Many genes are preferentially up-regulated by shade stimulus in the vasculature of Arabidopsis thaliana cotyledons, some of which are regulated by newly synthesized auxin in the mesophyll and/or epidermis.
Abstract
Plants sense and respond to light via multiple photoreceptors including phytochrome. The decreased ratio of red to far-red light that occurs under a canopy triggers shade-avoidance responses, which allow plants to compete with neighboring plants. The leaf acts as a photoperceptive organ in this response. In this study, we investigated how the shade stimulus is spatially processed within the cotyledon. We performed transcriptome analysis on microtissue samples collected from vascular and nonvascular regions of Arabidopsis (Arabidopsis thaliana) cotyledons. In addition, we mechanically isolated and analyzed the vascular tissue. More genes were up-regulated by the shade stimulus in vascular tissues than in mesophyll and epidermal tissues. The genes up-regulated in the vasculature were functionally divergent and included many auxin-responsive genes, suggesting that various physiological/developmental processes might be controlled by shade stimulus in the vasculature. We then investigated the spatial regulation of these genes in the vascular tissues. A small vascular region within a cotyledon was irradiated with far-red light, and the response was compared with that when the whole seedling was irradiated with far-red light. Most of the auxin-responsive genes were not fully induced by the local irradiation, suggesting that perception of the shade stimulus requires that a wider area be exposed to far-red light or that a certain position in the mesophyll and epidermis of the cotyledon be irradiated. This result was consistent with a previous report that auxin synthesis genes are up-regulated in the periphery of the cotyledon. Hence, auxin acts as an important intraorgan signaling factor that controls the vascular shade response within the cotyledon.
Plants under a canopy compete with neighboring plants for light by triggering various physiological responses, collectively known as shade-avoidance syndrome. For example, shade inhibits germination and promotes hypocotyl elongation, petiole elongation, hyponasty, and flowering (for review, see Franklin, 2008; Casal, 2013; Pierik and de Wit, 2014).
In the shade, plants are exposed to a relatively low red (R) to far-red (FR) light ratio because of the selective absorption of R light by plants in the upper canopy (Casal, 2013). Plants have a variety of photoreceptors with distinct spectral natures. Among them, phytochrome, which absorbs R (660 nm) and FR (730 nm) light, perceives the change in the R:FR ratio. Phytochrome photoconverts between the two forms, Pr and Pfr, and reaches the photoequilibrium state between these two forms. Thereby, the level of Pfr, which is biologically active, is reduced under low R:FR conditions to trigger the shade-avoidance response (Casal, 2013).
Among members of the phytochrome family, phytochrome B (phyB) is the major suppressor of the shade-avoidance response under high R:FR conditions. In addition, phyD and phyE contribute to the response (Franklin and Quail, 2010). The activated phytochromes bind to PHYTOCHROME INTERACTING FACTOR (PIF), which belongs to the basic helix-loop-helix transcription factor family and phosphorylate PIFs under high R:FR (Leivar and Quail, 2011). Conversely, phyB is converted back to the inactive Pr form under low R:FR, which leads to the accumulation of PIF proteins and the up-regulation of genes required for the shade response.
A variety of plant hormones have been proposed to be involved in the shade-avoidance response, including auxin (Tao et al., 2008), gibberellins (Beall et al., 1996), brassinosteroids (Kozuka et al., 2010), ethylene (Pierik et al., 2004), and jasmonate (Moreno et al., 2009), among others. Auxin is synthesized in two steps in the shade: TRP AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1; AT1G70560) first converts Trp to indole-3-pyruvic acid (Tao et al., 2008), and YUCCA (YUC) transforms indole-3-pyruvic acid into IAA, the major auxin (Mashiguchi et al., 2011; Won et al., 2011). The shade stimulus promotes the expression of YUC genes, including YUC2 (AT4G13260), YUC5 (AT5G43890), YUC8 (AT4G28720), and YUC9 (AT1G04180), which leads to hypocotyl and petiole elongation (Müller-Moulé et al., 2016). PIF4 (AT2G43010), PIF5 (AT3G59060) (Hornitschek et al., 2012), and PIF7 (AT5G61270) (Li et al., 2012) are responsible for the shade-induced YUC expression. Thereby, the level of auxin is increased by the shade stimulus. In addition to YUCs, the expression of PIN-FORMED (PIN) genes, which encode auxin efflux carriers, are also increased at a low R:FR (Devlin et al., 2003), probably to promote hypocotyl elongation (Keuskamp et al., 2010; Kohnen et al., 2016).
The transcriptome responses to the shade stimulus have been studied in different organs or parts of an organ. The responses of the leaf blade and petiole of Arabidopsis (Arabidopsis thaliana) rosette leaves to the shade were compared by DNA microarray analysis (Kozuka et al., 2010). More recently, the shade responses were compared between the cotyledons and shoot apex (Nito et al., 2015), and between cotyledons and the hypocotyl (Kohnen et al., 2016) by RNA sequencing (RNA-seq) analysis. Consistent with the well-accepted view that auxin biosynthesis is increased in response to the shade stimulus (Fletcher and Zalik, 1964; Casal, 2013), many auxin-responsive genes are up-regulated in different organs in somewhat different manners. However, the physiological consequence of auxin accumulation can vary depending on the organ (de Wit et al., 2015).
Spotlight irradiation has been adopted to investigate interorgan communication in the shade-avoidance response. For example, irradiation of cotyledons affects stem elongation in cucumber (Cucumis sativus; Black and Shuttleworth, 1974), whereas photoperception by the leaf and stem are both effective in mustard (Casal and Smith, 1988). Likewise, signals are transferred from the shoot to the root (Correll and Kiss, 2005; Van Gelderen et al., 2018). Furthermore, spotlight irradiation of cotyledons has been shown to induce hypocotyl elongation and the expression of many auxin-responsive genes in the hypocotyl (Tanaka et al., 2002; Procko et al., 2014; Nito et al., 2015). Similarly, local irradiation of the rosette leaf blade promotes petiole elongation (Kozuka et al., 2010), whereas leaf tip irradiation triggers leaf hyponasty (Michaud et al., 2017; Pantazopoulou et al., 2017).
Taken together, the shade stimulus is considered to increase local auxin production in photoperceptive organs, such as cotyledons and leaf blades, by up-regulating the auxin biosynthesis genes, YUCs. The newly produced auxin is then basipetally transported to the hypocotyl and petiole to induce elongation growth locally. However, there appear to be other layers of regulation. For example, local production of auxin in the hypocotyl itself also contributes to the hypocotyl elongation response (Kohnen et al., 2016). In addition, auxin metabolism in the target site plays a role in the regulation of elongation growth (Zheng et al., 2016). Hence, the shade stimulus appears to affect auxin in various ways in different parts of the plant.
Organs consist of multiple tissues. For instance, Endo et al. (2014) indicated that ∼77% of leaf mRNA is derived from mesophyll cells. Hence, analysis of the whole cotyledon mainly reflects the mesophyll response but not the other minor tissues such as the vasculature and epidermis. One approach to circumvent this problem is to isolate the tissue. The vasculature of Arabidopsis cotyledons has been isolated to demonstrate that FT (AT1G65480) is up-regulated in this tissue in response to the shade stimulus (Endo et al., 2005). More recently, the epidermal peel was used to examine the coregulation of auxin and brassinosteroid by the shade stimulus in Brassica hypocotyl (Procko et al., 2016).
Another approach to investigate intertissue communication is to locally express a certain component of light signal transduction, such as phytochrome itself, and observe the physiological consequence. Rousseaux et al. (1997) overexpressed phyA in different tissues to analyze leaf senescence and stem elongation responses to FR. Likewise, tissue-specific phyB-GFP-expressing lines have been used to determine the photoperceptive sites for the regulation of hypocotyl elongation, flowering, and stomatal development (Endo et al., 2005; Casson and Hetherington, 2014). In the hypocotyl gravitropism experiment, a variety of cell-specific phyB-expressing lines were generated to show that epidermal phyB promotes PIF degradation in the endodermis (Kim et al., 2016). Procko et al. (2016) generated cell-specific auxin-blocking lines to demonstrate the importance of auxin metabolism in the epidermis.
Despite the above efforts, it remains unclear how the shade signal is spatially processed within a certain organ such as the cotyledon. In this study, we first compared the transcriptome responses to the shade stimulus in vascular and nonvascular regions of the cotyledon and found that more genes, including many auxin-responsive genes, were up-regulated in the vasculature relative to the mesophyll and epidermis. We then examined the autonomy of the vascular response by spotlight irradiation. The results suggested that perception of the shade stimulus and auxin production in a wider area of the cotyledon was required for the vascular response.
RESULTS
Isolation of Micro Samples from Arabidopsis Cotyledons
To investigate the spatial regulation of the gene expression responses to the shade stimulus within cotyledons, micro samples were prepared from two distinct regions of cotyledons of 4-d-old Arabidopsis seedlings using a needle-based device (Kajiyama et al., 2015; Nito et al., 2015; Fig. 1). The MN (mesophyll-enriched, prepared with a needle) sample of ∼100 μm in diameter was excised from a nonvascular region in the cotyledon. The VN (vasculature-inclusive, prepared with a needle) sample was prepared in a similar manner from the central and more basal vascular region. Unfortunately the MN and VN samples were isolated from different locations in the cotyledon. Hence, a gene expressed in VN but not in MN might be expressed in mesophyll and/or epidermis in a specific position of the cotyledon rather than in the vasculature. However, the vasculature patterns varied between cotyledons and the mesophyll-enriched and vasculature-enriched tissue pieces could only be prepared from these locations at a reasonable success rate. To partially circumvent this problem, the vasculature was mechanically isolated (VS; vasculature, prepared by sonication) by ultrasonication from enzyme-treated cotyledons (Endo et al., 2005). Microscopy observation revealed that MN pieces mainly consisted of mesophyll and epidermis, whereas VN additionally contained vascular tissues. It was also confirmed that the mesophyll and epidermis were efficiently removed from VS.
