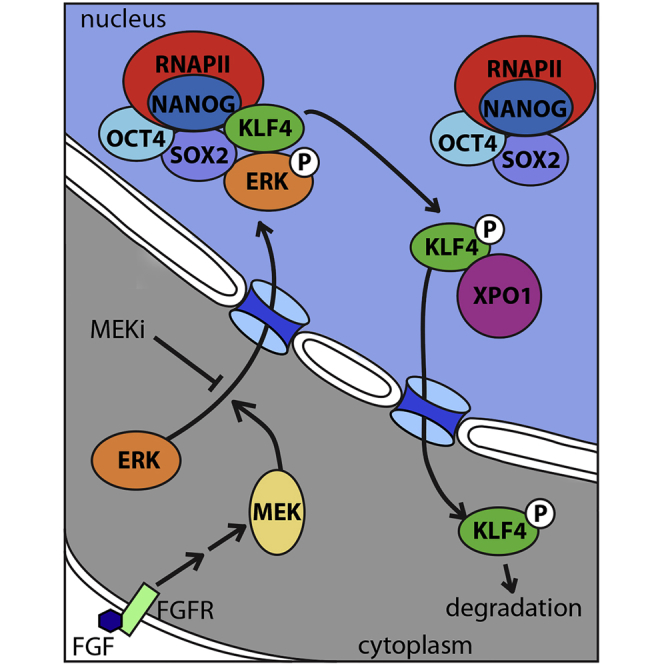
Summary
Cooperative action of a transcription factor complex containing OCT4, SOX2, NANOG, and KLF4 maintains the naive pluripotent state; however, less is known about the mechanisms that disrupt this complex, initiating exit from pluripotency. We show that, as embryonic stem cells (ESCs) exit pluripotency, KLF4 protein is exported from the nucleus causing rapid decline in Nanog and Klf4 transcription; as a result, KLF4 is the first pluripotency transcription factor removed from transcription-associated complexes during differentiation. KLF4 nuclear export requires ERK activation, and phosphorylation of KLF4 by ERK initiates interaction of KLF4 with nuclear export factor XPO1, leading to KLF4 export. Mutation of the ERK phosphorylation site in KLF4 (S132) blocks KLF4 nuclear export, the decline in Nanog, Klf4, and Sox2 mRNA, and differentiation. These findings demonstrate that relocalization of KLF4 to the cytoplasm is a critical first step in exit from the naive pluripotent state and initiation of ESC differentiation.
Keywords: KLF4, embryonic stem cell, nuclear export, pluripotency, XPO1, ERK, transcription factor, proximity ligation amplification
Graphical Abstract
Highlights
-
•
KLF4 nuclear export occurs as ESCs exit naive pluripotency
-
•
Active ERK and XPO1 interaction with KLF4 is required for nuclear export
-
•
KLF4 S132, an ERK phosphorylation site in KLF4, is required for nuclear export
-
•
Blocking KLF4 nuclear export prevents ESC differentiation
Dhaliwal and colleagues show that KLF4 is exported from the nucleus to initiate embryonic stem cell differentiation. KLF4 nuclear export is caused by FGF-MEK-ERK signaling whereby activated ERK phosphorylates KLF4, allowing interaction with nuclear export factor Xportin1. Blocking KLF4 nuclear export prevents embryonic stem cell exit from naive pluripotency and slows development of the embryo.
Introduction
Kruppel-like factor 4 (Klf4) is a transcriptional regulator of the naive pluripotent state sufficient to revert epiblast stem cells to naive embryonic stem cells (ESCs) (Bruce et al., 2007, Guo et al., 2009, Zhang et al., 2010). KLF4 forms a protein complex with the pluripotency master regulators OCT4 (POU5F1) and SOX2 and binds many of the same genomic regions as OCT4, SOX2, and NANOG (Chen et al., 2008, Gao et al., 2012, Wei et al., 2009). Leukemia inhibitory factor (LIF) can be used to maintain mouse ESCs in the pluripotent state, and Klf4 is a direct target of LIF signaling through activation of STAT3 (Bourillot et al., 2009, Hall et al., 2009, Niwa et al., 2009). In mouse ESCs, MAPK (mitogen-activated protein kinase) and GSK3 (glycogen synthase kinase 3) signaling promote differentiation; however, inhibition of both pathways through the use of two inhibitors (2i) supports self-renewal (Doble et al., 2007, Kunath et al., 2007, Ying et al., 2008). Mouse ESCs cultured in 2i media have been proposed to be the closest in vitro representation of pluripotent cells from pre-implantation embryos, referred to as naive pluripotency (Nichols and Smith, 2009). Media supplemented with 2i can support ESC self-renewal even in the absence of LIF; however, LIF is often included in the media to further support pluripotency maintenance (Nichols et al., 2009, Theunissen et al., 2011). The mechanisms of pluripotency exit, specifically how the pluripotency transcription factors are downregulated during differentiation, remain poorly understood.
Although KLF4 is involved in pluripotency maintenance in vitro, its expression has not been investigated in the pre-implantation mouse embryo; however, analysis of rhesus macaque embryos revealed nuclear KLF4 in all cells of pre-implantation blastocysts, including the pluripotent cells of the inner cell mass (ICM) (Harvey et al., 2009). Klf4 null mice survive early development, indicating that Klf4 is not required for embryogenesis, due to compensatory mechanisms involving other Klf factors (Jiang et al., 2008, Segre et al., 1999). Although not required during early mouse development, there may be a need for Klf4 downregulation upon differentiation, as overexpression of Klf4 has been shown to prevent differentiation of ESCs even in the absence of LIF (Zhang et al., 2010).
The mechanisms of pluripotency induction during reprogramming have been widely investigated, but less effort has been expended in understanding the mechanisms of pluripotency exit. As ESCs exit pluripotency, Klf4 and Nanog are the first transcription factors to show a decrease in transcript expression (Zhang et al., 2010). This, coupled with the observation that KLF4 protein binds the Nanog promoter and activates transcription, led to the idea that reduced Klf4 transcription is the trigger that causes exit from pluripotency (Zhang et al., 2010). To further investigate this mechanism, we monitored nuclear levels of KLF4 and NANOG protein in ESCs as they exit naive pluripotency. In contrast to the mechanism proposed above, our study revealed that pluripotency exit is initiated by an ERK-mediated interaction between KLF4 and the nuclear export factor XPO1 and the subsequent relocalization of KLF4 to the cytoplasm. Blocking KLF4 nuclear export by mutating the ERK phosphorylation site in the KLF4 protein or KLF4 nuclear export sequences (NES) required for interaction with XPO1 prevents exit from pluripotency. To investigate this mechanism in the embryo, we inhibited MEK signaling to prevent ERK activation; this resulted in a loss of KLF4/ERK2 and KLF4/XPO1 interaction and a disruption in KLF4 and NANOG downregulation in pre-implantation mouse blastocysts. These findings extend our proposed mechanism for controlling KLF4 nuclear levels to the pre-implantation embryo, demonstrating the significance of KLF4 nuclear export in regulating stem cell differentiation both in vitro and in vivo.
