328–4333. 10.1073/pnas.0530206100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Liu H., An C., Shi Y., Liu X., Yuan W., et al. (2013). Arabidopsis FRS4/CPD25 and FHY3/CPD45 work cooperatively to promote the expression of the chloroplast division gene ARC5 and chloroplast division. Plant J. 75 795–807. 10.1111/tpj.12240 [DOI] [PubMed] [Google Scholar]
- Gillaspy G. E. (2011). The cellular language of myo-inositol signaling. New Phytol. 192 823–839. 10.1111/j.1469-8137.2011.03939.x [DOI] [PubMed] [Google Scholar]
- Glynn J. M., Froehlich J. E., Osteryoung K. W. (2008). Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20 2460–2470. 10.1105/tpc.108.061440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A., Smith A. M. (2011). Starch and the clock: the dark side of plant productivity. Trends Plant Sci. 16 169–175. 10.1016/j.tplants.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Harmer S. L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60 357–377. 10.1146/annurev.arplant.043008.092054 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczian A., Bury E., Tscheuschler A., Kircher S., Toth R., et al. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15 2125–2130. 10.1016/j.cub.2005.10.042 [DOI] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Yang P., Lau O. S., Li G., Li J., et al. (2012). Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24 4590–4606. 10.1105/tpc.112.103994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M., Ringli C., Boylan M. T., Quail P. H. (1999). The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 13 2017–2027. 10.1101/gad.13.15.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M. E., Lisch D. R., Quail P. H. (2003). The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 34 453–471. 10.1046/j.1365-313X.2003.01741.x [DOI] [PubMed] [Google Scholar]
- Joly-Lopez Z., Bureau T. E. (2014). Diversity and evolution of transposable elements in Arabidopsis. Chromosome Res. 22 203–216. 10.1007/s10577-014-9418-8 [DOI] [PubMed] [Google Scholar]
- Joly-Lopez Z., Hoen D. R., Blanchette M., Bureau T. E. (2016). Phylogenetic and genomic analyses resolve the origin of important plant genes derived from transposable elements. Mol. Biol. Evol. 33 1937–1956. 10.1093/molbev/msw067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva A. A., Potokina E. K., Salina E. A. (2017). Features of Ppd-B1 expression regulation and their impact on the flowering time of wheat near-isogenic lines. BMC Plant Biol. 17(Suppl 1):172. 10.1186/s12870-017-1126-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Masuda T. (2016). Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 7:1811. 10.3389/fpls.2016.01811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin R. M. (2016). Tetrapyrrole signaling in plants. Front. Plant Sci. 7:1586. 10.3389/fpls.2016.01586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O. S., Deng X. W. (2010). Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 13 571–577. 10.1016/j.pbi.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Li D., Fu X., Guo L., Huang Z., Li Y., Liu Y., et al. (2016). FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113 9375–9380. 10.1073/pnas.1602960113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Siddiqui H., Teng Y., Lin R., Wan X. Y., Li J., et al. (2011). Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 13 616–622. 10.1038/ncb2219 [DOI] [PubMed] [Google Scholar]
- Li J., Li G., Gao S., Martinez C., He G., Zhou Z., et al. (2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22 3634–3649. 10.1105/tpc.110.075788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D. R., Feschotte C., Wang H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318 1302–1305. 10.1126/science.1146281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Wang H. (2004). Arabidopsis FHY3/FAR1 gene family and distinct roles of its members in light control of Arabidopsis development. Plant Physiol. 136 4010–4022. 10.1104/pp.104.052191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li Y., Zhong S. (2017). Interplay between light and plant hormones in the control of Arabidopsis seedling chlorophyll biosynthesis. Front. Plant Sci. 8:1433. 