The Arabidopsis pentatricopeptide repeat protein SOT5 has a critical role in both plastid rpl2 and trnK intron splicing and leaf development.
Abstract
Chloroplast biogenesis and development are highly complex processes requiring interaction between plastid and nuclear genomic products. Using a high-throughput screen for chloroplast biogenesis suppressors in Arabidopsis (Arabidopsis thaliana), we identified a suppressor of thf1 (sot5) that displays virescent and serrated leaves. Further characterization revealed that sot5 mutants are defective in leaf adaxial and abaxial polarity and act as enhancers of asymmetric leaves2. Map-based cloning identified SOT5 as a gene previously named EMB2279 that encodes a plastid-targeted pentatricopeptide repeat (PPR) protein with 11 PPR motifs. A G-to-A mutation in sot5 leads to a significant decrease in splicing efficiency, generating two additional mRNA variants. As reported previously, the sot5 null mutation is embryo lethal. SOT5 is predicted to bind to specific RNA sequences found in plastid rpl2 and trnK genes, and we found decreased splicing efficiency of the rpl2 and trnK genes in sot5 mutants. Together, our results reveal that the PPR protein SOT5/EMB2279 is required for intron splicing of plastid rpl2 and trnK, providing insights into the role of plastid translation in the coupled development between chloroplasts and leaves.
Chloroplast development, which is an essential event for the switching of plant growth from heterotrophy to autotrophy, relies on the coordinated expression of nuclear and plastid genes. Although significant progress has been made in the past few years toward understanding the molecular mechanisms underlying chloroplast development, it remains elusive how chloroplast gene expression is finely regulated in response to intracellular and environmental cues (Pogson et al., 2015; Chan et al., 2016). Since chloroplasts originated from endosymbiotic cyanobacteria and possess prokaryote-type circular DNA, the chloroplast remains the same machinery that carries out gene expression as prokaryotes (Jarvis and López-Juez, 2013). However, some novel events, such as RNA editing, intron splicing, and RNA cleavage and trimming (Stern et al., 2010), have evolved, making gene expression in chloroplasts distinct from that in prokaryotes.
In Arabidopsis (Arabidopsis thaliana) chloroplasts, 20 introns are distributed across 17 genes (de Longevialle et al., 2010); 19 of these introns belong to the group II family of introns, and only one, located in the trnL gene, belongs to group I. Original group I and II introns are large ribozymes that cleave phosphodiester bonds through two consecutive trans-esterification steps in a sequence-specific manner (Lehmann and Schmidt, 2003). The main differences between these two types of introns lie in their secondary structures and chemical reaction mechanisms for the first splicing step: group I introns are composed of 10 domains (P1–P10) that play a role in RNA folding for efficient splicing and ligation; group II introns have six domains (I–VI), each with distinct functions in intron self-splicing and/or retromobility (Lehmann and Schmidt, 2003; Dai et al., 2008; Pyle, 2016). The largest domain, domain I, functions as a structural scaffold and recognizes the exon positions for catalysis. Domains II and III enhance the catalysis of splicing. Domain IV, the most variable domain in different introns, contains the intron-encoded protein (IEP) sequence, which constitutes four different subdomains (reverse transcriptase, maturase, DNA binding, and endonuclease) that are required for RNA folding during splicing and intron retromobility after splicing. Chloroplast introns have no IEP sequence except for the trnK intron, which contains a limited coding sequence only for functional maturase (MatK; Zoschke et al., 2010) and the break point of the trans-splicing rps12-1 intron (Glanz and Kück, 2009). The highly conserved domain V contains the catalytic core that is essential for ribozyme activity. Domain VI contains the branch-point site adenosine required for the splicing reaction. Group II introns are further classified into several subclasses (IIA–IIF) that display distinct structural and functional differences (Pyle, 2016). Among the 19 group II introns in Arabidopsis chloroplasts, eight belong to group IIA and 11 belong to group IIB (de Longevialle et al., 2010). However, both group I and group II introns in plant organelles are degenerated and require additional trans-factors for efficient splicing in vivo (Hahn et al., 1998; de Longevialle et al., 2010). These trans-factors, including chloroplast IEP (MatK) and nucleus-encoded RNA-binding factors, are assembled with cis-elements (intron RNAs) into active ribonucleoprotein complexes for accurate and efficient self-splicing (Stern et al., 2010; Zoschke et al., 2010). The chloroplast matK gene was reported to be required for the splicing of seven group IIA introns, trnA, trnI, trnV, atpF, rpl2, rps12-2, and trnK (Zoschke et al., 2010).
In addition to the trans-factor encoded by matK, a number of nucleus-encoded proteins involved in chloroplast splicing events have been identified, particularly from maize (Zea mays), through genetic and biochemical approaches. Some of the trans-factors are required for splicing multiple introns, while others are required for specific introns (Stern et al., 2010). Chloroplast RNA Splicing1 (CRS1) and CRS2 (Jenkins et al., 1997) were the first reported trans-factors in maize. Whereas CRS1 contains three repeated chloroplast RNA splicing and ribosome maturation (CRM) domains and is essential solely for the splicing of the chloroplast atpF group II intron (Till et al., 2001; Keren et al., 2008), CRS2 is a homolog of bacterial peptidyl-tRNA hydrolase and required for the splicing of nine group II introns (Jenkins et al., 1997; Ostheimer et al., 2003). CRS2 forms a complex with CRS2-Associated Factor1 (CAF1) and/or CAF2, both of which are CRM proteins with two CRM domains, to facilitate the splicing of the CRS2-dependent introns (Ostheimer et al., 2003). The splicing specificity of CRS2/CAF-dependent introns is determined by CRM Family Member2 (CFM2) and CFM3 (Asakura et al., 2008). Two general splicing factors, an RNase III domain protein (RNC1) and WTF1 (WHAT IS THIS FACTOR1), were identified from CAF1- and/or CAF2-immunoprecipitated ribonucleoprotein particles by mass spectrometry (Watkins et al., 2007). RNC1, which contains two RNase III domains without RNA cleavage activity, and WTF1, which has the plant organelle RNA recognition domain (PORR), form heterodimers that splice most of chloroplast group II introns in a CAF1/CAF2-dependent or -independent manner (Kroeger et al., 2009). It remains unclear whether a core spliceosome, as in the nucleus, is present for the in vivo splicing of all group II introns in chloroplasts.
In addition to these general splicing factors, another group of proteins involved in chloroplast intron splicing events are pentatricopeptide repeat (PPR) proteins, which bind to their single-stranded RNA targets in a modular and base-specific fashion (Yin et al., 2013; Barkan and Small, 2014). For example, PPR4 and PPR5 are responsible for the splicing of the rps12 and trnG-UCC group II introns in maize, respectively (Schmitz-Linneweber et al., 2006; Beick et al., 2008); in Arabidopsis, ORGANELLE TRANSCRIPT PROCESSING70 (OTP70) is required for the splicing of the rpoC1 intron (Chateigner-Boutin et al., 2011), OTP51 for ycf3 intron 2 and several other group IIA introns as well (de Longevialle et al., 2008), THYLAKOID ASSEMBLY8 for ycf3 intron 2 and trnA (Khrouchtchova et al., 2012), EMBRYO DEFECTIVE2654 (EMB2654) for trans-splicing of rps12 intron 1 (Aryamanesh et al., 2017); in rice (Oryza sativa), WHITE STRIPE LEAF (WSL) is required for rpl2 intron splicing (Tan et al., 2014) and WSL4 for four group II introns, atpF, rpl2, rps12-2, and ndhA (Wang et al., 2017). Generally, PPR proteins are characterized by a one-repeat/one-nucleotide recognition mechanism (Prikryl et al., 2011; Yin et al., 2013). Thus, the binding specificity and affinity of a PPR protein to RNA targets are associated with the number of PPR repeats. It has been known that over 450 PPR proteins are present in Arabidopsis and divided into two subfamilies, designated P and PLS, according to the characteristics of the PPR motifs (Lurin et al., 2004). Most PPRs are involved in organellar gene expression, including RNA transcription and stability, RNA editing, RNA maturation, RNA translation, and RNA splicing (Schmitz-Linneweber and Small, 2008). However, the roles of most PPR proteins in RNA processing still remain unknown. It is thought that, during the group II intron splicing process, the intron-specific PPRs act in an initial sequence-specific binding step that alters RNA conformation ready for the recruitment of the relevant general splicing complex, such as the WTF/RNC1 complex (de Longevialle et al., 2010). This hypothesis still remains to be validated by experimental data.
Variegation mutants are valuable for the study of chloroplast biogenesis mechanisms. The study of suppressors of variegation mutants, such as var2 (variegation2) and thf1 (thylakoid formation1), has provided lots of insights into the mechanisms of chloroplast development (Yu et al., 2008; Wu et al., 2013, 2016; Hu et al., 2015; Ma et al., 2015). Here, we report another suppressor of thf1, namely sot5 (suppressor of thf1 5), whose gene encodes one of the Arabidopsis PPR proteins, EMB2279. We found that EMB2279/SOT5 is required specifically for plastid rpl2 and trnK intron splicing. The sot5 defects in plastid translation and leaf morphogenesis may indicate a role of plastid gene expression in regulating leaf development.
