The symbiotic receptor DMI2 in Medicago undergoes constitutive degradation by the proteasome, which is blocked upon rhizobia inoculation.
Abstract
Plants use receptor-like kinases to monitor environmental changes and transduce signals into plant cells. The Medicago truncatula (hereafter M. truncatula) DOES NOT MAKE INFECTIONS2 (DMI2) protein functions as a coreceptor of rhizobial signals to initiate nodule development and rhizobial infection during nitrogen-fixing symbiosis, but the mechanisms regulating DMI2 protein level and folding are still unknown. Here, we report that DMI2 protein abundance changes during nitrogen-fixing symbiosis. DMI2 accumulates in the nodules and is induced by rhizobia treatment through a posttranscriptional process. However, DMI2 induction is independent of the perception of Nod factor, a group of lipochitooligosaccharides secreted by rhizobia. The stability of the DMI2 protein is controlled by the proteasome pathway: in rhizobia-free environments, DMI2 is degraded by the proteasome, but during rhizobial infection, DMI2 is protected from the proteasome, resulting in protein accumulation. Furthermore, proteasome inhibitor-promoted accumulation of DMI2 protein in Medicago roots induces the expression of two early nodulation marker genes, supporting the hypothesis that DMI2 accumulation activates downstream symbiosis signaling. The extracellular region of DMI2 contains two malectin-like domains (MLDs) and a leucine-rich repeat. When conserved amino acids in the MLDs are mutated, DMI2 fails to restore nodule development in dmi2 mutants, and point-mutated MLD proteins are degraded constitutively, suggesting that the MLD may be vital for the accumulation of DMI2. Our findings suggest that legumes control nodule development through modulating the protein level of DMI2, revealing a layer of regulation in the interaction between plants and rhizobia in nitrogen-fixing symbiosis.
Plants form symbiotic relationships with surrounding microbes to gain access to nutrients in natural environments. Most land plants form beneficial interactions with arbuscular mycorrhizal (AM) fungi, developing mycorrhized roots, which provide phosphorous and micronutrients to plants in exchange for fixed carbon. A subset of plants, including plants of the legume family, develop nitrogen-fixing symbioses in specialized organs, or root nodules. This sophisticated symbiosis with rhizobia provides legumes with nitrogen fixed by rhizobia hosted inside plant cells of the nodules (Oldroyd et al., 2011).
Plants use receptors to discriminate between symbiotic and pathogenic microbes. In both AM and nitrogen-fixing symbiosis, plant roots detect the existence of beneficial microbes by a group of receptor-like kinases (RLKs). In nitrogen-fixing symbiosis, rhizobia secrete a group of lipochitooligosaccharides, or Nod factors, to initiate nodule development and symbiosis. In Medicago truncatula (hereafter Medicago), Nod factors are perceived by two RLKs: NFP (Nod Factor Perception) and LYK3 (LysM Domain-Containing Receptor-Like Kinase3), which are named NFR1 (Nod Factor Receptor1) and NFR5 in Lotus japonicus, respectively. The perception of Nod factors activates the common symbiosis signaling pathway, including root hair-associated calcium spiking, early nodulation gene activation, and cortical cell division (Schauser et al., 1999). DMI2 (DOES NOT MAKE INFECTIONS2, the name in M. truncatula)/SYMRK (the name in L. japonicus)/Nodulation Receptor Kinase (the name in Medicago sativa) is believed to interact with Nod factor receptors (Antolín-Llovera et al., 2014a). Mutations in DMI2 lead to the abortion of rhizobial infection at a very early stage (Endre et al., 2002; Stracke et al., 2002). DMI2 also is indispensable for AM symbiosis and plant-Frankia symbiosis, another independently evolved nitrogen-fixing symbiosis between certain plants and Frankia bacteria (Endre et al., 2002; Stracke et al., 2002; Gherbi et al., 2008), indicating a conserved role of DMI2 throughout the evolution of plant-microbe symbioses.
The DMI2 protein contains an intracellular kinase domain, a transmembrane domain, and the extracellular portion, including a region with antolinthree leucine-rich repeats (LRRs) and a malectin-like domain (MLD). In human cells, the single-domain protein Malectin functions in the endoplasmic reticulum (ER) lumen in protein quality control by binding to diglucosylated Glc2Man9GlcNAc2, a glycan composed of three glucoses, nine mannoses, and two GlcNAcs (Schallus et al., 2008). Utilizing its carbohydrate-binding activity, Malectin interacts directly with misfolded glycoproteins and inhibits their secretion (Qin et al., 2012).
While Malectin-like sequences are widespread among biological kingdoms, two features make MLD-containing proteins in plants unique: (1) their gene families are greatly expanded in plants, and (2) MLDs mostly occur in the extracellular portion of RLKs. A few MLD-containing RLKs have been shown to play vital roles in plant development, male-female interaction, disease resistance, and plant-microbe symbiosis (Endre et al., 2002; Boisson-Dernier et al., 2011; Hok et al., 2011; Haruta et al., 2014). Although the functions of MLDs have yet to be revealed, the position of MLDs in the extracellular portions of proteins points to the possibility that MLDs may be necessary for activating or deactivating the intracellular kinase domain through binding extracellular ligands. Interestingly, it has been reported that the MLD of SYMRK/DMI2 is cleaved constitutively, with or without rhizobia, and that the MLD-cleaved SYMRK/DMI2 protein outcompetes the full-length SYMRK/DMI2 in the interaction with Nod factor receptors (Antolín-Llovera et al., 2014b).
During nitrogen-fixing symbiosis, the host is required to provide nutrients to the rhizobia (Oldroyd et al., 2011). This burden on the plant has promoted the evolution of a sophisticated regulatory network controlling the scale and timing of nodule development (Oldroyd et al., 2011). Several reports show that overexpressing the full-length SYMRK/DMI2 or the intracellular kinase domain of SYMRK/DMI2 leads to spontaneous nodule formation even in the absence of rhizobia (Ried et al., 2014; Saha et al., 2014), suggesting that the protein level of DMI2 needs to be regulated precisely. SYMRK/DMI2 also has been reported to interact with two E3 ligases: SINA4 (SEVEN IN ABSENTIA4) and SIE3 (SYMRK-INTERACTING E3 UBIQUITIN LIGASE; Den Herder et al., 2012; Yuan et al., 2012). However, direct genetic evidence illustrating that these two E3 ligases affect the level of SYMRK/DMI2 in planta is still missing, and the dynamics of DMI2 protein levels during nitrogen-fixing symbiosis remain unknown.
