OsERS1, a glutamyl-tRNA synthetase, modulates early rice anther development through affecting metabolic homeostasis and redox status.
Abstract
Aminoacyl-tRNA synthetases (aaRSs) have housekeeping roles in protein synthesis, but little is known about how these aaRSs are involved in organ development. Here, we report that a rice (Oryza sativa) glutamyl-tRNA synthetase (OsERS1) maintains proper somatic cell organization and limits the overproliferation of male germ cells during early anther development. The expression of OsERS1 is specifically detectable in meristematic layer 2-derived cells of the early anther, and osers1 anthers exhibit overproliferation and disorganization of layer 2-derived cells, producing fused lobes and extra germ cells in early anthers. The conserved biochemical function of OsERS1 in ligating glutamate to tRNAGlu is enhanced by its cofactor aaRS OsARC. Furthermore, metabolomics profiling revealed that OsERS1 is an important node for multiple metabolic pathways, indicated by the accumulation of amino acids and tricarboxylic acid cycle components in osers1 anthers. Notably, the anther defects of the osers1 mutant are causally associated with the abnormal accumulation of hydrogen peroxide, which can reconstitute the osers1 phenotype when applied to wild-type anthers. Collectively, these findings demonstrate how aaRSs affect male organ development in plants, likely through protein synthesis, metabolic homeostasis, and redox status.
Reproductive cells in flowering plants link the dominant diploid sporophytic generation and the short haploid gametophytic generation. Male germ cells are produced within the sporophytic tissue called the anther (Ma, 2005). The anther primordium that forms from the floral meristem usually contains three layers: L1 to L3. L1 and L3 cells subsequently differentiate into the epidermis and connective tissue, respectively, whereas the L2 layer forms germ cells in the center of anther lobes and three surrounding supportive somatic cell layers. The early stages of anther development (stages 1–5) determine the formation of anther cell layers. At stage 2, the L2 layer undergoes rapid cell division and generates L2-derived (L2-d) cells. At stage 3, the central archesporial cells show a distinctive enlarged shape surrounded by the neighboring primary parietal cells (PPCs). At stage 4, PPCs form endothecium and secondary parietal cells (SPCs) via asymmetric cell division, and archesporial cells differentiate into sporogenous cells. Following this, the sporogenous cells continue to divide for subsequent meiosis and the SPC layer symmetrically divides to form the middle layer and tapetum at stage 5 (Ma, 2005; Wilson and Zhang, 2009; Zhang and Yang, 2014; Walbot and Egger, 2016).
One critical event in premeiotic anther development is rapid periclinal cell division and strict cell lineage specification. However, the underlying mechanism controlling this process remains largely unknown. Previous studies revealed the critical functions of a few regulators, such as receptor-like protein kinases, transcription factors, redox status, glycoprotein, and hormones, in specifying cell lineage and the fate of germ and somatic cells in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and maize (Zea mays; Wilson and Zhang, 2009; Zhang and Yang, 2014; Walbot and Egger, 2016).
Glutaredoxins (GRXs) are small oxidoreductases that modulate various cellular events and responses to oxidative stress (Holmgren, 2000). Rice MICROSPORELESS1 (MIL1) and maize Male Sterile Converted Anther1 (MSCA1) are homologs to two Arabidopsis GRXs, ROXY1 and ROXY2. These GRXs have been shown to regulate abaxial-adaxial anther lobe formation and the differentiation of microsporocytes, suggesting that GRXs trigger archesporial fate determination by controlling redox status. Further studies show that GRXs interact with TGA (TGACGTCA cis-element-binding protein) transcription factors to transcriptionally regulate targets (Chaubal et al., 2003; Xing and Zachgo, 2008; Hong et al., 2012b; Kelliher and Walbot, 2012; Yang et al., 2016).
Recent studies have shown the function of cell surface-localized leucine-rich repeat receptor-like kinases (LRR-RLKs) and their ligands, such as rice MULTIPLE SPOROCYTE1 (MSP1) and TAPETAL DETERMINANT1-LIKE A (OsTDL1A)/MIL2, in determining early anther cell fate. These are orthologous to Arabidopsis EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS and TAPETAL DETERMINANT1, respectively, and function in specifying anther cell identity and controlling cell numbers of tapetum and microsporocytes (Canales et al., 2002; Zhao et al., 2002, 2008; Nonomura et al., 2003; Yang et al., 2003; Hong et al., 2012a; Zhang and Yang, 2014). MSP1 and OsTDL1A were suggested to exert their effects in early anther cell differentiation via modulating redox status in rice (Yang et al., 2016).
In maize, early-stage anthers also require hypoxia to maintain proper division and determine cell fate. The maize GRX MSCA1 may maintain growth-generated hypoxic conditions to induce archesporial cells. Also, archesporial cells might silence mitochondria to lower the concentration of reactive oxygen species (ROS) and maintain reducing conditions (Kelliher and Walbot, 2012).
Aminoacyl-tRNA synthetases (aaRSs) are found in all protein-synthesizing organisms and can catalyze the ligation of amino acids and their cognate tRNA, an essential step for translation and amino acid dynamics (Ibba and Söll, 2000; Yamakawa and Hakata, 2010). Studies in mammals showed noncanonical and surprising roles of aaRSs in amino acid signaling, transcriptional control, and antiapoptosis (Guo et al., 2010). As housekeeping proteins, aaRSs are indispensable for plant development, but their detailed function in plant growth remains largely unclear. In Nicotiana benthamiana, silencing of Glutamyl-tRNA Synthetase (GluRS) and Seryl-tRNA Synthetase causes a severe yellowing phenotype in leaves due to significant reduction in the number of organelles (Kim et al., 2005). Mutation of a rice ValRS results in a reduced level of chlorophyll and abnormal chloroplast ribosome biogenesis (Wang et al., 2016). Among the 45 aaRSs in Arabidopsis, 21 are required for ovule development and embryogenesis (Berg et al., 2005). In addition, an Arabidopsis Cysteinyl-tRNA Synthetase (CysRS) expressed in the central cell of the female gametophyte may specify the fate of adjacent cells (Kägi et al., 2010). Noncanonical and surprising roles of aaRSs have been uncovered, such as amino acid signaling, transcriptional control, and antiapoptosis. Furthermore, eight members of aaRSs were found to exist in mammalian cytoplasm as a multitRNA synthetases complex (MSC) whose roles and mechanisms of formation are still unclear (Guo and Schimmel, 2013). However, the detailed role of aaRSs in plant organs and development is limited. Here, we report the critical function of the cytosol- and mitochondria-localized GluRS, OsERS1, in early anther cell proliferation and organization by affecting protein synthesis, amino acid-derived metabolism, and cellular redox status.
RESULTS
OsERS1 Limits L2-d Cell Proliferation and Maintains Anther Cell Organization
To explore the mechanisms underlying anther development in the cereal model plant rice, we identified a new male-sterile mutant, oryza sativa glutamyl-tRNA synthetase (osers1), because of the mutation in Glutamyl-tRNA Synthetase1 (see below). The osers1 mutant displayed no obvious phenotypic changes from the wild type in vegetative growth, inflorescence architecture, female reproductive organ, or seed shape (Supplemental Fig. S1, A and B). However, during the male reproductive stage, the mutant developed defective anthers that were pale and had fused lobes, resulting in the production of ∼20% viable pollen grains and an ∼10% seed-setting rate (Supplemental Fig. S1, C–F). The osers1 mutant showed a more tightly woven texture in the anther wall, and the pollen grains were shrunken and displayed rough, lumpy extine, in contrast to the wild type, which had a well-arranged cuticle on the exterior of the anther wall and mature pollen grains with smooth extine (Supplemental Fig. S1, G–N). All F1 plants of reciprocal crosses between osers1 and the wild type were normal, and the F2 offspring had an ∼3:1 segregation of normal versus abnormal anthers (116:42; χ2 = 0.58, P > 0.05), suggesting that the osers1 mutants contain a unifactorial and recessive mutation.
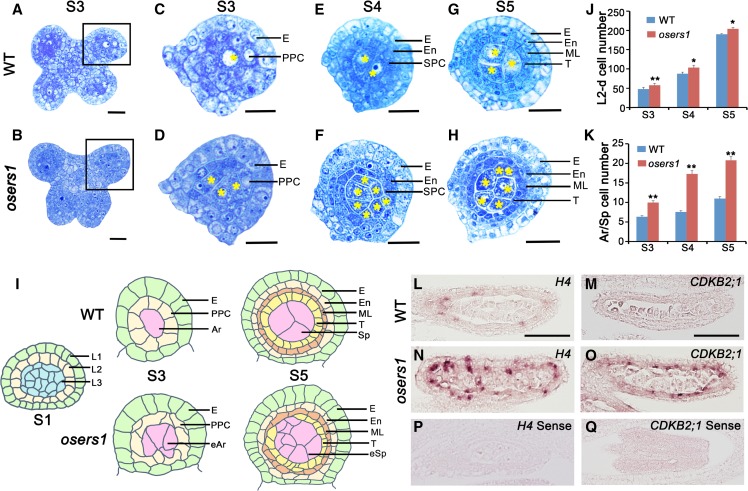
From stage 3 to 5 of anther development, ∼40% of osers1 anthers had adaxial fused lobes, which was distinct from the butterfly-shaped four lobes in wild-type anthers (Fig. 1, A and B; Supplemental Fig. S2, A–F and I). At stage 3, transverse section analysis showed that each wild-type anther formed ∼6.3 archesporial cells (n = 11) enringed by a single layer of PPCs, while each osers1 anther produced ∼10 archesporial-like cells (n = 12) enringed by PPCs organized in an irregular mosaic pattern (Fig. 1, C, D, I, and K). At stage 5, the wild-type archesporial cells divided further into ∼11 sporogenous cells (n = 15) surrounded by three circular inner layers derived from PPCs (Fig. 1, E, G, I, and K). However, each osers1 anther produced ∼20 sporogenous-like cells (n = 13), and ∼50% of the anthers had disordered wall layers with irregular boundaries (Fig. 1, F, H, I, and K; Supplemental Fig. S2I). Notably, the total number of somatic and germ cells derived from the L2 layer in the osers1 mutant was increased during stages 3 to 5 (Fig. 1J). Some sporogenous-like cells were ectopically embedded in subepidermal layers (Fig. 1I; Supplemental Fig. S2, G and H).
Figure 1.
