Abstract
Background/Aim: In this study, we determined the expression of selected circulating microRNAs (miRNA) and their potential roles as biomarkers in patients with atherosclerosis and a control group. Materials and Methods: In order to obtain insight into miRNA expression levels in atherosclerosis, we analyzed miRNA expression levels by real-time polymerase chain reaction (RT-PCR) in case (n=89) and healthy control (n=93) groups. Receiver operating characteristic curve analysis was performed to assess the diagnostic capability of miRNAs. Results: miRNA221 and miRNA221 expression levels were significantly lower in patients than controls (p=0.011 and p=0.004, respectively). Receiver operator curve analysis demonstrated that expression levels of miRNA221 [area under curve (AUC)=0.623, p=0.0086) and miRNA222 (AUC=0.654, p=0.0006) were significantly different between groups. There were positive correlations between miRNA122a and triglyceride (p=0.046) and very-low-density lipoprotein (p=0.029) levels. Conclusion: miRNA221 and miRNA222 could be convenient biomarkers for diagnosis of atherosclerosis.
Keywords: Atherosclerosis, biomarker, miRNA221, miRNA222, miRNA122a
Atherosclerosis is the predominant cause of mortality and morbidity in developed countries despite therapeutic improvements in interventional surgery and pharmacologic treatment strategies (1). According to World Health Organization (WHO) reports, 17.7 million people die each year because of cardiovascular diseases (CVD) (2). Thus, new strategies are needed for understanding underlying mechanisms of atherosclerosis to discover new therapeutic strategies and novel diagnostic biomarkers.
MicroRNAs (miRNAs) are endogenous, non-coding RNAs of 18-22 nucleotides in length that negatively regulate gene expression by interfering transcription or inhibit translation at the post-transcriptional level (3). It has been shown that 30% of protein-coding genes are controlled by miRNAs (4). Owing to their stable characteristics, miRNAs have emerged as potential biomarkers. Furthermore, miRNAs can easily be obtained from body fluids by non-invasive methods (5). They play crucial roles in diverse physiological and pathological cell process including proliferation, cell differentiation, migration, and apoptosis, as well as angiogenesis, cardiogenesis, endothelial and myocyte growth and lipoprotein metabolism (6,7). Although many studies showed relationships between miRNA processes and atherosclerosis, effective diagnostic therapeutic alternatives have not been validated yet.
In this study, we aimed to investigate the relationships between miRNA expressions and their potential therapeutic role as biomarkers in atherosclerosis.
Materials and Methods
Study population. Patient and control groups were selected after detailed clinical examinations at the Cardiovascular Surgery Departments of Marmara University and Derince Training and Research Hospital. The control group (n=93) consisted of individuals who presented for check-ups while the patients with atherosclerosis (n=89), defined as having coronary artery disease (CAD) were documented by angiography. Angiographic inclusion criteria were: ≥50% stenosis of at least one major coronary vessel because of atherosclerosis. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and the study protocol was approved by the Yeditepe University Medical Faculty Ethics Committee (file no: 03.06.2017/428) on research on humans. After obtaining informed consent of each participant, blood samples were collected in EDTA-containing tubes.
Sample preparation. Serum was obtained from whole blood samples which had been collected in 5 ml plain serum separator tubes. Serum samples were centrifuged at 1500 × g for 10 min then the specimen was decanted into a new sterile tube and frozen at −80˚C until miRNA assessments were conducted.
miRNA isolation from 200 μl serum was performed using a miRNeasy Serum/Plasma Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The optical density of miRNA isolated from samples was measured with NanoDrop2000 (Thermoscientific, Waltham, MA, USA). Reverse transcription from isolated miRNA samples to cDNA was performed with miScript II RT Kit (Qiagen). Transcripted miRNA concentration was evaluated and equalized using Qubit miRNA Assay Kit standard protocol on a Qubit 3.0 Fluorometer (Thermoscientific).
miRNA selection. Bioinformatics analysis of data compiled from several medical databases, including miRBase, TargetScanHuman, microrna.org and Embase, was used for selection of the miRNAs. The examined miRNAs (miRNA221, miRNA222, miRNA143, miRNA145, miRNA133a, miRNA 133b, miRNA 122a, miRNA 494, miRNA 499a) in this study were chosen to provide a comprehensive approach for atherosclerosis.