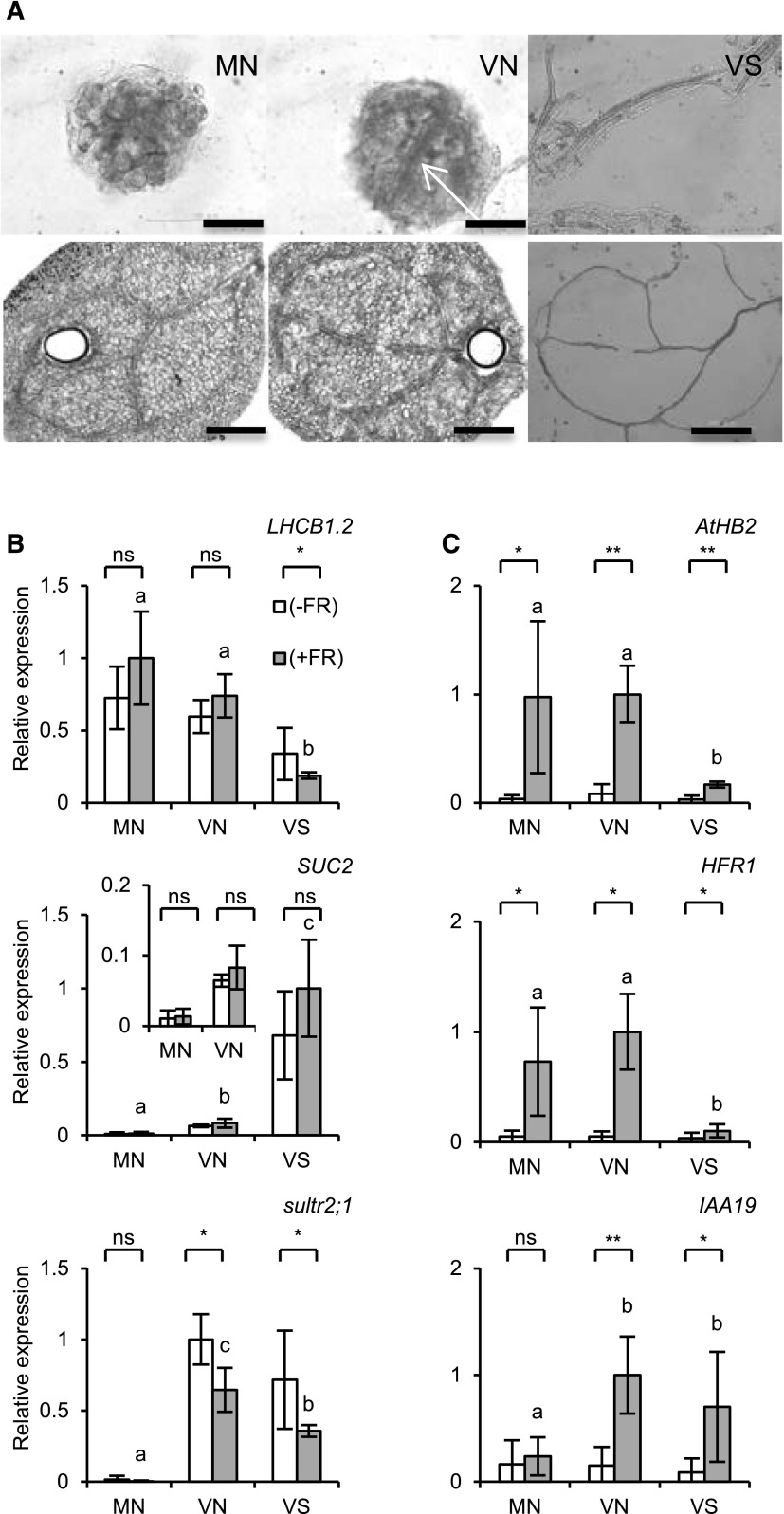
Figure 1.
Micro samples isolated from Arabidopsis cotyledons. A, Photographs of MN, VN, and VS micro samples collected from 4-d-old Arabidopsis cotyledons. MN and VN pieces of ∼100 μm in diameter (upper panel) were excised from the distal nonvascular and basal vascular regions of the cotyledon, respectively, with a needle-based device (lower panels). A hole much larger than the tissue piece was left because of the thickness of the needle cylinder wall. The arrow indicates the vasculature contained in VN. VS was isolated from whole cotyledons by enzyme treatment and ultrasonication. Bars = 50 and 200 μm in the upper and lower panels, respectively. B, Tissue marker gene expression measured by RT-qPCR in MN, VN, and VS. The 4-d-old seedlings were treated with (gray bar) or without (white bar) EOD-FR. LHBC1.2, SULTR2;1, and SUC2 are mesophyll, phloem and xylem parenchyma, and phloem companion cell markers, respectively. TUB2/TUB3 was used to standardize the results (for details, see “Materials and Methods”). Four microsample pieces were combined in a tube and subjected to cDNA synthesis (n ≥ 4, mean ± sd). Asterisks indicate significant differences for Student’s t test (**, P < 0.005; *, P < 0.05; ns, nonsignificant). Different letters denote significant differences between the FR-treated samples (P < 0.05, Student’s t test). The expression levels are shown in arbitrary units setting the highest value as 1. C, Gene expression responses of typical shade-responsive genes, ATHB2, HFR1, and IAA19, to EOD-FR. Experimental conditions and symbols are as described for B.
The expression levels of a housekeeping gene were then examined in those micro samples. In this work, the end-of-day FR (EOD-FR) treatment was employed to induce the shade-avoidance response (Kasperbauer, 1971; López-Juez et al., 1990). The 4-d-old seedlings were treated with or without EOD-FR of 50,000 μmol m−2 at the end of the fourth day of growth. The seedlings were further incubated for 2 h in darkness before the micro sample preparation. The cDNA sample was then synthesized from four micro pieces combined in a tube and subjected to reverse transcription-quantitative PCR (RT-qPCR) analysis after two rounds of amplification (Kajiyama et al., 2015). The expression levels of highly homologous housekeeping genes, TUBULIN BETA CHAIN2 (TUB2; AT5G62690) and TUB3 (AT5G62700) (Czechowski et al., 2005), were collectively assessed (TUB2/3 hereafter) with a common primer set. The TUB2/3 levels per unit cDNA varied to some extent and were higher in the order of VS, VN, and MN (Supplemental Fig. S1).
Expression of Tissue Marker and Shade-Responsive Genes in the Micro Samples
We examined the expression of tissue marker genes in the micro samples (Fig. 1). For this purpose, we employed TUB2/3 as an internal standard to estimate the total mRNA levels in the samples. However, the RT-qPCR and RNA-seq analysis (Supplemental Fig. S1) revealed that TUB2/3 expression per unit cDNA varied depending on the sample type (see below). Hence, we corrected the TUB2/3 values accordingly when they were used as internal controls (see “Materials and Methods” for details).
The expression of a mesophyll marker gene, LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN (LHCB1.2; AT1G29910; Susek et al., 1993), per unit cDNA was much higher in MN and VN than in VS regardless of the light condition (Fig. 1). The higher expression in MN and VN was consistent with the microscopy observation that both MN and VN contained mesophyll as a major component. Since the LHCB1.2 level was very similar between MN and VN, the mesophyll was suggested to be a major tissue even in VN. By contrast, LHCB1.2 was lower in VS, which is consistent with the observation that mesophyll was removed from VS (Fig. 1). However, VS might be contaminated with photosynthetic cells, although they were not clearly observed microscopically (Fig. 1) because LHCB1.2 expression was detected in VS to some extent.
The vascular marker gene, SUC-PROTON SYMPORTER2 (SUC2; AT1G22710), which is specifically expressed in phloem companion cells (Ivashikina et al., 2003), was expressed most highly in VS regardless of the light conditions (Fig. 1), which was not surprising because VS consisted almost entirely of vasculature whereas a large portion of VN was occupied by nonvascular tissues such as mesophyll and epidermis. Second, the expression was significantly higher in VN than in MN, indicating that the vasculature was indeed excluded from MN (Fig. 1). Unlike SUC2, another vascular marker, SULFATE TRANSPORTER2;1 (SULTR2;1; AT5G10180), which is expressed in both xylem parenchyma and phloem cells in leaves (Takahashi et al., 2000), was expressed equally in VN and VS despite the much higher proportion of vasculature in VS (Fig. 1). Hence, a substantial part of the xylem parenchyma might have been removed from VS probably during the ultrasonication treatment.
We then examined the expression of some of the known shade-responsive genes, such as ARABIDOPSIS THALIANA HOMEOBOX PROTEIN2 (ATHB2; AT4G16780), LONG HYPOCOTYL IN FAR-RED (HFR1; AT1G02340), and INDOLE-3-ACETIC ACID INDUCIBLE19 (IAA19; AT3G15540; Steindler et al., 1999; Devlin et al., 2003; Sessa et al., 2005) in the micro samples (Fig. 1). ATHB2 and HFR1 are transcription factors whose expression is directly regulated by PIFs (Hornitschek et al., 2012) to represent the primary action of phytochrome. The auxin-responsive IAA19 gene was chosen as a representative of the downstream response (Iglesias et al., 2017). The expression levels of ATHB2 and HFR1 were substantially increased by EOD-FR, especially in MN and VN. Since these patterns were very similar to that of LHCB1.2 (Fig. 1), ATHB2 and HFR1 appeared to be preferentially induced in the mesophyll (and/or epidermis) rather than the vasculature. By contrast, IAA19 expression was increased only in VN and VS by EOD-FR (Fig. 1). The pattern resembled that of SULTR2;1, indicating that IAA19 responded in vascular tissues (most likely in xylem parenchyma).
Initial Characterization of the RNA-Seq Data
To compare the transcriptome responses to EOD-FR in different tissues, MN, VN, and VS samples were prepared from cotyledons of 4-d-old seedlings for the RNA-seq analysis. For this purpose, six micro pieces were combined in a tube for cDNA synthesis. The samples were prepared in duplicate from seedlings treated with or without EOD-FR. After cDNA synthesis, the samples were subjected to two rounds of amplification (Kajiyama et al., 2015). The sample quality was checked using a Bioanalyzer to confirm the proper size distribution. We then confirmed that the tissue marker genes were expressed properly in those samples (Supplemental Fig. S2). Finally, those samples were subjected to an RNA-seq analysis and yielded 5.92 × 106 reads, of which 73% on average were mapped (4.36 × 106).
We first compared the read per million (RPM) values for representative housekeeping genes such as TUB2, TUB3, ACT2 (AT3G18780), and UBC9 (AT4G27960; Hong et al., 2010) in the 12 samples (Fig. 2; Supplemental Fig. S3A). As expected, the expression was not affected by EOD-FR. However, the levels varied depending on the sample type. The mean RPM values for TUB2, TUB3, and UBC9 were higher in the order of VS, VN, and MN, which was consistent with the RT-qPCR results for TUB2/3 (Supplemental Fig. S1). In contrast, ACT2 was higher in MN and VN than in VS. Hence, none of the tested housekeeping genes exhibited constant expression levels across all sample types.
