Tomato mitogen-activated protein kinase SlMPK1 negatively regulates high-temperature tolerance by regulating antioxidant defense, and the phosphorylation of substrate SlSPRH1 is involved in this pathway.
Abstract
High-temperature (HT) stress is a major environmental stress that limits plant growth and development. MAPK cascades play key roles in plant growth and stress signaling, but their involvement in the HT stress response is poorly understood. Here, we describe a 47-kD MBP-phosphorylated protein (p47-MBPK) activated in tomato (Solanum lycopersicum) leaves under HT and identify it as SlMPK1 by tandem mass spectrometry analysis. Silencing of SlMPK1 in transgenic tomato plants resulted in enhanced tolerance to HT, while overexpression resulted in reduced tolerance. Proteomic analysis identified a set of proteins involved in antioxidant defense that are significantly more abundant in RNA interference-SlMPK1 plants than nontransgenic plants under HT stress. RNA interference-SlMPK1 plants also showed changes in membrane lipid peroxidation and antioxidant enzyme activities. Furthermore, using yeast two-hybrid screening, we identified a serine-proline-rich protein homolog, SlSPRH1, which interacts with SlMPK1 in yeast, in plant cells, and in vitro. We demonstrate that SlMPK1 can directly phosphorylate SlSPRH1. Furthermore, the serine residue serine-44 of SlSPRH1 is a crucial phosphorylation site in the SlMPK1-mediated antioxidant defense mechanism activated during HT stress. We also demonstrate that heterologous expression of SlSPRH1 in Arabidopsis (Arabidopsis thaliana) led to a decrease in thermotolerance and lower antioxidant capacity. Taken together, our results suggest that SlMPK1 is a negative regulator of thermotolerance in tomato plants. SlMPK1 acts by regulating antioxidant defense, and its substrate SlSPRH1 is involved in this pathway.
High temperature (HT) is a major plant stress that disturbs cellular homeostasis and leads to severe retardation of crop growth and development, and even death. HT stress can be expected to become increasingly problematic as global warming leads to more adverse climatic changes (Tubiello et al., 2007). Plants have evolved a variety of response mechanisms to elevated temperatures (Kotak et al., 2007). Many HT-responsive genes have been identified, and altered gene expression plays an important role in plant HT tolerance (Larkindale and Vierling, 2008; Mittler et al., 2012). Tomato (Solanum lycopersicum) is one of the most important vegetable crops but is susceptible to HT stress, especially in greenhouse facilities. Nevertheless, the biological functions of most HT-responsive genes in tomato are largely unknown.
The MAPK cascades are one of the major and evolutionarily conserved signaling pathways by which extracellular stimuli are transduced into intracellular responses in eukaryotic cells. The basic modules consist of three interlinked protein kinase modules (MAPKKK-MAPKK-MAPK). The downstream targets of activated MAPKs can be transcription factors, protein kinases, or cytoskeleton-associated proteins (Nakagami et al., 2005). MAPK gene families show a similar complexity in different species, with 20 genes in Arabidopsis (Arabidopsis thaliana; Colcombet and Hirt, 2008), 17 genes in rice (Oryza sativa; Rohila and Yang, 2007), and 16 genes in tomato (Kong et al., 2012). In plants, MAPK cascades have been identified in signal transduction, including cell division, hormone responses, development, and disease resistance (Nakagami et al., 2005; Pitzschke et al., 2009; Meng and Zhang, 2013; Xu and Zhang, 2015). It is also well documented that MAPKs play key roles in the regulation of the plant’s response to abiotic stresses like drought, salinity, cold, ozone, and heavy metals (Nakagami et al., 2005; Sinha et al., 2011; Moustafa et al., 2014; Pitzschke, 2015; de Zelicourt et al., 2016; Zhao et al., 2017). By contrast, there are only a few reports concerning the involvement of MAPKs in the HT response (Evrard et al., 2013).
HT can up-regulate the expression of StMPK1 in potato (Solanum tuberosum; Blanco et al., 2006) and induce the activities of 46-kD HAMK in tobacco (Nicotiana tabacum) cells (Suri and Dhindsa, 2008) and AtMPK6 in Arabidopsis (Li et al., 2012). So far, however, little is known of the molecular mechanisms underlying the role of MAPK in response to HT. Li et al. (2012) showed that the AtMPK6-mediated activation of γVPE played an important role in HT-induced programmed cell death. It was also reported that AtMPK6 negatively regulates the HT response, and AtMPK6-phosphorylated HSFA2 might participate in the response (Evrard et al., 2013). However, the molecular roles of MAPK in response to HT need to be further elucidated. In the tomato genome, 16 MAPK genes, five MAPKK genes, and 89 MAPKKK genes have been identified (Kong et al., 2012; Wu et al., 2014). Studies of tomato MAPKs have focused mainly on biotic stresses. The tomato SlMPK1, SlMPK2, and SlMPK3 are activated upon stress responses caused by the wound-signaling peptide systemin, oligosaccharide elicitors, and aphids (Li et al., 2006; Kandoth et al., 2007; Stulemeijer et al., 2007). SlMKK2 is a key protein regulating immunity-associated programmed cell death in plants (Melech-Bonfil and Sessa, 2011; Oh et al., 2013). Recent studies showed that the silencing of tomato MPK1/2 by virus-induced gene silencing (VIGS) abolishes plant tolerance to heat, cold, and oxidative stress (Nie et al., 2013; Zhou et al., 2014; Lv et al., 2017). These results suggest an opposite function to AtMPK6, a close homolog to SlMPK1, under HT or cold stress in Arabidopsis (Li et al., 2012, 2017; Evrard et al., 2013; Zhao et al., 2017). Therefore, the function of SlMPK1 in abiotic stress resistance in tomato requires further molecular genetic evidence, especially in relation to HT stress.
Here, an HT-activated 47-kD MBP-phosphorylated protein (p47-MBPK) was identified as SlMPK1 using anion-exchange column purification and tandem mass spectrometry (MS/MS) analyses in tomato leaves. Furthermore, the possible molecular mechanisms underlying SlMPK1-mediated responses to HT were investigated. RNA interference (RNAi) silencing of SlMPK1 enhanced HT tolerance in transgenic tomato seedlings. Interestingly, analysis of the proteome using isobaric tags for relative and absolute quantification (iTRAQ) revealed that several proteins involved in antioxidant defense were significantly up-regulated in RNAi-SlMPK1 plants under HT stress. The RNAi-SlMPK1 plants possessed higher antioxidant defense capacity, while overexpression lines developed opposite phenotypes. In addition, a serine-proline-rich protein homolog (SlSPRH1) was shown to be a substrate of SlMPK1, with Ser-44 being the major phosphorylation site. When Ser-44 was mutated, the phosphorylation of SlSPRH1 by SlMPK1 was almost abolished, showing that Ser-44 phosphorylation is essential for the SlMPK1-mediated antioxidant defense involved in HT. Arabidopsis plants overexpressing SlSPRH1 were more sensitive to HT and possessed a lower antioxidant defense capacity. Our results reveal a potential involvement of SlMPK1 as part of the HT response in tomato and unravel the molecular mechanisms by which SlMPK1 negatively regulates the HT response.
RESULTS
HT-Activated p47-MBPK Is Identified as SlMPK1
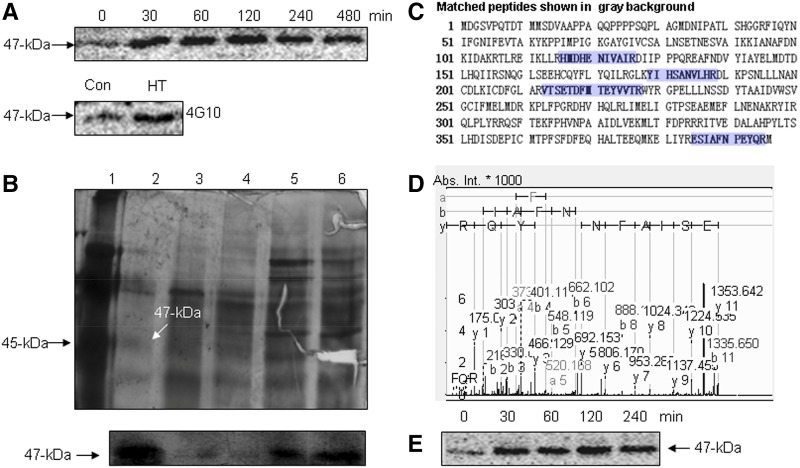
To search for MAPKs involved in the perception of HT signaling, an in-gel kinase activity assay was used. HT caused a significant increase in the activity of p47-MBPK (Fig. 1A). The p47-MBPK also was immunoprecipitated with the anti-pTyr monoclonal antibody 4G10, which has been widely used to demonstrate Tyr phosphorylation of MAPKs, an important characteristic of MAPKs. The results suggest that p47-MBPK is a MAPK-like protein. Katou et al. (2005) purified an elicitor-induced 51-kD MAPK, designated as StMPK1. We previously purified an abscisic acid (ABA)-activated p46-MAPK, identified as ZmMPK5 (Ding et al., 2009). To reveal the identity and function of the p47-MBPK, we first purified the kinase from HT-treated tomato leaves using different anion-exchange columns (Supplemental Fig. S1). The peak protein levels and kinase activities were confirmed in each step by silver staining and an in-gel kinase assay, respectively, revealing a 47-kD kinase in the poly-l-Lys-agarose column fractions with one band (Fig. 1B).
Figure 1.
Identification of HT-activated SlMPK1. A, HT-activated MBP kinase. The time courses of the induction of MBP kinase activities by HT (top) and the Tyr phosphorylation of HT-activated MBP kinase (bottom) are shown. B, Analysis of purification pools by gel electrophoresis (top) and in-gel kinase assay (bottom). Lane 1, Marker standards; lane 2, pooled fractions from the poly-l-Lys-agarose column; lane 3, pooled fractions from the Mono QTM 5/50 GL column; lane 4, pooled fractions from the Q-Sepharose HP column; lane 5, pooled fractions from the Phenyl-Sepharose FF column; lane 6, pooled fractions from the Q-Sepharose FF column. Proteins from different stages of purification were resolved on a 12% polyacrylamide gel containing SDS and stained with silver (top). The fractions were loaded onto a 12% SDS-polyacrylamide gel embedded with MBP, and an in-gel kinase assay was performed (bottom). C, Identification of SlMPK1 by MS/MS. A 47-kD band from the SDS-PAGE gel was cut, and the protein in-gel fragment was digested with trypsin followed by MALDI-TOF/TOF-MS/MS analyses. Proteins were identified by Mascot database searches. Matched peptides are shown in blue. D, MS/MS spectrum of the selected peptide (m/z 1,777.7845). Database matching indicated that this is a fragment (ESIAFNPEYQR) of SlMPK1. Abs. Int., Absolute intensity of the y axis. E, Immunoprecipitation kinase analysis of HT-induced SlMPK1. Total proteins were extracted from the HT-treated leaves at the indicated times, and SlMPK1 activity was measured.
To identify the p47-MBPK, the partly purified protein was silver stained on an SDS-PAGE gel and the band corresponding to 47 kD was in-gel digested with trypsin, followed by MALDI-TOF/TOF-MS/MS analysis. The search yielded a top score of 341 for Q7Y1Y6, SlMPK1 with four matched peptides, using the Mascot search engine (Fig. 1C). Furthermore, the selected tryptic peptide (m/z 1,777.7845) sequenced by MS/MS revealed an amino acid sequence of ESIAFNPEYQR, corresponding to the specific residues 385 to 395 of SlMPK1 (Fig. 1D). To further confirm the identity of p47-MBPK correlated with SlMPK1, a C-terminal-specific polyclonal antibody for SlMPK1 was prepared. The molecular mass of the immunoprecipitated protein kinase from HT-treated tomato leaves was 47 kD, and this protein was induced by HT stress (Fig. 1E). These results indicate that the HT-activated p47-MBPK is SlMPK1.