Real-time polymerase chain reaction (PCR) analysis of miRNA. Nine different custom primers for miRNA221-3p, miRNA222-3p, miRNA143-3p, miRNA145-3p, miRNA133a-3p, miRNA 133b-3p, miRNA 122a-3p, miRNA 494-3p and miRNA 499a-5p, which were embedded in PCR plate, were used to demonstrate miRNA expression levels (Qiagen). Three housekeeping assay genes, namely mirRNAU6, mirSNORD61, and SNORD68, were used for determine delta cycle threshold (ΔCt) values of each primers (Qiagen). Ct values were obtained by miScript SYBR Green PCR Kit (Qiagen) on an Applied Biosystems 7500 Fast Real-Time PCR instrument (Applied Biosystems, Foster City, CA, USA). The miRNA expression levels were calculated with Livak formula (2−ΔΔCт) (8). ΔCt, (2−ΔΔCt) and normalized expression levels [logn[2(−ΔΔCт)] were calculated using Microsoft Excel (Microsoft Corporation, Redmond, MA, USA).
Statistical analysis. Values are expressed as the mean±standard deviation (SD). Chi square and Fisher’s exact tests were used to compare demographic information. Student’s t-test was used to examine the significance of differences between the case and control groups. miRNA expression levels were analyzed by Student’s t-test. Correlations were determined using Pearson correlation test, and univariate linear regression analysis methods. Statistical analysis was performed using SPSS Ver. 23 software (IBMCorp., Armonk, NY, USA). The diagnostic value of circulating miRNAs determined by receiver operator curve (ROC) analysis using MedCalc version 15.0 (MedCalc Software, Ostend, Belgium) was used for ROC analysis with 95% confidence interval. Reported p-values are two sided and a significance level of p<0.05 was considered to indicate statistical significance.
Results
We determined the expression levels of circulating miRNAs in 89 patients with atherosclerosis and 93 healthy controls without evidence of CAD. The mean age of the patients and healthy controls was 58.97±13.79 and 57.07±9.80 years, respectively (p=0.08). There were no significant differences between two groups in terms of body mass index, body surface area, plasma cholesterol, triglyceride (TG), low-density lipoprotein and very low-density lipoprotein (VLDL). Regarding clinical risk factors, there were differences were observed in family history, diabetes and hypertension and high-density lipoprotein levels.
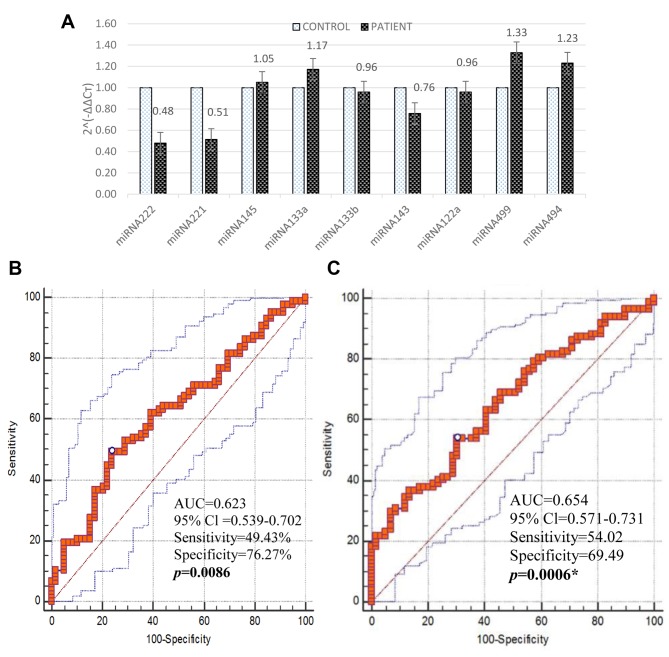
It was found that miRNA221, miRNA222, miRNA133b, miRNA143 and miRNA122a were down-regulated, whereas miRNA145, miRNA133a, miRNA494 and miRNA499a were up-regulated (Figure 1A). Logistic regression analysis of circulating miRNA expression levels in patients with atherosclerosis and control group are given in Table I. From the risk estimate analysis, altered levels of miRNA221 (p=0.011) and miRNA222 (p=0.004) were statistically significant.
