Abstract
The aim of this study was to evaluate the role of vimentin expression in the prognosis and progression of CRC. Meta-analysis was conducted to investigate the correlations between vimentin and prognosis and clinicopathological features in CRC. Literatures were searched by PubMed, Embase, ClinicalKey, CNKI, VIP, and WanFang databases. The Cancer Genome Atlas (TCGA) database was used to assess the association of vimentin expression with survival rate in CRC. Eleven reports with 1969 cases were included in the meta-analysis. The results showed that positive vimentin expression predicted a poor overall survival (OS) in the univariate analysis (HR: 2.087, 95%CI: 1.660-2.625) and multivariate analysis (HR: 1.633, 95%CI: 1.223-2.181). Vimentin overexpression also conferred worse disease-free survival (DFS) in the univariate analysis (HR: 2.069, 95%CI: 1.024-4.179) and multivariate analysis (HR: 2.802, 95%CI: 1.421-5.527). Moreover, upregulated vimentin is related to lymph node metastasis (OR: 2.288, 95%CI: 1.159-4.517), TNM stages (OR: 1.957, 95%CI: 1.333-2.873), and N stage (OR: 2.316, 95%CI: 1.482-3.620). Analysis of TCGA database indicated that elevated vimentin predicated a shorter OS (p=0.033). Our findings reveal that upregulated vimentin contributes to the progression and poor prognosis of CRC. Vimentin may be a prognostic biomarker and therapeutic target in patients with CRC.
1. Introduction
Colorectal cancer (CRC) is one of the most prevalent human malignancies and is considered as the fourth most common cause of cancer-related deaths worldwide [1, 2]. Although the overall incidence rate of CRC has declined, largely due to early clinical diagnosis and advanced therapies in developed countries, it is still very high in the East Asia [3]. Besides, the majority of patients with CRC suffer a poor clinical outcome, mainly because of unfavorable prognostic factors including distant/regional metastasis, local recurrence, and chemoresistance. Hence, increasing studies have focused on the molecular events related to these factors in CRC development, of which epithelial-mesenchymal transition (EMT) has received great attention in clinical research [4]. EMT is a dynamic process in which cells lose epithelial characteristics and acquire mesenchymal properties and is involved in the downregulation of epithelial markers and upregulation of mesenchymal markers [5, 6]. Vimentin is regarded as a sign of cell epithelial to mesenchymal conversion and seems to be one of the best indicators of EMT in tumorigenesis [7, 8]. Vimentin plays a vital role in the progression and prognosis of cancer via the EMT and the corresponding signaling pathways, which contributes to the tumorigenesis, metastasis, invasion, and therapeutic resistance of various tumors [9, 10]. Accumulating evidences have demonstrated that vimentin overexpression stimulates the metastasis and invasion of CRC [11–13]. However, its prognostic significance remains unclarified. A previous study suggested that vimentin could be a promising predictive marker for patients with stage III CRC [14], whereas a recent study indicated that vimentin was of no prognostic value for these patients [15].
To the best of our knowledge, so far there has been no systematic review on the prognostic significance of vimentin expression in CRC. Therefore, we conducted a study based on a meta-analysis and TCGA database to estimate the relations between vimentin and prognosis and progression in CRC.
2. Materials and Methods
2.1. Meta-Analysis
2.1.1. Search Strategy
Following the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) statements checklist, PubMed, Embase, ClinicalKey, CNKI, VIP, and WanFang databases were searched until Apr. 2018[16]. The search terms were as follows: (Vimentin or vim or vmt or vm or hel113 or ctrct30) and (colorectal or colon or rectum or colorectum) and (cancer or carcinoma or adenocarcinoma or tumor or neoplasm). No restrictions were placed on language. References of the retrieved and review articles were also screened by hand.
2.1.2. Selection Criteria
The included studies had to meet the following criteria: (1) patients with a pathological diagnosis of stage I to IV CRC who underwent radical surgery, (2) studies detected the level of vimentin protein or vimentin mRNA in the CRC tissues by immunohistochemistry (IHC) or real-time reverse transcription-polymerase chain reaction (qRT-PCR), (3) studies investigated the association of vimentin expression with the overall survival (OS), disease-free survival (DFS) or clinicopathological features such as age, gender, tumor size, differentiation, TNM stage, lymph node metastasis, and distant metastasis, (4) the survival data may be directly or indirectly obtained, and (5) when an author had several studies on the same patient population, only the most recent or largest sample article was included. The exclusion criteria in our meta-analysis included (1) titles, abstracts, systematic review, meta-analysis, case reports, letters, and conference data, (2) duplicated study, (3) studies used animals, cell lines, or others but not tumor tissues, (4) no effective data to estimate HR with its 95%CI, (5) studies combined vimentin with other markers to evaluate its clinical significance in CRC, or (6) a study with low quality.
2.1.3. Data Extraction
Data were extracted independently by two authors, and the inconsistent opinions were adjudicated by a third author. The information collected from each study are as follows: the first authors' last name, year of publication, countries, the study design, the number of patients, cancer site, the stage of cancer, the follow-up time, treatment, technology of detection, the value of cut-off, the type of survival analysis, and HR with 95%CI. Moreover, clinicopathological parameters were collected, including age, gender, tumor size, tumor site, serum CEA (carcinoembryonic antigen) level, differentiation, lymph node metastasis, distant metastasis, recurrence metastasis, lymphovascular invasion, venous invasion, TNM stages, T stage, and N stage.
2.1.4. Quality Assessment
The quality of each study was evaluated by the Newcastle-Ottawa Scale (NOS), which is a 9-star system containing the following three dimensions: the selection of cohorts, the comparability of cohorts, and the ascertainment of outcomes [17]. A study with 7-9 scores was classified as a high-quality study, whereas those with scores of 4–6 and 0-3 are moderate- and low-quality studies, respectively [18].
2.1.5. Statistical Analysis
The data analyses were performed using Comprehensive Meta-Analysis Software, v. 2.0 (CMA, Biostat, Englewood, NJ, USA). The prognostic value of vimentin expression in the CRC patients was estimated by summary HRs with 95%CIs. The HRs and 95%CIs were obtained directly from the univariate or multivariate survival analysis and indirectly from Kaplan–Meier survival curves as reported by Parmar [19]. Pooled ORs and corresponding 95%CIs were calculated to evaluate the relations between vimentin expression and the clinicopathological features, including age, gender, tumor size, tumor site, serum CEA level, differentiation, lymph node metastasis, distant metastasis, recurrence metastasis, lymphovascular invasion, venous invasion, TNM stages, T stage, and N stage. Heterogeneity was evaluated among studies by calculating the Q-statistic and I2 value. A significant heterogeneity was present among studies if a p value of <0.10 for the Q-test, the I2 value describes the percentage of variation across studies that are due to heterogeneity rather than chance, while an I2 of 0% indicates no observed heterogeneity, with 25% regarded as low, 50% as moderate, and 75% as high [20]. Furthermore, the random-effects model was used to provide more conservative pooled estimates [21]. Publication bias was assessed by constructing the funnel plots (there was no publication bias if the funnel plot was symmetric) and quantified using Begg's test [22] and Egger's test [23], in which a p-value<0.05 indicated the existence of potential publication bias. A sensitivity analysis was also performed to assess whether the combined estimates could have been markedly influenced by a single study, in which each study was omitted one by one and the analysis was repeated based on the remaining studies.