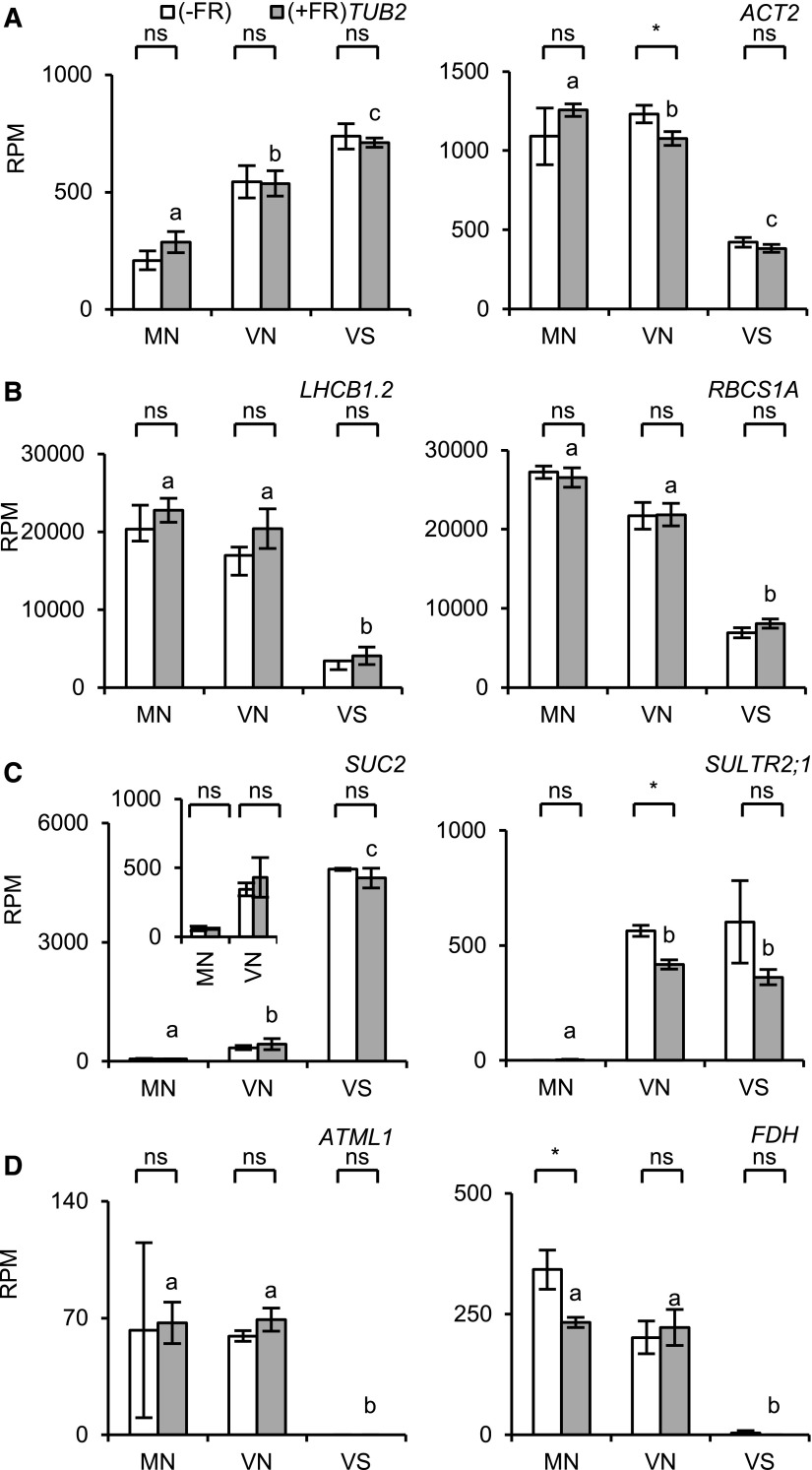
Figure 2.
Expression of housekeeping and tissue marker genes measured by RNA-seq in microsamples. The seedlings were light-treated as described for Figure 1 and subjected to RNA-seq analysis. The RPM values are shown. White bar, without EOD-FR; gray bar, with EOD-FR. The inset in C is a magnified view of SUC2 expression in MN and VN. Six microsample pieces were combined to synthesize the cDNA (n = 2, mean ± sd). Asterisks indicate significant differences for Student’s t test (*, P < 0.05; ns, nonsignificant). Different letters denote significant differences between the FR-treated samples (P < 0.05, Student’s t test). A, Housekeeping genes; B, mesophyll marker genes; C, vasculature marker genes; D, epidermis marker genes.
We then checked the tissue marker gene expression (Fig. 2, B–D; Supplemental Fig. S3, B–D). The mesophyll marker genes LHCB1.2. and RBCS1A (AT1G67090) were expressed at higher levels in MN and VN than in VS, as demonstrated by the RT-qPCR analysis (Fig. 1). However, the expression of LHCB2.3 (AT3G27690) was equally low in VN and VS, indicating that mesophyll tissues in MN and VN might have different characteristics, probably due to their positions in the cotyledon (Fig. 1).
Phloem marker genes, such as SUC2, SEOR1 (AT3G01680), and APL (AT1G79430) (Kondo et al., 2016; Yamaguchi et al., 2017), were expressed at much higher levels in VS and almost not at all in MN (Fig. 2; Supplemental Fig. S3D). Similarly, the xylem markers IRX3 (AT5G17420) and XCP1 (AT4G35350) were expressed most highly in VS (Supplemental Fig. S3C). Hence, the vasculature was highly concentrated in VS and efficiently excluded from MN. Strikingly, SULTR2;1 was equally expressed in VN and VS (Fig. 2), as shown by RT-qPCR (Fig. 1). We also checked the expression of the epidermis marker genes ATML1 (AT4G21750) and FDH (AT2G26250; Sessions et al., 1999; Takada et al., 2013), which were expressed at similar levels in MN and VN but not in VS (Fig. 2). Hence, the epidermis was equally included in MN and VN.
RNA-Seq Analysis of Shade-Signaling Genes
We checked the RPM values for the components of shade signal transduction (Casal, 2013) in the different sample types (Supplemental Fig. S4). Of five phytochrome genes, PHYA (AT1G09570), PHYB (AT2G18790), PHYC (AT5G35840), and PHYE (AT4G18130) were detected, whereas the expression of PHYD (AT4G16250) was very low in all the samples (<5). Basically, they were expressed ubiquitously in all the sample types. Hence, both the mesophyll/epidermis and vascular cells were suggested to be able to respond to the shade stimulus. As previously reported (Devlin et al., 2003), the expression levels of PHYA and PHYB were somewhat increased by FR treatment, at least in MN and VN. However, the expression of cryptochrome genes, CRY1 (AT4G08920) and CRY2 (AT1G04400), was more stable and not affected by EOD-FR.
Four PIF genes involved in shade avoidance, PIF3 (AT1G09530), PIF4, PIF5, and PIF7, were expressed in all sample types (Supplemental Fig. S4B). The COP1 (AT2G32950) and HY5 (AT5G11260) genes were also expressed ubiquitously (Supplemental Fig. S4D). None of those genes clearly responded to EOD-FR, although HY5 might be down-regulated to some extent in MN. In conclusion, major shade signaling components of shade signal transduction were expressed at similar levels in all sample types.
Selection of Light-Responsive Genes
To investigate the transcriptome response to EOD-FR (Supplemental Table S1), we selected genes that exhibited robust responses to EOD-FR. As a prefiltration step, we selected protein-coding genes that exhibited average RPM values >5 in at least one sample type (17,368 genes in total). We then chose genes that exhibited responses to EOD-FR in at least one sample type. The experiment was conducted in two light conditions (±FR) and on three samples (MN, VN, and VS; Supplemental Fig. S2). We selected genes with maximum expression among the three FR-treated conditions (+FR/MN, +FR/VN, and +FR/VS) that were more than 3 times higher than that in the three non-FR-treated conditions (-FR/MN, -FR/VN, and −FR/VS; Supplemental Fig. S5) to exclude genes that responded to EOD-FR in a certain sample type but were constitutively expressed at higher levels in other sample types. Among them, 241 genes that exhibited statistically significant responses (P < 0.1) were defined as up-regulated genes (Supplemental Table S2). In a similar manner, 209 down-regulated genes (1/3 > +FR/−FR, P < 0.1) were selected (Supplemental Table S3).
We performed a Gene Ontology (GO) enrichment analysis using the g:profiler website (http://biit.cs.ut.ee/gprofiler/; Reimand et al., 2016) to characterize the above 241 up-regulated gene sets (Supplemental Table S4). The enriched GO terms of lower orders (more specific) included “shade avoidance (GO:0009641),” “response to auxin (GO:0009733),” “cellular response to auxin stimulus (GO:0071365),” “auxin-activated signaling pathway (GO:0009734),” and “transcription factor activity, sequence-specific DNA binding (GO:0003700).” These terms were consistent with the general view that auxin plays an important role in the shade-avoidance response (Casal, 2013). Less characterized are the genes that are down-regulated by the shade stimulus. No GO term enrichment was observed for the 209 down-regulated gene set. As reported previously (Nito et al., 2015), down-regulation was expected to be more difficult to identify.
Clustering Analysis of Light-Responsive Genes
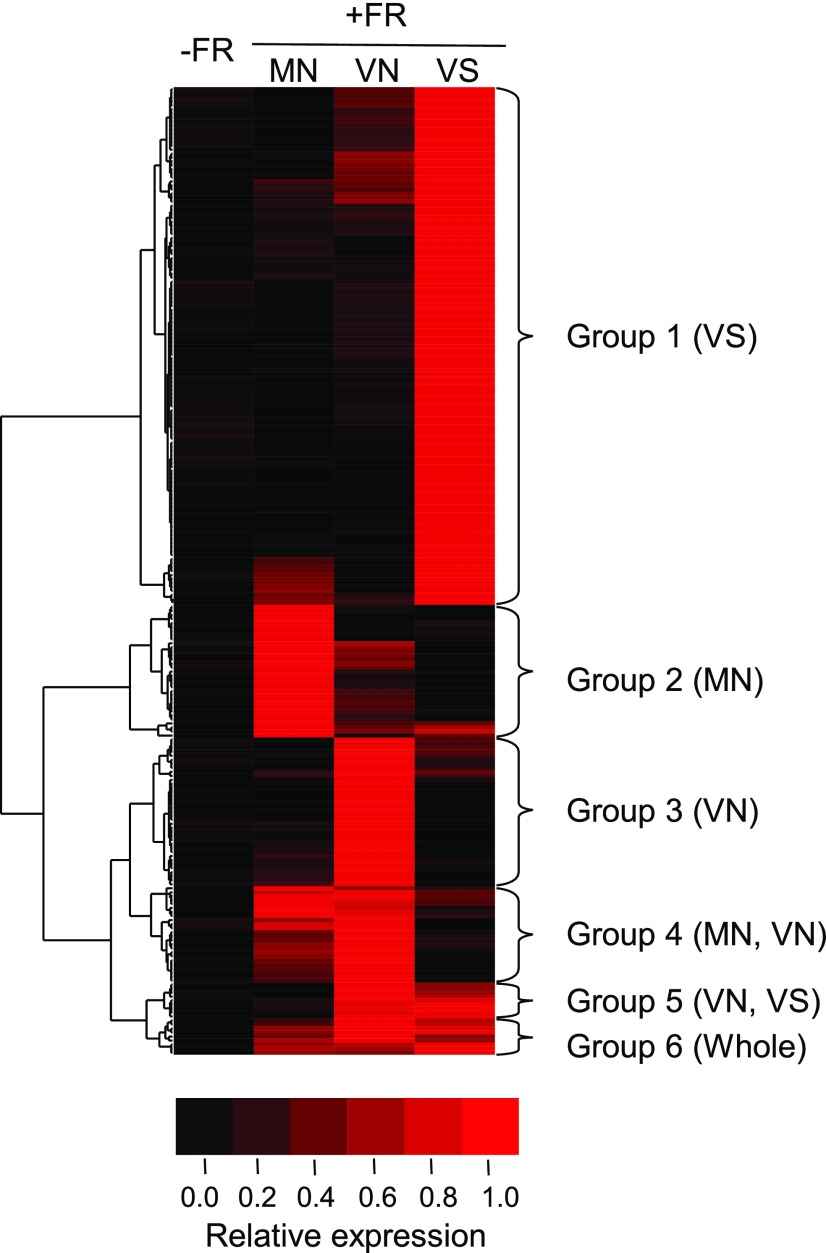
We performed cluster analysis of the above 241 up-regulated genes with Gene Cluster 3.0 (de Hoon et al., 2004). The genes were classified into six groups according to the response patterns in the different sample types (Fig. 3; Supplemental Table S2). Group 1 (VS) genes (129 genes) were up-regulated mainly in VS. This was the largest group accounting for 54% of the total up-regulated genes. Group 2 (MN) and Group 3 (VN) genes were preferentially expressed in MN and VN, respectively. They were the second largest, and each of them accounted for ∼15% of the up-regulated genes. The other three groups, Groups 4 (MN and VN), 5 (VN and VS), and 6 (whole), were more minor and accounted for approximately the remaining 15% of the genes in total.