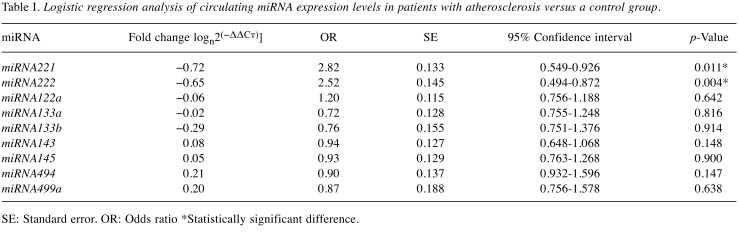
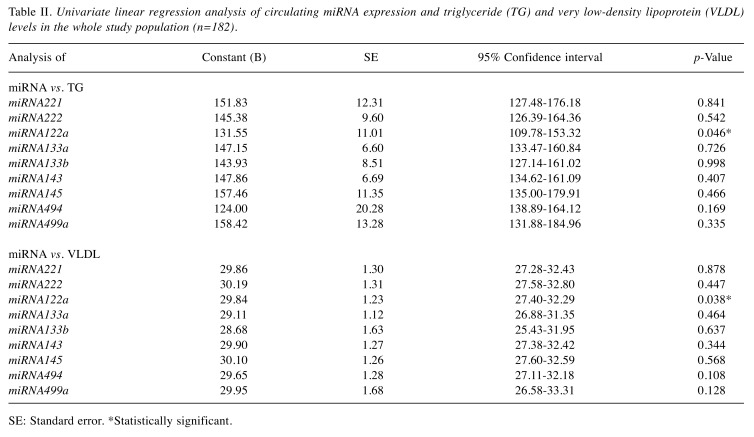
Figure 1. The expression of circulating miRNAs in patients with atherosclerosis. B, C: Receiver operator characteristic curves for linear regression of expression levels of circulating miRNA221 (B) and miRNA222 (C).
Table I. Logistic regression analysis of circulating miRNA expression levels in patients with atherosclerosis versus a control group.
SE: Standard error. OR: Odds ratio *Statistically significant difference.
Although there were statistically significant differences for only miRNA221 and miRNA222 expression levels, all miRNAs underwent ROC analysis to reveal results in detail. Area under the curve (AUC) results indicated that miRNA221 and miRNA222 not surprisingly achieved highest ROC values. miRNA221 reached a specificity of 49.43% and a sensitivity of 76.27% (AUC=0.623, p=0.0086) and miRNA222 had a specificity of 54.02% and a sensitivity of 69.49% (AUC=0.654, p=0.0006) (Figure 1B and C). Our results indicate that miRNA221 and miRNA222 could be eligible candidate biomarkers for diagnosis of atherosclerosis.
Determining lipid profiles and miRNA expression levels indicated that there was positive correlation between miRNA122a and TG (p=0.046) and VLDL (p=0.029) levels. Owing to limited study populations, these analyses were performed in all participants (n=182). Univariate linear regression analysis of miRNA expression and TG and VLDL levels are shown in Table II.
Table II. Univariate linear regression analysis of circulating miRNA expression and triglyceride (TG) and very low-density lipoprotein (VLDL) levels in the whole study population (n=182).