Here, we report that DMI2 protein levels are tightly regulated by legume hosts to properly respond to rhizobia infection. We find that, without rhizobia infection, DMI2 protein is constitutively degraded through the proteasome apparatus; during rhizobia infection, the DMI2 protein level is induced by blocking proteasome-mediated degradation. Meanwhile, if key amino acid residues in the DMI2-MLD are mutated, DMI2 is degraded constitutively, suggesting a key role of MLD in the regulation of DMI2 protein homeostasis. Taken together with the reports that overexpression of the DMI2/SYMRK kinase domain causes spontaneous nodulation (Ried et al., 2014; Saha et al., 2014), fine-tuning the protein level of DMI2 is critical for legumes to maximize the profit of nitrogen-fixing symbiosis at the lowest cost.
RESULTS
Rhizobia Induce DMI2 Protein at the Posttranscriptional Level
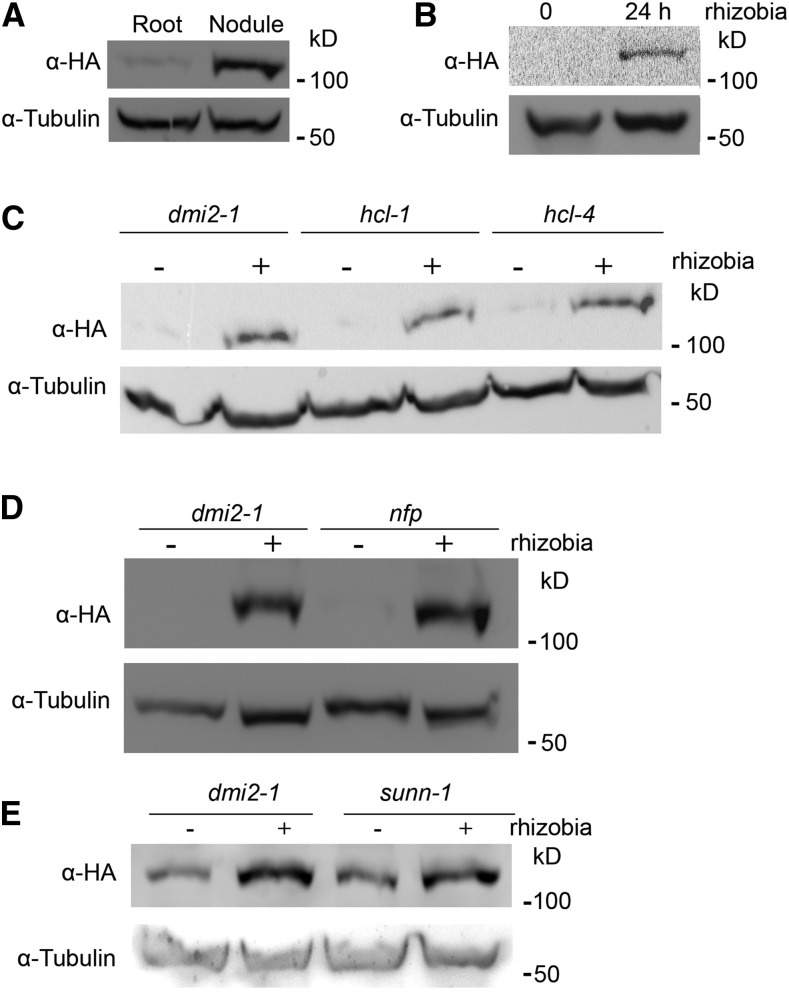
To determine the dynamics of DMI2 protein levels during nitrogen-fixing symbiosis, a stable transgenic Medicago dmi2-1 line was used. This line expresses the DMI2 genomic sequence driven by its native promoter; it is also fused to a dual affinity tag containing three copies of the hemagglutinin epitope (HA) and a single StrepII (ST); therefore, the resulting protein is named DMI2-HAST. The gDMI2:HAST construct complements the phenotype of dmi2 mutants, and the protein is easy to detect (Riely et al., 2013). After inoculating the transgenic lines with Sinorhizobium meliloti strain ABS7, we compared the protein accumulation of DMI2-HAST between nodules and untreated roots. The result showed that, before rhizobia treatment, the DMI2-HAST protein level in the roots was very low (Fig. 1A), which is consistent with previous reports that DMI2-HAST protein is almost undetectable (Riely et al., 2013). In nodules, the protein level of DMI2-HAST was much higher compared with rhizobia-free roots (Fig. 1A). Furthermore, by analyzing the expression of DMI2 in nodules and uninoculated roots in the M. truncatula gene expression atlas database (Benedito et al., 2008), we found that the transcription of DMI2 is not highly activated in whole nodules (Supplemental Fig. S1, A and B), which also is consistent with a previous report using northern-blot assays (Bersoult et al., 2005). These results suggest that the DMI2-HAST protein accumulates in the nodules through posttranscriptional regulation.
Figure 1.
Rhizobia treatment could induce the protein level of DMI2 in a Nod factor receptor-independent manner. A, DMI2-HAST protein accumulated in the nodules compared with uninoculated roots. Protein samples were taken from 14-d-old nodules and 4-week-old uninoculated roots. B, DMI2-HAST protein was induced to a much higher level 24 h after rhizobia treatment. Four-week-old Medicago roots were treated with S. meliloti strain ABS7 hemA:LacZ at the concentration of OD600 = 0.05 for 24 h. C and D, DMI2-HAST protein was induced to a similar level by rhizobia treatment in wild-type and Nod factor receptor mutant backgrounds. The gDMI2-HAST construct was transiently expressed in dmi2-1, hcl-1, and hcl-4, and transformed roots were treated with or without S. meliloti strain ABS7 hemA:LacZ. E, DMI2-HAST protein could be induced to a similar level by rhizobia in dmi2-1 and sunn-1 plants stably transformed with gDMI2-HAST. α-HA was used to detect the DMI2-HAST protein level, and tubulin was used as the loading control. The experiments were repeated five times with similar results.