2.2. Analysis of the Cancer Genome Atlas (TCGA) Database
We downloaded the data of 344 CRC cases on age, gender, race, tumor stage, survival information, and vimentin expression. Based on the median of vimentin expression, all cases were divided into high-expression (n=165) and low-expression groups (n=179). Survival rates were estimated by multivariate analysis. Cox regression analysis was used to perform the multivariate analysis, in which the confounding factors including age, gender, tumor stage, and race were adjusted. The results were considered statistically significant if p < 0.05.
3. Results
3.1. Meta-Analysis
3.1.1. Literature Search
The flow diagram of the literature search is shown in Figure 1. We searched 2704 titles or abstracts until the latest date of Apr. 2018, of which 18 articles were related to our research purpose. Finally, a total of 11 studies were included in this meta-analysis, and the main reasons for removing 7 studies from the remaining articles were as follows: 6 studies provided insufficient data for calculating HRs with 95%CIs [11, 13, 24–27], and 1 study retrieved patient data from the prospectively maintained hepato-pancreato-biliary database and its quality could not be assessed [28].
Figure 1.
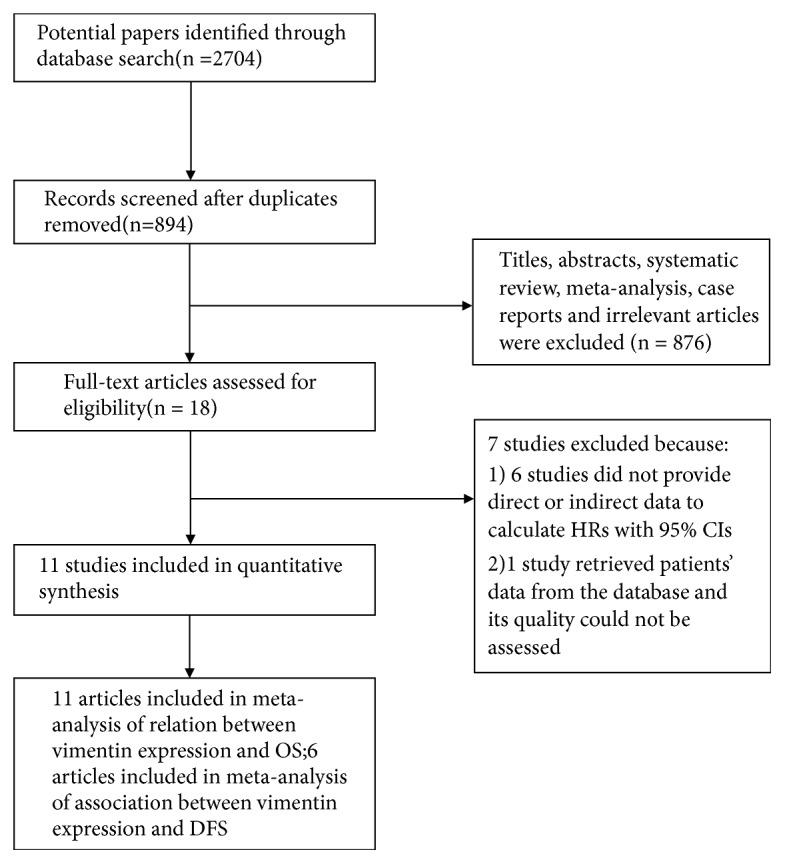
Flow diagram of the study selection in this meta-analysis. NOS: Newcastle-Ottawa-Scale; OS: overall survival; DFS: disease-free survival.
3.1.2. Study Characteristics and Quality Assessment
The main characteristics of the included studies are summarized in Tables 1 and 2. The type of study design was cohort study with 1969 cases. 7 studies were conducted in China [29–35], 2 studies were conducted in Japan [12, 14], and 2 studies were conducted in Korea [15, 36]. All of the included studies evaluated the correlation between vimentin expression and OS. Of these studies, 6 studies also assessed the association of vimentin expression with DFS [12, 14, 15, 29, 31, 34]. The quality scores of studies ranged from 7 to 9. Therefore, all of the included studies were high-quality studies (studies with a score⩾7), as shown in Table 1.
Table 1.
Characteristics of the included studies for the overall survival (OS) analysis.
| Study | Country and design | Case (N) |
Cancer site |
Stage | Follow-up (M) | Treatment | Technique | Cut off | Overall Survival HR (95% CI) |
NOS | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis |
Multivariate analysis |
||||||||||
| Ngan, 2007 | Japan; Cohort | 142 | CRC | II,III | 66 | resection, chemotherapy$ | IHC | >8.8% | 3.360(1.220-9.250)∧ | -& | 7 |
| Li, 2015 | China; Cohort | 41 | Colon cancer | I-IV | 64# | resection | IHC | scores≥2 | 6.670(1.420-31.380)∧ | - | 8 |
| Gao, 2014 | China; Cohort | 194 | CRC | I-IV | 52∗ | resection | IHC | >10% | 1.760(0.920-3.370)∧ | - | 7 |
| Yun, 2014 | Korea; Cohort | 409 | CRC | III | 80# | resection | IHC | scores≥1 | 1.000(0.507–1.975) | - | 8 |
| Xiao, 2015 | China; Cohort | 105 | CRC | I-IV | 81# | resection | IHC | Scores 2–3 | 2.038(1.142–3.795) | 1.573(0.792–2.133) | 8 |
| Liu, 2017 | China; Cohort | 203 | CRC | II | 80# | resection | IHC | scores≥6 | 2.092(1.058–4.135) | 2.028(1.021–4.029) | 8 |
| Toiyama, 2013 | Japan; Cohort | 181 | CRC | I-IV | 40∗ | resection | qRT-PCR and IHC | scores>2 | 2.540(1.470-4.380) | 1.470(0.790–2.750) | 8 |
| Huang, 2017 | China; Cohort | 117 | Colon cancer | II | 47∗ | resection, chemotherapy$ | IHC | Scores 2-4 | 1.790(0.240-13.320)∧ | 1.749(0.550–5.847) | 7 |
| Gao, 2015 | China; Cohort |
189 | CRC | I-IV | 52∗ | resection | IHC | >10% | 1.870(0.950-3.690)∧ | 1.905(1.066-3.407) | 7 |
| Wang, 2017 | China; Cohort | 102 | CRC | I-III | 56∗ | resection | IHC | scores>3 | - | 1.056(0.455–2.451) | 9 |
| ChOI, 2017 | Korea; Cohort | 286 | CRC | I-IV | 53 | resection, chemotherapy, and radiation therapy$ | IHC | >5% | 2.740(1.490-5.060)∧ | - | 7 |
CRC, colorectal cancer; qRT-PCR, real-time reverse transcription-polymerase chain reaction; IHC, immunohistochemistry; NOS, Newcastle-Ottawa Scale; N, the number of cases; M, months.
∗: median follow-up time.
#: maximum follow-up time.
$: adjuvant chemotherapy or radiation therapy after surgical resection.
&-: not available.
∧: data calculated from Kaplan–Meier survival curves.
Table 2.