Results
KLF4 Nuclear Export Occurs as ESCs Exit Naive Pluripotency
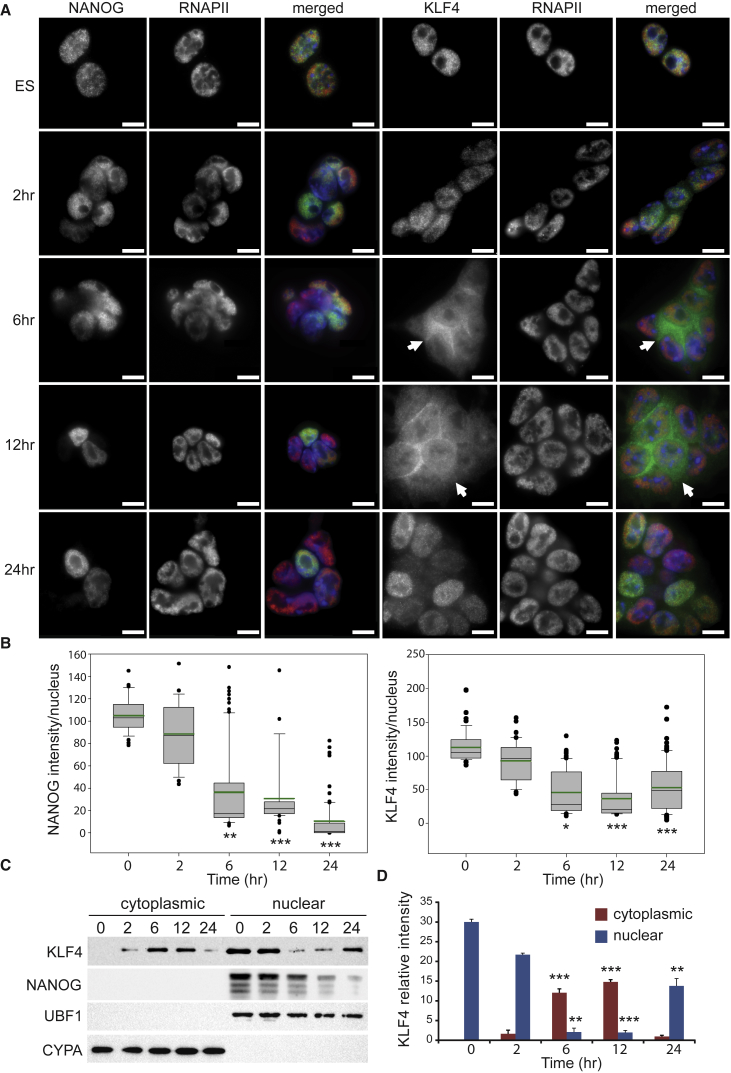
Levels of Klf4 and Nanog mRNA decline within the first 24 hr of pluripotency exit, and it has been suggested that reduced Klf4 transcription after removal of external maintenance factors is the trigger that initiates ESC differentiation; however, protein levels have not been investigated during this time frame (Zhang et al., 2010). We first confirmed that the decline in both Klf4 and Nanog mRNA occurred for ESCs maintained in LIF/2i after removal of these components, and observed a reduction in both Klf4 and Nanog mRNA at 6 hr (Figure S1A). To investigate changes in nuclear protein levels as ESCs exit naive pluripotency, we monitored subcellular localization of KLF4, OCT4, SOX2, and NANOG for 24 hr after removal of LIF/2i. In undifferentiated ESCs KLF4, OCT4, SOX2, and NANOG are located in the nucleus (Figures 1 and S1B). Quantification of immunofluorescence images from different times after differentiation induction indicated that SOX2 and OCT4 nuclear protein levels were maintained over the first 24 hr of ESC differentiation (Figures S1C and S1D), whereas NANOG and KLF4 exhibited dynamic behavior (Figure 1B). At 6 hr, a significant portion of KLF4 protein was localized in the cytoplasm (arrows in Figure 1A), suggesting that KLF4 nuclear export occurs as ESCs exit naive pluripotency. Over this same time period, NANOG nuclear protein levels decreased with a significant reduction first observed at 6 hr; however, NANOG was not observed in cytoplasm (Figures 1A and 1B). To obtain a more quantitative measure of the amount of cytoplasmic and nuclear KLF4, we separated ESCs into nuclear and cytoplasmic fractions and assayed them by immunoblot. This analysis confirmed the subcellular redistribution of KLF4; nuclear KLF4 was significantly reduced at 6 hr and cytoplasmic KLF4 was increased (Figures 1C and 1D). Interestingly, 24 hr after LIF/2i removal KLF4 was again predominantly nuclear, although at half the levels observed in undifferentiated ESCs. Total protein was also monitored, which confirmed the 50% decrease in KLF4 protein at 24 hr compared with undifferentiated ESCs (Figure S2A).
Figure 1.
KLF4 Nuclear Export Occurs as ESCs Exit Naive Pluripotency
(A) Immunofluorescence of ESCs cultured with LIF/2i and 2, 6, 12, and 24 hr after LIF/2i removal display KLF4 accumulation in the cytoplasm starting at 6 hr (arrows). Merged images display NANOG or KLF4 in green, RNAPII-S5P in red, and DAPI DNA stain in blue. Scale bars, 10 μm.
(B) Box-and-whisker plots display intensities per nucleus of NANOG (left) and KLF4 (right). Green line indicates the average intensity at each time point and black line indicates the median intensity. Boxes indicate interquartile range of intensity values and whiskers indicate the 10th and 90th percentiles; outliers are shown as black dots. Images were collected from at least three biological replicate samples and ≥100 nuclei were quantified for each. Statistical differences are indicated by ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) Immunoblot of nuclear and cytoplasmic fractions from ESCs cultured with LIF/2i (0) and 2, 6, 12, and 24 hr after LIF/2i removal. Cyclophilin A (CYPA) and the nucleolar protein upstream binding factor (UBF1) reveal purity of the cytoplasmic and nuclear fractions, respectively. KLF4 levels are increased in the cytoplasmic fraction at 6 hr and correspondingly decreased in the nuclear fraction. At 24 hr KLF4 was mainly located in the nuclear fraction.
(D) Quantification of relative intensity levels of KLF4 in immunoblots from three biological replicates. Statistical differences compared with the 0-hr values are indicated by ∗∗p < 0.01 and ∗∗∗p < 0.001. Error bars represent standard deviation.
See also Figure S1.
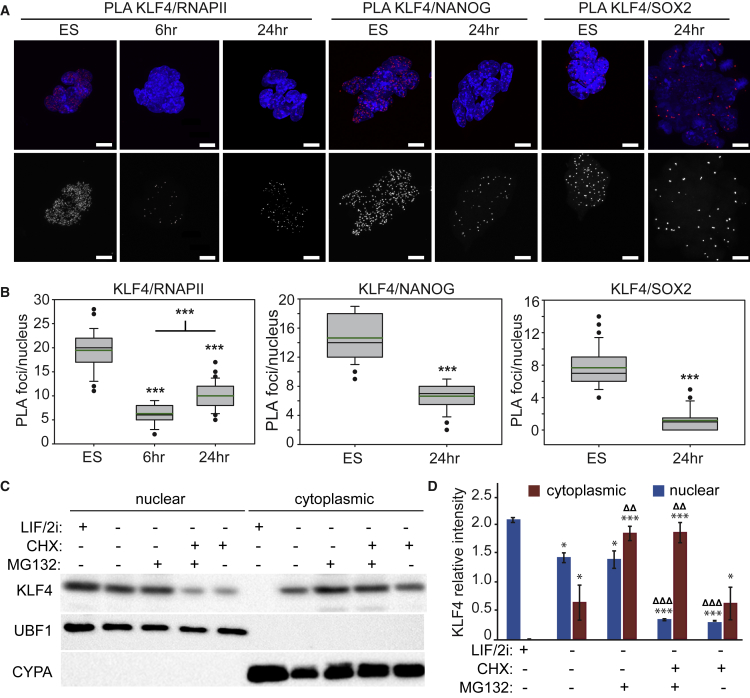
As KLF4 was observed to exit the nucleus at 6 hr but accumulate in the nucleus at 24 hr, we monitored nuclear protein complexes to determine whether this dynamic behavior disrupted KLF4 function in the nucleus. Proximity ligation amplification (PLA) (Fredriksson et al., 2002) was conducted to quantify the interaction between KLF4/RNAPII-S5P or pluripotency transcription factors in single nuclei. In undifferentiated ESCs, KLF4 interacts with RNAPII-S5P at about 20 nucleoplasmic foci (Figures 2A and 2B). As ESCs differentiate this is reduced to about 6 foci per nucleus at 6 hr; at 24 hr, on average 10 foci were detected, indicating that the reduction in nuclear KLF4 by half affects participation of KLF4 in polymerase complexes. In addition, interaction between KLF4/NANOG and NANOG/RNAPII-S5P (Figures 2A, 2B, and S2B) was reduced at 24 hr. This was expected as nuclear NANOG levels were dramatically reduced at 24 hr. Although SOX2 levels were found to be unchanged over the first 24 hr of ESC differentiation, and interaction of SOX2/RNAPII-S5P was unchanged (Figures S2A and S2B), interaction between KLF4/SOX2 was also reduced at 24 hr (Figure 2). Similarly, interaction between KLF4/OCT4 was reduced at 24 hr (Figure S2B). Together these data indicate that KLF4 function in nuclear transcription complexes is disrupted at 6 hr and only partially restored at 24 hr of differentiation.
Figure 2.
Dynamic Localization of KLF4 Alters Nuclear Complexes during Exit from Naive Pluripotency
(A) Proximity ligation amplification (PLA) indicating the interaction between KLF4/RNAPII-S5P, KLF4/NANOG, and KLF4/SOX2 in ESCs are disrupted 6 or 24 hr after LIF/2i withdrawal. Images shown are maximum-intensity projections. Scale bars, 10 μm.
(B) Box-and-whisker plots indicate the number of PLA foci per nucleus. Boxes indicate interquartile range of intensity values and whiskers indicate the 10th and 90th percentiles; outliers are shown as black dots. Images were collected from at least three biological replicate samples and ≥100 nuclei were quantified for each. Statistical differences are indicated by ∗∗∗p < 0.001.
(C) Immunoblot of nuclear and cytoplasmic fractions from ESCs after 24 hr LIF/2i withdrawal and 12 hr treatment with CHX or MG132.
(D) Quantification of relative intensity levels of KLF4 in immunoblots from three biological replicates reveals that the proteasome inhibitor MG132 prevents degradation of cytoplasmic KLF4 while the protein synthesis inhibitor cycloheximide (CHX) prevents accumulation of nuclear KLF4. Statistical differences compared with 0-hr values are indicated by ∗p < 0.05 and ∗∗∗p < 0.001; statistical differences between untreated 24-hr differentiated fractions (LIF/2i−) and CHX or MG132 treated fractions are indicated by ΔΔp < 0.01 and ΔΔΔp < 0.001. Error bars represent standard deviation. See also Figure S2.