To further investigate the protein level variation of DMI2 during rhizobia inoculation, we treated dmi2-1 gDMI2:HAST plant roots with rhizobia strain ABS7 and checked protein abundance during rhizobia infection at the earliest stage. Twenty-four hours post ABS7 strain treatment, the protein level of DMI2-HAST increased dramatically compared with untreated roots (Fig. 1B). Treating dmi2-1 gDMI2:HAST plants with one-half-strength basic nodulation medium, the liquid medium for rhizobia inoculation, Agrobacterium rhizogenes strain Arqua1, and S. meliloti strains Rm1021 and ABS7 showed that the DMI2-HAST protein level increased only during rhizobia inoculation (Supplemental Fig. S2A), indicating that the DMI2-HAST induction effect is specific to rhizobia. To establish how quickly the protein accumulates, we analyzed the protein level of DMI2-HAST at different time points post rhizobia treatment and found that, as early as 3 h after ABS7 inoculation, the DMI2-HAST protein level was already induced (Supplemental Fig. S2B). To rule out the possibility that the accumulation of DMI2-HAST was the result of transcriptional activation, we checked the expression level of DMI2 by reverse transcription-quantitative PCR (RT-qPCR) as well as by browsing the Medicago truncatula Gene Expression Atlas and the Medicago truncatula Genome Project version 4.0 database (Benedito et al., 2008; Krishnakumar et al., 2015). The RT-qPCR assay showed that the transcription of DMI2 was not induced substantially at 3, 6, 12, and 24 h after rhizobia inoculation, with microarray and RNA sequencing data showing similar results (Supplemental Fig. S3, A–C), which is consistent with previous reports that DMI2 transcripts are not induced significantly by rhizobia treatment in a northern-blot assay (Mirabella et al., 2005). Taking these results together, we conclude that rhizobia treatment induces the abundance of DMI2 protein by affecting posttranscriptional regulation at a very early stage of symbiosis.
It is reported that DMI2/SYMRK interacts directly with the Nod factor receptors to perceive Nod factors (Antolín-Llovera et al., 2014b). To determine whether the accumulation of DMI2-HAST protein in the presence of rhizobia is dependent on Nod factor receptors, we expressed gDMI2:HAST in hcl-1 and hcl-4 (two mutant alleles of the Medicago Nod factor receptor gene LYK3; Smit et al., 2007) as well as in dmi2-1. Similar to dmi2-1, 24 h post rhizobia treatment, the protein level of DMI2-HAST in hcl-1 and hcl-4 mutants accumulated to a much higher level compared with the control (Fig. 1C). We also performed this experiment with the mutant of another Nod factor receptor, NFP, and found similar results (Fig. 1D). In various experiments detecting the protein level changes of DMI2-HAST, the intensity of the DMI2-HAST bands in uninoculated Medicago roots ranged from very weak to hard to detect. It has been reported that, in hcl-1, hcl-4, and nfp mutants, the expression of DMI2 transcripts was identical to that in the wild type (Mirabella et al., 2005), indicating that the accumulation of DMI2-HAST protein in hcl-1, hcl-4, and nfp also is posttranscriptionally controlled.
To further confirm that the induction of DMI2 protein level by rhizobia was independent of Nod factor perception, we inoculated Medicago plants with wild-type S. meliloti RM1021 and two rhizobium mutant strains, RJW14 and JT210, which have defects in Nod factor synthesis (Wais et al., 2002). The protein level of DMI2 was induced to a similar level by RJW14 and JT210 compared with the wild-type strain (Supplemental Fig. S4, A and B). These results show that DMI2 protein accumulation induced by rhizobia is independent of Nod factor perception, suggesting the existence of another as-yet-unknown rhizobia signal.
SUNN (SUPER NUMERIC NODULES) is an LRR receptor kinase that functions in the shoot to regulate nodule numbers, and sunn mutants display a supernodulation phenotype (Penmetsa et al., 2003; Schnabel et al., 2005). Surprisingly, it was reported that overexpressing the DMI2/SYMRK kinase domain in sunn mutants decreases the number of nodules formed in these supernodulation mutants (Saha and DasGupta, 2015). To analyze whether SUNN could affect the induction of DMI2-HAST protein by rhizobia, we transformed gDMI2-HAST into dmi2-1 and sunn-1 backgrounds and checked the protein level of DMI2-HAST during ABS7 strain infection. The result showed that DMI2-HAST was induced to a similar level in sunn-1 and dmi2-1 mutant backgrounds (Fig. 1E), suggesting that the induction of DMI2 protein by rhizobia also is independent of SUNN.
DMI2 Protein Is Constitutively Degraded in Uninoculated Medicago Roots
The accumulation of DMI2 protein by rhizobia is regulated at the posttranscriptional level, indicating that protein stability regulation may play a role. To test this hypothesis, we performed a cell-free degradation assay using protein samples from dmi2-1 gDMI2-HAST roots grown in a sterile environment. DMI2-HAST protein from uninoculated roots was completely degraded within 2 h (Fig. 2A). To study which mechanism was responsible for the degradation of DMI2-HAST, several different proteolysis inhibitors were tested. While the protease inhibitor phenylmethylsulfonyl fluoride and a plant-specific protease inhibitor cocktail failed to rescue the protein level of DMI2, the proteasome inhibitor MG132 largely prevented DMI2-HAST protein degradation (Fig. 2A), showing that, in uninoculated roots, DMI2-HAST may be degraded in a proteasome-dependent manner.
Figure 2.
MG132 can block the degradation of DMI2 protein in uninoculated Medicago roots. A, MG132 could partially rescue DMI2-HAST protein level in a cell-free degradation assay. Cell-free DMI2-HAST protein samples were treated with different reagents for 2 h and subjected to western blot using α-HA. Tubulin was used to show equal loading. Start, Freshly extracted protein sample; Cocktail, plant-specific protease inhibitor mixture. B, MG132 treatment could induce the protein level of DMI2-HAST in vivo. Uninoculated Medicago roots were treated with 100 μm MG132, and the level of DMI2-HAST protein was detected. Experiments were repeated at least five times with similar results. C, MG132 could block the degradation of DMI2 independent of Nod factor reception. Samples were taken from dmi2-1, hcl-1, and hcl-4 plants expressing gDMI2-HAST and treated with or without 100 μm MG132 for 4 h. D and E, After MG132 treatment, the expression of NIN and ENOD11 was modestly induced in 2-week-old dmi2-1 gDMI2-HAST Medicago roots. Each dot represents a biological replicate. α-HA was used to detect the DMI2-HAST protein level, and tubulin was used as the internal control. Experiments were repeated two times with similar results.
The in planta protein degradation of DMI2-HAST was then examined by treating dmi2-1 gDMI2:HAST roots with MG132. Four hours post MG132 treatment, the accumulation of DMI2-HAST protein was enhanced significantly (Fig. 2B), showing that the inhibition of proteasome activity mimics the effect of rhizobia treatment. When treating hcl-1 and hcl-4 with MG132, the protein level of DMI2-HAST was similar to that in dmi2-1 plants, suggesting that the accumulation of DMI2 protein by MG132 treatment also is independent of Nod factor receptors (Fig. 2C). To rule out that MG132 treatment promoted the transcriptional activation of DMI2, we analyzed the dynamics of DMI2 transcripts during MG132 treatment by RT-qPCR. The transcript level of gDMI2:HAST was indistinguishable before and after MG132 treatment (Supplemental Fig. S5). These data show that MG132 can block the degradation of DMI2-HAST protein in uninoculated Medicago roots, suggesting that the DMI2-HAST protein is degraded through a proteasome apparatus in the absence of rhizobia.