SE: Standard error. *Statistically significant.
Discussion
miRNAs are thought to be novel negative gene regulators having crucial roles in biological and pathological processes such as proliferation, migration, oxidative stress and apoptosis. miRNAs show their effects not only by mRNA degradation but also translational repression of proteins. It has been shown that miRNAs participate in pathological processes including cancer, diabetes and obesity, as well as atherosclerosis (9). Circulating miRNAs have great interest as potential clinical biomarkers due to their highly stable and accessible features. Furthermore, miRNAs can easily be sampled from body fluids such as blood, saliva, urine with non-invasive methods (10).
There are several animal and in vitro studies regarding miRNAs as biomarkers in atherosclerosis (11), although few human data have been provided so far. The present study demonstrated that miRNA221 and miRNA222 expression levels were significantly lower in patients with atherosclerosis, and therefore could be eligible risk factors for atherosclerosis.
In the past decades, a great number of miRNAs have been recognized for their toll pathological processes of atherosclerosis. Because of the important role of vascular endothelial cells (VECs) on vascular system homoeostasis as regulators of proliferation, angiogenesis, platelet aggregation, cell adhesion, specific class of miRNAs were identified as affecting physiological regulation of VECs (11). miRNA221 and miRNA222 are members of same miRNA family, named antiangiogenic gene regulators, and are highly expressed in VECs. Decreased expression levels of miRNA221 and miRNA222 in serum samples demonstrated that limited cell proliferation had a crucial role in epithelial dysfunction in atherosclerosis. miRNA221 and miRNA222 modulate angiogenic activity of VECs via controlling proliferation factors such as human proto-oncogen c-KIT and receptor for stem cell factor (12). It has also been shown that miRNA221 and miRNA222 regulate c-KIT protein expressions at the post-transcriptional level without mRNA degradation, thus they can modulate proliferation of vascular cells (13). Liu et al. demonstrated interesting data regarding miRNA221 and miRNA222, they showed that these miRNAs have cell-specific functions. miRNA221 and miRNA222 have inverse effects on VECs and vascular smooth muscle cells (VSMCs). Gene expression of proto-oncogene c-KIT was very high in VECs while it was significantly lower in VSMCs. Therefore, altered miRNA221 and miRNA222 expression levels appear to have opposing effects on VSMCs and ECs (14). miRNA221 and miRNA222 have regulatory roles in VSMC proliferation in atherosclerosis (15). Zhang et al. demonstrated that patients with atherosclerosis had significantly lower miRNA221 and miRNA222 expression levels than healthy controls. They showed that down-regulated miRNA221 and miRNA222 expressions had crucial roles in the atherosclerotic process and suggested that these two miRNA could be potential candidate biomarkers for CAD (16).
The advantages presented by miRNAs show they are not only potential and useful biomarkers but also useful therapeutic targets. miRNA122a constitutes 70% of all miRNAs expressed in adult liver cells, and is therefore described as a mammalian liver-specific microRNA (17). Esau et al. demonstrated that miRNA122 plays important roles on lipid regulation by targeting six different mRNA which have direct or indirect effects on lipid metabolism. It has been shown that cholesterol synthesis rates decreased significantly in mice when miRNA122 was inhibited by antisense oligonucleotides. miRNA122 interference resulted in reduction of plasma cholesterol levels and hepatic fatty acid synthesis, while it increased in hepatic fatty-acid oxidation. Their results showed that miRNA122 could be a therapeutic target for lipid homoeostasis (18). Iliopoulos et al. reported that inhibition of miRNA122 had similar effects on lipid regulation in human hepatocyte-derived cell lines. Their data revealed an association between miRNA122 and lipogenic genes, up-regulation of miRNA122 inhibited gene regulation that cause TG accumulation in liver by increased lipogenesis and suppressed β-oxidation. Moreover, reduced expression levels of miRNA122 caused decrements in sterol regulatory element-binding protein 1c (SREBF-1c) gene, which regulates LDL receptor and cholesterol pathways in cell lines (19). In the current study, we demonstrated that miRNA122a positively correlated with lipid profiles. Complementary to our results, another study was performed by Song et al., with silencing of miRNA122 by antagomirs. Inhibition of miRNA122-stimulated bile acid synthesis, therefore serum cholesterol and TG levels decreased (20).