RESULTS
Identification and Characterization of the sot5 Mutant
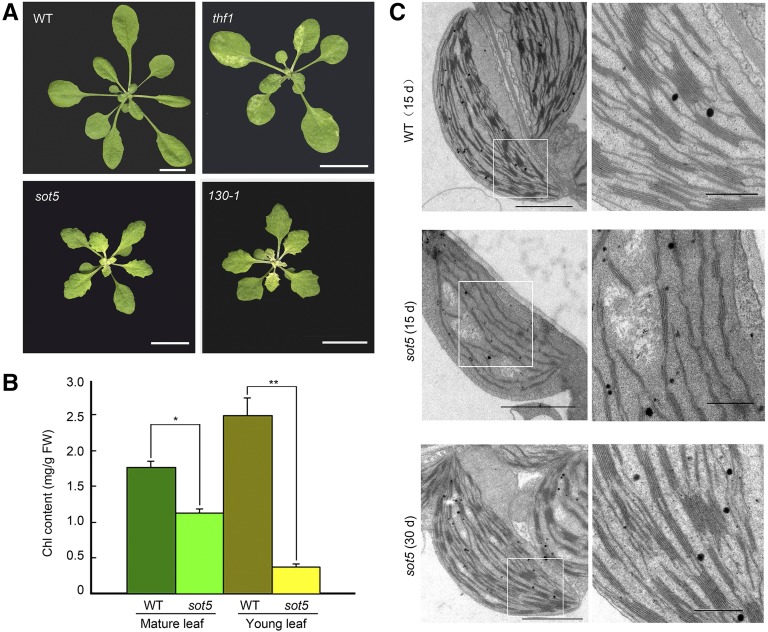
To investigate the molecular mechanisms by which THF1 mutations lead to defects in chloroplast development, we mutagenized thf1 seeds with ethyl methanesulfonate and screened for suppressors of thf1 leaf variegation, as reported previously (Wu et al., 2013, 2016; Hu et al., 2015; Ma et al., 2015). The sot5 single mutant was isolated from the F2 generation of the cross between a suppressor line, 130-1, and Columbia-0 (Col-0). Both sot5 and 130-1 displayed the same phenotype, such as virescent and serrated leaves (Fig. 1A). sot5 cotyledons were yellowish and contained about 74% of the chlorophyll (Chl) content of wild-type cotyledons (Supplemental Fig. S1, A and B). By contrast, young true leaves of sot5 were initially pale yellow but gradually became light green (Fig. 1A; Supplemental Fig. S1C). Total Chl analysis showed that sot5 had about 15% and 64% of the Chl content of the wild type in young and mature leaves, respectively (Fig. 1B). Transmission electron microscopy analysis of chloroplast ultrastructure demonstrated that chloroplasts in sot5 leaves were initially smaller and had fewer thylakoid membranes than those in wild-type leaves (Fig. 1C), but this difference was not apparent in mature leaves (Fig. 1C). Taken together, these results indicate that SOT5 is required for chloroplast development.
Figure 1.
Identification and characterization of the sot5 mutant. A, Phenotypes of 30-d-old wild-type (WT), thf1, sot5, and 130-1 plants. Bars = 1 cm. B, Chlorophyll contents of mature and young leaves from 45-d-old wild-type and sot5 plants. The data represent means ± sd of three biological replicates. Asterisks indicate significant differences between the wild type and sot5 (Student’s t test, *, P < 0.05 and **, P < 0.01). FW, Fresh weight. C, Ultrastructure of chloroplasts in the first wild-type and sot5 true leaves sampled from 15- or 30-d-old plants. The images at right are closeups of the boxes in the left images. Bars = 2 μm at left and 0.5 μm at right.
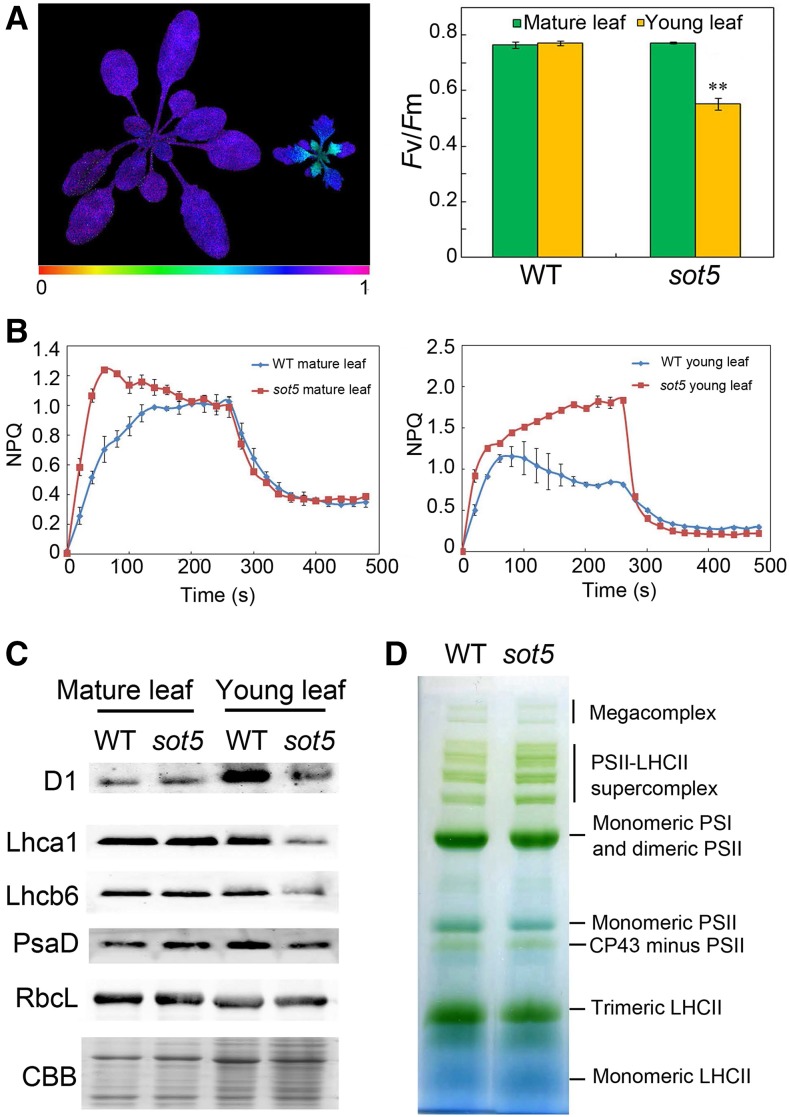
We also analyzed the photosynthetic capacity of sot5. The Fv/Fm value, which represents the maximum quantum yield of PSII photochemistry, was significantly lower in young leaves of sot5 than those of the wild type but was comparable in the mature leaves between sot5 and the wild type (Fig. 2, A and B). NPQ, which is an important photoprotective mechanism to dissipate excessive light energy absorbed by antennae as heat, increased more rapidly in sot5 than in the wild type in both mature and young leaves, whereas there was no obvious difference in NPQ relaxation in the dark between sot5 and the wild type (Fig. 2B), and young sot5 leaves maintained a higher level of NPQ than wild-type leaves in light (Fig. 2B). Immunoblot analysis showed that all photosynthetic proteins examined, such as D1, Lhca1, Lhcb6, PsaD, and RbcL, accumulated to apparently significantly lower levels in sot5 young leaves (Fig. 2C) compared with the wild type, but the difference of these proteins between the wild type and sot5 disappeared in mature leaves. Further analysis of photosynthetic complexes in sot5 leaves using blue-native (BN) PAGE assays showed that sot5 contained the same pattern of photosynthetic complexes as the wild type (Fig. 2D). Together, these results indicate that loss-of-function SOT5 reduces photosynthetic efficiency and capacity.
Figure 2.
Photosynthetic characterization of sot5 mutants. A, Analysis of Fv/Fm of 30-d-old wild-type (WT) and sot5 plants. The graph at right shows quantified Fv/Fm of the mature and young leaves shown at left (Chl fluorescence image). The data represent means ± sd (n = 6). Asterisks indicate a significant difference between the wild type and sot5 (Student’s t test, **, P < 0.05). B, Nonphotochemical quenching (NPQ) measurement of mature and young leaves from 30-d-old wild-type and sot5 plants. The data represent means ± sd (n = 6). C, Immunoblot analysis of photosynthetic proteins accumulated in mature and young leaves from 30-d-old wild-type and sot5 plants. Coomassie Brilliant Blue (CBB) staining is shown to check the difference in sample loading. D, BN-PAGE analysis of photosynthetic complexes in thylakoid membranes isolated from 30-d-old wild-type and sot5 plants. Each lane was loaded with 6 μg of Chl.
