RAPTOR1B positively regulates vegetative growth and development in Arabidopsis by controlling metabolic and hormonal integrity.
Abstract
Vegetative growth requires the systemic coordination of numerous cellular processes, which are controlled by regulatory proteins that monitor extracellular and intracellular cues and translate them into growth decisions. In eukaryotes, one of the central factors regulating growth is the serine/threonine protein kinase Target of Rapamycin (TOR), which forms complexes with regulatory proteins. To understand the function of one such regulatory protein, Regulatory-Associated Protein of TOR 1B (RAPTOR1B), in plants, we analyzed the effect of raptor1b mutations on growth and physiology in Arabidopsis (Arabidopsis thaliana) by detailed phenotyping, metabolomic, lipidomic, and proteomic analyses. Mutation of RAPTOR1B resulted in a strong reduction of TOR kinase activity, leading to massive changes in central carbon and nitrogen metabolism, accumulation of excess starch, and induction of autophagy. These shifts led to a significant reduction of plant growth that occurred nonlinearly during developmental stage transitions. This phenotype was accompanied by changes in cell morphology and tissue anatomy. In contrast to previous studies in rice (Oryza sativa), we found that the Arabidopsis raptor1b mutation did not affect chloroplast development or photosynthetic electron transport efficiency; however, it resulted in decreased CO2 assimilation rate and increased stomatal conductance. The raptor1b mutants also had reduced abscisic acid levels. Surprisingly, abscisic acid feeding experiments resulted in partial complementation of the growth phenotypes, indicating the tight interaction between TOR function and hormone synthesis and signaling in plants.
Eukaryotes have evolved sophisticated molecular and physiological mechanisms for their adaptation to changing environmental conditions. To be able to coordinate diverse environmental and nutritional inputs with the intricate metabolic pathways, several regulators controlling cellular growth behavior have evolved (Chantranupong et al., 2015). The Ser/Thr protein kinase Target of Rapamycin (TOR) is one of these central growth regulators, having been shown to positively control anabolism while repressing catabolic processes (Saxton and Sabatini, 2017). TOR, which is structurally and functionally conserved among all eukaryotes (Wullschleger et al., 2006; Saxton and Sabatini, 2017), was identified more than 25 years ago in a genetic screen for rapamycin resistance in the budding yeast Saccharomyces cerevisiae (Heitman et al., 1991). The inhibitory activity of the macrocyclic lactone rapamycin, which is an antimicrobial, immunosuppressive, and antitumor drug, is mediated through its direct interaction with the peptidyl cis/trans-isomerase FK506-binding protein 12 kD (FKBP12; Sehgal, 2003). The mode of action of this binary protein-drug complex, which binds to the FKBP12-rapamycin-binding domain of TOR, might be through steric repression of the TOR kinase activity or interference of the complex with the binding sites of essential TOR substrates (Baretić and Williams, 2014).
Two TOR genes have been identified in yeast, but only a single TOR has been identified in most other eukaryotes (Wullschleger et al., 2006). The presence of two TOR genes in yeast led to the discovery of two distinct TOR complexes, TORC1 and TORC2 (Loewith et al., 2002). Soon after the detection of the two TORCs in yeast, similar complexes were identified in mammals (Hara et al., 2002; Kim et al., 2002; Sarbassov et al., 2004) and in other species (Wullschleger et al., 2006). Interestingly, it turned out that only TORC1, which is composed of three core proteins, TOR, RAPTOR (Regulatory-Associated Protein of TOR), and LST8 (Lethal with Sec Thirteen8), was rapamycin sensitive; TORC2, which contains TOR, RICTOR (Rapamycin-Insensitive Companion of TOR), and LST8, was insensitive to the drug (Sarbassov et al., 2004). Based upon this essential difference, major progress in the understanding of the function and regulation of TORC1 was made in mammals and yeast (González and Hall, 2017; Saxton and Sabatini, 2017), while understanding of the rapamycin-insensitive TORC2 lagged behind. An ever-growing network of TORC1 interactions has been uncovered, providing evidence that TORC1 is a central regulator of anabolism, positively regulating all essential metabolic processes, including protein (Ma and Blenis, 2009), lipid (Lamming and Sabatini, 2013), and nucleotide metabolism (Ben-Sahra and Manning, 2017), while repressing catabolic processes like autophagy (Neufeld, 2010).
In Arabidopsis (Arabidopsis thaliana), a single TOR gene, two LST8 genes (LST8-1 and LST8-2), and two RAPTOR genes (RAPTOR1A and RAPTOR1B) have been annotated (Menand et al., 2002; Anderson et al., 2005; Deprost et al., 2005; Moreau et al., 2012), but Arabidopsis lacks a copy of RICTOR (Xiong and Sheen, 2014). Accordingly, it was concluded that the Arabidopsis genome encodes the rapamycin-sensitive TORC1, but the rapamycin-insensitive TORC2 seems to be absent. Unfortunately, Arabidopsis, like many other plant species, seems to be largely insensitive to the TORC1-inhibiting drug rapamycin, which was explained by sequence differences in FKBP12 in the plant lineage that render this protein unable to properly bind rapamycin (Xu et al., 1998; Menand et al., 2002; Sormani et al., 2007). This fact, in combination with the embryo lethality of tor null mutants (Menand et al., 2002; Ren et al., 2011), delayed the elucidation of TOR functions in photoautotrophic organisms compared with rapamycin-sensitive systems (Xiong and Sheen, 2014). This bottleneck led to the development of genetic approaches such as generating tor RNA interference (tor-RNAi) and tor artificial microRNA (amiR-tor) lines (Deprost et al., 2007; Xiong and Sheen, 2012; Caldana et al., 2013) as well as lines transformed with the yeast FKBP12, which confers rapamycin sensitivity in a transgenic background (Ren et al., 2012). In sum, these reverse genetic tools provided the basis for a number of studies that started to elucidate the function of TOR in plants. It was shown that TOR signaling in plants positively regulates several anabolic processes, including transcription and cell cycle processes (Xiong et al., 2013), ribosome biogenesis (Ren et al., 2011, 2012), translation (Ahn et al., 2011; Schepetilnikov et al., 2013), and central and lipid metabolism (Caldana et al., 2013), while catabolic processes, including autophagy, are inhibited by active TOR (Liu and Bassham, 2010; Ahn et al., 2011).
Still, little is known about the function and influence of RAPTOR, the essential TORC1-defining interaction partner of TOR. Interestingly, it was shown previously that even though both RAPTOR proteins show high sequence similarity to each other, only the knockout (KO) of RAPTOR1B results in significant defects of growth and development, while raptor1a T-DNA lines show almost no changes in their morphology and development (Anderson et al., 2005). In a previous study, we looked carefully at the morphology, germination behavior, and metabolic composition of raptor1b seeds, all of which were significantly altered compared with the wild type (Salem et al., 2017). To further deepen our understanding of how RAPTOR1B affects growth, development, and metabolism, we describe here the impact of the raptor1b mutation on the vegetative and reproductive growth of Arabidopsis plants. A detailed morphological and developmental phenotypic analysis of raptor1b T-DNA KO lines was performed and combined with physiological and molecular analyses. Our results revealed that RAPTOR1B is involved in most Arabidopsis developmental stages, from postembryonic growth to senescence. Accordingly, raptor1b plants exhibited a delay in the development of rosette leaves and primary root as well as defects in root hairs, delayed vegetative and reproductive growth, and delayed senescence. To better understand the physiological changes produced by mutation of RAPTOR1B, we comprehensively profiled the diurnal molecular changes in raptor1b lines compared with the wild type. The mutant exhibited significant changes in primary and secondary metabolism as well as changes in lipid and hormone levels. These significant molecular differences reflect the crucial roles of RAPTOR, a part of TORC1, in regulating growth, metabolism, and stress adaptation in plants.
RESULTS
RAPTOR1B Disruption Causes Developmental Delays
Unlike in most eukaryotes, which have only one gene for this protein, RAPTOR is encoded by two homologous genes in the Arabidopsis genome (RAPTOR1A [AT5G01770] and RAPTOR1B [AT3G08850]). Both genes are actively transcribed throughout development (Supplemental Fig. S1A). While the single KO of RAPTOR1A does not lead to visible phenotypes (Supplemental Fig. S1B), the double KO of RAPTOR1A and RAPTOR1B leads to arrested postembryonic growth and development (Anderson et al., 2005). Interestingly, the KO of RAPTOR1B led to substantial growth and developmental phenotypes, which seem to be partially compensated for by the increased expression of RAPTOR1A (Supplemental Fig. S1C).
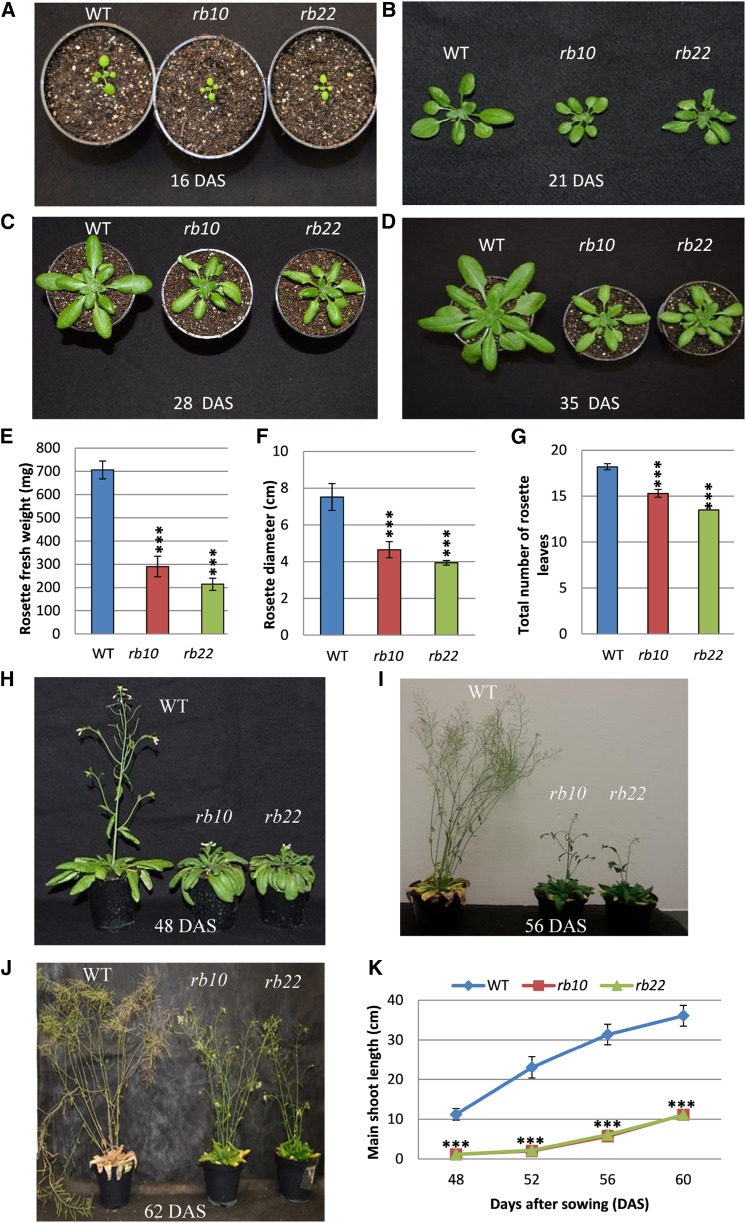
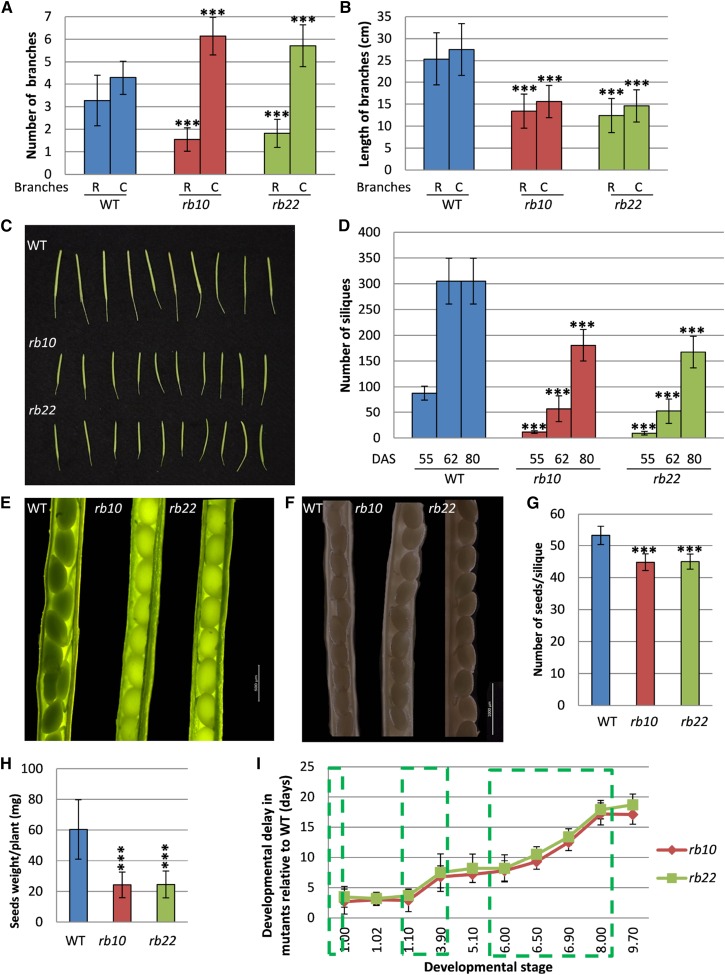
To better understand the impact of the raptor1b mutation on growth and development, we performed detailed physiological and developmental analyses of two independent raptor1b T-DNA mutants (rb10 and rb22) and compared them with their corresponding wild type (Columbia-0 [Col-0]). The most obvious phenotype of raptor1b plants is the significant delay and reduction in growth and development. This growth reduction starts with the germination of the seeds (Salem et al., 2017), which leads to delayed emergence of the first true leaves, followed by delayed and reduced growth of the whole rosette (Fig. 1, A–G; Supplemental Fig. S2, A and B). Similar to the perturbed vegetative growth, we also observed delayed and reduced growth throughout the reproductive growth phase. Mutant plants exhibited late production of visible flower buds, a delay in the opening of first flowers, and delayed bolting compared with the wild type (Fig. 1, H and I). Additionally, raptor1b plants demonstrated a significant reduction in the primary stem length. Mutant plants grew an approximately 1-cm-long stem 48 DAS, which elongated to 10 cm at 60 DAS, while wild-type plants already developed 10- and 40-cm-long inflorescences at the indicated time points (Fig. 1K). Furthermore, raptor1b plants lagged behind during the entire flowering period, including silique ripening and senescence stages. Accordingly, mutants started to produce their first flowers at 56 DAS, while wild-type plants had already completed the flowering stage at this time point. At 62 DAS, wild-type plants completed the senescence stage, while raptor1b plants only then ceased to flower (Fig. 1J). Moreover, raptor1b plants showed a defective shoot architecture, with a significantly increased number of axillary branches (Fig. 2A) and a significant reduction in axillary and secondary branch length at 80 DAS (Fig. 2B). In addition to effects on plant architecture, raptor1b plants reproducibly showed 10% abnormally developed flowers under long-day (LD) conditions, exhibiting increased numbers of petals, sepals, and/or stamens (Supplemental Fig. S2C). Along with this plethora of phenotypes, disruption of RAPTOR1B also caused a significant reduction in the number of siliques produced per plant accompanied by a significant decrease in seed/silique yield (Fig. 2, C–H).
