Salinity-induced endopolyploidy in Mesembryanthemum crystallinum plays a role in leaf development and the enlargement of epidermal bladder cells to increase their sodium storage capacity.
Abstract
Endopolyploidy occurs when DNA replication takes place without subsequent mitotic nuclear division, resulting in cell-specific ploidy levels within tissues. In plants, endopolyploidy plays an important role in sustaining growth and development, but only a few studies have demonstrated a role in abiotic stress response. In this study, we investigated the function of ploidy level and nuclear and cell size in leaf expansion throughout development and tracked cell type-specific ploidy in the halophyte Mesembryanthemum crystallinum. In addition to developmental endopolyploidy, we examined the effects of salinity stress on ploidy level. We focused specifically on epidermal bladder cells (EBC), which are modified balloon-like trichomes, due to their large size and role in salt accumulation. Our results demonstrate that ploidy increases as the leaves expand in a similar manner for each leaf type, and ploidy levels up to 512C were recorded for nuclei in EBC of leaves of adult plants. Salt treatment led to a significant increase in ploidy levels in the EBC, and these cells showed spatially related differences in their ploidy and nuclear and cell size depending on the positions on the leaf and stem surface. Transcriptome analysis highlighted salinity-induced changes in genes involved in DNA replication, cell cycle, endoreduplication, and trichome development in EBC. The increase in cell size and ploidy observed in M. crystallinum under salinity stress may contribute to salt tolerance by increasing the storage capacity for sodium sequestration brought about by higher metabolic activity driving rapid cell enlargement in the leaf tissue and EBC.
Endopolyploidy, also known as somatic polyploidy, occurs when DNA replication takes place in the absence of mitotic nuclear division in nongermline cells, such that the plant remains diploid but specific cells change their ploidy. This is in contrast to true polyploid organisms, such as autopolyploids and allopolyploids, in which the duplicated DNA content is inherited through the germline and perpetuated over subsequent generations in all cells (De Storme and Mason, 2014; Scholes and Paige, 2015). Endopolyploidy is present in animals but is particularly widespread among insects and plants (Edgar et al., 2014; Rangel et al., 2015; Leitch and Dodsworth, 2017), with an estimated 70% to 90% of angiosperms presenting endopolyploidy in at least one cell type (Joubès and Chevalier, 2000; Skaptsov et al., 2017). While embryonic, meristematic, and guard cells remain at the 2C DNA content, most other cell types, including those from roots, stems, leaves, flowers, fruits, and seeds, can undergo independent rounds of endoreduplication during development. This results in cells with a genome content that is orders of magnitude greater than the germline, most commonly ranging from 4C to 64C (Scholes and Paige, 2015). Endopolyploidy has long been recognized to play an important role in initiating and sustaining plant growth through cell expansion and is crucial for tissue formation, organ morphology, differentiation, and/or the function of specific cell types and the maintenance of cell identity (Lee et al., 2009; Bramsiepe et al., 2010; Chevalier et al., 2011). There also is evidence that plants employ endopolyploidy as an adaptation to stress tolerance. Plant genotypes or mutants that show higher cellular ploidy are more tolerant to stress, as was shown for UV-B and drought tolerance in Arabidopsis (Arabidopsis thaliana; Cookson et al., 2006; Gegas et al., 2014). It also has been demonstrated that endopolyploidy can be triggered in plants as a response to other environmental stresses, including high light and salinity (Schoenfelder and Fox, 2015; Scholes and Paige, 2015), as a means to improve fitness and survival under adverse conditions.
Mesembryanthemum crystallinum is an annual halophyte/xerophyte endemic to the coastline of South Africa that has been naturalized around the world in regions with a Mediterranean-type arid coastal environment. It shows facultative Crassulacean acid metabolism, switching from C3 photosynthesis to Crassulacean acid metabolism, following periods of water deficit or salinity stress and is an important emerging model plant that is widely used to understand the molecular mechanisms leading to adaptations to saline environments (Adams et al., 1998; Cushman and Bohnert, 2000; Barkla et al., 2002). One of these adaptations is the presence of modified, single-celled, nonglandular trichomes called epidermal bladder cells (EBC) that line the stem, leaves, and flower buds of the plant. These globular, balloon-like cells play an important role for the accumulation of sodium (Adams et al., 1998; Barkla et al., 2002; Agarie et al., 2007; Dassanayake and Larkin, 2017). The ability to isolate these cells by direct extraction of the cell sap, peeling of the leaf epidermis, or isolating single cells through freeze/thaw techniques has made them attractive for the analysis of single-cell-type systems biology (Barkla et al., 2012, 2016; Barkla and Vera-Estrella, 2015; Oh et al., 2015).
M. crystallinum shows endopolyploidy, and in earlier studies, ploidy levels up to 128C were reported in total leaf tissue of adult plants (De Rocher et al., 1990). In this study, our aim was to investigate the level of endopolyploidy as a function of leaf expansion throughout the development of the plant and in specialized cell types and to determine if salinity stress alters the developmentally programmed ploidy level. We also investigated the ploidy level in the large EBC on the leaf, stem, and flower bud, as these cells enlarge significantly with salt stress. To identify the transcriptomic signature and gain knowledge of the genes involved in DNA replication, endopolyploidy, and trichome development in EBC, the EBC transcriptome from control and salt-treated plants (Oh et al., 2015) was mined and compared with the published transcriptome from M. crystallinum root tissue (Tsukagoshi et al., 2015) that does not show high levels of endopolyploidy.
RESULTS
Developmental Endopolyploidy
To systematically measure developmental somatic polyploid variation in M. crystallinum tissues, including roots, leaves, stems, and leaf epidermal peels, nuclei samples were taken throughout plant development, beginning with the ungerminated seed through adult flowering plants. M. crystallinum has two distinct developmental stages: a juvenile stage and an adult stage (Adams et al., 1998). In the juvenile stage, which encompasses germination through 6 weeks of age, the first three sets of primary leaves develop at 1-week intervals and expand over a 3-week period (for an example of cotyledon expansion, see Supplemental Fig. S1); the fourth and fifth leaves expand over a 2-week period and then begin to senesce. At 6 weeks of age, the plant enters the adult growth stage, with growth along the primary axis terminating. At this stage, branching occurs, and side shoots develop above primary leaves 2 and 3 with branch leaves, followed by the development of flower bracts with bract leaves (Supplemental Fig. S1; Adams et al., 1998).
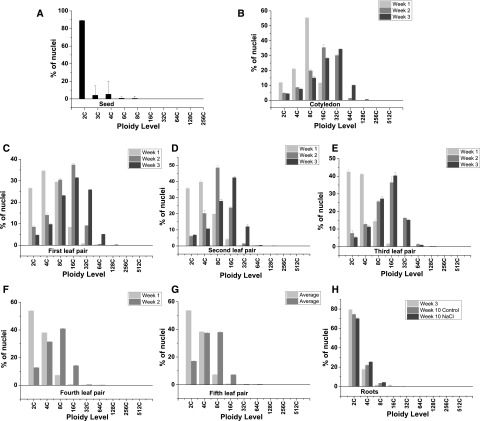
Flow cytometry analysis of M. crystallinum extracts revealed the presence of endopolyploid nuclei in all tissues studied (Figs. 1–4; Supplemental Figs. S2–S4). However, the degree of endoreduplication differed markedly between tissues and between developmental stages of the same tissue. In the ungerminated seed, the majority of the nuclei were 2C (88%), with only a small 4C peak detected (5.6%; Fig. 1A; Supplemental Fig. S2A). Small peaks corresponding to 3C and 6C nuclei also were detected in the seed and most likely correspond to the triploid endosperm. In the emerging cotyledon 7 d after germination (DAG), nuclei up to 16C were measured, with the majority being 8C (55%); however, by 14 DAG, the majority of the nuclei in the cotyledon were 16C (33%) and nuclei up to 64C were recorded (Fig. 1B; Supplemental Fig. S1B). In the cotyledon 21 DAG, nuclei of 128C were recorded, with the majority of the nuclei being 32C (34%; Supplemental Fig. S1B). During successive leaf development events in juvenile plants (up to 6 weeks), the number of 2C nuclei measured in week 1 of the emergence of each primary leaf, corresponding to 14, 21, 28, 35, and 42 DAG for the first, second, third, fourth, and fifth leaf pair, respectively, were seen to increase. In the first leaf pair, 26% of the nuclei were 2C, whereas 34% were 2C in the second leaf pair, 44% in the third leaf pair, 55% in the fourth leaf pair, and 54% in the fifth leaf pair, with ploidy up to 128C measured (Fig. 1, C–G, left; Supplemental Fig. S2). However, by the second week of development for all leaf pairs, the distribution had changed dramatically, with 2C nuclei representing only 9.2%, 7.1%, 4.3%, 12.8%, and 17.3% for the first, second, third, fourth, and fifth leaf pair, respectively, and in all cases 16C nuclei were the highest represented (Fig. 1, C–G, middle; Supplemental Fig. S2). The highest level of ploidy detected in juvenile plants was 256C in nuclei isolated from the third leaf pair at 3 weeks of development in 6-week-old plants (Fig. 1E, right; Supplemental Fig. S2E). This value is higher than the 128C reported by Adams et al. (1998).
Figure 1.
Average normalized nuclear count for each ploidy level detected in M. crystallinum juvenile plants. A, Seed. B, Cotyledon. C, First leaf pair. D, Second leaf pair. E, Third leaf pair. F, Fourth leaf pair. G, Fifth leaf pair. H, Roots. Leaves were measured over a 3-week period, as follows: week 1 (light gray bars), week 2 (medium gray bars), and week 3 (dark gray bars). Data are means ± se of three independent biological replicates.
Figure 4.
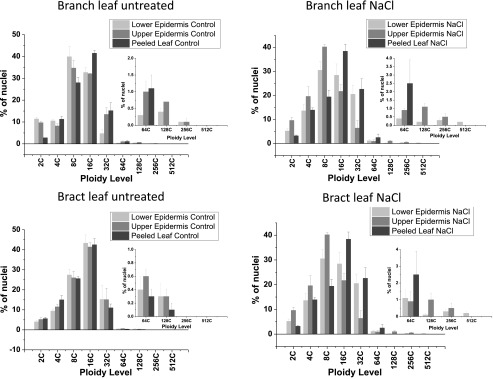
Bar graphs showing average normalized nuclear counts for each ploidy level in the branch or bract leaves from adult plants. Samples were as follows: peeled leaf (dark gray bars), upper epidermal peels (medium gray bars), and lower epidermal peels (light gray bars). Insets show expansion of the y axis to visualize high-ploidy nuclear counts. Data are means ± se of three independent biological replicates.