Suppression of SlMPK1 Increased HT Tolerance in Transgenic Plants
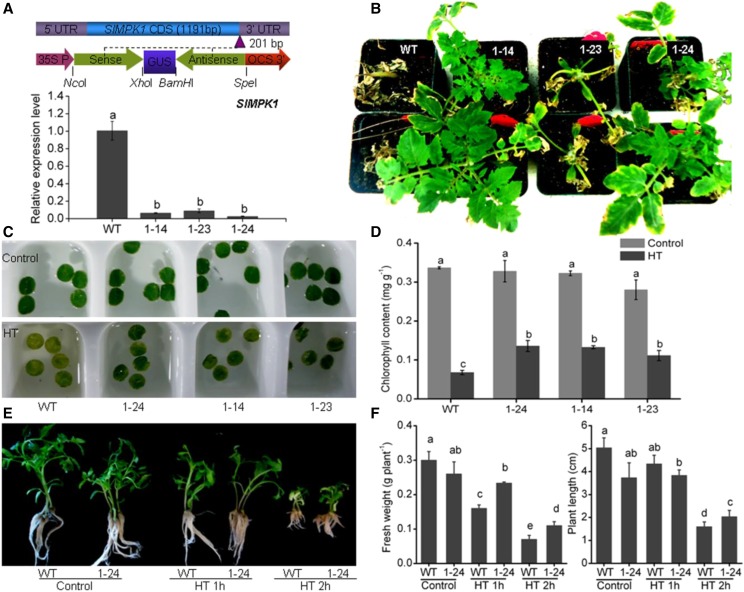
SlMPK1 (Solyc12g019460), homologous to AtMPK6, ZmMPK5, and OsMPK6, was first isolated from tomato seedlings by Holley et al. (2003). SlMPK1 can be activated by systemin and several oligosaccharide elicitors (Higgins et al., 2007). In this study, SlMPK1 activity was induced in HT-treated tomato leaves (Fig. 1, A and E). Given this result, we speculated that SlMPK1 might be involved in the regulation of the HT response. To test this hypothesis, RNAi was used to suppress the expression of SlMPK1 in transgenic tomato. Three independent RNAi lines (1-14, 1-23, and 1-24) were selected for HT tolerance testing at the seedling stage (Fig. 2A). The inhibition rate of SlMPK1 expression by RNAi was over 90%, but the expression of the homologous gene SlMPK2 was not changed (Supplemental Fig. S2B), indicating that SlMPK1 expression was suppressed specifically by RNAi in the transgenic plants. Under normal conditions, the growth of three RNAi lines was similar to that of wild-type plants (Supplemental Fig. S2A). When seedlings of the wild type and RNAi-SlMPK1 lines grown in soil were treated with HT, the RNAi lines showed more tolerance to HT (Fig. 2B). Consistent with these results, the chlorophyll content of leaf discs from HT-treated plants was lower in the wild type than in RNAi-SlMPK1 lines (Fig. 2, C and D). Similar results of growth inhibition also were observed in sterile seedlings of RNAi line 1-24 grown in Murashige and Skoog medium (Fig. 2, E and F). Taken together, these results demonstrate that SlMPK1 has a negative role in HT tolerance in tomato.
Figure 2.
Effects of SlMPK1 suppression on HT tolerance of tomato seedlings. A, Schematic diagram of the RNAi-SlMPK1 construct and the expression level of SlMPK1 in RNAi-SlMPK1 plants. The closed green arrows represent the partial sequence of SlMPK1 (top). UTR, Untranslated regions; CDS, coding sequence; 35S P, 35S promoter; OCS, OCS terminators. The restriction sites used are shown. The graph at bottom shows reverse transcription-quantitative PCR (RT-qPCR) analysis of the transcript levels of SlMPK1 of partial transgenic lines. The numbers indicate independent RNAi transgenic plants. B, Phenotypes of RNAi-SlMPK1 lines in nutritional bowl feeding matrix soils under HT. Seedlings of the wild type (WT) and RNAi-SlMPK1 lines 1-14, 1-23, and 1-24 at the five-leaf stage were subjected to 38°C/28°C for 3 d and then recovered at 25°C/20°C for 10 d. C, Phenotypes of leaf discs in RNAi-SlMPK1 lines under HT. The leaf discs from fourth fully expanded leaves were incubated at 45°C for 2 h, followed by recovery at 25°C/20°C for 2 d. D, Chlorophyll content of the leaf discs of RNAi-SlMPK1 transgenic plants under HT shown in C. Error bars represent sd values of three replicates. E, Phenotypes of RNAi-SlMPK1 line 1-24 in Murashige and Skoog medium under HT. F, Fresh weight and plant length of the wild-type and 1-24 plants shown in E. Statistical differences among the samples shown in A, D, and F are labeled with different letters according to the lsd test (P < 0.05, one-way ANOVA).
The expression of heat shock protein (HSP) and heat shock factor (HSF) genes serves as a master regulator of heat stress responses (HSR). To determine the relationship between SlMPK1 and HSR, we measured the transcript levels of HSFA2 and HSP101, the major heat-inducible HSF and HSP genes (Wu et al., 2017). Our results show that the expression of both genes did not change significantly in SlMPK1 RNAi lines when compared with the wild type at normal growth temperature or HT stress (Supplemental Fig. S2C).
Analysis of SlMPK1-Mediated Proteins under HT Conditions with the iTRAQ Proteomic Assay
To elucidate how SlMPK1 coordinately regulates HT tolerance, the proteins involved in the SlMPK1-mediated HT-responsive pathway were analyzed using iTRAQ. The RNAi line 1-24 was used for the proteomic assay. There was high reproducibility between the iTRAQ analyses according to the coefficient of variation (Supplemental Fig. S3, A and B). Data analysis detected a total of 2,258 and 2,433 proteins in the first and second iTRAQ analyses, respectively (Supplemental Fig. S3C). There were 1,787 proteins overlapping between two 8-plex iTRAQ analyses, and only proteins with consistent expression (fold change ratio > 1.2 or < 0.67, P < 0.05) in both 8-plex iTRAQ analyses were demonstrated to have statistical significance. Several HT-related proteins, such as HSP22.0, HSP101, ATHSP90.1, and HSP70b, were highly induced in both 8-plex iTRAQ analyses, suggesting that the HT treatment was effective (Supplemental Table S1). The results showed that the expression of only a small number of proteins changed in RNAi line 1-24 plants compared with the wild type under normal conditions (Supplemental Table S2). For example, PPC1 is a phosphoenolpyruvate carboxylase and is part of the adaptation of the plant to salt and drought (Sánchez et al., 2006).
Meanwhile, the expression of 48 proteins was significantly altered in the RNAi-SlMPK1 line 1-24 versus wild-type plants under HT conditions (Supplemental Table S3). An overview of functional protein networks affected in the RNAi-SlMPK1 line 1-24 under HT using STRING 9.0 is shown in Supplemental Figure S4. Molecular function enrichment analysis showed that 19 proteins have catalytic activities, including nine proteins with oxidoreductase activity (NRX1, THI1, GLU1, MDAR1, IMD2, SDR5, TKL1, HDS, and CRL1; Supplemental Table S4). In addition, CAT2 and AOR also have oxidoreductase activities that were not listed in the PANTHER version 12.0 of the tomato reference genome. Biological process enrichment categorizes these proteins into different metabolic processes, including protein folding (e.g. CHAPERONIN20 [CPN20]), amino acid biosynthetic process (e.g. MS1), lipid metabolic process (e.g. ANNAT4), and translation (e.g. emb2394). Interestingly, if we used Bonferroni correction for functional enrichment, oxidoreductase activity (GO:0016491) was the only significantly enriched molecular function (Supplemental Table S5), indicating that oxidoreductase activity-mediated processes are one of the main processes in the SlMPK1-mediated response to HT. Twenty proteins having significant consistency of expression in at least three of four biological replicates are shown in Table I, including seven oxidoreductases (NRX1, THI1, GLU1, MDAR1, IMD2, CAT2, and AOR).
Table I. Proteins abundant in the leaves of RNAi-SlMPK1 tomato line 1-24 compared with the wild type under HT conditions using iTRAQ.
| UniProt Identifier | Gene Identifiera | Nameb | Ic | II | III | IV | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 116:114d | Pd | 121:118 | P | 116:114 | P | 121:118 | P | |||
| Q9M5A8 | Solyc07g042250 | CPN20 | 2.2284 | 0.00445 | 1.6144 | 0.04330 | 1.7567 | 0.08371 | 2.3335 | 0.02852 |
| C5IU71 | Solyc05g052600 | SBPASE | 2.2080 | 4.37E-05 | 1.3183 | 0.00801 | 1.6749 | 0.51661 | 5.8076 | 7.05E-09 |
| K4BAE6 | Solyc02g082760 | CAT2 | 1.9770 | 0.00157 | 3.1333 | 0.00022 | 1.3062 | 0.64461 | 1.8707 | 0.00136 |
| K4B1S1 | Solyc01g103450 | CPHSC70-2 | 1.9767 | 3.24E-05 | 1.8880 | 0.01473 | 0.7379 | 0.09048 | 4.1687 | 6.57E-08 |
| K4DAD5 | Solyc11g069790 | CPN60A | 1.8030 | 0.00306 | 2.7797 | 0.00334 | 1.5754 | 0.02498 | 1.8880 | 0.00038 |
| K4CUF4 | Solyc09g065270 | RRF | 1.7539 | 0.00311 | 1.5276 | 0.02924 | 1.7311 | 0.50232 | 1.9770 | 0.04506 |
| K4BW77 | Solyc05g005460 | NRX1 | 1.6444 | 0.03503 | 1.5704 | 0.03808 | 1.2022 | 0.04712 | 1.3062 | 0.160419 |
| K4CH99 | Solyc07g064160 | THI1 | 1.6293 | 0.01993 | 4.6132 | 0.00785 | 0.9376 | 0.20589 | 1.9589 | 0.00549 |
| K4BHA1 | Solyc03g063560 | GLU1 | 1.6144 | 6.86E-05 | 1.2942 | 0.04700 | 1.2803 | 0.95947 | 1.5704 | 1.68E-06 |
| K4CEJ1 | Solyc07g043320 | 1.6144 | 0.00725 | 1.0186 | 0.655478 | 2.3768 | 0.00037 | 1.2474 | 0.02173 | |
| Q672Q6 | Solyc02g079950 | PSBQ | 1.5276 | 0.00833 | 1.4723 | 0.00914 | 1.3908 | 0.06042 | 2.8054 | 0.00419 |
| K4AYG3 | Solyc01g087730 | PRPL1 | 1.4860 | 0.01086 | 1.7865 | 0.00430 | 0.50583 | 0.01430 | 2.0512 | 0.02356 |
| K4BW79 | Solyc05g005480 | AOR | 1.4859 | 0.00513 | 1.5996 | 0.083761 | 2.2909 | 0.00182 | 2.2699 | 0.03217 |
| Q5NE20 | Solyc02g086820 | CA1 | 1.4454 | 0.01144 | 1.8197 | 0.03724 | 0.3162 | 0.01374 | 2.5586 | 0.00038 |
| K4B1F9 | Solyc01g102310 | 1.4191 | 0.00155 | 1.1482 | 0.263537 | 1.6904 | 0.01809 | 1.5849 | 0.01383 | |
| K4CQW8 | Solyc09g009390 | MDAR1 | 1.3062 | 0.03410 | 1.0765 | 0.993515 | 2.0137 | 0.03218 | 1.3727 | 0.01575 |
| K4AV63 | Solyc01g028810 | CPN60B2 | 1.2942 | 0.00080 | 2.6062 | 1.04E-05 | 0.7178 | 0.46698 | 2.2080 | 0.00105 |
| K4BX77 | Solyc05g009030 | IMD2 | 1.2706 | 0.02061 | 1.6293 | 0.03579 | 1.5560 | 0.99387 | 2.0324 | 0.02828 |
| K4BMN4 | Solyc03g120850 | CPN60B1 | 1.2482 | 0.03575 | 2.2284 | 0.00645 | 1.0765 | 0.55860 | 3.0479 | 0.00764 |
| K4B0D9 | Solyc01g097520 | ANNAT4 | 0.2089 | 0.00403 | 0.7516 | 0.488531 | 0.6546 | 0.04915 | 0.2378 | 0.00022 |
The UniProt identifier entries were obtained via UniProt (http://www.uniprot.org), and the gene identifiers were translated using UniProt’s ID Mapping. bThe name of the detected protein. cI and II indicate two biological replicates using an iTRAQ 8-plex, and III and IV indicate another two replicates in another iTRAQ 8-plex. dThese values were calculated as the ratio of 116 and 121 label to 114 and 118 label, respectively. 116 and 121 represent RNAi-SlMPK1 line 1-24 under HT, and 114 and 118 represent the wild type. Only proteins identified with more than one peptide and quantitation results with P < 0.05 were considered to be statistically different from unity. The proteins were considered to be differentially expressed if their iTRAQ ratios were greater than 1.2 or less than 0.67. The raw data are shown in Supplemental Table S3. Proteins with significantly different protein ratios (m/w) in at least three of four biological replicates are shown here. Significant changes are indicated by boldface type.
The expression of HT-related protein-coding genes such as CPN20 and CPN60A1 was highly up-regulated in HT-treated wild-type plants, suggesting that the HT treatment was effective. The expression pattern of some protein-coding genes, such as CAT2, CPN20, CPN60A1, CPN60B1, and CPN60B2, was consistent with the variations in protein abundance (Supplemental Fig. S5). Although the expression of the HSR master regulator genes HSFA2 and HSP101 did not change, the expression of several heat-inducible genes, such as CPN60A, CPN20, and CPHSC70-2, increased in the SlMPK1 RNAi lines 1-14 and 1-24 under HT stress (Supplemental Fig. S5).