OsERS1 is essential for cell proliferation and organization in early rice anther development. A to H, Cytological comparison of anther development between the wild type (WT) and the osers1 mutant. S3 to S5 indicate anther developmental stages. A and B show anthers in stage 3, C and D show magnifications of the boxed regions in A and B, respectively, E and F show anther lobes in stage 4, and G and H show anther lobes in stage 5. Yellow asterisks indicate archesporial cells at S3 and sporogenous cells at S4 to S5. Bars = 15 μm. I, Diagrams of anther development from stages 1 to 5 in the wild type and the osers1 mutant. Disorganization of somatic cell layers and excess archesporial/sporogenous-like cells are shown in the mutant. J and K, Quantification of cell overproliferation in osers1 anther. J, Average L2-d cell counts of anther cross sections. K, Average archesporial/sporogenous cell counts of the anther cross sections. All error bars represent sd. S3 wild type, n = 11; S4 wild type, n = 13; S5 wild type, n = 15; S3 osers1, n = 12; S4 osers1, n = 14; S5 osers1, n = 13. Student’s t test: *, P < 0.05 and **, P < 0.01. Ar, Archesporial cell; E, epidermis; eAr, excess archesporial-like cell; En, endothecium; eSp, excess sporogenous-like cell; ML, middle layer; Sp, sporogenous cell; T, tapetum. L to Q, In situ hybridization of cell division marker genes in stage 4 anthers. Bars = 50 μm.
The increased numbers of anther cells in the osers1 mutant prompted us to compare cell division activity between the wild type and the osers1 mutant. In situ hybridization showed stronger expression signals for HISTONE4 (H4), a cell-division marker for the S-phase of mitosis, and CYCLIN-DEPEDENT KINASE B2-1 (CDKB2;1), a marker of the G2/M phase transition (Umeda et al., 1999), in both somatic and germ cells of osers1 lobes, compared with the wild type (Fig. 1, L–Q). Reverse transcription-quantitative PCR (RT-qPCR) analysis also showed increased expression of H4 and CDKB2;1 in osers1 anthers (Supplemental Fig. S3B). Due to the lack of marker genes for cell lineage during early anther development, we chose MSP1 to examine the relationship of the MSP1 pathway and OsERS1. In situ hybridization showed similar distribution patterns of MSP1 in both wild-type and osers1 anthers: that is, preferential accumulation on anther wall cells rather than in central sporogenous cells (Supplemental Fig. S3A). RT-qPCR showed no significant difference in the expression of MSP1, as well as in that of two other genes involved in anther development, MIL1 and MIL2, between wild-type and osers1 anthers (Supplemental Fig. S3B). Unlike osers1, mutations of MSP1, MIL2, and MIL1 led to misidentified anther wall cell layers and complete male sterility (Nonomura et al., 2003; Hong et al., 2012a,b). These results indicate that the cell properties are still maintained in osers1 anthers, and OsERS1 is strongly implicated in cell proliferation and organization in the rice anther.
OsERS1 Encodes a Glutamyl-tRNA Synthetase
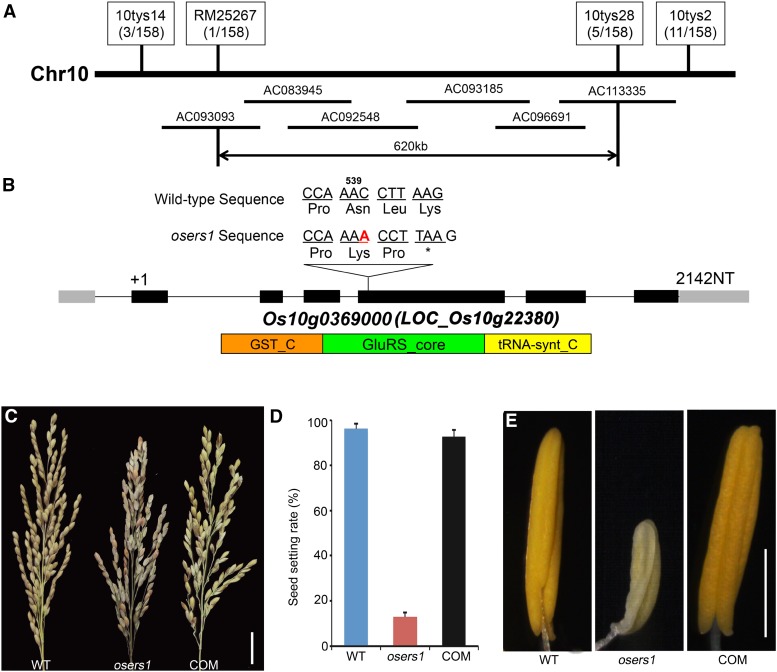
Using map-based cloning combined with the whole-genome sequencing-based MutMap method (Abe et al., 2012; Lü et al., 2015), we identified the mutated OsERS1 gene as having an insertion of A after A539 in the LOC_Os10g22380 coding region (also known as Os10g0369000; see “Materials and Methods”); this resulted in a premature stop codon in the open reading frame (Fig. 2, A and B). The identity of OsERS1 was confirmed by the complementary assay, in which the male-sterile phenotype in the osers1 mutant was rescued by the expression of a genomic fragment that contained the wild-type OsERS1 gene (Fig. 2, C–E).
Figure 2.
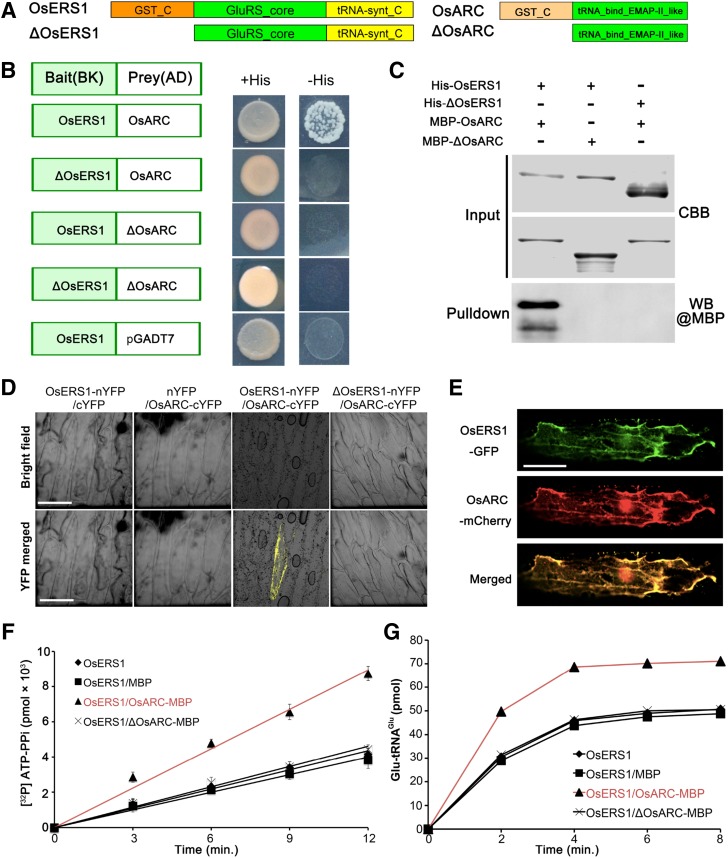
Molecular identification of OsERS1. A, Fine-mapping of OsERS1 to chromosome 10. The molecular markers used for mapping are indicated. Numbers in parentheses indicate the number of recombinants. AC numbers are rice genomic DNA accession numbers of bacterial artificial chromosome clones. B, Schematic representation of OsERS1 and functional domains. The red A indicates an insertion after A539 in oser1. NT, Nucleotides. Black rectangles represent exons. The asterisk means a stop codon. GST_C, Glutathione S-transferase C-terminal domain; GluRS_core, catalytic core domain of glutamyl-tRNA synthetase; tRNA-synt_C, tRNA synthetases class I anticodon binding domain. C to E, Complementation of the osers1 mutant. C, Inflorescences with mature seeds. D, Average seed-setting rates in panicles (n = 20 for each set). Error bars represent sd. E, Mature anthers. COM, Complementation; WT, wild type. Bars = 2 cm in C and 1 mm in E.
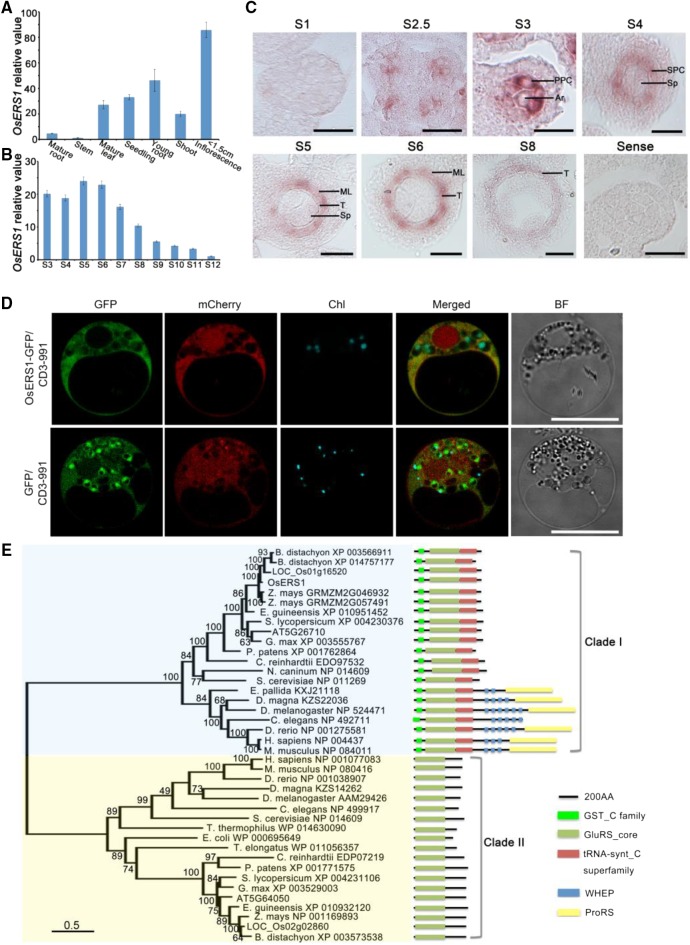
OsERS1 encodes a putative glutamyl-tRNA synthetase (GluRS) that contains an N-terminal GST_C (C terminus of GST) domain, a glutamyl-tRNA synthetase catalytic core, and a C-terminal tRNA-synt_C domain (Fig. 2B). RT-qPCR showed that OsERS1 was expressed in the wild type at low levels in mature organs but at high levels in young tissues, including seedlings and young inflorescences (Fig. 3A). In the anther, OsERS1 transcript levels were relatively high during stages 3 to 6 and decreased in later stages (Fig. 3, B and C). In situ hybridization detected OsERS1 mRNA in presumptive archesporial cells and neighboring L2-d cells before stage 3. During stages 3 to 5, when anther cells undergo rapid cell division, OsERS1 mRNA was detected in PPC and archesporial cells of stage 3, the SPC and sporogenous cells of stage 4, and the tapetum, middle layer, and sporogenous cells of stage 5. The expression of OsERS1 in germ cells decreased significantly after stage 5 (Fig. 3C; Supplemental Fig. S4A). The rice genome has two OsERS1 homologs, LOC_Os01g16520 and LOC_Os02g02860, but these homologs had low expression in the anther and did not appear to have altered expression in the osers1 mutant, suggesting that the phenotype of the osers1 mutant was caused by aberrant OsERS1 in the anther as opposed to the two homologs (Supplemental Fig. S4B). Considering that protein synthesis is essential in the cytosol, chloroplast, and mitochondria, we performed protein localization studies in rice protoplast cells and onion (Allium cepa) cells by coexpressing OsERS1-eGFP and the mitochondrial marker ScCOX4-mCherry (carried by vector CD3-991; Nelson et al., 2007). Our results showed that OsERS1 is localized mainly in the cytoplasm and is partially shared by the mitochondria (Fig. 3D; Supplemental Fig. S4C).