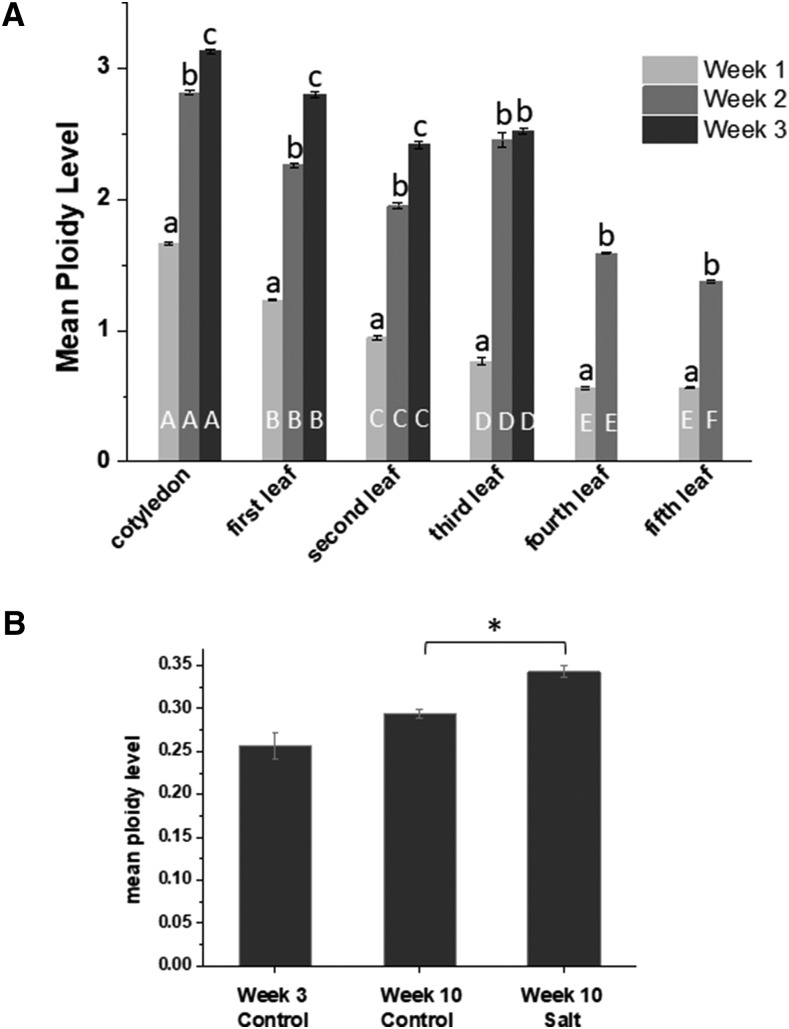
Mean ploidy level was calculated from Equation 1 (see “Materials and Methods”) for all developmental stages in the juvenile plant, and the average mean ploidy level of three biological replicates is reported in Figure 2. This value takes into consideration the number of nuclei of each represented ploidy level multiplied by the number of endoreduplication cycles needed to reach the corresponding ploidy level (Barow and Meister, 2003). As the emergence and expansion of the cotyledon, first, second, third, fourth, and fifth leaf pairs followed, in all cases, maximal ploidy level increased significantly with the age of the leaf (Fig. 2); however, the mean ploidy level at each week of leaf development was highest in the cotyledon and then decreased significantly with each subsequent leaf pair in the juvenile plants.
Figure 2.
Mean ploidy levels in cotyledons, leaves, and roots from M. crystallinum. A, Mean ploidy was calculated as shown in “Materials and Methods” from cotyledons, leaf pair 1, leaf pair 2, leaf pair 3, leaf pair 4, and leaf pair 5 over a 3- or 2-week period, as follows: week 1 (light gray bars), week 2 (medium gray bars), and week 3 (dark gray bars). Data are means ± se of three independent biological replicates. Columns with the same letter are not significantly different in Duncan’s multiple range test (P < 0.05), with white uppercase letters denoting the comparison with developmental stages with each week and black lowercase letters comparing values for the weeks within each development stage. B, Mean ploidy levels in young and old roots from control or salt-treated plants. Significance in Duncan’s multiple range test (P < 0.05) is indicated by the asterisk.
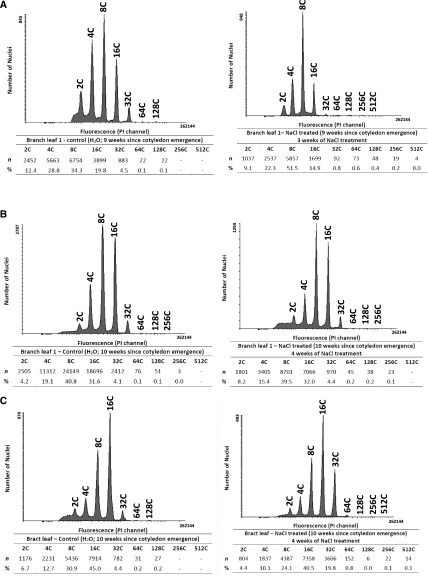
In adult plants, ploidy was measured in the first branch leaf at 9 and 10 weeks following germination. These adult leaves are smaller than the late juvenile leaves (Supplemental Fig. S1B). Similar to ploidy levels measured in juvenile plants, leaves progressed from having a predominance of 8C ploidy to showing increased levels of 16C ploidy in week 2 of development (Fig. 3, A and B, left). Ploidy changes in adult leaves were not as dramatic as those observed in the large juvenile leaves, which also showed higher rates of expansion (Fig. 1; Supplemental Fig. S2).
Figure 3.
Flow cytometry analysis of endopolyploidy in M. crystallinum adult plants. Propidium iodide (PI) was used to stain nuclei in the chopped plant material from branch leaf of 9-week-old plants (A), branch leaf of 10-week-old plants (B), and bract leaf (C). Plants were untreated (left) or salt treated (right). Acquisition was performed on a BD FACS Canto II flow cytometer, and data were analyzed using Flowing Software version 2.5.1. The histogram is representative of three independent experiments with biological replicates from three different plants. Mean ploidy levels in the samples shown were calculated as described in “Materials and Methods.”
Ploidy also was measured in bract leaves, which are small leaves that develop below the flower bud in adult plants (Supplemental Fig. S1B). In these leaves, nuclei from 2C to 128C were detected, with the greatest percentage of the nuclei being 16C (Fig. 3C).
In root tissue, the ploidy level remained low relative to the leaf. The majority of nuclei were 2C, and the highest ploidy level reached was 32C in the young roots (Fig. 1H; Supplemental Fig. S1H). The mean ploidy level showed no significant variation with plant age (Fig. 2B).
Ploidy Levels in M. crystallinum EBC
M. crystallinum EBC are present on the upper and lower epidermis of the leaves as well as on the stems of the developing plant and enlarge during leaf expansion (Supplemental Fig. S1C; Adams et al., 1998; Oh et al., 2015). The largest EBC are detected on the lower surface of the leaf (Supplemental Fig. S1C), on the stem, and on the flower bud (Oh et al., 2015). These cells are present on untreated plants but swell significantly when plants are treated with salt and can reach diameters of 1.6 mm (Table I).
Table I. Estimation of ploidy and nuclear and cell size in the EBC from leaves, stems, and flower buds.
| Tissue | Treatment | Mean Nuclear Area | Nuclear Volume | No. | Estimated Ploidy from Fluorescence Microscopy | Flow Cytometry Size Estimate of Ploidy | Bladder Cell Area |
|---|---|---|---|---|---|---|---|
| µm2 | µm3 | µm2 | |||||
| Bract leaf lower epidermis EBC | Water | 925 ± 79 | 117,842 | 21 | 1,024C | 128C | 28,577 ± 629 |
| Salt | 1,815 ± 33 | 323,894 | 133 | 2,048C | 256C | 36,818 ± 861 | |
| Bract leaf upper epidermis EBC | Water | 956 ± 52 | 123,816 | 20 | 1,024C | 128C | 18,641 ± 342 |
| Salt | 978 ± 19 | 128,114 | 162 | 1,024C | 128C | 21,128 ± 454 | |
| Flower bud EBC | Water | 1,230 ± 87 | 180,695 | 25 | 1,024C | 128C | 433,456 ± 15,707 |
| Salt | 9,406 ± 154 | 3,821,167 | 35 | 32,768C | 65,536C | 2,208,355 ± 99,677 | |
| Stem EBC | Water | 1,238 ± 46 | 182,461 | 26 | 1,024C | 128C | 42,111 ± 884 |
| Salt | 6,586 ± 105 | 2,238,830 | 28 | 8,192C | 2,048C | 62,677 ± 1,969 |
To measure ploidy in the EBC and compare values with those obtained from nuclei isolated from the whole leaf and peeled leaf, epidermal peels were made using the branch leaf and the bract leaf. In the epidermal peels from the untreated branch leaf, ploidy up to 256C was measured in both lower and upper epidermal peels, a level not detected when whole-leaf extractions were employed (Fig. 3; Supplemental Fig. S3), and more 128C nuclei were detected than were detected for the whole leaf (Supplemental Fig. S3, A–C, untreated). In the branch leaf tissue, with upper and lower epidermis removed, ploidy of only up to 64C was measured, with the majority of the nuclei measuring 16C (Supplemental Fig. S3A, untreated). Compared with the peeled leaf, epidermal peels showed a higher percentage of 2C nuclei, which can be attributed to the higher representation of guard cells in the epidermal peels. Both upper and lower epidermal peels also had a higher number of nuclei with 8C ploidy level than the peeled leaf (Fig. 3). When bract leaf tissue was analyzed, ploidy up to 256C was detected in the epidermal peels from the untreated leaf, and similar to the branch leaf, the highest ploidy detected in the peeled leaf minus the epidermis was 64C (Fig. 4, untreated; Supplemental Fig. S4, A–C).
Salinity-Induced Endopolyploidy in Leaves and EBC
In order to determine if M. crystallinum also showed salt stress-induced polyploidy in addition to developmental endopolyploidy in leaf tissue, we measured ploidy levels in tissue isolated from salt-treated adult plants and compared those with data obtained from untreated plants. Measurement of ploidy in the whole branch leaf and bract leaf from adult salt-treated plants showed increases in the maximum C value detected (Figs. 3 and 4). Highest ploidy levels increased from 128C in untreated whole leaves up to 512C in salt-treated leaves, indicating at least one, and in some cases two, additional rounds of endoreduplication. In root tissue, while there was no increase in maximum ploidy level detected with salt treatment (Supplemental Fig. S2H), mean ploidy level showed a significant increase between salt-treated and untreated root tissue (Fig. 2).
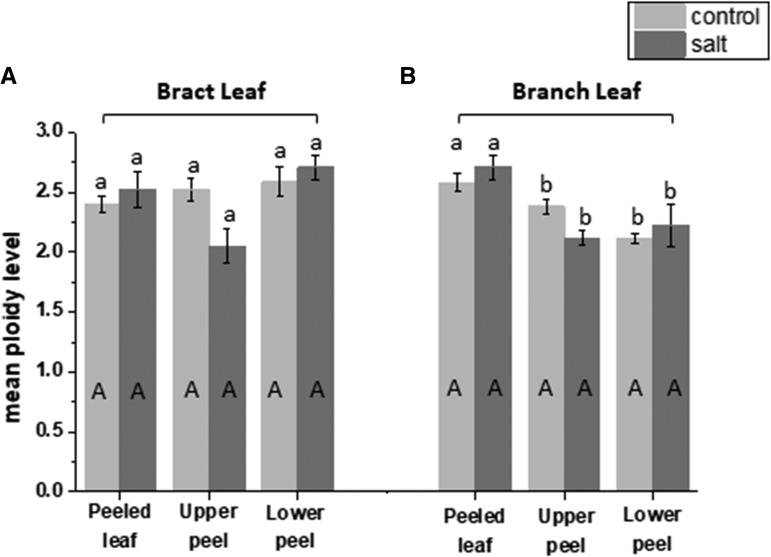
To measure the contribution of EBC to the salt-induced increase in ploidy level reached in the leaves of adult plants, epidermal peels of the upper and lower sides of the branch and bract leaf were compared with the values obtained in the peeled leaf. In the peeled branch leaf, salt treatment resulted in a significant increase in 32C and 64C nuclei compared with those detected in the peeled leaf from untreated plants. However, the highest ploidy detected in both control and salt-treated mesophyll cells was 128C (Supplemental Figs. S3A and S4A). When ploidy in the epidermal peels from salt-treated branch leaves was compared with that in epidermal peels of untreated leaves, ploidy up to 512C was detected in the lower epidermis of treated leaves compared with only 256C in the corresponding peels from the untreated leaves, and considerably more 256C nuclei were observed in the lower epidermis from salt-treated plants (Fig. 3; Supplemental Fig. S3, B and C). In the bract leaf, there was a much larger proportion of 32C and 64C nuclei in the peeled leaf from the salt-treated plant, and as in the branch leaf, the highest ploidy in both treated and untreated leaves was 64C (Fig. 4B; Supplemental Fig. S4A). In the epidermal peels from this leaf, the highest levels of ploidy were measured in the lower epidermal peels from the salt-treated tissue (512C), and only a few nuclei detected in the untreated epidermal peels were higher than 128C (Fig. 4; Supplemental Fig. S4, B and C). However, when measurements of mean ploidy level were calculated, there was no statistical difference between treatments (Fig. 5), despite the detection of higher ploidy levels in the salt-treated samples. These results indicate that salt stress induces at least one extra round of endoreduplication than that observed in the untreated tissue of the same age and that the high ploidy levels (greater than 64C) detected in the whole leaf reflects the contribution of the ploidy level from the EBC.