Up-Regulation of Antioxidant Defense in the SlMPK1-Suppressed Lines under HT
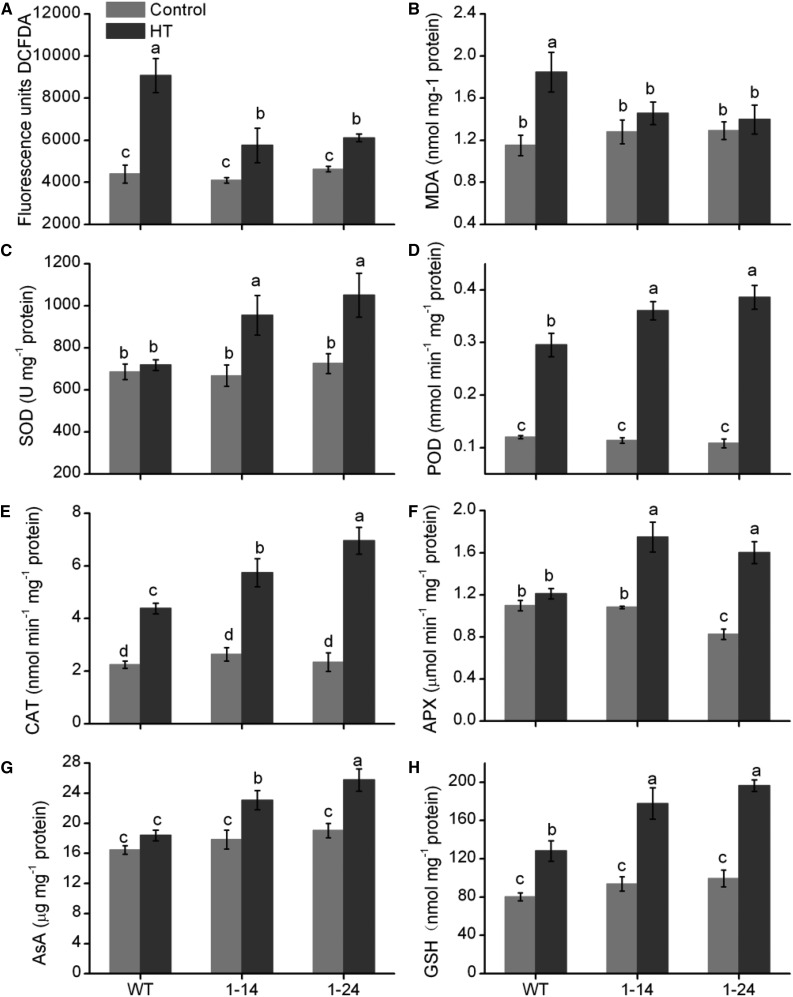
The iTRAQ analysis indicated that the abundance of many proteins involved in cell redox homeostasis increases in the RNAi-SlMPK1 line 1-24 under HT conditions. Plant catalases are part of the major reactive oxygen species (ROS) scavenging network. For example, a decrease in the activity of the catalase CAT2 in the Arabidopsis cat2-1 mutant correlates with a greater accumulation of H2O2 and higher oxidative damage in leaves (Bueso et al., 2007). MDAR1 is a monodehydroascorbate reductase crucial for maintaining a reduced pool of ascorbate (AsA), a major antioxidant and radical scavenger in plants (Eltayeb et al., 2007). The increased oxidoreductase levels in the RNAi-SlMPK1 plants under HT should be reflected in increased antioxidant capacity and decreased levels of ROS. Under HT conditions, the level of H2O2 in RNAi-SlMPK1 lines was lower than in wild-type plants (Fig. 3A). Lipid peroxidation was estimated by determining malondialdehyde (MDA) content and showed results similar to those of H2O2 (Fig. 3B).
Figure 3.
Response of antioxidant defense to HT in RNAi-SlMPK1 plants. H2O2 accumulation (A), the accumulation of the membrane lipid peroxidation product MDA (B), the activities of SOD (C), POD (D), CAT (E), and APX (F), and the content of the nonenzymatic antioxidant AsA (G) and GSH (H) are shown in the wild type (WT) and RNAi-SlMPK1 line 1-24 under control (25°C) or HT (42°C) conditions for 4 h. Error bars represent sd values of three replicates. Statistical differences among the samples are labeled with different letters according to the lsd test (P < 0.05, one-way ANOVA). DCFDA, 2′,7′-Dichlorofluorescin diacetate.
The generation of ROS is limited or scavenged by antioxidant enzymes like superoxide dismutase (SOD), guaiacol peroxidase (POD), CAT, and ascorbate peroxidase (APX) as well as by nonenzymatic antioxidants such as AsA and glutathione (GSH; Mittler, 2002). There were significant differences in the activity of APX and the accumulation of AsA and GSH between RNAi-SlMPK1 lines and the wild type (Fig. 3). Concomitantly with the increase in the levels of proteins like CAT2 and MDAR1, our physiological analysis revealed a significant increase in the activities of enzymes involved in AsA-GSH metabolism and regulation in the RNAi-SlMPK1 plants. Therefore, the enhanced HT tolerance observed in RNAi-SlMPK1 plants might be related to the activation of the antioxidant defense system, which seems to be the result of de novo synthesis and the activation of enzymes.
Overexpression of SlMPK1 Decreased HT Tolerance in Transgenic Plants
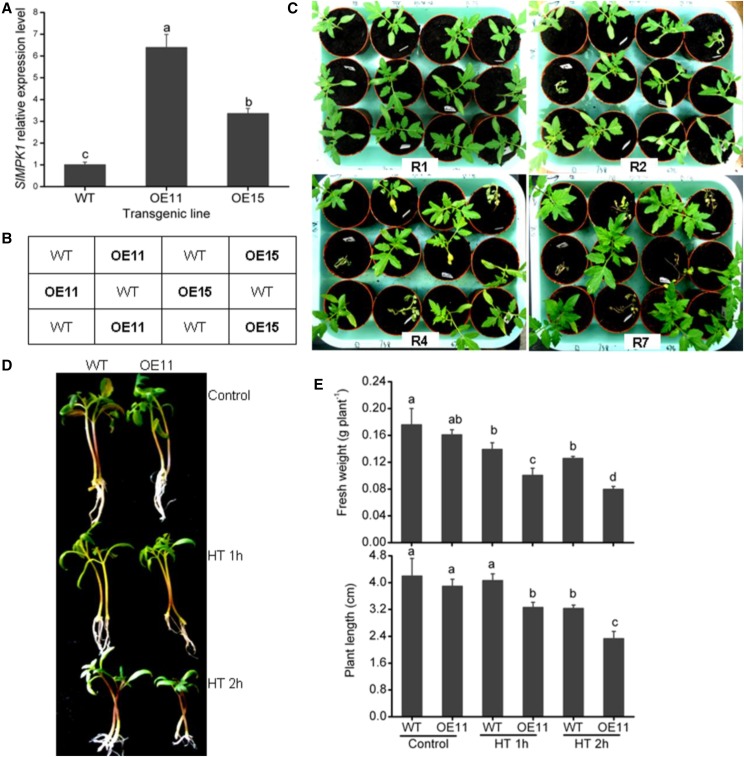
Genetic and physiological analyses of the RNAi-SlMPK1 line suggest that SlMPK1 plays a negative role in the regulation of HT resistance in tomato. We predicted that overexpression of SlMPK1 might result in decreased HT tolerance. To test this hypothesis, transgenic plants were generated by overexpressing the full-length SlMPK1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Two SlMPK1-overexpressing (OE) lines showed significantly increased SlMPK1 expression (Fig. 4A), indicating that SlMPK1 was overexpressed successfully. We first analyzed the tolerance of transgenic tomato to HT stress and found that SlMPK1-OE lines were nearly dead after HT stress, while all the wild-type plants survived (Fig. 4C). Furthermore, the SlMPK1-OE lines exhibited more severe growth inhibition than wild-type plants in Murashige and Skoog medium under HT conditions (Fig. 4D). Statistical analysis showed that the SlMPK1-OE transgenic plants suffered significantly more suppression of shoot growth than wild-type plants after 2 h of HT stress (Fig. 4E). These results suggest that overexpression of SlMPK1 has a negative effect on HT tolerance. We also determined H2O2 level, MDA content, and two enzyme activities in SlMPK1-OE transgenic plants under HT conditions. Under HT conditions, the accumulation of H2O2 and MDA in SlMPK1-OE transgenic plants was higher than in wild-type plants (Supplemental Fig. S6). By contrast, the activities of antioxidant enzymes in SlMPK1-OE transgenic plants were lower than in the wild-type plants.
Figure 4.
Effects of SlMPK1 overexpression on the HT tolerance of tomato seedlings. A, Expression levels of SlMPK1 in SlMPK1-OE plants. RT-qPCR analysis of transcript abundance of SlMPK1 in two independent OE transgenic plants is shown. B, Chart of the material combination of plants in C. C, Phenotypes of SlMPK1-OE lines in nutritional bowl feeding matrix soils under HT. Seedlings of the wild type (WT) and SlMPK1-OE lines OE11 and OE15 at the three-leaf stage were subjected to 38°C/28°C for 1 d and then recovery at 25°C/20°C for 1 d (R1), 2 d (R2), 4 d (R4), and 7 d (R7). D, Phenotypes of SlMPK1-OE transgenic line OE11 under HT. Detached plants were planted in a new Murashige and Skoog medium and grown at 25°C/20°C for 3 d and then incubated at 45°C for 1 or 2 h, followed by recovery at 25°C/20°C for 7 d. E, Fresh weight and plant length of the wild type and SlMPK1-OE transgenic line OE11 in Murashige and Skoog medium under HT as shown in D. Statistical differences among the samples are labeled with different letters according to the lsd test (P < 0.05, one-way ANOVA).
SlMPK1 Interacts with SlSPRH1
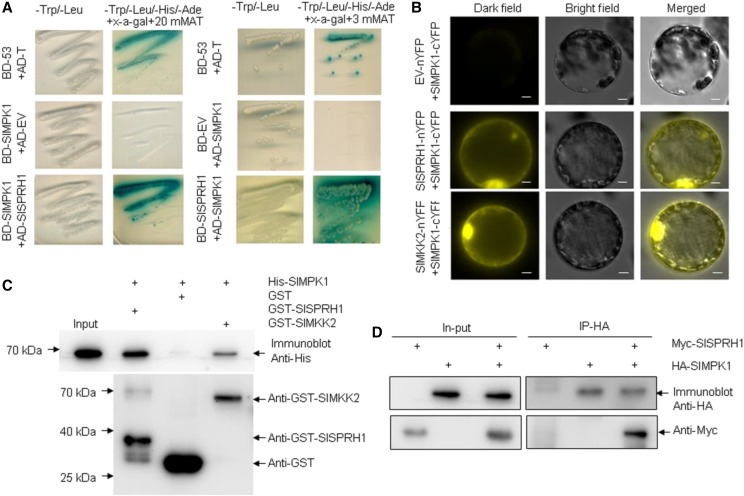
MAPKs play important roles in various cellular processes by binding to many types of proteins, such as substrates, other protein kinases, protein phosphatases, cytoskeletal proteins, and transcription factors (Tanoue and Nishida, 2003). To identify downstream targets of SlMPK1, we screened a tomato cDNA library using SlMPK1 as the bait in a yeast two-hybrid (Y2H) assay. The screen identified Solyc06g053700 as a candidate SlMPK1-interacting protein. Solyc06g053700 is a Ser-Pro-rich protein homolog of unknown function, homologous to Arabidopsis At1g04330 (fragment). We tentatively named this protein SlSPRH1. Cotransformation of SlSPRH1 with SlMPK1 confirmed their interaction in yeast (Fig. 5A). SlMKK2 has been reported to interact with SlMPK1 (Kandoth et al., 2007) and was used as a positive control in this study. The results showed that SlMPK1 interacts with SlSPRH1 in a bimolecular fluorescence complementation (BiFC) assay (Fig. 5B). To provide further evidence for such interaction, we analyzed the interaction in vitro using pull-down assays. Using the immobilization of recombinant SlSPRH1 fusion protein on GST Sepharose beads, we found that GST-SlSPRH1, but not GST alone, was able to pull down His-SlMPK1 in vitro (Fig. 5C). Furthermore, the SlMPK1-SlSPRH1 interaction was confirmed by coimmunoprecipitation (Co-IP) experiments in vivo on Nicotiana benthamiana leaves. Immunoprecipitates of transiently expressed SlMPK1-HA in leaves transformed with SlMPK1-HA and SlSPRH1-myc were found to contain SlSPRH1 (Fig. 5D). Taken together, these data show that SlMPK1 can interact with SlSPRH1 in vitro and in vivo.
Figure 5.