Figure 3.
Cluster analysis of the up-regulated genes. The 241 up-regulated genes were clustered using Cluster 3.0 software into six groups according to the response patterns in the different sample types. For this analysis, the average expression levels of +FR/MN, +FR/VN, +FR/VS, and −FR (data from all three tissue types were combined) were calculated and normalized by setting the highest value to 1. Brighter red indicates higher expression.
To characterize each group, we performed GO enrichment analysis (Reimand et al., 2016). Among the six groups, only Groups 1 (VS), 3 (VN), and 4 (MN, VN) exhibited enrichment (Supplemental Table S4). Terms related to auxin were enriched in Group 1 (VS) and 3 (VN), whereas Group 4 (MN and VN) was enriched with the term “response to far-red light (GO: 0010218).” We combined the vascular groups, Groups 1 (VS) and 5 (VN and VS), and subjected the combined group to GO term enrichment analysis and confirmed the enrichment of auxin-related GO terms (Supplemental Table S4). Group 3 (VN) was not included in the vascular group because the Group 3 genes could be expressed in the mesophyll/epidermis in a specific position of the cotyledon rather than in the vasculature.
We also examined the classification of known shade-responsive genes in the above analysis (Supplemental Table S5). The 53 genes were reported in at least three out of five literatures as genes up-regulated by shade (Nito et al., 2015). They were up-regulated in different types of samples such as whole seedlings and rosette leaf blades. Among those 53 core shade-responsive genes, 22 were found in the present up-regulated gene set. Of these, 16 genes (72%) were classified into Group 2 (MN) or 4 (MN and VN), indicating that previously known shade-responsive genes tended to be expressed in the mesophyll/epidermis. This tendency was confirmed by a similar analysis with 98 core up-regulated genes reported by Sellaro et al. (2017). Among 11 overlapping genes, nine belonged to the mesophyll/epidermis group consisting of Groups 2 (MN), 4 (MN and VN), and 6 (whole), whereas only one belonged to the vascular group (Groups 1 and 5). Consistent with this view, the GO term “shade avoidance (GO: 0009641)” was enriched in the mesophyll/epidermis group (Supplemental Table S4).
Although no particular GO term was enriched in the 209 down-regulated genes, we attempted to classify them by cluster analysis (Supplemental Fig. S6). Among them, 143 genes (68% of down-regulated genes) were preferentially expressed and down-regulated by EOD-FR in VS. We also observed several other groups with different response patterns. However, no GO term was enriched in any of those groups. Hence, the significance of the down-regulated genes remained unclear.
Confirmation of the RNA-Seq Results by RT-qPCR
To evaluate the reliability of the RNA-seq analysis, 22 genes were selected from each group and examined by RT-qPCR. Those genes included nine core shade-responsive genes, five auxin-related genes that showed strong responses to the shade (>10-fold), five abscisic acid-related genes, and three cell wall-related genes. Samples were freshly prepared (n = 3 or 4) in addition to the original RNA-seq samples (n = 2). Proper expression levels of TUB2/3 and tissue marker genes were confirmed after the first and second round of cDNA amplification, respectively. Among the 22 genes selected from the 241 up-regulated genes (Supplemental Table S2), 17 genes exhibited a significant light response (P < 0.05; Fig. 4; Supplemental Fig. S7). Three genes apparently responded to EOD-FR, although the response was not statistically significant; the deviations were too large in several conditions to draw any conclusion for the remaining two genes.
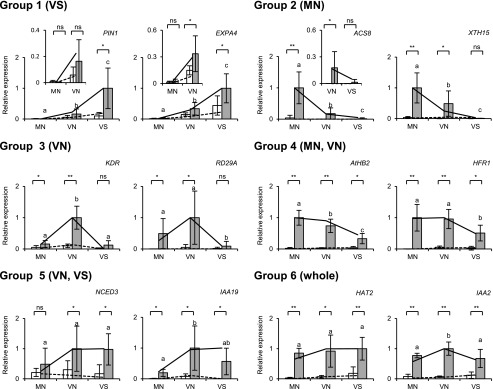
Figure 4.
Confirmation of the RNA-seq results by RT-qPCR. Representative genes were selected from each of the six groups shown in Figure 3 and examined by RT-qPCR. The samples included the original samples for RNA-seq (n = 2) and freshly prepared samples (n = 3 or 4). The seedlings were light-treated as described in Figure 1. TUB2/TUB3 was used to standardize the results (mean ± sd). Asterisks indicate significant differences for Student’s t test (**, P < 0.005; *, P < 0.05; ns, nonsignificant). Different letters denote significant differences between the FR-treated samples (P < 0.05, Student’s t test). White bar, without EOD-FR; gray bar, with EOD-FR. The insets are magnified views of the expression in MN and VN, or VN and VS. RNA-seq results (Supplemental Table S2) with (solid line) and without EOD-FR (broken line) are shown for comparison. The expression levels are shown in arbitrary units setting the highest value to 1.
The tissue-specific response patterns were generally reproducible (Fig. 4; Supplemental Fig. S7). Five genes representing Group 1 (VS), including three auxin-responsive genes (Goda et al., 2008), LAX2 (AT2G21050), IAA30 (AT3G62100), and IAA1 (AT4G14560), were induced most strongly in VS, excluding IAA1. The response of PIN1 (AT1G73590) in particular was observed almost exclusively in VS. Genes representing Groups 3 (VN) and 5 (VN and VS) were also confirmed to be up-regulated preferentially in the respective sample types, including KDR (AT1G26945; Group 3), RD29A (AT5G52310; Group 3), NCED3 (AT3G14440; Group 5), and IAA19 (Group 5). Likewise, the pattern was also reproduced for the genes mainly induced in the mesophyll/epidermis, including ACS8 (AT4G37770; Group 2), XTH15 (AT4G14130; Group 2), ATHB2 (Group 4), HFR1 (Group 4), IAA29 (AT4G32280; Group 4), GH3.3 (AT2G23170; Group 4), HAT2 (AT5G47370; Group 6), and IAA2 (AT3G23030; Group 6).
Expression Patterns of Auxin-Related Genes in the RNA-Seq Data
Through the GO term enrichment analysis, we found that there were many auxin-related genes among the up-regulated genes (Supplemental Table S4). Hence, we expanded our analysis and checked the expression levels of all genes reported as auxin-related (Iglesias et al., 2017) in the RNA-seq data (Supplemental Fig. S8). About half of the auxin synthesis genes were upregulated by shade in the different sample types, whereas the other half were down-regulated. Some genes, such as YUC8 and YUC9, increased in MN, while YUC3 (AT1G04610) and YUC4 (AT5G11320) increased in VN and VS, respectively. Some of the auxin metabolism genes, including GH3s, were up-regulated mainly in VN, whereas the others did not show clear shade responses. These GH3 genes were known to conjugate excess IAA with several amino acids by homeostasis (Staswick et al., 2005). With respect to the auxin transport genes, up-regulated, down-regulated, and nonregulated genes were observed almost equally.
We also checked the expression of TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB) genes, AUXIN RESPONSE FACTOR (ARF) genes, AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) genes, and SMALL AUXIN UP RNA (SAUR) genes separately. TIR1/AFBs are components of the ubiquitin E3 ligase, which binds to auxin and promotes degradation of IAAs (Parry et al., 2009; Calderón Villalobos et al., 2012). IAAs suppress the auxin response by inhibiting ARFs, which promote the expression of auxin-responsive genes (Chapman and Estelle, 2009). SAURs were originally identified as auxin-inducible small RNAs (McClure and Guilfoyle, 1987). The subfamily SAUR19-24 was reported to promote cell expansion by preventing inactivation of the plasma membrane proton pump (Spartz et al., 2014).
Most of the TIR1/AFB and ARF genes did not respond to shade. Interestingly, many ARF genes were expressed mainly in VS. About half of the IAA genes, including IAA19 and IAA29, were up-regulated by shade mainly in VN or VS. However, about one-third of the IAAs were down-regulated, albeit less prominently. In the case of SAURs, almost an equal number of up-regulated and down-regulated genes were found. It was noteworthy that up-regulation of SAURs was observed more in VN and VS than in MN. In summary, both up- and down-regulations were observed in auxin-related genes. Consistent with the GO term enrichment analysis (Supplemental Table S4), the up-regulation was observed more in VN and VS than in MN.
RT-qPCR Analysis of YUC Gene Expression
The RNA-seq analysis demonstrated that the sites of auxin synthesis and responses did not necessarily match (Supplemental Fig. S8). To gain further insights into this phenomenon, we reexamined the expression of auxin-synthesis YUC genes in the different sample types by RT-qPCR (Supplemental Fig. S9). As reported previously (Müller-Moulé et al., 2016), YUC8 and YUC9 were induced by the shade stimulus. They responded preferentially in MN as suggested by the RNA-seq analysis. Since the expression in VN was much lower than that in MN, the position within the cotyledon might be important for their expression. This result is consistent with the reports that the GUS reporter gene expression is uniformly observed in the periphery of the rosette leaf (Chen et al., 2016; Müller-Moulé et al., 2016).
In addition, we examined YUC2, YUC3, Y UC4, and YUC5 expression because YUC2, YUC3, and YUC5 have been reported to be up-regulated by shade (Tao et al., 2008; Li et al., 2012; Nito et al., 2015; Goyal et al., 2016; Kohnen et al., 2016; Müller-Moulé et al., 2016), while YUC4 responded to shade in this RNA-seq analysis (Supplemental Fig. S8). However, no clear shade response was observed by the RT-qPCR analysis (Supplemental Fig. S9). In summary, the mesophyll/epidermis appeared to be a major site of shade-induced auxin biosynthesis in this study, although the auxin response was more prominent in the vasculature.