Figure 3.
Expression pattern and phylogenetic analysis of OsERS1. A and B, RT-qPCR data showing relative expression levels of OsERS1 in various tissues (A) and anthers at various stages (B). For B, total RNA was extracted from whole rice flowers before stage 6 and from anthers after stage 7. All error bars represent sd (n = 3). C, In situ hybridization of OsERS1 in the wild-type anther at different stages. A sense probe was used as the control. Ar, Archesporial cell; ML, middle layer; Sp, sporogenous cell; T, tapetum. Bars = 15 μm. D, Subcellular localization of OsERS1 in rice protoplasts. GFP was used as a control. Vector CD3-991 containing the mitochondria-specific protein ScCOX4 fused with mCherry served as the organelle marker. Chl, Chloroplast; BF, bright field. Bars = 10 μm. E, Neighbor-joining tree of OsERS1 and its homologs showing the progressive recruitment of novel functional domains. AA, Amino acids; GST_C, C terminus of GST; GluRS_core, glutamyl-tRNA synthetase catalytic core; tRNA-synt_C, tRNA synthetase class I anticodon binding domain; ProRS, prolyl-tRNA synthetase.
Phylogenetic analysis of 40 OsERS1 homologs from 21 species from diverse kingdoms clearly divided these proteins into two clades: the eukaryote-specific GluRSs (clade I) and the prokaryote-originated GluRSs (clade II; Fig. 3E; Supplemental Table S1). Intriguingly, most eukaryotes contain both types. Clade II GluRS contained only the catalytic domain, while clade I GluRSs showed more complex structures. The progressive appearance of novel domains, such as GST_C, tRNA-synt_C, repeats of the WHEP domain (initially discovered in human tryptophanyl-tRNA synthetase [TrpRS; W], histidyl-tRNA synthetase [H], and GluRS-prolyl-tRNA synthetase [ProRS; EP]), and chimeric GluRS-ProRS in clade I, indicated multiple domain-recruiting events during evolution (Gao et al., 2010). In grass crops such as rice and maize, there seemed to be a duplication of GST_C containing GluRS, which we did not observe in other species (Fig. 3E), indicating that GluRS might have an expanded function for evolutionary adaption in higher organisms.
OsERS1 Functions as a Ligase between Glu and tRNAGlu
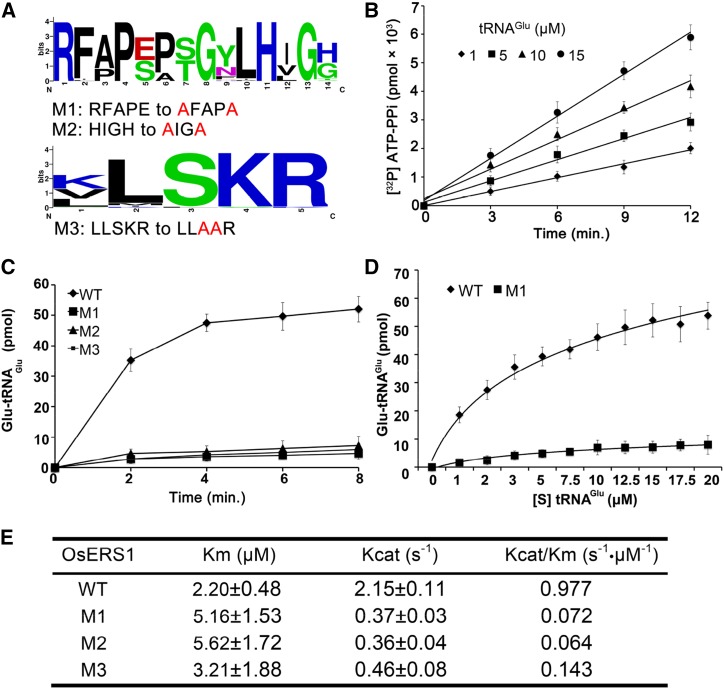
The synthesis of glutamyl-tRNA is a two-step reaction: activation of Glu by ATP and glutamylation of tRNAGlu. In the catalytic domain, GluRS has a core active site (RFAPE in OsERS1) that binds Glu and two key motifs called HIGH and KMSKS (LLSKR in OsERS1) that are involved in ATP binding and tRNA binding, respectively (Freist et al., 1997; Ibba and Söll, 2000). Sequence alignment showed that these three motifs are highly conserved in the 40 GluRS homologs (Fig. 4A; Supplemental Fig. S5).
Figure 4.
OsERS1 functions as a ligase of Glu and tRNAGlu. A, Conserved sites in the catalytic domain of OsERS1 used for site-directed mutagenesis. The figure was generated from 40 homologs of OsERS1 by WebLogo (http://weblogo.berkeley.edu). B, ATP-PPi exchange rate in the presence of increased concentrations of tRNAGlu. C, tRNAGlu aminoacylation by wild-type (WT) and mutant (M1–M3) OsERS1. D, Kinetics of tRNAGlu aminoacylation by wild-type OsERS1 and OsERS1 mutant M1 under increased tRNAGlu concentrations. Wild-type OsERS1, R2 = 0.99; OsERS1 mutant M1, R2 = 0.97. E, Kinetic constants determined by tRNAGlu charging assays. Kinetics of the OsERS1 and OsERS1 M1 to M3 aminoacylation reaction was measured using varied concentrations of tRNAGlu, and reactions were conducted for 10 min under standard conditions. Reciprocal initial velocity was plotted against the reciprocal tRNAGlu concentration according to Lineweaver-Burk to calculate the corresponding Km values. All error bars represent sd (n = 3).
To confirm the function of OsERS1 in glutamyl-tRNA synthesis, we performed an ATP-pyrophosphate (PPi) exchange assay, which showed that OsERS1 has PPi exchange activity and that its reaction rate is proportional to the concentration of tRNAGlu (Fig. 4B). An aminoacylation assay revealed that OsERS1 has the catalytic activity of aminoacylation of tRNAGlu with Glu (Fig. 4, C and D), yielding a Km value (tRNAGlu as substrate) of 2.2 μm and a Kcat/Km value of 0.977 s–1 μm–1 (Fig. 4E). OsERS1’s affinity for tRNAGlu is similar to that of the Thermus thermophilus GluRS but lower than that of Rattus spp. liver and Escherichia coli (Freist et al., 1997). Also, OsERS1’s Km value is lower than that of wheat (Triticum aestivum) GluRSs, possibly because the latter function as dimers (Thomes et al., 1983). However, it is still not clear whether GluRS can form dimers in rice.
Mutations of the three conserved motifs of OsERS1s, Arg-209 and Glu-213 in the active site region, His-219 and His-222 in the HIGH motif, and Ser-442 and Lys-443 in the KMSKS motif (Fig. 4A; Supplemental Fig. S5), caused significantly reduced glutamylation activity and enzyme efficiency (Kcat/Km; Fig. 4, C–E). These results suggest that OsERS1 has catalytic activity in tRNAGlu glutamylation and that the RFAPE, HIGH, and KMSKS motifs are required for OsERS1’s enzymatic function.
The Cofactor OsARC Stimulates the Catalytic Activity of OsERS1
The GST_C domain of GluRS is noncatalytic because it lacks thioredoxin, but it was shown to link the enzyme to protein complexes (Dixon et al., 2002). In yeast and mammals, GluRS/GluRS-ProRS interacts with the aaRS cofactor Arc1p/p18 to form a complex and modulate enzyme activities (Galani et al., 2001; Graindorge et al., 2005; Gao et al., 2010). Rice has a single homolog of yeast Arc1p, OsARC (encoded by LOC_Os01g60660), which contains a heterogeneous GST_C domain (Fig. 5A). To examine whether OsERS1 can form a complex with OsARC, we conducted yeast two-hybrid and in vitro pull-down assays; these confirmed that OsERS1 can interact with OsARC and that the interaction requires the GST_C domains (Fig. 5, B and C;. Bimolecular fluorescence complementation (BiFC) showed that only the cells expressing full-length OsERS1-nYFP and OsARC-cYFP displayed yellow fluorescence signals (Fig. 5D), verifying this interaction in vivo. OsERS1-eGFP and OsARC-mCherry were highly colocalized when coexpressed in onion cells (Fig. 5E), supporting our hypothesis that OsERS1 and OsARC can form a complex. Moreover, compared with that of OsERS1 alone or OsERS1-ΔOsARC (truncated OsARC protein without the GST_C domain; Fig. 5, F and G), the OsERS1-OsARC complex had ∼2.5 times the ATP-PPi exchange rate of Glu and ∼1.4 times the tRNAGlu aminoacylation efficiency in the plateau stage, indicating an enhancement effect promoted by OsARC. In addition, the GluRS and cofactor pairs are conserved components of MSCs in various organisms (Laporte et al., 2014). However, the structure and composition of plant MSC have not been dissected.
Figure 5.