Figure 1.
Vegetative and reproductive growth phenotypes of Col-0 and two independent raptor1b mutants. A to D, Delayed and reduced vegetative growth phenotypes of raptor1b (rb10 and rb22) for seeds grown on soil at 16 d after sowing (DAS; A), 21 DAS (B), 28 DAS (C), and 35 DAS (D). Col-0 was used as the wild type (WT). E to G, Rosette biomass (E), rosette diameter (F), and number of rosette leaves (G) of raptor1b compared with wild-type plants grown under LD conditions at 28 DAS. H to J, Reproductive growth phenotypes for wild-type and raptor1b plants grown on soil at 48 DAS (H), 56 DAS (I), and 62 DAS (J). K, Main shoot length of raptor1b compared with the wild type at different time points after sowing. Data represent means ± sd for 20 biological replicates from at least two independent experiments. Asterisks indicate significant differences from the wild type under the same condition (***, P < 0.001, Student’s t test).
Figure 2.
Defective reproductive growth of raptor1b. A, Shoot branching of raptor1b mutants compared with wild-type (WT) plants at 80 DAS. B, Shoot length of raptor1b compared with wild-type plants at 80 DAS. C, Cauline branches; R, rosette branches. C, Siliques of raptor1b compared with the wild type. D, Number of siliques of raptor1b at 55 to 80 DAS. E and F, Seeds of raptor1b from young (E) and mature (F) siliques. Bars = 500 and 1,000 µm for green and mature siliques, respectively. G, Number of seeds per silique in raptor1b. H, Seed yield in raptor1b. I, Delayed growth of raptor1b. A stepwise delay is seen in different growth stages of raptor1b relative to the wild type. Green boxes mark delay increases during different stages of growth, as defined in Supplemental Figure S1D. Data represent means ± sd for 20 biological replicates from at least two independent experiments. Asterisks indicate significant differences from the wild type under the same condition (***, P < 0.001, Student’s t test).
Growth Delays Are Especially Pronounced during Distinct Developmental Transitions in raptor1b Lines
The observed growth delay in raptor1b plants compared with the wild type may either linearly increase throughout the life cycle or occur in a stepwise manner at specific developmental stages. To address this, plants were again grown side by side, and the time required to reach certain developmental stages was scored. For this purpose, we made use of the well-described nomenclature characterizing the different developmental stages of Arabidopsis from germination to senescence (Boyes et al., 2001; Supplemental Fig. S2D).
As can be seen from the line plot in Figure 2I, the developmental delay did not occur linearly but, rather, occurred stepwise. The initial delay happened during the first 7 d of the germination phase and resulted in a delay of 3 to 4 d, which was stably maintained until the setting of the 10th rosette leaf after 21 d (stage 1.10; Figs. 1, A and B, and 2I; Supplemental Fig. S2D). Following this stable growth phase of approximately 3 weeks, a second delay of an additional 3 to 4 d was introduced. This delay emerged during the completion of rosette growth, namely between the juvenile and adult rosette stages (stage 1.10 and the beginning of stage 3.90, rosette growth achieved; Figs. 1D and 2I; Supplemental Fig. S2D). Subsequent to this delay phase, a delay free week of growth ensued before the last and longest retardation. This additional delay of approximately 10 d occurred during the completion of flowering (stages 6.50 and 6.90) and silique ripening (stage 8.00; Figs. 1, H and I, and 2I; Supplemental Fig. S2D). Once this last delay was over, the raptor1b plants proceeded to senesce delay free (Figs. 1J and 2I; Supplemental Fig. S2D).
Taken together, these results suggest that the reduction of functional RAPTOR1B is not equally important throughout the entire life cycle of Arabidopsis but specifically affects the major developmental transitions, namely the onset of germination, the transition from juvenile vegetative to adult vegetative, and the entire flowering stage (Supplemental Table S1).
The Morphology and Anatomy of raptor1b Leaves Is Altered Significantly
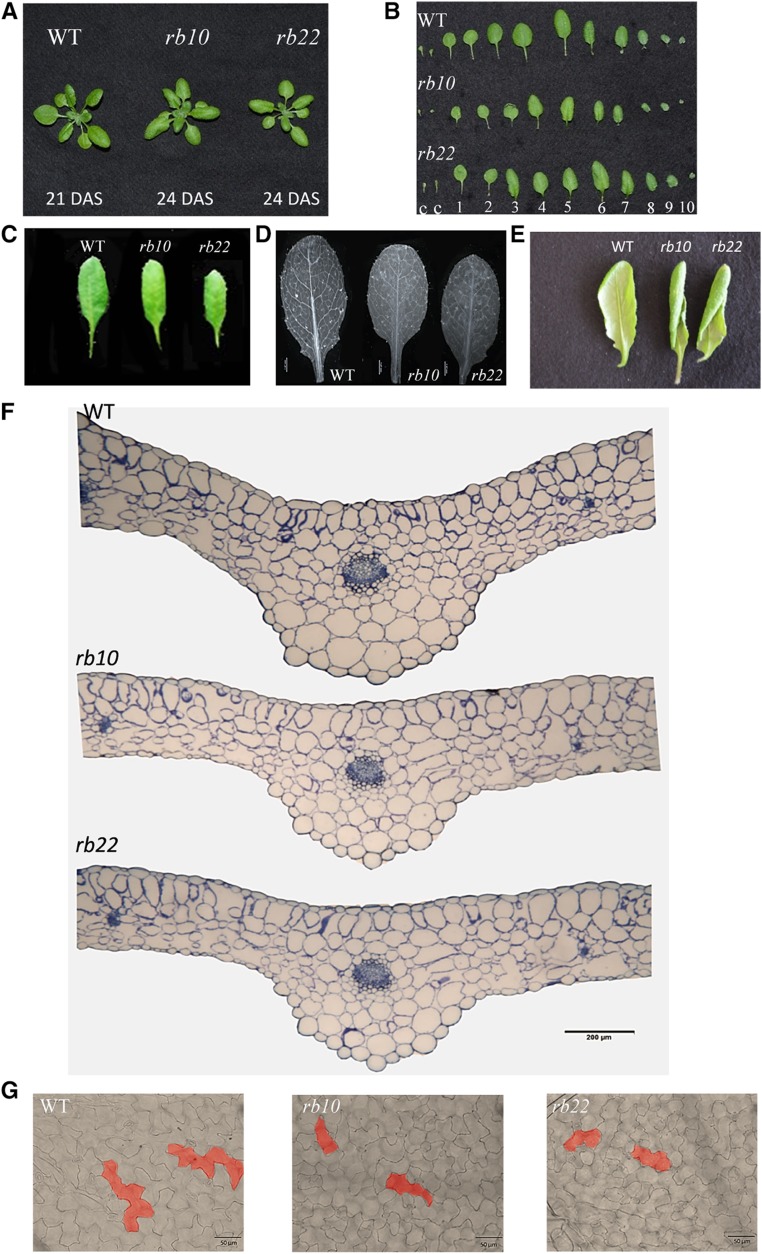
Interestingly, even though the collected data revealed that the vegetative growth of raptor1b plants was delayed (Supplemental Fig. S3A; Supplemental Tables S2 and S3), leaf and rosette size and shape did not seem to be macroscopically altered. To better understand if these macroscopic observations were manifested at the microscopic level, we analyzed size-matched raptor1b rosette leaves at the same developmental stage (stage 1.10, 10-rosette-leaves stage; Fig. 3, A–C). In contrast to the nearly identical leaf size, we observed that raptor1b leaves exhibited an altered leaf-vein phenotype with significantly decreased subbranching of the veins (Fig. 3D). This vein phenotype was accompanied by strong leaf curling in several raptor1b rosette leaves, whereby leaves started to turn from the adaxial to the abaxial surface followed by twisting around the lengthwise axis, compared with the generally flat-shaped wild-type leaves (Fig. 3E; Supplemental Fig. S3B).
Figure 3.
Analysis of the reduced leaf growth phenotype in raptor1b. A, Developmental stage- and size-matched rosettes of the wild type (WT) and raptor1b at the 10-rosette-leaves stage. B, Individual leaves of raptor1b compared with the wild type from developmentally matched rosettes (10 rosette leaves). Two cotyledons (c) and true leaves are numbered in order of their emergence. C, Comparison of the same leaf (leaf 5) of raptor1b and the wild type at the same developmental stage (10 rosette leaves). D, Reduced vein-density phenotype in raptor1b compared with the wild type. Bar = 1.47 mm. E, Leaf-curling phenotype in raptor1b. The abaxial side of leaves shows the start of curling. F, Cross sections of leaves of the wild type and raptor1b stained with Toluidine Blue. Bar = 200 µm. G, More-and-smaller-cells phenotype in adaxial epidermal cells of raptor1b. Bars = 50 µm. Leaf sections and cell size analyses were done on rosette leaf 5 at the same developmental stage of producing 10 rosette leaves.
Leaf cross sections showed that raptor1b rosette leaves were significantly thinner, both in the midrib and the lamina regions, than the corresponding wild-type leaves at the same developmental stage (stage 1.10; Fig. 3F; Supplemental Fig. S4, A–D). Additionally, the microscopic leaf sections demonstrated that the palisade cells in the lamina region of raptor1b leaves exhibited deformations and defects in their cellular organization compared with the tightly packed and elongated palisade cells of the wild type (Fig. 3F; Supplemental Fig. S4, C–E). The mesophyll cells of the raptor1b leaves also evolved deformed cell shapes with large intercellular spaces, compared with the generally rounded and loosely packed wild-type mesophyll cells (Fig. 3F; Supplemental Fig. S4, C–E). In addition to these cellular phenotypes, raptor1b leaves contained regularly structured but slightly smaller vascular bundles in the midrib region (Supplemental Fig. S4F).
Consistent with our observation of the slightly smaller cells of the vasculature, our microscopic analysis disclosed that raptor1b leaves, albeit almost size matched to the wild type, had a significant reduction in average cell size, meaning that the cell number per leaf must have increased. This phenotype was true for upper and lower epidermal cells as well as for mesophyll cells (Fig. 3G; Supplemental Fig. S5, A–C; Supplemental Table S4). The increased number of epidermal cells was consequently accompanied by an increase in the number of trichomes (Supplemental Fig. S5D) and stomatal cells (Supplemental Table S4). Interestingly, besides their decreased size, both cell types developed no morphological changes (Supplemental Fig. S5E). These results suggested that raptor1b cells have a defect in cell elongation but not necessarily in cell proliferation. This setting allows the plant to maintain leaf identity by compensating for decreased cell size with an increased cell number.