Spotlight Treatment of the Cotyledon
As described above, the sites of auxin synthesis and responses did not appear to match. To gain insights into this phenomenon, we conducted spotlight irradiation experiments, which were successfully applied in a previous work to analyze interorgan communications between cotyledons and the shoot apex (Nito et al., 2015). First, we confirmed that phytochrome was converted from Pfr to Pr by FR spotlight irradiation only in the irradiated area (Supplemental Fig. S10). For this purpose, we used the Bpro transgenic plant, which expresses a phyB-GFP protein under the control of its own promoter (Endo et al., 2005). The phyB-GFP forms speckles in the nucleus upon photoactivation (Kircher et al., 2002; Nagatani, 2004), whereas the speckles disappear upon activation by EOD-FR (Nito et al., 2015)
The right side of the cotyledon of 4-d-old Bpro seedlings grown under white light was treated with an FR spotlight guided through an acrylic fiber with a diameter of 250 μm. As a control, the whole seedling was irradiated with FR. After 30 to 40 min of incubation in the dark, the seedlings were subjected to microscopy observation. We confirmed that phyB-GFP speckles disappeared only in the irradiated area but not in the nonirradiated area on the other side (Supplemental Fig. S10). The rate of nuclei with speckles within the nonirradiated area was 78.6%, but it dropped to 3.3% in the irradiated area.
The Response of Auxin-Responsive Genes to Spotlight Irradiation
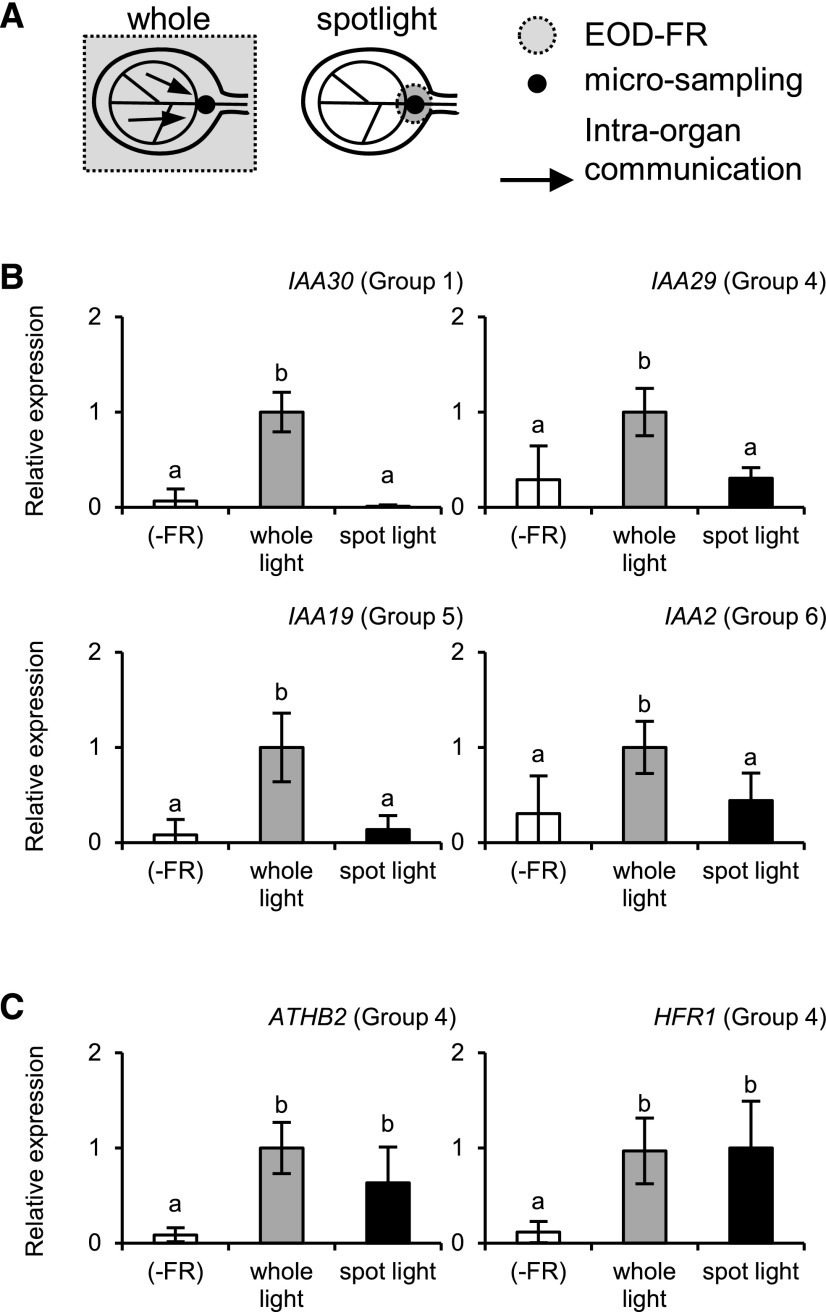
To examine how auxin-responsive genes respond to local FR irradiation, the vascular region in the cotyledon was treated with an FR spotlight, and the VN micro sample was collected from the irradiated area (Fig. 5). As a control, the whole seedling was irradiated. In this experiment, we examined six typical auxin-responsive genes, IAA1, IAA30, IAA29, GH3.3, IAA19, and IAA2 (Supplemental Table S2). Four genes were induced more efficiently in VN by whole-seedling irradiation and only weakly by local spotlight irradiation (Fig. 5). In the case of IAA1 and GH3.3, the experimental error was too large to draw a clear conclusion. By contrast, ATHB2 and HFR1, with auxin-independent responses (Li et al., 2012; Nito et al., 2015), were fully induced even by local irradiation (Fig. 5). Hence, the shade responses of the auxin-responsive genes in the vascular region were controlled by the global perception of the shade stimulus rather than the local perception.
Figure 5.
Gene expression responses to spotlight EOD-FR irradiation in the wild type (accession Landsberg erecta). A, Diagram explaining the spotlight and whole-seedling irradiation of a cotyledon. The VN micro sample was excised from the irradiated area for RT-qPCR analysis. B and C, Gene expression responses to the whole-seedling (gray bar) or spotlight (black bar) EOD-FR irradiation in VN. Seedlings not treated with EOD-FR were used as controls (white bar). The seedlings were light-treated as described for Figure 1, except that two micro sample pieces were combined in a tube for cDNA synthesis. TUB2/TUB3 was used to standardize the results. The expression levels are shown in arbitrary units setting the highest value to 1 (n ≥ 4, mean ± sd). Different letters denote significant differences between the samples (P < 0.05, Student’s t test).
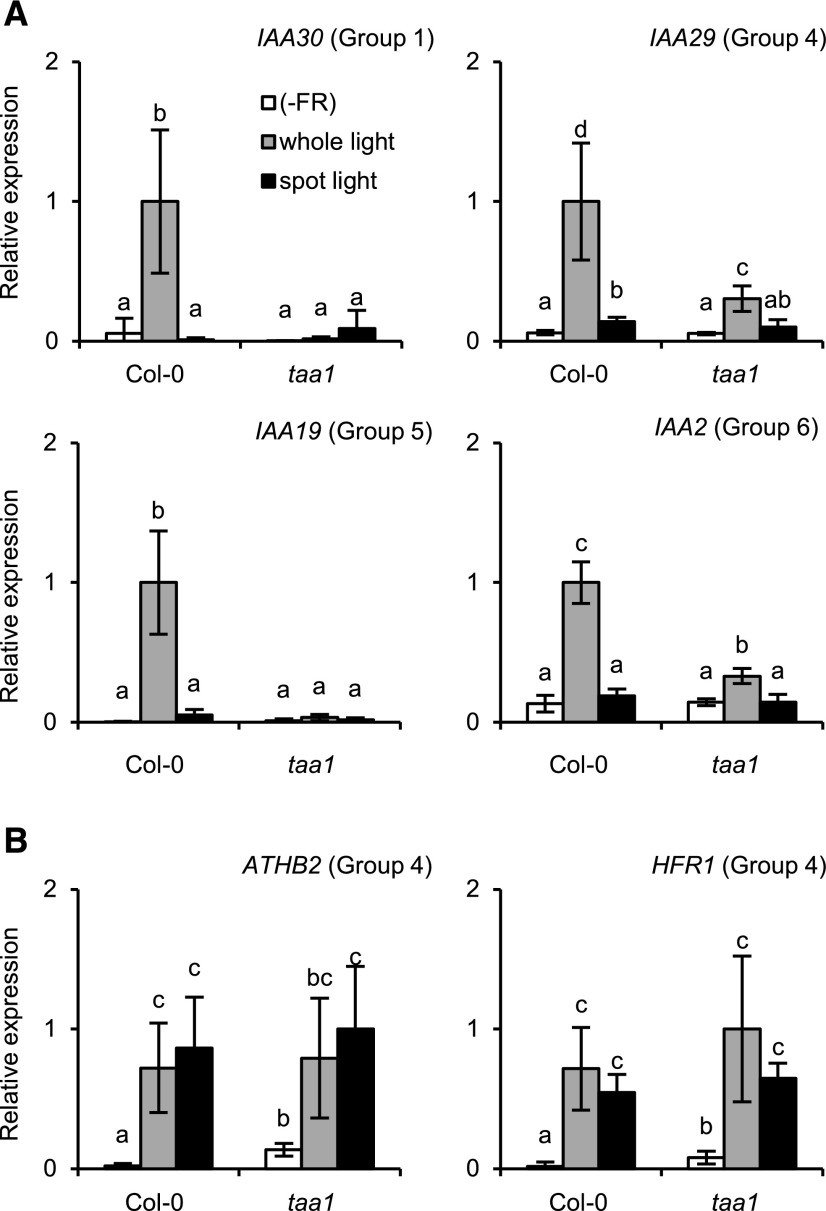
To further confirm the involvement of auxin in the above responses, we conducted the same experiment in the taa1 mutant, which is deficient for the shade-induced increase in auxin biosynthesis (Tao et al., 2008), and the respective wild type, Col-0. All four auxin-responsive genes were induced only by whole-seedling irradiation in the wild type (Fig. 6). The responses were greatly reduced in the taa1 mutant, indicating that their responses were auxin dependent. In addition, the ATHB2 and HFR1 responses were confirmed to be auxin independent (Fig. 6). We also confirmed that expression of the tissue marker genes was not affected by the light treatment (Supplemental Fig. S11).
Figure 6.
Gene expression responses to EOD-FR in the taa1 mutant and respective wild type (accession Col-0). The cDNA samples were prepared and analyzed as described for Figure 5, except that four micro sample pieces were combined in a tube for cDNA synthesis. The bar colors are as described for Figure 5. TUB2/TUB3 was used to standardize the results. The expression levels are shown in arbitrary units setting the highest value to 1 (n ≥ 3, mean ± sd). Different letters denote significant differences between the samples (P < 0.05, Student’s t test). A, Auxin-responsive genes; B, AtHB2 and HFR1.
The above observation implied that auxin transport might be involved in the regulation of auxin-responsive genes by shade. Hence, we examined the effects of an auxin transport inhibitor, 1-N-naphthylphtalamic acid (NPA; Teale and Palme, 2018), on the local induction of the auxin-responsive genes. First, we confirmed that NPA inhibited the promotion of hypocotyl elongation by shade (Supplemental Fig. S12A). In this experiment, NPA solution was added to the medium solidified with agar 24 h before the EOD-FR treatment. We then examined the responses of IAA19 and IAA29 to EOD-FR in the presence of NPA, but no inhibitory effect was observed (Supplemental Fig. S12B). To exclude the possibility that NPA did not penetrate into the cotyledon by the above method, we vacuum-infiltrated the NPA solution into detached cotyledons and exposed them to EOD-FR (Supplemental Fig. S12C). However, the result was the same. Hence, the NPA-sensitive auxin transport mechanism might not be required for the response.