The complex of OsERS1 and OsARC enhances OsERS1’s catalytic activity. A, Domain structures of OsERS1 and OsARC and their truncations. B, OsERS1 and OsARC interact in yeast. Interaction is verified by growth of the yeast strain in selective medium. C, Pull-down assays between His-OsERS1 and MBP-OsARC in vitro. The precipitated MBP-OsARC was detected by α-MBP antibody. MBP-tagged ΔOsAR and His-tagged ΔOsERS1 were used as a control to show the interaction in a GST_C domain-dependent manner. CBB, Coomassie Brilliant Blue; WB, western blot. D, BiFC assays in onion epidermal cells showing the interaction between OsERS1 and OsARC. cYFP, C-terminal split of YFP; nYFP, N-terminal split of YFP. Bars = 50 μm. E, Colocalization of OsERS1-eGFP and OsARC-mCherry in onion epidermal cells. Bar = 50 μm. F, Influence of the OsERS1-OsARC complex on ATP-PPi exchange. A total of 15 μm tRNAGlu was used as the substrate. G, Influence of the OsERS1-OsARC complex on tRNAGlu aminoacylation. All error bars represent sd (n = 3).
Mutation of OsERS1 Leads to Metabolic Alterations in Anthers
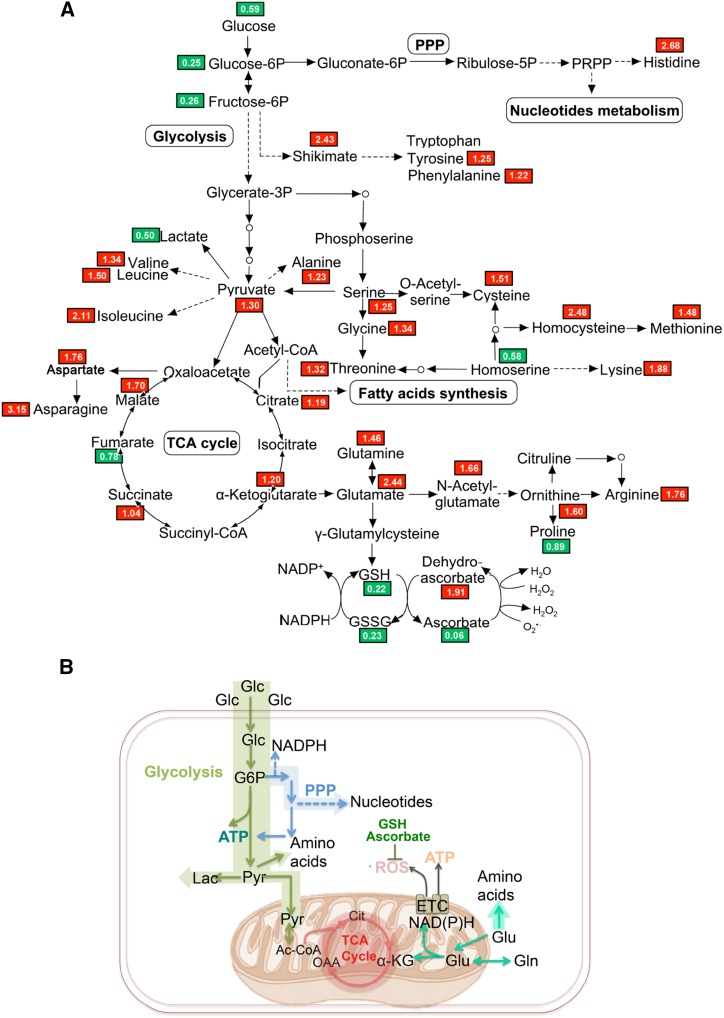
The substrate for OsERS1, Glu, occupies a central position that links key metabolic pathways (Newsholme et al., 2003). Loss of function of OsERS1 may cause an accumulation of free Glu that is not used for protein synthesis in the anther. To test this possibility, we conducted nontargeted metabolomics profiling in wild-type and osers1 anthers. A total of 297 metabolites were identified, covering all major primary and several secondary metabolism pathways (Supplemental Table S2). Consistent with the deficiency in ligating Glu with tRNAGlu, the osers1 mutant had 2.44 times the level of Glu compared with the wild type. With the exception of glutathione in both reduced (GSH) and oxidized (GSSG) status, the levels of Glu family members, including Gln, α-ketoglutarate (α-KG), His, and Arg, were increased in the osers1 mutant. One Glu derivative, α-KG, also is an intermediate of the tricarboxylic acid cycle and links the metabolism of amino acids and carbohydrates (Jeong et al., 2013). As a result, most metabolites of the tricarboxylic acid cycle, such as oxaloacetate (OAA) and malate, were increased in the osers1 mutant (Fig. 6A; Supplemental Fig. S6B; Supplemental Table S2). This is also consistent with changes in the expression of genes involved in Glu metabolism and the tricarboxylic acid cycle, such as genes encoding Gln synthetases (GS2 and GS3), Glu dehydrogenase (GDH1), and oxoglutarate dehydrogenase (ODH2; Supplemental Table S3). Besides the Glu family, the Leu, Ser, and Asp families derived from pyruvate, phosphoglycerate, and OAA also were increased in the osers1 mutant. We propose that increased levels of amino acids in the mutant may have been caused by the accumulation of Glu, OAA, and pyruvate, which are important precursors for amino acid synthesis (Fig. 6; Supplemental Fig. S6A; Supplemental Table S2).
Figure 6.
Metabolic rewiring in the osers1 mutant. A, The identified important metabolites are mapped onto a simplified metabolic network. Red and green blocks indicate metabolites with higher and lower average levels, respectively, in the osers1 mutant compared with the wild type. All values are averages of three independent trials. B, Putative metabolism in osers1 anthers. Major metabolic pathways are shown, including glycolysis, pentose phosphate pathway (PPP), tricarboxylic acid (TCA) cycle, and potential ROS producers and scavengers. Ac-CoA, Acetyl-coenzyme A; Cit, citrate; ETC, electron transport chain; α-KG, α-ketoglutarate; Lac, lactate; Pyr, pyruvate.
In addition, we detected the expression level of 12 aaRS genes highly expressed in reproductive organs. Most of the aaRS genes showed subtle expression changes between the wild type and the osers1 mutant, except for CysRS1 and TrpRS2, which had lower expression in the osers1 mutant (Supplemental Fig. S7), implying that the amino acid changes in the mutant are caused mainly by the loss of function of OsERS1. By contrast, we observed decreased levels of glycolysis intermediates, including Glc, Glc-6-P, and Fru-6-P, in the osers1 mutant (Fig. 6; Supplemental Fig. S6B); this indicates a higher consumption of these compounds in the osers1 mutant. Decreased Glc supply and less lactate production suggest that pyruvate was likely used by the tricarboxylic acid cycle and for amino acid biosynthesis. Increased expression of PPP genes and the accumulation of purine and pyrimidine indicate that more intermediates of glycolysis were used to support nucleotide biosynthesis in osers1 anthers (Supplemental Fig. S6C; Supplemental Tables S2 and S3). Metabolic analysis showed abnormal fatty acid synthesis in the osers1 mutant, such as cutin and wax, which is consistent with the scanning electron microscopic analysis of osers1 anther wall and pollen grain (Supplemental Fig. S1, G–N, and S6D; Supplemental Table S2).
Overall, the osers1 anthers show more active amino acid and carbohydrate metabolism with aerobic characteristics, such as an active tricarboxylic acid cycle and less lactate production. The increased Glu family amino acids, as well as those derived from OAA and pyruvate, contribute to the significant changes in biosynthesis of the amino acid pathway in osers1 anthers (Fig. 6B; Supplemental Fig. S6A; Supplemental Table S2). Notably, Glu may provide mitochondrial anaplerosis to the tricarboxylic acid cycle to stimulate mitochondrial activities, which produces ROS as an inevitable by-product. Consistent with this, decreased cellular antioxidants (GSH/GSSG and ascorbate) suggest a disturbed redox status in osers1 anthers (Fig. 6B). Corresponding with the activated cell division in osers1 anthers, more carbohydrates are consumed in glycolysis to generate energy and provide more intermediates for the PPP, which enhances the synthesis of nucleotides and macromolecules. These results reveal that the metabolic homeostasis is disturbed in osers1 anthers and that OsERS1 plays key roles in amino acid-related metabolism.
Altered Redox Status Affects Early Anther Development
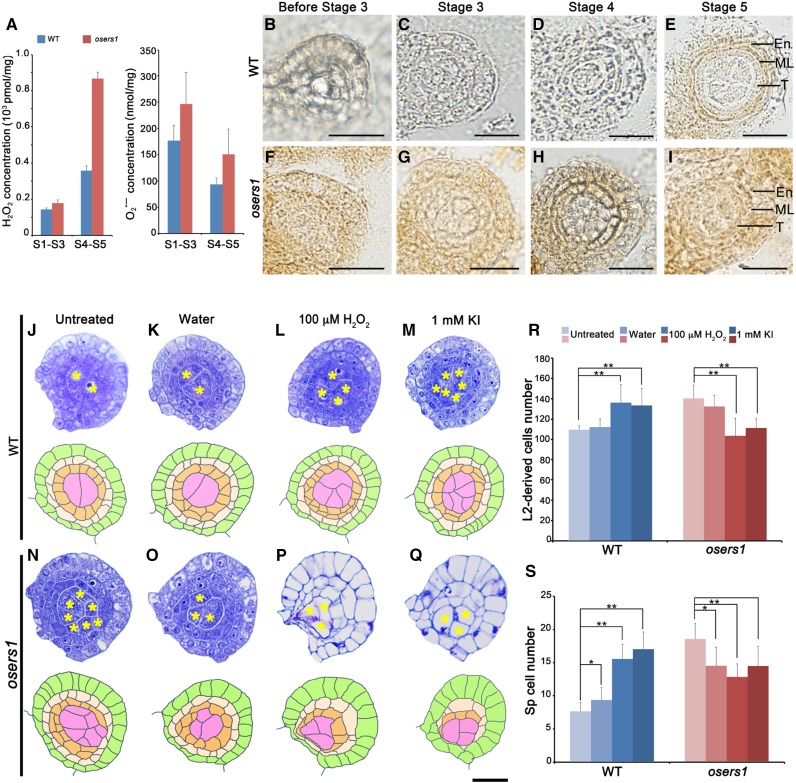
Global metabolomics profiling suggested that ROS might be produced by mitochondrial activities in the osers1 mutant. To verify this, we first measured hydrogen peroxide (H2O2), a stable ROS molecule implicated in cell proliferation and differentiation (Møller et al., 2007; Tsukagoshi et al., 2010). Fluorometric quantitation assays showed that H2O2 content in the osers1 mutant at stages 4 to 5 was ∼900 pmol mg−1, twice that in wild-type anthers at the same stage (Fig. 7A, left). Also, the precursor of H2O2, superoxide radical, was increased slightly in the osers1 mutant (Fig. 7A, right), which contributed to the redox status change in the osers1 mutant. In addition, 3,3′-diaminobenzidine (DAB) staining of wild-type anthers showed that the H2O2 signal was preferentially present in middle layer and endothecium cells until stage 5 (Fig. 7, B–E). However, osers1 anthers displayed detectable H2O2 in the anther primordium before stage 3 (Fig. 7F) and strong H2O2 signals in anther cells at stages 4 to 5 (Fig. 7, G–I). In later stages, even though the distribution of H2O2 was similar in both wild-type and osers1 anther wall layers, stronger H2O2 signals were seen in osers1 anther wall layers (Supplemental Fig. S8). The lower expression levels of genes encoding ascorbate peroxidases in the mutant (Supplemental Table S3) may have contributed to the accumulation of ROS.