Figure 5.
Mean ploidy levels calculated in a peeled leaf, the upper epidermal peel, and the lower epidermal peel of bract and branch leaves taken from untreated (light gray bars) or salt-treated (dark gray bars) M. crystallinum plants. Mean ploidy was calculated as shown in “Materials and Methods” from peeled leaves, upper leaf peels, and lower leaf peels from both bract leaves (A) and branch leaves (B). Data are means ± se of three independent biological replicates. Columns with the same letter are not significantly different in Duncan’s multiple range test (P < 0.05), with uppercase letters denoting the effects of NaCl treatment and lowercase letters referring to the comparison of different tissues.
Nuclear Size and Ploidy Level
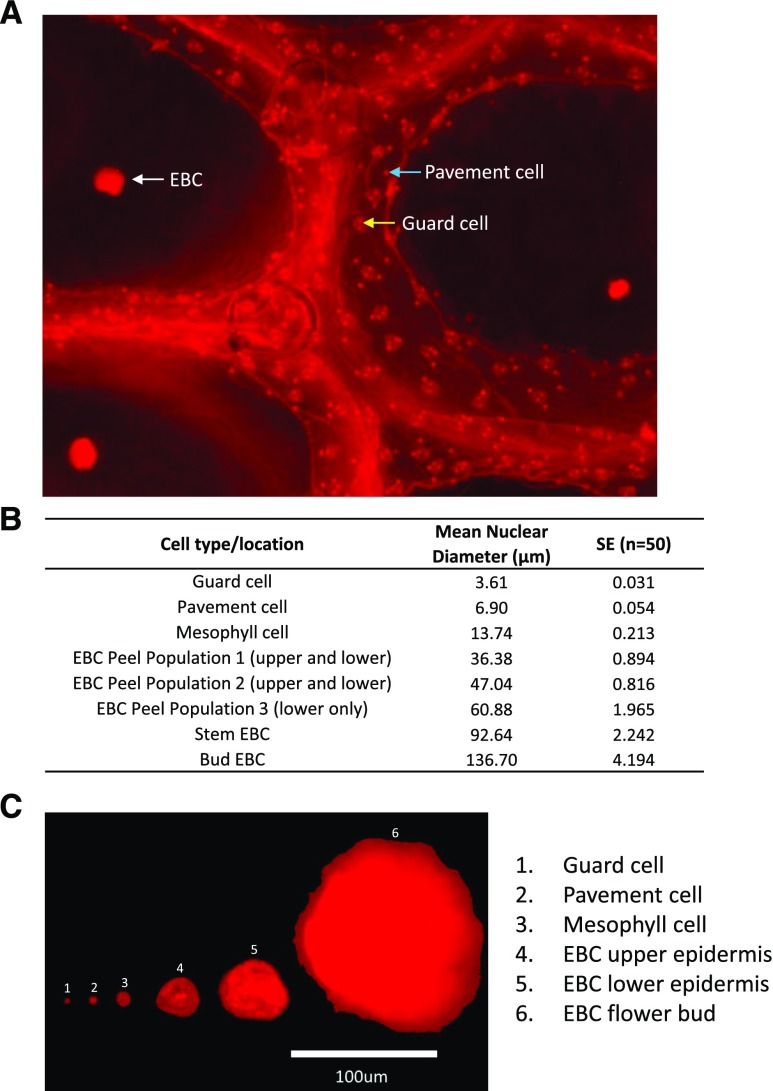
When measuring ploidy in the epidermal peels by flow cytometry, it was apparent that the number of nuclei detected appeared to be lower than that expected from the number of EBC present on the tissue. This raised the possibility that the highly polyploid nuclei were too large for the flow cell and/or were not passing through the 100-μm filter used to filter cellular debris prior to flow cytometry. This would result in an underestimation of the ploidy level in these cells and result in incorrect reporting of mean ploidy level in this tissue. To determine the size of the nuclei in the epidermal peels, as well as within other cells of the leaf, we labeled tissue with PI, and nuclei were visualized using fluorescence microscopy. A range of different-sized nuclei were detected and measured in the leaf tissue and assigned to specific cell types, where possible (Fig. 6, A and B). In epidermal peels, we identified several populations of nuclei based on their mean nuclear diameter, as indicated in Figure 6B. In upper peels, EBC nuclei with an average of 36 and 47 μm were detected, and in lower peels, an additional group with an average of 60 μm was identified, which was not observed in the upper peel. The largest leaf nuclei were detected in the epidermal peels from the lower surface of leaves from salt-treated plants. However, stems and flower buds from salt-treated plants, which have bigger EBC than those found on the leaves (Fig. 6), possessed the largest nuclei found in the plant, with diameters of up to 136 μm detected in the EBC of flower buds (Fig. 6C). These findings suggested that flow cytometry data significantly underestimated the EBC ploidy level due to the physical constraints of the method. In order to estimate the level of ploidy in the large nuclei that were observed by fluorescence microscopy in the leaf but could not be measured by flow cytometry, two approaches were taken and are described in detail in “Materials and Methods.” For the first approach, we associated the nuclear volume determined by fluorescence microscopy with a known C value for specific cell types and then used this information to estimate the C value in the large EBC from their area, assuming a perfect sphere (Table I). From this method, ploidy levels of up to 2,048C were estimated for nuclei from the EBC of bract leaf lower epidermis from salt-treated plants, up to 8,192C for nuclei in the EBC of the stem from salt-treated plants, and up to 32,768C for nuclei from the EBC of flower buds from salt-treated plants, representing 10, 12, and 15 rounds of endoreduplication, respectively (Table I).
Figure 6.
Fluorescence microscopy of nuclei in different M. crystallinum cell types from salt-treated plants. A, Lower epidermal peels were stained in PI solution for 5 min in the dark, and a representative fluorescence microscopy image is shown. The white arrow indicates an EBC nucleus, the yellow arrow indicates the nucleus in a guard cell, and the blue arrow indicates a pavement cell nucleus. B, Table showing calculated nuclear diameters for selected cell types. Data are means ± se of 50 measurements per cell type. C, Composed figure showing the relative nuclear size of representative nuclei from different cell types.
In the second approach, spherical particles with known diameters were included in the flow cytometry nuclei suspension. This allowed for within-suspension nuclei, already gated into ploidy groups based on their relative fluorescence, to be assigned a size value for the forward scatter property (FSC), which is used here as a proxy for nuclei diameter. The association between nuclei diameter and the endoreduplication index (ERI) then could be extrapolated and enabled an ERI estimation for the bladder cells too large to run through the flow cytometer (Supplemental Fig. S5).
Using these values, we estimated the ploidy in salt-treated plants of up to 256C in the leaf, 2,048C in the stem, and 65,536C in the extremely large nuclei of the flower bud (136 μm), representing seven, 10, and 16 rounds of endoreduplication, respectively (Table I). Therefore, rather than a single round of endoreduplication being induced by salinity (from 256C to 512C), as detected initially solely from the flow cytometry data, up to six additional rounds would account for the values estimated in salt-treated EBC of the flower bud.
Transcriptomic Response Associated with Endoreplication in M. crystallinum EBC in Response to Salt Stress
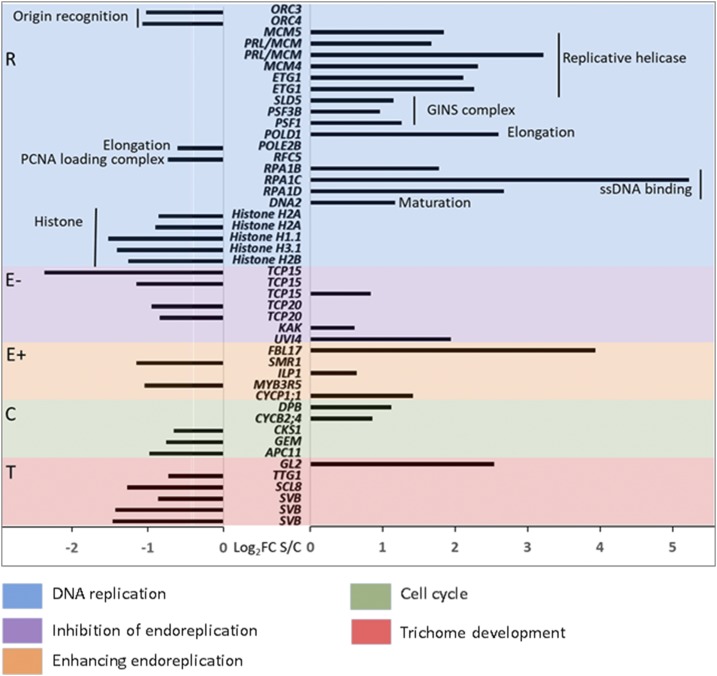
Endopolyploidy involves the tight control of molecular mechanisms that initiate and then maintain endoreplication in the cell, allowing endocycling cells to replicate their DNA during the synthesis (S) phase but arresting progress to the mitosis phase, cycling instead between the S and gap (G) phases. Down-regulation of mitotic cyclins and mitotic cyclin-dependent kinase (CDK) activity play a key role in this regulation (De Veylder et al., 2011). However, endopolyploid cells also must regulate genes involved in DNA replication (De Veylder et al., 2011). In order to assess the transcriptomic signature that may define the higher ploidy level observed in EBC under salt stress and gain a further understanding of the underlying processes, we used an EBC cell type-specific RNA-sequencing (RNA-seq) replicated data set (Oh et al., 2015), generated from control and salt-treated plants (200 mm NaCl for 2 weeks) of the same developmental age (8 weeks). We selected a focal set of genes (see “Materials and Methods”) associated with DNA replication (R), positive regulation of endoreplication (E+), negative regulation of endoreplication (E−), cell cycle (C), and trichome development (T). This focal set of genes is referred to as RECT genes in the following analyses for the letters indicated above (Fig. 7; Supplemental Table S1). It should be noted that many of the genes considered for this analysis may fit in more than one category, but to avoid overlaps and redundancy, we categorized them in the order listed. We also selected a number of additional genes involved in nuclear size, cytoskeletal components, and cell expansion that do not fit into our RECT subset but may be important for EBC alterations under salt stress, and these are analyzed and discussed separately.
Figure 7.
Fold change (log2) of significantly differentially expressed genes associated with endoreplication in the M. crystallinum bladder cell transcriptome in response to 200 mm NaCl. Significantly different expression was evaluated at an adjusted P < 0.05 and false discovery rate = 0.01.
Of the 165 genes in the RECT gene set, 43 genes (39%) show significantly differential expression between salt-treated and control EBC, as compared with only 22% of the total transcripts in the EBC transcriptome (8,285 out of 37,341; Oh et al., 2015). Particularly striking were the changes in transcripts associated with DNA replication (R in Fig. 7 and Supplemental Table S1). Many of the genes associated with the replicative helicase and GINS complex were significantly up-regulated in the EBC from salt-treated plants, while histones were down-regulated.
A Comparison of Transcriptome Responses Associated with Endoreplication between EBC and Root Transcriptomes
Our results indicate a much higher level of endoreplication in EBC compared with that measured in roots (Fig. 1). To test whether the EBC transcriptome carries a transcriptomic signature supporting the striking ploidy difference, we compared the normalized expression of RECT genes between the M. crystallinum EBC and root transcriptomes (Oh et al., 2015; Tsukagoshi et al., 2015). Supplemental Tables S2 and S3 give the ordered list of RECT genes in their respective functional categories, their normalized expression between the EBC and root transcriptomes, and the transcripts that show significantly different expression in response to salt stress. It should be noted that we identified differential expression based on a within-tissue comparison, as these two transcriptomes were created from plants grown in different growth environments and were sampled at two different developmental stages (Oh et al., 2015; Tsukagoshi et al., 2015). We did not focus on the general salt response in these two tissues. Despite the differences in growth conditions and development stages, the root tissues consistently represent mostly 2C cells and the mature EBC consistently represent a higher ploidy level than 2C (Fig. 1). Therefore, we only contrast the overall outcome of the normalized RECT gene expression between roots and EBC that is likely responsible for the difference in the ploidy status of the tissues where the ploidy remains significantly different between the two tissues regardless of developmental age, growth conditions, and salt treatments.