10.3389/fpls.2017.01433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xie Y., Wang H., Ma X., Yao W., Wang H. (2017). Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 29 2269–2284. 10.1105/tpc.17.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Qin G., Zhang J., Liang Y., Song Y., Zhao M., et al. (2011). D-myo-inositol-3-phosphate affects phosphatidylinositol-mediated endomembrane function in Arabidopsis and is essential for auxin-regulated embryogenesis. Plant Cell 23 1352–1372. 10.1105/tpc.111.083337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Tian T., Lin R., Deng X. W., Wang H., Li G. (2016). Arabidopsis FHY3 and FAR1 regulate light-induced myo-inositol biosynthesis and oxidative stress responses by transcriptional activation of MIPS1. Mol. Plant 9 541–557. 10.1016/j.molp.2015.12.013 [DOI] [PubMed] [Google Scholar]
- Ma L., Xue N., Fu X., Zhang H., Li G. (2017). Arabidopsis thaliana FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 (FAR1) modulate starch synthesis in response to light and sugar. New Phytol. 213 1682–1696. 10.1111/nph.14300 [DOI] [PubMed] [Google Scholar]
- Makarova K. S., Aravind L., Koonin E. V. (2002). SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem. Sci. 27 384–386. 10.1016/S0968-0004(02)02140-0 [DOI] [PubMed] [Google Scholar]
- Maple J., Moller S. G. (2007). Plastid division: evolution, mechanism and complexity. Ann. Bot. 99 565–579. 10.1093/aob/mcl249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C. R., Salome P. A., Michael T. P. (2002). The Arabidopsis circadian system. Arabidopsis Book 1:e0044. 10.1199/tab.0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac A. C., Terry M. J. (2002). Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 32 549–559. 10.1046/j.1365-313X.2002.01443.x [DOI] [PubMed] [Google Scholar]
- Meng P. H., Raynaud C., Tcherkez G., Blanchet S., Massoud K., Domenichini S., et al. (2009). Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS One 4:e7364. 10.1371/journal.pone.0007364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S. Y., Froehlich J. E., Osteryoung K. W. (2006). PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18 2517–2530. 10.1105/tpc.106.045484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T., Nielsen E. (2011). Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14 489–497. 10.1016/j.pbi.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Nilsson L., Muller R., Nielsen T. H. (2007). Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 30 1499–1512. 10.1111/j.1365-3040.2007.01734.x [DOI] [PubMed] [Google Scholar]
- Oakenfull R. J., Davis S. J. (2017). Shining a light on the Arabidopsis circadian clock. Plant Cell Environ. 40 2571–2585. 10.1111/pce.13033 [DOI] [PubMed] [Google Scholar]
- Oliver K. R., McComb J. A., Greene W. K. (2013). Transposable elements: powerful contributors to angiosperm evolution and diversity. Genome Biol. Evol. 5 1886–1901. 10.1093/gbe/evt141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R. C., Huang S. C., Song L., Lewsey M. G., Bartlett A., Nery J. R., et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165 1280–1292. 10.1016/j.cell.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K. W., Pyke K. A. (2014). Division and dynamic morphology of plastids. Annu. Rev. Plant Biol. 65 443–472. 10.1146/annurev-arplant-050213-035748 [DOI] [PubMed] [Google Scholar]
- Ouyang X., Li J., Li G., Li B., Chen B., Shen H., et al. (2011). Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23 2514–2535. 10.1105/tpc.111.085126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter A., Inigo S., Fernandez-Calvo P., Heyndrickx K. S., Dhondt S., Shi H., et al. (2017). The transcriptional repressor complex FRS7-FRS12 regulates flowering time and growth in Arabidopsis. Nat. Commun. 8:15235. 10.1038/ncomms15235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrig J., Yu Z., Chae K. S., Kim J. H., Han K. H., Fischer R. (2017). The Aspergillus nidulans Velvet-interacting protein, VipA, is involved in light-stimulated heme biosynthesis. Mol. Microbiol. 105 825–838. 10.1111/mmi.13739 [DOI] [PubMed] [Google Scholar]