Figure 7.
Redox status controls L2-d cell division and organization. A, Measurement of H2O2 and superoxide radicals (O2•−) in young spikelets. All error bars represent sd (n = 3). S1–S3, Stages 1 to 3; S4–S5, stages 4 to 5. B to I, DAB staining analysis for H2O2 in anther lobes. En, Endothecium; ML, middle layer; T, tapetum; WT, wild type. Bars = 15 μm. J to Q, Cross sections (top rows) and diagrams (bottom rows) of wild-type and osers1 anther lobes of stage 4 under treatments. Yellow asterisks indicate sporogenous cells. Bar = 15 μm. R and S, Average counts of L2-d cells (R) and sporogenous(-like) cells (S) in cross sections from anthers of stage 4 under the indicated treatments. All error bars represent sd (wild type, n = 16; wild type-water, n = 12; wild type-H2O2, n = 18; wild type-KI, n = 17; osers1, n = 14; osers1-water, n = 13; osers1-H2O2, n = 16; osers1-KI, n = 15). Student’s t test: *, P < 0.05 and **, P < 0.01.
To test whether the increased level of H2O2 in the osers1 mutant causes the abnormal division and patterning of L2-d cells, we injected 100 μm H2O2 and 1 mm of the ROS-removal reagent potassium iodide (KI) into young inflorescences of the wild type and the osers1 mutant (Supplemental Fig. S9). Transverse section observations revealed that each stage 4 wild-type anther treated with water showed a slight increase in the number of sporogenous-like cells, from 7.5 (n = 16) to 9.2 (n = 12), which may have been triggered by wounding (Fig. 7, J, K, and S). Remarkably, the number of L2-d cells in each wild-type anther treated with H2O2 increased from 109 (n = 16) in untreated anthers to 135.4 (n = 18), and sporogenous-like cell numbers increased from 7.5 to 15.4. Therefore, treatment with 100 μm H2O2 in the wild type led to overproliferation of L2-d cells, ectopic sporogenous-like cells, and disordered parietal cell layers, phenocopying the defects of the osers1 mutant (Fig. 7, L, R, and S). Wild-type anthers treated with KI also showed increases in the number of L2-d cells to 132.7 and sporogenous-like cells to 16.8 (n = 17), while parietal cells were organized differently compared with those treated with H2O2 (Fig. 7, M, R, and S). Compared with that of the untreated anther (139.3 L2-d cells and 18.5 sporogenous-like cells; n = 14), in the osers1 mutant, each anther treated with water showed a slight decrease of L2-d cell numbers to 131.5 and sporogenous-like cell numbers to 14.5 (n = 13; Fig. 7, N, O, R, and S). However, each osers1 anther treated with H2O2 and KI showed a reduced number of L2-d cells and sporogenous-like cells (H2O2, 102.9 and 12.7, respectively [n = 16]; KI, 110.5 and 14.5, respectively [n = 15]). Aside from changes in cell numbers, we observed highly vacuolated cells in impaired osers1 anthers (Fig. 7, P–S). We reasoned that elevations of ROS by H2O2 injection and neutralization of ROS by KI injection both disturbed ROS homeostasis, which may contribute to repressed cell division, and that the osers1 anthers appear vulnerable to changes in redox status. Together, these results suggest that changed redox status disturbs early cell division in rice anthers.
DISCUSSION
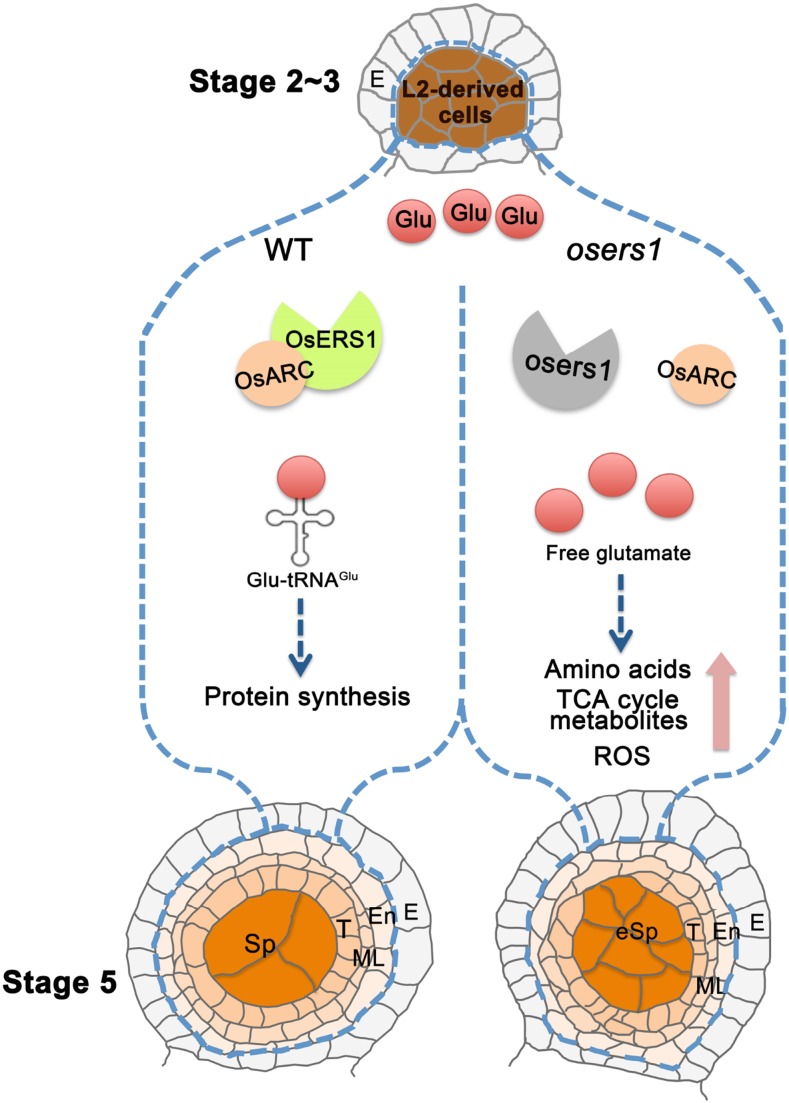
Early anther development is a complex event characterized by rapid cell division and fate specification to support the development of functional anther layers that are properly organized within the anther (Zhang and Yang, 2014; Walbot and Egger, 2016). In this study, we revealed that OsERS1, a glutamyl-tRNA synthetase, functions in early anther cell division and patterning, likely through maintaining physiological homeostasis of amino acids and carbohydrate metabolism as well as redox status (Fig. 8).
Figure 8.
Working model for OsERS1 function in rice anthers. In rice anthers, L2-d cells generate somatic endothecium, middle layer, tapetum, and germinal sporogenous cells. OsERS1 is expressed specifically in L2-d cells at high levels during stage 2 to 3 and later in cells differentiated from L2-d cells. In the wild type (WT), OsERS1 combined with OsARC are required for protein synthesis and to maintain L2-d cell number and patterning during early anther development. Loss of function of OsERS1 resulted in the accumulation of free Glu and changes in amino acids and their derived metabolic pathways. Subsequently, ROS produced by metabolic activities affected redox status and cell behavior, leading to changes in anther development. E, Epidermis; En, endothecium; eSp, excess sporogenous-like cell; ML, middle layer; Sp, sporogenous cell; T, tapetum; TCA, tricarboxylic acid.
Our genetic and cytological data demonstrate that loss of function of OsERS1 results in fused anther lobes with supernumerary L2-d cells and disarranged anther wall cell layers. In osers1 anthers, L2-d cell division was accelerated, but the L1 layer remained normal and formed a rigid enclosure, which aggravated the disarranged cell layers and created abaxial fused lobes. In spite of its putative housekeeping function, expression of OsERS1 shows spatial-temporal characteristics rather than a constitutive pattern. In the anther, OsERS1 transcripts are initially expressed in cells of presumptive lobes at the four primordial corners. Then, OsERS1 mRNA is detected in both parietal and germ cells, preferentially in those undergoing periclinal division or newly formed cell layers (Fig. 3C). After stage 5, the germ cells develop into microspores via meiosis, and the abundance of OsERS1 mRNA in germ cells decreased to a very low level, indicating that OsERS1 mainly affects early cell division and patterning. This expression pattern is in agreement with the phenotype of increased L2-d cells and disarranged cell layers in the osers1 mutant.
Phenotypically, mutations of the LRR-RLK-ligand pair MSP1-MIL2 or the GRX MIL1 lead to misidentified anther wall cell layers and complete male sterility (Nonomura et al., 2003; Zhao et al., 2008; Hong et al., 2012a,b), while the osers1 anther still produces a low proportion of viable pollen grains (Supplemental Fig. S1). Anther defects in the osers1 mutant occur earlier than those in the reported rice mutants with abnormal early anther development, such as gamyb-4, udt1 (undeveloped tapetum1), and tip2 (tdr interacting protein2; Jung et al., 2005; Liu et al., 2010; Fu et al., 2014). We showed that there are no significant changes in the expression of MSP1, MIL2, and MIL1 in the osers1 mutant compared with the wild type, and previous studies showed that OsERS1 has no obviously altered expression in msp1, mil2, gamyb-4, udt1, or tip2 mutants (Jung et al., 2005; Aya et al., 2009; Liu et al., 2010; Fu et al., 2014; Yang et al., 2016). Therefore, we propose that OsERS1 plays an independent role with known regulators in early anther development. Although loss of function of OsERS1 causes abnormal anther morphogenesis and decreased pollen vitality, the mutant still can form a small number of seeds with normal shape, suggesting that fertilization is not completely disturbed. However, we cannot exclude the possibility that female gametophyte development also is affected.