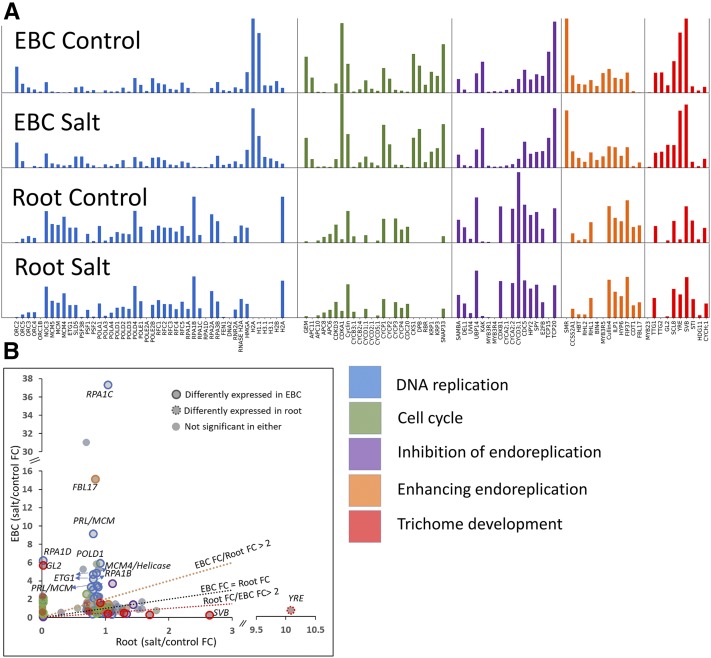
All functional categories show differences in expression between EBC and roots (Fig. 8A). There are more distinct differences in the overall pattern between the tissues than differences between control and salt-treated samples in the same tissue (Fig. 8A). More RECT genes are regulated significantly differently in response to salt stress in EBC than in roots. Our EBC transcriptome had 8,285 out of 37,341 transcripts significantly differentially expressed in response to salt stress (Oh et al., 2015), compared with only 53 differentially expressed transcripts out of 53,516 in roots (Tsukagoshi et al., 2015). There are 43 significantly differentially expressed RECT genes in EBC compared with only one differentially expressed gene detected in the root transcriptome in response to salt stress (Fig. 8B; Supplemental Table S3).
Figure 8.
Differential responses of selected genes to salt stress in root and EBC transcriptomes. A, Normalized expression values within a category scaled from 0 to 1 on the y axis. B, Fold change (FC) between salt and control treatments.
Is Polyploidy a Common Occurrence in Halophytes with EBC?
To determine if high levels of endopolyploidy are a common feature of plants with bladder-type specialized trichomes, the nuclei in the leaves of salt-treated Chenopodium quinoa were measured. Like M. crystallinum, C. quinoa has EBC on the surface of the leaves; however, they are considerably smaller, do not enlarge over time, and accumulate very little salt (Orsini et al., 2011). C. quinoa is a natural tetraploid (Jarvis et al., 2017), and measurement of polyploidy in leaves showed maximum levels of only 8C from adult 200 mm salt-treated plants (Supplemental Fig. S4), suggesting the absence of high levels of endopolyploidy in this plant.
DISCUSSION
In plants, most cell types undergo one or two rounds of endoreduplication, with more specialized cell types commonly exhibiting higher ploidy levels (Leitch and Dodsworth, 2017). Highly metabolically active cells appear to have the highest levels of endopolyploidy measured, as has been demonstrated in endosperm and suspensor cells of the seeds of Arum maculatum and Phaseolus coccineus (24,567C and 8,192C, respectively; Leitch and Dodsworth, 2017), suggesting that polyploidy is associated with the most active and specialized cells (Cookson et al., 2006). However, the role of endopolyploidy is still far from clear, and there are surprisingly few biological functions that are specifically attributed to endopolyploidy (Fox and Duronio, 2013).
In M. crystallinum, endopolyploidy up to 256C was measured in the leaf tissue of juvenile plants and up to 2,048C was estimated in leaf tissue of adult untreated plants. This appears to be developmentally programmed, increasing with leaf age and leaf number (Figs. 1–5), seemingly in line with cotyledon/leaf expansion (Supplemental Fig. S1). In Arabidopsis, leaf growth has been linked to increased endopolyploidy (Massonnet et al., 2011), and cell size increases proportionally with the increase in ploidy level, although to levels much lower than seen in M. crystallinum (Tsukaya, 2013). Previous results from M. crystallinum also showed changes in ploidy with leaf number; however, in that study, the highest ploidy detected was only 64C (De Rocher et al., 1990). This difference between our study and that earlier report can be attributed to the pore size of the filters used to remove cellular debris from the nuclear extractions prior to flow cytometry. In the study of De Rocher et al. (1990), filters of 60 μm followed by 10 μm were employed to filter the nuclei (Galbraith et al., 1983), and many of the larger nuclei would have been unable to pass through the mesh. In contrast, in our study, we discovered that large nuclei were not passing through the standard filters (30 and 60 μm), so we utilized filters with a pore size of 100+ μm for our analysis. Despite this, the largest nuclei detected by fluorescence microscopy were unable to pass through these filters or were subsequently too large for the flow cell (Fig. 6; Table I).
In addition to developmentally regulated endopolyploidy, M. crystallinum also showed salinity-induced polyploidy with at least two (and up to possibly six) additional rounds of endoreduplication measured in nuclei isolated from adult salt-treated plants (200 mm NaCl; Figs. 2–4; Table I). Increasing evidence links endopolyploidy with higher stress tolerance in plants as a strategy to allow cellular transcriptional and metabolic optimization to mitigate stress effects (Scholes and Paige, 2015). In cucumber (Cucumis sativus), a 15-d UV-B irradiation induced a doubling of the ploidy of a small number of epidermal cells from 2C to 4C and an increase in cell size. Additionally, these cells showed increased peroxidase activity and altered levels of phenolic compounds (Yamasaki et al., 2007, 2010). In Arabidopsis, accessions showing UV-B resistance also showed higher rounds of endoreduplication, with nuclei in leaf cells of the Kondara UV-B-tolerant accession showing 64C in comparison with 32C in the UV-B-sensitive Columbia-0 accession (Gegas et al., 2014). However, despite these lines having larger leaves, they did not show increased pigment levels, and UV-B irradiation was not shown to induce endoreduplication. Salinity stress has been shown to alter the ploidy level measured in root cells of sorghum (Sorghum bicolor ‘610’) plants. Only 1.3% of cells in control plants showed nuclei with DNA contents higher than 4C, while 41.2% of the cells in salt-treated plants were higher than 4C and nuclei up to 32C were recorded in some of the samples (Ceccarelli et al., 2006); however, the maximum ploidy level in the roots of salt-treated plants was highly variable between cells. Also in that study, no salt-induced alterations in ploidy levels were observed in shoot tissue, indicating that the response was root specific. In M. crystallinum, small but significant salinity-induced ploidy levels were observed in root tissue, and the low ploidy level measured in this tissue may reflect that roots do not sequester sodium (Barkla et al., 2002).
Trichomes typically are thought to show elevated levels of endopolyploidy, and on average, mature trichomes show a DNA content of between 16C and 32C (De Rocher et al., 1990; Melaragno et al., 1993; Hülskamp, 2004; Skaptsov et al., 2017); however, several Arabidopsis trichome mutants have C values reported up to 64C (Hülskamp, 2004). EBC, which line the leaves, stems, and flower buds of both salt-treated and untreated M. crystallinum plants, are considered large modified balloon-like trichomes (Adams et al., 1998; Oh et al., 2015) and may contribute to the polyploidy measured in the whole leaf. In order to determine the ploidy level of EBC, epidermal peels were made of the abaxial and adaxial leaf epidermis and nuclei were extracted and ploidy was compared with the level of the peeled leaf. In adult plants, ploidy levels of up to 64C were measured in the peeled leaf (Figs. 3 and 4), whereas ploidy levels in the epidermal peels reached values as high as 512C (Fig. 4). The highest ploidy values were measured in epidermal peels of salt-treated plants, and differences were observed between the upper and lower epidermal peels, with lower epidermal peels showing a level of ploidy higher than that of the upper peel. This is correlated with the increased size of EBC from the lower peels relative to the upper peels (Supplemental Fig. S1). Differences in ploidy level depending on leaf surface orientation also have been observed for epidermal cells of Phaseolus vulgaris primary leaves (Kinoshita et al., 2008), where, under high light, upper epidermal pavement cells showed higher levels of endoreduplication than were shown by lower epidermal pavement cells, with more 4C than 2C nuclei. The pavement cells with the higher ploidy also had increased cell size. However, this result is opposite what was observed in the EBC of M. crystallinum in relation to upper epidermis and lower epidermis ploidy variation (Fig. 4), and the ploidy difference was confined to pavement cells in P. vulgaris, as no increased DNA synthesis was observed in either guard cells or trichomes (Kinoshita et al., 2008).
A positive relationship between cell size, nuclear size, and ploidy level has been demonstrated in animal cells (Fox and Duronio, 2013; for review, see Schoenfelder and Fox, 2015) and in various endoreduplicating plant tissues. This includes the metaxylem in maize (Zea mays) roots, Aloe arborea (Agavaceae), and Zebrina pendula (Commelinaceae; List, 1963), the endosperm in sorghum (Kladnik et al., 2006), the pericarp in tomato (Solanum lycopersicum; Bourdon et al., 2012), and in species encompassing a wide variation in endopolyploidy (Barow and Meister, 2003). As the nuclear DNA content undergoes exponential amplification through endoreduplication, the nuclear volume is expected to increase accordingly. Increases in nuclear volume with increased polyploidy were clearly represented in EBC (Table I; Fig. 5), with the largest nuclei being detected in the largest EBC located on the flower bud, having an estimated ploidy level of 32,768C to 65,536C and a nuclei diameter of close to 136 μm.
To understand the role of gene expression in the regulation of cellular mechanisms influencing developmental and salt-induced ploidy levels in M. crystallinum EBC, including DNA replication, endoreduplication, cell and nuclear enlargement, and trichome development, we assembled a focal set of RECT genes for specific analysis.
The basal levels of transcripts in the RECT category R (DNA replication) in roots are higher compared with those in EBC. This is likely due to the higher number of cells in roots, including the root tips that are actively dividing, compared with EBC (Fig. 8A; Supplemental Table S2). All the complexes that coordinately regulate DNA replication are maintained at a relatively higher level in roots than in EBC, except for histone-coding transcripts. The absence of several histone-coding transcripts in the root may suggest incomplete representation of histone genes in the M. crystallinum reference root transcriptome. Therefore, the transcriptomic signature for DNA replication as an overall process does not appear to be significantly higher in EBC compared with roots. Nevertheless, when we checked the significantly differentially expressed genes in response to salt stress within each tissue, many of the genes associated with DNA replication showed a higher up-regulation in EBC than in roots (Fig. 8B; Supplemental Table S3). For example, the magnitude of response to salt stress in EBC by RNA polymerase I and III subunit C (RPA1C), DNA polymerase Δ1 (POLD1), prolifera (PRL; two copies), minichromosome maintenance replicative helicase (MCM4), and E2F target gene1 (ETG1; two copies) is much higher than in roots (Fig. 8B). These transcripts contribute to the replicative helicase, elongation, and single-strand DNA-binding complexes integral to the DNA replication process (Forsburg, 2004; Shultz et al., 2007; Takahashi et al., 2010) and are important for polyploidization (Guo et al., 2014). Figure 8A additionally illustrates how complexes in DNA replication are expressed coordinately in a single tissue while maintaining relative basal levels of individual transcripts within a complex. The findings that salt-treated EBC have increased levels of replication fork components (MCMs, GINs, single-stranded DNA-binding protein) but decreased initiation (ORC) and elongation complex components, as well as decreased histone expression, collectively suggest that mature salt-treated EBC have arrested the cell cycle in late S-phase with unresolved replication forks.