Interactions between SlMPK1 and SlSPRH1. A, Y2H assay of interactions between SlMPK1 and SlSPRH1. SlMPK1 (as bait) was cloned into pGBKT7 (BD) and SlSPRH1 (as prey) was cloned into pGADT7 (AD). AD-T and BD-p53 was used as a positive control, and AD-empty vector (EV) and SlMPK1-BD was used as a negative control. Simultaneously, SlSPRH1 (as bait) was cloned into pGBKT7 (BD) and SlMPK1 (as prey) was cloned into the pGADT7 (AD). AD-SlMPK1 and BD-EV was used as a negative control. B, BiFC assay for detecting molecular interactions between SlMPK1 and proteins (SlMKK2 or SlSPRH1) transiently coexpressed in tobacco leaf protoplasts. SlMPK1 was fused with the C terminus of YFP, and SlMKK2 and SlSPRH1 were fused with the N terminus of YFP. The images were obtained from the YFP channel, the differential interference contrast channel, and a merged image of the two channels. The positive control was SlMKK2-nYFP/SlMPK1-cYFP, and the negative control was EV-nYFP and SlMPK1-cYFP. Bars = 50 μm. C, SlMPK1 interacts physically with SlSPRH1. His-tagged SlMPK1 was incubated with immobilized GST or GST-tagged SlSPRH1. Beads were washed, fractionated by 12% (w/v) SDS-PAGE, and subjected to immunoblot analysis using an antibody against His (top) or GST (bottom). Immobilized GST was used as a negative control, and GST-tagged SlMKK2DD was used as a positive control. D, In vivo Co-IP assay of the SlMPK1 and SlSPRH1 interaction in tobacco leaves. Crude lysates precleared by protein A-Sepharose beads (In-put) were immunoprecipitated (IP) with anti-HA antibody and then detected with anti-HA and anti-Myc antibodies for SlMPK1-HA and SlSPRH1-Myc, respectively.
SlMPK1 Phosphorylates SlSPRH1
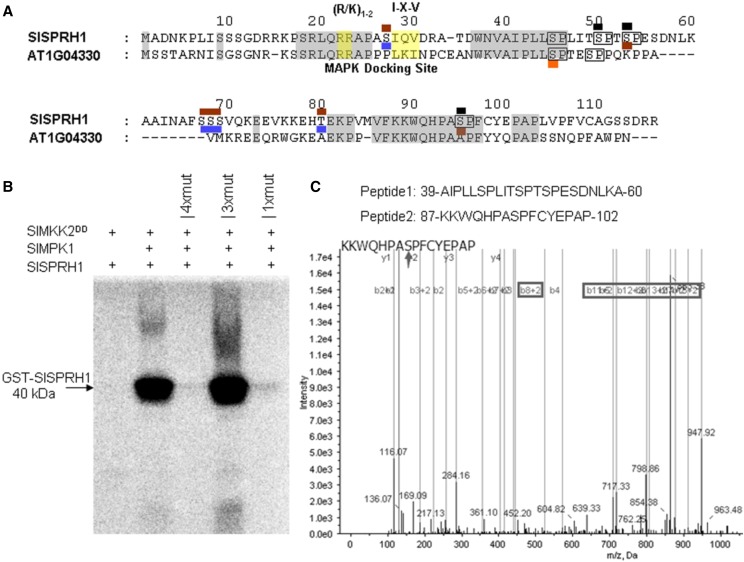
All MAPK-interacting proteins use a signature motif known as the D motif [-(R/K)1-2-(X)2-6-L/I/V-X-L/I/V-] to interact with MAPKs (Tanoue et al., 2000). A search shows one D motif (RRAPASIQV) located in the N terminus of SlSPRH1 (Fig. 6A). To identify the SlMPK1 phosphorylation sites on SlSPRH1, recombinant SlSPRH1 was phosphorylated in vitro by SlMPK1. MS analysis identified several phosphopeptides (Fig. 6; Supplemental Fig. S7). Ser-52 and Ser-94 were found to be phosphorylated exclusively in the SlMPK1-treated SlSPRH1. Other sites, like Ser-27 and Thr-79, were found to be phosphorylated in both the control and the kinase-treated SlSPRH1, which implies that Ser-27 and Thr-79 phosphorylation results from the activity of an Escherichia coli kinase (Li et al., 2017). However, the two phosphorylated sites Ser-44 and Ser-49 in SlSPRH1 were not identified by MS. To further confirm these SlMPK1-phosphorylated sites, two peptides were synthesized and tested in vitro as substrates for SlMPK1. MS analysis of the peptides showed that Ser-49, Ser-52, and Ser-94 were phosphorylated exclusively by SlMPK1 (Fig. 6A; Supplemental Fig. S7). While the Ser-49 phosphorylation site was found only in the peptide, the Ser-44 phosphorylation site was not detected by MS.
Figure 6.
Phosphorylation of SlSPRH1 by SlMPK1. A, Identification of phosphorylation sites of SlMPK1-targeted SlSPRH1 using liquid chromatography (LC)-MS/MS analysis. The amino acid sequence of SlSPRH1 and Arabidopsis homologous At1g04330 phosphorylation sites are marked. The potential MAPK target sites (S/T-P) are boxed in the SlSPRH1 sequence, where S/T-P sites detected as being phosphorylated are marked in brown in the SlMPK1-treated SlSPRH1 protein and blue in the background SlSPRH1 protein and those detected as being phosphorylated by AtMPK6 are shown in orange (Palm-Forster et al., 2012). Partial MS/MS spectra are shown in Supplemental Figure S7. B, In vitro phosphorylation of purified wild-type and mutant SlSPRH1 substrate by SlMPK1. His-tagged SlMPK1 (His-SlMPK1) and GST-tagged SlSPRH1 (GST-SlSPRH1) were used in phosphorylation reactions with 32P labeling. 4xmut indicates all identified Ser residues, Ser-44, Ser-49, Ser-52, and Ser-94, changed to Ala, 3xmut indicates all Ser residues changed to Ala except Ser-44, and 1xmut indicates only Ser-44 changed to Ala. The constitutive-active form of SlMKK2DD was used to activate SlMPK1 and as a negative control for unspecific phosphorylation of SlSPRH1 protein. Phosphorylated SlSPRH1 was visualized by a Typhoon phosphor imager after gel electrophoresis. The experiment was repeated at least three times with similar results. C, MS analysis of synthetic peptides phosphorylated in vitro by SlMPK1. Synthetic peptides were incubated with His-SlMPK1 and GST-SlMKK2DD in a kinase reaction, which was then centrifugally filtered through a Millipore 5-kD cutoff filter. The filtrate was determined by LC-MS/MS. The arrow indicates the phosphorylation site of the peptide. The phosphorylated sites are marked in black in A.
To further characterize the phosphorylation of SlSPRH1, His-tagged SlMPK1 and GST-tagged SlSPRH1 were purified and used in kinase assays in vitro. SlSPRH1 was phosphorylated in the presence of recombinant SlMPK1 activated by SlMPKK2DD (Fig. 6B). Sequence analysis indicated that there are four potential phosphorylation sites (with a Ser or Thr residue followed by a Pro residue, S/T-P motif) in SlMPK1 (Ser-44, Ser-49, Ser-52, and Ser-94; Fig. 6A). The phosphorylation sites Ser-49, Ser-52, and Ser-94 were confirmed by MS. Previously, Ser-44 in At1g04330, homologous to Ser-44 in SlSPRH1, was examined as the major target of AtMPK6 (Palm-Forster et al., 2012). To identify the phosphosites of SlSPRH1, Ala exchange of the potential phosphosites was performed. Changes of all four corresponding residues to Ala in SlSPRH1 (4хmut) almost completely abolished SlSPRH1 phosphorylation (Fig. 6B). We also mutated SlSPRH1 at the three predicted phosphosites except Ser-44 (3xmut) and found no effect on the phosphorylation of SlSPRH1. By contrast, when only the Ser-44 site was mutated (1хmut), phosphorylation of SlSPRH1 was almost abolished (Fig. 6B). The fact that the phosphosite Ser-44 was not detected by MS might be a limitation of the MS technique. These results indicate that SlSPRH1 is a substrate of SlMPK1, that Ser-44, Ser-49, Ser-52, and Ser-94 are phosphorylation sites, and that Ser-44 is a phosphosite conserved between the two homologous proteins (Supplemental Fig. S8) and is the most preferred site in SlSPRH1 for phosphorylation by SlMPK1.
Overexpression of SlSPRH1 Decreased HT Tolerance in Transgenic Plants
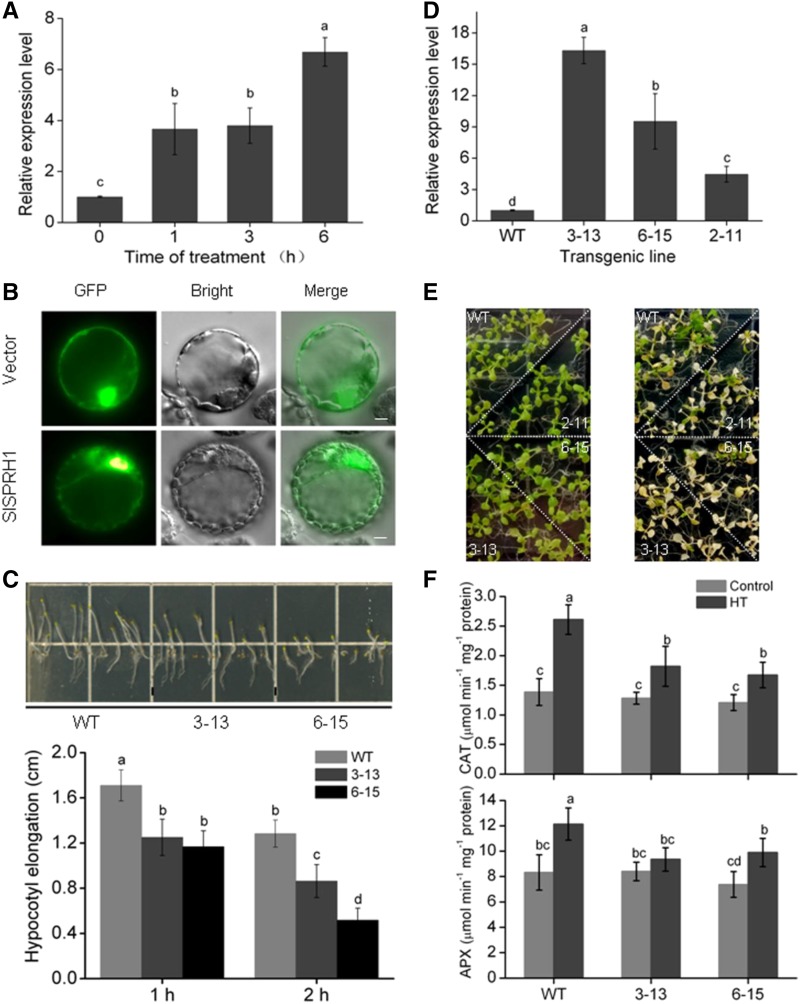
To investigate the role of SlSPRH1 in HT stress, we first measured the expression of SlSPRH1 under HT stress. HT significantly increased the expression of SlSPRH1 in tomato leaves (Fig. 7A). At the same time, SlMPK1 expression also was induced by HT (Supplemental Fig. S10). The BiFC results showed that the interaction between SlMPK1 and SlSPRH1 was observed in both nucleus and cytoplasm (Fig. 5B). To further confirm the localization of SlSPRH1, a 35S-SlMPK1-GFP construct and a 35S-GFP control were produced, transformed into N. benthamiana (Fig. 7B), and visualized by fluorescence microscopy. GFP fluorescence from SlSPRH1-GFP was observed in the nucleus and cytoplasm but predominantly in the nucleus. To further explore the molecular function of SlSPRH1, full-length SlSPRH1 was heterologously expressed in Arabidopsis plants. Hypocotyl elongation is known to be inhibited by HT and has been used as a parameter for HT tolerance (Queitsch et al., 2000). The hypocotyl length and growth of HT-stressed, SlSPRH1-expressing plants were reduced relative to the wild type when plants were grown in darkness (Fig. 7C). Transgenic Arabidopsis seedlings grown on solid agar Murashige and Skoog medium plates treated with 42°C for 1.5 h were almost completely dead after recovery for 10 d (Fig. 7E). These results suggest that the heterologous expression of SlSPRH1 has a negative effect on HT tolerance in Arabidopsis.
Figure 7.
SlSPRH1 negatively regulates Arabidopsis tolerance to HT stress. A, SlSPRH1 expression is induced by HT. RT-qPCR analyses of transcript abundance of SlSPRH1 is shown in leaves of wild-type tomato plants during HT (42°C) for 0, 1, 3, and 6 h. B, Subcellular localization of SlSPRH1. The SlSPRH1-GFP protein and GFP, driven by CaMV 35S, were transformed separately into tobacco protoplast and visualized by fluorescence microscopy. Images were taken in representative cells expressing GFP (top) or SlSPRH1-GFP fusion protein (bottom) under bright field (middle) or dark field (left). The merged images are shown at right. Bars = 50 μm. C, Hypocotyl length of wild-type (WT) and SlSPRH1-expressing Arabidopsis lines in one-half-strength Murashige and Skoog medium under HT stress. After germination for 3 d, the plates covered with foil were subjected to HT treatment at 45°C for 1 and 2 h, followed by vertical culturing in the dark for 6 d. D, Expression of SlSPRH1 in SlSPRH1-expressing lines. RT-qPCR analyses of the transcript abundance of SlSPRH1 is shown in the three independent transgenic plants. ACTIN was used as an internal control. E, Phenotypes of SlSPRH1-expressing transgenic lines 3-13, 6-15, and 2-11 under HT stress. Ten-day-old Arabidopsis seedlings in one-half-strength Murashige and Skoog medium (left) were subjected to HT treatment at 42°C for 1.5 h, followed by recovery for 10 d (right). F, Response of antioxidant enzymes to HT in the SlSPRH1-expressing plants. Two-week-old wild-type and transgenic line 3-13 and 6-15 plants were treated with HT for 3 h. The activities of CAT and APX were determined. Error bars represent sd values of three replicates. Statistical differences among the samples are labeled with different letters according to the lsd test (P < 0.05, one-way ANOVA).