Characteristics of the included studies for the disease-free survival (DFS) analysis.
| Study | Country and design |
Case (N) |
Cancer site | Stage | Follow-up (M) | Treatment | Technique | Cut off | Disease-free Survival HR (95% CI) |
NOS | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis |
Multivariate analysis |
||||||||||
| Ngan, 2007 | Japan; Cohort | 142 | CRC | II,III | 66 | resection, chemotherapy$ | IHC | >8.8% | 2.810(1.220-6.480)∧ | 3.450(1.650–7.220) | 7 |
| Li, 2015 | China; Cohort | 41 | Colon cancer | I-IV | 64# | resection | IHC | scores≥2 | 12.080(2.350-62.000)∧ | 11.636(3.093-43.770) | 8 |
| Yun, 2014 | Korea; Cohort | 409 | CRC | III | 80# | resection | IHC | scores≥1 | 0.769(0.419–1.413) | -& | 8 |
| Liu, 2017 | China; Cohort | 203 | CRC | II | 80# | resection | IHC | scores≥6 | 1.847(1.007–3.386) | 2.032(1.106–3.734) | 8 |
| Toiyama, 2013 | Japan; Cohort | 181 | CRC | I-IV | 40∗ | resection | qRT-PCR and IHC | scores>2 | 2.130(0.730-6.170)∧ | - | |
| Wang, 2017 | China; Cohort | 102 | CRC | I-III | 56∗ | resection | IHC | scores>3 | - | 1.409(0.667–2.975) | 9 |
CRC, colorectal cancer; IHC, immunohistochemistry; NOS, Newcastle-Ottawa Scale; N, the number of cases; M, months.
∗: median follow-up time.
#: maximum follow-up time.
$: adjuvant chemotherapy after surgical resection.
&-: not available.
∧: data calculated from Kaplan–Meier survival curves.
3.1.3. Association of Vimentin Expression and OS
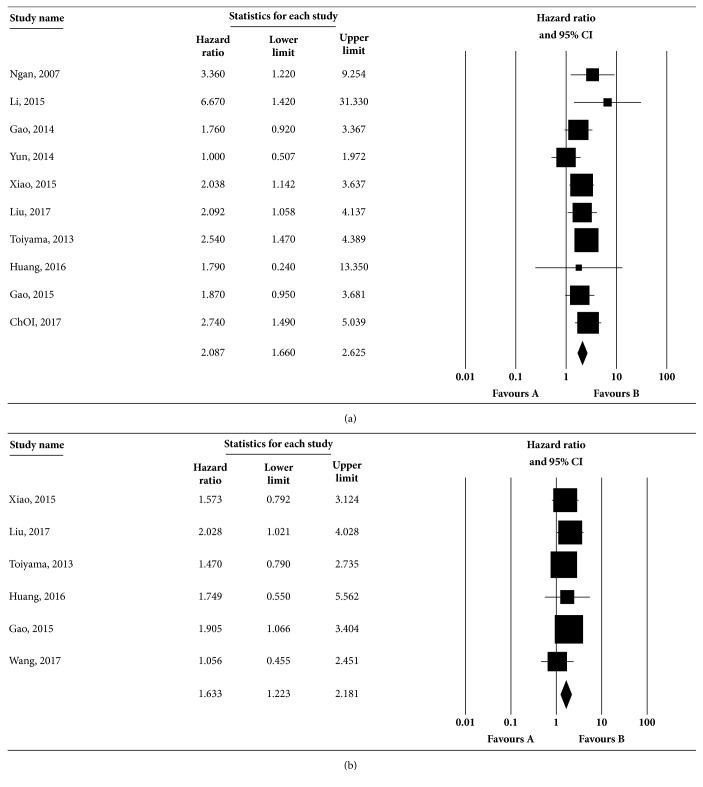
A total of 11 studies investigated the significance of vimentin expression in the OS of CRC, of which 10 studies performed the univariate analysis, and 6 studies performed the multivariate analysis. In the univariate analysis, the heterogeneity was not statistically significant (Q=9.180, I2=1.966%, p=0.421), and the results of the pooled HR showed that positive vimentin expression predicted a poor OS (HR: 2.087, 95%CI: 1.660-2.625) based on the random-effects model, as shown in Figure 2(a). Moreover, the multivariate analysis indicated the relation between positive vimentin expression and an unfavorable OS (HR: 1.633, 95%CI: 1.223-2.181, Figure 2(b)) by the random-effects model. There was not significant heterogeneity among the studies (Q=1.818, I2=0%, p=0.874). In sensitivity analyses, no great fluctuation was observed in the pooled results when one study was ruled out in univariate or multivariate analyses (Fig S1A and B), which suggested that the results of this meta-analysis were reliable. Begg and Egger tests were conducted to evaluate the publication bias of the included studies. No evident publication bias was detected based on the symmetric distribution of funnel plot and P values in Begg (p=0.655 in univariate analysis, Fig S2A, p=0.851 in multivariate analysis, Fig S2B) and Egger tests (p=0.454 in univariate analysis, Fig S2A, p=0.562 in multivariate analysis, Fig S2B). The subgroup analyses were also conducted to verify the above findings, and the detailed results are summarized in Table 3.
Figure 2.
Association between vimentin expression and OS rate in CRC by univariate (a) and multivariate (b) analyses. Studies were combined using the random-effects model. (a) The pooled HR for OS was 2.087 (95% CI: 1.660-2.625; p for heterogeneity = 0.421, I2= 1.966%). (b) The pooled HR for OS was 1.633 (95% CI: 1.223-2.181; p for heterogeneity =0.874, I2=0%). The square boxes indicate study-specific estimates. The size of each box reflects the study's weight in the analysis, and the horizontal lines represent 95% CIs. The diamond represents the pooled HRs and 95% CI. The p value < 0.1 indicated the existence of heterogeneity among studies.
Table 3.
Subgroup analysis of HR in overall survival (OS) by univariate and multivariate analyses.
| Variables | Study (N) | Heterogeneity test | HR (95%CI) | p | ||
|---|---|---|---|---|---|---|
| Q | I2 (%) | p | ||||
| Overall survival (U) | 10 | 9.180 | 1.966 | 0.421 | 2.087(1.660-2.625) | 0.000∗ |
| Country | ||||||
| China | 6 | 2.536 | 0.000 | 0.771 | 2.041(1.476-2.823) | 0.000∗ |
| Japan | 2 | 0.227 | 0.000 | 0.634 | 2.717(1.638-4.506) | 0.000∗ |
| Korea | 2 | 4.688 | 78.670 | 0.030 | 1.740(1.083-2.796) | 0.022∗ |
| Sample | ||||||
| ≤200 | 7 | 3.683 | 0.000 | 0.719 | 2.241(1.667-3.012) | 0.000∗ |
| >200 | 3 | 4.873 | 58.956 | 0.087 | 1.842(1.242-2.732) | 0.002∗ |
| Cancer site | ||||||
| CRC | 8 | 6.950 | 0.000 | 0.434 | 2.038(1.619-2.565) | 0.000∗ |
| Colon | 2 | 1.034 | 3.255 | 0.309 | 4.088(1.200-13.928) | 0.024∗ |
| Year | ||||||
| 2014-2017 | 8 | 7.517 | 6.882 | 0.377 | 1.938(1.500-2.505) | 0.000∗ |
| Before 2014 | 2 | 0.227 | 0.000 | 0.634 | 2.706(1.672-4.378) | 0.000∗ |
| Overall survival (M) | 6 | 1.818 | 0.000 | 0.874 | 1.633(1.223-2.181) | 0.001∗ |
| Country | ||||||
| China | 5 | 1.677 | 0.000 | 0.795 | 1.681(1.213-2.331) | 0.002∗ |
| Japan | 1 | - | - | - | - | - |
| Sample | ||||||
| ≤200 | 5 | 1.353 | 0.000 | 0.852 | 1.559(1.133-2.144) | 0.006∗ |
| >200 | 1 | - | - | - | - | - |
| Cancer site | ||||||
| CRC | 5 | 1.804 | 0.000 | 0.772 | 1.626(1.206-2.191) | 0.001∗ |
| Colon | 1 | - | - | - | - | - |
| Year | ||||||
| 2014-2017 | 5 | 1.677 | 0.000 | 0.795 | 1.681(1.213-2.331) | 0.002∗ |
| Before 2014 | 1 | - | - | - | - | - |
N, the number of the included studies; U, univariate analysis; M, multivariate analysis; CRC, colorectal cancer.