Both the MCM and the RPA proteins also have been shown to play roles in triggering DNA damage repair mechanisms in response to stalled replication forks (Harper and Elledge, 2007; Hu et al., 2016). To examine this possibility in the EBC, we analyzed the expression patterns of putative orthologs of 22 genes that have been shown to be induced by a variety of genotoxic agents (Supplemental Table S4; Yi et al., 2014). Of the genes in this set that were represented by one or more putative orthologs in the transcriptome assembly, three genes were significantly up-regulated in EBC, but the expression of most was unchanged by salt treatment, and two of these genes were significantly down-regulated (Supplemental Table S4). Additionally, while several Arabidopsis siamese-related (SMR) CDK inhibitor genes are up-regulated by exposure to DNA-damaging agents, the only M. crystallinum SMR gene showing a significant change upon salt treatment was down-regulated (Fig. 6). One interpretation of these results is that the endoreplicated salt-treated EBC may have arrested the cell cycle at the inter-S phase checkpoint with unresolved replication forks, but without activating the DNA damage checkpoint. Perhaps this checkpoint is lacking in these highly endoreplicated cells, or alternatively, the replication forks remain poised to be reactivated.
Several positive markers of endoreplication are maintained at a constitutively higher level in EBC than in roots (Fig. 8A). These include CCS52A1 (an activating subunit of the anaphase-promoting complex [APC]), known for its role in promoting endoreplication in both shoots and roots (Breuer et al., 2009; Kasili et al., 2010; Baloban et al., 2013; Takahashi et al., 2013), HOBBIT, which codes for another subunit of the APC (Blilou et al., 2002), and RHL2 (brassinosteroid-insensitive5 [BIN5]), a DNA topoisomerase. SMR, myb domain protein 3R5 (MYB3R5), and BIN4 (MIDGET) were not detected in roots (Fig. 8A; Supplemental Table S2), while SMR in EBC was detected at the highest normalized expression in the subset of genes searched under positive regulators of endoreplication (Fig. 7). SMR genes are ubiquitous in all plant lineages and are found routinely in roots and shoots (Kasili et al., 2011; Kumar et al., 2015). In contrast, constitutive expression of positive regulators of endoreplication, FKBP12 interacting protein of 37 kD (FIP37), chromatin licensing and DNA replication factor1 (CDT1), and F box-like17 (FBL17), were over 3-fold higher in roots than in EBC (Fig. 8A; Supplemental Table S2), consistent with their established roles in root meristem cell proliferation and subsequent development (Caro et al., 2007a; Noir et al., 2015; Shen et al., 2016). These genes are down-regulated in the roots upon salt stress, when salinity would most likely hinder root growth.
It is somewhat surprising that only a handful of these major known regulators of endoreplication showed increased expression in salt-treated EBC; however, it is noteworthy that many are expressed in both control and salt-treated EBC at levels apparently comparable to those of endoreplicated Arabidopsis tissues. Both control and salt-treated EBC have stopped dividing and likely express sufficient levels of these regulators of entry into endoreplication. Two subunits of topoisomerase V1, BIN4 and root hairless1 (RHL1), and FBL17 are the exception to this pattern, being up-regulated in the EBC under salt stress. BIN4 and RHL1 are required for endoreplication (Breuer et al., 2007) and are known for their up-regulation under environmental stress due to the additional function of topoisomerase VI as an integrator of reactive oxygen species stress signals. FBL17 plays a key role in the G1/S transition, degrading CDK inhibitors that prevent the activation of E2F by CDKs (Zhao et al., 2012). The almost 15-fold up-regulation of FBL17 in salt-treated EBC makes it the most highly up-regulated gene in the EBC transcriptome (Figs. 6 and 7B; Supplemental Table S3). The Arabidopsis FBL17 gene contains E2F-binding sites, creating a positive feedback loop that drives S phase entry. Endoreplication is almost completely absent in loss-of-function fbl17 mutants, and growth is greatly impaired (Noir et al., 2015). Not surprisingly, no root transcripts associated with positive regulators of endoreplication were significantly up-regulated under salt stress.
Identifying the significance of negative regulators of endoreplication in regulating higher ploidy levels is compounded by the fact that many proteins associated with the cell cycle form multiple complexes that can have antagonistic functions determined by the complex and not by the individual protein. This makes it difficult to interpret the net dominant process associated with differential expression of single transcripts, except when multiple members of a complex show coexpression that supports coregulation. For example, CDKB1 is highly active in dividing cells and CDKB protein abundance follows the transcription pattern (Inzé and De Veylder, 2006). In M. crystallinum, CDKB1 transcripts are expressed more than 50 times higher in roots compared with EBC, and A- and D-type cyclins that form a complex with CDKB1 (Inzé and De Veylder, 2006) also are expressed over 15-fold higher in roots than in EBC (Fig. 8A; Supplemental Table S3). Up-regulation of these genes would maintain cell division in root tissue. Negative regulators of the APC/cyclosome (C) complex, SAMBA (Eloy et al., 2012), DP-E2F-like1 (DEL1; Vlieghe et al., 2005), and ultraviolet-B-insensitive4 (UVI4; Heyman et al., 2017), together with ubiquitin-specific protease14 (UBP14; Xu et al., 2016), which would suppress endoreplication, also are more highly expressed in roots than in EBC.
Several other transcripts, including high ploidy2 (HPY2; Huang et al., 2009; Zhang et al., 2010), spindly (SPY; Cui et al., 2014), and cell cycle Ser/Thr-protein kinase5 (CDC5; Lin et al., 2007), important for cell proliferation in root development and known for their negative regulation of the endocycle, also are maintained at higher levels in roots than in EBC (Fig. 8A; Supplemental Table S3).
Negative regulators of endoreplication (E−), kaktus (KAK), UVI4, and the MYB domain protein MYB3R1, show significant increases in transcript levels in EBC when treated with salt. This may be explained in light of their pleiotropic tissue-specific roles in flavonoid pathway regulation coupled to trichome development (Haga et al., 2011; Patra et al., 2013) and response to stress (Hase et al., 2006; Cai et al., 2011). Other negative regulators of endoreplication genes, teosinte branched1/cycloidea/proliferating cell factor15 (TCP15) and (TCP20), are found at relatively high abundance in EBC (Fig. 8A; Supplemental Table S2) but are significantly down-regulated upon salt stress (Supplemental Table S3). In Arabidopsis, cellular oxidative stress is known to inactivate the TCP proteins, and this is mirrored transcriptionally by the down-regulation of TCP15 (Viola et al., 2013). Salt stress is known to induce many oxidative stress response pathways in the M. crystallinum EBC transcriptome (Oh et al., 2015). Overall, the highly polyploid M. crystallinum EBC maintains a low basal level of negative regulators of endoreplication, in contrast to the root transcriptome, which maintains a high level of genes in this category required for the high level of mitotic activity.
Among the genes involved in the cell cycle considered for this analysis (Fig. 8A, green bars; Supplemental Table S3), several APC/C complex core units, APC6, APC8, and APC10 (Heyman and De Veylder, 2012), are constitutively maintained more than 2- to 68-fold higher in roots than in EBC. The CDK-activating kinases CDKD;3 (Takatsuka et al., 2015), CYCB3;1 (Inzé and De Veylder, 2006), and CDC20;1 (Kevei et al., 2011) also are found at more than 2- to 20-fold higher in roots than in EBC. This difference can be explained by the mitotically active state of the root tissue compared with EBC.
The SMR proteins, which are positive regulators of endoreplication, are known to interact with CYC-CDK complexes to promote endoreplication in Arabidopsis trichomes (Kumar et al., 2015). As noted before, multiple copies of SMR transcripts are expressed in EBC, with some showing very high relative expression compared with other positive regulators. SMRs also are known to interact with CYCB2;4 (Van Leene et al., 2010), an underexplored member of the cyclin B family, which was found at a much higher basal level in EBC compared with roots (Supplemental Table S2). Additionally, GL2-expression modulator (GEM), a regulator of GL2 (Caro et al., 2007b), and GL2, a transcription factor involved in trichome development, branching, and positioning as well as nonhair cell differentiation in the root epidermis (Ishida et al., 2008; Lin and Aoyama, 2012), are both found at higher constitutive levels in EBC than in roots. The GEM protein not only regulates GL2 expression but also binds to CDT1 and has been proposed as a link between development and DNA replication (Caro et al., 2007a).
Plant stress signaling often leads to growth and developmental responses through the modulation of cell cycle genes (De Veylder et al., 2007; for review, see Komaki and Sugimoto, 2012). In response to salt stress, the EBC transcriptome shows six significantly differentially regulated genes out of 24 considered under the cell cycle category (Fig. 7; Supplemental Table S3). Among these, DPB and CKS1 are present at relatively high levels in EBC, with DPB significantly up-regulated with salt, as has been shown for this gene in other plants (Ma et al., 2014), while CKS1 is significantly down-regulated by salt (Supplemental Tables S2 and S3), which contrasts with evidence in other plants under abiotic stress (Zhao et al., 2013; Biermans et al., 2015; Tamirisa et al., 2017). We could not identify representative DPB and CKS1 genes in the root transcriptome.
Trichome development is tightly coupled with endoreplication (Hülskamp, 2004; Yang and Ye, 2013; Orr-Weaver, 2015). Most genes regulating trichome development are not unique to trichomes, but their tissue-specific regulation in trichomes can provide meaningful insight into the overall cellular state and ploidy level. Out of 13 genes considered in this category, we observed five genes in EBC and one gene in roots significantly regulated in response to salt (Supplemental Table S3).
Trichome initiation activators, GL2 and transparent testa glabra2 (TTG2), which encode HD-Zip and WRKY transcription factors, are both found in EBC transcriptomes at higher basal levels than in roots (Supplemental Table S1). Upon salt stress, GL2 shows an almost 6-fold increase in expression in EBC with no change in root expression. GL2 has been shown to be positively regulated by MYB23 and TTG1 (Szymanski et al., 1998; Lin and Aoyama, 2012) and negatively regulated by GEM (Caro et al., 2007a). MYB23 (a paralog of GL1 with functional redundancy; Kirik et al., 2005) and TTG1 (a WD40 protein) also are found at higher basal levels in EBC than in roots (Fig. 8A; Supplemental Table S3), and GEM is significantly down-regulated in EBC upon salt treatment. GEM, with the interaction of GL2 and TTG1, also regulates histone acetylation and methylation, providing a link to the epigenetic control of cell division and growth (Caro et al., 2007a).
Collectively, these results suggest that multiple regulators required for EBC identity, endoreplication, and development are expressed coordinately through the action of downstream regulators.
Yap1p response element (YRE/CER3/WAX2/FLP1) and shavenbaby (SVB) are two of the most highly abundant transcripts in EBC, at basal levels of 250- and 15-fold higher in EBC than in roots, respectively (Fig. 8A; Supplemental Table S3). YRE is a wax biosynthesis pathway gene involved in cuticle formation and required for trichome development and also is expressed in lateral root primordia (Kurata et al., 2003), while SVB is important for normal trichome development, although its function is unknown (Marks et al., 2009). YRE expression does not change significantly in response to salt stress in EBC, possibly due to its already constitutively high expression, but shows a significant 15-fold up-regulation in roots (Fig. 8B; Supplemental Table S3). The stress-specific response of YRE in roots exemplifies the functional significance of cuticle development in roots in stress adaptation (Lee and Suh, 2015), yet to allow rapid cell division and expansion in root lateral primordia, YRE is found at a very low basal expression level (Fig. 8A; Supplemental Table S2). SVB shows the highest relative expression among all RECT genes in both EBC and roots.
Stichel (STI) is the only trichome development gene that shows a higher basal level in roots compared with EBC (Supplemental Table S2). STI, a positive regulator of trichome branching (Ilgenfritz et al., 2003), may not play a significant role in the formation of globe-shaped unbranched EBC.