Recently, there have been several reports showing that overexpressing the kinase domain or the full-length DMI2/SYMRK protein can induce a spontaneous nodulation phenotype in legume plants (Ried et al., 2014; Saha et al., 2014). To evaluate whether MG132-induced DMI2 protein accumulation carries any biological significance, we checked the expression of ENOD11 (EARLY NODULIN11) and NIN (NODULE INCEPTION PROTEIN), two marker genes of early nodule development (Schauser et al., 1999), in dmi2-1 and dmi2-1 gDMI2:HAST roots after MG132 treatment. As shown in Figure 2, D and E, 4 h after MG132 treatment in dmi2-1 gDMI2:HAST roots, the expression of NIN and ENOD11 was induced to a higher level compared with dmi2-1 mutants. We conclude that DMI2 protein accumulation may be able to partially activate downstream nodulation signaling.
DMI2 Protein Is Protected from Degradation in Inoculated Roots
To further test the effect of MG132 on DMI2 protein in inoculated Medicago roots, S. meliloti ABS7-inoculated dmi2-1 gDMI2-HAST roots were treated with MG132 for 4 h. Compared with untreated roots, the protein level of DMI2-HAST was not increased further (Supplemental Fig. S6), showing that MG132 has little impact on DMI2 protein level in the presence of rhizobia, which indicates that, in inoculated Medicago roots, proteasome-mediated degradation of DMI2 has already been blocked (Fig. 2B). Thus, during nitrogen-fixing symbiosis, DMI2 protein is protected from proteasome-mediated degradation.
MLD Is Vital for DMI2 Function
In the extracellular region, DMI2 has an LRR and an MLD. To gain more insights about the MLD, we aligned the MLD with human Malectin protein using a homology-modeling method (http://swissmodel.expasy.org/; Biasini et al., 2014). While DMI2-MLD was described originally as a Malectin-like sequence, we found that it actually contains two tandem matches to the human Malectin A domain, and each match has about 140 amino acids (Supplemental Fig. S7, A and B). The human Malectin protein is reported to bind Glc2Man9GlcNAc2 and regulate protein folding (Schallus et al., 2008). However, very little is known about the function of DMI2-MLD or any MLD in plants.
To study the function of DMI2-MLD, we aligned the amino acid sequence of DMI2-MLD with its close homologs in the plant kingdom. The results show that DMI2-MLD is conserved among its homologs in dicots (Supplemental Fig. S8), suggesting that MLD may have a conserved role. To gain further insights into the function of DMI2-MLD, we performed site-directed mutagenesis. We analyzed the following point mutations: DMI2C39D, DMI2Y95A, DMI2F111A, DMI2Y164A, DMI2F271A, DMI2Y291A, and DMI2F294A, where the numbers indicate the locations of these residues in the full-length DMI2 protein, including the signal peptide sequence (Supplemental Fig. S9, A and B). We targeted these amino acids because they were conserved among the homologs included in the alignment (Supplemental Fig. S8), pointing to a greater possibility that these point mutations may affect the function of DMI2-MLD. Among them, Y95A, Y164A, and Y291A may be in the predicted ligand-binding pockets (Supplemental Fig. S9A; Schallus et al., 2008).
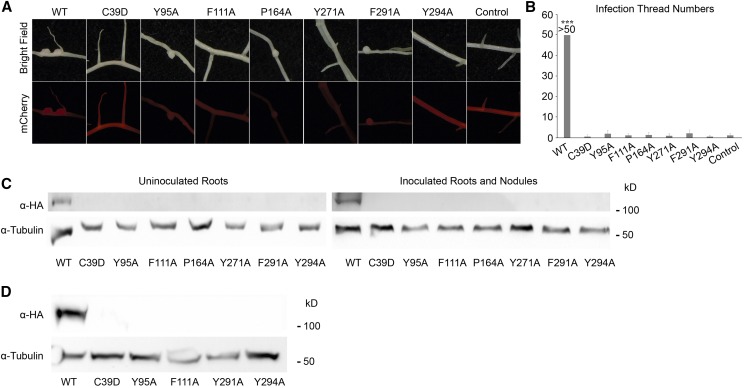
To investigate whether mutating the conserved amino acids in MLD would affect the function of the DMI2 protein, we introduced wild-type gDMI2-HAST and the seven MLD point-mutated versions into dmi2-1 mutant plants using the hairy root transformation method. As shown in Table I, 14 d after rhizobia inoculation, dmi2-1 plants expressing wild-type gDMI2-HAST generated many nodules, while dmi2-1 roots expressing MLD point mutants had few nodules, and even fewer of which were pink, suggesting a failure in nitrogen fixation (Fig. 3A). Some point mutations, such as DMI2C39D, had no nodules at all (Table I). These results show that, regarding sufficient nodule development, MLD is vital for the proper function of DMI2.
Table I. Summary of the complementation assay using wild-type and MLD point mutation versions of gDMI2-HAST.
While dmi2-1 plants transformed with wild-type gDMI2-HAST could generate a large number of pink nodules, roots expressing gDMI2-HAST versions with amino acid substitutions in MLD were impaired in nodule development and generated very few pink nodules. Control, Plants transformed with empty vector. Transformed roots were selected based on the mCherry fluorescence marker of the gDMI2-HAST construct. Experiments were repeated more than three times with similar results.
| Parameter | Wild Type | C39D | Y95A | F111A | P164A | Y271A | F291A | Y294A | Control |
|---|---|---|---|---|---|---|---|---|---|
| Plants in total | 13 | 14 | 11 | 12 | 13 | 14 | 12 | 11 | 14 |
| Transformed plants | 13 | 13 | 11 | 12 | 13 | 14 | 12 | 11 | 14 |
| Plants with nodules | 13 | 0 | 9 | 4 | 9 | 1 | 5 | 3 | 0 |
| Nodule numbers | >300 | 0 | 29 | 4 | 17 | 1 | 19 | 4 | 0 |
| Pink nodules | >300 | 0 | 5 | 0 | 0 | 1 | 2 | 0 | 0 |
Figure 3.