Root Length and Root Meristem Size Are Reduced in raptor1b Plants
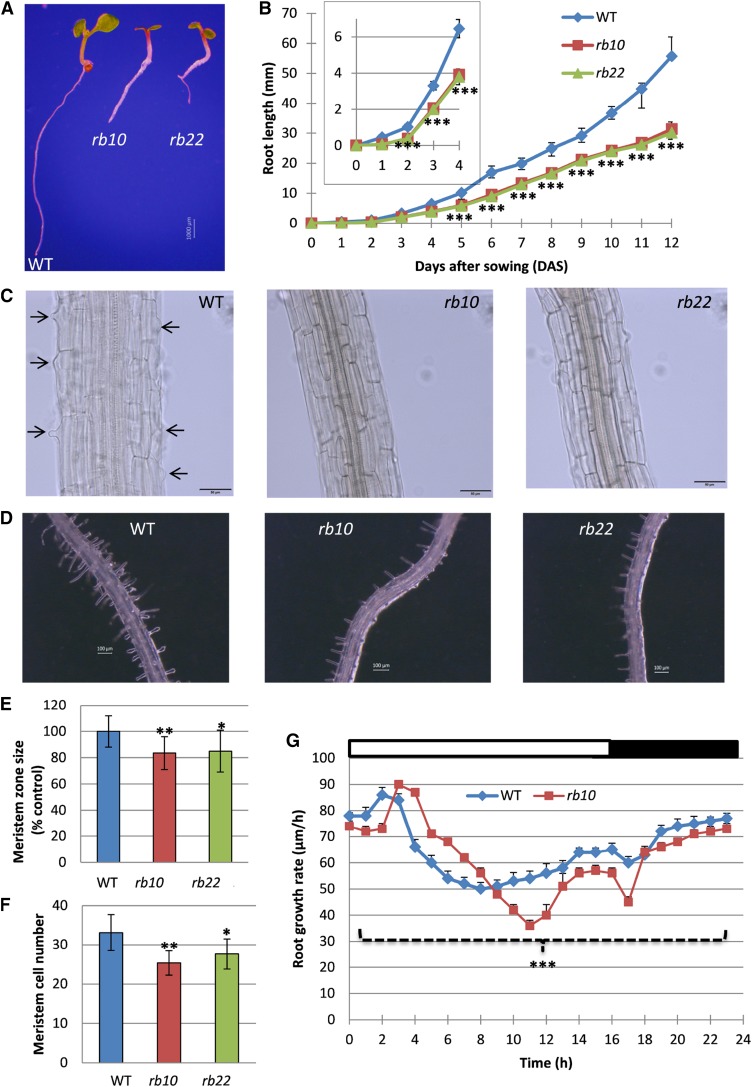
In light of the growth phenotypes, especially the reduced size of raptor1b leaf cells, we inspected the raptor1b root phenotype. This was especially interesting because three recent publications have reported that the chemical or genetic inhibition of the TOR complex leads to reduced root cell size in combination with restricted meristematic zone size (Ren et al., 2012; Montané and Menand, 2013; Xiong et al., 2013). To analyze the roots, wild-type and raptor1b plants were grown for 12 d on vertical agar plates with root length scored every day. As expected from the aboveground phenotypes obtained in soil-grown plants, the roots of the agar-grown plants displayed delayed and reduced growth over time (Fig. 4, A and B; Supplemental Fig. S6A). Looking at the day-to-day growth within the first 12 DAS, we determined that the root length reduction in the mutants compared with the wild type increased continuously over time (Fig. 4B).
Figure 4.
Root phenotypes of raptor1b. A, Seedlings of raptor1b and the wild type (WT) grown on 0.5× Murashige and Skoog (0.5× MS) agar plates at 7 DAS. Bar = 1,000 µm. B, Primary root length of raptor1b. Root length was monitored from seedlings grown on 0.5× MS agar plates for 12 DAS. The inset shows root length for 0 to 4 DAS. C, Delayed initiation of root hairs of raptor1b at 7 DAS. Black arrows indicate the initiation of root hairs in the wild type. Bars = 50 µm. D, Root hair growth of raptor1b compared with the wild type at 12 DAS. Bars = 100 µm. E and F, Meristem zone size (E) and meristem cell number (F) of raptor1b at 12 DAS. G, Root growth rate of the wild type and raptor1b. Root elongation was measured continuously over a period of 10 d for seedlings grown on 0.5× MS agar plates. White and black bars on the top indicate light and dark periods, respectively. Data represent means ± sd for 20 biological replicates for B, E, and F and for six biological replicates for G. Asterisks indicate significant differences between the wild type and raptor1b under the same condition (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, Student’s t test). The 0-d time point in B refers to seeds that were imbibed for 3 d at 4°C.
A closer inspection of raptor1b roots further indicated that, similar to previous observations in TOR-inhibited plants (Ren et al., 2012; Montané and Menand, 2013; Xiong and Sheen, 2013), mutation of RAPTOR1B led to a delay in root hair initiation and to a significant decrease in root hair numbers (Fig. 4, C and D). Interestingly, the reduced root hair size and number are fully complemented over time, as we have shown by comparing wild-type and raptor1b roots grown for 7 and 12 d (Supplemental Fig. S6B).
In addition to the root hair phenotype, it was also reported that chemical inhibition of TOR by either rapamycin (Ren et al., 2012; Xiong et al., 2013) or AZD-8055 (Montané and Menand, 2013) leads to decreases in meristem size and activity. These results were clearly confirmed by our analysis, revealing that meristem size and also the number of meristem cells, in stage-matched raptor1b and wild-type seedlings, were reduced significantly. This indicates that the disruption of RAPTOR1B, similar to the inactivation of TOR, leads to repressions in meristem activity, differentiation, and expansion (Fig. 4, E and F; Supplemental Fig. S6C).
These root growth phenotypes of raptor1b mutants prompted us to conduct a detailed, continuous root growth analysis. To this end, root elongation kinetics of wild-type and raptor1b plants were recorded using noninvasive video imaging (Yazdanbakhsh and Fisahn, 2012). As shown in Figure 4G, upon illumination, a pronounced transient increase in root growth rate emerged in the wild type, consistent with previous data (Yazdanbakhsh et al., 2011). This maximal root elongation rate occurred with a delay of 1 h in raptor1b lines (Fig. 4G). Surprisingly, between 4 and 8 h after the onset of illumination, the elongation rates of raptor1b roots transiently exceeded those of the wild type before decreasing continuously below the wild-type level. Derived from this clearly deviating growth behavior of the mutants (Fig. 4G), the total root elongation rates of the raptor1b plants were reduced significantly, accounting for the decreased root growth depicted in Figure 4B.
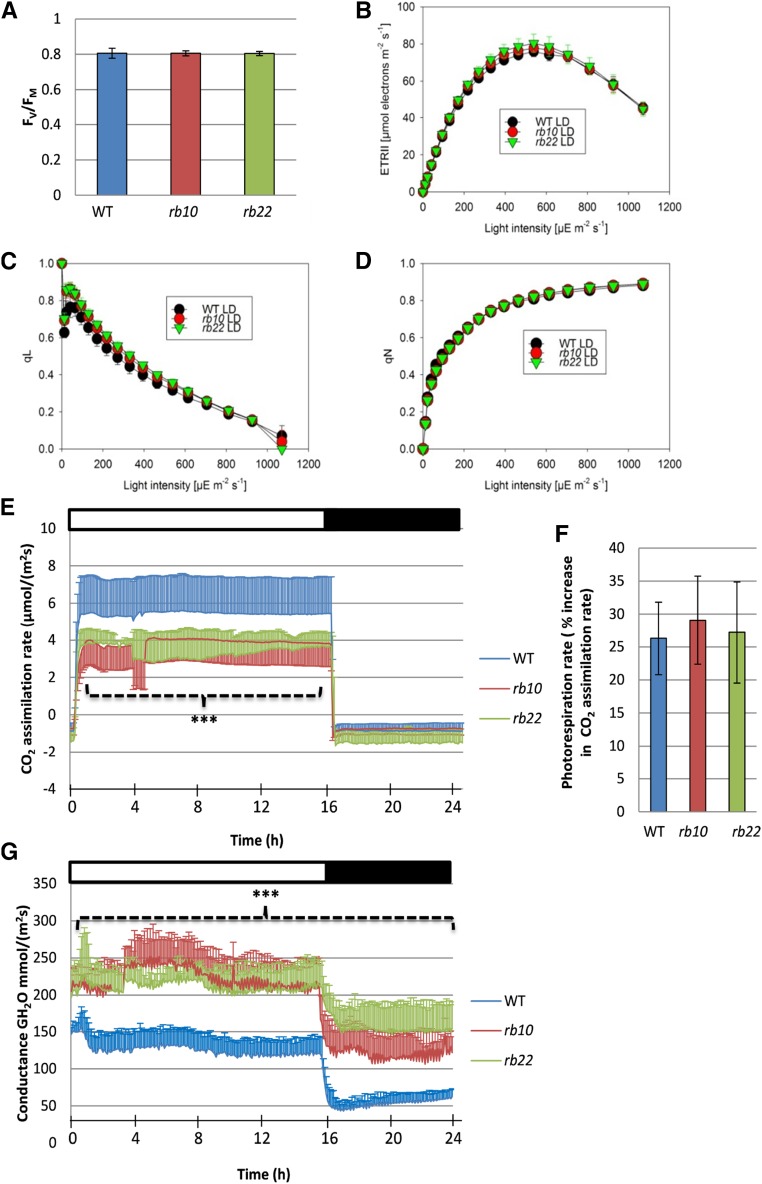
The Photosynthetic Electron Transport Rate Remains Unaffected, While the CO2 Assimilation Rate Is Altered in raptor1b Plants
The delayed vegetative growth observed in the raptor1b plants suggested that their photosynthetic activity might be down-regulated. Therefore, we measured in vivo chlorophyll fluorescence parameters in developmentally matched (stage 1.10) rosette leaves of wild-type and raptor1b plants (Fig. 5). Application of a saturating light pulse to dark-adapted plants provided an estimate of the maximum quantum efficiency of PSII (Fv/Fm). Wild-type and raptor1b plants exhibited almost identical Fv/Fm (Fig. 5A). We also investigated the photosynthetic performance of wild-type and raptor1b plants under increasing light intensities, subsequent to dark adaption. Light-response curves indicated indistinguishable linear electron transport rates (ETRs) between raptor1b and wild-type plants (Fig. 5B). Additionally, no difference between wild-type and raptor1b plants could be detected in PSII acceptor site reduction upon progressively increased illumination (qL; Fig. 5C). Similarly, light-response curves of nonphotochemical quenching (qN), which increased with light intensity, were almost identical for wild-type and raptor1b plants (Fig. 5D).
Figure 5.
Photosynthetic efficiency and gas-exchange measurements of the wild type (WT) and raptor1b. A, Fv/Fm of wild-type and raptor1b leaves. B to D, Light-response curves of linear ETR (B), qL (C), and qN (D) of the wild type and raptor1b. E, Net CO2 assimilation rate of raptor1b compared with the wild type. F, Assimilation rate under nonphotorespiratory conditions of the wild type and raptor1b. G, Stomatal conductance of raptor1b compared with the wild type. Measurements were performed on rosette leaves from soil-grown plants at the same developmental stage under LD conditions. Plants were dark adapted for 30 min before the commencement of chlorophyll fluorescence imaging at the beginning of the day, 1 h after illumination. Data represent means ± sd for five biological replicates. Asterisks indicate significant differences from the wild type under the same condition (***, P < 0.001, Student’s t test). White and black bars on the top of the graphs in E and G indicate light and dark periods, respectively.
The results derived from the chlorophyll fluorescence imaging analysis were complemented by extraction and quantification of photosynthetic pigments. These measurements also did not reveal significant changes in total chlorophyll or total carotenoid contents between wild-type and raptor1b plants (Supplemental Fig. S7, A–C), suggesting that the photosynthetic apparatus is likely unaltered between the two genotypes.
To deepen the investigation of the photosynthetic performance of raptor1b leaves, we analyzed whether the CO2 assimilation rate in wild-type and raptor1b leaves was unaltered, similar to the light-driven electron transport. Contrary to the almost identical ETRs, the CO2 assimilation rates of raptor1b leaves exhibited a significant reduction when measured at ambient CO2 (350 µL L−1) and a growth light intensity of 150 µE m–2 s–1, while no difference was observed for respiratory CO2 release during the night (Fig. 5E).
Since CO2 uptake rates decreased significantly in raptor1b plants while the ETR was maintained, we reasoned that photorespiration, as a putative mechanism for the acceptance of photosynthetic electrons, could be increased and, therefore, explain our observations. Therefore, we compared CO2 assimilation rates under atmospheric oxygen as well as under microaerobic conditions, the latter expected to fully repress photorespiration and result in an increase in net CO2 assimilation. Unexpectedly, the increase in net assimilation under nonphotorespiratory conditions was similar in wild-type and raptor1b plants (Fig. 5F).