∗: the value of p<0.05 indicates statistical significance.
&-: not available.
3.1.4. Association of Vimentin Expression and DFS
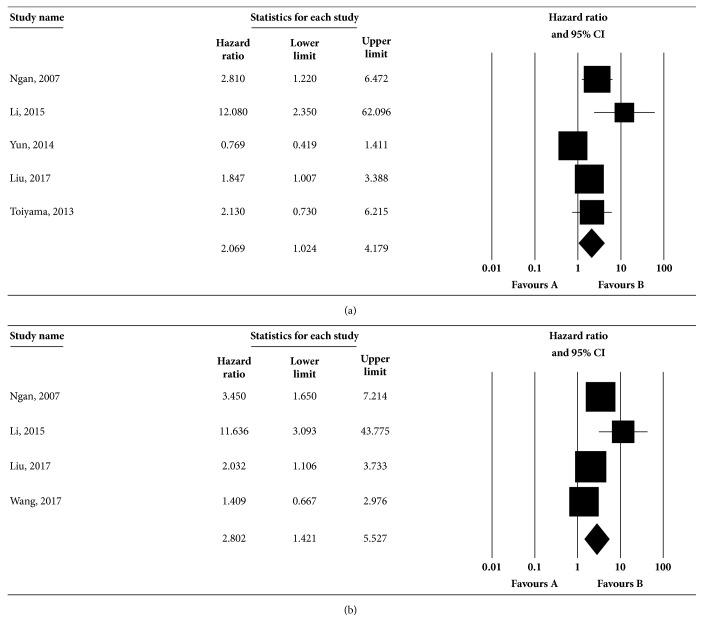
Six papers provided data on the effect of vimentin expression on the DFS of CRC. Among these studies, 5 studies conducted the univariate analysis, and 4 studies conducted the multivariate analysis. The univariate analysis showed that the combined HR was 2.069 (95%CI: 1.024-4.179, Figure 3(a)) via the random-effects model with substantial heterogeneity (Q=13.668, I2=70.734%, p=0.008); the multivariate analysis indicated that the estimated effect was HR 2.802 (95%CI: 1.421-5.527, Figure 3(b)) based on the random-effects model with potential heterogeneity (Q=8.604, I2=65.133%, p=0.035). The result of sensitivity test in univariate analysis showed that there was no statistical significance in the pooled HRs of the remaining studies by omission of Ngan, 2007, Li, 2015, Liu, 2017 and Toiyama, 2013 (Fig S1C), which were not consistent with the combined estimates. Therefore, the four studies might be the sources of significant heterogeneity. However, excluding any single study did not affect the result of DFS in multivariate analysis (Fig S1D), which needs further discussion. Publication bias was estimated by Begg and Egger test. There was no indication of publication bias based on the symmetric distribution of funnel plot and P values in Begg (p=0.142 in univariate analysis, Fig S2C, p=0.497 in multivariate analysis, Fig S2D) and Egger tests (p=0.119 in univariate analysis, Fig S2C, p=0.217 in multivariate analysis, Fig S2D).
Figure 3.
Association between vimentin expression and DFS rate in CRC by univariate (a) and multivariate (b) analyses. Studies were combined using the random-effects model. (a) The pooled HR for DFS was 2.069 (95% CI: 1.024-4.179; p for heterogeneity =0.008, I2=70.734%). (b) The pooled HR for DFS was 2.802 (95% CI: 1.421-5.527; p for heterogeneity =0.035, I2=65.133%). The square boxes indicate study-specific estimates. The size of each box reflects the study's weight in the analysis, and the horizontal lines represent 95% CIs. The diamond represents the pooled HRs and 95% CI. The p value < 0.1 indicated the existence of heterogeneity among studies.
3.1.5. Correlations between Vimentin and Clinicopathological Characteristics
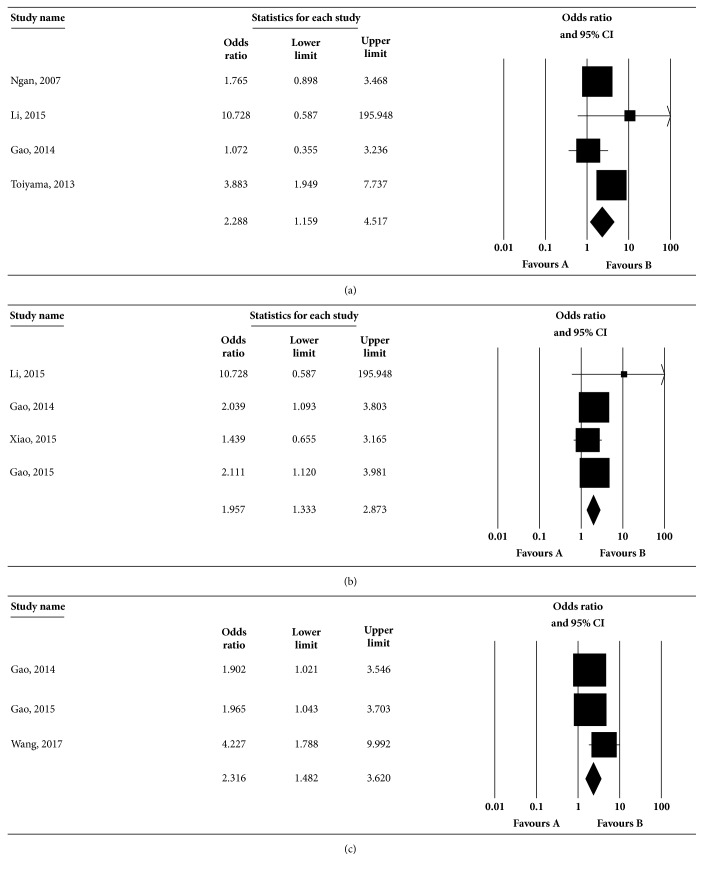
Ten studies were included to estimate the association between vimentin expression and clinicopathological characteristics in CRC. Table 4 showed the combined ORs of vimentin in various parameters. The summary results suggested that positive vimentin expression was related to lymph node metastasis (OR: 2.288, 95%CI: 1.159-4.517, Figure 4(a)), TNM stages (OR: 1.957, 95%CI: 1.333-2.873, Figure 4(b)), and N stage (OR: 2.316, 95%CI: 1.482-3.620, Figure 4(c)). No significant correlation was observed between vimentin and other clinicopathological characteristics (p>0.05). The results of the sensitivity analysis indicated that the summary results were not influenced by excluding any one study in each clinicopathological characteristic (data not shown). There was no significant publication bias in the majority of the clinicopathological characteristics except venous invasion and N stage (p<0.05, data not shown).