The observation that KLF4 protein became predominantly nuclear again at 24 hr led us to investigate whether this was newly synthesized KLF4 or the same pool of protein that was exported at 6 hr. To investigate this we treated ESCs undergoing differentiation for 12 hr, when KLF4 is almost exclusively cytoplasmic (Figure 1D), for a further 12 hr with the protein synthesis inhibitor cycloheximide (CHX) in the presence or absence of the proteasome inhibitor MG132. Immunoblotting revealed that KLF4 protein levels in the nuclear fraction are significantly reduced at 24 hr when cells are treated with CHX, indicating that KLF4 accumulation in the nucleus at 24 hr requires protein synthesis (Figures 2C and 2D). The significantly increased amount of KLF4 in the cytoplasmic fraction of the MG132-treated cells indicates that KLF4 exported to the cytoplasm undergoes proteosomal degradation.
ERK Activation and Interaction with KLF4 Is Coincident with KLF4 Nuclear Export
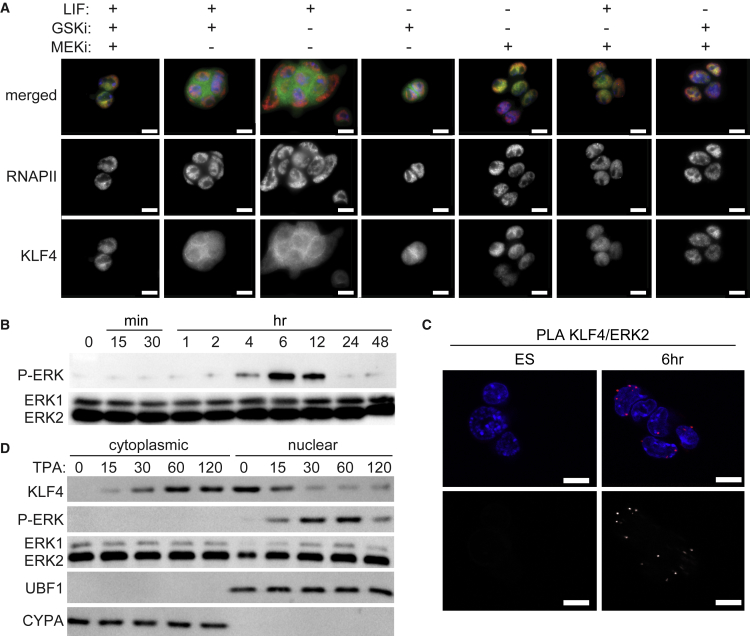
As KLF4 was exported to the cytoplasm 6 hr after removal of LIF/2i, we investigated the mechanisms involved by removal of individual media components (LIF, MEK inhibitor, GSK inhibitor) for 6 hr followed by KLF4 subcellular localization by immunofluorescence (Figure 3A). Removal of the MEK inhibitor resulted in relocalization of KLF4 to the cytoplasm; however, when the MEK inhibitor remained in the media, KLF4 was localized to the nucleus. Stimulation of the fibroblast growth factor receptor (FGFR) by FGF4 causes activation of the MEK-ERK pathway and is required for ESC differentiation (Kunath et al., 2007). KLF4 nuclear export after removal of MEK inhibition was blocked by an FGFR inhibitor, indicating that FGFR activation is required for this process (Figure S3A). Together these observations suggest that ERK activation plays a central role in mediating ESC differentiation by controlling the subcellular localization of KLF4.
Figure 3.
Active ERK2 Interacts with KLF4 Initiating KLF4 Nuclear Export
(A) Immunofluorescence images of ESCs cultured for 6 hr with the indicated components: LIF, MEKi (MEK1 inhibitor, PD0325901), and GSKi (GSK3 inhibitor, CHIR99021). Merged images display KLF4 in green, RNAPII-S5P in red, and DAPI in blue. Scale bars, 10 μm.
(B) Immunoblot for activated ERK1/2 (pTEpY) and total ERK1/2 in ESCs cultured with LIF/2i (0), 15 and 30 min, 1, 2, 4, 6, 12, 24, and 48 hr after LIF/2i removal.
(C) Proximity ligation amplification (PLA) for KLF4/ERK2 displays the interaction between ERK2 and KLF4 6 hr after LIF/2i withdrawal but not in ESCs maintained in LIF/2i. Images shown are single optical sections. Merged images display DAPI in blue and PLA in red. Bottom images display grayscale PLA signal. Scale bars, 10 μm.
(D) Immunoblot for ESCs treated with TPA and sampled at the indicated time in minutes. With TPA treatment, ERK phosphorylation and KLF4 nuclear export occur more rapidly starting at 15 min. Cyclophilin A (CYPA) and the nucleolar protein upstream binding factor (UBF1) reveal purity of the cytoplasmic and nuclear fractions, respectively.
See also Figure S3.
To determine whether ERK activation was coincident with KLF4 nuclear export, we monitored total and active ERK1/2 by immunoblot (Figure 3B). Phosphorylated active ERK (pTEpY) was detected 4–6 hr after removal of LIF/2i when cytoplasmic KLF4 was observed. ERK phosphorylation was no longer observed at 24 hr when KLF4 again accumulates in the nucleus. Upon investigation of the subcellular localization of ERK2 in ESCs, we observed phosphorylated ERK in the nucleus and increased overlap of ERK2 with KLF4 at 6 hr (Figures S3B and S3C). The concurrence between KLF4 nuclear export, ERK2 phosphorylation, and nuclear accumulation suggested that active nuclear ERK2 promotes KLF4 nuclear export. To investigate whether ERK2 directly interacts with KLF4, we conducted PLA to identify proximity between KLF4 and ERK2. In undifferentiated ESCs, ERK2 remains inactive and KLF4/ERK2 PLA revealed no interaction between these two proteins; however, after 6 hr of differentiation, when phosphorylated ERK2 accumulates in the nucleus, interaction between KLF4 and ERK2 was observed (Figure 3C).
As ESCs maintained in LIF/2i are prevented from differentiating by inhibiting MEK-ERK signaling and the release of this inhibition may be linked to KLF4 nuclear export, we sought to determine whether KLF4 nuclear export would occur during pluripotency exit for ESCs maintained in LIF. We observed KLF4 nuclear export at 12 hr, which was delayed compared with cells maintained in LIF/2i (Figure S3D). MEK-ERK signaling is often rapid, peaking within minutes. To determine whether MEK-ERK signaling and KLF4 nuclear export can occur more rapidly in ESCs, we treated undifferentiated cells with TPA (12-O-tetradecanoylphorbol-13-acetate), an upstream activator of the MEK-ERK pathway (Figure 3D). TPA treatment caused ERK phosphorylation and KLF4 nuclear export at 15–30 min, indicating that this response can be rapid. In addition, a more dramatic decrease in both Klf4 and Nanog transcript levels was observed 2 hr after LIF/2i removal in the presence of TPA (Figure S3E).
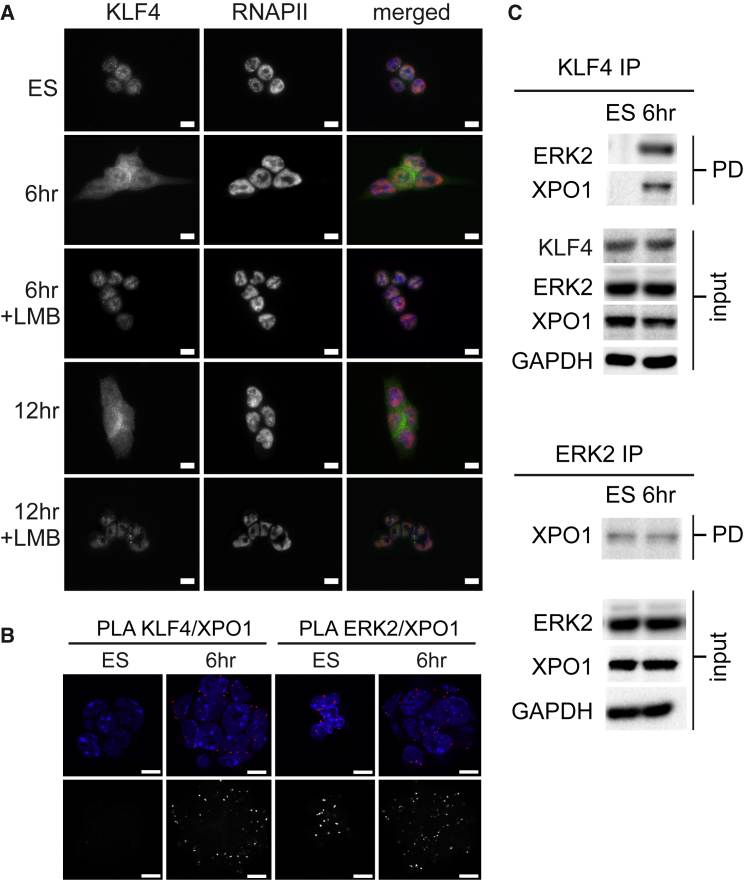
KLF4 Nuclear Export Occurs through an XPO1-Mediated Nuclear Export Mechanism
To investigate the involvement of Xportin1 (XPO1) in KLF4 nuclear export, we differentiated ESCs in the presence or absence of the XPO1-mediated nuclear export inhibitor leptomycin B (LMB) (Figure 4A). LMB was found to block KLF4 nuclear export, as KLF4 remained nuclear. XPO1-mediated nuclear export occurs when XPO1 binds to a cargo protein NES. PLA for KLF4/XPO1 revealed an interaction between KLF4 and XPO1 only in differentiating ESCs (Figure 4B). ERK2 and XPO1 were also shown to interact by PLA; however, signals were observed in both ESCs and 6-hr cells. Interactions between KLF4/ERK2, KLF4/XPO1, and ERK2/XPO1 were validated by co-immunoprecipitation (Figure 4C). ERK2 is unphosphorylated in undifferentiated cells; however, we identified interaction with XPO1 under these conditions, suggesting that continuous export of ERK2 by XPO1 prevents nuclear accumulation of unphosphorylated ERK2; indeed this has been described in both Xenopus and rat cells where ERK nuclear export depends on XPO1 and the MEK NES (Adachi et al., 2000). In addition, we found that MEK interacts with XPO1 in both undifferentiated and 6-hr differentiated ESCs; however, KLF4 does not interact with MEK in either condition (Figure S4), despite the observation that both MEK and KLF4 interact with XPO1 in differentiating ESCs, indicating that export of KLF4 is independent of the MEK NES in ESCs.