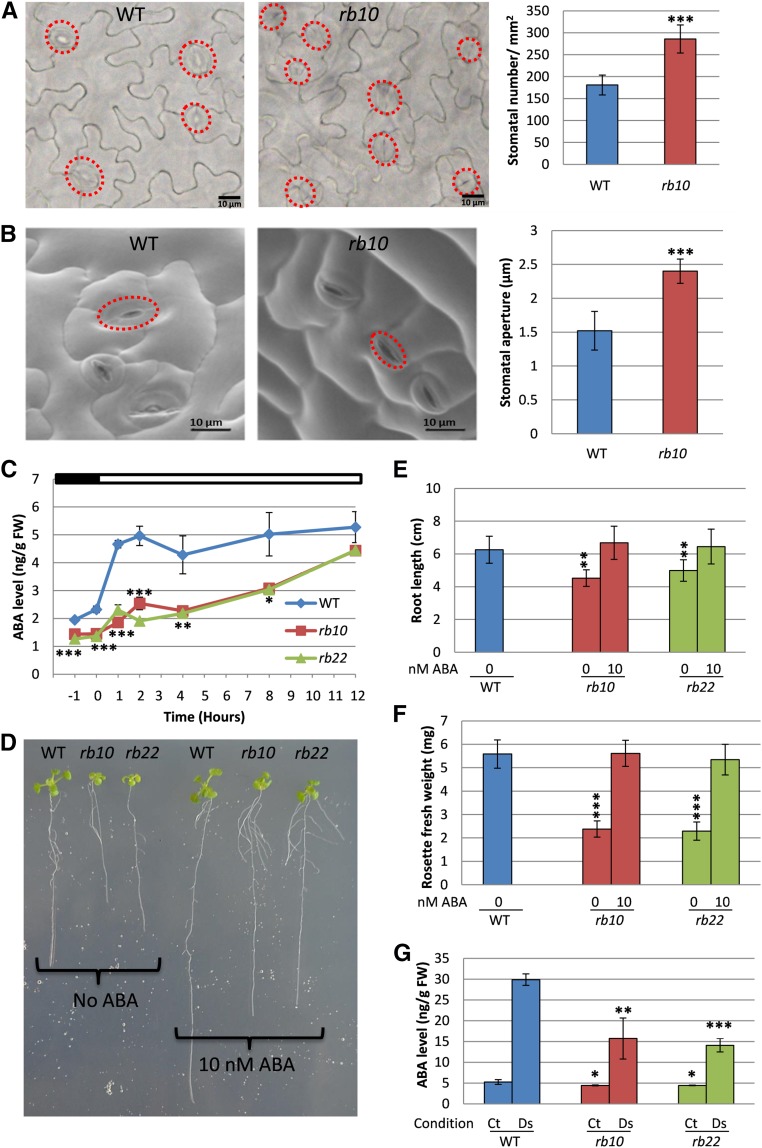
Interestingly, gas-exchange measurements revealed highly elevated stomatal conductance rates throughout the entire diurnal cycle in both raptor1b lines (Fig. 5G). This increased stomatal conductance, especially in the dark period, suggests insufficient stomatal closure in raptor1b plants. To validate this assumption, we further analyzed stomatal size and aperture. As expected from the conductance measurements, the microscopic analysis revealed that raptor1b leaves showed, next to the higher densities of smaller stomata (Fig. 6A), significantly increased stomatal aperture (Fig. 6B).
Figure 6.
raptor1b shows phenotypes of abscisic acid (ABA) deficiency. A and B, Stomatal number (A) and stomatal aperture size (B) in raptor1b. C, Changes in ABA level in raptor1b compared with the wild type (WT) before and after illumination. D to F, Exogenous application of ABA on raptor1b. D, Seedlings of raptor1b treated with ABA and grown for the same time as wild-type seedlings without ABA. E and F, Effects of ABA treatment on raptor1b root growth defect (E) and rosette biomass (F). G, Effects of drought stress on ABA in raptor1b. Samples were harvested 12 h after illumination from nontreated plants (control [Ct]) and after induction of drought stress (Ds) for 6 d. Microscopic analysis was performed on rosette leaf 5 from the wild type and raptor1b at the same developmental stage (10 rosette leaves) from plants that were grown on soil under LD growth conditions. At least 200 and 50 stomata were analyzed for stomatal number and stomatal aperture size, respectively. Red circles in A and B indicate positions of stomata. For ABA extraction, the rosettes of raptor1b and the wild type were harvested at the same developmental stage (10 rosette leaves) from plants cultivated on soil under normal-light LD growth conditions. The first time point (‒1) represents samples that were harvested 1 h before illumination, while 0 h represents samples harvested just before illumination. The time points 1, 2, 4, 8, and 12 h represent samples that were harvested in the light period at 1, 2, 4, 8, and 12 h after the onset of illumination, respectively. Data represent means ± sd for five biological replicates. Asterisks indicate significant differences between the wild type and raptor1b under the same condition (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, Student’s t test). White and black bars on the top of the graph in C indicate light and dark periods, respectively. FW, Fresh weight.
The Levels of ABA Are Decreased Significantly in raptor1b Plants
Increased stomatal conductance and aperture have been reported frequently to be caused by decreased ABA levels within the leaves (Assmann, 2003; Cutler et al., 2010; Munemasa et al., 2015). Absolute quantification of ABA levels in developmentally matched rosette leaves of the wild type and raptor1b was performed on samples that were harvested during a day/night cycle. Perfectly in line with the permanently increased stomatal conductance (Figure 5G), ABA levels in raptor1b leaves were reduced significantly throughout the entire diurnal time course (Fig. 6C).
In contrast to the negative effect of ABA on seed germination (Finkelstein, 2013; Salem et al., 2017), ABA synthesis mutants develop smaller leaves than their respective wild type. These mutants can be complemented during vegetative growth by ABA feeding (LeNoble et al., 2004; Barrero et al., 2005). Accordingly, we tested if the suppressed growth phenotype of raptor1b lines can be complemented by the external application of low concentrations of ABA. The application of 10 nm ABA rescued rosette growth as well as root expansion to wild-type control levels (Fig. 6. D–F).
Based on the observation of the reduced ABA levels, we asked whether raptor1b lines might have a basic limitation in the synthesis of this hormone or if the reduced levels are due to differential regulation of the synthesis pathway. To address this question, we subjected leaves of wild-type and raptor1b plants at the 10-rosette-leaves stage to drought stress, a well-known abiotic stress that induces ABA synthesis (Assmann, 2003; Cutler et al., 2010; Munemasa et al., 2015). Indeed, raptor1b responded to drought stress by producing higher ABA concentrations (Fig. 6G), even though ABA levels were clearly lower than those observed in drought-stressed wild-type plants.
Carbon Partitioning Is Altered Significantly in raptor1b Leaves
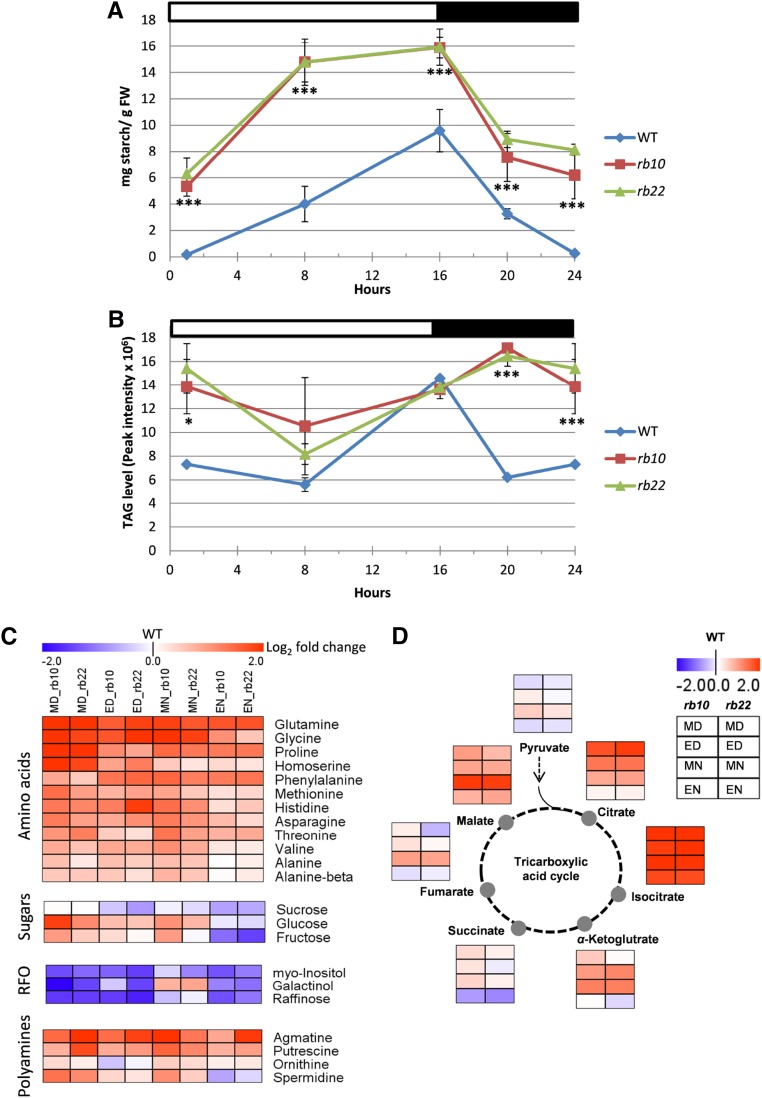
To gain more insight into the delayed and changed growth phenotypes, we asked how these alterations impact the main carbon sinks, namely cell walls, total proteins, and carbon storage (starch and triacylglycerols [TAGs]). As a starting point, transitory starch, the main carbon storage molecule in plants (Streb and Zeeman, 2012), was measured throughout the diurnal growth cycle in raptor1b and wild-type leaves. These measurements indicated that the levels of starch were continuously higher in mutant plants compared with the wild type, even though the absolute level of starch accumulation and degradation within raptor1b and wild-type plants was almost identical, with a value of approximately 10 mg g−1 fresh weight (Fig. 7A). A striking difference between the two genotypes was the starting concentration, almost no starch in the wild type and approximately 6 mg g−1 fresh weight in raptor1b plants, which made raptor1b appear like one of the previously described starch-excess mutants (Kötting et al., 2005; Comparot-Moss et al., 2010; Santelia et al., 2011). At the same time, the accumulation and degradation pattern of starch during the day and night was clearly different between the wild type and raptor1b. Starch almost linearly accumulated in the wild type during the day and was degraded again throughout the night, but its accumulation was biphasic in raptor1b plants. During the first half of the light phase, starch accumulated at a high rate, while in the second half, the accumulation seemed to almost cease (Fig. 7A). Similarly, the degradation of starch was significantly more rapid in the first half of the night while flattening out completely in the second half.
Figure 7.
Influence of raptor1b mutation on carbon storage and primary metabolites. A, and B, Diurnal changes in starch and TAG in raptor1b relative to the wild type (WT). FW, Fresh weight. C, Heat map representing the changes of amino acids, sugars, raffinose family of oligosaccharides (RFOs), and polyamines in raptor1b compared with the wild type. D, Changes in tricarboxylic acid cycle intermediates upon raptor1b mutation. For extraction, rosettes of raptor1b and the wild type were harvested at different time points from plants at the same developmental stage (10 rosette leaves). Plants were cultivated on soil under normal-light LD growth conditions. For heat maps, fold changes were calculated for each compound by dividing the average level of each independent raptor1b line by the average level of the wild type at the same time point. The log2 fold change in metabolite abundance was used to generate heap maps. This allowed more precise comparison by having one value for the wild type (0). The log2 fold changes provide the intensity of the color, according to the scale in the legend. Metabolite levels at each time point are presented in a color scale (blue = decrease, red = increase, white = zero, the wild-type value). Data represent means ± sd for five biological replicates. Asterisks indicate significant differences between the wild type and raptor1b under the same condition (***, P < 0.001, Student’s t test). White and black bars on the top of the graphs in A and B indicate light and dark periods, respectively. ED, End of the day; EN, end of the night; MD, middle of the day; MN, middle of the night.
In addition to the massive increase in the primary chloroplastic carbon storage, we asked if the raptor1b mutation also led to similar changes in the diurnal accumulation of cytosolic carbon storage, namely TAGs. TAGs, which are the major lipid-based carbon storage, are synthetized in the endoplasmic reticulum and stored in lipid droplets (Li-Beisson et al., 2013). Similar to the starch phenotype, raptor1b lines exhibited more of a shift in their accumulation pattern during the day than an absolute change in the TAG concentration (Fig. 7B). Intriguingly, raptor1b plants showed a delayed increase in TAG levels from the middle of the day to the middle of the night, before decreasing steeply until the end of the night. By contrast, wild-type TAGs demonstrated more rapid accumulation, starting already at middle of the day with a maximum at end of the day. Once the light was switched off, wild-type plants showed a steep decrease in TAGs until middle of the night and no change until end of the night (Fig. 7B).