The decreased HT tolerance in SlMPK1-OE tomato plants might be related to a down-regulation of the antioxidant defense system. To determine whether the overexpression of SlSPRH1 has a similar effect, the enzyme activities were measured in SlSPRH1-expressing Arabidopsis plants under HT conditions. The activities of APX, CAT, and SOD were lower in SlMPK1-expressing transgenic plants than in wild-type plants, while the MDA level was higher under HT conditions (Fig. 7F; Supplemental Fig. S9). These results suggest that the negative effect of SlSPRH1 heterologous expression on HT tolerance might be related to a decrease in antioxidant defenses.
Phosphorylation of Ser-44 in SlSPRH1 Is Involved in SlMPK1-Mediated Antioxidant Defense under HT
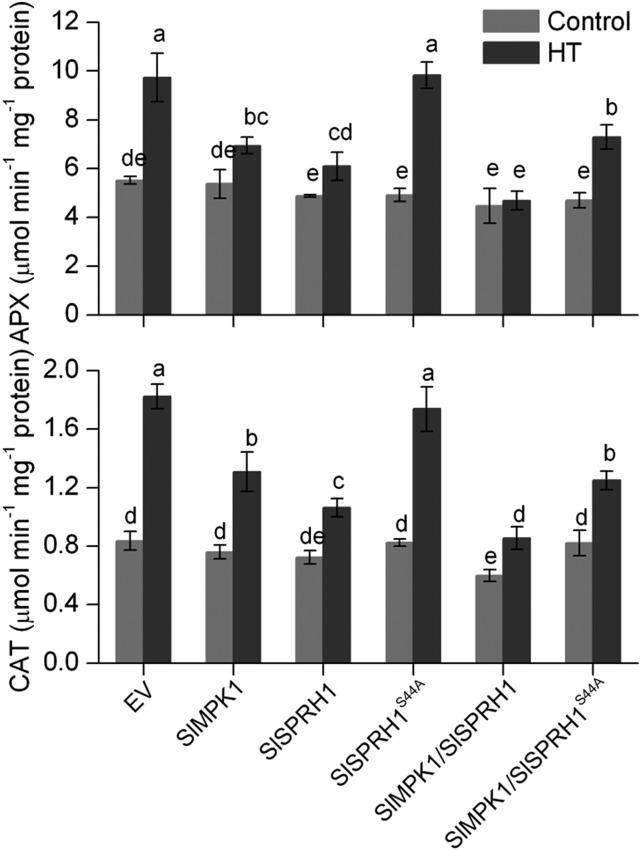
To further determine whether SlSPRH1 acts downstream of SlMPK1 in the regulation of antioxidant defense under HT, the SlSPRH1 mutant (SlSPRH1S44A) alone or SlMPK1 and SlSPRH1S44A simultaneously were transiently expressed in Arabidopsis mesophyll protoplasts. APX and CAT are two major H2O2-scavenging enzymes of antioxidant defense. The activities of APX and CAT were lower in protoplasts expressing SlMPK1 or SlSPRH1 and even lower in protoplasts coexpressing SlMPK1 and SlSPRH1 (Fig. 8). However, enzyme activities in protoplasts expressing SlSPRH1S44A or SlMPK1 and SlSPRH1S44A were the same as those harboring empty vector or SlMPK1, respectively. This results suggests that the mutation S44A (SlSPRH1S44A) cancels the inhibition of enzyme activities and that SlMPK1-mediated phosphorylation of SlSPRH1 at Ser-44 regulates antioxidant defense under HT.
Figure 8.
Effects of Ser-44 mutation on SlMPK1-mediated antioxidant defense under HT stress. Arabidopsis protoplasts transiently expressing SlMPK1 alone, SlSPRH1 alone, SlSPRH1S44A alone, SlMPK1 and SlSPRH1simultaneously, SlMPK1 and SlSPRH1S44A simultaneously, or empty vector (EV) were kept at 25°C for 1 h and then kept at 36°C for 3 h. The activities of APX and CAT were measured as described in “Materials and Methods.” Values are means ± sd of three different experiments. Statistical differences among the samples are labeled with different letters according to the lsd test (P < 0.05, one-way ANOVA).
DISCUSSION
SlMPK1 Is a Negative Regulator of HT Responses in Tomato
The most extensively studied plant MAPKs are Arabidopsis AtMPK6, AtMPK3, and AtMPK4, all of which are activated by a diversity of stimuli, including abiotic stresses, pathogens, and oxidative stress (Pitzschke et al., 2009; Pitzschke, 2015; de Zelicourt et al., 2016). The Arabidopsis AtMPK6 and its functional orthologs in other species have been shown to be involved in integrating and transducing several signal stimuli for appropriate cellular responses to various stresses (Pitzschke et al., 2009; Kumar and Kirti, 2010; Xu and Zhang, 2015; Lv et al., 2017). It has been well established that MPK6 is a positive regulator of defense responses in plants (Nakagami et al., 2005; Pitzschke et al., 2009; Sinha et al., 2011; Moustafa et al., 2014; Pitzschke, 2015; de Zelicourt et al., 2016). For example, overexpression of OsMAPK5 in rice transgenic plants increased tolerance, while suppression led to hypersensitivity to various stresses, including salt, drought, and cold (Xiong and Yang, 2003). However, other studies showed that MPK6 also plays a negative role in defense responses. Silencing WIPK and SIPK in tobacco enhances basal resistance against Tobacco mosaic virus but breaks N-mediated resistance (Kobayashi et al., 2010). Cotton (Gossypium hirsutum) GhMPK6a, a homolog of MPK6, negatively regulates osmotic tolerance and bacterial infection in transgenic N. benthamiana (Li et al., 2013). Recently, two research groups reported that an MPK3/6 cascade negatively regulates freezing tolerance (Li et al., 2017; Zhao et al., 2017). Interestingly, positive and negative roles in regulating defense responses have been reported recently for soybean (Glycine max) MPK6 (Liu et al., 2014). So far, it is unclear how MAPKs respond and adapt to HT stress. As MAPKs constitute a large gene family, the characterization of more HT-responsive MAPKs will provide a better understanding of the roles of individual members in the stress signaling network. There are only two reports showing that an mpk6 Arabidopsis mutant displays higher HT tolerance (Li et al., 2012; Evrard et al., 2013). We previously purified an ABA-activated p46-MAPK in maize (Zea mays), identified as ZmMPK5 (Ding et al., 2009). In this study, we purified a p47-MBPK using anion-exchange columns and identified it as SlMPK1 using MS/MS (Fig. 1; Supplemental Fig. S1). This study focused on SlMPK1, an ortholog of AtMPK6, NtSIPK, and OsMPK6. Recent studies showed that the silencing of tomato SlMPK1 by VIGS abolishes plant tolerance to heat, cold, and oxidative stress (Nie et al., 2013; Zhou et al., 2014; Lv et al., 2017). According to our results, silencing of SlMPK1 in transgenic tomato enhances HT tolerance (Fig. 2), whereas overexpression of SlMPK1 leads to decreased HT tolerance (Fig. 4). These contrasting results may be related to differences in HT treatments. In our study (long-term trial), the seedlings of wild-type and RNAi-SlMPK1 transgenic tomato plants were subjected to 38°C/28°C (day/night) for 3 d, then recovered at 25°C/20°C for 10 d. After that, the phenotype of wild-type and RNAi-SlMPK1 transgenic tomato plants is visible, and RNAi-SlMPK1 tomato plants grow better than wild-type plants (Fig. 2B). In their treatment (short-term trial), after 42°C for 10 h, the plants did not display a phenotype, and physical indicators were used to study the roles of the tomato MPK gene (Nie et al., 2013). Therefore, the results of gene function studies using VIGS need to be further verified by stable genetic plant material.
SlMPK1 Is Involved in HT Responses via Regulating HT-Responsive Proteins, Including Antioxidant Defense Proteins
To better understand the molecular basis of SlMPK1-mediated HT tolerance in tomato, we first compared the protein abundance profiles of RNAi-SlMPK1 and wild-type plants under normal or HT conditions. Our results suggest that only 10 proteins show significant changes in abundance in the RNAi-SlMPK1 line 1-24 compared with the wild type under normal conditions (Supplemental Table S2). OPR3 is a peroxisome 12-oxophytodienoate-10,11-reductase and is required for jasmonate synthesis involving the MKK2 pathway (Brader et al., 2007). Ariga et al. (2015) showed that AtCSP41B-overexpressing transgenic Arabidopsis lines exhibited heat and salinity stress tolerance and concluded that the maintenance of CSP41B expression under abiotic stresses may alleviate photoinhibition and improve survival under such stresses. ASR (Q2QJT5) is an ABA stress ripening protein related to drought and salt tolerance not found in Arabidopsis (Fischer et al., 2011). AOR is an alkenal/one oxidoreductase, and its homologous protein (AT1G23740) was shown to be down-regulated in the absence of ATMPK6 (mpk6, R6-7) using quantitative proteomics (Miles et al., 2009). Here, AOR (Solyc05g005480) was down-regulated in the RNAi-SlMPK1 line 1-24 compared with the wild type. Therefore, the results suggest that SlMPK1 suppression improves HT tolerance in RNAi-SlMPK1 tomato plants.
Interestingly, a number of stress-responsive proteins were altered in the RNAi-SlMPK1 line 1-24 versus wild-type plants under HT conditions. These proteins showed diverse functions, such as protein folding, amino acid biosynthetic process, lipid metabolic process, translation, and oxidation-reduction process (Supplemental Tables S3 and S4). The protein-coding gene transcripts noted by others as HT regulated are, not surprisingly, HSPs/molecular chaperones (CPN60A1, CPN60B1, CPN60B2, CPN20, and CPHSC70-2). CPN60 activity is most abundant in the developing green tissues and might be involved in chloroplast biogenesis and plastid division (Ahsan et al., 2010). Salvucci (2008) demonstrated that CPN60B plays a role in acclimating photosynthesis to HT, possibly by protecting Rubisco activase from thermal denaturation. Chloroplast CPN20 is a cochaperonin of CPN60. Moreover, Arabidopsis stromal CPHTC70s are essential for plant development and important for thermotolerance in germinating seeds (Su and Li, 2008). Consistent with these results, the chlorophyll content of RNAi line 1-24 was less affected by HT stress than that of wild-type leaves (Fig. 2, C and D). These results suggest that SlMPK1-mediated CPN60 accumulation may protect the chloroplast proteins from HT-induced thermal aggregation or denaturation. Moreover, SBPASE is a Calvin cycle enzyme and functions in photosynthetic carbon fixation. The list also includes three ribosomal family proteins (Solyc09g065270, Solyc01g087730, and Solyc12g100160).
Oxidoreductase activity (GO:0016491) was the only significantly enriched molecular function (Supplemental Table S5), and seven oxidoreductases (CAT2, MDAR1, NRX1, THI1, GLU1, IMD2, and AOR) showed significant consistency of expression in at least three of the four biological replicates (Table I). This result suggests that the oxidoreductase activity-mediated oxidation-reduction process is one of the main processes in the SlMPK1-mediated response to HT. One important response mechanism to HT in plants is the antioxidant defense machinery, including both enzymatic and nonenzymatic antioxidants that work in concert to scavenge HT-caused ROS and protect plant cells from oxidative stress (Mittler, 2002; Kotak et al., 2007; Miller et al., 2010). In the preliminary proteomic analysis, the protein levels of CAT2 and MDAR1 were up-regulated in the RNAi-SlMPK1 line 1-24 compared with the wild type under HT conditions (Table I). In addition, high levels of activity of four enzymes (SOD, POD, CAT, and APX) were detected in the RNAi-SlMPK1 lines under HT conditions (Fig. 3). Both AsA and GSH act as important redox buffers in the AsA-GSH cycle. In this study, the contents of AsA and GSH also were higher in the RNAi-SlMPK1 line 1-24 than in the wild type under HT stress. These results suggest that RNAi-SlMPK1 lines possess a more efficient antioxidant network than the wild type. This is corroborated by the accumulation of lower levels of H2O2 in the RNAi-SlMPK1 lines. As an excess of H2O2 results in oxidative stress in plants, our results indicate that SlMPK1 suppression decreases HT-induced oxidative damage, consistent with the reduced levels of MDA, a parameter for determining membrane lipid peroxidation. An earlier study showed that the ectopic expression of GhMPK6a in N. benthamiana reduced drought and salt tolerance, with elevated MDA and ROS content relative to wild-type plants (Li et al., 2013). Indeed, it has been reported that CPN20 mediates FeSOD activity independent of its cochaperonin role in Arabidopsis chloroplasts, supporting a common role for CPN20 in the activation of FeSOD for oxidative stress protection and chloroplast development (Kuo et al., 2013). It is conceivable that the SlMPK1-mediated antioxidant system is effective for removing the ROS produced under HT conditions. In this regard, SlMPK1-OE transgenic plants displayed lower tolerance to HT, with low levels of enzyme activities and high levels of H2O2 and MDA (Fig. 4; Supplemental Fig. S6). These results indicate that SlMPK1 confers resistance to HT-induced oxidative stress by regulating key enzyme activities. There were no significant differences in the antioxidant system between the wild type and transgenic lines (RNAi or OE lines) under normal conditions. However, the transgenic plants showed altered activities of antioxidant enzymes after HT stress, suggesting a function for SlMPK1 as a negative regulator in HT stress through regulation of the antioxidant system. NRX1, THI1, GLU1, IMD2, and AOR are five proteins involved in the oxidation-reduction process. NRX1 has thioredoxin-disulfide reductase activity. GLU1 is a ferredoxin-dependent Glu synthase. IMD2 is a 3-isopropylmalate dehydrogenase involved in Leu biosynthesis. THI1 is a thiamine (vitamin B1) synthase with a dual function in thiamine biosynthesis and mitochondrial DNA damage tolerance. Furthermore, thiamine confers enhanced tolerance to oxidative stress in Arabidopsis (Tunc-Ozdemir et al., 2009). Recent studies suggested that THI1 may play roles in plant abiotic stress responses such as sugar deprivation, high salinity, drought, hypoxia, and oxidative stress (Li et al., 2016a). AOR is a NADPH-dependent reductase involved in the detoxification of reactive carbonyls (α,β-unsaturated carbonyl compounds) to protect chloroplast function in plants (Yamauchi et al., 2011). Taken together, our results suggest that SlMPK1-mediated HT tolerance in plants might be related to the balance of cellular redox homeostasis.