Figure 4.
KLF4 Nuclear Export Occurs through an XPO1-Mediated Mechanism
(A) Immunofluorescence images of ESCs cultured for 6 and 12 hr without LIF/2i in the presence of 5 μg/mL leptomycin B (LMB), which inhibits XPO1-mediated nuclear export. Merged images display KLF4 in green, RNAPII-S5P in red, and DAPI in blue. Scale bars, 10 μm.
(B) Proximity ligation amplification (PLA) displays the interaction between KLF4/XPO1 and XPO1/ERK2 in ESCs and 6 hr after LIF/2i withdrawal. Images shown are single optical sections. Merged images display DAPI in blue and PLA in red. Bottom images display the grayscale PLA signal. Scale bars, 10 μm.
(C) KLF4 and ERK2 immunoprecipitation (IP) for undifferentiated ESCs and ESCs 6 hr after LIF/2i removal, probed with anti-ERK2 and anti-XPO1 (PD, pull-down). GAPDH levels reveal equal loading of the input.
See also Figure S4.
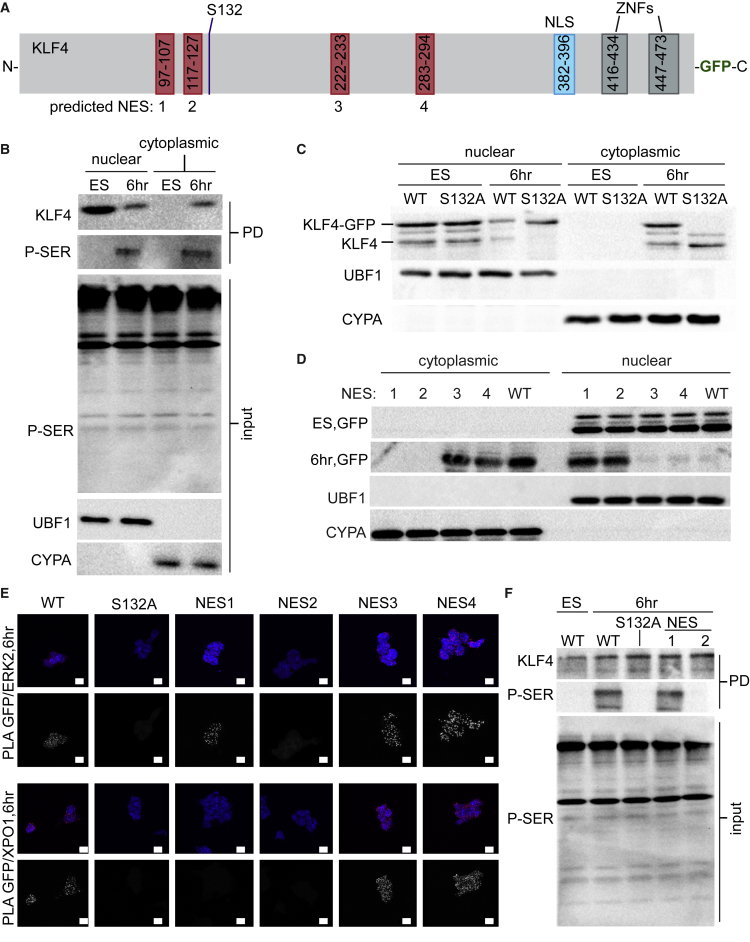
KLF4 Nuclear Export Requires Both Nuclear Export Sequences and Phosphorylation of S132
As KLF4 nuclear export was initiated by interaction of KLF4 with active ERK2, and KLF4 has been shown to be phosphorylated by ERK at S132 (described as S123 in Kim et al. (2012) as their construct lacked the first 9 amino acids of the endogenous KLF4 protein), we next investigated the role of S132 in KLF4 nuclear export (Figure 5). To determine whether KLF4 is phosphorylated prior to nuclear export, we immunoprecipitated KLF4 from nuclear and cytoplasmic fractions of undifferentiated and 6-hr differentiated ESCs and subjected it to immunoblotting using an anti-phosphoserine antibody (Figure 5B). The presence of a phosphoserine band in both the cytoplasmic and nuclear fractions at 6 hr suggests that activated ERK2 interacts with KLF4 and phosphorylates KLF4 at a serine residue. PLA for ERK2/RNAPII-S5P revealed that at 6 hr, when active ERK2 enters the nucleus, it interacts with active RNAPII (Figure S5A) which could facilitate interaction with, and phosphorylation of, KLF4 associated with active RNAPII (Figure 2).
Figure 5.
The KLF4 ERK Phosphorylation Site S132, NES1, and NES2 Are Required for KLF4 Nuclear Export
(A) Schematic of mouse KLF4 depicting predicted nuclear export signals (NES), nuclear localization signal (NLS), ERK phosphorylation site S132, and zinc fingers (ZNFs).
(B) Immunoprecipitated KLF4 protein from nuclear and cytoplasmic fractions of ESCs cultured with LIF/2i and 6 hr after LIF/2i removal, probed with anti-KLF4 and anti-phosphoserine (P-SER; PD, pull-down). Cyclophilin A (CYPA) and the nucleolar protein upstream binding factor (UBF1) reveal purity of the cytoplasmic and nuclear fractions, respectively.
(C) Nuclear and cytoplasmic fractions prepared from KLF4-GFP ES lines indicate that wild-type (WT) KLF4-GFP (top band) but not KLF4S132A-GFP is exported to the cytoplasm 6 hr after LIF/2i removal. Endogenous KLF4 (bottom band) is exported to the cytoplasm in both cases.
(D) Nuclear and cytoplasmic fractions prepared from WT KLF4-GFP and KLF4-GFP NES mutants (1–4). Anti-GFP immunoblot indicated that NES1 and NES2 are required for nuclear export of KLF4 after removal of LIF/2i.
(E) Proximity ligation amplification (PLA) for GFP/ERK2 and GFP/XPO1 indicated that the interaction between KLF4/ERK2 was disrupted by S132 and NES2 mutations. The interaction between KLF4/XPO1 was disrupted by S132, NES1, and NES2 mutations. Images shown are maximum-intensity projections. Scale bars, 10 μm.
(F) Anti-phosphoserine immunoblot of GFP-Trap revealed that S132A and NES2 mutations disrupted KLF4 serine phosphorylation.
See also Figure S5.
To investigate the role of S132 in KLF4 nuclear export, we generated ESC lines that stably express wild-type (WT) KLF4-GFP, an S132 mutant (KLF4(S132A)-GFP), or a phosphomimetic (KLF4(S132D)-GFP). Endogenous KLF4 and WT KLF4-GFP were observed to exit the nucleus at 6 hr (Figure 5C). By contrast, KLF4(S132A)-GFP remains in the nuclear fraction, indicating that S132 is required for nuclear export. In addition, the phosphomimetic KLF4(S132D)-GFP is exported to the cytoplasm in undifferentiated ESCs, suggesting that phosphorylation of S132 is sufficient for KLF4 nuclear export (Figure S5B). Nuclear export occurs when XPO1 binds a cargo protein NES; to investigate this we mutated four predicted NES in KLF4-GFP (Figure 5A). Immunoblot with anti-GFP revealed that mutation of NES1 or NES2 inhibited KLF4 export to the cytoplasm, whereas mutation of NES3 or NES4 had no effect (Figure 5D).