MLD is required for DMI2 protein to function properly in the early nodule development process and to stabilize full-length DMI2 proteins in plants. A, Images showing representative nodules in dmi2-1 plants transforming wild-type (WT) gDMI2-HAST and MLD point mutation versions of gDMI2-HAST. Although some point mutation versions could generate nodules, most of them were white and small. mCherry fluorescence was used to select the transformed roots. B, dmi2-1 plants expressing gDMI2-HAST containing point mutations in MLD had few infection threads in a quantitative assay. The number of infection threads in dmi2-1 plants expressing wild-type gDMI2-HAST was estimated to be more than 50. More than 10 transformed plants were used for each line. Data represent means and sd. Asterisks show a significant difference (***, P < 0.001, Student’s t test). C, DMI2 protein containing amino acid substitutions in MLD is unstable in Medicago plants, with or without rhizobia inoculation. In dmi2-1 gDMI2-HAST plants, DMI2-HAST protein could be seen, while the MLD amino acid substitution version proteins were totally undetectable. D, MG132 treatment could not rescue the constitutive degradation of DMI2 protein containing amino acid substitutions. dmi2-1 plants expressing wild-type gDMI2-HAST and MLD point mutation versions were treated with 100 μm MG132, and the protein level was tested. Experiments were repeated three times with similar results.
To enter legume roots, rhizobia normally have to penetrate plant cells through a plant-derived tubular structure at infected root hair cells, known as the infection thread (Oldroyd et al., 2011). It has been shown that, in dmi2/symrk mutants, the development of the infection thread is blocked (Endre et al., 2002; Stracke et al., 2002). To find out whether blocking the function of MLD could affect the role of DMI2 in infection thread formation, we checked the infection thread phenotype of dmi2-1 mutants transformed with wild-type gDMI2-HAST and the seven MLD point-mutated versions of gDMI2-HAST. Three days after rhizobia inoculation, infection threads could be seen using the microscope in dmi2-1 mutants transformed with wild-type gDMI2-HAST. In contrast, there were few infection thread-like structures in the plants transformed with gDMI2-HAST containing point mutations in MLD (Supplemental Fig. S10). Counting the numbers of infection threads, we found that dmi2-1 roots transformed with wild-type gDMI2-HAST could produce large numbers of infection threads, but the roots transformed with gDMI2-HAST with point mutations in MLD had very few infection threads (Fig. 3B). These results show that MLD is necessary for DMI2 protein to function properly in infection thread formation. Taken together, the proper function of MLD is necessary for DMI2 protein to play a fundamental role in nodule development at the very early stage.
MLD Is Required for the Homeostasis of DMI2
Since the protein level of DMI2 is important for its proper function in nodule development, we studied whether the amino acid substitutions in MLD would affect the protein level of DMI2. In Medicago roots expressing either wild-type gDMI2-HAST or MLD point mutations of gDMI2-HAST, the protein could be detected in plants transformed with wild-type sequence, but the plants transformed with gDMI2-HAST containing amino acid substitutions in MLD could not generate detectable DMI2-HAST protein, with or without rhizobia inoculation (Fig. 3C). To rule out the possibility that the disappearance of the MLD point-mutated protein is regulated at the transcriptional level, we checked the transcripts of wild-type gDMI2-HAST and the MLD point mutation versions using RT-qPCR. We found that transgenic plants could generate comparable amounts of wild-type gDMI2-HAST and MLD point mutation transcripts (Supplemental Fig. S11), albeit the expression level varied due to variations in hairy root transformation. These results show that the proper function of MLD is critical for the precise regulation of DMI2 protein abundance at the posttranscriptional stage.
To find out whether the degradation of MLD amino acid substitution versions of DMI2 protein is dependent on the proteasome apparatus, we treated dmi2-1 plants expressing wild-type gDMI2-HAST and MLD point mutation versions with MG132 and checked DMI2 levels before and after MG132 treatment. As shown in Figure 3D, 4 h after MG132 treatment, the protein level of wild-type full-length DMI2-HAST was strongly induced, while MLD point mutation versions could not accumulate the protein. This result shows that the degradation of DMI2 containing point mutations in MLD could not be rescued by inhibiting proteasome activity with MG132. Considering that the human Malectin protein binds to carbohydrate and functions in ER quality control (Schallus et al., 2008; Qin et al., 2012), it is possible that the MLD is required for the proper folding of DMI2 in the ER; blocking MLD function may activate ER quality control signaling and result in the degradation of DMI2.
As MLD is indispensable for the function of DMI2 in Medicago, we further investigated the origin of MLD and whether it is conserved in the plant kingdom. We constructed a phylogenetic tree of DMI2 in dicots, monocots, and basal angiosperm species and found that MLD is not present in monocot homologs of DMI2, but the protein from Amborella trichopoda, a basal angiosperm species, has an MLD (Supplemental Fig. S12). On the other hand, we examined the phylogenetic tree of the closest DMI2 paralog in M. truncatula, Medtr7g057170. To our surprise, the orthologs of this gene from every species had an MLD. These results suggest that MLD is important for the function of DMI2 in dicots and basal angiosperms; however, in monocot plants, the MLD is missing, indicating that there may be other proteins functioning together with DMI2 to perform the function of MLD (Supplemental Fig. S12).
DISCUSSION
Rhizobia Inoculation Can Block the Protein Degradation of DMI2
Localized to the plasma membrane, RLKs are able to detect environmental changes through extracellular domains and transduce the signals into cells through their intracellular catalytic domains (Lemmon and Schlessinger, 2010). To ensure the proper activation of RLKs during development and stress-response processes, the protein levels of RLKs must be kept in check by plants to avoid runaway intracellular responses. Here, we report that the protein level of the M. truncatula symbiosis receptor DMI2 is regulated at the posttranslational level to modulate the proper response to rhizobia inoculation.
DMI2 plays a key role regulating plant-microbe symbiosis, as it is required for the symbiosis between plants and AM fungi, plant-actinomycete nitrogen-fixing symbiosis, and legume plant-rhizobia nitrogen-fixing symbiosis (Gherbi et al., 2008). Furthermore, DMI2 displays protein level increases coinciding with rhizobia inoculation. When plants are grown in the absence of rhizobia, the DMI2 protein is kept at a very low level (Fig. 1, A and B); during rhizobia infection, through a currently unknown mechanism, rhizobia block the degradation of DMI2, resulting in increased protein levels. As a consequence, accumulated DMI2 induces plant roots to start the process of nodule development. This is consistent with previous reports that, when the DMI2/SYMRK kinase domain or full-length protein is overexpressed in legume roots, plants will generate spontaneous nodules without rhizobia infection (Ried et al., 2014; Saha et al., 2014).
We also found that MG132 treatment caused accumulation of the protein level of DMI2 in uninoculated Medicago roots (Fig. 2, A and B), indicating that the turnover of DMI2 in the absence of rhizobia infection is through proteasome-mediated protein degradation. More importantly, MG132 treatment mimicked the effect of rhizobia inoculation with respect to activating NIN and ENOD11 (Fig. 2, D and E). In inoculated roots, MG132 treatment did not increase the protein level of DMI2 (Supplemental Fig. S6), suggesting that, after rhizobia inoculation, DMI2 protein is already protected from proteasome degradation. The robust induction of DMI2 protein levels by MG132 also suggests that MG132 treatment can be used for further characterization of the DMI2 protein. For example, it may be used to find DMI2-interacting partners, especially the substrates of the DMI2 kinase domain.