In conclusion, our results demonstrated potential data for association between miRNA221, miRNA222 and atherosclerosis. Although metabolic effects of miRNA221 and miRNA222 were vague, altered expression levels in serum of patients with atherosclerosis indicated that they could be candidate novel circulating biomarkers for diagnosis. Besides, expression level of miRNA122a has a significant impact on serum TG and VLDL levels, because of positive correlations between them. These results should be supported with more detailed future studies.
Conflicts of Interest
The Authors declare that there are no financial disclosures or conflict of interest associated with this study.
References
- 1.Wang T, Palucci D, Law K, Yanagawa B, Yam J, Butany J. Atherosclerosis: pathogenesis and pathology. Diag Histopathol. 2012;18:461–467. [Google Scholar]
- 2.http://www.who.int/cardiovascular_diseases/en/ [Google Scholar]
- 3.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Luo X, Lu Y, Yang B. miRNAs at the heart of the matter. J Mol Med. 2008;86(7):771–783. doi: 10.1007/s00109-008-0341-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C, Pei F, Zhu X, Duan DD, Zeng C. Circulating microRNAs as novel and sensitive biomarkers of acute myocardial Infarction. Clin Biochem. 2012;45(10-11):727–732. doi: 10.1016/j.clinbiochem.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santovito D, Mezzetti A, Cipollone F. MicroRNAs and atherosclerosis: new actors for an old movie. Nutr Metab Cardiovasc Dis. 2012;22(11):937–943. doi: 10.1016/j.numecd.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Wojciechowska A, Braniewska A, Kozar-Kamińska K. MicroRNA in cardiovascular biology and disease. Adv Clin Exp Med. 2017;26(5):865–874. doi: 10.17219/acem/62915. [DOI] [PubMed] [Google Scholar]
- 8.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 9.Santovito D, Mezzetti A, Cipollone F. MicroRNAs and atherosclerosis: new actors for an old movie. Nutr Metab Cardiovasc Dis. 2012;22(11):937–943. doi: 10.1016/j.numecd.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79(4):581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 12.Rippe C, Blimline M, Magerko KA, Lawson BR, LaRocca TJ, Donato AJ, Seals DR. MicroRNA changes in human arterial endothelial cells with senescence: relation to apoptosis, eNOS and inflammation. Exp Gerontol. 2012;47(1):45–51. doi: 10.1016/j.exger.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108(9):3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Cheng Y, Yang J, Xu L, Zhang C. Cell-specific effects of miR-221/222 in vessels: molecular mechanism and therapeutic application. J Mol Cell Cardiol. 2012;52(1):245–255. doi: 10.1016/j.yjmcc.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104(4):476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Shao S, Geng H, Yu Y, Wang C, Liu Z, Yu C, Jiang X, Deng Y, Gao L, Zhao J. Expression profiles of six circulating microRNAs critical to atherosclerosis in patients with subclinical hypothyroidism: a clinical study. J Clin Endocrinol Metab. 2014;99(5):E766–774. doi: 10.1210/jc.2013-1629. [DOI] [PubMed] [Google Scholar]
- 17.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may down-regulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1(2):106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 18.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid Res. 2010;51(6):1513–1523. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song KH, Li T, Owsley E, Chiang JY. A putative role of micro RNA in regulation of cholesterol 7alpha-hydroxylase expression in human hepatocytes. J Lipid Res. 2010;51(8):2223–2233. doi: 10.1194/jlr.M004531. [DOI] [PMC free article] [PubMed] [Google Scholar]