We asked if two of the major carbon sinks of actively growing plants, namely the protein concentration and cell wall sugars, induced more pronounced adaptations upon RAPTOR1B disruption. To address this question, we measured the monosaccharide composition of plant cell wall polysaccharides after acidic hydrolysis and the total protein content of the cell. We did not detect any significant changes in the monosaccharides or crystalline cellulose, nor in the absolute protein content in raptor1b lines (Supplemental Fig. S8, A–C). This finding suggests that the timing of these processes seems to be changed but the absolute levels were not altered. This result was particularly unexpected, because cell sizes within the analyzed leaves were clearly modulated, indicating that cell wall expansion might be influenced.
RAPTOR1B Disruption Decreases S6 Kinase Activity and Has a Significant Impact on the Leaf Proteome
The standard measure for TOR kinase activity is to determine the phosphorylation state of the ribosomal protein S6 kinase (S6K), one of the main downstream targets of TOR in mammals (Pearson et al., 1995) and also in Arabidopsis (Schepetilnikov et al., 2011, 2013; Xiong and Sheen, 2012; Xiong et al., 2013). In this study, we used a previously developed microtiter plate-based assay to indirectly determine the activity of TOR by measuring the activity of the S6K toward a synthetic substrate (Sun et al., 2016). Since we observed a nonlinear effect of the RAPTOR1B mutation on Arabidopsis development (Fig. 2I), we decided to initially compare the influence of the RAPTOR1B KO on S6K activity at different developmental stages. Surprisingly, the results indicated that the kinase activity was substantially different in rosette leaves derived from plants at differential developmental stages (Supplemental Fig. S8D). Interestingly, S6K activity obtained from rosette leaves of plants harvested at the 10-rosette-leaves stage showed the strongest reduction of more than 70%, while leaves from full-grown rosettes or rosettes at the reproductive stage were almost wild type like in S6K activity (Supplemental Fig. S8D).
Due to the fact that the S6K activity was lowest at the 10-rosette-leaves stage, we decided to determine the impact of this repressed TOR activity on the total proteome. To address this issue, we performed a global analysis of the leaf proteome in the wild type and raptor1b. A total of 3,143 proteins were unambiguously identified (Supplemental Data Set S1), of which 477 proteins were significantly up- or down-regulated at least in one of the raptor1b T-DNA KO lines compared with the wild type. To perform protein data analysis and visualization, we used the Cytoscape (Shannon et al., 2003) and MAPMAN (Thimm et al., 2004) tools to elucidate biological processes that might be regulated at the protein level. The MAPMAN representation of the proteomic data indicated that raptor1b had significant changes in proteins assigned to several categories of metabolism, including starch and Suc, glycolysis, gluconeogenesis, cell wall, the tricarboxylic acid cycle, photosynthesis, signaling, redox regulation, nucleotide, hormone, amino acid, protein, lipid, and secondary metabolism (Supplemental Fig. S9A; Supplemental Data Set S1).
Proteins involved in TAG degradation, degradation of glucosinolates, jasmonic acid metabolism, RNA processing, DNA synthesis, protein synthesis, and defense against biotic stress were down-regulated significantly. Conversely, proteins involved in the Calvin-Benson cycle, starch synthesis, amino acid metabolism, secondary metabolism, and antioxidant machinery were up-regulated significantly (Supplemental Fig. S9B; Supplemental Data Set S1).
Influence of RAPTOR1B Disruption on the Levels of Primary Metabolites
The results obtained from the carbon partitioning and proteomic study indicated that raptor1b mutants should show modulations in primary, secondary, and lipid metabolism. This prompted us to perform global metabolic profiling to reveal if the RAPTOR1B mutation was associated with significant changes in metabolism. Based on a previously developed method (Salem et al., 2016), we extracted several metabolites associated with different compound classes. In the first step, we analyzed the primary metabolites derived from gas chromatography-mass spectrometry-based measurements (Salem et al., 2016).
Similar to the accumulation of storage reserves, we observed massive diurnal changes in multiple primary metabolites in raptor1b compared with the wild type (Supplemental Data Set S2). Interestingly, even though the total protein content did not change, the raptor1b lines accumulated significantly higher amounts of amino acids (Fig. 7C). For instance, the levels of aliphatic amino acids, such as Ala, Ile, Leu, and Val, and aromatic amino acids, such as Phe, were increased strongly in raptor1b, especially at end of the night. Additionally, the levels of other amino acids, such as Asn, Gln, Gly, Pro, and Thr, increased significantly (Fig. 7C; Supplemental Fig. S10A).
Furthermore, raptor1b plants displayed adaptation in sugar concentrations. For instance, the level of the disaccharide Suc was significantly lower in raptor1b lines, while the levels of Glc and Fru were increased strongly (Fig. 7C; Supplemental Fig. S10A). Moreover, the levels of tricarboxylic acid cycle intermediates, such as citrate, isocitrate, 2-oxoglutarate, fumarate, and malate, increased in raptor1b (Fig. 7D). Other primary metabolites, such as myoinositol, galactinol, and raffinose of the raffinose family of oligosaccharides, also decreased significantly, while some components of the polyamine pathway, such as putrescine, agmatine, and spermidine, were increased strongly in raptor1b (Fig. 7C).
Nitrogen Uptake and Fixation Are Altered in raptor1b Lines
The observed significant changes in C partitioning and the C/N balance in storage and primary metabolism in raptor1b mutants prompted us to analyze the influence of RAPTOR1B disruption on nitrogen uptake and fixation. We initially measured the absolute concentrations of nitrate and ammonium by ion chromatography (Watanabe et al., 2013). Both nitrate and ammonium were increased significantly in raptor1b lines, indicating higher availability for the synthesis of the increased amino acids (Supplemental Fig. S10B). To validate if these increased concentrations were based on higher availability or uptake, or simply due to decreased activity of the reducing enzymes, we determined the enzyme activities of the nitrogen-fixing enzymes. Interestingly, the activities of the key nitrogen-assimilating enzymes nitrate reductase and nitrite reductase increased significantly in raptor1b leaf tissue, while that of Gln synthetase, the main enzyme in nitrogen fixation into amino acids (Coruzzi, 2003), was unchanged in one mutant line, with the other line showing slight reduction (Supplemental Fig. S10B).
The Main Structural Lipid Classes Are Changed Only Slightly in the raptor1b Mutation
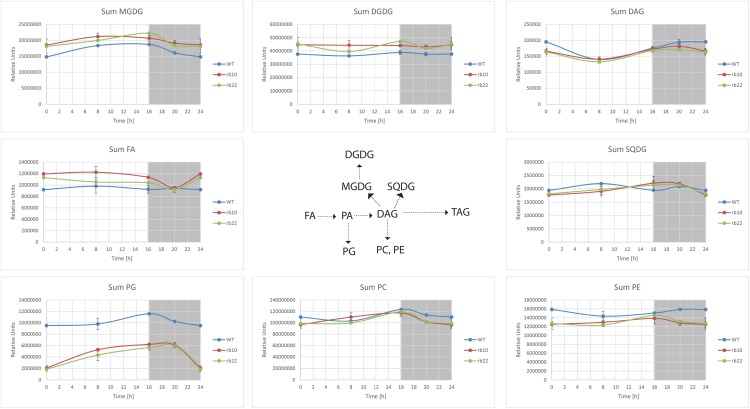
Next, we asked whether the raptor1b mutation led to changes in the composition of structural lipids, which are required for building the major organelles (Li-Beisson et al., 2013). To answer this question and to obtain more information about the regulation of lipids by RAPTOR1B, we performed comprehensive lipid profiling in raptor1b and wild-type lines using rosette leaves of plants that were stage matched (Salem et al., 2016). Aside from the previously mentioned significant increase in TAGs (storage lipids), smaller but still significant changes in the structural lipids were detected in raptor1b lines (Fig. 8; Supplemental Fig. S11; Supplemental Data Set S3). For plastidic glycolipids, raptor1b leaves expressed slightly higher levels of MGDG and DGDG compared with the wild type, while the total levels of PG and SQDG were decreased significantly. Especially PG levels showed that this phosphoglycerolipid class was decreased in raptor1b at all time points (Fig. 8; Supplemental Fig. S11; Supplemental Data Set S3).
Figure 8.
Influence of raptor1b mutation on lipid metabolism. For lipid extraction, the rosettes of raptor1b and the wild type (WT) were harvested from plants at the same developmental stage (10 rosette leaves). Plants were cultivated on soil under normal-light LD growth conditions. DAG, Diacylglyceride; DGDG, digalactosyldiacylglycerol; FA, fatty acid; MGDG, monogalactosyldiacylglycerol; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; SQDG, sulfoquinovosyldiacylglycerol.
The total amounts of the main endomembrane lipids, namely PC and PE, were partially decreased in raptor1b, while the levels of the less abundant phosphatidylserine and ceramides were almost unaffected (Fig. 8; Supplemental Fig. S11; Supplemental Data Set S3). Similarly, we did not see changes in the levels of DAG, which can act as intermediates in lipid biosynthetic pathways (Li-Beisson et al., 2013).
Regulation of Secondary Metabolites by RAPTOR1B
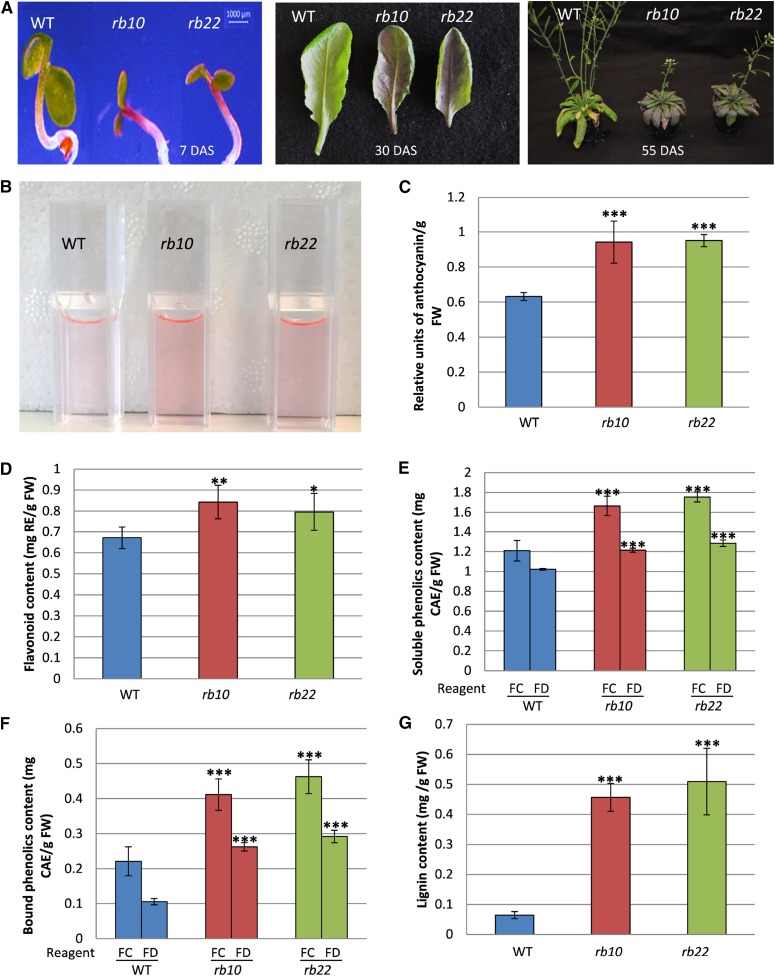
A regularly observed phenotype of raptor1b is the accumulation of purple pigmentation, suggesting increased levels of anthocyanins in young seedlings and small rosette leaves, but also in bolted plants (Fig. 9A). This visual phenotype was analyzed by extracting and quantifying the total anthocyanins from wild-type and raptor1b tissue (Fig. 9, B and C). Apart from the anthocyanins, total flavonoids and other phenolic secondary metabolites, such as free soluble and cell wall-bound phenolics, including lignin, were measured spectrophotometrically. All of these specialized compound classes showed significant increases in their levels in raptor1b lines (Fig. 9, D–G).
Figure 9.
Influence of raptor1b mutation on phenolics accumulation. A, Visible accumulation of anthocyanin in young raptor1b seedlings (7 DAS), rosette leaves (30 DAS), and mature plants (55 DAS). B, Anthocyanins extracted from raptor1b compared with the wild type (WT). C to G, Total anthocyanin (C), flavonoids (D), soluble phenolics (E), cell wall-bound phenolics (F), and total lignin (G) in raptor1b compared with the wild type. For extraction, the rosettes of raptor1b and the wild type were harvested from plants at the same developmental stage (10 rosette leaves). Plants were cultivated on soil under normal-light LD growth conditions. Data represent means ± sd for five biological replicates. Asterisks indicate significant differences between the wild type and raptor1b under the same condition (***, P < 0.001, Student’s t test). CAE, Chlorogenic acid equivalent; FC, Folin-Ciocalteu reagent; FD, Folin-Denis reagent; FW, fresh weight; RE, rutin equivalent.