SlMPK1 Cascades Involved in the HT Response
MAPKs play important roles in various cellular processes by binding to many types of proteins (Tanoue and Nishida, 2003). Our results indicate that SlMPK1 may be a negative regulator of HT signaling by suppressing the ROS-scavenging pathway in tomato. What is the SlMPK1 pathway in response to HT? As a first step, we analyzed proteomic data and found that several SlMPK1-mediated homologous proteins also were observed in the Arabidopsis MAPKKK double mutant anp2anp3 (Takáč et al., 2014). A comparative proteomic analysis of anp2anp3 revealed an overabundance of core enzymes such as SOD, DHAR1, and the FeSOD1-associated regulatory protein CPN20, which ensure favorable cellular redox conditions as well as accelerated defense against the overproduction of ROS (Takáč et al., 2014). In this study, many proteins controlling the oxidation-reduction process also were discovered (Supplemental Table S6). There are about 80 putative MAPKKKs in Arabidopsis and 89 MAPKKKs in tomato, making up the most complex and the largest group of MAPK pathway components. However, few MAPKKKs have been functionally characterized. At1g73660 encoding a putative MAPKKK was reported to negatively regulate salt tolerance in Arabidopsis (Gao and Xiang, 2008). SlMKK9 (Solyc03g097920), with homology to Arabidopsis AtMKK9, was identified to interact with SlMPK1 using Y2H assays in our laboratory (data not shown). Loss of MKK9 activity in Arabidopsis reduces salt sensitivity, indicating that MKK9 negatively regulates salt stress (Xu et al., 2008). MKK9 might be an upstream component of SlMPK1 in response to HT. Recently, the MKK4/5-MPK3/6 cascade was found to negatively regulate cold responses (Zhao et al., 2017). Therefore, we can infer that there may be one or more MAPKKK-MKK cascades upstream of SlMPK1 negatively responding to HT signaling, which still requires further investigation.
The multifunctionality of MPK6 is likely to be conferred by its different substrates. Until now, however, only a few substrates have been reported with functional data (Guo et al., 2016). To gain insight into the interactions downstream of SlMPK1, we used Y2H assays to screen a tomato cDNA library and obtained a set of candidate proteins. Our data demonstrate that SlSPRH1 is a substrate of SlMPK1. SlSPRH1 has four Ser-Pro motifs. SlSPRH1 interacts with SlMPK1 in Y2H, BiFC, in vitro pull-down, and Co-IP assays (Fig. 5). SlSPRH1 is a protein of unknown function, homologous to Arabidopsis At1g04330 (Palm-Forster et al., 2012). Recently, it was also reported as PH2 and shown to be involved in pathogen defense (Palm-Forster et al., 2017). The first putative phosphorylation site within the primary protein sequence (Ser-44) is modified predominantly by AtMPK6 and is the same site in the conserved region (Ser-44) of SlSPRH1. The Prox-Ser-Pro (PxSP) motif has been touted previously as a high-stringency site for MAPK-directed phosphorylation. AtMPK6 can phosphorylate ERF10 and ACS6 on a Ser residue within a PxSP motif (Stulemeijer et al., 2007). The predicted PxSP motif matches for Ser-52 and Ser-94 in SlSPRH1. Here, we provide MS data supporting that Ser-49, Ser-52, and Ser-94 can be phosphorylated by SlMPK1 (Fig. 6; Supplemental Fig. S7). In vitro phosphorylation assays together with site-directed mutagenesis of the phosphorylated sites indicate that SlSPRH1 can be phosphorylated by the activated SlMPK1. Although the Ser-44 phosphorylation site was not found by MS/MS, the site-directed mutagenesis results indicate that Ser-44 is the most preferred site in SlSPRH1 for phosphorylation by SlMPK1. This residue (in the motif AIPLLSP) is conserved in closely related homologs of dicotyledonous plants but not in monocotyledonous plants (Supplemental Fig. S8). Phosphorylation of MAPK substrates often affects protein stability or turnover rates. Phosphorylation by MPK4 increases the stability of MYB75 (Li et al., 2016b). By contrast, phosphorylation by MPK3 decreases the stability of WRKY46 (Sheikh et al., 2016). Palm-Forster et al. (2017) showed that the double phosphosite mutant of PH2 displayed enhanced stability compared with the wild type. Therefore, we speculate that the phosphorylation of Ser-44 by SlMPK1 may affect SlSPRH1 stability, which, in turn, affects downstream signals. The motif AIPLLSP may act as a phosphorylated substrate for MAPK to play an important role in plant growth and development. Whether SlSPRH1 is mostly phosphorylated on the Ser-44 site after SlMPK1 activation in vivo remains to be established.
To explore the molecular function of SlSPRH1 in HT responses, transgenic Arabidopsis plants expressing SlSPRH1 were generated. SlSPRH1-expressing plants showed shorter growth and lower antioxidant defense capacity than wild-type plants under HT stress (Fig. 7; Supplemental Fig. S9). These results suggest that SlSPRH1 has a negative effect on HT tolerance. Furthermore, we predicted that the phosphorylation of site Ser-44 might be involved in the SlMPK1-mediated HT response. The mutation S44A (SlSPRH1S44A) blocked SlMPK1-mediated inhibition in protoplasts under HT (Fig. 8). These data suggest that SlSPRH1 acts downstream of SlMPK1 in the regulation of antioxidant defenses under HT. Future studies will be required to determine how the phosphorylation of S44A by SlMPK1 affects the function of the SlMPK1-SlSPRH1 cascade in tomato plants.
In summary, our data demonstrate that SlMPK1 responds to HT and plays a negative role in HT tolerance through regulating antioxidant defenses. SlSPRH1 is a phosphorylation substrate of SlMPK1, and the phosphorylation at Ser-44 by SlMPK1 is essential for SlMPK1-mediated responses to HT. These results suggest a possible molecular mechanism involving SlMPK1 in HT responses and provide insight into the MAPK cascade in tomato plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Tomato (Solanum lycopersicum ‘OFSN’), RNAi-SlMPK1 lines, SlMPK1-OE lines, tobacco (Nicotiana tabacum), and Arabidopsis (Arabidopsis thaliana Columbia-0 ecotype background, wild-type and SlSPRH1-OE lines) were used in this study. The germinated tomato seeds were sown in plastic pots containing compost soil mix and grown under a 14-h-light/10-h-dark photoperiod at 25°C/20°C (day/night) with 70% relative humidity. For the sterile culture, seeds were germinated on Murashige and Skoog medium under the same conditions. Nicotiana benthamiana seedlings were grown under a 16-h-light/8-h-dark photoperiod at 25°C with 70% relative humidity in a growth chamber. About 1- to 1.5-month-old N. benthamiana seedlings were used for transient expression. Arabidopsis plants were grown under a 16-h-light/8-h-dark photoperiod at 23°C on one-half-strength Murashige and Skoog plates.
Partial Purification of p47-MBPK and Identification by MS
The purification process was carried out as described by Ding et al. (2009) with slight modifications. Tomato leaves treated with 42°C for 2 h were harvested, frozen, and stored at −80°C. Frozen tomato leaves (1,300 g) were ground to a fine powder in the presence of liquid N2 and mixed with 1,000 mL of extraction buffer. The crude homogenate was centrifuged at 23,000g for 1 h, and the resulting supernatant fraction was brought to 30% (NH4)2SO4 saturation. The pellets were then dissolved in buffer A for ultracentrifugation, and the supernatant was loaded onto a Sephadex G 25 M column for desalting. All chromatographic runs were carried out on the AKTA Purifier 100 system and the AKTA Prime System (GE Healthcare). The fractions were loaded onto the following columns: Q-Sepharose FF column, Phenyl-Sepharose FF column, Q-Sepharose HP column, Mono QTM 5/50 GL column, and poly-l-Lys-agarose column (Supplemental Methods S1). Proteins from different stages of purification were resolved on a 12% polyacrylamide gel containing SDS and stained with silver. The 47-kD band was excised and digested with trypsin, and the tryptic digest was analyzed by LC-MS/MS. Proteins were identified using MS/MS ion search of the Mascot search engine (Matrix Science).
Generation of Transgenic Plants
For the RNAi construct, SlMPK1 (Solyc12g019460) was analyzed in the Sol Genomics Network (https://solgenomics.net/search/unigene.pl?unigene_id=576603). The RNAi-SlMPK1 construct was made by introducing a fragment targeting the 201-bp 3′ untranslated region of SlMPK1 (containing XhoI, SpeI, NcoI, and BamHI restriction sites) into pGSA1285 vector (Adams-Phillips et al., 2004). For the tomato overexpression construct, the full-length cDNA of SlMPK1 was amplified with specific primers containing BamHI and SacI restriction sites and inserted into binary vector pBI121 driven by the CaMV 35S promoter as described previously (Meli et al., 2010). RNAi and overexpression of SlMPK1 in the putative lines were examined by RT-qPCR using specific primers (Supplemental Methods S1). Positive tomato T0 plants were transplanted to plastic pots containing a compost-soil mix and grown in the greenhouse for the collection of T0 seeds. A kanamycin spraying test was used in the genetic segregation analysis (Weide et al., 1989), and single-copy homozygous T1 seeds were used for further study. To generate SlSPRH1-expressing plants, the full-length cDNA of SlSPRH1 was inserted into binary vector pBI121. The constructs were introduced into Arabidopsis (Columbia-0 ecotype) by the floral dip method (Clough and Bent, 1998). Homozygous T3 seeds were harvested for further analysis.
HT Stress
For the HT tolerance assay, seedlings planted in matrix soils or sterile culture were used. Seedlings of the wild type and RNAi-SlMPK1 lines 1-14, 1-23, and 1-24 in the matrix soils at the five-leaf stage were subjected to 38°C/28°C (day/night) for 3 d and then recovered at 25°C/20°C for 10 d. The RNAi-SlMPK1 transgenic line 1-24 in Murashige and Skoog medium was used. After surface sterilization at 25°C, seeds were allowed to germinate on Murashige and Skoog medium at 25°C/20°C for 10 d. Seedlings of the same size were chosen and cut at the bottom of the hypocotyls using a sharp blade (the height of the detached plantlets was kept at 2 cm). The detached plantlets were moved to a new Murashige and Skoog medium and grown at 25°C/20°C for another 3 d, incubated at 45°C for 1 or 2 h, and then followed by recovery at 25°C/20°C for 14 d.
Seedlings of the wild type and the SlMPK1-OE lines OE11 and OE15 in the matrix soils at the three-leaf stage were subjected to 38°C/28°C (day/night) for 1 d and then recovery at 25°C/20°C for 7 d. For the sterile seedling treatment, seedlings of the same size were chosen and cut at the bottom of the hypocotyls using a sharp blade when two cotyledons were expanded completely. The detached plantlets were moved to a new Murashige and Skoog medium and grown at 25°C/20°C for 3 d, incubated at 45°C for 1 or 2 h, and then followed by recovery at 25°C/20°C for 7 d.