In addition to the RECT genes discussed above, transcripts for genes that control cell size, nuclear size, and morphology were analyzed in the EBC transcriptome, as large increases in cell size and the size of the nucleus in highly polyploid cells were measured (Fig. 5; Table I). Little nuclei (Linc) genes have been shown to control nuclear size and morphology in Arabidopsis (Dittmer et al., 2007) and are homologous to Nuclear Matrix Constituent Protein1 (NMCP1), a large nuclear coiled-coil protein localized to the nuclear periphery that showed domain homology to lamins and was first identified in Daucus carota (Kimura et al., 2014). Arabidopsis linc1 and linc2 mutants not only showed reductions in the nuclear size of epidermal cells but also reduced counts of polyploid levels, resulting in a mean ploidy level decrease from an average of 7.3C to 5.5C (Dittmer et al., 2007), suggesting a role in endopolyploidy as well as nuclear size. Significant up-regulation of a Linc1 gene was observed in EBC transcripts from salt-treated samples (Supplemental Table S1), and NMCP1 proteins have been shown to be induced by salt stress in Vigna spp. (Vincent et al., 2007).
The P-type cyclins CYCP1;1 and CYCP3;1 were found at much higher levels in the root transcriptome than in EBC (Supplemental Table S2), but CYCP1;1 was significantly up-regulated in EBC with salt treatment. P-type cyclins may link cell division to the nutritional status of a cell (Torres Acosta et al., 2004) and play a role in cell size determination, as deletion of a P-type cyclin with homology to AtCYCP1 from the malaria parasite Plasmodium berghei resulted in a significant reduction in oocyte size (Roques et al., 2015).
Rapid expansion of the bladder cells in the EBC from salt-treated cells would require a flexible and loose cell wall. Significant down-regulation of multiple trichome birefringence genes in EBC from salt-treated plants (Supplemental Table S1), which have been shown to be involved in the O-acetylation of cell wall polysaccharides (Gille and Pauly, 2012), may contribute to cell wall loosening. Arabidopsis trichome birefringence mutants have lower levels of crystalline cellulose in trichome cell walls, altered pectin composition in trichomes and stems, and swollen epidermal cells (Bischoff et al., 2010).
CONCLUSION
In M. crystallinum, the rapid expansion of cells during leaf development is due to a substantial increase in ploidy, beginning in emerging leaves, to higher than 1,024C in leaves of adult plants. Salt treatment leads to a further increase in developmentally programmed ploidy, and this is particularly illustrated in the EBC of the flower bud, with nuclei of greater than 100 μm in diameter being estimated at a ploidy of 32,768C or higher. Transcripts and proteins involved in endopolyploidy are shown to be regulated by salt in the EBC, suggesting environmental as well as developmental cues. It has been proposed that increased ploidy helps to mitigate stress damage, and the increases in cell volume and ploidy observed in M. crystallinum under salinity may contribute to tolerance by increasing the store size for sodium sequestration. This study shows that M. crystallinum is an outstanding model for studying endopolyploidization and its physiological role in the plant in relationship to both developmental and environmental stress tolerance.
At the molecular level, the increased knowledge of plant replication, endoduplication, and cell cycle regulation through comparison of tissue- and species-specific transcriptomes provides clues to understand the basis of endoreduplication control in EBC and plants in general.
MATERIALS AND METHODS
Plant Material
Mesembryanthemum crystallinum and Chenopodium quinoa were grown from seed in potting mix consisting of one-third sphagnum peat moss (TEAM; Theriault and Hachey), one-third vermiculite No. 2 (Auspearl), and one-third perlite P400 medium (Auspearl). Potting mix was supplemented with slow-release osmocote (5 g L−1; 16-9-12; Scotts Australia) and micronutrients (Micromax; 1 g L−1; Scotts Australia) in 15-cm-diameter, 1-L pots in a greenhouse under natural irradiation and photoperiod from October 2016 to March 2017. Temperature during the experiments in the greenhouse fluctuated from a minimum of 15°C at night to a maximum of 35°C during a short period on the hottest days, with average daytime temperatures around 28°C. NaCl treatment (200 mm) was initiated 6 weeks after germination for a period of 14 d. For treatments, plants were either watered daily with tap water (untreated) or treated with NaCl (200 mm). Seeds of M. crystallinum were from inbred lines derived from material originally collected by Winter et al. (1978). C. quinoa was grown from seed kindly provided by Sergey Shabala (University of Tasmania) and is used in this study as a comparator halophyte.
For each sample, three biological replicates, being individual plants, were grown.
Extraction of Nuclei
Approximately 20 mg of leaf tissue with or without epidermis or 5 mg of epidermal peel from the upper or lower leaf was removed from the leaf, and nuclei were extracted from this tissue. Tissue was chopped with a new sharp stainless steel razor blade for 90 s in a sterile petri dish on ice in 250 or 100 µL of cold (4°C) Galbraith’s nuclei extraction buffer (Galbraith, 1990; 45 mm MgCl2, 20 mm MOPS, 30 mm sodium citrate, and 0.05% [v/v] Triton X-100, pH 7). This suspension was then filtered through a 100-µm nylon mesh filter before being stained with 50 µg mL−1 PI and left to incubate on ice, in darkness, for 5 min (Galbraith et al., 1997).
Flow Cytometry
For flow cytometry analysis, each experiment was replicated three times by taking leaf tissue from three different plants for each age/treatment/leaf pair and isolating nuclei from those samples. All three biological replicates were analyzed on the same day.
Samples were analyzed using a BD FACS Canto II flow cytometer (BD Biosciences). PI that was bound to nucleic acids was excited using the 488-nm blue laser with the emission spectra captured using the 556-nm LP mirror with a 585/42-nm filter. Each sample was run for 5 min at a low flow rate to increase resolution and sensitivity and analyzed using Flowing Software version 2.5.1 (Perttu Terho, Centre for Biotechnology). To reduce interference from debris and other cellular particles, events were gated on SSC and PI fluorescence to distinguish between noise and fluorescence signals from the nucleus. Flow data were plotted on a logarithmic scale so that peak height was directly proportional to the number of nuclei of the corresponding ploidy level. Nuclei population peaks were selected with percentage coefficient of variation below 10%, and the values corresponded to the expected incremental doubling of fluorescence from 2C to 512C.
The mean endoreduplication cycles per nucleus in each sample were calculated using the following equation (Barow and Meister, 2003):
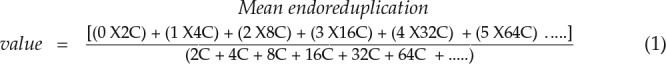 |
(1) |
Fluorescence Microscopy
Leaves, stems, or flower buds were either peeled or sectioned using forceps or a razor blade, respectively, and tissue was incubated in a 0.1% (v/v) Triton X-100 (pH 7) solution containing 50 µg mL−1 PI at room temperature in darkness for 15 min. Nuclei from the large epidermal bladder cells of the stem and buds were extracted by plunge freezing whole buds or stems in liquid nitrogen. Large individual bladder cells were then separated using forceps and placed directly on a microscope slide to thaw on top of a 1-µL drop of 1 mg mL−1 PI. This freeze/thaw step disrupts the cell wall, allowing for the large cell to become freely suspended in the PI solution for easy uptake and visualization. A Nikon E600 fluorescence microscope with G-2A filter cube was used to visualize nuclei fluorescence for all tissue and nuclei preparations. Images were captured with an Olympus DP73 camera using CellSens software and, when necessary, as for the measurement of nuclear diameter and volume, were further analyzed in ImageJ (Abramoff et al., 2004).
Estimation of Ploidy in Large EBC
Two separate approaches were used to allow the estimation of ploidy level in large EBC. For the first, we used nuclei of known identity whose ploidy was measured by flow cytometry (i.e. guard cells, pavement cells, etc.) and calculated the volume of those nuclei from fluorescence microscopy using ImageJ software (Abramoff et al., 2004) to associate nuclear volume with C value. The volumes of the large PI-stained EBC nuclei from leaf epidermis, stem, and flower bud were then estimated from their area, assuming a perfect sphere, and the ploidy levels were extrapolated (Table I).
In the second approach, the pollen of M. crystallinum, which are spherical with a mean diameter of 31.9 ± 0.3 µm (n = 50), and BD FACS size standard beads (3 µm) were added to the nuclei suspension. The FSC values of these two standards gated the 2C to 16C nuclei populations, with the 32C population sitting slightly larger than the pollen FSC. From this, the 2C to 64C nuclei populations were assigned a nucleus diameter based on FSC, and a standard curve was made to relate ERI with nuclei diameter (Supplemental Fig. S6).
Selection of Target Genes
To investigate the transcriptomic signals of endoreplication in M. crystallinum EBC, we created a custom set of focal genes for our current analyses because a single Gene Ontology (GO) term could not capture the majority of genes associated with DNA replication and endoreplication processes. First, we selected genes annotated under the GO term DNA endoreplication and added genes associated with DNA replication, cell cycle processes, and trichome development, as EBC are considered modified trichomes. Because of the lag time in updating GO annotations to mirror published research that add specific or novel functions to proteins, the literature was searched for additional genes that had identified roles critical to endoreplication. The reference transcriptome developed for EBC (Oh et al., 2015) was then searched to mine for putative orthologs of the selected Arabidopsis (Arabidopsis thaliana) genes in M. crystallinum using reciprocal BLAST. We included matches that had an e-value less than 10−5 and genes represented by contigs that had a length above 300 bp. From previously published research, we knew that BLAST was an ineffective tool to search for SMR genes, one of the hallmark gene families in endoreplication (Kumar et al., 2015). Therefore, we used a custom program developed in our laboratory (Kumar et al., 2015) to search for putative SMR genes in the nonmodel plants and manually checked alignments with known SMR genes before using the putative genes in this analysis. We categorized each gene into the following functional categories: DNA replication, positive regulation of endoreplication, negative regulation of endoreplication, cell cycle processes, trichome development, and other genes indirectly associated with endoreplication, to follow each category in different tissue/species (Supplemental Table S2).
Significantly Differently Expressed Genes in M. crystallinum
The expression values for selected genes (Supplemental Table S1) were taken from RNA-seq reads analyzed by Oh et al. (2015) from 8-week-old plants treated for 2 weeks with 200 mm NaCl. We used an adjusted P cutoff value of 0.05 and a false discovery rate value of 0.01 for uniquely mapped reads normalized based on the methods described by Oh et al. (2015) to detect significantly differently expressed genes between control and salt-treated EBC replicated samples.
The differently expressed genes in M. crystallinum roots were extracted by aligning RNA-seq reads to the reference root transcriptome published by Tsukagoshi et al. (2015), which used 5-d-old seedlings treated with 140 to 500 mm NaCl for 24 h on Murashige and Skoog plates with 1% [w/v] Suc. We used Bowtie (version 1.1.2; Langmead et al., 2009) to identify uniquely mapped reads with a seed alignment length of 50 bp and no more than one mismatch allowed in the seed alignment between the Illumina read and the reference transcriptome. To create biological replicates of the reference root transcriptome for a comparable analysis to our EBC transcriptome, we combined the 0 and 140 mm salt treatments as two biological replicates for the control condition, given the minimal changes observed between the low- and zero-salt treatments in the root transcriptomes based on a principal component analysis test (Tsukagoshi et al., 2015). For the salt-treated response, we used 250 and 500 mm salt treatments as two biological replicates (Tsukagoshi et al., 2015). The control and salt-treated root samples were then analyzed for differentially expressed genes using DESeq (Anders and Huber, 2010) similarly conducted for the EBC transcriptomes to detect significantly different expression.