We next investigated the effect of KLF4 mutation on interaction with ERK2 and XPO1 (Figure 5E). Similar to results for the interaction of endogenous KLF4 with ERK2 and XPO1, we found no interactions in undifferentiated ESCs (Figure S5C). Cells differentiated for 6 hr displayed interaction between WT KLF4-GFP and ERK2 as well as XPO1 (Figure 5E). Mutation of KLF4 at S132 or NES2, immediately upstream of S132, disrupted the interaction of KLF4-GFP with ERK2, whereas mutation of NES1, 3, or 4 had no effect. The interaction between KLF4-GFP and XPO1 was disrupted when S132, NES1, or NES2 was mutated, whereas mutation of NES3 or NES4 had no effect. To evaluate the effect of specific mutations on the phosphorylation of KLF4, we immunoprecipitated cell lysates with GFP-Trap and conducted immunoblotting using anti-phosphoserine (Figure 5F). Serine phosphorylation at 6 hr was disrupted by mutation of S132 or NES2. Together, these data indicate that NES1, NES2, and S132 are required for the interaction of KLF4 with XPO1 and KLF4 nuclear export.
To rule out the possibility that mutant KLF4-GFP was not able to participate in nuclear complexes, we investigated interaction with RNAPII-S5P by PLA, which revealed that all KLF4-GFP mutants participated in transcriptional complexes to a similar degree (Figure S5D). To determine whether KLF4 nuclear localization is important for interaction with ERK2, we generated an ESC line stably expressing a KLF4 nuclear localization mutant, KLF4(NLS)-GFP. GFP/ERK2 PLA in these cells revealed no interaction between KLF4(NLS)-GFP and ERK2 after 6 hr, indicating that localization to the nucleus is important for this interaction (Figure S5D). Together these data indicate the S132 ERK phosphorylation site, NES1, and NES2 are together required for KLF4 nuclear export. Furthermore, mutation of these residues does not appear to interfere with the participation of KLF4 in active RNAPII transcriptional complexes.
Inhibiting KLF4 Nuclear Export Delays Differentiation of ESCs
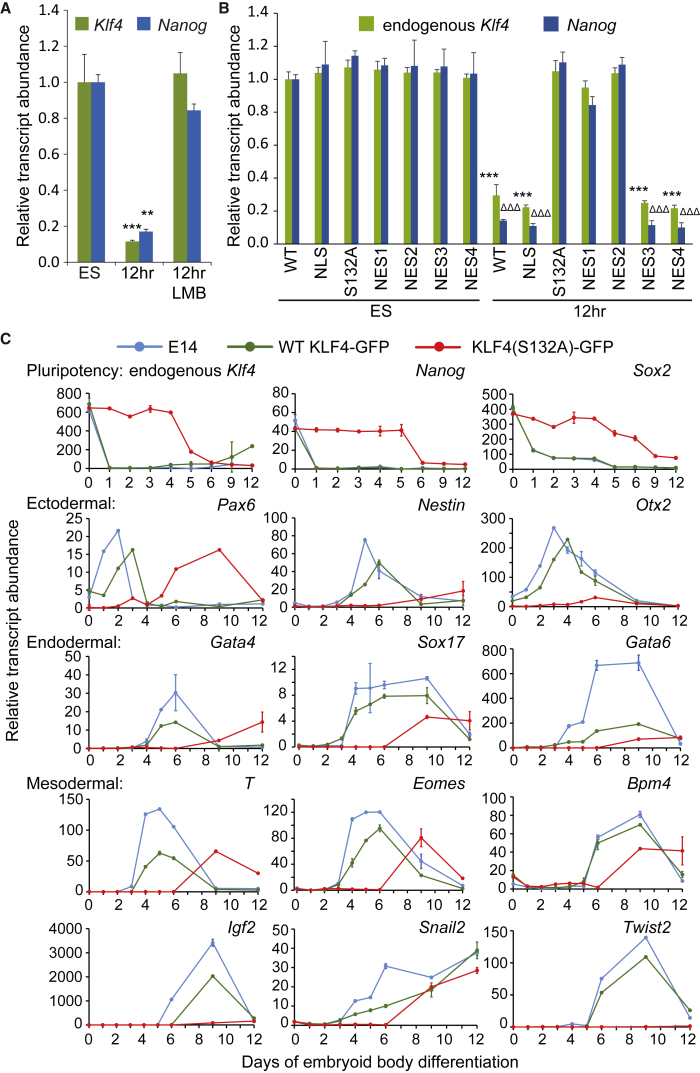
Nanog downregulation upon ESC differentiation has been suggested to occur due to a decrease in Klf4 transcription, which was assumed to be followed by a subsequent decrease in KLF4 protein levels (Zhang et al., 2010). By contrast, our data reveal that KLF4 is exported to the cytoplasm at this time, suggesting that this export causes Nanog and Klf4 downregulation at the transcriptional level. To determine whether nuclear export is required for the downregulation of Nanog and Klf4, we monitored transcript abundance by qRT-PCR in undifferentiated and 12 hr differentiated ESCs in the presence or absence of the nuclear export inhibitor LMB. The presence of LMB blocked KLF4 nuclear export and the early decrease in both Klf4 and Nanog transcripts observed after 12 hr of differentiation (Figure 6A). In the same samples, no change in the amount of Sox2 or Oct4 transcripts was observed (Figure S6A).
Figure 6.
KLF4 Nuclear Export Inhibition Delays Exit from Naive Pluripotency and Differentiation of Embryoid Bodies
(A) Treatment with 5 μg/mL LMB prevents downregulation in Klf4 and Nanog transcripts that normally occurs 12 hr after LIF/2i withdrawal. Average data from three biological replicates are normalized to levels observed in undifferentiated ESCs. Statistical differences are indicated by ∗∗p < 0.01 and ∗∗∗p < 0.001. Error bars represent standard deviation.
(B) Expression of endogenous Klf4 and Nanog is maintained after 12 hr of differentiation in KLF4(S132A)-GFP, KLF4(NES1)-GFP, and KLF4(NES2)-GFP mutants but not in wild-type (WT) KLF4-GFP, KLF4(NLS)-GFP, KLF4(NES3)-GFP, and KLF4(NES4)-GFP mutants. Average data from three biological replicates are normalized to the levels observed in undifferentiated ESCs expressing WT KLF4-GFP. Statistical differences from the undifferentiated ESC levels for endogenous Klf4 transcript levels are indicated by ∗∗∗p < 0.001, and Nanog transcript levels are indicated by ΔΔΔp < 0.001. Error bars represent standard deviation.
(C) Relative gene expression analysis for pluripotency and germ layer markers in untransfected ESCs (E14), WT KLF4-GFP, and KLF4(S132A)-GFP transfected ESCs differentiated to embryoid bodies for 12 days reveals that expression of KLF4(S132A)-GFP delays ESC differentiation. Data shown are averages of three qRT-PCR replicates for each time point. Error bars represent standard deviation.
See also Figure S6.
To investigate the effect of inhibiting nuclear export of KLF4 specifically, we analyzed relative transcript levels of Nanog and endogenous Klf4 in KLF4-GFP-expressing cells. Blocking KLF4 nuclear export by mutating S132, NES1, or NES2 prevents the decline in transcript levels for Nanog and endogenous Klf4 normally observed at 12 hr but does not affect Oct4 and Sox2 levels (Figures 6B and S6B). In addition, inhibiting KLF4 nuclear export by mutation of S132 prevents the reduction in KLF4/RNAPII-S5P interaction observed for WT KLF4 at both 6 and 24 hr (Figure S6C). By contrast, expression of WT KLF4-GFP or the NLS, NES3, and NES4 mutants, all of which exit the nucleus, or in the case of the NLS mutant are resident in the cytoplasm, did not prevent the decline in transcript levels for Nanog and endogenous Klf4.
Next we investigated the effect of blocking KLF4 nuclear export on ESC differentiation. We observed that alkaline phosphatase (AP) activity was maintained 5 days after withdrawal of LIF/2i in cells expressing KLF4(S132A)-GFP but not WT KLF4-GFP (Figure S6D). To evaluate the effect of blocking KLF4 nuclear export on the specification of germ layers, we differentiated ESCs to embryoid bodies (EBs) (Figure 6C). qRT-PCR revealed that expression of the pluripotency markers Nanog, Klf4, and Sox2 was maintained in KLF4(S132A)-GFP EBs for 5 days, whereas WT KLF4-GFP EBs displayed a reduction in the expression of these genes similar to the pattern displayed by untransfected ESCs. In addition, the onset of expression for all germ layer markers was delayed by 4–5 days in KLF4(S132A)-GFP EBs compared with WT KLF4-GFP and untransfected embryonic day 14 (E14) ESCs. Together these data indicate that inhibiting KLF4 nuclear export in ESCs maintains naive pluripotency in the absence of external signals (LIF/2i) and delays differentiation to endoderm, mesoderm, and ectoderm by about 5 days.