Table 4.
Analysis of relationships between vimentin and clinicopathological variables in CRC.
| Clinical pathological variable | Study (N) | Pooled OR (95%CI) |
p value | Heterogeneity test | ||
|---|---|---|---|---|---|---|
| Q | I2 (%) | p | ||||
| Age (≤60/>60) | 6 | 0.964(0.742-1.253) | 0.786 | 1.368 | 0.000 | 0.928 |
| Tumor size (>5cm/≤5cm) | 5 | 0.782(0.581-1.053) | 0.105 | 2.582 | 0.000 | 0.630 |
| Gender (male/female) | 10 | 0.943(0.761-1.169) | 0.594 | 5.800 | 0.000 | 0.760 |
| Tumour site (Colon/Rectum) | 5 | 0.929(0.690-1.250) | 0.626 | 3.835 | 0.000 | 0.429 |
| CEA level (>5ng/ml/≤5 ng/ml) | 3 | 0.937(0.650-1.352) | 0.728 | 1.559 | 0.000 | 0.459 |
| Differentiation (poor/well or mod) | 8 | 0.954(0.407-2.234) | 0.914 | 53.357 | 86.881 | 0.000 |
| Lymph node metastasis (present/absent) | 4 | 2.288(1.159-4.517) | 0.017∗ | 5.717 | 47.520 | 0.126 |
| Distant metastasis (present/absent) | 2 | 4.122(0.820-20.730) | 0.086 | 2.551 | 60.807 | 0.110 |
| Recurrence (present/absent) | 2 | 3.726(0.215-64.509) | 0.366 | 5.993 | 83.314 | 0.014 |
| Lymphovascular invasion (present/absent) | 4 | 1.244(0.736-2.103) | 0.415 | 6.367 | 52.883 | 0.095 |
| Venous invasion (present/absent) | 3 | 1.180(0.220-6.328) | 0.847 | 3.912 | 48.880 | 0.141 |
| TNM stages (III-IV/I-II) | 4 | 1.957(1.333-2.873) | 0.001∗ | 1.973 | 0.000 | 0.578 |
| T stage (T3–T4/T1–T2) | 3 | 0.756(0.520-1.098) | 0.142 | 1.242 | 0.000 | 0.537 |
| N stage (N1–N2/N0) | 3 | 2.316(1.482-3.620) | 0.000∗ | 2.515 | 20.477 | 0.284 |
N, the number of the included studies; CEA, carcinoembryonic antigen; CRC, colorectal cancer.
∗: the value of p<0.05 indicates statistical significance.
Figure 4.
Correlations between vimentin expression and clinicopathological characteristics. Studies were combined using the random-effects model. (a) The pooled OR for lymph node metastasis was 2.288 (95% CI: 1.159-4.517; p for heterogeneity =0.126, I2=47.52%). (b) The pooled OR for TNM stages was 1.957 (95% CI: 1.333-2.873; p for heterogeneity =0.578, I2=0%). (c) The pooled OR for N stage was 2.316 (95% CI: 1.482-3.620; p for heterogeneity =0.284, I2=20.477%). The square boxes indicate study-specific estimates. The size of each box reflects the study's weight in the analysis, and the horizontal lines represent 95% CIs. The diamond represents the pooled ORs and 95% CI. The p value < 0.1 indicated the existence of heterogeneity among studies.
3.2. Analysis of the Cancer Genome Atlas (TCGA) Database
The association between vimentin expression and prognosis of CRC was also evaluated from TCGA data. The result suggested that high vimentin expression indicated a shorter OS compared with low-expression group (p=0.033, Figure 5).
Figure 5.
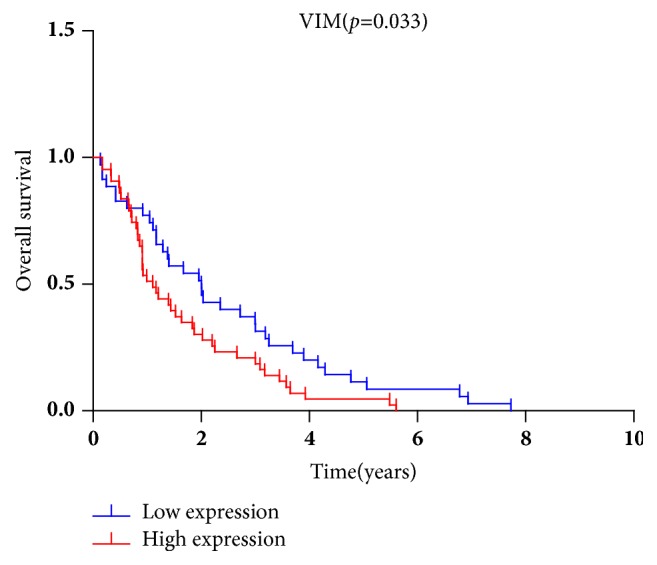
Increased vimentin expression was associated with reduced OS in CRC. The data on survival and vimentin expression downloaded from TCGA database indicated 165 cases with vimentin upregulation and 179 cases with vimentin downregulation. Survival rates were estimated using Cox regression analysis. The p value <0.05 was considered statistically significant.
4. Discussion
A total of 11 cohort studies on the relation between vimentin expression and the prognosis of CRC were included in this review. To our knowledge, this report is the first meta-analysis combined with TCGA database to evaluate the value of vimentin in predicting the progression and prognosis of CRC. The results of meta-analysis suggested that positive vimentin expression predicted a poorer OS in both univariate and multivariate analyses. In the univariate analysis of DFS, the combined HR indicated that the association of positive vimentin expression with the shorter survival in CRC. Moreover, in the multivariate analysis of DFS, in which confounding factors are adjusted, we found that vimentin could be a significant prognostic factor. The potential heterogeneities existed in the two analyses of DFS. The main reasons are as follows: (1) excluding Ngan, 2007 [14], Li, 2015 [29], Liu, 2017 [31], and Toiyama, 2013 [12], influenced the pooled HR of DFS in univariate analysis; (2) the results of publication bias showed that the funnel plots were not symmetric in univariate and multivariate analyses, which may be the sources of significant heterogeneities among studies; (3) the survival data were obtained by calculation based on survival curves in three studies [12, 14, 29], which may result in an inaccurate HR of DFS. The results of the current meta-analysis also indicated that upregulated vimentin correlated well with lymph node metastasis, advanced TNM stages, and N stage. Moreover, substantial heterogeneities were observed in differentiation, distant metastasis, recurrence, and lymphovascular invasion, which may depend on the differences in country, cancer stage, and sample size among the included studies. The analysis from TCGA database indicated that elevated vimentin expression predicted a shorter OS.