Leaf Morphology and Cell Proliferation Were Altered in sot5
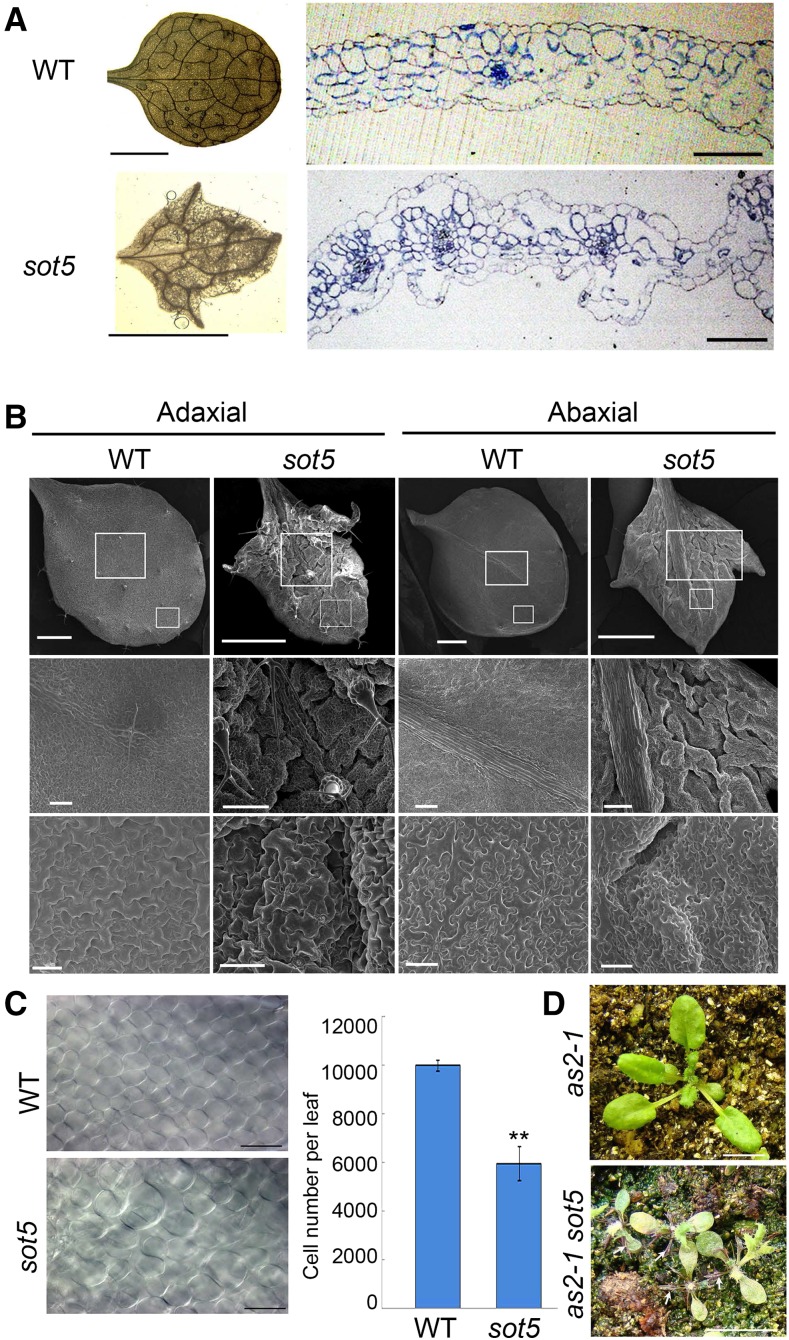
In addition to photosynthesis-related defects, sot5 mutants displayed leaf morphology defects. The first true leaf of sot5 had serrations and simplified vascular bundles (Fig. 3A), and cross sections of the first leaf showed that the epidermis of sot5 was not as smooth as that of the wild type and that the air spaces between mesophyll cells were larger in sot5 than in the wild type (Fig. 3A). In addition, the anatomical identity of palisade and spongy cells disappeared in sot5 leaves (Fig. 3A), indicating that the leaf adaxial-abaxial polarity was altered in sot5 mutants. Using scanning electron microscopy to examine the structure of abaxial and adaxial cells in sot5 and wild-type leaves, we found that both adaxial and abaxial sides of sot5 leaves were severely wrinkled and that adaxial epidermal cells resembled abaxial ones (Fig. 3B), suggesting reduced adaxial cell identity in the sot5 mutant. Since sot5 leaves are small and their mesophyll cells are loosely arranged, we hypothesized that the number of mesophyll cells was reduced in sot5. As expected, the number of mesophyll cells in sot5 was about 60% of the wild-type value (Fig. 3C), indicating that cell proliferation is inhibited in sot5 leaves. Together, these results indicate that SOT5 plays an important role in establishing abaxial-adaxial leaf polarity.
Figure 3.
Leaf morphology and adaxial-abaxial polarity are altered in sot5. A, Morphology and cross section of the first true leaves from 25-d-old wild-type (WT) and sot5 plants. Bars = 5 mm at left and 100 µm at right. B, Scanning electron microscopy analysis of adaxial and abaxial surfaces of the first true leaves from 25-d-old wild-type and sot5 plants. Enlargements of the larger and smaller squares in the top row are shown in the middle and bottom rows, respectively. Bars = 1 mm in the top row, 200 µm in the middle row, and 50 µm in the bottom row. C, Comparison of palisade cell size (left) and number (right) from the first true leaves of 25-d-old wild-type and sot5 plants. The data represent means ± sd of 10 leaves. Asterisks indicate a significant difference between the wild type and sot5 (Student’s t test, ** P < 0.05). Bars = 50 µm. D, sot5 enhances the leaf adaxial-abaxial polarity of as2-1. White arrows indicate filamental leaves. Bars = 1 cm.
In Arabidopsis leaf development, ASYMMETRIC LEAVES1 (AS1) and AS2 form an epigenetic repressor complex that plays a critical role in repressing abaxial-determining genes and promotes adaxial polarity formation (Husbands et al., 2015). AS1 or AS2 loss-of-function mutants exhibit mild abaxialized leaves, such as lotus-like leaves (Xu et al., 2003). Previous work has demonstrated that the Arabidopsis mutants with altered leaf cell proliferation and leaf adaxial-abaxial patterning (asymmetric leaves1/2 enhancer7, elongator, and scabra1 [sca1]) can enhance the as2 phenotype, resulting in narrow and even needle-like (abaxialized) leaves (Yuan et al., 2010; Xu et al., 2012; Mateo-Bonmatí et al., 2015). To genetically test whether leaf polarity is altered in sot5 mutants, we generated double sot5 as2 mutants and found that they displayed many narrow or needle-like leaves (Fig. 3D), supporting our hypothesis that sot5 is an enhancer of as2. Together, these results indicate that SOT5 has a role in promoting leaf adaxial identity formation.
SOT5 Encodes a Chloroplast-Localized PPR Protein
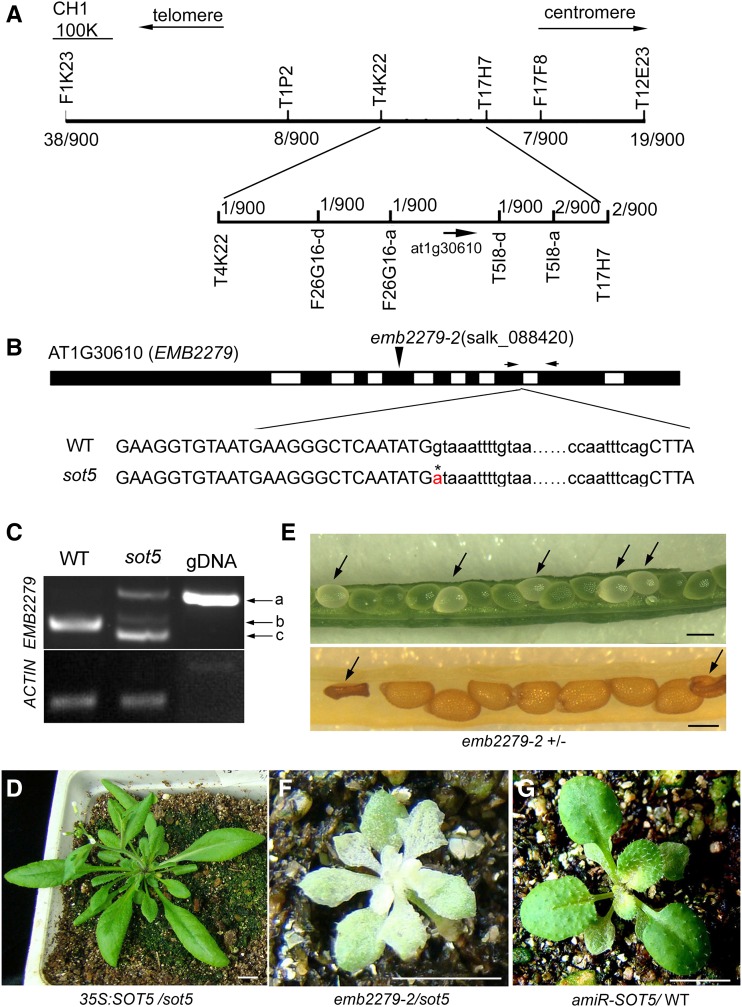
Genetic analysis showed that the sot5 phenotype was controlled by a single nuclear recessive mutation. To clone sot5, we produced an F2 population from the cross between 130-1 and Landsberg erecta (Ler) for map-based cloning. Using 900 recombinant plants, we mapped the sot5 gene on the upper arm of chromosome 1 within an ∼70-kb region between F26G16-a and T5I8-d markers, in which there are 20 annotated genes (Fig. 4A). Bioinformatic analysis revealed that, among these 20 candidates, six genes encode chloroplast-localized proteins, one of which is a PPR protein (AT1G30610), named EMB2279. DNA sequencing showed a G-to-A substitution at the first nucleotide of the seventh intron in AT1G30610 (Fig. 4B). Reverse transcription (RT)-PCR analysis using the primer pair shown in Figure 4B detected three variants of the transcripts in sot5 but only one in the wild type (Fig. 4C). Sequencing of these three transcripts revealed that the longest transcript (a in Fig. 4C) contained the seventh intron, the middle transcript (b in Fig. 4C) was the same as the wild type, and the shortest transcript (c in Fig. 4C) lacked 22-bp nucleotides at the 3′ end of the seventh exon. The 22-bp nucleotide deletion resulted in a truncated form of SOT5. Thus, the sot5 point mutation leads to a low efficiency of the intron splicing and missplicing of the exon. By contrast, we did not detect any alternative splicing events in the seventh intron of the SOT5 gene in the wild type, which is contrary to recorded data in TAIR, where two transcriptional variants with or without the seventh intron of the SOT5 gene are described. Based on our data, SOT5 encodes only one protein with 978 amino acid residues.