- Rubio V., Linhares F., Solano R., Martin A. C., Iglesias J., Leyva A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15 2122–2133. 10.1101/gad.204401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Zhu D., Li J., Rubio V., Zhou Z., Shen Y., et al. (2008). Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol. Cell 31 607–613. 10.1016/j.molcel.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salojarvi J., Smolander O. P., Nieminen K., Rajaraman S., Safronov O., Safdari P., et al. (2017). Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 49 904–912. 10.1038/ng.3862 [DOI] [PubMed] [Google Scholar]
- Siddiqui H., Khan S., Rhodes B. M., Devlin P. F. (2016). FHY3 and FAR1 act downstream of light stable phytochromes. Front. Plant Sci. 7:175. 10.3389/fpls.2016.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. M., Zeeman S. C., Smith S. M. (2005). Starch degradation. Annu. Rev. Plant Biol. 56 73–98. 10.1146/annurev.arplant.56.032604.144257 [DOI] [PubMed] [Google Scholar]
- Stirnberg P., Zhao S., Williamson L., Ward S., Leyser O. (2012). FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J. 71 907–920. 10.1111/j.1365-313X.2012.05038.x [DOI] [PubMed] [Google Scholar]
- Streb S., Zeeman S. C. (2012). Starch metabolism in Arabidopsis. Arabidopsis Book 10:e0160. 10.1199/tab.0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Ji Q., Huang Y., Jiang Z., Bao M., Wang H., et al. (2013). FAR-RED ELONGATED HYPOCOTYL3 and FAR-RED IMPAIRED RESPONSE1 transcription factors integrate light and abscisic acid signaling in Arabidopsis. Plant Physiol. 163 857–866. 10.1104/pp.113.224386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Wang W., Chen D., Ji Q., Jing Y., Wang H., et al. (2012). Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24 1984–2000. 10.1105/tpc.112.097022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valluru R., Van den Ende W. (2011). Myo-inositol and beyond–emerging networks under stress. Plant Sci. 181 387–400. 10.1016/j.plantsci.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Wang H., Deng X. W. (2002). Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 21 1339–1349. 10.1093/emboj/21.6.1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang H. (2015a). Multifaceted roles of FHY3 and FAR1 in light signaling and beyond. Trends Plant Sci. 20 453–461. 10.1016/j.tplants.2015.04.003 [DOI] [PubMed] [Google Scholar]
- Wang H., Wang H. (2015b). Phytochrome signaling: time to tighten up the loose ends. Mol. Plant 8 540–551. 10.1016/j.molp.2014.11.021 [DOI] [PubMed] [Google Scholar]
- Wang W., Tang W., Ma T., Niu D., Jin J. B., Wang H., et al. (2016). A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J. Integr. Plant Biol. 58 91–103. 10.1111/jipb.12369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska A., Vlad F., Sirichandra C., Redko Y., Jammes F., Valon C., et al. (2008). An update on abscisic acid signaling in plants and more. Mol Plant 1 198–217. 10.1093/mp/ssm022 [DOI] [PubMed] [Google Scholar]
- Wattebled F., Dong Y., Dumez S., Delvalle D., Planchot V., Berbezy P., et al. (2005). Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol. 138 184–195. 10.1104/pp.105.059295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam G. C., Johnson E., Peng J., Carol P., Anderson M. L., Cowl J. S., et al. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5 757–768. 10.1105/tpc.5.7.757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Xu F., Zhang Y., Lin J., Song C., Fang X. (2015). Fine mapping and candidate gene analysis of a novel PANICLE AND SPIKELET DEGENERATION gene in rice. Euphytica 206 793–803. 10.1007/s10681-015-1525-x [DOI] [Google Scholar]
- Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., et al. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. U.S.A. 106 21431–21436. 10.1073/pnas.0907670106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Hare P. D., Yang S. W., Zeidler M., Huang L. F., Chua N. H. (2005). FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J. 43 356–370. 10.1111/j.1365-313X.2005.02453.x [DOI] [PMC free article] [PubMed] [Google Scholar]