Comparison of the Shade Responses in Different Positions within the Cotyledon
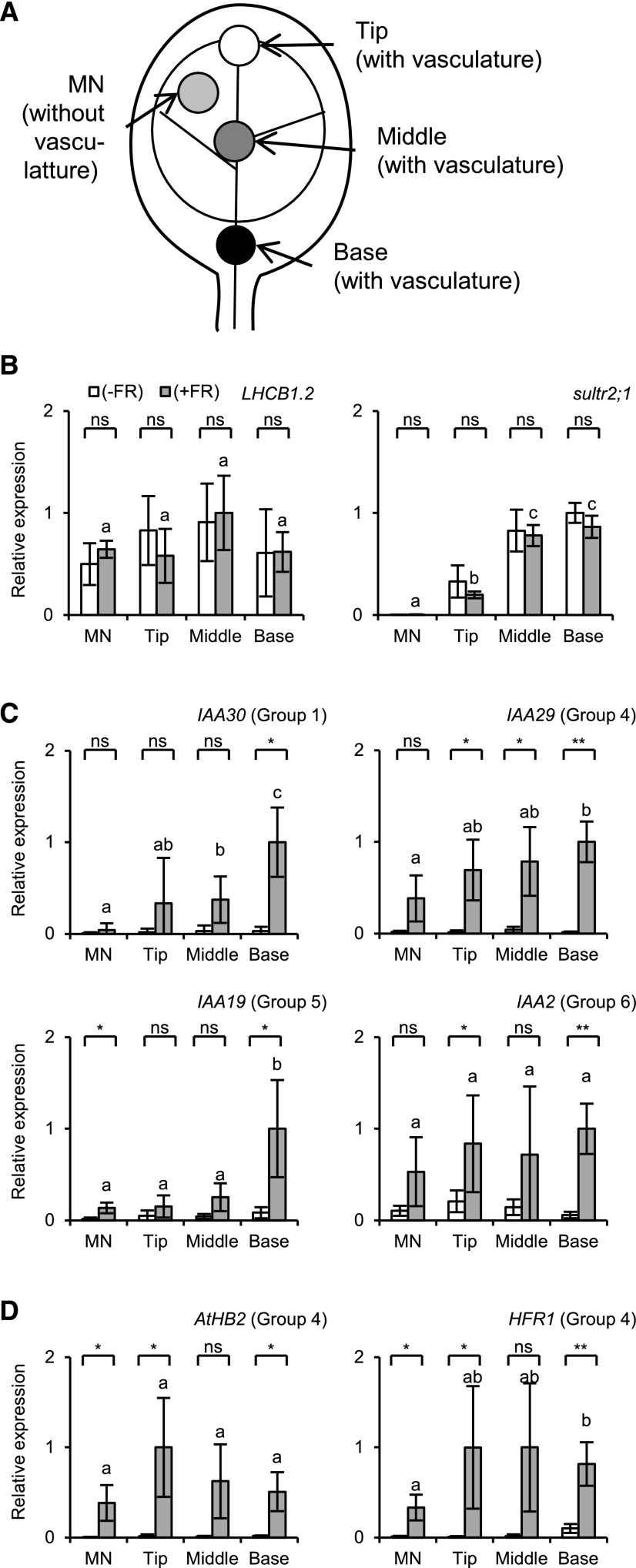
The extent of the response for some genes might be determined by the tissue type and also by the position in the cotyledon (see the sections “Initial Characterization of the RNA-Seq Data,” “Clustering Analysis of Light-Responsive Genes,” and “RT-qPCR Analysis of YUC Gene Expression”). Hence, we examined how the responses differed depending on the position. Whole seedlings were treated with EOD-FR and the micro samples containing vasculature tissues were prepared from the tip, middle, and basal parts of the cotyledon (Fig. 7). The MN samples were included as a control. To confirm the sample quality, two tissue marker genes were first examined. The mesophyll marker gene (LHCB1.2) was evenly detected along the proximal-distal axis, whereas the xylem parenchyma marker gene (sultr2;1) increased toward the basal position (Fig. 7).
Figure 7.
Gene expression responses along the proximal-distal axis in the cotyledon. A, Diagram explaining the location of microsampling. The micro samples containing vasculature tissue were excised with the needle-based device for the RT-qPCR analysis. B to D, Relative expression of tissue marker genes (B), nonautonomous genes (C), and autonomous genes (D). The cDNA samples were prepared and analyzed as for Figure 1, except that six micro sample pieces were combined in a tube for cDNA synthesis. White bar, without EOD-FR; gray bar, with EOD-FR. TUB2/TUB3 was used to standardize the results. The expression levels are shown in arbitrary units setting the highest value to 1 (n ≥ 3, mean ± sd). Asterisks indicate significant differences for Student’s t test (**, P < 0.005; *, P < 0.05; ns, nonsignificant). Different letters denote significant differences between the FR-treated samples (P < 0.05, Student’s t test).
Then, we determined the expression levels of several shade-responsive genes in these samples. The expression of IAA30 (Group 1; VS) and IAA19 (Group 5; VN and VS) increased toward the base (Fig. 7). This gradient matched with that of xylem parenchyma marker expression. Hence, these genes might be specifically expressed in the vasculature. It is noteworthy here that this pattern is almost opposite to that of YUC8 and YUC9 (Chen et al., 2016; Müller-Moulé et al., 2016; Supplemental Fig. S8). In contrast to the above two genes, IAA2 (Group 6; whole) and IAA29 (Group 4; MN and VN) were expressed more evenly. Hence, the expression patterns of the auxin response genes differed depending on the gene. Two autonomous genes, AtHB2 and HFR1 (Fig. 5), were induced at similar levels in all samples (Fig. 7), indicating that at least some of the primary responses are equally induced regardless of the position in the cotyledon.
DISCUSSION
Preparation and Initial Characterization of Micro Samples
In this study, the method used to prepare cDNA samples for the RNA-seq analysis from microtissue pieces (Kajiyama et al., 2015) was successfully adopted to compare the gene expression responses in vascular and nonvascular regions in cotyledons. The results of the mesophyll marker gene expression analysis (Figs. 1 and 2; Supplemental Fig. S3B) indicated that mesophyll/epidermis mRNA constituted a major part of the total mRNA in MN and VN. Likewise, the vasculature marker analysis demonstrated that the vasculature was highly concentrated in VS, which was included as one component of VN and was completely excluded from MN.
Multiple tissue marker genes provided consistent results (Figs. 1 and 2; Supplemental Fig. S3). However, certain markers exhibited exceptional patterns. The vascular marker SULTR2;1 was expressed at similar levels in VN and VS, although the vasculature was highly concentrated in VS compared with VN. This discrepancy was probably because xylem parenchyma, which is one of the sites of SULTR2;1 expression (Takahashi et al., 2000), was partially excluded in VS. Unlike the other mesophyll markers, the expression of LHCB2.3 was low in VN compared with MN, which may suggest that mesophyll cells in more distal parts of the cotyledon (MN) might differ in some features from those in more basal parts (VS).
It might be more direct to analyze the isolated tissues to investigate the tissue-specific gene expression. However, tissue isolation takes longer and may damage the tissue. Indeed, a xylem parenchyma domain expressing SULTR2;1 appeared to be damaged and partially excluded from VS during isolation (Figs. 1 and 2). The needle-based method can provide more intact tissues within a short time, although the resultant samples usually consist of multiple tissues. In this work, these two methods were employed to complement each other.
Transcriptome Responses to the Shade Stimulus in Different Tissues
Transcriptome responses to the shade stimulus have been studied first at the whole-seedling level (Devlin et al., 2003; Salter et al., 2003) and more recently in different organs (Kozuka et al., 2010; Nito et al., 2015; Kohnen et al., 2016; Pantazopoulou et al., 2017). Meta-analysis on available transcriptome data has been reported recently (Sellaro et al., 2017). Thereby, the importance of interorgan communication in the response has been demonstrated. Taking advantage of the novel method to analyze microtissue pieces (Nito et al., 2015), we expanded the scope to intraorgan communications within cotyledons in this study.
The up-regulated genes that were identified included reasonable numbers of known shade-responsive genes. Among the 53 major shade-induced genes (Nito et al., 2015), 22 were found in this list, which was not surprising because MN and VN contained large proportions of mesophyll/epidermis that should have been a major component of previous samples such as whole seedlings. In the previous work, more shade-induced genes were found in the shoot apex, including the vasculature, than in cotyledons (106 versus 31 genes; Nito et al., 2015). Likewise, 57.3% of the 241 up-regulated genes were preferentially responsive in the vasculature (Groups 1 and 5; Supplemental Table S2). Hence, the vasculature might be a major site of the shade response.
The above result is intriguing because the vasculature plays a critical role in long-distance signaling (Notaguchi and Okamoto, 2015). In addition, various types of genes that were presumed to be involved in biological processes, such as vascular development, cell wall synthesis/maintenance, and abiotic stress responses, were found in the vascular group (Groups 1 and 5). For example, SODIUM HYDROGEN EXCHANGER4 (NHX4; AT3G06370; Group 1; VS) encodes a Na+/K+ antiporter. It is noteworthy that the vacuole NHX family, of which NHX4 is a member, affects cell expansion (Claussen et al., 1997; Bassil et al., 2011; McCubbin et al., 2014). Another example is FLP1 (Group 1; VS). Overexpression of FPF1, a close homolog of FLP1, causes a constitutive shade-avoidance response (Wang et al., 2014). EXPANSIN A4 (EXPA4; Group 1; VS) encodes an α-expansin, which mediates acid-induced wall loosening (Cosgrove, 2015). Nevertheless, its expression is apparently not affected by auxin (Goda et al., 2008). In summary, the possibility arises that various physiological processes are controlled by shade in the vasculature.
It is noteworthy that phytochromes and PIFs were expressed approximately equally in all the sample types (Supplemental Fig. S4). Hence, we checked how PIF target genes responded in different sample types. For this purpose, we defined 264 core PIF target genes (Supplemental Table S6) that were reported to be PIF-binding genes in the literature (Hornitschek et al., 2012; Oh et al., 2012; Zhang et al., 2013; Pfeiffer et al., 2014). Interestingly, 12 core PIF target genes, including HFR1, ATHB2, GA2OX6, IAA29, and CKX5, were found in the mesophyll group (Groups 2 and 4; 57 genes in total; Supplemental Table S2). Only five core PIF target genes were identified among the 138 vascular genes (Groups 1 and 5). Hence, the primary shade responses appeared to be more prominent in the mesophyll/epidermis than in the vasculature. This is not surprising because the vasculature should be shaded by the surrounding green tissue, which would make the vasculature not suitable for shade detection.
As discussed above, the responses observed in the vasculature might not be directly regulated by phytochrome. Indeed, the vascular response appeared to be regulated at least in part by auxin synthesized in the surrounding tissues (see below). However, the genes that were up-regulated in the vasculature may not all be auxin dependent (Supplemental Table S2). Typical examples are FLP1 and EXPA4 (AT2G39700; Fig. 4), which are excluded from the list of auxin-responsive genes (Goda et al., 2008) and not associated with the GO term “response to auxin (GO:0009733).” Since the vasculature consists of functionally divergent tissues, such as phloem, companion cells, and xylem parenchyma, the response could be regulated in various ways according to the subtissue types. This feature should be explored in future studies.