To further deepen the understanding of the observed changes in the total levels of these secondary metabolites, and additionally to assess the status of other secondary metabolite classes in raptor1b, we performed ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) metabolic profiling on the polar fraction using our comprehensive, sequential extraction method (Salem et al., 2016).
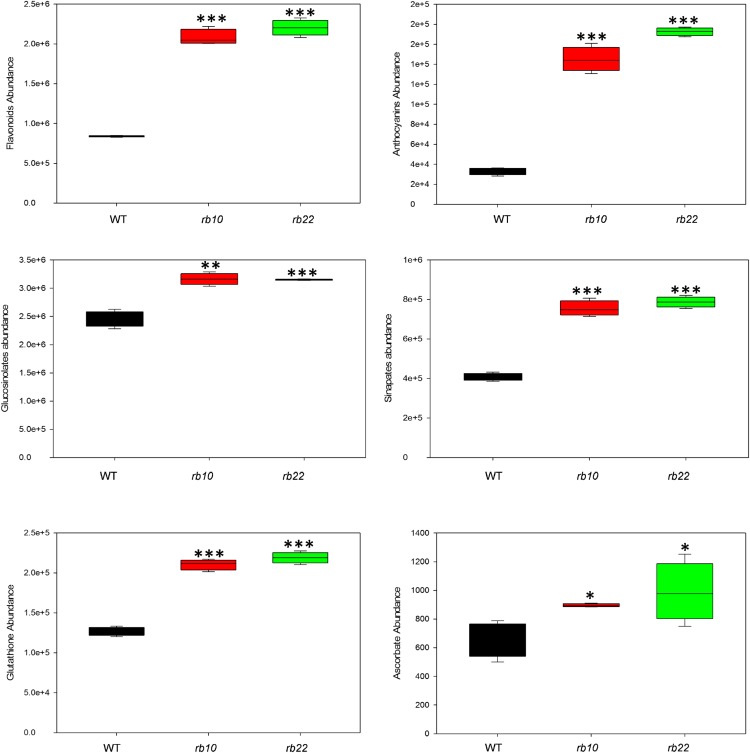
These measurements confirmed that flavonoids and anthocyanins were increased significantly in raptor1b mutants (Fig. 10). A detailed overview of the specific species can be found in Supplemental Data Set S4. Additionally, we found that sinapate esters (Milkowski and Strack, 2010) as well as glucosinolate levels (Halkier and Gershenzon, 2006) increased significantly in raptor1b. Moreover, we detected significant accumulation of antioxidant compounds including reduced and oxidized glutathione, as well as scopoletin (Fig. 10; Supplemental Fig. S12; Supplemental Data Set S4). These results suggest that mutation of RAPTOR1B led to massive modulation of secondary metabolism by heavily inducing the accumulation of stress-related, protective metabolites.
Figure 10.
Influence of raptor1b mutation on secondary metabolism. The sum of the peak intensities of detected flavonoids, anthocyanins, glucosinolates, and sinapates, as well as the intensities of glutathione and ascorbate, are shown. For extraction, the rosettes of raptor1b and the wild type (WT) were harvested from plants at the same developmental stage (10 rosette leaves). Plants were cultivated on soil under normal-light LD growth conditions. Data represent means ± sd for five biological replicates. Asterisks indicate significant differences between the wild type and raptor1b under the same condition (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, Student’s t test).
Reactive Oxygen Species Production and Autophagy Are Induced in raptor1b Plants
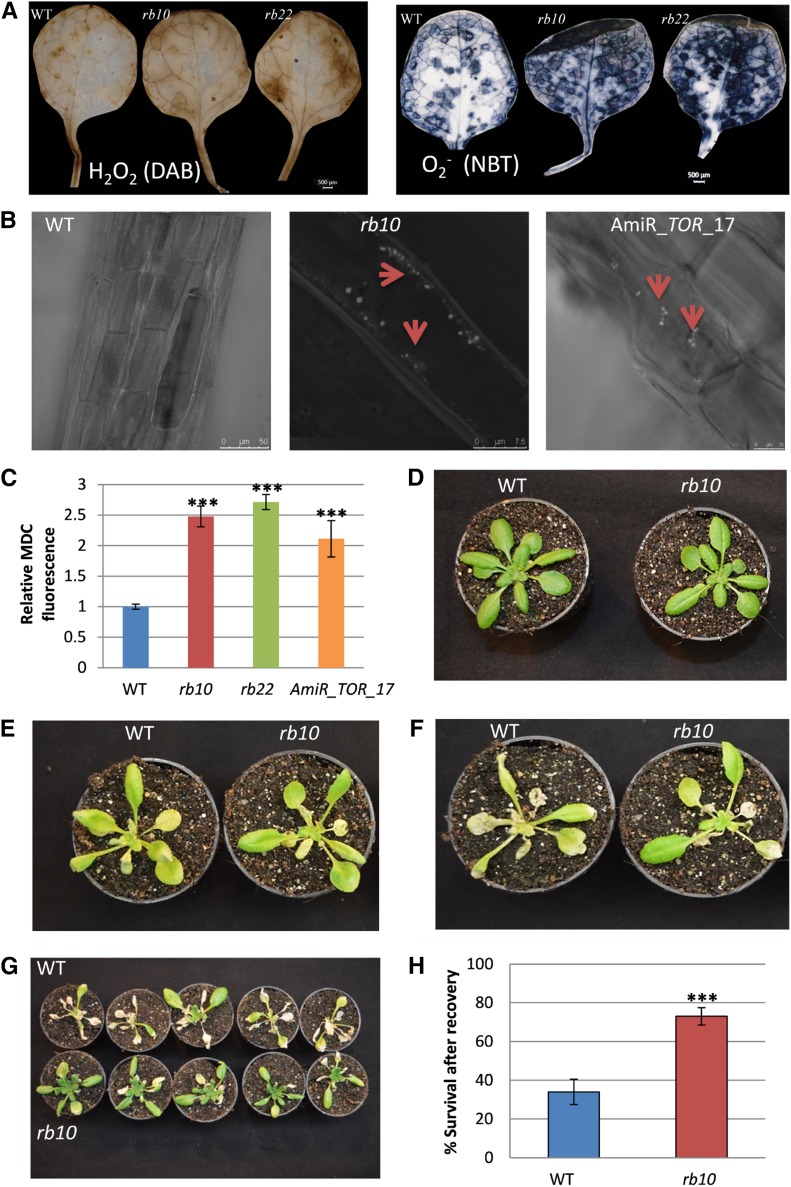
As mentioned in the previous section, raptor1b plants accumulated stress-related primary and secondary metabolites. Several phenol-derived metabolites, such as flavonoids and anthocyanins, have been reported frequently as reactive oxygen species (ROS) scavengers in plants exposed to stressful conditions (Fini et al., 2011; Lepistö and Rintamäki, 2012). Excess light, as well as other environmental cues like nutrient excess or depletion, can be key stimuli for stress-related anthocyanin production in green plants (Steyn et al., 2002; Das et al., 2011). To investigate if the increase in these stress-induced metabolites is connected to ROS levels, we determined the levels of common ROS, including superoxide (O2•−) and hydrogen peroxide (H2O2), in wild-type and raptor1b leaves. As expected from the metabolic signatures, we found that the amounts of both H2O2 and O2•− were increased significantly in raptor1b plants (Fig. 11A).
Figure 11.
Influence of raptor1b mutation on ROS, autophagy, and tolerance to carbon starvation. A, Histochemical staining of ROS in rosette leaves. The levels of H2O2 and O2•− were assessed by 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining, respectively. Bars = 500 µm. B, MDC-labeled autophagosomes observed by fluorescence microscopy in roots of 10-d-old seedlings of the wild type (WT), raptor1b, and the TOR-repressed line amiR-TOR-17. Red arrowheads indicate the positions of autophagosomes. C, MDC-derived fluorescence of roots was quantified by confocal microscopy following raptor1b mutation or TOR repression relative to the wild type. D to H, Effects of raptor1b on the survival of fixed-carbon starvation. The wild type and raptor1b at the same developmental stage (D) were exposed to 12 d of dark treatment (E), then allowed to recover for 4 d (F) and 7 d (G) after dark treatment. Survival rate (H) was scored after 7 d of recovery from fixed-carbon starvation. Data represent means ± sd for five biological replicates for C and 20 biological replicates for H from at least two independent experiments. Asterisks indicate significant differences between the wild type and raptor1b under the same condition (***, P < 0.001, Student’s t test).
Previous reports indicated that the induction of ROS can be linked to the induction of autophagy (Pérez-Pérez et al., 2012), which is connected to the inhibition of TOR in Arabidopsis (Liu and Bassham, 2010; Pu et al., 2017) and Chlamydomonas reinhardtii (Pérez-Pérez et al., 2010). Therefore, we asked if ROS accumulation in raptor1b mutants is associated with the induction of autophagy. To test this hypothesis, we quantified autophagosomes, the hallmark of autophagy, using the acidotropic dye monodansylcadaverine (MDC), which selectively labels these subcellular structures (Liu and Bassham, 2010). To benchmark our analysis strategy, we used our previously developed estradiol-inducible amiR lines of AtTOR (amiR_TOR_17 and amiR_TOR_9; Caldana et al., 2013), which induce autophagy. In accordance with previous results, these lines showed a clear increase in autophagosomes (Liu and Bassham, 2010; Pu et al., 2017; Fig. 11B).
Next, we repeated the analysis using the rb10 lines. Visible inspection of MDC-labeled roots clearly disclosed increased quantities of autophagosomes in raptor1b plants compared with the wild type (Fig. 11B). In addition to the KO of RAPTOR1B in rb10 plants and the inducible repression of TOR, we additionally analyzed estradiol-induced amiR lines of AtRAPTOR1B (amiR_RAPTOR1B) and compared their autophagosome phenotype with the wild type and amiR lines of RAPTOR1A using another amiR line (amiR_RAPTOR1A). We observed that MDC-labeled autophagosomes were rarely detectable in wild-type and amiR_RAPTOR1A plants, but similar to the rb10 RAPTOR1B KO plants, amiR_RAPTOR1B lines showed a strong increase in autophagosomes (Supplemental Fig. S13). To statistically illustrate these results, we quantified the total fluorescence derived from confocal images. As can be seen from Figure 11C, these results depict significantly higher fluorescence levels in raptor1b and TOR-repressed lines compared with their wild type, confirming that activation of autophagy is induced upon the repression of RAPTOR1B but not RAPTOR1A.
raptor1b Plants Are More Tolerant to Carbon Starvation Conditions
Our results indicated that raptor1b plants accumulate higher levels of storage materials, such as starch and TAG (Fig. 7, A and B), accompanied by a significant induction of autophagy (Fig. 11, A–C). Accordingly, we hypothesized that these mutants, in contrast to autophagy mutants (Chung et al., 2010; Suttangkakul et al., 2011), should be able to tolerate nutrient deprivation conditions better than their respective wild type. To test this hypothesis, the effects of fixed-carbon starvation were evaluated by a greening-after-extended-darkness assay (Choy et al., 2008; Chung et al., 2010). For this purpose, wild-type and raptor1b plants were subjected to 12 d of darkness after they reached the 10-rosette-leaves developmental stage. The plants were then allowed to recover for 1 week, and their survival rate was scored. As predicted, raptor1b developed a visibly better ability to regreen and produce new leaves after the 12-d carbon starvation and, hence, recovered from the extended darkness (Fig. 11, D–G). Additionally, mutant plants also evolved a significantly higher survival rate after recovery compared with the wild type (Fig. 11H), indicating that they are more tolerant to carbon starvation conditions.
DISCUSSION
RAPTOR Links TOR Signaling to Cell Elongation
Our detailed phenotypical and biochemical analysis of the raptor1b mutants revealed multiple developmental, growth, and molecular phenotypes. One of these phenotypes was the significant decrease in the number and length of root hairs (Fig. 4, C and D). This root hair phenotype was shown in conditionally repressed TOR RNAi lines (Xiong and Sheen, 2012) as well as in plants transformed with a copy of the yeast FKBP12 gene, which made the otherwise rapamycin-insensitive Arabidopsis plants sensitive to high (10 µm) concentrations of the TOR-repressing drug (Ren et al., 2012). Similarly, this phenotype was confirmed in Arabidopsis plants treated with several ATP-competitive TOR inhibitors (Montané and Menand, 2013).
The formation of root hairs is based on the extension of root epidermal cells, a process dependent upon the synthesis and rearrangement of cell wall components (Grierson et al., 2014). Previous studies have already shown that the transcriptional down-regulation of cell wall-related genes, especially extensins (Moreau et al., 2012; Caldana et al., 2013; Xiong et al., 2013; Dong et al., 2015), could be associated to this phenotype. In this study, we provide evidence that the receptor-like protein kinase FERONIA (FER) is decreased significantly in raptor1b plants. The KO of FER has been shown to cause severe root hair defects (Duan et al., 2010) in addition to globally reducing cell elongation during vegetative growth (Guo et al., 2009). The main target of FER in root hair development has been shown to be the Rho-related GTPase of Plants2 (ROP2), which localizes to the root hair tip and drives, possibly with other members of the ROP family (Molendijk et al., 2001), cell elongation in a pH-dependent and ROS-mediated process (Grierson et al., 2014). Interestingly, two recent studies (Li et al., 2017; Schepetilnikov et al., 2017) also showed that ROP2 is associated with auxin-dependent TOR activation and signaling in Arabidopsis.