We used the sum of the reads per kilobase of transcript per million mapped reads (RPKM) values from each reference transcript to represent the expression of the associated reference gene model if different transcript copy numbers were present in the two transcriptomes to follow gene expression patterns across tissues in Figure 8A. We then normalized the expression value for each gene within a category to assess the expression pattern across categories between EBC and root samples. The expression of each gene was divided by the sum expression for a given category and normalized to the mean expression of that category (Supplemental Table S2). We added a value of 1 (numerically insignificant expression) to all normalized expression values to avoid having undefined expression due to 0 RPKM observed for some of the genes in one tissue type when values were converted to log2. Finally, all values were scaled between 0 and 1 to visualize expression on a linear scale, with 1 being the highest expression value and 0 being the lowest expression in each category. These values were plotted using Matplotlib, a graphing tool (Hunter, 2007), in a custom python script (Fig. 8A).
For comparison between significantly differently expressed genes that have variable numbers of transcript copies in roots and EBC (Fig. 8B), we used one-to-one reciprocal BLAST to identify the identical transcripts between the two tissues. For transcripts that had missing copies in one tissue, we added an artificial value of 0-fold change with an adjusted P value of 1 (not significant) to represent the remaining expression in the relevant tissue (Supplemental Table S3).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. M. crystallinum cotyledon expansion, stem branch anatomy, and leaf cross section.
Supplemental Figure S2. Flow cytometry analysis of endopolyploidy in M. crystallinum juvenile plants.
Supplemental Figure S3. Flow cytometry analysis of endopolyploidy in M. crystallinum epidermal peels and peeled branch leaf tissue.
Supplemental Figure S4. Flow cytometry analysis of endopolyploidy in M. crystallinum epidermal peels and peeled bract leaf tissue.
Supplemental Figure S5. Standard curve used to estimate the ERI of large-diameter nuclei from the EBC.
Supplemental Figure S6. Flow cytometry analysis of endopolyploidy in C. quinoa.
Supplemental Table S1. RECT gene expression in M. crystallinum EBC in response to salt treatment.
Supplemental Table S2. M. crystallinum RECT gene expression in EBC and roots in response to salinity.
Supplemental Table S3. Normalized fold change of RECT genes between EBC and root transcriptomes.
Supplemental Table S4. Normalized expression of selected DNA damage-responsive genes in response to salt treatment in epidermal bladder cells.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We acknowledge discussions with Hans Bohnert for igniting our interest in endopolyploidy in M. crystallinum. Thanks to Sergey Shabala for C. quinoa seeds, Carolyn Raymond for statistical analysis and Alicia Hidden for horticultural assistance. The high-performance computing service at Louisiana State University is acknowledged for facilitating the comparative transcriptomic analyses.
Footnotes
This work was funded by an SCU seed grant to B.J.B., NSF grant 1615782 to J.C.L., and NSF grant 1616827 and the Next-Generation BioGreen21 Program (grant no. PJ011379) of the Rural Development Administration, Republic of Korea, to M.D.
Articles can be viewed without a subscription.
References
- Abramoff MD, Magalhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton Int 11: 36–42 [Google Scholar]
- Adams P, Nelson DE, Yamada S, Chmara W, Jensen RG, Bohnert HJ, Griffiths H (1998) Growth and development of Mesembryanthemum crystallinum (Aizoaceae). New Phytol 138: 171–190 [DOI] [PubMed] [Google Scholar]
- Agarie S, Shimoda T, Shimizu Y, Baumann K, Sunagawa H, Kondo A, Ueno O, Nakahara T, Nose A, Cushman JC (2007) Salt tolerance, salt accumulation, and ionic homeostasis in an epidermal bladder-cell-less mutant of the common ice plant Mesembryanthemum crystallinum. J Exp Bot 58: 1957–1967 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloban M, Vanstraelen M, Tarayre S, Reuzeau C, Cultrone A, Mergaert P, Kondorosi E (2013) Complementary and dose-dependent action of AtCCS52A isoforms in endoreduplication and plant size control. New Phytol 198: 1049–1059 [DOI] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R (2015) Single cell-type comparative metabolomics of epidermal bladder cells from the halophyte Mesembryanthemum crystallinum. Front Plant Sci 6: 435 10.3389/fpls.2015.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Camacho-Emiterio J, Pantoja O (2002) Na+/H+ exchange in the halophyte Mesembryanthemum crystallinum L. is associated with cellular sites of Na+ storage. Funct Plant Biol 29: 1017–1024 [DOI] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Pantoja O (2012) Protein profiling of epidermal bladder cells from the halophyte Mesembryanthemum crystallinum. Proteomics 12: 2862–2865 10.1002/pmic.201200152 [DOI] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Raymond C (2016) Single-cell-type quantitative proteomic and ionomic analysis of epidermal bladder cells from the halophyte model plant Mesembryanthemum crystallinum to identify salt-responsive proteins. BMC Plant Biol 16: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barow M, Meister A (2003) Endopolyploidy in seed plants is differently correlated to systemics, organ, life strategy and genome size. Plant Cell Environ 26: 571–584 [Google Scholar]
- Biermans G, Horemans N, Vanhoudt N, Vandenhove H, Saenen E, Van Hees M, Wannijn J, Vangronsveld J, Cuypers A (2015) Arabidopsis thaliana seedlings show an age-dependent response on growth and DNA repair after exposure to chronic γ-radiation. Environ Exp Bot 109: 122–130 [Google Scholar]
- Bischoff V, Nita S, Neumetzler L, Schindelasch D, Urbain A, Eshed R, Persson S, Delmer D, Scheible WR (2010) TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol 153: 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Frugier F, Folmer S, Serralbo O, Willemsen V, Wolkenfelt H, Eloy NB, Ferreira PC, Weisbeek P, Scheres B (2002) The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev 16: 2566–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon M, Pirrello J, Cheniclet C, Coriton O, Bourge M, Brown S, Moïse A, Peypelut M, Rouyère V, Renaudin JP, et al. (2012) Evidence for karyoplasmic homeostasis during endoreduplication and a ploidy-dependent increase in gene transcription during tomato fruit growth. Development 139: 3817–3826 [DOI] [PubMed] [Google Scholar]
- Bramsiepe J, Wester K, Weinl C, Roodbarkelari F, Kasili R, Larkin JC, Hülskamp M, Schnittger A (2010) Endoreplication controls cell fate maintenance. PLoS Genet 6: e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Stacey NJ, West CE, Zhao Y, Chory J, Tsukaya H, Azumi Y, Maxwell A, Roberts K, Sugimoto-Shirasu K (2007) BIN4, a novel component of the plant DNA topoisomerase VI complex, is required for endoreduplication in Arabidopsis. Plant Cell 19: 3655–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Kawamura A, Ichikawa T, Tominaga-Wada R, Wada T, Kondou Y, Muto S, Matsui M, Sugimoto K (2009) The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 21: 2307–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Tian S, Liu C, Dong H (2011) Identification of a MYB3R gene involved in drought, salt and cold stress in wheat (Triticum aestivum L.). Gene 485: 146–152 [DOI] [PubMed] [Google Scholar]
- Caro E, Castellano MM, Gutierrez C (2007a) A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature 447: 213–217 [DOI] [PubMed] [Google Scholar]
- Caro E, Castellano MM, Gutierrez C (2007b) GEM, a novel factor in the coordination of cell division to cell fate decisions in the Arabidopsis epidermis. Plant Signal Behav 2: 494–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli M, Santantonio E, Marmottini F, Amzallag GN, Cionini PG (2006) Chromosome endoreduplication as a factor of salt adaptation in Sorghum bicolor. Protoplasma 227: 113–118 [DOI] [PubMed] [Google Scholar]
- Chevalier C, Nafati M, Mathieu-Rivet E, Bourdon M, Frangne N, Cheniclet C, Renaudin JP, Gévaudant F, Hernould M (2011) Elucidating the functional role of endoreduplication in tomato fruit development. Ann Bot 107: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson SJ, Radziejwoski A, Granier C (2006) Cell and leaf size plasticity in Arabidopsis: what is the role of endoreduplication? Plant Cell Environ 29: 1273–1283 [DOI] [PubMed] [Google Scholar]
- Cui H, Kong D, Wei P, Hao Y, Torii KU, Lee JS, Li J (2014) SPINDLY, ERECTA, and its ligand STOMAGEN have a role in redox-mediated cortex proliferation in the Arabidopsis root. Mol Plant 7: 1727–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3: 117–124 [DOI] [PubMed] [Google Scholar]
- Dassanayake M, Larkin JC (2017) Making plants break a sweat: the structure, function, and evolution of plant salt glands. Front Plant Sci 8: 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rocher EJ, Harkins KR, Galbraith DW, Bohnert HJ (1990) Developmentally regulated systemic endopolyploid in succulents with small genomes. Science 250: 99–101 [DOI] [PubMed] [Google Scholar]
- De Storme N, Mason A (2014) Plant speciation through chromosome instability and ploidy change: cellular mechanisms, molecular factors and evolutionary relevance. Curr Plant Biol 1: 10–33 [Google Scholar]
- De Veylder L, Beeckman T, Inzé D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A (2011) Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 16: 624–634 [DOI] [PubMed] [Google Scholar]
- Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ (2007) LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 19: 2793–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Zielke N, Gutierrez C (2014) Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat Rev Mol Cell Biol 15: 197–210 [DOI] [PubMed] [Google Scholar]
- Eloy NB, Gonzalez N, Van Leene J, Maleux K, Vanhaeren H, De Milde L, Dhondt S, Vercruysse L, Witters E, Mercier R, et al. (2012) SAMBA, a plant-specific anaphase-promoting complex/cyclosome regulator is involved in early development and A-type cyclin stabilization. Proc Natl Acad Sci USA 109: 13853–13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. (2004) Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev 68: 109–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ (2013) Endoreplication and polyploidy: insights into development and disease. Development 140: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW. (1990) Isolation and flow cytometric characterization of plant protoplasts. Methods Cell Biol 33: 527–547 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 10.1126/science.220.4601.1049 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Lambert GM, Macas J, Dolezel J (1997) Analysis of nuclear DNA content and ploidy in higher plants. Curr Protoc Cytometry 2: 7.6.1–7.6.22 [DOI] [PubMed] [Google Scholar]
- Gegas VC, Wargent JJ, Pesquet E, Granqvist E, Paul ND, Doonan JH (2014) Endopolyploidy as a potential alternative adaptive strategy for Arabidopsis leaf size variation in response to UV-B. J Exp Bot 65: 2757–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Pauly M (2012) O-Acetylation of plant cell wall polysaccharides. Front Plant Sci 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Wu Z, Song J, Jiang F, Wang Z, Deng S, Walker VK, Zhou S (2014) Juvenile hormone-receptor complex acts on mcm4 and mcm7 to promote polyploidy and vitellogenesis in the migratory locust. PLoS Genet 10: e1004702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga N, Kobayashi K, Suzuki T, Maeo K, Kubo M, Ohtani M, Mitsuda M, Demura T, Nakamura K, Jürgens G, et al. (2011) Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol 157: 706–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Hase Y, Trung KH, Matsunaga T, Tanaka A (2006) A mutation in the uvi4 gene promotes progression of endo-reduplication and confers increased tolerance towards ultraviolet B light. Plant J 46: 317–326 [DOI] [PubMed] [Google Scholar]
- Heyman J, De Veylder L (2012) The anaphase-promoting complex/cyclosome in control of plant development. Mol Plant 5: 1182–1194 [DOI] [PubMed] [Google Scholar]