Figure 4.
SOT5 encodes EMB2279, a PPR protein. A, Map-based cloning of SOT5. SOT5 was mapped between two markers, F26G16-a and T5I8-d, on chromosome 1. The molecular markers used for fine-mapping are shown. Numbers indicate the recombinant sot5 mutants in 900 F2 individuals from crosses between sot5 (Col-0 ecotype) and Ler. B, Model for the SOT5 (AT1G30610) gene structure. The flanking sequences of the seventh intron are shown below the gene model. The G-to-A point mutation is labeled with an asterisk and red font in sot5 at the first position of the seventh intron. The position of the T-DNA insertion in emb2279-2 (SALK_088420) is shown above the gene model. The arrows show the primers used for RT-PCR. WT, Wild type. C, RT-PCR analysis of SOT5 transcripts in Col-0 and sot5 by using the primer pair shown in B. There are three bands, designated a, b, and c, in sot5. Genomic DNA (gDNA) was used as a temperate control, while the expression of ACTIN was used as a positive control. D, Overexpression of SOT5 cDNA complemented the phenotype of sot5. Bar = 1 cm. E, The null mutation emb2279-2 is embryo lethal. The top image shows the albino seeds (arrows) in a young emb2279-2 heterozygote silique. The bottom image shows the aborted seeds (arrows) in an older emb2279-2 heterozygote silique. Bars = 1 mm. F, Allelic test of sot5 and emb2279-2. The F1 plant of sot5/emb2279-2 appears albino and is seedling lethal. Bar = 1 cm. G, Knockdown lines of SOT5 by the artificial microRNA technique exhibit a leaf virescent phenotype. Bar = 1 cm.
To confirm the map-based cloning result, we transformed the SOT5 cDNA under the control of the cauliflower mosaic virus 35S promoter into the sot5 mutant. All the sot5 phenotypes were complemented by overexpressing SOT5 (Fig. 4D), confirming that SOT5 encoding the 978-amino acid polypeptide is functional. A previous study reported that the null mutation emb2279-2 is embryo lethal (Lurin et al., 2004; Cushing et al., 2005), causing embryo development to stop at the globular stage (Tzafrir et al., 2003). To determine whether emb2279 and sot5 are alleles of the same gene, we first identified emb2279-2 (SALK_088420) from the Arabidopsis Biological Resource Center (ABRC) stock. Indeed, about 25% of the seeds were albino and aborted in the emb2279-2 heterozygote siliques (Fig. 4E), confirming the embryo-lethal phenotype of emb2279. We then crossed sot5 with the emb2279-2 heterozygote. We found that half of the F1 plants were almost albino and half of them were wild type like when they were cultured on one-half-strength Murashige and Skoog medium. The albino plants could turn a bit green when growing on the medium but could not survive after transfer to the soil, since their leaves were severely pale (Fig. 4F). Furthermore, we generated SOT5 knockdown lines by an artificial microRNA technique. Transgenic plants with low expression levels of SOT5 showed a virescent phenotype (Fig. 4G). Taken together, our data suggest that sot5 is a weak allele of emb2279.
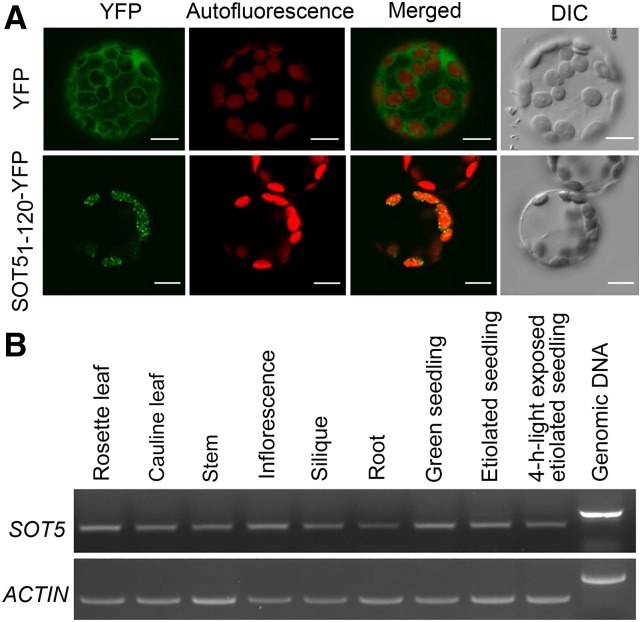
Although SOT5/EMB2279 was predicted to target chloroplasts, no evidence supports this prediction. To test this, we made a construct in which the first N-terminal 120 amino acids containing the transit peptide of SOT5 were fused to yellow fluorescent protein (YFP) driven by the cauliflower mosaic virus 35S promoter and transformed it into protoplasts. Confocal microscopy analysis showed that YFP signal was totally overlapped with Chl autofluorescence (Fig. 5A), indicating that SOT5 is localized in chloroplasts. RT-PCR analysis showed that SOT5 was expressed ubiquitously in all examined tissues (Fig. 5B): in 5-d-old green or etiolated seedlings and in etiolated seedlings that had been exposed to light for 4 h (Fig. 5B). These results indicate that SOT5 functions in both green and nongreen tissues.
Figure 5.
Subcellular localization of SOT5 and tissue expression patterns of SOT5. A, Microscopy analysis of the YFP fusion protein SOT51-120-YFP transiently expressed in Arabidopsis protoplasts. The green fluorescence of SOT51-120-YFP was overlapped with chloroplast autofluorescence in merged images. DIC, Differential interference contrast microscopy. Bars = 10 µm. B, RT-PCR analysis of SOT5 expression in various tissues and seedlings. Total RNA was extracted from rosette and cauline leaves, stems, inflorescences, siliques, and roots of 40-d-old plants. Total RNA was extracted from 5-d-old seedlings grown in light or darkness or etiolated seedlings exposed to light for 4 h. Genomic DNA was used as a template control, and ACTIN expression was used as a loading control of RNA.