In agreement with the root hair phenotype, which was driven by defective cell elongation, we similarly confirmed a TOR-specific cell size phenotype in the leaf tissue (Fig. 3G; Supplemental Fig. S5). As we have shown previously for the amiR repression of TOR (Caldana et al., 2013), the KO of RAPTOR1B led to smaller epidermal and mesophyll cells (Fig. 6A; Supplemental Fig. S5). Contrary to the results using the inducible amiR system, where we only observed the smaller cell size in smaller leaves without employing developmental stage matching (Caldana et al., 2013), the RAPTOR1B T-DNA lines allowed us to additionally look at the impact on organ size at different growth stages. Accordingly, we were able to show that the phenotype of smaller cells did not necessarily lead to smaller leaves, since mutant plants were compensating for the reduced cell size with increased cell number (Fig. 6, A and B). Therefore, these results place RAPTOR/TOR upstream of cell elongation without affecting cell division.
raptor1b Shows Delays during Developmental Stage Transitions
To set up a fair comparison system for the wild type and raptor1b, we determined the time required for each genotype to reach distinct, well-defined developmental stages (Boyes et al., 2001). Contrary to our initial expectation, the mutation of RAPTOR1B did not result in a linear developmental delay due to continuous growth repression but, instead, accumulated delays during three distinct developmental phases (Fig. 2I). The first delay, of approximately 2 to 3 d, was accumulating during the germination process, a result we described in great detail in a study of the RAPTOR1B mutation and its impact on seed development (Salem et al., 2017). The second delay, of an additional 7 d, which mostly affected vegetative growth, was obtained between the 10-rosette-leaves stage and the termination of rosette growth (3.90). The last and longest delay, of an additional 10 d, was obtained during the beginning of flower development (5.1 and 6.0) and silique ripening (8.0), leading to a total delay of approximately 15 to 20 d (Fig. 2I).
Looking in more detail at these developmental stages, we did see a transition delay for all the main transitions within the life cycle of a flowering plant, including germination, heterotrophic to autotrophic, juvenile vegetative to adult vegetative, and adult vegetative to reproductive (Huijser and Schmid, 2011). The proper function of RAPTOR/TOR seems to be required in each of these steps. The proper timing of these transitions is controlled by multiple factors, including endogenous signals and environmental conditions (Huijser and Schmid, 2011). Prominent among these signals are nutrients, including sugar (Yu et al., 2013) and nitrogen-containing compounds (Vidal et al., 2014), in addition to hormones (Gazzarrini et al., 2004) and microRNAs (Huijser and Schmid, 2011). A recent study in Arabidopsis has already linked TOR and sugar signals to the heterotrophic-to-autotrophic transition (Xiong et al., 2013), while our analysis of seed development indicated that the germination process of raptor1b was delayed significantly due to high ABA levels (Salem et al., 2017). It was now interesting to integrate our data and determine which other factors could influence the transitions from juvenile to adult and also the transition from adult to reproductive stage in raptor1b. Contrary to the high-ABA phenotype detected in raptor1b seeds (Salem et al., 2017), a constitutive low-ABA phenotype throughout the day was detected in raptor1b leaves, indicating that different developmental stages also can result in differential hormonal phenotypes in the mutant (Fig. 6C). Accordingly, it was also not surprising that the hormonal phenotype was not based on the inability of raptor1b to produce higher ABA levels if drought stress was applied (Fig. 6G). These results indicate that, depending on the context, the loss of RAPTOR1B had different impacts on the phenotypic outcome. One factor that was shown to strongly modulate ABA levels in plants, especially in seeds, is the B3 domain transcription factor FUSCA3 (FUS3; Gazzarrini et al., 2004). Interestingly, FUS3 also was shown to be a positive regulator of miR156 (Wang and Perry, 2013), which negatively regulates the phase transition from juvenile to adult (Guo et al., 2017) and, further, the adult-to-reproductive state (Huijser and Schmid, 2011). Recently, it was demonstrated that FUS3 interacts physically with SNF Kinase Homolog10 (KIN10; Tsai and Gazzarrini, 2012). KIN10 is the catalytic subunit of the cellular energy sensor SnRK1 (Snf1-related protein kinase; Broeckx et al., 2016) and phosphorylates FUS3, which seems to stabilize the transcription factor by an unknown mechanism (Tsai and Gazzarrini, 2012). Our proteomic data indicate that KIN10, which can stabilize the FUS3 protein, was reduced by almost 50% (Supplemental Fig. S9B; Supplemental Data Set S1), providing a rationale for the decreased ABA levels and also for the increased levels of miR156, which would explain the delay in phase transitions.
Recent data showed that AKIN10, as a component of SnRK1, interacts directly with RAPTOR and phosphorylates it at an unknown position (Nukarinen et al., 2016). A similar scenario has been described in mammalian cells, where AMP kinase, the ortholog of SnRK1 in animals, has been shown to bind and phosphorylate RAPTOR within the TOR complex and, thus, repress TOR complex activity (Gwinn et al., 2008). Taken together, these results suggest that the RAPTOR-containing TOR complex and SnRK1 might have antagonistic functions and that active SnRK1 represses TOR directly. Still, the decrease of AKIN10 protein levels in the raptor1b background might indicate that the direct antagonistic function also relies on the presence of a functional RAPTOR within the TOR complex.
Photosynthetic Electron Transport Is Not Affected in raptor1b, But CO2 Fixation Is Reduced Significantly
Photosynthesis provides the energy and the reduction equivalents for carbon fixation and other anabolic processes, which serve as the basis for biomass accumulation and growth (Smith and Stitt, 2007). Accordingly, it was speculated that the repression of RAPTOR1B, which resulted in significantly reduced and delayed growth (Figs. 1–4), might lead to decreased photosynthetic efficiency and reduced carbon assimilation. Transcription data in Arabidopsis indicate that reduced TOR signaling represses photosynthetic genes (Ren et al., 2012; Dong et al., 2015), while the repression of either S6K or RAPTOR in rice (Oryza sativa) leads to massively reduced synthesis of photosynthetic pigments, thylakoid lipids, and photosynthesis rates (Sun et al., 2016).
In accordance with the results from the rice study (Sun et al., 2016), we also detected a significant reduction of S6K activity in raptor1b plants (Supplemental Fig. S8D), indicating that the KO of RAPTOR1B leads to reduced TOR activity. Nevertheless, we did not detect any changes in the levels of chlorophyll or other photosynthetic pigments (Supplemental Fig. S7). Similarly, we did not detect relevant changes in the concentration and the composition of the main thylakoid lipids, namely the glycolipids MGDG, DGDG, and SQDG (Fig. 8). Based on these contradicting observations, we were not surprised that the linear electron transport in raptor1b was not altered compared with the wild type. The light-response curves of chlorophyll fluorescence parameters (ETR, qL, and qN) and the levels of dark respiration and photorespiration were largely unaltered between wild-type and mutant leaves (Fig. 5). Therefore, these data suggest that, contrary to the results obtained in rice (Sun et al., 2016), the primary reactions of photosynthesis are not affected significantly by the raptor1b mutation in Arabidopsis. This result was further supported by the proteomic profiles of proteins associated with photosynthetic reactions (Supplemental Data Set S1), where only a few singular changes could be detected. Accordingly, the question arises of the underlying causes for these major differences between the rice study and our data. On the one hand, this could indicate that there is a functional difference in RAPTOR and S6K activity between monocot and dicot plants. Alternatively, these severe phenotypic differences could possibly also be related to the balance between survival and lethality observed in the initial RAPTOR studies (Anderson et al., 2005; Deprost et al., 2005). While the first study showed that raptor1b lines were embryo lethal (Deprost et al., 2005), the study by Anderson et al. (2005) obtained plants with severe growth phenotypes but otherwise fully vital, indicating that these lines are highly sensitive to the applied growth conditions. Accordingly, it also may be that the different phenotypic responses between Arabidopsis and rice might be related to the different growth conditions of rice and Arabidopsis. Nevertheless, the rice phenotype seems to be more similar to the phenotype obtained after the repression of TOR, since the down-regulation of TOR leads to severe chlorosis and, finally, to lethality (Deprost et al., 2007; Caldana et al., 2013). This observation, therefore, clearly implies that the Arabidopsis RAPTOR1B mutation is partially complemented by the second homolog, RAPTOR1A, especially since double mutants are essentially lethal (Anderson et al., 2005).
In contrast to the unaltered photosynthetic electron transport rates, we found significantly reduced CO2 assimilation rates in raptor1b (Fig. 5E). This observation could not be explained by a decrease in stomatal conductance, since conductance was constitutively elevated in the mutants (Fig. 5G). The increased conductance also was accompanied by a larger number of stomata per leaf (Fig. 6A) in addition to a larger aperture of these stomata (Fig. 6B). This phenotype could be explained by the low ABA concentration in the raptor1b mutant throughout the entire day (Fig. 6C), since ABA is required for the efficient control and closure of stomata (Finkelstein, 2013). As a consequence of the increased stomatal opening, transpiration rates were quite high in these plants, which leads to decreased water use efficiency linked to a decreased turgor pressure and reduced cell elongation (Ache et al., 2010). Taken together, the reduced ABA levels explain a number of observed phenotypes, including smaller epidermal and mesophyll cells due to lower turgor pressure and, as a consequence, altered leaf structure and organization (Fig. 3; Supplemental Fig. S4). Accordingly, ABA treatment of raptor1b plants complemented the growth repression (Fig. 6D), indicating that the other low-ABA phenotypes possibly could be reverted by external ABA treatment.
raptor1b Shows a Starch-Excess Phenotype
Starch is the largest carbon-storage compound in Arabidopsis, accumulating during the day to serve as a carbon resource to promote growth during the night (Smith and Stitt, 2007). Half of the CO2 fixed in the Calvin-Benson cycle is actually stored as transitory starch (Zeeman and Ap Rees, 1999). Accordingly, the partitioning of fixed carbon between starch storage and its utilization for biomass accumulation determine, at least in part, the growth rate of the plant (Sulpice et al., 2009).
Previous studies in genetically or chemically repressed TOR plants (Deprost et al., 2007; Caldana et al., 2013; Dobrenel et al., 2013), as well as in mutants of the TOR complex component lst8 (Moreau et al., 2012), confirmed this negative correlation by showing significantly higher starch concentrations in plants of reduced size. These results, at first glance, were in full agreement with the results obtained for raptor1b mutants, where starch concentrations were almost twice as high compared with the wild type (Fig. 7A). However, a major difference became obvious when looking at these results in more detail. Starch data for the raptor1b mutants were collected at five time points throughout an entire day (Fig. 7A) and, thus, contrary to the previous TOR-related studies, where only a single time point was sampled, provided information on diurnal starch distribution. raptor1b plants not only contained higher concentrations in the middle or at the end of the day compared with the wild type but also contained higher concentrations at the end of the night (Fig. 7A). Accordingly, the diurnal starch concentration did not decrease to wild-type levels, indicating that raptor1b plants have a starch-excess phenotype. In previous studies, it was reported that starch excess was not only characterized by higher starch concentrations at the end of the day but also by continuously accumulating starch concentrations between subsequent days (Streb and Zeeman, 2012). Therefore, contrary to the previous assumptions, it cannot be concluded that raptor1b plants accumulate more starch during the day than wild-type plants, since both genotypes showed an increase of ∼10 mg starch g−1 fresh weight during the 16-h light phase (Fig. 7A). Nevertheless, due to the fact that CO2 assimilation in raptor1b mutants was reduced (Fig. 5E), while the absolute amount of fixed starch was the same as in the wild type, it can be concluded that significantly lower amounts of fixed carbon were partitioned to anabolic processes. In agreement with this assumption, the amount of Suc, which directly fuels from carbon fixation into anabolic growth processes (Smith and Stitt, 2007), was decreased significantly in raptor1b mutants (Fig. 7C).
A similar starch-excess phenotype was described for the akin10/11 double mutant (Baena-González et al., 2007) of the catalytic subunits of SnRK1. This result, at first glance, is a little confusing, since the function of SnRK1, the energy sensor of the cell, was suggested to be antagonistic to that of a functional TOR-RAPTOR complex (Baena-González and Hanson, 2017). Still, the similarity in the starch phenotypes seems to be in agreement with the observation that the amount of AKIN10 was repressed significantly in raptor1b mutants (Supplemental Fig. S9B). A possible reason for these overlapping starch phenotypes might emerge from the fact that AKIN10 colocalizes and phosphorylates RAPTOR (Nukarinen et al., 2016), which might provide the necessary entry point for the antagonistic, negative-feedback regulation between the TOR complex and SnRK1. To further elucidate these relationships, additional results will be required.