We confirmed the in vitro activity of OsERS1 in catalyzing the ligation between Glu and tRNAGlu. OsERS1 is dominantly localized in the cytosol and partially shared by mitochondria. The product, glutamyl-tRNA, is an essential substrate for protein synthesis. Compared with the prokaryotic (E. coli) GluRS (Freist et al., 1997), OsERS1 possesses a relatively lower affinity to tRNAGlu. One explanation is that the additional GST_C domain may change space conformation and reduce substrate affinity. However, the interaction between OsERS1 and its cofactor OsARC overcomes this disadvantage by enhancing the activity of OsERS1 for synthesizing glutamyl-tRNA (Fig. 5). In the early anther, loss of function of OsERS1 may impair translational processes in the cells that normally accumulate high OsERS1. However, the osers1 anthers maintain partial fertility and produce viable pollen, suggesting that other components may remedy the absence of OsERS1 to maintain protein synthesis in the anther.
Metabolic status is associated with tumorigenesis, stem cell renewal, pluripotency maintenance, and differentiation (Cairns et al., 2011; Schuster et al., 2014; Sperber et al., 2015; Ito and Ito, 2016). In plants, glycolysis and mitochondrial metabolism activate the Arabidopsis root meristem via Glc-target of rapamycin signaling (Xiong et al., 2013). In maize, the anther primordium is more likely glycolysis dependent, as the enclosed niche provides hypoxic conditions for archesporial cells (Kelliher and Walbot, 2012). We speculate that the hypoxia status specifying cell identity and cell division must be well controlled by the specific metabolic pathway within anther cells. In this study, we showed that, as an important metabolic regulatory node, OsERS1 is involved in intracellular Glu dynamics, and Glu could refuel the tricarboxylic acid cycle that occupies a central position in multiple metabolic pathways. Within this context, the Arabidopsis cytosolic GluRS is an important target of 14-3-3 proteins in carbon/nitrogen metabolism and sugar-sensing pathways. GluRS is degraded when sugar starvation occurs in order to release Glu and support cell survival (Cotelle et al., 2000; Coruzzi and Zhou, 2001). In this work, we showed that the mutation of OsERS1 disturbs the homeostasis of the metabolic network, as evidenced by increased amounts of amino acids, tricarboxylic acid components, and active aerobic metabolism. We speculate that such a metabolic shift acts as a signal to remove the limitation on cell proliferation in the anther lobe. Consistent with this, some mutants of 21 Arabidopsis aaRSs showed defective female gametogenesis and embryo development but no obvious defect during male gametogenesis except for in the ova9 mutant, which had a mutation in GlnRS (Berg et al., 2005). However, how these specific aaRSs are involved in reproductive development remains unclear. Although we observed no obvious change of other aaRS genes in the osers1 mutant (Supplemental Fig. S7), we cannot rule out the function of other aaRSs in male reproduction.
ROS exert dose-dependent impacts on cells. A lower or moderate basal level of ROS is essential for cell proliferation and differentiation, but higher levels of ROS are cytotoxic, causing cell damage and death (Sena and Chandel, 2012; Schieber and Chandel, 2014; Mittler, 2017). In rice anthers, the ROS molecule H2O2 is initially accumulated in the middle layer at stage 5; this may act as a signal that triggers cell differentiation (Zhang and Yang, 2014; Yang et al., 2016). In this study, we observed the production of H2O2 in osers1 anthers as early as the formation of archesporial cells, which may cause the overproliferation of anther cells and disorganized cell layers (Fig. 7). In support, disruption of the redox status in wild-type anthers by injecting low concentrations of H2O2 resulted in a mimic of the phenotype of the osers1 mutant (Fig. 7). Similarly, in the shoot apical meristem of Arabidopsis, elevating H2O2 dramatically enlarges meristem size by increasing cell numbers in the peripheral zone (Zeng et al., 2017). In maize, treatment with a high concentration of H2O2 (1 mm) on the anther inhibits cell division in both somatic and germ cells (Kelliher and Walbot, 2012), demonstrating multifaceted regulatory aspects of ROS in cell division. The study in maize also indicates that the anther primordium may use glycolysis as a major energy source, rather than through Glc oxidation in mitochondria to avoid ROS pressure and maintain genomic integrity (Kelliher and Walbot, 2012). In this study, we demonstrated that increased ROS causes the overproliferation of early anther cells in the mutant, suggesting that reductive redox status is essential to maintain normal cell division and organization during rice anther development.
In summary, we discuss the role of a conserved glutamyl-tRNA synthetase, OsERS1, in limiting anther cell division and maintaining cell patterning (Fig. 8). The catalytic activity of OsERS1 in ligating Glu to tRNAGlu can be enhanced by its cofactor OsARC. Loss of function of OsERS1 causes changes in a variety of metabolites, such as the accumulation of amino acids and tricarboxylic acid cycle intermediates, and leads to aerobic metabolic features in the mutant. Together with the H2O2 assays, we concluded that OsERS1 maintains normal early anther cell division and patterning, likely through affecting protein synthesis, amino acid homeostasis, and redox status in rice. Our study reveals how the aaRSs control the cell division and organ development derived from meristematic cells in plants.
MATERIALS AND METHODS
Materials, Plant Growth, and Phenotypic Analysis
The osers1 mutant was isolated from a rice (Oryza sativa ssp. japonica) mutant library in the cv 9522 background (Chen et al., 2006). Rice plants were grown in the artificial atmospheric chambers on the Waite Campus, University of Adelaide, and in a paddy field at Shanghai Jiao Tong University. Plants and flowers were photographed with a Nikon E995 digital camera and a Motic K400 dissecting microscope. Photography of plant materials and observation of anthers were done as described by Li et al. (2006) and imaged with a Nikon Ni-E optical microscope and a Philips XL20 scanning electron microscope. Cell counts were determined based on cross sections from more than 10 anthers, and statistical analysis included all four lobes of whole anthers.
Map-Based Cloning and Complementation of the Mutant
For fine-mapping, we identified 158 mutants from an F2 population that was generated from a cross between the osers1 mutant (japonica) and cv 9311 (indica). To map the OsERS1 locus, we used bulked segregation analysis and developed insertion-deletion molecular markers based on the sequence differences between rice cultivars, described in the National Center for Biotechnology Information. The OsERS1 gene was initially located to chromosome 10 between two insertion-deletion molecular markers, 10tys14 and 10tys2, and finer mapping narrowed down the gene to the region between RM25267 and 10tys28. Mapping combined with genome sequencing was performed using the previously published method (Abe et al., 2012; Lü et al., 2015).
For functional complementation, an 8.3-kb genomic DNA fragment containing the entire OsERS1 coding region, a 3.2-kb upstream sequence, and a 1.7-kb downstream sequence was amplified from a BAC clone and subcloned through EcoRI and BstElI into the binary vector pCAMBIA1301 (p1301), which carries a hygromycin resistance marker, to generate p1301-OsERS1. We induced calli from young panicles of the homogenous osers1 mutant and transformed the calli with Agrobacterium tumefaciens EHA105, which carried the p1301-OsERS1 plasmid. At least 25 independent transgenic lines were obtained after PCR genotyping using the primers listed in Supplemental Table S4.
RT-qPCR
We extracted total RNA from plants using the TRIZOL reagent (Invitrogen), following the manufacturer’s instructions. cDNA was synthesized from 4 μg of total RNA using a reverse transcriptase kit (Bio-Rad). SYBR Green Mix (Kapa) was used for RT-qPCR on a QuantStudio Flex 6 (Life Technologies) machine. The level of ACTIN2 mRNA was used as the internal control. For expression analysis of OsERS1, aaRSs, metabolism-related genes, or anther developmental genes, we denatured the DNA at 95°C for 10 min, followed by 42 to 45 cycles at 95°C for 10 s, 56°C for 15 s, and 72°C for 15 s. The RT-qPCR results shown are from representatives of experiments repeated at least three times with similar results. Primer sequences are listed in Supplemental Table S4.
RNA in Situ Hybridization
We amplified OsERS1-, MSP1-, H4-, and CDKB2;1-specific fragments of the exon region (OsERS1, 70–391 bp; MSP1, 3,462–3,841 bp; H4, full length; CDKB2;1, 415–739 bp) by PCR using specific primers fused with the T7 promoter (sequences are listed in Supplemental Table S4). Digoxigenin-labeled antisense and sense probes were transcribed using an in vitro transcription kit (Roche) according to the manufacturer’s instructions. In situ hybridization was performed according to a published protocol (Zeng et al., 2017).
Subcellular Localization
35Spro::OsERS1-eGFP (full-length OsERS1 cDNA fused with eGFP) was generated in pCAMBIA1301 and expressed in rice protoplasts and onion (Allium cepa) epidermal cells using PEG-mediated transfections and a particle delivery system, respectively. The construction of the organelle marker (CD3-991) was described by Nelson et al. (2007). For rice protoplast cells, stem tissues of 14-d-old seedlings were processed following the protocol described by Zhang et al. (2011). For onion epidermal cells, the onion was cut into 2- × 2-cm squares and placed on a plate for 4 h with Murashige and Skoog medium containing 40 g L−1 d-mannitol. For transformation, 5 μg of each plasmid DNA was coated with 1-μm gold particles (Bio-Rad) and delivered into onion epidermal cells using a PDS-1000/He particle delivery system (Bio-Rad). Fluorescence microscopy was performed using a Leica TCS SP5 laser scanning confocal microscope. Fluorescence signals for mCherry (excitation, 587 nm; emission, 580–630 nm; 15% power), eGFP (excitation, 483 nm; emission, 500–530 nm; 15% power), and chloroplasts (excitation, 504 nm; emission, 650–750 nm; 15% power) were detected with a 20× (dry) objective for onion cells and a 63× (oil) objective for protoplast cells.
Phylogenetic Analysis and Motif Alignment
We constructed a phylogenetic tree with aligned full-length sequences of homologs of OsERS1. MEGA (version 7.0) and the neighbor-joining methods were used with a p-distance model and pairwise deletion and bootstrap (Gap Ktuple, 2; length penalty, 5; Window, 4; Diagonals, 4; 1,000 replicates). The maximum parsimony method of MEGA also was used to support the neighbor-joining tree using the default parameter. Amino acid sequences from regions 199 to 228 and 434 to 454 in OsERS1 were used for motif alignment by MEGA.