SOT5 Is Required for Splicing of the Plastid rpl2 and trnK Introns
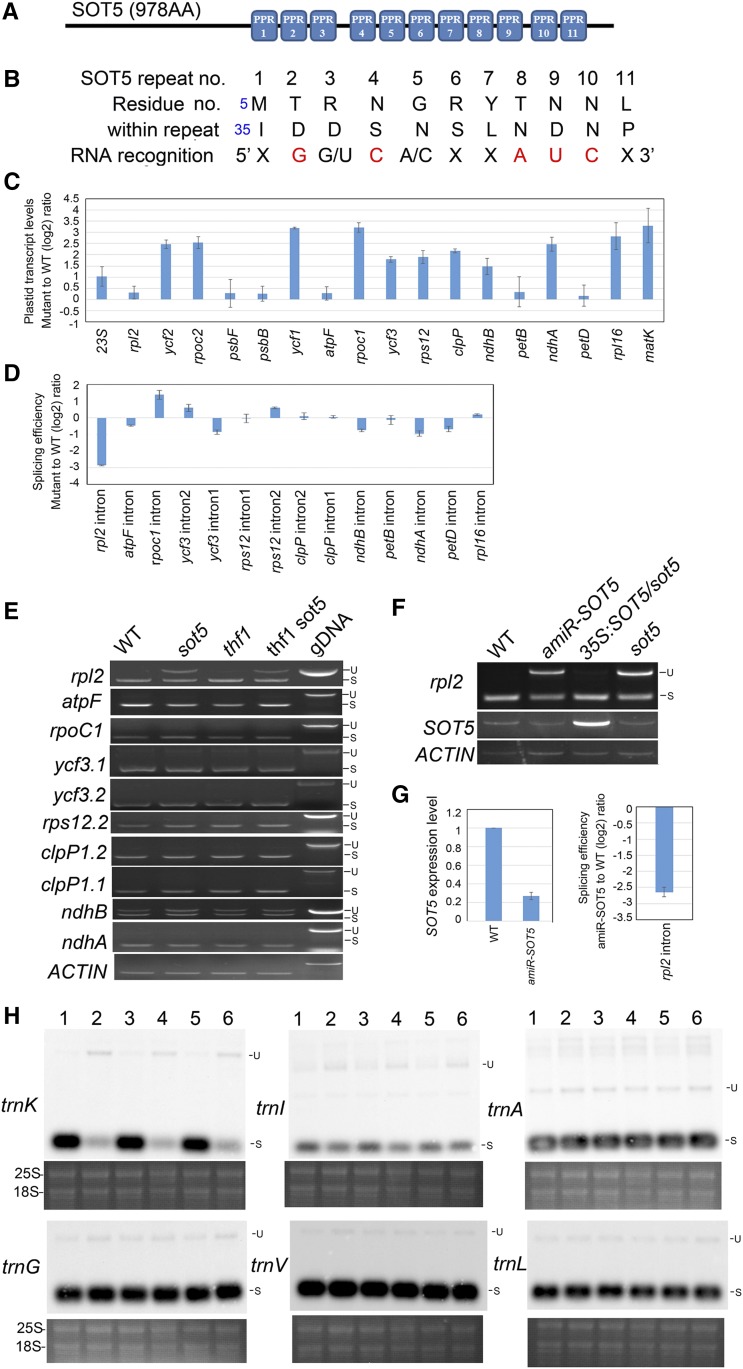
SOT5 is a P-subfamily PPR protein composed of 978 amino acid residues. ScanProsite (https://prosite.expasy.org/) analysis showed that SOT5 has 11 PPR motifs at the C terminus and no other function-known domains at the N terminus (Fig. 6A). BLAST searches demonstrated that SOT5 is a single-copy gene in Arabidopsis and is conserved in monocots and dicots, but not in Bryophyta, Lycopodiophyta, Chlorophyta, and Rhodophyta (http://plants.ensembl.org/Arabidopsis_thaliana/Gene/Compara_Ortholog?db=core;g=AT1G30610;r=1:10846513-10850724). The 11-PPR motifs of SOT5 also are present in the mosses Physcomitrella patens and Marchantia polymorpha, in the fern Selaginella moellendorffii, but not in green algae (Supplemental Fig. S2).
Figure 6.
SOT5 is required for the splicing of the plastid rpl2 and trnK introns. A, Schematic SOT5 protein containing 11 PPR motifs. B, Predicted 11 ribonucleotides targeted by the 11 PPR motifs shown in A. In each repeat, the two key amino acid residues and their nucleotide targets are shown. X indicates unpredictable; two nucleotides with a slash mean optional; the red nucleotides are precisely predicted. C, RT-qPCR analysis of relative expression levels of the plastid genes containing the predicted target sequences or group IIA introns in sot5. Three biological replicates were analyzed. D, Splicing efficiency analysis of 14 plastid introns in sot5 by RT-qPCR. Three biological replicates were analyzed. E, RT-PCR analysis of intron retention in eight plastid genes. Genomic DNA (gDNA) was used as a template control. S, Spliced; U, unspliced. F, RT-PCR analysis of rpl2 intron splicing in wild-type (WT), amiR-SOT5/sot5, 35S:SOT5/sot5, and sot5 plants. G, Analysis of SOT5 transcript levels and splicing efficiency of rpl2 in wild-type and amiR-SOT5 plants by RT-qPCR. H, Northern-blot analysis of plastid trn genes that contain introns in different genotypes. Lanes 1 to 6 indicate the wild type, sot5, thf1, thf1 sot5, 35S:SOT5/sot5, and amiR-SOT5, respectively. The ethidium bromide-stained gel under each blot is shown as a loading control. Two biological replicates were analyzed, and one representative result is shown.
Since PPR proteins participate in RNA processing, such as RNA splicing, editing, maturation, and stabilization, through directly recognizing specific RNA sequences, we propose that the target of SOT5 also might be conserved in higher plants. To identify the targets of SOT5, we first predicted its binding target, the single-stranded RNAs, according to the PPR code and previously identified single-stranded RNAs targeted by different PPRs (Barkan et al., 2012; Yin et al., 2013; Yagi et al., 2014). Our analysis revealed that the 11 ribonucleotides targeted by the 11 PPR motifs of SOT5 are XG(G/U)C(A/C)XXAUCX, with X representing any nucleotide that cannot be precisely predicted and nucleotides in parentheses being optional (Fig. 6B). We then searched the plastome using this potential SOT5-binding sequence and found that 14 plastid genes, ATCG00860 (ycf2.1), ATCG01280 (ycf2.2), ATCG00170 (rpoC2), ATCG00950 (23S.1 rRNA), ATCG01180 (23S.2 rRNA), ATCG00030 (trnK), ATCG01130 (ycf1.2), ATCG00100 (trnG), ATCG00360 (ycf3), ATCG00570 (psbF), ATCG00680 (psbB), ATCG00830.1 (rpl2.1), ATCG01310 (rpl2.2), and ATCG01100 (ndhA), contain the sequence (Supplemental Table S1). Based on these data, we designed primers flanking the binding sites and analyzed the expression levels of these candidate genes by reverse transcription-quantitative PCR (RT-qPCR). None of these genes were down-regulated significantly in sot5 (Fig. 6C), but some NEP genes, such as rpoc2, ycf1, and ycf2, were up-regulated in sot5 (Fig. 6C). These results indicate that SOT5 is not required for the RNA stability of these candidate genes.
Among these candidate genes, rpl2, ndhA, trnK, and trnG contain the group II intron. Therefore, we investigated whether SOT5 was involved in the intron splicing of these chloroplast genes. RT-qPCR analysis showed that the intron splicing efficiency for only rpl2 was reduced significantly in sot5 (Fig. 6D). Further RT-PCR analysis using the primers flanking the group II intron showed that the precursor levels of rpl2 transcripts were accumulated in sot5 and thf1 sot5 but not in the wild type and thf1, but no difference was detected in transcript levels of other intron-containing genes between the wild type and sot5 (Fig. 6E). Consistently, the levels of unspliced rpl2 transcripts accumulated in the SOT5 knockdown line (amiR-SOT5) but were restored to wild-type levels in complementary lines (35S:SOT5/sot5; Fig. 6, F and G). Since mature tRNAs are small and have strong secondary structure and base modifications, they are difficult to detect precisely by RT-PCR (de Longevialle et al., 2008). To assess mature tRNA and precursor levels, we employed northern-blot analysis. Our data demonstrated that only trnK unspliced precursors accumulated significantly in sot5, sot5 thf1, and amiR-SOT5, and trnK spliced mature products were reduced dramatically in these genotypes compared with the wild type and 35S:SOT5/sot5 (Fig. 6H), suggesting that SOT5 mutations result in the reduced splicing efficiency of trnK. We were unable to verify the binding site of SOT5 in vitro, because the recombinant SOT5 protein was not expressed in prokaryotic and eukaryotic systems.
Since the matK gene, which is required for the splicing of seven group IIA introns in trnA, trnI, trnV, atpF, rpl2, rps12-2, and trnK (Zoschke et al., 2010), is located in the trnK intron, it is possible that lowered trnK intron splicing efficiency in sot5 would affect the level of matK transcripts and subsequently affect the intron splicing of all group IIA genes. Moreover, decreased rpl2 and trnK might affect plastid protein translation, leading to a decrease in MatK activity in sot5. However, our results demonstrated that the relative level of matK transcripts was enhanced in sot5 (Fig. 6C), probably due to the higher accumulation of trnK precursors in sot5, and the splicing efficiency of atpF, rps12-2, trnA, trnI, and trnV was not affected significantly by SOT5 mutations (Fig. 6, C and H). These results suggest that, in sot5 mutants, MatK activity is high enough to maintain the splicing of these introns, and the reduced splicing efficiency of rpl2 and trnK should be caused directly by SOT5 mutations.
SOT5 Mutations Also Suppress the Leaf Variegation of var2 and Inhibit Plastid rRNA Processing
Since thf1 leaf variegation is attributed to a low level of var2 (Zhang et al., 2009), we tested whether sot5 can suppress the variegation phenotype of var2. Genetic analysis showed that var2 sot5 double mutants displayed a similar phenotype to sot5 mutants (Fig. 7A), suggesting that sot5 is a suppressor of var2. This is consistent with the previously reported results that a decrease of plastid translation efficiency can suppress leaf variegation (Yu et al., 2008; Ma et al., 2015; Wu et al., 2016).
Figure 7.