Involvement of Auxin in the Shade-Avoidance Responses in the Cotyledon
As many as 20 genes related to auxin were found in the up-regulated genes (Supplemental Table S2). They were enriched in the vascular groups, Groups 1 (VS) and 5 (VNVS), indicating that the auxin-related shade response was enhanced in the vasculature. It should be noted here that some well-known shade-responsive genes were among them, such as IAA1 (Group 1) and IAA19 (Group 5). This finding was not surprising because whole-seedling or organ samples contain vasculature, albeit at low ratios. It is noteworthy that not all auxin-responsive genes were preferentially up-regulated in the vasculature. Genes such as IAA29 (Group 4) and GH3.3 (Group 4) were up-regulated preferentially in the mesophyll/epidermis (Supplemental Fig. S7).
Increased expression of YUC genes, such as YUC2, YUC5, YUC8, and YUC9, in the shade has been proposed to be responsible for the up-regulation of auxin-responsive genes in response to shade stimulus (Li et al., 2012). The shade response was confirmed for YUC8 and YUC9 in this study (Supplemental Fig. S9). Importantly, their expression was most prominent in MN, much weaker in VN, and not detectable in VS, indicating that they were expressed in a specific part of the mesophyll/epidermis. It is intriguing that GUS reporter expression driven by the YUC8 and YUC9 promoters is induced more strongly in the leaf margin in slightly older Arabidopsis plants (Müller-Moulé et al., 2016). More recently, photoperception by the rosette leaf tip has been shown to promote leaf hyponasty through the action of auxin (Michaud et al., 2017; Pantazopoulou et al., 2017). Hence, auxin might be synthesized more actively in the distal and/or marginal ends of the cotyledon.
Nonautonomous Regulation of Auxin-Responsive Genes
In contrast to YUC expression, the auxin responses were observed more prominently in the vascular region (Supplemental Table S4; Supplemental Fig. S8). Furthermore, some of the IAA responses increased toward the base of the cotyledon (Fig. 7), which was opposite to the expression patterns of YUC8 and YUC9 (Supplemental Fig. S9). Hence, the site of auxin synthesis and responses did not to match in the cotyledon. Consistent with this view, the local irradiation of the VN region was ineffective to induce the expression of IAA30, IAA29, IAA19, and IAA2 (Figs. 5 and 6). This result implies that YUC genes should be up-regulated globally or at least in a specific area of the cotyledon to elicit the response. We speculate that the auxin synthesized in the mesophyll/epidermis is transported and accumulated in the vasculature, especially at the basal end of the cotyledon. This view is consistent with current knowledge that auxin is loaded into the vasculature from surrounding cells in young leaves for long-distance transport (Marchant et al., 2002; Notaguchi and Okamoto, 2015). However, it remains possible that not only auxin levels but also the sensitivity to auxin might be altered in the vasculature to up-regulate the expression of auxin-responsive genes. To answer the question ultimately, we would need to measure the auxin levels in different parts of the cotyledon, which is very difficult at present.
The previous studies have shown that auxin transport is an important element of auxin response to shade (Keuskamp et al., 2010; Michaud et al., 2017; Pantazopoulou et al., 2017). PIN proteins are auxin efflux carriers that facilitate auxin transport (Blilou et al., 2005; Wisniewska et al., 2006). The pin3pin4pin7 loss-of-function mutant fails to promote hypocotyl elongation in response to shade (Kohnen et al., 2016). Interestingly, PIN3 and PIN7 expression increases in response to shade at the whole-seedling level (Devlin et al., 2003). Hence, polar auxin transport was envisaged to be involved in the shade response within the cotyledon. To examine this possibility, a well-known auxin transport inhibitor, NPA (Teale and Palme, 2018), was employed. However, NPA did not affect the responses of IAA19 and IAA29 in VN despite the fact that it inhibited hypocotyl elongation (Supplemental Fig. S12). A similar result that induction of SAUR22 in cotyledons and the hypocotyl remains robust in the pin3pin4pin7 mutant has been reported previously (Kohnen et al., 2016). Hence, auxin movement by an NPA-insensitive mechanism might contribute to the response.
CONCLUSION
As mentioned previously, auxin links spatially separated organs, such as the leaf and hypocotyl, in shade responses (Tanaka et al., 2002; Procko et al., 2014; Kohnen et al., 2016). This analysis extends this view regarding the role of auxin in intraorgan communication. The shade stimulus promotes auxin synthesis in the photoperceptive mesophyll/epidermis. The synthesized auxin is then accumulated in the vasculature, probably through the action of some auxin carriers in the cotyledon. The vasculature then facilitates the long-distance transport of auxin to target organs such as the hypocotyl. Phytochromes are envisaged to regulate or modify various aspects of auxin throughout this process.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Most experiments were performed with Arabidopsis (Arabidopsis thaliana) accession Landsberg erecta. The Bpro-7 transgenic plant, in which the phyB-GFP fusion protein was expressed under the control of the PHYB authentic promoter (Endo et al., 2005), was used for nuclear phyB speckle observation. In addition, the taa1/sav3 mutant (Tao et al., 2008) and its background accession Columbia (Col-0) were used for the spotlight experiments. Seeds were sown on Murashige and Skoog medium (pH 5.5–6.5) solidified with 0.8% (w/v) agar and supplemented with 0.5% (w/v) sugar and 0.2% vitamin B5. Seeds were incubated at 4°C in the dark for 2 d to induce germination. Seedlings were grown for 4 d at 23°C in the chamber under short-day cycles (8 h light/16 h dark) before the EOD-FR treatment. A fluorescent lamp FL20SS EX-N/18 TT (Mitsubishi) supplied the white light (120 μmol m−2 s−1).
Light Treatments
EOD-FR was supplied at the end of the fourth day of the growth period to induce the shade-avoidance response (Kasperbauer, 1971; López-Juez et al., 1990). Whole seedlings were treated with FR (2 min, 427 μmol m−2 s−1) using an FR light-emitting diode panel ISL-150×150-FR (CCS) except for the spotlight experiment. For the spotlight experiments, the cotyledon was treated with a spotlight of 1,370 μmol m−2 s−1 for 4 s. The spotlight was from an FR light-emitting diode bulb VSF741N1 (Alpha-one), on top of which was attached an acrylic fiber with a diameter of 250 μm (Nito et al., 2015). As controls, whole seedlings were treated with a bare FR light-emitting diode bulb (1,000 μmol m−2 s−1, 5 s). The light intensity was measured with a quantum sensor LI-250A (LI-COR). The FR-treated seedling was incubated in the dark for 2 h before sample collection unless otherwise stated.
Micro Sample Collection and Observation
Micro samples were excised from different parts of Arabidopsis cotyledons with a needle-based device (Nito et al., 2015). The vasculature was isolated from Arabidopsis cotyledons using a previous method (Endo et al., 2014) with modifications. For this purpose, 20 cotyledons were cut and placed in a 1.5-mL tube containing 300 μL enzyme solution (0.75% Cellulase Onozuka R-10 [Yakult], 0.25% Macerozyme R-10 [Yakult], 0.4 m mannitol, 5 mm MES-KOH, pH 5.6, and 8 mm CaCl2). The cotyledons were then ultrasonicated for 45 s to remove mesophyll and epidermal tissues. The bare vasculature was collected with tweezers.
For bright-field microscopy observation, a BX51 microscope (Olympus) equipped with a CCD camera DP30BW (Olympus) was used. The DP manager imaging software (Olympus) was used to take the pictures. For observation of phyB-GFP, a confocal microscope FV1000-D (Olympus) and FV10-ASW software (Olympus) were used. The phyB-GFP fluorescence was observed 30 to 40 min after the FR treatment. The z axis sections (3-μm intervals) were stacked to encompass the epidermis and the top layer of the mesophyll.
cDNA Synthesis
cDNA was synthesized and amplified as previously described (Nito et al., 2015). The micro sample pieces were frozen in 0.5 μL RLC lysis buffer (Qiagen) and homogenized with a pestle. After reverse transcription, cDNA was amplified by PCR and purified twice with AMPure XP beads (Beckman Coulter). After the second-round PCR amplification, cDNA was purified using the Wizard SV gel and PCR cleanup system (Promega). The cDNA concentration was measured using the Qubit dsDNA HS assay kit (Life Technologies) and Qubit 2.0 Fluorometer (Life Technologies).
RNA-Seq Analysis
The RNA-seq library was prepared with 1 ng cDNA using the KAPA HyperPlus Kit (KAPA Biosystems) following the manufacturer’s instructions. The quality was checked using a Bioanalyzer (Agilent Technologies). The library was sequenced using a HiSeq 2500 sequencer (single end, 50 bp; Illumina). Sequenced reads were preprocessed, mapped, and quantified according to a previously described pipeline (Kamitani et al., 2016) with the following modifications. Gene models in TAIR10 were used as reference sequences.
RT-qPCR Analysis
The RT-qPCR analysis was performed using the Faststart Essential DNA Green Master (Roche) and LightCycler 96 system (Roche). A 1-ng aliquot of the amplified cDNA (1 mg/mL) was mixed with 6.2 μL PCR-grade water, 7.5 μL Faststart Essential DNA Green Master 2× concentrated, 0.3 μL primer (FW-RV), and 1 μL cDNA and subjected to the RT-qPCR reaction. The PCR conditions were as follows: 95°C for 10 min, 45 cycles of 95°C for 20 s, 55°C for 10 s, and 72°C for 10 s, and finally 95°C for 10 s, 65°C for 60 s, and 97°C for 1 s for melting.
As an internal standard, the expression levels of the TUB2/TUB3 genes, which were collectively detected with a primer set common to these genes, were determined. Since the relative TUB2/TUB3 expression levels per 1 ng of cDNA were 1, 1.75, and 2.18 in MN, VN, and VS, respectively, on the basis of the RT-qPCR analysis (Supplemental Fig. S1), the TUB2/TUB3 values were corrected based on these ratios as an internal control. The primer sequences are shown in Supplemental Table S7.
NPA Treatment
For the hypocotyl elongation and gene expression assays, NPA solution at 25 μm (<0.1% DMSO in water) was applied onto the agar plate 24 h before the EOD-FR treatment. Alternatively, the 25 μm NPA solution was vacuum-infiltrated into the detached cotyledons 1 h before the EOD-FR treatment. Vacuum infiltration was performed by soaking cotyledons in the NPA solution in a microtube and repeatedly evacuating the tube using a syringe attached on top of it.
Accession Numbers
The RNA-seq sequence data are available in the DDBJ Sequenced Read Archive under accession number DRA006771.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Relative TUB2/TUB3 expression levels in different sample types.
Supplemental Figure S2. Marker gene expression in cDNA samples subjected to RNA-seq analysis.
Supplemental Figure S3. Expression of additional housekeeping and tissue marker genes measured by RNA-seq.