To measure the hypocotyl length of Arabidopsis, the seeds were plated in rows on one-half-strength Murashige and Skoog medium, and the plates were covered with foil. After 2 d of cold treatment (4°C), the foil-wrapped plates were placed in a vertical position at 23°C for 3 d. Wrapped plates were then subjected to HT at 45°C for 1 or 2 h. After that, the plates were incubated in a vertical position at 23°C under light or dark for another 6 d, and the hypocotyl length was measured. For Arabidopsis growth, 10-d-old seedlings in one-half-strength Murashige and Skoog medium were subjected to HT treatment at 42°C for 1.5 h, followed by recovery for 10 d.
iTRAQ Proteomic Assay
Protein Extraction and Digestion
Total protein extraction from leaf tissue of three plants grown in different pots of each treatment (wild-type and RNAi-SlMPK1 1-24 plants treated with normal conditions or 42°C for 4 h) was performed according to the method of Gong et al. (2014). iTRAQ labeling is shown in Supplemental Figure S3, and SCX fractionation and MS analysis are as described (Supplemental Methods S1).
Protein Identification
The MS/MS spectra were extracted and analyzed with ProteinPilot software (version 4.5; AB SCIEX) searching against a UniProt tomato protein database (35,812 proteins, updated in September 2017). Detected protein threshold [Unused ProtScore (Conf)] was 0.05 (10%), and the FDR Analysis tab was checked. All identified proteins had an Unused Protscore of greater than 1.3 (which corresponds to proteins identified with greater than 95% confidence), as calculated by the software, and a global false discovery rate of 1% or less determined at the protein level by the PSPEP algorithm. To be considered as differentially expressed, proteins were required to have P < 0.05, as calculated by the software. The coefficient of variation also was calculated for each iTRAQ study. For protein abundance ratios measured using iTRAQ, fold change greater than 1.2 or less than 0.67 was considered significant. Only proteins that were identified in two 8-plex iTRAQ analyses were included. Protein Ontology classification was performed using the PANTHER classification system (http://pantherdb.org, PANTHER Overrepresentation Test, release 20170413; Gene Ontology tomato database, released October 24, 2017).
Y2H Screening
The desired genes were cloned into the pGBKT7 vectors and transformed into the Y2HGold yeast strain (Clontech). Then, competent cells of a single clone from the bait transformant (Y2HGold) were transformed with the cDNA library plasmids constructed with HT-stressed tomato seedlings. The clones were selected on SD/−Trp/−Leu/−His/−Ade/+AbA/+20 mm 3-Amino-1,2,4-Triazole (3-AT) medium (self-activation of SlMPK1 was effectively suppressed at 20 mm 3-AT), and blue colonies were considered to be potential positive clones. These clones were tested subsequently by PCR for the library plasmids. The full-length coding sequences of SlMPK1 and SlSPRH1 were cloned into the pGBKT7 and pGADT7 vectors, respectively. The bait and prey constructs were cotransformed into the Y2HGold yeast strain using the lithium acetate method. Then, transformants were selected on SD/−Trp/−Leu medium. The colonies were inoculated onto SD/−Trp/−Leu and SD/−Trp/−Leu/−His/−Ade/+AbA/+X-α-gal/+20 mm 3-AT media, grown at 30°C for 2 to 3 d, and photographed. To further confirm the interaction, the full-length coding sequences of SlSPRH1 and SlMPK1 were cloned into the pGBKT7 and pGADT7 vectors, respectively. pGBKT7-53 and pGADT7-T were used as positive controls.
BiFC
Full-length coding sequences of SlMPK1 and other genes without a stop codon were cloned into the pSPYCE and pSPYNE vectors, respectively. These constructs were transferred into Agrobacterium tumefaciens EHA105. Then, cultures were resuspended in a freshly made solution containing 0.5% Glc, 50 mm MES-KOH, pH 5.6, 10 mm MgCl2, and 0.1 mm acetosyringone. The OD600 of the cell suspensions was adjusted to 0.5, incubated at room temperature in darkness for 2 h before being mixed at a 1:1 ratio, and infiltrated into the leaves of 5-week-old N. benthamiana. Protoplasts were extracted after 40 h of dark culture. Fluorescent signal was detected with a fluorescence microscope (Axio Observer Z1; Zeiss). Coexpression of SlMPK1-cYFP with SlMPKK2-nYFP was used as a positive control.
Recombinant Protein, in Vitro Pull-Down, and in Vitro Phosphorylation Assays
The DNA fragment encoding full-length SlMPK1 was cloned into pCzn I vector with a 6× His tag at the N terminus of the protein. His-SlMPK1 was overexpressed by adding 0.5 mm isopropylthio-β-galactoside and purified from Escherichia coli BL21 (Plyss) cells by nickel-agarose beads. A constitutive-active form of SlMKK2 was developed by replacing the conserved Ser/Thr residues Thr-215 and Ser-221 with Asp (SlMKK2T215D/S221D, referred to hereafter as SlMKK2DD). DNA fragments encoding full-length SlMKK2DD or SlSPRH1 were cloned into pGEX-4T-1 vector with a GST tag at the N terminus of the protein. GST-SlMKK2DD and GST-SlSPRH1 proteins were overexpressed by adding 0.5 mm isopropylthio-β-galactoside and purified from E. coli Arctic Express cells by Glutathione Sepharose 4B beads. In vitro phosphorylation assays were performed as described previously (Pérez-Salamó et al., 2014) with slight modifications. In vitro protein-protein interaction assays (pull down) was shown in Supplemental Methods S1.
In Vivo Co-IP Assay
Co-IP was carried out as described by Gou et al. (2015) with slight modifications. The HA and Myc tags are present in the pSPYCE-35S and pSPYNE-35S vector system (Schütze et al., 2009; Gou et al., 2015), so pSPYCE-SlMPK1 and pSPYNE-SlSPRH1 can be immunoprecipitated using anti-HA or anti-Myc antibody. A. tumefaciens GV3101 harboring each of the two SlMPK1-HA and SlSPRH1-Myc constructs was infiltrated solely or coinfiltrated into the abaxial side of 4-week-old N. benthamiana leaves using a 1-mL needleless syringe according to http://www.bio-protocol.org/e95 (Supplemental Methods S1).
Protein Phosphorylation Site Determination by LC-MS/MS
GST-SlSPRH1 was phosphorylated by His-SlMPK1 in a kinase reaction (25 mm Tris-HCl, pH 7.5, 1 mm EGTA, 20 mm MgCl2, 1 mm DTT, and 1× phosphatase inhibitor) in the presence of 200 µm ATP and GST-SlMKK2DD at room temperature for 1 h. As a control, a parallel reaction without kinases (His-SlMPK1 and GST-SlMKK2DD) was included. After Coomassie Blue staining, GST-SlSPRH1 protein bands were excised, destained, and dried completely. Tryptic digests were analyzed on an LC-MS system consisting of Easy-nLC 1000 (Thermo Fisher Scientific) coupled to a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Acquired raw data files were processed using Proteome Discoverer software (version 2.1; Thermo Fisher Scientific) and Sequest HT database search engines against a tomato protein database (Supplemental Methods S1).
Synthetic Peptide Phosphorylation Site Determination by MS/MS
Five micrograms of each peptide (AIPLLSPLITSPTSPESDNLKA or KKWQHPASPFCYEPAP, synthesized by Ontores Biotechnology) was incubated with His-SlMPK1 and GST-SlMKK2DD in a kinase reaction (25 mm Tris-HCl, pH 7.5, 1 mm EGTA, 20 mm MgCl2, 1 mm DTT, 200 µm ATP, and 1× phosphatase inhibitor) at room temperature for 1 h. The reaction buffer was centrifugally filtered through a Millipore 5-kD cutoff filter at 9,000g at 4°C. The filtrate was purified and determined by LC-MS/MS.
Subcellular Localization
To determine the subcellular localization of SlSPRH1, SlMPK1 was cloned into the pEGAD vector containing the GFP reporter gene to produce the fusion construct pEGAD-SlMPK1-GFP under the control of 35S. The fusion construct and the control vector (pEGAD) were transferred separately into A. tumefaciens strain EHA105 by heat shock and infiltrated into the leaves of 5-week-old N. benthamiana. Protoplasts were extracted after 40 h of dark culture, and fluorescent signals were detected using a fluorescence microscope (Axio Observer Z1; Zeiss).
Arabidopsis Mesophyll Protoplast Transient Expression
SlMPK1, SlSPRH1, or SlSPRH1S44A was cloned into the pUC19 vector with two 35S enhancer promoters. Protoplast isolation and transfection were carried out as described previously by Yoo et al. (2007). For transient expression assays, isolated protoplasts were transfected with pUC19-35S-SlMPK1, pUC19-35S-SlSPRH1, or pUC19-35S-SlSPRH1S44A plasmid alone or cotransfected with pUC19-35S-SlMPK1 and pUC19-35S-SlSPRH1 or pUC19-35S-SlSPRH1S44A by polyethylene glycol-mediated transformation at 25°C and then kept at 36°C for 3 h. Total proteins were extracted for enzyme activity determination.
Accession Numbers
Accession numbers are as follows: SlSPRH1 (Solyc06g053700) and SlMPK1 (Solyc12g019460).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Elution profiles of protein concentration and kinase activity from each chromatography step.
Supplemental Figure S2. Expression level of HT-responding genes under HT stress.
Supplemental Figure S3. iTRAQ proteomics analysis of SlMPK1-mediated response to HT.
Supplemental Figure S4. Prediction of functional protein networks involved in SlMPK1-mediated response to HT stress using STRING 9.0 applied to proteomic data.
Supplemental Figure S5. RT-qPCR analysis of transcript levels of genes coding SlMPK1-mediated proteins under HT.
Supplemental Figure S6. Response of antioxidant defense to HT in the SlMPK1-OE plants.
Supplemental Figure S7. Summary of MS-based mapping of in vitro phosphorylated residues in SlSPRH1 by SlMPK1.
Supplemental Figure S8. Multiple sequence alignments of the conserved motif (AIPLLSP) in plants.
Supplemental Figure S9. The response of antioxidant enzymes to HT in SlSPRH1-OE plants.
Supplemental Figure S10. The expression level of SlMPK1 under HT.
Supplemental Table S1. Proteins abundant in the tomato leaves of the wild type under HT stress.
Supplemental Table S2. Proteins abundant in the tomato leaves of RNAi-SlMPK1 line 1-24 as compared with the wild type under normal conditions.
Supplemental Table S3. Proteins abundant in the tomato leaves of RNAi-SlMPK1 line 1-24 as compared with the wild type under HT conditions.
Supplemental Table S4. Functional enrichment of identified proteins involved in SlMPK1-mediated response to HT with no Bonferroni correction.
Supplemental Table S5. Functional enrichment of identified proteins involved in SlMPK1-mediated response to HT with Bonferroni correction.
Supplemental Table S6. Similar proteins in the RNAi-SlMPK1 line 1-24 and the anp2anp3 mutant.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31101092 and 31761130073), by the China Postdoctoral Science Foundation-funded project (grant nos. 20110491464 and 2012T50520), by the Natural Science Foundation of the Higher Education Institutions of Jiangsu Province (grant no. 10KJB180010), by the Jiangsu Government Scholarship for Overseas Studies (grant no. JS-2012-012), and by a research grant from Fujian Agriculture and Forestry University (KXGH17005).
Articles can be viewed without a subscription.