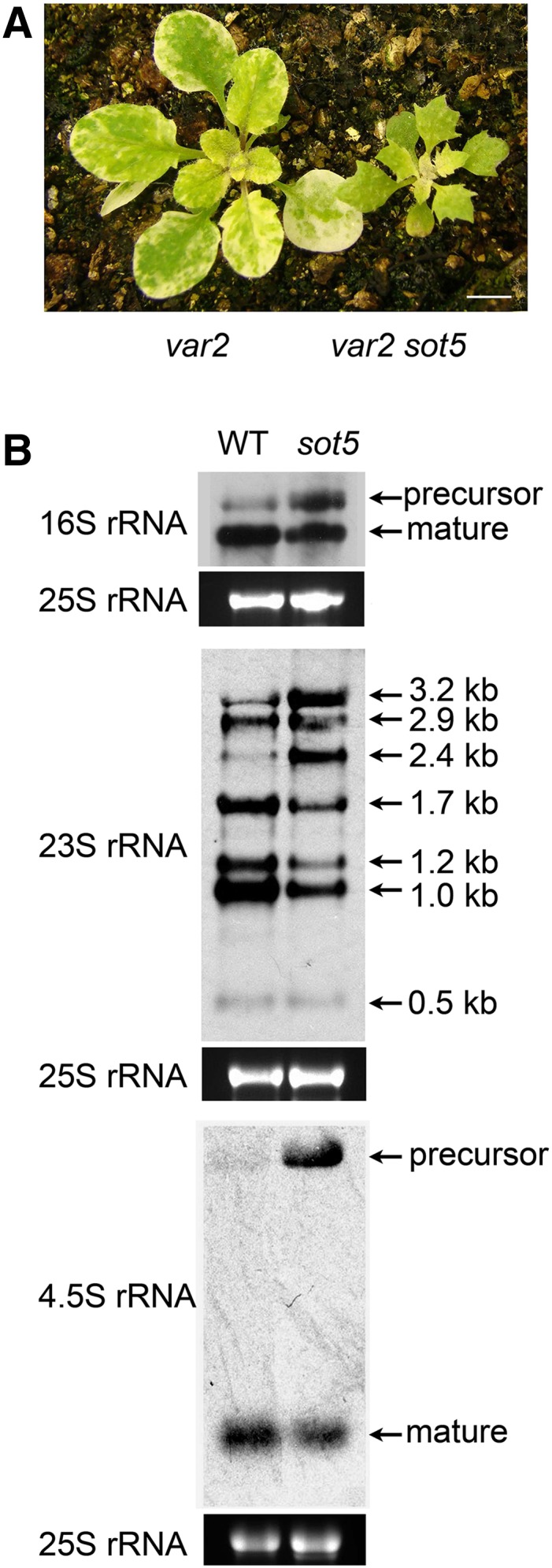
Loss-of-function sot5 suppresses leaf variegation of var2 and inhibits plastid rRNA processing. A, Phenotypes of 30-d-old var2 and var2 sot5 plants. sot5 also can suppress the leaf variegation phenotype of var2. Bar = 1 cm. B, Northern-blot analysis of plastid rRNA processing in the wild type (WT) and sot5. Ethidium bromide staining of the 25S rRNA is shown below each blot to indicate differences in gel loading.
It was reported previously that mutants deficient in plastid translation display a reduced efficiency of plastid rRNA processing (Yu et al., 2008; Ma et al., 2015; Wu et al., 2016). Based on this, we investigated the effect of sot5 mutations on rRNA processing. Northern-blot analysis showed that levels of mature 16S, 23S, and 4.5S rRNA were reduced in sot5 compared with the wild type, whereas their precursors overaccumulated (Fig. 7B), similar to our previously reported mutants, prpl11-2 and prps9 (Supplemental Fig. S3), which were defective in plastid ribosomal protein large subunit 11 and ribosomal protein small subunit 9, respectively (Wu et al., 2013; Ma et al., 2015). These results indicate that plastid rRNA processing is linked to ribosome biogenesis and that the sot5 mutant is indeed defective in ribosome biogenesis and protein translation in plastids due to the reduced splicing of rpl2 and trnK genes. The reduced efficiency of plastid rRNA processing may be the indirect effect of a translation defect in sot5. However, since 23S rRNA also was predicted to be the target of SOT5, we cannot rule out the possibility that SOT5 also can bind directly to 23S rRNA and facilitate its processing.
DISCUSSION
A Role of SOT5 in rpl2 and trnK Intron Splicing
In this study, we identified a PPR protein, SOT5/EMB2279, that is involved in the splicing of the plastid rpl2 and trnK introns. Since both Rpl2 and trnK are essential for plastid protein translation, complete disruption of SOT5 would lead to embryo lethality in Arabidopsis, similar to previously reported results for EMB2279 (Lurin et al., 2004; Cushing et al., 2005). The sot5 mutant contains a point mutation that leads to a low efficiency of the seventh intron splicing and missplicing of the seventh exon, resulting in a hypomorphic allele of emb2279. In TAIR, the SOT5 locus (AT1G30610) is recorded to produce two types of mRNA depending on whether the seventh intron is removed or not: the longer transcript encodes a 1,006-amino acid-long peptide with 10 PPR motifs, whereas the shorter one encodes a 978-amino acid-long peptide with 11 PPR motifs. However, we only detected the shorter transcript, which lacks 84 bp compared with the longer one, indicating that SOT5 has 11 PPR motifs. Consistent with the previous reports (Lurin et al., 2004; Cushing et al., 2005), total disruption of SOT5 leads to embryo lethality in Arabidopsis, whereas knocking down SOT5 expression through the artificial microRNA technique or partially knocking out the function of SOT5 by a point mutation resulted in a virescent phenotype. These results are consistent with the fact that most of the suppressors for thf1 and var2 leaf variegation are mutants defective in the plastid translation system (Miura et al., 2007; Yu et al., 2008; Ma et al., 2015; Wu et al., 2016).
It is generally recognized that binding sites of PPR proteins that are involved in intron splicing are located in the intron (Schmitz-Linneweber et al., 2006; Beick et al., 2008; de Longevialle et al., 2008, 2010), which is helpful for maintaining RNA conformation and the subsequent recruitment of general splicing factors to facilitate intron splicing. Consistently, the predicted SOT5-binding site is located in the intron of trnK. In contrast, the predicted SOT5-binding site is located in the exon of rpl2, although there is a shorter motif containing eight nucleotides that overlapped with the predicted SOT5-binding site in the rpl2 intron (523–530; Supplemental Table S1). Based on this, it is possible that SOT5 can facilitate both rpl2 intron splicing and transcript stabilization. Considering that the plastid rpl2 intron was independently lost in several species, such as in the Caryophyllales and Lythraceae (Downie et al., 1991; Gu et al., 2016), it will be interesting to examine whether the role of SOT5 homologs in these species is associated with rpl2 transcript stabilization. In addition, we cannot exclude the possibility that the binding of SOT5 to the rpl2 exon also is required for intron splicing. This hypothesis is consistent with the notion that the specific pattern of OTP51 action cannot be explained by an intron feature, because no conserved RNA motif was found in the ycf3.2, atpF, trnV, and trnK group IIA introns (de Longevialle et al., 2008). Whether PPR protein-binding sites can be located in an exon needs to be investigated further in the future.
Retrograde Signaling from Plastids to the Nucleus Regulates Leaf Development
Besides the leaf virescent phenotype, sot5 displays multiple defects in leaf development, such as serrated leaves and less vascular bundles (Fig. 3). It is well documented that abnormal leaf morphology occurs frequently in mutants with dysfunctional chloroplasts: for example, in DEFECTIVE CHLOROPLAST AND LEAVES, functioning in plastid rRNA maturation (Keddie et al., 1996; Bellaoui et al., 2003); SCA3, encoding a plastidic RpoTp RNA polymerase (Hricová et al., 2006); SCA1, encoding a plastid-type ribosomal protein S5 (Mateo-Bonmatí et al., 2015); ENF2, encoding a chloroplast-localized and functionally unknown protein, with low similarity to the bacterial polyamine transporters PotD and PotF (Tameshige et al., 2013); and ANGULATA7, encoding a DnaJ-like zinc finger domain protein localized in chloroplasts (Muñoz-Nortes et al., 2017). It has been suggested that the effect of chloroplast development on leaf morphology is associated with improper cell proliferation and expansion during leaf development (Mateo-Bonmatí et al., 2015). Recent studies showed that the serrated leaves and abnormal vascular development in initiation factor3, encoding a plastid translation initiation factor, were linked to altered auxin homeostasis and auxin-regulated pathways (Zheng et al., 2016); however, it remains unknown how chloroplast gene expression affects auxin biosynthesis and/or distribution. Our study also demonstrated that the layer of vertically elongated palisade cells beneath the adaxial epidermis was missing in sot5 (Fig. 3), suggesting that adaxial domain formation is inhibited in sot5 leaf primordia. Furthermore, the as2 sot5 double mutant had more severe abaxialized filamentous leaves, which were similar to those of sca1 as2 mutants. Both sca1 and sot5 are defective in a plastid ribosomal subunit (Mateo-Bonmatí et al., 2015), indicating that plastid translation may trigger a retrograde signal that regulates leaf polarity development. The sot5 mutant provides ideal material in which to investigate the molecular mechanisms by which leaf development is feedback regulated by plastid gene expression for improved photosynthesis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was used as the wild type in this study. Single mutants used in this study were described previously: thf1-1 (Huang et al., 2006), var2 (Yu et al., 2004) and as2-1 (Xu et al., 2003). The emb2279-2 (SALK_088420) mutant that was reported previously (Cushing et al., 2005) was obtained from the ABRC stock center (https://abrc.osu.edu/) and was genotyped according to the guidelines described there. Double mutants were identified in F2 generations derived from crosses between single mutants by the PCR-based genotyping procedure. Seeds were surface sterilized by 75% (v/v) ethanol and stratified at 4°C for 3 d and then sown onto one-half-strength Murashige and Skoog medium with 1% Suc, or seeds were sown directly into soil and grown in a phytotron with long-day conditions (16 h of light/8 h of dark) and light intensity (100 μmol photons m−2 s−1) at 22°C.