Recombinant Protein Purification
Wild-type and mutant OsERS1 were cloned into the pET32(a) vector for expression in the BL21 (DE3) Escherichia coli strain. Expression of the wild type and its variants was induced in Luria-Bertani broth with 0.5 mm isopropyl β-d-1-thiogalactopyranoside for 4 h at 23°C. We purified the proteins with Ni-NTA affinity resin (Qiagen) according to standard procedures and dialyzed against storage buffer containing 50 mm Tris-HCl (pH 7.5), 100 mm NaCl, 10 mm 2-mercaptoethanol, 3 mm MgCl2, and 50% (v/v) glycerol. The coding sequences of OsARC and ΔOsARC (deletion of GST_C) were fused to the N terminus of maltose-binding protein (MBP) in the vector pMAL2c-x. We expressed the fusion proteins in E. coli (strain BL21) and purified them using amylose resin (New England Biolabs) according to the manufacturer’s instructions. Supplemental Table S4 lists the primer sequences for the constructs.
tRNA Transcription and Purification
The DNA fragment covering the T7 promoter and the tRNAGlu gene (http://gtrnadb.ucsc.edu/#eukarya; chr1.trna78-GluTTC; 5′-GCCCCTATCGTCTAGTGGTTCAGGACATCTCTCTTTCAAGGAGGCAGCGGGGATTCGACTTCCCCTGGGGGTA-3′) was synthesized as a double-stranded molecule by Invitrogen, phosphorylated, and ligated into pUC19 (precleaved by EcoRI and BamHI) to construct the plasmid pUC19-tRNAGlu. T7 in vitro transcription was carried out at 37°C in a reaction mixture containing 40 mm Tris-HCl (pH 8), 22 mm MgCl2, 1 mm spermidine, 5 mm DTT, 0.5% (v/v) Triton X-100, 60 to 80 ng μL−1 tDNA template, 5 mm NTP (each), 0.8 units μL−1 RNase inhibitor, 20 mm GMP, 500 units μL−1 T7 RNA polymerase, and 1 unit mL−1 pyrophosphatase for 3 h. Subsequently, 5 units mL−1 DNase I (RNase I free) was added and incubated for 1 h to digest the transcription template. We loaded the transcripts onto a 15% (w/v) PAGE-8M urea gel with 1 mm thickness and 40 cm length and ran the gel at a constant 100 W for 4 h. The tRNA band was cut from the gel and eluted with 0.5 m NaAc (pH 5.2) at room temperature (three times), ethanol precipitated at 20°C after two phenol/chloroform extractions, and dissolved in 5 mm MgCl2. The tRNA was denatured at 80°C for 5 min and slowly cooled to room temperature (Fang et al., 2014).
ATP-PPi Exchange and Aminoacylation
The ATP-PPi exchange reaction mixture contains 100 mm Tris-HCl (pH 7.8), 12 mm MgCl2, various concentrations of tRNAGlu, 5 mm l-Glu, 4 mm ATP, 2 mm [32P]tetrasodium pyrophosphate (PerkinElmer), 0.1 mg mL−1 BSA, and 100 nm His-OsERS1 with or without 100 nm MBP-OsARC (or MBP-ΔOsARC, deleted for GST_C). The formation of [32P]ATP at 37°C was followed at various time intervals in 9-μL aliquots. Samples at specific time points were terminated using 1 mL of quenching buffer containing 2% (w/v) activated charcoal, 3.5% (v/v) HClO4, and 50 mm tetrasodium pyrophosphate. The charcoal suspension was put through a Whatman GF/A filter, washed four times with 5 mL of water, and rinsed with 10 mL of 100% ethanol. The charcoal powder on the filters was dried, and the synthesized [32P]ATP was quantified using a scintillation counter (Beckman Coulter; Chen et al., 2000).
We performed aminoacylation reactions in a mixture of 100 mm Tris-HCl (pH 7.8), 30 mm KCl, 12 mm MgCl2, 2 mm DTT, 4 mm ATP, 20 mm [3H]Glu (15 Ci mm−1; PerkinElmer), 15 μm tRNAGlu, and 100 nm His-OsERS1 with or without 100 nm MBP-OsARC (or MBP-ΔOsARC, deleted for GST_C) at 37°C. We used 0.5 to 20 μm tRNAGlu for kinetic constant analyses of OsERS1 and its variants (mutations of three key motifs). Reactions were initiated with the addition of the enzyme and were conducted in a 37°C heat block. Aliquots (9 μL) were taken at different time points and quenched on Whatman filter pads that had been presoaked with 5% (w/v) TCA. The pads were washed three times for 10 min each time with cold 5% (w/v) TCA and once with cold 100% ethanol; the washed pads were then dried. Radioactivity was quantified in the scintillation counter. The mixture of OsERS1 and OsARC was incubated at room temperature for 30 min to allow complexation before ATP-PPi and aminoacylation assays (Chen et al., 2001).
Yeast Two-Hybrid, Pull-Down, BiFC, and Colocalization Assays
We performed yeast two-hybrid experiments according to the manufacturer’s instructions (Clontech). The coding sequences of OsERS1 and ΔOsERS1 (deleted for GST_C) were fused in frame with the GAL4 DNA-binding domain of the bait vector pGBKT7. The coding sequences of OsARC and ΔOsARC were cloned in the prey vector pGADT7. Each bait-prey pair was cotransformed into the yeast strain AH109. To analyze protein-protein interactions using His auxotrophy assays, yeast colonies were patched in duplicate onto His– and His+ plates and kept at 30°C for 2 to 3 d.
Amylose resin beads containing MBP, MBP-OsARC, and MBP-ΔOsARC were incubated with His-OsERS1 in pull-down buffer (15 mm HEPES-NaOH at pH 7.9, 50 mm potassium glutamate, 5 mm magnesium chloride, 5% glycerol, 0.1% Nonidet P-40, 1 μg μL−1 BSA, and 1 mm DTT). We rotated the mixture at 4°C for 1 h and washed the beads five times with wash buffer (15 mm HEPES-NaOH at pH 7.9, 50 mm potassium glutamate, 5 mm magnesium chloride, and 0.1% Nonidet P-40). Proteins were eluted from the beads by boiling in SDS-PAGE sample buffer, and the eluate was analyzed by immunoblot detected with anti-MBP (1:1,000 dilution) antibodies.
The cDNAs of OsERS1, ΔOsERS1, and OsARC were cloned individually into pSAT1-nEYFP-N1 and pSAT1-cEYFP-C1-B vectors that contain either N- or C-terminal eYFP fragments. The method for BiFC assays using onion epidermal cells is described above (“Subcellular Localization”). YFP fluorescence was imaged at an excitation wavelength of 514 nm and emission wavelengths of 525 to 546 nm.
For colocalization between OsERS1 and OsARC1, we constructed 35Spro::OsERS1-eGFP and 35Spro::OsARC-mCherry by cloning cDNA sequences of OsERS1 and OsARC into p1301. The method for colocalization assays using onion epidermal cells is described above (“Subcellular Localization”).
Metabolomics Analysis
Over 120 mg of rice anther from wild-type and osers1 plants was collected, frozen with liquid nitrogen, and stored at –80°C until metabolomics analysis. We ground samples into a fine powder and analyzed methanol extracts from a 40-mg sample using the analytical platform composed of ultra-HPLC-tandem mass spectrometry and gas chromatography-mass spectrometry. METABOLON performed the experiments and raw data analysis (http://www.metabolon.com). Metabolites were named after standard references (http://www.kegg.jp).
ROS Measurement and H2O2 Staining
We collected rice spikelets from wild-type and osers1 plants and sorted them according to spikelet length. Samples were ground into a fine powder and processed according to the instructions of the Hydrogen Peroxide Assay Kit (abcom; catalog no. ab102500). Fluorometric readings were collected from a POLARstar OPTIMA device (BMG Labtech). Three independent experiments were performed to calculate mean values and sd. Aliquots of sample powder were suspended in 65 mm potassium phosphate buffer (pH 7.8) to extract superoxide radicals. Following a modified method (Wang and Jiao, 2000), the reaction mixture (0.5 mL of extract, 0.7 mL of potassium phosphate buffer, and 0.3 mL of 10 mm hydroxylammonium chloride) was incubated at 25°C for 60 min. Then, 0.5 mL of the above reaction mixture was added to 0.5 mL of 19 mm sulfanilic acid and 0.5 mL of 1% α-naphthylamine. After a 20-min coloring reaction at 25°C, the optical density was determined at 530 nm in a spectrophotometer. The linear standard curve was determined by 0 to 5 μm NaNO2 reacted with sulfanilic acid and α-naphthylamine.
Freshly collected anthers were carefully immersed in the fixative solution O.C.T. Compound (Sakura) and frozen in liquid nitrogen. Embedded fresh tissues were sectioned into 9-μm-thick sections using a Leica CM1800 cryostat. Production of H2O2 was performed by incubating sectioned anthers in the DAB solution (Vector Laboratories) for 3 to 4 min, optimized from the manufacturer’s instructions, and photographed with a Nikon Ni-E optical microscope.
Anther Treatments
We removed the outermost whorls of leaves from wild-type and osers1 stems to locate the inflorescences. Next, 1 mm KI and 100 μm H2O2 solutions were freshly prepared and injected through leaf whorls using an 18-gauge needle into the airspace about 5 cm above the first stem node, where the young inflorescence was developed (Supplemental Fig. S9). Water injection served as a control. After 72 h of treatment, rice spikelets were fixed, embedded in resin, and sectioned as described above.
Accession Numbers
Accession numbers are as follows: OsERS1, LOC_Os10g22380; OsERS2, LOC_Os01g16520; OsERS3, LOC_Os02g02860; OsARC, LOC_Os01g60660; MSP1, LOC_Os01g68870; H4, LOC_Os09g38020; CDKB2;1, LOC_Os08g40170; AlaRS3, LOC_Os10g10244; ArgRS2, LOC_Os05g07030; AsnRS1, LOC_Os01g27520; AspRS3, LOC_Os02g46130; CysRS1, LOC_Os03g04960; GlnRS1, LOC_Os01g09000; GlyRS3, LOC_Os08g42560; LysRS2, LOC_Os03g38980; MetRS2, LOC_Os06g31210; ProRS2, LOC_Os12g25710; ThrRS2, LOC_Os08g19850; and TrpRS2, LOC_Os12g35570.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic analysis of the osers1 mutant.