- Heyman J, Polyn S, Eekhout T, De Veylder L (2017) Tissue-specific control of the endocycle by the anaphase promoting complex/cyclosome inhibitors UVI4 and DEL1. Plant Physiol 175: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Cools T, De Veylder L (2016) Mechanisms used by plants to cope with DNA damage. Annu Rev Plant Biol 67: 439–462 [DOI] [PubMed] [Google Scholar]
- Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, Shi S, Wang J, Wang Y, Xie Q, et al. (2009) The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J 60: 666–678 [DOI] [PubMed] [Google Scholar]
- Hülskamp M. (2004) Plant trichomes: a model for cell differentiation. Nat Rev Mol Cell Biol 5: 471–480 [DOI] [PubMed] [Google Scholar]
- Hunter JD. (2007) Matplotlib: a 2D graphics environment. Comput Sci Eng 9: 90–95 [Google Scholar]
- Ilgenfritz H, Bouyer D, Schnittger A, Mathur J, Kirik V, Schwab B, Chua NH, Jürgens G, Hülskamp M (2003) The Arabidopsis STICHEL gene is a regulator of trichome branch number and encodes a novel protein. Plant Physiol 131: 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Jarvis DE, Ho YS, Lightfoot DJ, Schmöckel SM, Li B, Borm TJA, Ohyanagi H, Mineta K, Michell CT, Saber N, et al. (2017) The genome of Chenopodium quinoa. Nature 542: 307–312 [DOI] [PubMed] [Google Scholar]
- Joubès J, Chevalier C (2000) Endoreduplication in higher plants. Plant Mol Biol 43: 735–745 [DOI] [PubMed] [Google Scholar]
- Kasili R, Walker JD, Simmons LA, Zhou J, De Veylder L, Larkin JC (2010) SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics 185: 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasili R, Huang CC, Walker JD, Simmons LA, Zhou J, Faulk C, Hülskamp M, Larkin JC (2011) BRANCHLESS TRICHOMES links cell shape and cell cycle control in Arabidopsis trichomes. Development 138: 2379–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei Z, Baloban M, Da Ines O, Tiricz H, Kroll A, Regulski K, Mergaert P, Kondorosi E (2011) Conserved CDC20 cell cycle functions are carried out by two of the five isoforms in Arabidopsis thaliana. PLoS ONE 6: e20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Fujino K, Ogawa K, Masuda K (2014) Localization of Daucus carota NMCP1 to the nuclear periphery: the role of the N-terminal region and an NLS-linked sequence motif, RYNLRR, in the tail domain. Front Plant Sci 5: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita I, Sanbe A, Yokomura EI (2008) Difference in light-induced increase in ploidy level and cell size between adaxial and abaxial epidermal pavement cells of Phaseolus vulgaris primary leaves. J Exp Bot 59: 1419–1430 [DOI] [PubMed] [Google Scholar]
- Kirik V, Lee MM, Wester K, Herrmann U, Zheng Z, Oppenheimer D, Schiefelbein J, Hulskamp M (2005) Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132: 1477–1485 [DOI] [PubMed] [Google Scholar]
- Kladnik A, Chourey PS, Pring DR, Dermastia M (2006) Development of the endosperm of Sorghum bicolor during the endoreduplication-associated growth phase. J Cereal Sci 43: 209–215 [Google Scholar]
- Komaki S, Sugimoto K (2012) Control of the plant cell cycle by developmental and environmental cues. Plant Cell Physiol 53: 953–964 [DOI] [PubMed] [Google Scholar]
- Kumar N, Harashima H, Kalve S, Bramsiepe J, Wang K, Sizani BL, Bertrand LL, Johnson MC, Faulk C, Dale R, et al. (2015) Functional conservation in the SIAMESE-RELATED family of cyclin-dependent kinase inhibitors in land plants. Plant Cell 27: 3065–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T, Kawabata-Awai C, Sakuradani E, Shimizu S, Okada K, Wada T (2003) The YORE-YORE gene regulates multiple aspects of epidermal cell differentiation in Arabidopsis. Plant J 36: 55–66 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Suh MC (2015) Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep 34: 557–572 [DOI] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ (2009) Endoreplication: polyploidy with purpose. Genes Dev 23: 2461–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch I, Dodsworth S (2017) Endopolyploidy in plants. eLS
- Lin Q, Aoyama T (2012) Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: update on the roles of the classic regulator. J Integr Plant Biol 54: 729–737 [DOI] [PubMed] [Google Scholar]
- Lin Z, Yin K, Zhu D, Chen Z, Gu H, Qu LJ (2007) AtCDC5 regulates the G2 to M transition of the cell cycle and is critical for the function of Arabidopsis shoot apical meristem. Cell Res 17: 815–828 [DOI] [PubMed] [Google Scholar]
- List A. (1963) Some observations on DNA content and cell and nuclear volume growth in the developing xylem cells of certain higher plants. Am J Bot 50: 320 [Google Scholar]
- Ma TY, Li ZW, Zhang SY, Liang GT, Guo J (2014) Identification and expression analysis of the E2F/DP genes under salt stress in Medicago truncatula. Genes Genomics 36: 819–828 [Google Scholar]
- Marks MD, Wenger JP, Gilding E, Jilk R, Dixon RA (2009) Transcriptome analysis of Arabidopsis wild-type and gl3-sst sim trichomes identifies four additional genes required for trichome development. Mol Plant 2: 803–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet C, Tisné S, Radziejwoski A, Vile D, De Veylder L, Dauzat M, Granier C (2011) New insights into the control of endoreduplication: endoreduplication could be driven by organ growth in Arabidopsis leaves. Plant Physiol 157: 2044–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noir S, Marrocco K, Masoud K, Thomann A, Gusti A, Bitrian M, Schnittger A, Genschik P (2015) The control of Arabidopsis thaliana growth by cell proliferation and endoreplication requires the F-box protein FBL17. Plant Cell 27: 1461–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Barkla BJ, Vera-Estrella R, Pantoja O, Lee SY, Bohnert HJ, Dassanayake M (2015) Cell type-specific responses to salinity: the epidermal bladder cell transcriptome of Mesembryanthemum crystallinum. New Phytol 207: 627–644 [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL. (2015) When bigger is better: the role of polyploidy in organogenesis. Trends Genet 31: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini F, Accorsi M, Gianquinto G, Dinelli G, Antognoni F, Ruiz Carrasco KB, Martinez EA, Alnayef M, Marotti I, Bosi S, et al. (2011) Beyond the ionic and osmotic response to salinity in Chenopodium quinoa: functional elements of successful halophytism. Funct Plant Biol 38: 818–831 [DOI] [PubMed] [Google Scholar]
- Patra B, Pattanaik S, Yuan L (2013) Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J 74: 435–447 [DOI] [PubMed] [Google Scholar]
- Rangel J, Strauss K, Seedorf K, Hjelmen CE, Johnston JS (2015) Endopolyploidy changes with age-related polyethism in the honey bee, Apis mellifera. PLoS ONE 10: e0122208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roques M, Wall RJ, Douglass AP, Ramaprasad A, Ferguson DJP, Kaindama ML, Brusini L, Joshi N, Rchiad Z, Brady D, et al. (2015) Plasmodium P-type cyclin CYC3 modulates endomitotic growth during oocyst development in mosquitoes. PLoS Pathog 11: e1005273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder KP, Fox DT (2015) The expanding implications of polyploidy. J Cell Biol 209: 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes DR, Paige KN (2015) Plasticity in ploidy: a generalized response to stress. Trends Plant Sci 20: 165–175 [DOI] [PubMed] [Google Scholar]
- Shen L, Liang Z, Gu X, Chen Y, Teo ZWN, Hou X, Cai WM, Dedon PC, Liu L, Yu H (2016) N(6)-Methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev Cell 38: 186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz RW, Tatineni VM, Hanley-Bowdoin L, Thompson WF (2007) Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol 144: 1697–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaptsov MV, Lomonosova MN, Kutsev MG, Smirnov SV, Shmakov AI (2017) The phenomenon of endopolyploidy in some species of the Chenopodioideae (Amaranthaceae). Botany Letters 164: 47–53 [Google Scholar]
- Szymanski DB, Jilk RA, Pollock SM, Marks MD (1998) Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125: 1161–1171 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Quimbaya M, Schubert V, Lammens T, Vandepoele K, Schubert I, Matsui M, Inzé D, Berx G, De Veylder L (2010) The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. PLoS Genet 6: e1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Kajihara T, Okamura C, Kim Y, Katagiri Y, Okushima Y, Matsunaga S, Hwang I, Umeda M (2013) Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr Biol 23: 1812–1817 [DOI] [PubMed] [Google Scholar]
- Takatsuka H, Umeda-Hara C, Umeda M (2015) Cyclin-dependent kinase-activating kinases CDKD;1 and CDKD;3 are essential for preserving mitotic activity in Arabidopsis. Plant J 82: 1004–1017 [DOI] [PubMed] [Google Scholar]
- Tamirisa S, Vudem DR, Khareedu VR (2017) A cyclin dependent kinase regulatory subunit (CKS) gene of pigeonpea imparts abiotic stress tolerance and regulates plant growth and development in Arabidopsis. Front Plant Sci 8: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres Acosta JA, de Almeida Engler J, Raes J, Magyar Z, De Groodt R, Inzé D, De Veylder L (2004) Molecular characterization of Arabidopsis PHO80-like proteins, a novel class of CDKA;1-interacting cyclins. Cell Mol Life Sci 61: 1485–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Suzuki T, Nishikawa K, Agarie S, Ishiguro S, Higashiyama T (2015) RNA-seq analysis of the response of the halophyte, Mesembryanthemum crystallinum (ice plant) to high salinity. PLoS ONE 10: e0118339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. (2013) Does ploidy level directly control cell size? Counterevidence from Arabidopsis genetics. PLoS ONE 8: e83729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, et al. (2010) Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent D, Ergül A, Bohlman MC, Tattersall EA, Tillett RL, Wheatley MD, Woolsey R, Quilici DR, Joets J, Schlauch K, et al. (2007) Proteomic analysis reveals differences between Vitis vinifera L. cv. Chardonnay and cv. Cabernet Sauvignon and their responses to water deficit and salinity. J Exp Bot 58: 1873–1892 [DOI] [PubMed] [Google Scholar]
- Viola IL, Güttlein LN, Gonzalez DH (2013) Redox modulation of plant developmental regulators from the class I TCP transcription factor family. Plant Physiol 162: 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GTS, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inzé D, De Veylder L (2005) The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol 15: 59–63 [DOI] [PubMed] [Google Scholar]
- Winter K, Lüttge U, Winter E, Troughton JH (1978) Seasonal shift from C3 photosynthesis to Crassulacean Acid Metabolism in Mesembryanthemum crystallinum growing in its natural environment. Oecologia 34: 225–237 10.1007/BF00345168 [DOI] [PubMed] [Google Scholar]
- Xu Y, Jin W, Li N, Zhang W, Liu C, Li C, Li Y (2016) UBIQUITIN-SPECIFIC PROTEASE14 interacts with ULTRAVIOLET-B INSENSITIVE4 to regulate endoreduplication and cell and organ growth in Arabidopsis. Plant Cell 28: 1200–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Noguchi N, Mimaki K (2007) Continuous UV-B irradiation induces morphological changes and the accumulation of polyphenolic compounds on the surface of cucumber cotyledons. J Radiat Res (Tokyo) 48: 443–454 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Shimada E, Kuwano T, Kawano T, Noguchi N (2010) Continuous UV-B irradiation induces endoreduplication and peroxidase activity in epidermal cells surrounding trichomes on cucumber cotyledons. J Radiat Res 51: 187–196 [DOI] [PubMed] [Google Scholar]
- Yang C, Ye Z (2013) Trichomes as models for studying plant cell differentiation. Cell Mol Life Sci 70: 1937–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi D, Alvim Kamei CL, Cools T, Vanderauwera S, Takahashi N, Okushima Y, Eekhout T, Yoshiyama KO, Larkin J, Van den Daele H, et al. (2014) The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell 26: 296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Qi Y, Yang C (2010) Arabidopsis SUMO E3 ligase AtMMS21 regulates root meristem development. Plant Signal Behav 5: 53–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FY, Hu F, Zhang SY, Wang K, Zhang CR, Liu T (2013) MAPKs regulate root growth by influencing auxin signaling and cell cycle-related gene expression in cadmium-stressed rice. Environ Sci Pollut Res Int 20: 5449–5460 [DOI] [PubMed] [Google Scholar]
- Zhao X, Harashima H, Dissmeyer N, Pusch S, Weimer AK, Bramsiepe J, Bouyer D, Rademacher S, Nowack MK, Novak B, et al. (2012) A general G1/S-phase cell-cycle control module in the flowering plant Arabidopsis thaliana. PLoS Genet 8: e1002847. [DOI] [PMC free article] [PubMed] [Google Scholar]