Map-Based Cloning, Plasmid Construction, and Transformation
Suppressors were identified by screening M2 seeds that were mutagenized by 0.1% (w/v) ethyl methanesulfonate in the thf1 genetic background. The suppressor line was crossed to Ler, and the F2 seedlings were used as mapping populations. The mutation was mapped using simple sequence length polymorphism markers, and information is available at TAIR (http://www.arabidopsis.org). The SOT5/EMB2279 coding DNA sequence (CDS) and the CDS for the first 120 amino acids including transit peptide sequence (fragment CDS) were cloned into the pENTR SD/D-TOPO entry vector (Invitrogen). The SOT5/EMB2279 CDS was then recombined into the pGWB2 destination vector (Nakagawa et al., 2007). Artificial microRNA of SOT5 was built according to Schwab et al. (2006). The destination vectors were transformed into Col-0 or sot5 mutants according to the method described by Clough and Bent (1998). For transient subcellular localization analysis, the coding sequence of the first 120 amino acids including transit peptide sequence was recombined into the p2GWY7 vector (http://gateway.psb.ugent.be/vector/show/p2GWY7/search/index/) and transformed into the Arabidopsis protoplasts according to the method of Wu et al. (2009).
Analyses of Chl Content, Chl Fluorescence, and Chloroplast Ultrastructure
Total Chl was extracted with 80% acetone at 4°C for 24 h in darkness. The levels of Chl a and Chl b were determined as described by Porra et al., 1989. Chl fluorescence was analyzed using Imaging PAM 101 (Walz), and Fv/Fm values were determined according to the manufacturer’s instructions. For chloroplast ultrastructure analysis, the first true leaves of 15- and 30-d-old plants were sampled and fixed in a solution of 4% glutaraldehyde, then processed, embedded, and viewed via transmission electron microscopy as described (Zhou et al., 2009).
Protein Extraction, Immunoblot, and BN-PAGE Analyses
Protein extraction, immunoblot, and BN-PAGE analyses were performed as described previously (Huang et al., 2013).
Microscopy
Thin-section specimens and leaf samples for scanning electron microscopy were prepared according to our previous report (Huang et al., 2006). Samples for measuring and observing leaf mesophyll cells were processed as described by Xue et al. (2015).
RNA Isolation and RT-qPCR
Total RNA was extracted from leaves of 25-d-old plants using an RNeasy plant mini kit (Qiagen), according to the manufacturer’s instructions. Downstream DNase I treatment and RT-PCR steps were conducted as described previously by Huang et al. (2013). Quantitative and semiquantitative RT-PCR of chloroplast genes was carried out using the gene-specific primers described previously by de Longevialle et al. (2008) and designed by ourselves (Supplemental Table S2). Three biological repeats were analyzed, and each sample was run in triplicate. The data set was normalized using ACTIN2 as a reference. The method to quantify the transcript level and splicing efficiency of plastid genes was described previously (de Longevialle et al., 2008).
Northern-Blot Analysis
For northern-blot analysis of plastid tRNAs, the following biotinylated oligomer probes, which were described previously by de Longevialle et al. (2008), were synthesized by Thermo Fisher Scientific: trnK, 5′-AAAGCCGAGTACTCTACCGTTG-3′; trnL, 5′-TGGGGATAGAGGGACTTGAAC-3′; trnG, 5′-CATCGTTAGCTTGGAAGGCTAAGGG-3′; trnI, 5′-CTACCACTGAGCTAATAGCCC-3′; trnV, 5′-GTGTAAACGAGGTGCTCTACCTAA-3′; and trnA, 5′-GGACTCGAACCGCTGACATCCGCC-3′. Each probe sequence is in its exon. Total RNAs used for northern blot were extracted from 15-d-old seedlings according to the manufacturer’s instructions (Promega Denaturing Solution; Z5651). A total of 1 to 2 μg of RNA was run on 2% agarose/formaldehyde gels, transferred to positively charged nylon membranes (Roche; 11417240001), and hybridized with biotinylated oligomer probes at 42°C. The North2South Chemiluminescent Hybridization and Detection Kit (Thermo Fisher Scientific; 17097) was used for the downstream steps according to the kit instructions. Northern-blot analysis of plastid rRNAs was performed as described previously (Wu et al., 2016).
Prediction of SOT5-Binding Sequences
SOT5-binding sequences were predicted as described previously (Barkan et al., 2012; Yin et al., 2013; Yagi et al., 2014; Shen et al., 2015, 2016). In brief, each PPR motif coordinates with one RNA base, and the combinations of the fifth and 35th amino acids (known as the PPR code) within each motif confer RNA-specific binding. The PPR motif sequences from SOT5 were used to search against the previously defined RNA recognition database for PPRs, leading to the following predicted recognition sequence: XG(G/U)C(A/C)XXAUCX, where X indicates a nucleotide that cannot be predicted precisely. The Patmatch tool (https://www.arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl) was used to predict the binding sequences according to the SOT5 PPR code, XGKCMXXATCX (where X means any base, K means G or T, and M means A or C) by searching TAIR10 genes (+introns, +UTRs; DNA) database, with other parameters at default settings (Supplemental Table S1).
Accession Numbers
Sequence data from this article can be found in the TAIR data libraries under the following accession numbers: EMB2279 (AT1G30610.2), rpl2.1 (ATCG00830.1), rpl2.2 (ATCG01310.1), trnK (ATCG00030.1), ycf2.1 (ATCG00860.1), ycf2.2 (ATCG01280.1), rpoc2 (ATCG00170.1), 23S.1 (ATCG00950.1), 23S.2 (ATCG01180.1), ycf1.2 (ATCG01130.1), ycf3 (ATCG00360.1), psbF (ATCG00570.1), psbB (ATCG00680.1), ndhA (ATCG01100.1), trnG (ATCG00100.1), trnA (ATCG00940.1 and ATCG01190.1), trnI (ATCG00930.1 and ATCG01200.1), trnV (ATCG00450.1), and trnL (ATCG00400.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic comparison between wild-type and sot5 seedlings.
Supplemental Figure S2. The PPR motifs of SOT5 are conserved in higher plants.
Supplemental Figure S3. Northern-blot analysis of plastid rRNA processing in sot5, prpl11, and prps9 mutants.
Supplemental Table S1. Plastid genes that are predicted to be targets of SOT5.
Supplemental Table S2. Primer used in this study.
Acknowledgments
We thank the ABRC for providing the emb2279-2 (SALK_088420) allele; X. Gao, J. Li, and Z. Zhang for sample preparation for scanning and transmission electron microscopy; W. Zhou for help with northern blots; and J. Xue for help with leaf mesophyll cell observation.
Footnotes
This study was supported by grants from the Chinese 973 Program (2015CB910900) to P.Y. and W.H., the Chinese National Special Grant for Transgenic Crops (2016ZX08009003-005) and the Shanghai Engineering Research Center of Plant Germplasm Resources (17DZ2252700) to J.H., and the National Natural Science Foundation of China (30900093) to W.H.
Articles can be viewed without a subscription.