Although the mechanisms coupling ligand recognition in the extracellular parts and the activation of intracellular catalytic domains are surprisingly diverse (Lemmon and Schlessinger, 2010), there are reports that ligand binding induces the protein levels of RLKs. For instance, it is widely known that, in human cells, insulin treatment can increase the intracellular protein abundance of insulin receptors (Lemmon and Schlessinger, 2010). In Arabidopsis (Arabidopsis thaliana), FLS2 (FLAGELLIN SENSING2) is an LRR RLK that functions as a receptor for bacterial flagellin protein or flg22, a 22-amino acid active peptide derivative (Gómez-Gómez and Boller, 2000). FLS2 can be ubiquitinated by two related U-box E3 ligases, PUB12 and PUB13, without the presence of pathogen (Lu et al., 2011). The ubiquitination and subsequent degradation of FLS2 protects plant cells from the harm of the excessive activation of defense response. These findings indicate that the induction of RLK protein levels by their ligands may be a widespread phenomenon.
DMI2 Protein Level Is at the Center of Regulating Nodule Development and Rhizobial Initiation
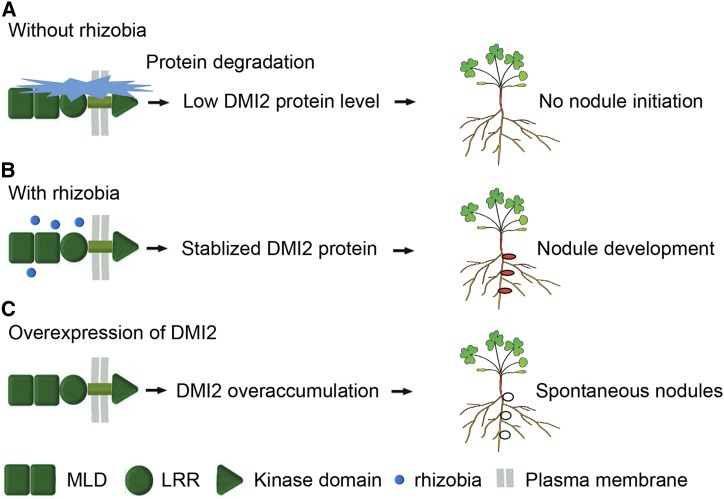
It is broadly known that, when growing in environments with abundant nitrogen, legume plants do not make nodules, despite the presence of rhizobia. Similarly, when nodulating plants are provided with bioavailable nitrogen, the plant will shut down the nodules immediately in order to save energy and resources. Hypernodulation mutants, which generate many more nodules than the wild type, are reported to have severe growth defects, suggesting that excessive nodules have negative effects on the fitness of plants (Schnabel et al., 2005). Understanding how plants maintain the ability to make the right number of nodules at the right time is a great interest of the research community. Our results show that the DMI2 protein accumulates during rhizobial inoculation. Taken together with reports that overexpressing the kinase domain or full-length DMI2/SYMRK results in spontaneous nodulation or a hypernodulation phenotype in legumes (Ried et al., 2014; Saha et al., 2014), we conclude that the protein level of DMI2/SYMRK is a master determinate signal of nodule development (Fig. 4).
Figure 4.
Plants control nodule development through fine regulation of DMI2 protein level. A, Without the presence of rhizobia, DMI2 protein is constitutively degraded by the proteasome apparatus and the nodule development signaling pathway is suppressed. B, Rhizobia inoculation could block the degradation of DMI2 protein, and subsequently, stabilized DMI2 could initiate nodule development signaling; thus, plants could develop functional nodules. C, When full-length DMI2 protein or the kinase domain is overexpressed, overaccumulated DMI2 protein can activate the nodule development signaling pathway without the presence of rhizobia, leading to a spontaneous nodulation phenotype. Red ellipses in B indicate functional nodules, and the white ellipses in C represent spontaneous nodules.
Our results show that the induction of DMI2 protein is independent of Nod factor reception (Fig. 1C), pointing to the possibility that there is another hidden signaling pathway used by rhizobia to regulate DMI2 protein levels, likely through blocking protein degradation. It has been reported that DMI2/SYMRK interacts with several E3 ligases in L. japonicus, like SINA4 and SIE3 (Den Herder et al., 2012; Yuan et al., 2012). As these reports have shown that SINA4 and SIE3 could ubiquitinate DMI2/SYMRK, our results provide the necessary evidence that the protein level of DMI2/SYMRK is altered during rhizobia infection in a proteasome-dependent manner in vivo. To truly confirm that SINA4 and SIE3 are responsible for DMI2/SYMRK protein turnover, expressing DMI2/SYMRK in sina4 and sie3 mutants to investigate DMI2/SYMRK protein dynamics is necessary.
Since DMI2 also is required for the symbiosis between plants and AM fungi and Frankia bacteria, it will be interesting to find out whether the protein level of DMI2 is induced by AM treatment. We would predict that this is the case, because we found that the induction of DMI2 protein by rhizobia is independent of Nod factor perception (Fig. 1C). Furthermore, any differences in DMI2 protein dynamics between rhizobia and AM infection might be related to how plants distinguish these two different symbionts, even though they utilize a shared symbiosis signaling pathway.
MLD Is Required for the Proper Folding of DMI2 Protein
The extracellular part of DMI2 contains an MLD domain and an LRR. Considering that the MLDs reside at the extracellular domain of RLKs in plants, it is speculated that it may directly recognize some microbial signals or signals generated from plant-microbe interaction and, subsequently, control the proper activation of the kinase domain. We found that the MLD is at least required for the proper folding of DMI2 protein because, after we introduced amino acid substitutions into the MLD, plants could not generate full-length DMI2 protein even in the presence of the proteasome inhibitor MG132 or rhizobia inoculation (Fig. 3, C and D). Human Malectin protein is reported to function in the ER (Schallus et al., 2008). Through its affinity to carbohydrate molecules, human Malectin protein could bind to misfolded proteins and activate ER quality control signaling (Qin et al., 2012). From our results, we speculate that DMI2-MLD also may function in the ER quality control process to guide the proper folding and successful secretion of the protein of which MLD itself is a part. The exact ligand for DMI2-MLD has yet to be found, but if this hypothesis is true, the likely scenario is that DMI2-MLD could bind to some carbohydrate ligands similar to diglucosylated Glc2Man9GlcNAc2 during protein folding in the ER lumen (Schallus et al., 2008). A carbohydrate microarray is required to find the exact ligand of DMI2-MLD.