Vimentin is a major component of the intermediate filament (IF) family and is involved in maintaining the cellular integrity and stability [37]. Increasing studies investigated the prognostic roles of vimentin expression and its clinicopathological significance in cancer [38–41]. However, the results of the published studies were inconsistent. A recent study indicated that vimentin overexpression in the invasive front of CRC significantly correlated with poor OS (p=0.028) [36], which is similar to our findings. Besides, a novel study based on computational modeling also supported this conclusion and identified vimentin as a valuable biomarker for CRC [42]. However, Yun et al. revealed that vimentin failed to indicate a significant association with prognosis in CRC [15]. The contradictions between the published studies may result from the differences in sample size, CRC stage, and the study design. Moreover, the prognosis-indicative role of vimentin was shown in other cancers and diseases. Nakashima et al. found that vimentin expression was markedly upregulated in micropapillary components of lung adenocarcinomas and it predicted adverse clinical outcome [43]. Tian et al. investigated the prognostic role of E-cadherin and vimentin expression in various subtypes of soft tissue leiomyosarcomas (LMS). They suggested that the patients with the gain of E-cadherin and loss of vimentin expression represented favorable trend of survival. Furthermore, the two markers might serve as good biomarkers of the LMS clinical outcome [44]. Our findings also showed that the overexpression of vimentin was associated with lymph node metastasis, advanced TNM stages, and N stage, whereas no significant relation was observed between upregulated vimentin and age, tumor size, gender, tumor site, serum CEA level, differentiation, distant metastasis, recurrence metastasis, lymphovascular invasion, venous invasion, and T stage. The increasing evidences suggested that high vimentin expression correlated well with the clinicopathological characteristics in other cancers such as cholangiocarcinoma (CCA), lung cancer, and liver cancer [45–47]. The findings were consistent with the results of our study. Hence, vimentin expression played a crucial role in the progression and prognosis of CRC.
Additionally, other markers could influence the progression and prognosis of cancer through regulating vimentin expression. A recent study reported that vimentin overexpression and the EMT were induced by PLAGL2 via Wnt/β-catenin signaling pathway in CRC, which stimulated the migration and invasion of tumor cells and may validate our findings that vimentin was related to lymph node metastasis in CRC [48]. The EMT of CRC were inhibited by loss of BMI-1 in inflammatory microenvironment through TLR4/MD-2/MyD88-mediated NF-κB signaling, which was beneficial to the prognosis of CRC [49]. Moreover, the expression of EMT-associated genes could be regulated by microRNAs. A recently published study indicated that miR-194 significantly upregulated vimentin expression in CRC, which resulted in cell migration and promoted the development of CRC [50]. MiR-375 inhibited the invasion and metastasis of CRC via targeting SP1 and regulating EMT-associated genes [51]. The EMT and metastasis in CRC were also suppressed by long noncoding RNA LINC01133 directly binding to SRSF6 [52]. In addition, the EMT of CRC cells was regulated by the renin angiotensin system, where vimentin expression was reduced by the blocker of renin angiotensin system peptide ANG II type 1 receptor (AT1R) and thus inhibited the metastasis and invasion of CRC [53]. Because of the significantly prognostic role of vimentin, researchers focused on its effect on the treatment of cancer. Lahat et al. showed that vimentin was a novel anticancer therapeutic target by mice xenograft studies [54]. Subsequently, cancer cell biologists turned their attention to reprogramming cancer stem cells to normal stem cells. Hugwil reported that a human monoclonal antibody, CLN-IgG, recognized vimentin expressing on the cell surface of the malignant tumor to reprogram cancer stem cells to normal organogenesis and thereby suppressed the progression of cancer [55]. In recent years, most anticancer products and drugs exerted their functions effectively by acting on multiple anticancer molecular targets. A recent study showed that cyclometalated gold (III) complexes realized its anticancer effect by the specific engagement with multiple cellular targets including vimentin [56]. Some anticancer drugs could inhibit migration and invasion of tumor cells by regulating EMT-associated genes expression [57, 58].
There are limited studies on the role of vimentin in the progression and prognosis of CRC patients and the results are inconsistent among studies. Thus, we first conducted a study with meta-analysis and TCGA database to investigate the value of vimentin in predicting the progression and prognosis of CRC. Compared with a single study with small sample size, a meta-analysis can provide a more stable result and make a more convincing conclusion because it summarizes the single sample data and combines all the existing evidences. Furthermore, we applied TCGA database to verify the prognostic value of vimentin in CRC, which includes the complete and updated data and thus could make the conclusions more reliable.
Similar to all studies, the present study has several limitations. First, the number of the included studies and sample size are smaller (a total of 11 included articles with 1969 cases). When the number of the studies is smaller than 10 in subgroup analysis, the power of publication bias test is declined and the combined results are unstable [59]. Thus, prospective studies with large sample sizes are needed to confirm the value of vimentin in predicting the progression and prognosis of CRC. Second, six studies did not directly provide the survival data, and the HRs of OS and DFS were obtained by calculation according to survival curves, which may cause an inaccurate pooled HR [14, 29, 30, 33, 35, 36]. Moreover, we could not perform subgroup analysis in assessing the relations between vimentin and clinicopathological characteristics due to the smaller included studies. Finally, the positive vimentin expression was defined based on cut-off value in each study, and the values were inconsistent among the included studies. Therefore, the heterogeneities may exist among the studies, which may weaken the reliability of the combined results and influence the conclusions.
This is the first study with meta-analysis and TCGA database to demonstrate that positive vimentin expression predicted a poor OS and DFS in both univariate and multivariate analyses. Additionally, upregulated vimentin was related to lymph node metastasis, advanced TNM stages, and N stage. In summary, high vimentin expression contributes to the progression and poor outcome of CRC patients. Vimentin may be a promising biomarker for survival prediction and a potential target for the treatment strategies in patients with CRC. In the future, our findings should be confirmed by more well-designed cohort or experimental studies.
Acknowledgments
The authors would like to thank the laboratory members Xiaojuan Zhao, Ziqing Zhu, and Qiqi Li for discussion. This work was supported by the National Natural Science Foundation of China (Grant no. 31401081).
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Le Du performed the literature search, extracted data for analysis, and wrote the article. Jingchuan Li conducted the literature search, extracted data for analysis, and revised the manuscript. Lei Lei extracted data for analysis and revised the manuscript. Hongjuan He and Erfei Chen conducted the meta-analysis. Jing Dong revised the manuscript. Jin Yang had the idea for the article, extracted data for analysis, and revised the manuscript. All authors read and approved the manuscript.
Supplementary Materials
The sensitivity analyses and publication bias of the included studies.