MEK-ERK Signaling during Embryogenesis Downregulates KLF4 and NANOG
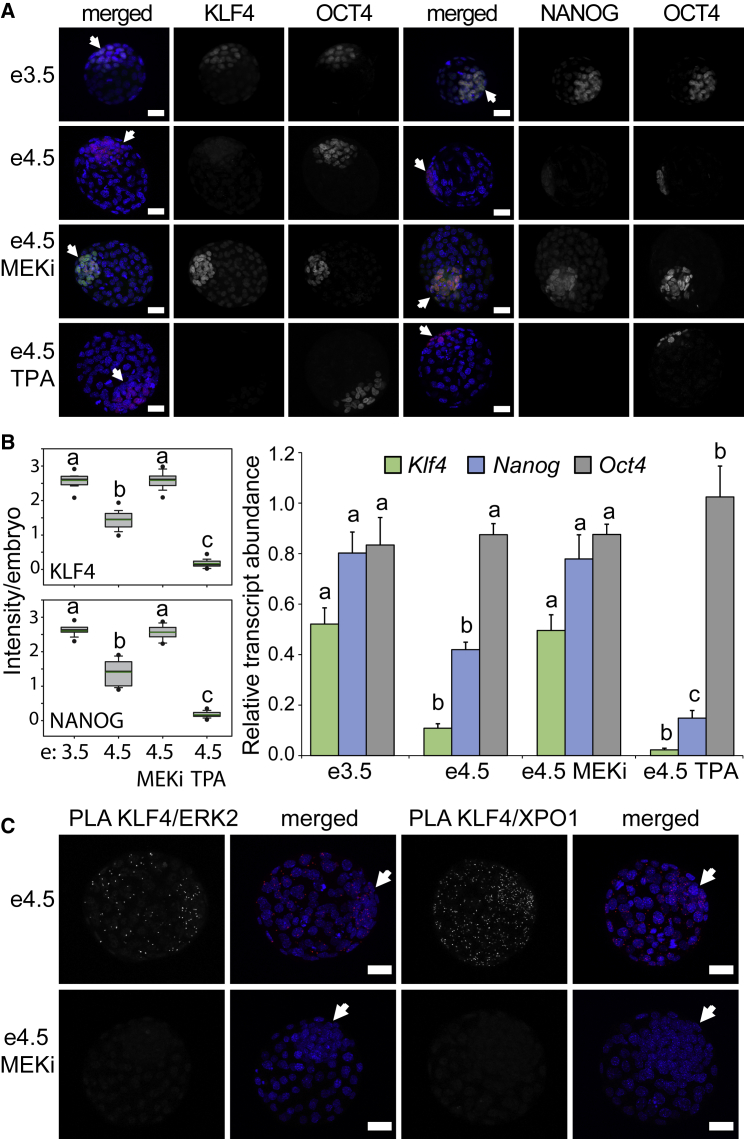
In the embryo, FGF-MEK-ERK signaling occurs between E3.25 and E4.5 and is required for embryo development (Yamanaka et al., 2010). To investigate the effect of MEK-ERK signaling on KLF4 and NANOG expression, we monitored protein and mRNA levels in mouse embryos (Figure 7A). In E3.5 blastocysts, immunofluorescence for KLF4 and NANOG revealed more intense signal in the OCT4-positive ICM (arrows). To evaluate changes in protein levels, we quantified images from at least 30 embryos in each group (Figure 7B). At E4.5, KLF4 and NANOG protein levels were both significantly reduced compared with E3.5 levels, and this reduction was inhibited by treatment with the MEK inhibitor. TPA treatment, by contrast, caused a further reduction in the levels of both KLF4 and NANOG protein to nearly undetectable levels. Similar to the changes in protein levels, mRNA for both Klf4 and Nanog was reduced at E4.5 compared with E3.5, and this reduction was prevented by treatment with the MEK inhibitor (Figure 7B). No decrease in OCT4 protein or mRNA levels was detected in these experiments (Figures 7 and S7A).
Figure 7.
KLF4 Downregulation in the Inner Cell Mass of Mouse Blastocysts Depends on MEK Activation
(A) Immunofluorescence images of mouse blastocysts at E3.5 and E4.5 maintained for 24 hr in control conditions, MEKi (MEK1 inhibitor, PD0325901) treated, or TPA treated. Blastocysts at E3.5 display nuclear KLF4 and NANOG in the OCT4-positive ICM (arrows). Blastocysts at E4.5 display more diffuse and lower KLF4 and NANOG levels in the ICM. Blastocysts at E4.5 maintained for 24 hr in MEKi display higher KLF4 and NANOG expression in the ICM. Blastocysts at E4.5 treated with TPA display low levels of KLF4 and NANOG, whereas OCT4 levels remain high. Images shown are maximum-intensity projections. Merged images display DAPI in blue, KLF4 or NANOG in green, and OCT4 in red. Scale bars, 25 μm.
(B) Quantification of immunofluorescence images from ≥30 embryos in each group reveal significant changes in KLF4 and NANOG protein levels (left). qRT-PCR from three separate pools of embryos at each stage reveals significant differences in Klf4 and Nanog mRNA levels (right). Error bars represent standard deviation. Statistical differences (p < 0.05) between different treatments for each gene are indicated by different letters.
(C) Proximity ligation amplification (PLA) for KLF4/ERK2 and KLF4/XPO1 displays the interaction between KLF4, ERK2 and XPO1 in E4.5 blastocysts maintained in control conditions but not in blastocysts pre-treated with MEKi for 24 hr. The interactions were observed in both the ICM (arrows) and trophectoderm. Images shown are maximum-intensity projections. Merged images display DAPI in blue and PLA signal in red. Scale bars, 25 μm.
See also Figure S7.
As treatment with the MEK inhibitor blocked the downregulation of KLF4, we investigated the interaction of KLF4 with ERK2 or XPO1 in mouse blastocysts using the PLA assay. XPO1 and ERK2 are present throughout the embryo as confirmed by immunofluorescence (Figure S7B). The localization of XPO1 was not affected by the MEK inhibitor, whereas ERK2 shifted from nuclear localization under control conditions to a cytoplasmic localization for embryos maintained in the MEK inhibitor, consistent with a shift to inactive ERK2 under these conditions. Similar to our observations in cultured ESCs, PLA for KLF4/ERK2 or KLF4/XPO1 revealed that interaction between these proteins in blastocysts requires MEK-ERK signaling (Figure 7C). Interestingly we found that the signal was not restricted to the ICM but occurred throughout the blastocyst, indicating that active ERK2, KLF4, and XPO1 interact in the trophectoderm as well as the ICM. Although MEK inhibition does not cause as dramatic an increase in the levels of KLF4 in the trophectoderm cells as observed in the ICM, a notable increase was observed compared with untreated E4.5 embryos (Figure 7A). We did note that in some early blastocysts (E3), KLF4 protein was localized in all nuclei including the NANOG-positive ICM (arrow) and the NANOG-negative trophectoderm (asterisk in Figure S7C), suggesting that KLF4 is initially expressed in most cells and later downregulated in the trophectoderm. Considered together, these findings indicate that, as observed in ESCs, ERK activation is required for the reduction in KLF4 nuclear protein levels in the embryo, which involves XPO1-mediated nuclear export. Furthermore, this drop in KLF4 protein is coincident with a reduction in Klf4 and Nanog transcript levels as well as NANOG protein levels.
Discussion
Although differentiation of ESCs occurs over several days, we identified significant changes in nuclear environment occur within 6 hr of differentiation, primarily KLF4 nuclear export, which we show initiates exit from the naive pluripotent state. The pluripotent state is regulated by an interconnected network of transcription factors of which OCT4, SOX2, NANOG, and KLF4 play a central role in pluripotency maintenance through binding to many of the same enhancer regions to regulate gene expression (Chen et al., 2008, Chen et al., 2012, Zhou et al., 2014). Our study reveals that KLF4 followed by NANOG are the first proteins of this network to be removed from nuclear transcription complexes and that this is initiated by export of KLF4 from the nucleus, mediated by XPO1 in response to ERK-mediated phosphorylation of KLF4. Interaction between KLF4, ERK2, and XPO1 also occurs in pre-implantation blastocysts leading to decreased KLF4 and NANOG, and blocking this process during EB formation delays exit from naive pluripotency by 5 days. These findings reveal that XPO1-mediated KLF4 nuclear export initiates exit from the naive pluripotent state both in vitro and in the pre-implantation embryo.
KLF4 and NANOG display reduced occupancy in nuclear protein complexes associated with transcription within 24 hr of LIF/2i removal. In undifferentiated ESCs, NANOG functions as part of a complex with OCT4 and SOX2, and together these three proteins bind the same DNA consensus motif (Chen et al., 2008, Gao et al., 2012). KLF4 also binds many of the same enhancer regions and activates similar genes, although it binds a separate DNA consensus motif (Chen et al., 2008, Moorthy et al., 2017). It has been proposed that pluripotency is maintained by the precise balance of the pluripotency transcription factors (Loh and Lim, 2011); our data indicate that this balance is first disrupted by export of KLF4 from the nucleus, followed by reduced levels of nuclear NANOG, causing the removal of these two transcription factors from nuclear complexes.