Supplemental Figure S4. Phytochrome and light-signaling gene expression measured by RNA-seq.
Supplemental Figure S5. A diagram explaining how the light-responsive genes were selected.
Supplemental Figure S6. Cluster analysis of down-regulated genes.
Supplemental Figure S7. Additional RT-qPCR confirmation results.
Supplemental Figure S8. Cluster analysis of auxin-related genes.
Supplemental Figure S9. Expression of auxin biosynthesis genes.
Supplemental Figure S10. Evaluation of spotlight irradiation with a phyB-GFP-expressing line.
Supplemental Figure S11. Tissue marker gene expression in VN micro samples collected from cotyledons treated with a spotlight.
Supplemental Figure S12. Effects of NPA on the hypocotyl and gene expression responses.
Supplemental Table S1. RPM values for all genes analyzed in this study.
Supplemental Table S2. List of the 241 up-regulated genes.
Supplemental Table S3. List of the 209 down-regulated genes.
Supplemental Table S4. List of GO terms enriched in the gene groups.
Supplemental Table S5. Responses of 53 core shade-responsive genes
Supplemental Table S6. List of PIF1, PIF3, PIF4, and PIF5 target genes.
Supplemental Table S7. List of primers used in this study.
Acknowledgments
We thank Dr. Joanne Chory at the Salk Institute for kindly providing the taa1 mutant seeds.
Footnotes
This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology, Japan, through a Grant-in-Aid for Scientific Research on Innovative Areas (no. 22120002) (to A.N.), by Japan Society for the Promotion of Science KAKENHI Grant JP 15H04389 (to A.N.), by Japan Science and Technology Agency CREST Grant JPMJCR15O2 (to A.J.N.), and by Japan Society for the Promotion of Science KAKENHI Grants JP16H06171 and JP16H01473 (A.J.N.).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Bassil E, Tajima H, Liang YC, Ohto MA, Ushijima K, Nakano R, Esumi T, Coku A, Belmonte M, Blumwald E (2011) The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23: 3482–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall FD, Yeung EC, Pharis RP (1996) Far-red light stimulates internode elongation, cell division, cell elongation, and gibberellin levels in bean. Can J Bot 74: 743–752 [Google Scholar]
- Black M, Shuttleworth JE (1974) The role of the cotyledons in the photocontrol of hypocotyl extension in Cucumis sativus L. Planta 117: 57–66 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64: 403–427 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Smith H (1988) The loci of perception for phytochrome control of internode growth in light-grown mustard: Promotion by low phytochrome photoequilibria in the internode is enhanced by blue light perceived by the leaves. Planta 176: 277–282 [DOI] [PubMed] [Google Scholar]
- Casson SA, Hetherington AM (2014) phytochrome B Is required for light-mediated systemic control of stomatal development. Curr Biol 24: 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M (2009) Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43: 265–285 [DOI] [PubMed] [Google Scholar]
- Chen L, Tong J, Xiao L, Ruan Y, Liu J, Zeng M, Huang H, Wang JW, Xu L (2016) YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J Exp Bot 67: 4273–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen M, Lüthen H, Blatt M, Böttger M (1997) Auxin-induced growth and its linkage to potassium channels. Planta 201: 227–234 [Google Scholar]
- Correll MJ, Kiss JZ (2005) The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol 46: 317–323 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2015) Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol 25: 162–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA (2003) A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol 133: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M, Ljung K, Fankhauser C (2015) Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol 208: 198–209 [DOI] [PubMed] [Google Scholar]
- Endo M, Nakamura S, Araki T, Mochizuki N, Nagatani A (2005) Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17: 1941–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Shimizu H, Nohales MA, Araki T, Kay SA (2014) Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RA, Zalik S (1964) Effect of light quality on growth and free indoleacetic acid content in Phaseolus vulgaris. Plant Physiol 39: 328–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA. (2008) Shade avoidance. New Phytol 179: 930–944 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Goyal A, Karayekov E, Galvão VC, Ren H, Casal JJ, Fankhauser C (2016) Shade promotes phototropism through phytochrome B-controlled auxin production. Curr Biol 26: 3280–3287 [DOI] [PubMed] [Google Scholar]
- Hong SM, Bahn SC, Lyu A, Jung HS, Ahn JH (2010) Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol 51: 1694–1706 [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, Xenarios I, Fankhauser C (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Iglesias MJ, Sellaro R, Zurbriggen MD, Casal JJ (2017) Multiple links between shade avoidance and auxin networks. J Exp Bot 69: 213–228 [DOI] [PubMed] [Google Scholar]
- Ivashikina N, Deeken R, Ache P, Kranz E, Pommerrenig B, Sauer N, Hedrich R (2003) Isolation of AtSUC2 promoter-GFP-marked companion cells for patch-clamp studies and expression profiling. Plant J 36: 931–945 [DOI] [PubMed] [Google Scholar]
- Kajiyama T, Fujii A, Arikawa K, Habu T, Mochizuki N, Nagatani A, Kambara H (2015) Position-specific gene expression analysis using a microgram dissection method combined with on-bead cDNA library construction. Plant Cell Physiol 56: 1320–1328 [DOI] [PubMed] [Google Scholar]
- Kamitani M, Nagano AJ, Honjo MN, Kudoh H (2016) RNA-Seq reveals virus-virus and virus-plant interactions in nature. FEMS Microbiol Ecol 92: fiw176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperbauer MJ. (1971) Spectral distribution of light in a tobacco canopy and effects end-of-day light quality on growth and development. Plant Physiol 47: 775–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LACJ, Peeters AJM, Pierik R (2010) Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA 107: 22740–22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Song K, Park E, Kim K, Bae G, Choi G (2016) Epidermal phytochrome B inhibits hypocotyl negative gravitropism non-cell-autonomously. Plant Cell 28: 2770–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Adám E, Schäfer E, Nagy F (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnen MV, Schmid-Siegert E, Trevisan M, Petrolati LA, Sénéchal F, Müller-Moulé P, Maloof J, Xenarios I, Fankhauser C (2016) Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28: 2889–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Nurani AM, Saito C, Ichihashi Y, Saito M, Yamazaki K, Mitsuda N, Ohme-Takagi M, Fukuda H (2016) Vascular cell induction culture system using Arabidopsis leaves (VISUAL) reveals the sequential differentiation of sieve element-like cells. Plant Cell 28: 1250–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuka T, Kobayashi J, Horiguchi G, Demura T, Sakakibara H, Tsukaya H, Nagatani A (2010) Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol 153: 1608–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E, Nagatani A, Buurmeijer WF, Peters JL, Furuya M, Kendrick RE, Wesselius JC (1990) Response of light-grown wild-type and aurea-mutant tomato plants to end-of day far-red light. J Photochem Photobiol B 4: 391–405 [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T (1987) Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol 9: 611–623 [DOI] [PubMed] [Google Scholar]
- McCubbin T, Bassil E, Zhang S, Blumwald E (2014) Vacuolar Na+/H+NHX-type antiporters are required for cellular K+ homeostasis, microtubule organization and directional root growth. Plants (Basel) 3: 409–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud O, Fiorucci AS, Xenarios I, Fankhauser C (2017) Local auxin production underlies a spatially restricted neighbor-detection response in Arabidopsis. Proc Natl Acad Sci USA 114: 7444–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JE, Tao Y, Chory J, Ballaré CL (2009) Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc Natl Acad Sci USA 106: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Moulé P, Nozue K, Pytlak ML, Palmer CM, Covington MF, Wallace AD, Harmer SL, Maloof JN (2016) YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. PeerJ 4: e2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. (2004) Light-regulated nuclear localization of phytochromes. Curr Opin Plant Biol 7: 708–711 [DOI] [PubMed] [Google Scholar]
- Nito K, Kajiyama T, Unten-Kobayashi J, Fujii A, Mochizuki N, Kambara H, Nagatani A (2015) spatial regulation of the gene expression response to shade in Arabidopsis seedlings. Plant Cell Physiol 56: 1306–1319 [DOI] [PubMed] [Google Scholar]
- Notaguchi M, Okamoto S (2015) Dynamics of long-distance signaling via plant vascular tissues. Front Plant Sci 6: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulou CK, Bongers FJ, Küpers JJ, Reinen E, Das D, Evers JB, Anten NPR, Pierik R (2017) Neighbor detection at the leaf tip adaptively regulates upward leaf movement through spatial auxin dynamics. Proc Natl Acad Sci USA 114: 7450–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M (2009) Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA 106: 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH (2014) Combinatorial complexity in a transcriptionally centered signalling hub in Arabidopsis. Mol Plant 7: 1598–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, de Wit M (2014) Shade avoidance: phytochrome signalling and other aboveground neighbour detection cues. J Exp Bot 65: 2815–2824 [DOI] [PubMed] [Google Scholar]
- Pierik R, Cuppens MLC, Voesenek LACJ, Visser EJW (2004) Interactions between ethylene and gibberellins in phytochrome-mediated shade avoidance responses in tobacco. Plant Physiol 136: 2928–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Crenshaw CM, Ljung K, Noel JP, Chory J (2014) Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol 165: 1285–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, Chory J (2016) The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev 30: 1529–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J (2016) g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res 44: W83–W89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux MC, Ballaré CL, Jordan ET, Vierstra RD (1997) Directed overexpression of PHYA locally suppresses stem elongation and leaf senescence responses to far-red radiation. Plant Cell Environ 20: 1551–1558 [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683 [DOI] [PubMed] [Google Scholar]
- Sellaro R, Pacín M, Casal JJ (2017) Meta-analysis of the transcriptome reveals a core set of shade avoidance genes in Arabidopsis. Photochem Photobiol 93: 692–702 [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I (2005) A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Weigel D, Yanofsky MF (1999) The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J 20: 259–263 [DOI] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM (2014) SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Takada S, Takada N, Yoshida A (2013) ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development 140: 1919–1923 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K (2000) The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J 23: 171–182 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Nakamura S, Mochizuki N, Nagatani A (2002) Phytochrome in cotyledons regulates the expression of genes in the hypocotyl through auxin-dependent and -independent pathways. Plant Cell Physiol 43: 1171–1181 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale W, Palme K (2018) Naphthylphthalamic acid and the mechanism of polar auxin transport. J Exp Bot 69: 303–312 [DOI] [PubMed] [Google Scholar]
- Van Gelderen K, Kang C, Pierik R (2018) Light signalling, root development and plasticity. Plant Physiol 176: 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fan S, Song M, Pang C, Wei H, Yu J, Ma Q, Yu S (2014) Upland cotton gene GhFPF1 confers promotion of flowering time and shade-avoidance responses in Arabidopsis thaliana. PLoS One 9: e91869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertová D, Brewer PB, Ruzicka K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi YL, Suzuki R, Cabrera J, Nakagami S, Sagara T, Ejima C, Sano R, Aoki Y, Olmo R, Kurata T, et al. (2017) Root-knot and cyst Nematodes activate procambium-associated genes in Arabidopsis roots. Front Plant Sci 8: 1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH (2013) A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novák O, Chen W, Ljung K, Noel JP, Chory J (2016) Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants 2: 16025. [DOI] [PMC free article] [PubMed] [Google Scholar]