References
- 1.Fitzmaurice C., Hamavid H., Dicker D., Allen C., Naghavi M. The Global Burden of Non-Hodgkin Lymphoma. Clinical Lymphoma, Myeloma & Leukemia. 2015;15:p. S70. doi: 10.1016/j.clml.2015.07.142. [DOI] [Google Scholar]
- 2.He H., Lei L., Chen E., et al. The screening of the functional microRNA binding site SNPs in sporadic colorectal cancer genes. Cancer Biology & Therapy. 2017;18(6):407–413. doi: 10.1080/15384047.2017.1323584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA: A Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Gurzu S., Silveanu C., Fetyko A., Butiurca V., Kovacs Z., Jung I. Systematic review of the old and new concepts in the epithelial-mesenchymal transition of colorectal cancer. World Journal of Gastroenterology. 2016;22(30):6764–6775. doi: 10.3748/wjg.v22.i30.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serrano-Gomez S. J., Maziveyi M., Alahari S. K. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Molecular Cancer. 2016;15(1, article 18) doi: 10.1186/s12943-016-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiery J. P., Sleeman J. P. Complex networks orchestrate epithelial-mesenchymal transitions. Nature Reviews Molecular Cell Biology. 2006;7(2):131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 7.Their J. P. Epithelial-mesenchymal transitions in tumor progression. Nature Reviews Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 8.Zeisberg M., Neilson E. G. Biomarkers for epithelial-mesenchymal transitions. The Journal of Clinical Investigation. 2009;119(6):1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heerboth S., Housman G., Leary M., et al. EMT and tumor metastasis. linical and Translational Medicine. 2015;4(1, article 6) doi: 10.1186/s40169-015-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato M., Shames D. S., Hasegawa Y. Emerging evidence of epithelial-to-mesenchymal transition in lung carcinogenesis. Respirology. 2012;17(7):1048–1059. doi: 10.1111/j.1440-1843.2012.02173.x. [DOI] [PubMed] [Google Scholar]
- 11.Rashed H. E., Hussein S., Mosaad H., et al. Prognostic significance of the genetic and the immunohistochemical expression of epithelial-mesenchymal-related markers in colon cancer. Cancer Biomarkers. 2017;20(1):107–122. doi: 10.3233/CBM-170034. [DOI] [PubMed] [Google Scholar]
- 12.Toiyama Y., Yasuda H., Saigusa S., et al. Increased expression of slug and vimentin as novel predictive biomarkers for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis. 2013;34(11):2548–2557. doi: 10.1093/carcin/bgt282. [DOI] [PubMed] [Google Scholar]
- 13.Tan J. L., Chen Q., Qiao S. S., Qin Q. Y., Deng M. L. Li. Jr, et al. Expression and clinical significance of vimentin in colorectal adenocarcinoma. Journal of Guangxi Medical University. 2016:829–831. [Google Scholar]
- 14.Ngan C. Y., Yamamoto H., Seshimo I., et al. Quantitative evaluation of vimentin expression in tumour stroma of colorectal cancer. British Journal of Cancer. 2007;96(6):986–992. doi: 10.1038/sj.bjc.6603651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun J.-A., Kim S.-H., Hong H. K., et al. Loss of E-cadherin expression is associated with a poor prognosis in stage III colorectal cancer. Oncology (Switzerland) 2014;86(5-6):318–328. doi: 10.1159/000360794. [DOI] [PubMed] [Google Scholar]
- 16.Moher D., Shamseer L., Clarke M. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Systematic Reviews. 2015;4(1, article 1) doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European Journal of Epidemiology. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 18.Yang L., Liu B., Huang B., et al. A functional copy number variation in the WWOX gene is associated with lung cancer risk in Chinese. Human Molecular Genetics. 2013;22(9):1886–1894. doi: 10.1093/hmg/ddt019. [DOI] [PubMed] [Google Scholar]
- 19.Parmar M. K. B., Torri V., Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statistics in Medicine. 1998;17(24):2815–2834. doi: 10.1002/(SICI)1097-0258(19981230)17:24<2815::AID-SIM110>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 20.Guyatt G. H., Oxman A. D., Kunz R., et al. GRADE guidelines: 7. Rating the quality of evidence—inconsistency. Journal of Clinical Epidemiology. 2011;64(12):1294–1302. doi: 10.1016/j.jclinepi.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Ioannidis J. P., Patsopoulos N. A., Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. British Medical Journal. 2007;335(7626):914–916. doi: 10.1136/bmj.39343.408449.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Begg C. B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 23.Egger M., Smith G. D., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. British Medical Journal. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaca A., Govender P., Locketz M., Naidoo R. The role of miRNA-21 and epithelial mesenchymal transition (EMT) process in colorectal cancer. Journal of Clinical Pathology. 2017;70(4):331–336. doi: 10.1136/jclinpath-2016-204031. [DOI] [PubMed] [Google Scholar]
- 25.Lapertosa G., Baracchini P., Fulcheri E., Tanzi R. Prognostic value of the immunocytochemical detection of extramural venous invasion in Duke's C colorectal adenocarcinomas. The American Journal of Pathology. 1989;135(5):939–945. [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y., Jiang H. S., Ma L. J., Liu S. Q., Jin Z. J. Xu Q.Clinical significance and correlation of RhoGDI2 and EMT-related proteins expression in colorectal carcinoma tissues. Chin J Lab Diagn. 2015:1465–1469. [Google Scholar]
- 27.Zhang C., Xu J. H., Liu T., Cui H. Clinical of signal transduction and activators of transcription 3,E-cadherin in colon cancer. Chin J Gastrointest Surg. 2011;14:202–205. [PubMed] [Google Scholar]
- 28.Lau L. F., Murone C., Williams D. S., et al. Metabolic response evaluation for colorectal liver metastases and correlation to pathologic response and tumour markers. ANZ Journal of Surgery. 2018;88(3):E108–E113. doi: 10.1111/ans.13680. [DOI] [PubMed] [Google Scholar]
- 29.Li L. L. Expression and Clinical Significance of E-Canderin and Vimentin in Han and Uigur in Xinjiang Area of Colon Cancer. Xinjiang Medical University; 2015. [Google Scholar]
- 30.Gao Z.-H., Lu C., Wang Z.-N., et al. ILEI: A novel marker for epithelial-mesenchymal transition and poor prognosis in colorectal cancer. Histopathology. 2014;65(4):527–538. doi: 10.1111/his.12435. [DOI] [PubMed] [Google Scholar]
- 31.Liu L.-G., Yan X.-B., Xie R.-T., Jin Z.-M., Yang Y. Stromal expression of vimentin predicts the clinical outcome of stage II colorectal cancer for high-risk patients. Medical Science Monitor. 2017;23:2897–2905. doi: 10.12659/MSM.904486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao S., Liu L., Lu X., Long J., Zhou X., Fang M. The prognostic significance of bromodomain PHD-finger transcription factor in colorectal carcinoma and association with vimentin and E-cadherin. Journal of Cancer Research and Clinical Oncology. 2015;141(8):1465–1474. doi: 10.1007/s00432-015-1937-y. [DOI] [PubMed] [Google Scholar]