Downregulation of Klf4 has been shown to precede Nanog downregulation and is the first observed change in expression of pluripotency-associated genes (Zhang et al., 2010); however, when investigating KLF4 protein we observed that KLF4 is exported from the nucleus concurrently with the downregulation in Klf4 and Nanog transcription and that blocking KLF4 nuclear export blocks this downregulation. KLF4 and NANOG bind to enhancer regions upstream of the Nanog promoter and have been shown to be required for Nanog transcription (Sanchez-Castillo et al., 2014, Zhang et al., 2010). Similarly, there are a number of binding sites for KLF4 and NANOG at enhancers downstream of Klf4, required for Klf4 transcription (Pradeepa et al., 2016, Sanchez-Castillo et al., 2014). After 24 hr of differentiation, we observed accumulation of KLF4 in the nucleus at half of the levels observed in undifferentiated ESCs; however, Klf4 and Nanog transcript levels were not found to increase (Figure S1A). The inability of nuclear KLF4 at 24 hr to induce de novo transcription of Klf4 and Nanog could be due to the reduced interaction of KLF4 with other transcriptional complexes (RNAPII and OCT4/SOX2) and due to reduced NANOG in the nucleus at 24 hr, both of which may prevent cooperative action of the complex and subsequent activation of Klf4 and Nanog transcription.
XPO1 is often involved in constitutive nuclear export to maintain the cytoplasmic accumulation of proteins, which can enter the nucleus constitutively. In an ESC-specific context, ERK2 interacts with XPO1 irrespective of its activation state, consistent with the idea that continual export of the inactive MEK/ERK complex maintains the cytoplasmic levels of these proteins. KLF4, by contrast, only interacts with XPO1 after ERK activation occurs. Furthermore, we showed that S132, the ERK phosphorylation site, as well as two predicted NES are required for this interaction and nuclear export, indicating that it is the S132 phosphorylated form of KLF4 that interacts with XPO1. In support of this, disruption of S132, either by direct mutation or mutation of NES2 that prevented phosphorylation of S132, disrupted the interaction of KLF4 with XPO1. NES1 was also required for the interaction of KLF4 and XPO1; however, mutation of this site did not affect KLF4 serine phosphorylation, suggesting that NES1 is the functional NES that supports the direct interaction of KLF4 and XPO1, whereas mutation of NES2 indirectly affects the interaction of KLF4 and XPO1 by preventing KLF4 phosphorylation at S132. These findings indicate that in the absence of S132 phosphorylation, NES1 is masked and not available for interaction with XPO1.
Differentiation impairment observed in ESCs expressing S132A mutant KLF4 revealed that nuclear export of KLF4 is critical for exit from the pluripotent state and subsequent differentiation of ESCs to specific germ layers. Our findings are different from those of Zhang et al. (2010) who found that overexpression of WT KLF4 prevents differentiation; this difference may be due to expression levels, as in our cells KLF4-GFP was expressed at levels comparable with endogenous KLF4 and did not prevent differentiation. In mouse pre-implantation blastocysts, MEK-ERK signaling downregulates KLF4 and NANOG, similar to our observations in ESCs; this mechanism involves the interaction between KLF4/ERK2 and KLF4/XPO1. Although Klf4 is not required for early embryogenesis in the mouse, as embryos lacking Klf4 survive to birth due to compensatory mechanisms involving other Klf factors, our in vitro differentiation experiments revealed that expression of constitutively nuclear KLF4 in ESCs prevents pluripotency exit and differentiation. This suggests that a failure in KLF4 downregulation could result in phenotypic consequences during early development. Our finding that KLF4 is the first of the pluripotency factors to be removed from transcription-associated nuclear complexes, through active ERK- and XPO1-dependent nuclear export, indicates KLF4 nuclear export as a critical first step in exiting the naive pluripotent state and committing to differentiation.
Experimental Procedures
Embryonic Stem Cell Culture
Mouse ESCs (E14TG2a; ATCC [CRL-1821]), were maintained in feeder-free conditions on 0.1% gelatin in DMEM supplemented with 15% (v/v) fetal bovine serum (FBS), 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 2 mM GlutaMAX, 0.1 mM 2-mercaptoethanol, 1,000 U/mL LIF, 3 μM CHIR99021 (GSK3β inhibitor, Biovision) and 1 μM PD0325901 (MEK inhibitor, Invivogen); referred to as LIF/2i medium. The differentiation medium contained the same components with the exception of LIF/2i. For FGFR inhibition, the MEK inhibitor was replaced with 25 nM PD173074 (Selleckchem). For TPA (NEB) treatment, cells were cultured overnight in low serum (0.2%) ESC medium, followed by treatment with 200 nM TPA in the absence of LIF/2i. For ESCs maintained in LIF alone, cells were cultured in ESC medium containing LIF without 2i for 8–10 passages before removal of LIF to induce differentiation. For evaluation of AP activity, cells were seeded at 250 cells/well in a 12-well plate and allowed to differentiate for 5 days. AP staining (Millipore) was performed following the manufacturer’s instructions. EBs were formed by the hanging-drop method; 1,000 cells were suspended in 20-μL droplets of ESC medium without LIF/2i (Ohnuki and Kurosawa, 2013). EBs were transferred to 0.1% gelatin on day 4, and collected until day 12; RNA was isolated for gene expression analysis by qRT-PCR.
Immunofluorescence and Proximity Ligation Amplification
Cells were fixed for 20 min in neutral buffered 10% formalin, blocked, and permeabilized for 30 min with 10% FBS in 0.1% Triton X-100 in PBS, both at room temperature. Cells were incubated sequentially with primary antibodies in antibody buffer (0.2% FBS, 0.1% Triton X-100 in PBS). After three PBST (0.1% Tween 20 in PBS) washes, cells were incubated in secondary antibodies. Cells were counterstained in DAPI, then washed twice with PBS and once in distilled water. Coverslips were mounted onto glass microscope slides using Vectashield (Vector Labs). Images were collected using a 100× magnification objective lens and analyzed to determine the intensity per nucleus (area defined by the DAPI counterstain) using VOLOCITY 6.0.1. A t test was performed to analyze significant differences in the mean intensity data of differentiating ESCs as compared with undifferentiated ESCs (0 hr). Controls using no primary antibody were conducted for all secondary antibodies, which revealed that there was no non-specific binding of the secondary antibodies. All immunofluorescence experiments were carried out on at least three biological replicate samples. Antibodies used are listed in Table S1.
PLA was conducted using Duolink (Sigma-Aldrich) following the manufacturer's instructions. Images were collected using a Leica TCS SP8 and a 63× magnification objective lens. The number of PLA foci per nucleus was quantified using Imaris 7.1 by manual 3D masking of nuclei in ESC colonies defined by the DAPI signal. All PLA experiments were carried out on at least three biological replicate samples. A t test was performed to evaluate significant differences in foci number per nucleus.
Immunoblot and Co-Immunoprecipitation
Cells were separated into nuclear and cytoplasmic fractions as previously described (Dhaliwal and Mitchell, 2016, Mitchell et al., 2012). Protein was extracted using RIPA buffer containing protease inhibitor complete EDTA free (Roche) and phosphatase inhibitor cocktail (Millipore) and quantified using bicinchoninic acid (Thermo Fisher Scientific). Protein samples were analyzed by SDS-PAGE (15% Bis-Tris resolved with a 5% stacking gel). Blots were incubated with primary antibodies (Table S1) followed by horseradish peroxidase-conjugated secondary antibodies. Blots were quantified by relative intensity using background correction from adjacent regions. At least three biological replicates were analyzed for each experiment, and statistical differences were determined by t test.
For immunoprecipitation of protein, fractions or total lysates in RIPA were incubated overnight with the appropriate antibody and then incubated overnight with a 50:50 mixture of protein A and protein G Dynabeads. Beads were washed three times with non-denaturing lysis buffer (20 mM Tris-HCl, 137 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA, 1 mM PMSF, and proteinase inhibitors), twice with PBS, eluted in loading buffer, and analyzed by SDS-PAGE. To immunoprecipitate KLF4-GFP, we used GFP-Trap (Chromotek) according to the manufacturer's protocol.
Real-Time qPCR
Total RNA was purified using TRIzol (Thermo Fisher Scientific). Following a DNaseI digestion to remove DNA, total RNA was reverse transcribed with random primers using the iScript cDNA synthesis kit (Bio-Rad). Gene expression was monitored by qPCR using genomic DNA to generate standard curves. Gapdh expression was used to normalize expression values. Three biological replicates were analyzed for each experiment, and significant differences in expression were determined by t test. Primers used are listed in Table S2.
Expression of KLF4 Mutants
The nuclear localization signal (NLS) was predicted using NLStradamus (Nguyen Ba et al., 2009) and NES were predicted using Wregex (Prieto et al., 2014). A mouse KLF4-GFP vector (RG206691) was obtained from Origene and subjected to site-directed mutagenesis (SDM) (QuikChange Lightning, Agilent Technologies). Primers for SDM are indicated in Table S3. Sequence-confirmed plasmids were transfected by electroporation and cells were selected with 400 μg/mL G418. The cells were sorted by fluorescence-activated cell sorting, and individual clones selected and maintained in 50 μg/mL G418 to obtain KLF4-GFP-positive clones. Expression of each KLF4-GFP construct was confirmed by immunoblot (Figure S5E).