By showing the protein-level dynamics of DMI2 during rhizobia inoculation and the altered protein behavior of DMI2 with amino acid substitutions in the MLD, we show the probable function of MLD and present a previously unidentified signaling pathway of how rhizobia affect DMI2 protein levels.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Medicago truncatula ecotype A17 was used for all the experiments in this study. Plants were grown under 16 h of light/8 h of dark at 22°C. Agrobacterium rhizogenes strain Arqua1 was used for plant hairy root transformation, and the procedure was conducted as described previously (Boisson-Dernier et al., 2001). Transgenic roots were selected based on antibiotic resistance against kanamycin at the concentration of 15 μg mL−1 in Fahraeus medium for 10 d.
Sinorhizobium meliloti strain ABS7 hemA:LacZ was used for rhizobia inoculation. Fresh overnight rhizobia culture was centrifuged and suspended in one-half-strength basic nodulation medium to a concentration of OD600 = 0.05. Five milliliters of liquid rhizobia culture was used per Medicago plant. Nodulation and infection thread phenotypes were checked 14 and 7 d post inoculation, respectively; for protein sample collection, tissues were collected at different time points, as indicated in the figure legends.
Evaluation of Symbiotic Phenotypes
To analyze the nodule development phenotype, 14 d after ABS7 hemA:LacZ inoculation, plants were harvested and total and pink nodules were counted. For infection thread analysis, 7 d post ABS7 hemA:LacZ inoculation, Medicago roots were stained for β-galactosidase activity as described previously (Pan et al., 2016). Root samples were filled with staining buffer (0.5 m sodium phosphate buffer, pH 7.2, 10% (v/v) Triton X-100, 100 mm potassium ferrocyannide, and 100 mm potassium ferricyanide) containing 2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, then incubated at 37°C overnight. Infection threads were checked, and photographs were taken using a Nikon E200 microscope. The number of infection threads was counted on at least 10 individual transformed plants for each construct.
Plasmids and Vectors
pKGW-RR::gDMI2-HAST was used to express wild-type gDMI2-HAST in plants stably and transiently (Riely et al., 2013). To make point mutations in the MLD, an overlapping PCR method was used to generate desired amino acid substitutions in pENTER::gDMI2-HAST (Heckman and Pease, 2007). After sequencing to confirm that the sequences were correct, the gDMI2-HAST sequences containing various point mutations in the MLD were introduced into the pKGW-RR vector using an LR recombination kit (Invitrogen). Then, the plasmids containing the point mutations were transformed into A. rhizogenes strain Arqua1 by electrotransformation.
Protein Extraction and Immunoblot
To extract proteins from uninoculated roots, inoculated roots, and nodules of Medicago plants, plant tissues were collected at the time points indicated in the figure legends and put into liquid nitrogen immediately. After grinding the tissues into a fine powder, proteins were extracted using native extraction buffer 1 (50 mm Tris-MES, pH 8, 0.5 m Suc, 1 mm MgCl2, 10 mm EDTA, and 5 mm DTT) with or without the protease inhibitor cocktail (Sigma-Aldrich; Liu et al., 2010). Extracted proteins were boiled for 5 min with 6× SDS sampling buffer and stored at −20°C.
For immunoblots, various protein samples were loaded onto a 12% (w/v) acrylamide SDS running gel, and the protein samples were transferred from the gel to nitrocellulose membranes (Adventec) by electroblotting. After blocking the membrane with milk, the membrane was subjected to incubation with 1:500 diluted anti-HA antibody (New England Biolabs) and 1:4,000 diluted anti-α-tubulin antibody (Life Sciences). The secondary antibodies were anti-rat and anti-mouse, respectively (Life Sciences). As the secondary antibodies were conjugated with horseradish peroxidase, the membrane was treated with the detection reagent from Thermo Scientific (products 1859707 and 1859678). The bands on the membrane were visualized via a G-box machine (New England Biogroup).
Cell-Free Degradation Assay and MG132 Treatment
Cell-free degradation assays were performed following previous reports (Spoel et al., 2009). Extracted protein samples were split into individual centrifuge tubes using equal amounts, different reagents were added into the tubes as indicated (MG132 concentration, 40 μm mL−1), and the tubes were shaken slowly at room temperature for 2 h. Individual samples were boiled with 6× SDS sampling buffer and subjected to immunoblot with anti-HA and anti-tubulin antibodies.
For MG132 in vivo treatments, 2-week-old Medicago plants were washed clean and put into distilled water containing 100 μm mL−1 MG132 at 22°C. Root samples were collected at different time points during MG132 treatment as indicated.
RNA Extraction and RT-qPCR
Total RNA extraction of rhizobia-inoculated or MG132-treated Medicago roots was carried out using TRIzol (Invitrogen) following the manufacturer’s instructions. To eliminate possible DNA contamination, extracted RNA was treated with the Turbo DNA-free kit (Life Science Technologies) using the manufacturer’s instructions. The iScript cDNA synthesis kit (Bio-Rad) was used for second chain synthesis following the manufacturer’s instructions step by step.
RT-qPCR was performed on the Eppendorf Mastercycler ep Realplex system. PCR was conducted using ExTaq polymerase (Takara) with SYBR Green dye (Thermo Fisher Scientific) in a 20-μL total volume: distilled, deionized water, 9.9 μL; 10× ExTaq buffer, 2 μL; 2.5 mm deoxyribonucleotide triphosphate mix, 2 μL; 10 μm primer mix, 2 μL; SYBR Green dye, 2 μL; template cDNA, 2 μL; ExTaq polymerase, 0.1 μL. For real-time PCR, annealing temperature was set at 60°C and elongation time was 30 s. When the PCR was finished, the melting curve was analyzed to rule out possible nonspecific amplification, and the result represented was the mean threshold cycle value of three technical replicates. Three biological replicates were used for each sample. PROTODERMAL FACTOR2 (locus identifier Medtr6g084690) was used as the internal control.
Protein Domain Analysis and Sequence Alignments
The gene expression analysis using existing databases was divided into two groups: for microarray analysis, the data were obtained from the Medicago truncatula Gene Expression Atlas (http://mtgea.noble.org/v3/; Benedito et al., 2008); for RNA sequencing analysis, the Medicago truncatula Genome Project version 4.0 (http://medicago.jcvi.org/medicago/index.php) and the symbimics Web site (https://iant.toulouse.inra.fr/symbimics/) were used (Young et al., 2011; Roux et al., 2014).
For protein structure prediction and conserved residue analysis, the SWISS-MODEL database (http://swissmodel.expasy.org/) was used, and the DMI2-MLD domain structure was predicted based on the structure of the human Malectin A domain. The positions of conserved amino acids were mapped to the predicted structure in the same program (Biasini et al., 2014).