- 33.Huang Z., Ai Z., Li N., et al. Over expression of galectin-3 associates with short-term poor prognosis in stage II colon cancer. Cancer Biomarkers. 2017;17(4):445–455. doi: 10.3233/CBM-160661. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Wu Z., Hu L. Epithelial-Mesenchymal Transition Phenotype, Metformin, and Survival for Colorectal Cancer Patients with Diabetes Mellitus II. Gastroenterology Research and Practice. 2017;2017 doi: 10.1155/2017/2520581.2520581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai H., Xie S. Determination of emission factors from motor vehicles under different emission standards in China. Journal of Peking University: Natural Science Edition. 2010;03:319–326. [Google Scholar]
- 36.Choi E., Bae J. S., Kang M. J., et al. Expression of epithelial-mesenchymal transition and cancer stem cell markers in colorectal adenocarcinoma: Clinicopathological significance. Oncology Reports. 2017;38(3):1695–1705. doi: 10.3892/or.2017.5790. [DOI] [PubMed] [Google Scholar]
- 37.Satelli A., Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cellular and Molecular Life Sciences. 2011;68(18):3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuyuhiro Y., Yashiro M., Noda S., et al. Clinical significance of vimentin-positive gastric cancer cells. Anticancer Reseach. 2010;30(12):5239–5244. [PubMed] [Google Scholar]
- 39.Jin H., Morohashi S., Sato F., et al. Vimentin expression of esophageal squamous cell carcinoma and its aggressive potential for lymph node metastasis. Journal of Biomedical Research. 2010;31(2):105–112. doi: 10.2220/biomedres.31.105. [DOI] [PubMed] [Google Scholar]
- 40.Hu L., Lau S. H., Tzang C.-H., et al. Association of Vimentin overexpression and hepatocellular carcinoma metastasis. Oncogene. 2004;23(1):298–302. doi: 10.1038/sj.onc.1206483. [DOI] [PubMed] [Google Scholar]
- 41.McInroy L., Määttä A. Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochemical and Biophysical Research Communications. 2007;360(1):109–114. doi: 10.1016/j.bbrc.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 42.Bukhari S., Mokhdomi T. A., Chikan N. A., et al. Affinity proteomics led identification of vimentin as a potential biomarker in colon cancers: Insights from serological screening and computational modelling. Molecular BioSystems. 2015;11(1):159–169. doi: 10.1039/c4mb00506f. doi: 10.1039/c4mb00506f. [DOI] [PubMed] [Google Scholar]
- 43.Nakashima H., Jiang S.-X., Sato Y., et al. Prevalent and up-regulated vimentin expression in micropapillary components of lung adenocarcinomas and its adverse prognostic significance. Pathology International. 2015;65(4):183–192. doi: 10.1111/pin.12257. [DOI] [PubMed] [Google Scholar]
- 44.Tian W., Wang G., Yang J., Pan Y., Ma Y. Prognostic role of E-cadherin and Vimentin expression in various subtypes of soft tissue leiomyosarcomas. Medical Oncology. 2013;30(1, article no. 401) doi: 10.1007/s12032-012-0401-y. [DOI] [PubMed] [Google Scholar]
- 45.Saentaweesuk W., Araki N., Vaeteewoottacharn K., et al. Activation of Vimentin is Critical to Promote a Metastatic Potential of Cholangiocarcinoma Cells. Oncology Research : Featuring Preclinical and Clinical Cancer Therapeutics. 2017;25 doi: 10.3727/096504017X15009778205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye Z., Zhang X., Luo Y., et al. Prognostic values of vimentin expression and its clinicopathological significance in non-small cell lung cancer: A meta-analysis of observational studies with 4118 cases. PLoS ONE. 2016;11(9) doi: 10.1371/journal.pone.0163162.e0163162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshikawa M., Morine Y., Ikemoto T., et al. Elevated preoperative serum cea level is associated with poor prognosis in patients with hepatocellular carcinoma through the epithelial-mesenchymal transition. Anticancer Reseach. 2017;37(3):1169–1175. doi: 10.21873/anticanres.11430. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y., Guo P., Zhu Z., et al. Pleomorphic adenoma gene like-2 induces epithelial-mesenchymal transition via Wnt/β-catenin signaling pathway in human colorectal adenocarcinoma. Oncology Reports. 2017;37(4):1961–1970. doi: 10.3892/or.2017.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye K., Chen Q., Sun Y., Lin J., Xu J. Loss of BMI-1 dampens migration and EMT of colorectal cancer in inflammatory microenvironment through TLR4/MD-2/MyD88-mediated NF-κB signaling. Journal of Cellular Biochemistry. 2018;119(2):1922–1930. doi: 10.1002/jcb.26353. [DOI] [PubMed] [Google Scholar]
- 50.Cai H.-K., Chen X., Tang Y.-H., Deng Y.-C. MicroRNA-194 modulates epithelial–mesenchymal transition in human colorectal cancer metastasis. OncoTargets and Therapy. 2017;10:1269–1278. doi: 10.2147/OTT.S125172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui F., Wang S., Lao I., et al. miR-375 inhibits the invasion and metastasis of colorectal cancer via targeting SP1 and regulating EMT-associated genes. Oncology Reports. 2016;36(1):487–493. doi: 10.3892/or.2016.4834. [DOI] [PubMed] [Google Scholar]
- 52.Kong J., Sun W., Li C., et al. Long non-coding RNA LINC01133 inhibits epithelial–mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Letters. 2016;380(2):476–484. doi: 10.1016/j.canlet.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen L., Ager E. I., Neo J., Christophi C. Regulation of colorectal cancer cell epithelial to mesenchymal transition by the renin angiotensin system. Journal of Gastroenterology and Hepatology. 2016;31(10):1773–1782. doi: 10.1111/jgh.13307. [DOI] [PubMed] [Google Scholar]
- 54.Lahat G., Zhu Q.-S., Huang K.-L., et al. Vimentin is a novel anti-cancer therapeutic target; insights from In Vitro and In Vivo mice xenograft studies. PLoS ONE. 2010;5(4) doi: 10.1371/journal.pone.0010105.e10105 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Hugwil A. V. The meaning of the anti-cancer antibody CLN-IgG (Pritumumab) generated by human×human hybridoma technology against the cyto-skeletal protein, vimentin, in the course of the treatment of malignancy. Medical Hypotheses. 2013;81(3):489–495. doi: 10.1016/j.mehy.2013.05.037. [DOI] [PubMed] [Google Scholar]
- 56.Fung S. K., Zou T., Cao B., et al. Cyclometalated Gold(III) Complexes Containing N-Heterocyclic Carbene Ligands Engage Multiple Anti-Cancer Molecular Targets. Angewandte Chemie International Edition. 2017;56(14):3892–3896. doi: 10.1002/anie.201612583. [DOI] [PubMed] [Google Scholar]
- 57.Lai Y.-J., Tai C.-J., Wang C.-W., et al. Anti-Cancer Activity of Solanum nigrum (AESN) through Suppression of Mitochondrial Function and Epithelial-Mesenchymal Transition (EMT) in Breast Cancer Cells. Molecules (Basel, Switzerland) 2016;21(5) doi: 10.3390/molecules21050553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu J., Yang H., Zhou X., Wang H., Gong L., Tang C. Bisdemethoxycurcumin suppresses migration and invasion of highly metastatic 95D lung cancer cells by regulating E-cadherin and vimentin expression, and inducing autophagy. Molecular Medicine Reports. 2015;12(5):7603–7608. doi: 10.3892/mmr.2015.4356. [DOI] [PubMed] [Google Scholar]
- 59.Sterne J. A. C., Gavaghan D., Egger M. Publication and related bias in meta-analysis: power of statistical tests and prevalence in the literature. Journal of Clinical Epidemiology. 2000;53(11):1119–1129. doi: 10.1016/s0895-4356(00)00242-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sensitivity analyses and publication bias of the included studies.