Blastocyst Collection, Immunostaining, and Gene Expression
CD-1 females 6–8 weeks old were injected intraperitoneally with 5 IU of pregnant mare serum gonadotropin (Prospec, HOR-272) at 4:30 pm followed by 5 IU of human chorionic gonadotropin (Prospec, HOR-250) 46 hr later and mated with CD-1 males. Females were euthanized by cervical dislocation and day 3–3.5 embryos were flushed out in M2 medium (Millipore, MR-015-D). Embryos were either subsequently fixed or cultured in a humidified incubator at 37°C with 5% CO2 in potassium-supplemented simplex optimized medium (KSOM) (Millipore, MR-106-D), KSOM with 0.5 μM MEK inhibitor (PD0325901, Invivogen) for 24 hr, or KSOM for 24 hr followed by 25 min with 200 nm TPA.
Following fixation in neutral buffered 10% formalin (Sigma-Aldrich) for 15 min, blastocysts were rinsed in PBS and permeabilized in 0.3% Triton X-100 for 15 min. Blocking was done in 10% goat serum and 0.1% Triton X-100 in PBS for 1 hr at room temperature. Antibodies (Table S1) were diluted in 5% goat serum and 0.1% Triton X-100 in PBS. Blastocysts were washed in PBS/0.05% Tween 20. Counterstaining was done with 0.1 μg/mL DAPI in PBS and embryos were mounted in 90% glycerol. All PLA experiments were carried out on at least ten embryos. PLA-positive control was detection of the core RNAPII RPB1 together with detection of the S5P CTD of RPB1; the negative control was the KLF4 primary antibody alone (Figure S7B). All immunofluorescence experiments were conducted on at least 30 embryos. Images were collected with a Leica TCS SP8 at 40×. The immunofluorescence images were analyzed for average staining intensities of KLF4, OCT4, and NANOG using IMARIS.
For gene expression analysis, ≥15 embryos were pooled for each condition. The cDNA was made using a SuperScript III CellsDirect cDNA Synthesis Kit (Thermo Fisher) and gene expression was monitored by qPCR with genomic DNA used to generate standard curves. Gapdh expression was used to normalize expression values. Three biological replicates were analyzed for each experiment, and significant differences in expression were determined by ANOVA. Primers used are listed in Table S2.
All animal experiments were approved by the University Animal Care Committee (UACC) at the University of Toronto and the Bioscience Local Animal Care Committee (LACC).
Author Contributions
J.A.M. and N.K.D. conceived and designed the experiments. N.K.D. performed all experiments with the following assistance: K.M. collaborated on the embryo experiments, and N.K.D., S.D., and H.T.E.J. generated KLF4-GFP mutant constructs. N.K.D. and J.A.M. analyzed the data and wrote the manuscript, which was approved by all co-authors.
Acknowledgments
We would like to thank all the members of the Mitchell lab for helpful discussions. This work was supported by the Canadian Institutes of Health Research (FRN 111214 and 153186), the Canada Foundation for Innovation, and the Ontario Ministry of Research and Innovation (operating and infrastructure grants held by J.A.M.). Studentship funding was provided by Ontario Graduate Scholarships held by N.K.D.
Published: March 8, 2018
Footnotes
Supplemental Information includes seven figures and three tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.02.007.
Supplemental Information
References
- Adachi M., Fukuda M., Nishida E. Nuclear export of MAP kinase (ERK) involves a MAP kinase kinase (MEK)-dependent active transport mechanism. J. Cell Biol. 2000;148:849–856. doi: 10.1083/jcb.148.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourillot P.Y., Aksoy I., Schreiber V., Wianny F., Schulz H., Hummel O., Hubner N., Savatier P. Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells. 2009;27:1760–1771. doi: 10.1002/stem.110. [DOI] [PubMed] [Google Scholar]
- Bruce S.J., Gardiner B.B., Burke L.J., Gongora M.M., Grimmond S.M., Perkins A.C. Dynamic transcription programs during ES cell differentiation towards mesoderm in serum versus serum-freeBMP4 culture. BMC Genomics. 2007;8:365. doi: 10.1186/1471-2164-8-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Morris Q., Mitchell J.A. Enhancer identification in mouse embryonic stem cells using integrative modeling of chromatin and genomic features. BMC Genomics. 2012;13:152. doi: 10.1186/1471-2164-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Dhaliwal N.K., Mitchell J.A. Nuclear RNA isolation and sequencing. Methods Mol. Biol. 2016;1402:63–71. doi: 10.1007/978-1-4939-3378-5_7. [DOI] [PubMed] [Google Scholar]
- Doble B.W., Patel S., Wood G.A., Kockeritz L.K., Woodgett J.R. Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev. Cell. 2007;12:957–971. doi: 10.1016/j.devcel.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gustafsdottir S.M., Ostman A., Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- Gao Z., Cox J.L., Gilmore J.M., Ormsbee B.D., Mallanna S.K., Washburn M.P., Rizzino A. Determination of protein interactome of transcription factor Sox2 in embryonic stem cells engineered for inducible expression of four reprogramming factors. J. Biol. Chem. 2012;287:11384–11397. doi: 10.1074/jbc.M111.320143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Harvey A.J., Armant D.R., Bavister B.D., Nichols S.M., Brenner C.A. Inner cell mass localization of NANOG precedes OCT3/4 in rhesus monkey blastocysts. Stem Cells Dev. 2009;18:1451–1458. doi: 10.1089/scd.2009.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Chan Y.S., Loh Y.H., Cai J., Tong G.Q., Lim C.A., Robson P., Zhong S., Ng H.H. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Kim M.O., Kim S.H., Cho Y.Y., Nadas J., Jeong C.H., Yao K., Kim D.J., Yu D.H., Keum Y.S., Lee K.Y. ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4. Nat. Struct. Mol. Biol. 2012;19:283–290. doi: 10.1038/nsmb.2217. [DOI] [PubMed] [Google Scholar]
- Kunath T., Saba-El-Leil M.K., Almousailleakh M., Wray J., Meloche S., Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- Loh K.M., Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell. 2011;8:363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Mitchell J.A., Clay I., Umlauf D., Chen C.Y., Moir C.A., Eskiw C.H., Schoenfelder S., Chakalova L., Nagano T., Fraser P. Nuclear RNA sequencing of the mouse erythroid cell transcriptome. PLoS One. 2012;7:e49274. doi: 10.1371/journal.pone.0049274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy S.D., Davidson S., Shchuka V.M., Singh G., Malek-Gilani N., Langroudi L., Martchenko A., So V., Macpherson N.N., Mitchell J.A. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 2017;27:246–258. doi: 10.1101/gr.210930.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba A.N., Pogoutse A., Provart N., Moses A.M. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Niwa H., Ogawa K., Shimosato D., Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Ohnuki Y., Kurosawa H. Effects of hanging drop culture conditions on embryoid body formation and neuronal cell differentiation using mouse embryonic stem cells: optimization of culture conditions for the formation of well-controlled embryoid bodies. J. Biosci. Bioeng. 2013;115:571–574. doi: 10.1016/j.jbiosc.2012.11.016. [DOI] [PubMed] [Google Scholar]
- Pradeepa M.M., Grimes G.R., Kumar Y., Olley G., Taylor G.C., Schneider R., Bickmore W.A. Histone H3 globular domain acetylation identifies a new class of enhancers. Nat. Genet. 2016;48:681–686. doi: 10.1038/ng.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto G., Fullaondo A., Rodriguez J.A. Prediction of nuclear export signals using weighted regular expressions (Wregex) Bioinformatics. 2014;30:1220–1227. doi: 10.1093/bioinformatics/btu016. [DOI] [PubMed] [Google Scholar]
- Sanchez-Castillo M., Ruau D., Wilkinson A.C., Ng F.S., Hannah R., Diamanti E., Lombard P., Wilson N.K., Gottgens B. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 2014;43:D1117–D1123. doi: 10.1093/nar/gku895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre J.A., Bauer C., Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Theunissen T.W., van Oosten A.L., Castelo-Branco G., Hall J., Smith A., Silva J.C. Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr. Biol. 2011;21:65–71. doi: 10.1016/j.cub.2010.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Yang Y., Zhang P., Andrianakos R., Hasegawa K., Lyu J., Chen X., Bai G., Liu C., Pera M. Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem Cells. 2009;27:2969–2978. doi: 10.1002/stem.231. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y., Lanner F., Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Andrianakos R., Yang Y., Liu C., Lu W. Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J. Biol. Chem. 2010;285:9180–9189. doi: 10.1074/jbc.M109.077958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.Y., Katsman Y., Dhaliwal N.K., Davidson S., Macpherson N.N., Sakthidevi M., Collura F., Mitchell J.A. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 2014;28:2699–2711. doi: 10.1101/gad.248526.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.