Supplemental Figure S2. Cytological analysis of wild-type and osers1 anthers.
Supplemental Figure S3. Expression of genes that encode regulators of early anther development in osers1 anthers.
Supplemental Figure S4. Molecular characteristics of OsERS1.
Supplemental Figure S5. Alignment of conserved motifs among GluRSs.
Supplemental Figure S6. Shift of major metabolic pathways in osers1 anthers.
Supplemental Figure S7. RT-qPCR detection of expression levels of aaRSs genes.
Supplemental Figure S8. DAB staining analysis for H2O2 in anther lobes.
Supplemental Figure S9. Chemical treatments to anthers.
Supplemental Figure S10. Full scan figures of the gel and western blot.
Supplemental Table S1. Alignment of GluRS amino acid sequences.
Supplemental Table S2. Heat map of metabolite changes between the wild type and the osers1 mutant.
Supplemental Table S3. RT-qPCR detection of the expression changes of genes in metabolic pathways between the wild type and the osers1 mutant.
Supplemental Table S4. Sequences of primers used in this study.
Acknowledgments
We thank Zhijing Luo and Mingjiao Chen for performing osers1 mutant screens and rice crosses; Enduo Wang and Dr. Yong Wang (Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences) for help with enzyme activity measurements; Chi-kuang Wen (Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences) for providing the CD3-991 vector; Dr. Sheng Quan, Dr. Guorong Qu, Qian Luo, and Jin Zhou (Shanghai Jiao Tong University) for support in metabolomics analysis; Dr. Gwenda Mayo (Adelaide Microscopy) for technical support; Jianping Hu (Michigan State University) for valuable comments; Deborah Devis (University of Adelaide) for revision of the article; and Diane Mather (University of Adelaide) for advice and comments.
Footnotes
The contributions of D.Z. and G.L. were supported by Startup Funding from the School of Agriculture, Food, and Wine, University of Adelaide, and the contributions of X.Y. by a Beacon Scholarship, University of Adelaide. This work was supported by the National Key Research and Development Program of China (2016YFD0100804); the Innovative Research Team, Ministry of Education, and 111 Project (grant no. B14016); and Australia-China Science and Research Fund Joint Research Centre grant ACSRF48187.
Articles can be viewed without a subscription.
References
- Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, et al. (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30: 174–178 [DOI] [PubMed] [Google Scholar]
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M, Rogers R, Muralla R, Meinke D (2005) Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J 44: 866–878 [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW (2011) Regulation of cancer cell metabolism. Nat Rev Cancer 11: 85–95 [DOI] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H (2002) EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol 12: 1718–1727 [DOI] [PubMed] [Google Scholar]
- Chaubal R, Anderson JR, Trimnell MR, Fox TW, Albertsen MC, Bedinger P (2003) The transformation of anthers in the msca1 mutant of maize. Planta 216: 778–788 [DOI] [PubMed] [Google Scholar]
- Chen JF, Guo NN, Li T, Wang ED, Wang YL (2000) CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry 39: 6726–6731 [DOI] [PubMed] [Google Scholar]
- Chen JF, Li T, Wang ED, Wang YL (2001) Effect of alanine-293 replacement on the activity, ATP binding, and editing of Escherichia coli leucyl-tRNA synthetase. Biochemistry 40: 1144–1149 [DOI] [PubMed] [Google Scholar]
- Chen L, Chu HW, Yuan Z, Pan AH, Liang WQ, Huang H, Shen MS, Zhang DB, Chen L (2006) Isolation and genetic analysis for rice mutants treated with 60Co γ-ray. J Xiamen Univ 45: 81–85 [Google Scholar]
- Coruzzi GM, Zhou L (2001) Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects.’ Curr Opin Plant Biol 4: 247–253 [DOI] [PubMed] [Google Scholar]
- Cotelle V, Meek SE, Provan F, Milne FC, Morrice N, MacKintosh C (2000) 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J 19: 2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R (2002) Plant glutathione transferases. Genome Biol 3: S3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang ZP, Wang M, Ruan ZR, Tan M, Liu RJ, Zhou M, Zhou XL, Wang ED (2014) Coexistence of bacterial leucyl-tRNA synthetases with archaeal tRNA binding domains that distinguish tRNALeu in the archaeal mode. Nucleic Acids Res 42: 5109–5124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freist W, Gauss DH, Söll D, Lapointe J (1997) Glutamyl-tRNA sythetase. Biol Chem 378: 1313–1329 [PubMed] [Google Scholar]
- Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W (2014) The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26: 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani K, Grosshans H, Deinert K, Hurt EC, Simos G (2001) The intracellular location of two aminoacyl-tRNA synthetases depends on complex formation with Arc1p. EMBO J 20: 6889–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graindorge JS, Senger B, Tritch D, Simos G, Fasiolo F (2005) Role of Arc1p in the modulation of yeast glutamyl-tRNA synthetase activity. Biochemistry 44: 1344–1352 [DOI] [PubMed] [Google Scholar]
- Guo M, Schimmel P (2013) Essential nontranslational functions of tRNA synthetases. Nat Chem Biol 9: 145–153 10.1038/nchembio.1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Yang XL, Schimmel P (2010) New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol 11: 668–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. (2000) Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid Redox Signal 2: 811–820 [DOI] [PubMed] [Google Scholar]
- Hong L, Tang D, Shen Y, Hu Q, Wang K, Li M, Lu T, Cheng Z (2012a) MIL2 (MICROSPORELESS2) regulates early cell differentiation in the rice anther. New Phytol 196: 402–413 [DOI] [PubMed] [Google Scholar]
- Hong L, Tang D, Zhu K, Wang K, Li M, Cheng Z (2012b) Somatic and reproductive cell development in rice anther is regulated by a putative glutaredoxin. Plant Cell 24: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M, Söll D (2000) Aminoacyl-tRNA synthesis. Annu Rev Biochem 69: 617–650 [DOI] [PubMed] [Google Scholar]
- Ito K, Ito K (2016) Metabolism and the control of cell fate decisions and stem cell renewal. Annu Rev Cell Dev Biol 32: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, et al. (2013) SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 23: 450–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kägi C, Baumann N, Nielsen N, Stierhof YD, Gross-Hardt R (2010) The gametic central cell of Arabidopsis determines the lifespan of adjacent accessory cells. Proc Natl Acad Sci USA 107: 22350–22355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Walbot V (2012) Hypoxia triggers meiotic fate acquisition in maize. Science 337: 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Lee JY, Cho HS, Lee SS, Ha HJ, Kim S, Choi D, Pai HS (2005) Inactivation of organellar glutamyl- and seryl-tRNA synthetases leads to developmental arrest of chloroplasts and mitochondria in higher plants. J Biol Chem 280: 37098–37106 [DOI] [PubMed] [Google Scholar]
- Laporte D, Huot JL, Bader G, Enkler L, Senger B, Becker HD (2014) Exploring the evolutionary diversity and assembly modes of multi-aminoacyl-tRNA synthetase complexes: lessons from unicellular organisms. FEBS Lett 588: 4268–4278 [DOI] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Bao W, Liang W, Yin J, Zhang D (2010) Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development. J Integr Plant Biol 52: 670–678 [DOI] [PubMed] [Google Scholar]
- Lü Y, Cui X, Li R, Huang P, Zong J, Yao D, Li G, Zhang D, Yuan Z (2015) Development of genome-wide insertion/deletion markers in rice based on graphic pipeline platform. J Integr Plant Biol 57: 980–991 [DOI] [PubMed] [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2017) ROS are good. Trends Plant Sci 22: 11–19 [DOI] [PubMed] [Google Scholar]
- Møller IM, Jensen PE, Hansson A (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58: 459–481 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Newsholme P, Procopio J, Lima MMR, Pithon-Curi TC, Curi R (2003) Glutamine and glutamate: their central role in cell metabolism and function. Cell Biochem Funct 21: 1–9 [DOI] [PubMed] [Google Scholar]
- Nonomura K, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N (2003) The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15: 1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber M, Chandel NS (2014) ROS function in redox signaling and oxidative stress. Curr Biol 24: R453–R462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C, Gaillochet C, Medzihradszky A, Busch W, Daum G, Krebs M, Kehle A, Lohmann JU (2014) A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev Cell 28: 438–449 [DOI] [PubMed] [Google Scholar]
- Sena LA, Chandel NS (2012) Physiological roles of mitochondrial reactive oxygen species. Mol Cell 48: 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H, et al. (2015) The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol 17: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomes JC, Ratinaud MH, Julien R (1983) Dimeric glutamyl-tRNA synthetases from wheat: kinetic properties and functional structures. Eur J Biochem 135: 479–484 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Umeda M, Umeda-Hara C, Yamaguchi M, Hashimoto J, Uchimiya H (1999) Differential expression of genes for cyclin-dependent protein kinases in rice plants. Plant Physiol 119: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V, Egger RL (2016) Pre-meiotic anther development: cell fate specification and differentiation. Annu Rev Plant Biol 67: 365–395 [DOI] [PubMed] [Google Scholar]
- Wang SY, Jiao H (2000) Scavenging capacity of berry crops on superoxide radicals, hydrogen peroxide, hydroxyl radicals, and singlet oxygen. J Agric Food Chem 48: 5677–5684 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang C, Zheng M, Lyu J, Xu Y, Li X, Niu M, Long W, Wang D, Wang H, et al. (2016) WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiol 170: 2110–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ZA, Zhang DB (2009) From Arabidopsis to rice: pathways in pollen development. J Exp Bot 60: 1479–1492 [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S (2008) ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J 53: 790–801 [DOI] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J (2013) Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Hakata M (2010) Atlas of rice grain filling-related metabolism under high temperature: joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol 51: 795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Qian X, Chen M, Fei Q, Meyers BC, Liang W, Zhang D (2016) Regulatory role of a receptor-like kinase in specifying anther cell identity. Plant Physiol 171: 2085–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SL, Xie LF, Mao HZ, Puah CS, Yang WC, Jiang L, Sundaresan V, Ye D (2003) Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15: 2792–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Dong Z, Wu H, Tian Z, Zhao Z (2017) Redox regulation of plant stem cell fate. EMBO J 36: 2844–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yang L (2014) Specification of tapetum and microsporocyte cells within the anther. Curr Opin Plant Biol 17: 49–55 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DZ, Wang GF, Speal B, Ma H (2002) The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, de Palma J, Oane R, Gamuyao R, Luo M, Chaudhury A, Hervé P, Xue Q, Bennett J (2008) OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J 54: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]