The phylogenic tree was built using the MEGA 7.0 program under the developer’s instruction; all the sequences used are summarized in “Supplemental Data.”
Accession Numbers
Sequence data from this article can be found in Supplemental Table S1. The GenBank accession numbers for the genes used to build the phylogenetic tree also are presented there.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression of DMI2 in nodules compared with roots.
Supplemental Figure S2. Induction of DMI2 by rhizobia.
Supplemental Figure S3. DMI2 expression with rhizobia or Nod factor treatment.
Supplemental Figure S4. DMI2 level induction by rhizobia strains with defects in Nod factor synthesis.
Supplemental Figure S5. DMI2-HAST expression with MG132 treatment.
Supplemental Figure S6. DMI2 levels in rhizobia-inoculated Medicago roots treated with MG132.
Supplemental Figure S7. Predicted protein structure of two human Malectin A matches in DMI2-MLD.
Supplemental Figure S8. Alignment of DMI2-MLD with its closest plant homologs.
Supplemental Figure S9. Positions of amino acid substitutions in the DMI2-MLD protein sequence.
Supplemental Figure S10. Representative images showing infection thread development in wild-type and mutated MLD plants.
Supplemental Figure S11. DMI2-HAST expression in wild-type and mutated MLD plants.
Supplemental Figure S12. MLD presence or absence in DMI2 orthologs across species.
Supplemental Table S1. Accession numbers.
Acknowledgments
We thank Douglas Cook from the University of California, Davis, for sharing the gDMI2-HAST-pKGWRR plasmid and the dmi2-1 gDMI2-HAST Medicago seeds. We thank Sharon Long from Stanford University for the hcl-1, hcl-4, and nfp mutant seeds and Julia Frugoli from Clemson University for the sunn-1 seeds. We thank Alice Cheung, Heng-Ming Wu, and Dr. Minsoo Kim from the University of Massachusetts, Amherst, for providing critical comments on the article.
Footnotes
This project is funded by the USDA National Institute of Food and Agriculture AFRI award 2015-67013-22915.
Articles can be viewed without a subscription.
References
- Antolín-Llovera M, Petutsching EK, Ried MK, Lipka V, Nürnberger T, Robatzek S, Parniske M (2014a) Knowing your friends and foes: plant receptor-like kinases as initiators of symbiosis or defence. New Phytol 204: 791–802 [DOI] [PubMed] [Google Scholar]
- Antolín-Llovera M, Ried MK, Parniske M (2014b) Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr Biol 24: 422–427 [DOI] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Bersoult A, Camut S, Perhald A, Kereszt A, Kiss GB, Cullimore JV (2005) Expression of the Medicago truncatula DM12 gene suggests roles of the symbiotic nodulation receptor kinase in nodules and during early nodule development. Mol Plant Microbe Interact 18: 869–876 [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42: W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Kessler SA, Grossniklaus U (2011) The walls have ears: the role of plant CrRLK1Ls in sensing and transducing extracellular signals. J Exp Bot 62: 1581–1591 [DOI] [PubMed] [Google Scholar]
- Den Herder G, Yoshida S, Antolín-Llovera M, Ried MK, Parniske M (2012) Lotus japonicus E3 ligase SEVEN IN ABSENTIA4 destabilizes the symbiosis receptor-like kinase SYMRK and negatively regulates rhizobial infection. Plant Cell 24: 1691–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kaló P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Gherbi H, Markmann K, Svistoonoff S, Estevan J, Autran D, Giczey G, Auguy F, Péret B, Laplaze L, Franche C, et al. (2008) SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc Natl Acad Sci USA 105: 4928–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2: 924–932 [DOI] [PubMed] [Google Scholar]
- Hok S, Danchin EG, Allasia V, Panabières F, Attard A, Keller H (2011) An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant Cell Environ 34: 1944–1957 [DOI] [PubMed] [Google Scholar]
- Krishnakumar V, Kim M, Rosen BD, Karamycheva S, Bidwell SL, Tang H, Town CD (2015) MTGD: the Medicago truncatula Genome Database. Plant Cell Physiol 56: e1. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J (2010) Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabella R, Hartog M, Franken C, Geurts R, Bisseling T (2005) Expression pattern of DMI genes in Medicago nodules. In Wang Y.-P., Lin M., Tain Z.-X., Elmerich C., Newton W. E., eds, Biological Nitrogen Fixation, Sustainable Agriculture and the Environment. Springer, pp 153–155 [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Pan H, Oztas O, Zhang X, Wu X, Stonoha C, Wang E, Wang B, Wang D (2016) A symbiotic SNARE protein generated by alternative termination of transcription. Nat Plants 2: 15197. [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Frugoli JA, Smith LS, Long SR, Cook DR (2003) Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol 131: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin SY, Hu D, Matsumoto K, Takeda K, Matsumoto N, Yamaguchi Y, Yamamoto K (2012) Malectin forms a complex with ribophorin I for enhanced association with misfolded glycoproteins. J Biol Chem 287: 38080–38089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried MK, Antolín-Llovera M, Parniske M (2014) Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases. eLife 3: e03891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely BK, Larrainzar E, Haney CH, Mun JH, Gil-Quintana E, González EM, Yu HJ, Tricoli D, Ehrhardt DW, Long SR, et al. (2013) Development of tools for the biochemical characterization of the symbiotic receptor-like kinase DMI2. Mol Plant Microbe Interact 26: 216–226 [DOI] [PubMed] [Google Scholar]
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrère S, Sallet E, Courcelle E, Moreau S, et al. (2014) An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 77: 817–837 [DOI] [PubMed] [Google Scholar]
- Saha S, DasGupta M (2015) Does SUNN-SYMRK crosstalk occur in Medicago truncatula for regulating nodule organogenesis? Plant Signal Behav 10: e1028703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Dutta A, Bhattacharya A, DasGupta M (2014) Intracellular catalytic domain of symbiosis receptor kinase hyperactivates spontaneous nodulation in absence of rhizobia. Plant Physiol 166: 1699–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallus T, Jaeckh C, Fehér K, Palma AS, Liu Y, Simpson JC, Mackeen M, Stier G, Gibson TJ, Feizi T, et al. (2008) Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol Biol Cell 19: 3404–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, de Carvalho-Niebel F, Duc G, Frugoli J (2005) The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol 58: 809–822 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al. (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Wais RJ, Keating DH, Long SR (2002) Structure-function analysis of nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis. Plant Physiol 129: 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Zhu H, Gou H, Fu W, Liu L, Chen T, Ke D, Kang H, Xie Q, Hong Z, et al. (2012) A ubiquitin ligase of symbiosis receptor kinase involved in nodule organogenesis. Plant Physiol 160: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]