References
- Aryamanesh N, Ruwe H, Sanglard LVP, Eshraghi L, Bussell JD, Howell KA, Small I, des Francs-Small CC (2017) The pentatricopeptide repeat protein EMB2654 is essential for trans-splicing of a chloroplast small ribosomal subunit transcript. Plant Physiol 173: 1164–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Bayraktar OA, Barkan A (2008) Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA 14: 2319–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I (2012) A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet 8: e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol 28: 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaoui M, Keddie JS, Gruissem W (2003) DCL is a plant-specific protein required for plastid ribosomal RNA processing and embryo development. Plant Mol Biol 53: 531–543 [DOI] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, des Francs-Small CC, Delannoy E, Kahlau S, Tanz SK, de Longevialle AF, Fujii S, Small I (2011) OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J 65: 532–542 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cushing DA, Forsthoefel NR, Gestaut DR, Vernon DM (2005) Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 221: 424–436 [DOI] [PubMed] [Google Scholar]
- Dai L, Chai D, Gu SQ, Gabel J, Noskov SY, Blocker FJ, Lambowitz AM, Zimmerly S (2008) A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol Cell 30: 472–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle AF, Hendrickson L, Taylor NL, Delannoy E, Lurin C, Badger M, Millar AH, Small I (2008) The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J 56: 157–168 [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Small ID, Lurin C (2010) Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol Plant 3: 691–705 [DOI] [PubMed] [Google Scholar]
- Downie SR, Olmstead RG, Zurawski G, Soltis DE, Soltis PS, Watson JC, Palmer JD (1991) Six independent losses of the chloroplast DNA RPL2 intron in dicotyledons: molecular and phylogenetic implications. Evolution 45: 1245–1259 [DOI] [PubMed] [Google Scholar]
- Glanz S, Kück U (2009) Trans-splicing of organelle introns: a detour to continuous RNAs. BioEssays 31: 921–934 [DOI] [PubMed] [Google Scholar]
- Gu C, Tembrock LR, Johnson NG, Simmons MP, Wu Z (2016) The complete plastid genome of Lagerstroemia fauriei and loss of rpl2 intron from Lagerstroemia (Lythraceae). PLoS ONE 11: e0150752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D, Nickelsen J, Hackert A, Kuck U (1998) A single nuclear locus is involved in both chloroplast RNA trans-splicing and 3′ end processing. Plant J 15: 575–581 [Google Scholar]
- Hricová A, Quesada V, Micol JL (2006) The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol 141: 942–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Zhu Y, Wu W, Xie Y, Huang J (2015) Leaf variegation of Thylakoid Formation1 is suppressed by mutations of specific σ-factors in Arabidopsis. Plant Physiol 168: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pi L, Liang W, Xu B, Wang H, Cai R, Huang H (2006) The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell 18: 2479–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Chen Q, Zhu Y, Hu F, Zhang L, Ma Z, He Z, Huang J (2013) Arabidopsis thylakoid formation 1 is a critical regulator for dynamics of PSII-LHCII complexes in leaf senescence and excess light. Mol Plant 6: 1673–1691 [DOI] [PubMed] [Google Scholar]
- Husbands AY, Benkovics AH, Nogueira FTS, Lodha M, Timmermans MCP (2015) The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell 27: 3321–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Jenkins BD, Kulhanek DJ, Barkan A (1997) Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keddie JS, Carroll B, Jones JDG, Gruissem W (1996) The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J 15: 4208–4217 [PMC free article] [PubMed] [Google Scholar]
- Keren I, Klipcan L, Bezawork-Geleta A, Kolton M, Shaya F, Ostersetzer-Biran O (2008) Characterization of the molecular basis of group II intron RNA recognition by CRS1-CRM domains. J Biol Chem 283: 23333–23342 [DOI] [PubMed] [Google Scholar]
- Khrouchtchova A, Monde RA, Barkan A (2012) A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA 18: 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger TS, Watkins KP, Friso G, van Wijk KJ, Barkan A (2009) A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc Natl Acad Sci USA 106: 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K, Schmidt U (2003) Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol 38: 249–303 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Wu W, Huang W, Huang J (2015) Down-regulation of specific plastid ribosomal proteins suppresses thf1 leaf variegation, implying a role of THF1 in plastid gene expression. Photosynth Res 126: 301–310 [DOI] [PubMed] [Google Scholar]
- Mateo-Bonmatí E, Casanova-Sáez R, Quesada V, Hricová A, Candela H, Micol JL (2015) Plastid control of abaxial-adaxial patterning. Sci Rep 5: 15975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura E, Kato Y, Matsushima R, Albrecht V, Laalami S, Sakamoto W (2007) The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 19: 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Nortes T, Pérez-Pérez JM, Ponce MR, Candela H, Micol JL (2017) The ANGULATA7 gene encodes a DnaJ-like zinc finger-domain protein involved in chloroplast function and leaf development in Arabidopsis. Plant J 89: 870–884 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Ostheimer GJ, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A (2003) Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J 22: 3919–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Ganguly D, Albrecht-Borth V (2015) Insights into chloroplast biogenesis and development. Biochim Biophys Acta 1847: 1017–1024 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectrometry. Biochim Biophys Acta 975: 384–394 10.1016/S0005-2728(89)80347-0 [DOI] [Google Scholar]
- Prikryl J, Rojas M, Schuster G, Barkan A (2011) Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci USA 108: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM. (2016) Group II intron self-splicing. Annu Rev Biophys 45: 183–205 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A (2006) A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18: 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Wang X, Liu Y, Li Q, Yang Z, Yan N, Zou T, Yin P (2015) Specific RNA recognition by designer pentatricopeptide repeat protein. Mol Plant 8: 667–670 [DOI] [PubMed] [Google Scholar]
- Shen C, Zhang D, Guan Z, Liu Y, Yang Z, Yang Y, Wang X, Wang Q, Zhang Q, Fan S, et al. (2016) Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat Commun 7: 11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Tameshige T, Fujita H, Watanabe K, Toyokura K, Kondo M, Tatematsu K, Matsumoto N, Tsugeki R, Kawaguchi M, Nishimura M, et al. (2013) Pattern dynamics in adaxial-abaxial specific gene expression are modulated by a plastid retrograde signal during Arabidopsis thaliana leaf development. PLoS Genet 9: e1003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Tan Z, Wu F, Sheng P, Heng Y, Wang X, Ren Y, Wang J, Guo X, Zhang X, et al. (2014) A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol Plant 7: 1329–1349 [DOI] [PubMed] [Google Scholar]
- Till B, Schmitz-Linneweber C, Williams-Carrier R, Barkan A (2001) CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D (2003) The Arabidopsis SeedGenes Project. Nucleic Acids Res 31: 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ren Y, Zhou K, Liu L, Wang J, Xu Y, Zhang H, Zhang L, Feng Z, Wang L, et al. (2017) WHITE STRIPE LEAF4 encodes a novel P-type PPR protein required for chloroplast biogenesis during early leaf development. Front Plant Sci 8: 1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins KP, Kroeger TS, Cooke AM, Williams-Carrier RE, Friso G, Belcher SE, van Wijk KJ, Barkan A (2007) A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19: 2606–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis Sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Zhu Y, Ma Z, Sun Y, Quan Q, Li P, Hu P, Shi T, Lo C, Chu IK, et al. (2013) Proteomic evidence for genetic epistasis: ClpR4 mutations switch leaf variegation to virescence in Arabidopsis. Plant J 76: 943–956 [DOI] [PubMed] [Google Scholar]
- Wu W, Liu S, Ruwe H, Zhang D, Melonek J, Zhu Y, Hu X, Gusewski S, Yin P, Small ID, et al. (2016) SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J 85: 607–621 [DOI] [PubMed] [Google Scholar]
- Xu D, Huang W, Li Y, Wang H, Huang H, Cui X (2012) Elongator complex is critical for cell cycle progression and leaf patterning in Arabidopsis. Plant J 69: 792–808 [DOI] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H (2003) Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130: 4097–4107 [DOI] [PubMed] [Google Scholar]
- Xue J, Luo D, Xu D, Zeng M, Cui X, Li L, Huang H (2015) CCR1, an enzyme required for lignin biosynthesis in Arabidopsis, mediates cell proliferation exit for leaf development. Plant J 83: 375–387 [DOI] [PubMed] [Google Scholar]
- Yagi Y, Nakamura T, Small I (2014) The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J 78: 772–782 [DOI] [PubMed] [Google Scholar]
- Yin P, Li Q, Yan C, Liu Y, Liu J, Yu F, Wang Z, Long J, He J, Wang HW, et al. (2013) Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504: 168–171 [DOI] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR (2004) The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J 37: 864–876 10.1111/j.1365-313X.2003.02014.x [DOI] [PubMed] [Google Scholar]
- Yu F, Liu X, Alsheikh M, Park S, Rodermel S (2008) Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 20: 1786–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Luo D, Li G, Yao X, Wang H, Zeng M, Huang H, Cui X (2010) Characterization of the AE7 gene in Arabidopsis suggests that normal cell proliferation is essential for leaf polarity establishment. Plant J 64: 331–342 [DOI] [PubMed] [Google Scholar]
- Zhang L, Wei Q, Wu W, Cheng Y, Hu G, Hu F, Sun Y, Zhu Y, Sakamoto W, Huang J (2009) Activation of the heterotrimeric G protein alpha-subunit GPA1 suppresses the ftsh-mediated inhibition of chloroplast development in Arabidopsis. Plant J 58: 1041–1053 10.1111/j.1365-313X.2009.03843.x [DOI] [PubMed] [Google Scholar]
- Zheng M, Liu X, Liang S, Fu S, Qi Y, Zhao J, Shao J, An L, Yu F (2016) Chloroplast translation initiation factors regulate leaf variegation and development. Plant Physiol 172: 1117–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Cheng Y, Yap A, Chateigner-Boutin AL, Delannoy E, Hammani K, Small I, Huang J (2009) The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J 58: 82–96 [DOI] [PubMed] [Google Scholar]
- Zoschke R, Nakamura M, Liere K, Sugiura M, Börner T, Schmitz-Linneweber C (2010) An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci USA 107: 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]