References
- Adams-Phillips L, Barry C, Kannan P, Leclercq J, Bouzayen M, Giovannoni J (2004) Evidence that CTR1-mediated ethylene signal transduction in tomato is encoded by a multigene family whose members display distinct regulatory features. Plant Mol Biol 54: 387–404 [DOI] [PubMed] [Google Scholar]
- Ahsan N, Donnart T, Nouri MZ, Komatsu S (2010) Tissue-specific defense and thermo-adaptive mechanisms of soybean seedlings under heat stress revealed by proteomic approach. J Proteome Res 9: 4189–4204 [DOI] [PubMed] [Google Scholar]
- Ariga H, Tanaka T, Ono H, Sakata Y, Hayashi T, Taji T (2015) CSP41b, a protein identified via FOX hunting using Eutrema salsugineum cDNAs, improves heat and salinity stress tolerance in transgenic Arabidopsis thaliana. Biochem Biophys Res Commun 464: 318–323 [DOI] [PubMed] [Google Scholar]
- Blanco FA, Zanetti ME, Casalongué CA, Daleo GR (2006) Molecular characterization of a potato MAP kinase transcriptionally regulated by multiple environmental stresses. Plant Physiol Biochem 44: 315–322 [DOI] [PubMed] [Google Scholar]
- Brader G, Djamei A, Teige M, Palva ET, Hirt H (2007) The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis. Mol Plant Microbe Interact 20: 589–596 [DOI] [PubMed] [Google Scholar]
- Bueso E, Alejandro S, Carbonell P, Perez-Amador MA, Fayos J, Bellés JM, Rodriguez PL, Serrano R (2007) The lithium tolerance of the Arabidopsis cat2 mutant reveals a cross-talk between oxidative stress and ethylene. Plant J 52: 1052–1065 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413: 217–226 [DOI] [PubMed] [Google Scholar]
- de Zelicourt A, Colcombet J, Hirt H (2016) The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci 21: 677–685 [DOI] [PubMed] [Google Scholar]
- Ding H, Zhang A, Wang J, Lu R, Zhang H, Zhang J, Jiang M (2009) Identity of an ABA-activated 46 kDa mitogen-activated protein kinase from Zea mays leaves: partial purification, identification and characterization. Planta 230: 239–251 [DOI] [PubMed] [Google Scholar]
- Eltayeb AE, Kawano N, Badawi GH, Kaminaka H, Sanekata T, Shibahara T, Inanaga S, Tanaka K (2007) Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 225: 1255–1264 [DOI] [PubMed] [Google Scholar]
- Evrard A, Kumar M, Lecourieux D, Lucks J, von Koskull-Döring P, Hirt H (2013) Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. PeerJ 1: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer I, Camus-Kulandaivelu L, Allal F, Stephan W (2011) Adaptation to drought in two wild tomato species: the evolution of the Asr gene family. New Phytol 190: 1032–1044 [DOI] [PubMed] [Google Scholar]
- Gao L, Xiang CB (2008) The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant Mol Biol 67: 125–134 [DOI] [PubMed] [Google Scholar]
- Gong B, Zhang C, Li X, Wen D, Wang S, Shi Q, Wang X (2014) Identification of NaCl and NaHCO3 stress responsive proteins in tomato roots using iTRAQ-based analysis. Biochem Biophys Res Commun 446: 417–422 [DOI] [PubMed] [Google Scholar]
- Gou M, Zhang Z, Zhang N, Huang Q, Monaghan J, Yang H, Shi Z, Zipfel C, Hua J (2015) Opposing effects on two phases of defense responses from concerted actions of HEAT SHOCK COGNATE70 and BONZAI1 in Arabidopsis. Plant Physiol 169: 2304–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Feng P, Chi W, Sun X, Xu X, Li Y, Ren D, Lu C, Rochaix JD, Leister D, et al. (2016) Plastid-nucleus communication involves calcium-modulated MAPK signalling. Nat Commun 7: 12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins R, Lockwood T, Holley S, Yalamanchili R, Stratmann JW (2007) Changes in extracellular pH are neither required nor sufficient for activation of mitogen-activated protein kinases (MAPKs) in response to systemin and fusicoccin in tomato. Planta 225: 1535–1546 [DOI] [PubMed] [Google Scholar]
- Holley SR, Yalamanchili RD, Moura DS, Ryan CA, Stratmann JW (2003) Convergence of signaling pathways induced by systemin, oligosaccharide elicitors, and ultraviolet-B radiation at the level of mitogen-activated protein kinases in Lycopersicon peruvianum suspension-cultured cells. Plant Physiol 132: 1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth PK, Ranf S, Pancholi SS, Jayanty S, Walla MD, Miller W, Howe GA, Lincoln DE, Stratmann JW (2007) Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc Natl Acad Sci USA 104: 12205–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou S, Yoshioka H, Kawakita K, Rowland O, Jones JDG, Mori H, Doke N (2005) Involvement of PPS3 phosphorylated by elicitor-responsive mitogen-activated protein kinases in the regulation of plant cell death. Plant Physiol 139: 1914–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Seo S, Hirai K, Yamamoto-Katou A, Katou S, Seto H, Meshi T, Mitsuhara I, Ohashi Y (2010) Silencing of WIPK and SIPK mitogen-activated protein kinases reduces tobacco mosaic virus accumulation but permits systemic viral movement in tobacco possessing the N resistance gene. Mol Plant Microbe Interact 23: 1032–1041 [DOI] [PubMed] [Google Scholar]
- Kong F, Wang J, Cheng L, Liu S, Wu J, Peng Z, Lu G (2012) Genome-wide analysis of the mitogen-activated protein kinase gene family in Solanum lycopersicum. Gene 499: 108–120 [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Kumar KRR, Kirti PB (2010) A mitogen-activated protein kinase, AhMPK6 from peanut localizes to the nucleus and also induces defense responses upon transient expression in tobacco. Plant Physiol Biochem 48: 481–486 [DOI] [PubMed] [Google Scholar]
- Kuo WY, Huang CH, Liu AC, Cheng CP, Li SH, Chang WC, Weiss C, Azem A, Jinn TL (2013) CHAPERONIN 20 mediates iron superoxide dismutase (FeSOD) activity independent of its co-chaperonin role in Arabidopsis chloroplasts. New Phytol 197: 99–110 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Vierling E (2008) Core genome responses involved in acclimation to high temperature. Plant Physiol 146: 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Wang M, Wu XM, Chen DH, Lv HJ, Shen JL, Qiao Z, Zhang W (2016a) THI1, a thiamine thiazole synthase, interacts with Ca2+-dependent protein kinase CPK33 and modulates the S-type anion channels and stomatal closure in Arabidopsis. Plant Physiol 170: 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ding Y, Shi Y, Zhang X, Zhang S, Gong Z, Yang S (2017) MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev Cell 43: 630–642.e4 [DOI] [PubMed] [Google Scholar]
- Li Q, Xie QG, Smith-Becker J, Navarre DA, Kaloshian I (2006) Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling cascades. Mol Plant Microbe Interact 19: 655–664 [DOI] [PubMed] [Google Scholar]
- Li S, Wang W, Gao J, Yin K, Wang R, Wang C, Petersen M, Mundy J, Qiu JL (2016b) MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28: 2866–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang L, Wang X, Zhang W, Hao L, Chu X, Guo X (2013) Cotton GhMPK6a negatively regulates osmotic tolerance and bacterial infection in transgenic Nicotiana benthamiana, and plays a pivotal role in development. FEBS J 280: 5128–5144 [DOI] [PubMed] [Google Scholar]
- Li Z, Yue H, Xing D (2012) MAP Kinase 6-mediated activation of vacuolar processing enzyme modulates heat shock-induced programmed cell death in Arabidopsis. New Phytol 195: 85–96 [DOI] [PubMed] [Google Scholar]
- Liu JZ, Braun E, Qiu WL, Shi YF, Marcelino-Guimarães FC, Navarre D, Hill JH, Whitham SA (2014) Positive and negative roles for soybean MPK6 in regulating defense responses. Mol Plant Microbe Interact 27: 824–834 [DOI] [PubMed] [Google Scholar]
- Lv X, Ge S, Jalal Ahammed G, Xiang X, Guo Z, Yu J, Zhou Y (2017) Crosstalk between nitric oxide and MPK1/2 mediates cold acclimation-induced chilling tolerance in tomato. Plant Cell Physiol 58: 1963–1975 [DOI] [PubMed] [Google Scholar]
- Melech-Bonfil S, Sessa G (2011) The SlMKK2 and SlMPK2 genes play a role in tomato disease resistance to Xanthomonas campestris pv. vesicatoria. Plant Signal Behav 6: 154–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meli VS, Ghosh S, Prabha TN, Chakraborty N, Chakraborty S, Datta A (2010) Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc Natl Acad Sci USA 107: 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Miles GP, Samuel MA, Ranish JA, Donohoe SM, Sperrazzo GM, Ellis BE (2009) Quantitative proteomics identifies oxidant-induced, AtMPK6-dependent changes in Arabidopsis thaliana protein profiles. Plant Signal Behav 4: 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 10.1111/j.1365-3040.2009.02041.x [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 10.1016/S1360-1385(02)02312-9 [DOI] [PubMed] [Google Scholar]
- Mittler R, Finka A, Goloubinoff P (2012) How do plants feel the heat? Trends Biochem Sci 37: 118–125 [DOI] [PubMed] [Google Scholar]
- Moustafa K, AbuQamar S, Jarrar M, Al-Rajab AJ, Trémouillaux-Guiller J (2014) MAPK cascades and major abiotic stresses. Plant Cell Rep 33: 1217–1225 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10: 339–346 [DOI] [PubMed] [Google Scholar]
- Nie WF, Wang MM, Xia XJ, Zhou YH, Shi K, Chen Z, Yu JQ (2013) Silencing of tomato RBOH1 and MPK2 abolishes brassinosteroid-induced H2O2 generation and stress tolerance. Plant Cell Environ 36: 789–803 [DOI] [PubMed] [Google Scholar]
- Oh CS, Hwang J, Choi MS, Kang BC, Martin GB (2013) Two leucines in the N-terminal MAPK-docking site of tomato SlMKK2 are critical for interaction with a downstream MAPK to elicit programmed cell death associated with plant immunity. FEBS Lett 587: 1460–1465 [DOI] [PubMed] [Google Scholar]
- Palm-Forster MAT, Eschen-Lippold L, Lee J (2012) A mutagenesis-based screen to rapidly identify phosphorylation sites in mitogen-activated protein kinase substrates. Anal Biochem 427: 127–129 [DOI] [PubMed] [Google Scholar]
- Palm-Forster MAT, Eschen-Lippold L, Uhrig J, Scheel D, Lee J (2017) A novel family of proline/serine-rich proteins, which are phospho-targets of stress-related mitogen-activated protein kinases, differentially regulates growth and pathogen defense in Arabidopsis thaliana. Plant Mol Biol 95: 123–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Salamó I, Papdi C, Rigó G, Zsigmond L, Vilela B, Lumbreras V, Nagy I, Horváth B, Domoki M, Darula Z, Medzihradszky K, Bögre L, et al. (2014) The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol 165: 319–334 10.1104/pp.114.237891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A. (2015) Modes of MAPK substrate recognition and control. Trends Plant Sci 20: 49–55 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohila JS, Yang Y (2007) Rice mitogen-activated protein kinase gene family and its role in biotic and abiotic stress response. J Integr Plant Biol 49: 751–759 [Google Scholar]
- Salvucci ME. (2008) Association of Rubisco activase with chaperonin-60beta: a possible mechanism for protecting photosynthesis during heat stress. J Exp Bot 59: 1923–1933 [DOI] [PubMed] [Google Scholar]
- Sánchez R, Flores A, Cejudo FJ (2006) Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress. Planta 223: 901–909 [DOI] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C (2009) Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol 479: 189–202 [DOI] [PubMed] [Google Scholar]
- Sheikh AH, Eschen-Lippold L, Pecher P, Hoehenwarter W, Sinha AK, Scheel D, Lee J (2016) Regulation of WRKY46 transcription factor function by mitogen-activated protein kinases in Arabidopsis thaliana. Front Plant Sci 7: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha AK, Jaggi M, Raghuram B, Tuteja N (2011) Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal Behav 6: 196–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stulemeijer IJE, Stratmann JW, Joosten MHAJ (2007) Tomato mitogen-activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol 144: 1481–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li HM (2008) Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol 146: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri SS, Dhindsa RS (2008) A heat-activated MAP kinase (HAMK) as a mediator of heat shock response in tobacco cells. Plant Cell Environ 31: 218–226 [DOI] [PubMed] [Google Scholar]
- Takáč T, Šamajová O, Vadovič P, Pechan T, Košútová P, Ovečka M, Husičková A, Komis G, Šamaj J (2014) Proteomic and biochemical analyses show a functional network of proteins involved in antioxidant defense of the Arabidopsis anp2anp3 double mutant. J Proteome Res 13: 5347–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T, Nishida E (2003) Molecular recognitions in the MAP kinase cascades. Cell Signal 15: 455–462 [DOI] [PubMed] [Google Scholar]
- Tanoue T, Adachi M, Moriguchi T, Nishida E (2000) A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol 2: 110–116 [DOI] [PubMed] [Google Scholar]
- Tubiello FN, Soussana JF, Howden SM (2007) Crop and pasture response to climate change. Proc Natl Acad Sci USA 104: 19686–19690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunc-Ozdemir M, Miller G, Song L, Kim J, Sodek A, Koussevitzky S, Misra AN, Mittler R, Shintani D (2009) Thiamin confers enhanced tolerance to oxidative stress in Arabidopsis. Plant Physiol 151: 421–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide R, Koornneef M, Zabel P (1989) A simple, nondestructive spraying assay for the detection of an active kanamycin resistance gene in transgenic tomato plants. Theor Appl Genet 78: 169–172 [DOI] [PubMed] [Google Scholar]
- Wu J, Wang J, Pan C, Guan X, Wang Y, Liu S, He Y, Chen J, Chen L, Lu G (2014) Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS ONE 9: e103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JR, Wang LC, Lin YR, Weng CP, Yeh CH, Wu SJ (2017) The Arabidopsis heat-intolerant 5 (hit5)/enhanced response to aba 1 (era1) mutant reveals the crucial role of protein farnesylation in plant responses to heat stress. New Phytol 213: 1181–1193 [DOI] [PubMed] [Google Scholar]
- Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang S (2015) Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci 20: 56–64 [DOI] [PubMed] [Google Scholar]
- Xu J, Li Y, Wang Y, Liu H, Lei L, Yang H, Liu G, Ren D (2008) Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J Biol Chem 283: 26996–27006 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Hasegawa A, Taninaka A, Mizutani M, Sugimoto Y (2011) NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J Biol Chem 286: 6999–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhao C, Wang P, Si T, Hsu CC, Wang L, Zayed O, Yu Z, Zhu Y, Dong J, Tao WA, et al. (2017) MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev Cell 43: 618–629.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Xia XJ, Zhou YH, Shi K, Chen Z, Yu JQ (2014) RBOH1-dependent H2O2 production and subsequent activation of MPK1/2 play an important role in acclimation-induced cross-tolerance in tomato. J Exp Bot 65: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]