Induction of Autophagy and Altered Nitrogen Metabolism Are Basic Signatures of raptor1b
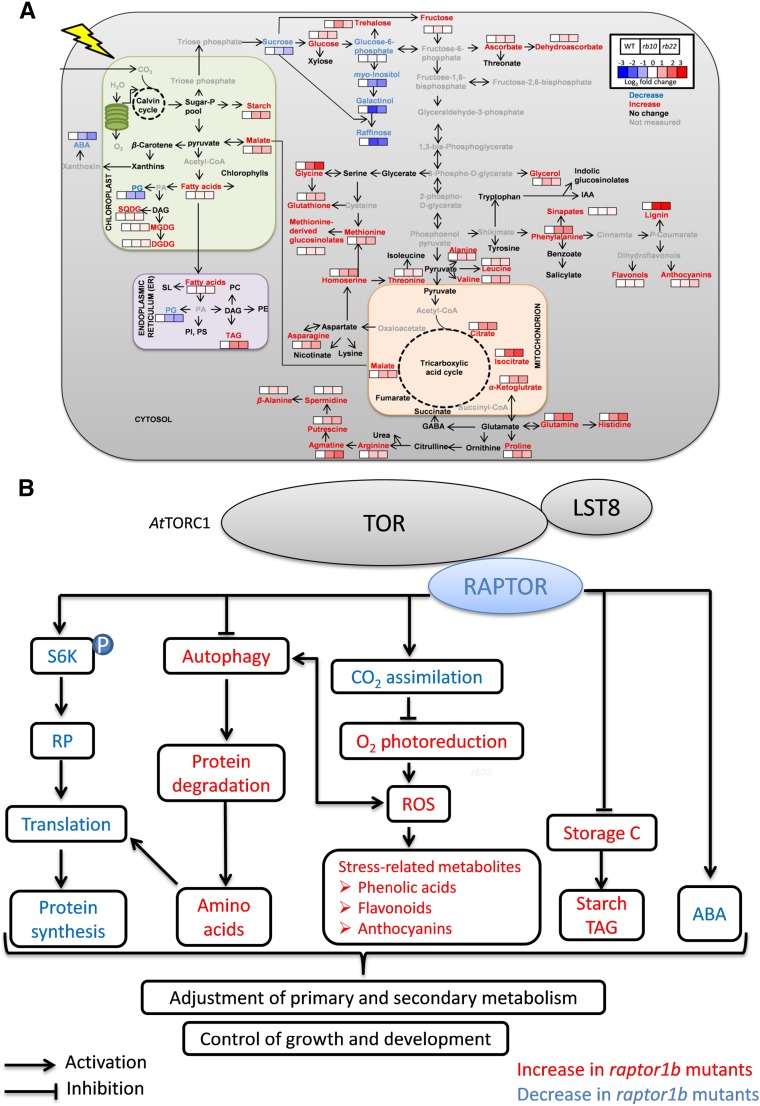
Beyond the starch-excess phenotype, we observed major changes in primary and secondary metabolism in raptor1b (Fig. 7C; Supplemental Figs. S10A and S12; Supplemental Data Sets S2 and S4). These changes are systematically summarized, in comparison with the lipid changes, in Figure 12A. The most obvious pattern in this overview is characterized by the massive increase in amino acids and secondary metabolites. The accumulation of the majority of amino acids and tricarboxylic acid cycle intermediates in raptor1b is consistent with the described phenotypes for the lst8 mutation (Moreau et al., 2012) and TOR repression (Deprost et al., 2007; Ren et al., 2012; Caldana et al., 2013). The increased amino acid levels were discussed previously to have several possible causes (Caldana et al., 2013), such as the repression of protein biosynthesis and the induction of proteolysis by autophagy.
Figure 12.
Mutation of RAPTOR1B leads to changes of growth and metabolism in Arabidopsis. A, raptor1b shows significantly changed compounds, including lipids, metabolites, and hormones. The log2 fold changes of metabolite abundance in raptor1b relative to the wild type at the end of the day are shown by color scale, blue and red for significant decrease and increase (P < 0.05, Student’s t test), respectively. Metabolites with blue, red, and black characters represent significant decrease, increase, and no change, respectively. Metabolites with gray characters were not measured. For simplicity, only some of the main pathways are shown. Solid arrows represent single or multiple reactions. GABA, γ-Aminobutyric acid; IAA, indole-3-acetic acid; PI, phosphatidylinositol; PS, phosphatidylserine; SL, sphingolipids. B, Model for the influence of TOR/RAPTOR inhibition on signaling networks, growth, and metabolism in Arabidopsis. TOR as a central regulator controls plant growth and metabolism through the phosphorylation of regulatory targets. Arrows indicate activation, and bar-headed lines indicate inhibition.
Decreased protein biosynthesis has been demonstrated at multiple levels after TOR repression in animal systems (Ma and Blenis, 2009) and also in plants (Deprost et al., 2007; Sormani et al., 2007; Ahn et al., 2011; Ren et al., 2011, 2012; Schepetilnikov et al., 2011, 2013; Dobrenel et al., 2016). These protein biosynthesis targets were, among others, translation initiation, measured by decreased polysomal loading (Deprost et al., 2007; Sormani et al., 2007; Ahn et al., 2011; Schepetilnikov et al., 2011, 2013; Dobrenel et al., 2016), as well as ribosomal biogenesis, measured by decreased synthesis of rRNA (Ren et al., 2011, 2012). Since we did not measure polysome loading or rRNA concentrations, the evidence of repressed protein biosynthesis in our data mostly comes from the significant decrease in S6K1 activity (Supplemental Fig. S8D), which has been shown to control translation (Dobrenel et al., 2016), and also from the significant decreases in the levels of several ribosomal proteins (Supplemental Fig. S9).
One alternative mechanism, which could lead to an increased accumulation of amino acids, comes from the described increase in catabolic activity after TOR repression. The connection between TOR and autophagy and protein degradation has been well established in multiple animal systems (Dunlop and Tee, 2014), with more evidence being gathered for this interaction in plants (Díaz-Troya et al., 2008). TOR is a repressor of autophagy in C. reinhardtii (Pérez-Pérez and Crespo, 2010) and Arabidopsis (Liu and Bassham, 2010). Our study, therefore, is in full agreement with these TOR-related results; moreover, we also confirm the results of a very recent study showing that the mutation of RAPTOR1B induces autophagy (Pu et al., 2017), namely by demonstrating the induction of autophagosomes in the raptor1b background (Fig. 11, B and C; Supplemental Fig. S13). In addition to the autophagosome phenotype, we show that raptor1b plants are significantly more resistant to fixed-carbon starvation (Fig. 11, D–H), which is a common hallmark of plants with high autophagy activity (Chung et al., 2010). Additionally, our data are in full agreement with a recent study using etiolated seedlings as a model for carbon starvation, where it was shown that the mutation of essential autophagy genes leads to decreased amounts of amino acids and several secondary metabolites, including glucosinolates and flavonoids (Avin-Wittenberg et al., 2015). This result, which is opposite to our phenotype of the raptor1b mutants, where almost all secondary metabolites were increased (Figs. 9, 10, and 12A), might illustrate that the induction of autophagy by TOR repression leads to a redirection of nutrients into secondary metabolism.
CONCLUSION
In conclusion, the physiological and molecular phenotypes observed in raptor1b support the fundamental roles of TOR signaling in balancing plant growth and development via the modulation of cellular metabolism. This seems to be achieved through diverse mechanisms, including a selective regulation of hormonal balance and the direct regulation of essential biological processes controlling anabolic and catabolic metabolism. Figure 12B summarizes these major findings in a simplified model, which awaits further studies to fill the gaps with better mechanistic understanding.
MATERIALS AND METHODS
Plant Material, Cultivation, and Growth Conditions
Detailed phenotypic analysis was performed on two raptor1b T-DNA KO lines (SALK_101990 and SALK_022096), which were abbreviated rb10 and rb22, respectively. These raptor1b mutants were described in our previous report (Salem et al., 2017). Mutants of RAPTOR1A (raptor2-1; SALK_043920) have been described before (Anderson et al., 2005; Deprost et al., 2005). These mutants were compared with wild-type Arabidopsis (Arabidopsis thaliana) of the ecotype Col-0. For plate-based phenotypic analysis, seeds (wild-type Col-0 and raptor1b T-DNA KO lines) were surface sterilized before being sown on plates containing 0.5× MS salts and 1% (v/v) agar. The plates were arranged vertically in racks before being transferred to a cold room (4°C) in darkness for 3 d to synchronize germination. The plates were then transferred to phytotrons with a 16/8-h light/dark cycle (LD). The average light intensity was maintained at 120 µmol m‒2 s‒1. The day/night temperature and relative humidity were maintained at 20°C/16°C and 60%/75%, respectively. For phenotypic analysis on soil under photoautotrophic conditions, seeds were stratified in water at 4°C in darkness for 3 d before being sown on soil in pots. LD growth conditions were maintained at a 16/8-h light/dark cycle. The average light intensity was maintained at 150 µmol m‒2 s‒1. The day/night temperature and relative humidity were 20°C/16°C and 60%/75%, respectively. For C-starvation treatment, wild-type and raptor1b plants were transferred, after they reached the 10-rosette-leaves developmental stage, to extended darkness for 12 d. The survival percentage was scored after the plants were allowed to recover for 1 week under LD conditions. For hormone treatment, seeds of the wild type and raptor1b were germinated on 0.5× MS medium containing 1% (w/v) agar for 7 d, and then the 1-week-old seedlings were transferred to new plates with or without 10 nm ABA for another 10 d. For drought stress, wild-type and raptor1b plants were grown under standard LD conditions until they reached the same developmental stages (10 rosette leaves). After this, watering was stopped for 6 d. Samples (rosette leaves) were harvested at the end of the day from untreated control and drought-treated plants.
RNA Extraction and Reverse Transcription-Quantitative PCR Analysis of RAPTOR1A
Young seedlings (10 d old) of the wild type and raptor mutants were harvested and snap frozen in liquid nitrogen. Total RNA was extracted from 50 mg of tissue using NucleoSpin RNA plant lysis buffer according to the manufacturer’s instructions (Macherey-Nagel). Total RNA (300 ng) was used as a template to perform reverse transcription reactions using the RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific). Reverse transcription-quantitative PCR assays were performed using the Maxima SYBR Green/ROX qPCR Master Mix (2X) kit (Thermo Scientific) according to the manufacturer’s protocol. Transcript levels were determined using the following primers: RAPTOR1A_qRTf (5′-ATGCAAAGTGAATATGGAGTTGGG-3′) and RAPTOR1A_qRTr (5′-GCGTAAATTCTATTTGGCAAAGCC-3′).
Microscopic Analysis of Cell Size and Number
Images of individual leaves and young seedlings were captured using a Leica stereomicroscope (MZ 12.5, DC DFC 420, Soft Imaging System; LAS). For the analysis of meristem size, roots of 12-DAS seedlings were mounted in chloral hydrate solution (Perilli and Sabatini, 2010) and examined with a microscope. For the measurement of cell size and number, rosette leaf 5 was chosen from both the wild type and raptor1b at the same developmental stage (10 rosette leaves) with five biological replicates for each genotype. The leaves were bleached using a solution of 12.5% (v/v) acetic acid in ethanol, then the samples were washed using ethanol in different proportions and finally examined in 1 m KOH. Photographs were taken from at least six different positions, covering the whole lamina. For measurements of cell size and number, images were captured using the Olympus Epi-Florescence Microscope BX-51, DC View III (5 MP), Soft Imaging System (CellSens). The number of cells per fixed area (0.14 mm2) was counted from at least 36 sections for each genotype. The average cell area was calculated after measuring at least 200 cells for each genotype.
Microscopic Analysis of Leaf Structure
Freshly harvested fifth leaves were fixed in fixative buffer (4% [w/v] paraformaldehyde and 0.25% [w/v] glutaraldehyde in PBS, pH 7.4) for 1 h under vacuum at room temperature before a 24-h incubation in fresh fixative buffer at 4°C. The leaves were washed with PBS three times (15 min each) before dehydration in a graded ethanol series and embedding in Technovit 7100 historesin (Heraeus-Kulzer). The leaf sections (3 µm thick), obtained with a microtome (Fully Automated Rotary Microtome Leica RM2265; Leica Microsystems), were stained with 0.05% (w/v) Toluidine Blue O (in 100 mm phosphate buffer, pH 7.2) for 1 min or 10% (v/v) aqueous Lugol solution for 2 min. Images were captured using the Olympus Epi-Florescence Microscope BX-51, DC View III (5 MP), Soft Imaging System (CellSens).
For the measurement of stomatal aperture, rosette leaves (leaf 5) from plants grown under LD conditions were detached from the wild type and raptor1b at the same developmental stage (10 rosette leaves). The detached leaves were frozen rapidly in liquid nitrogen before examination with a tabletop microscope (TM3030Plus; Hitachi High-Technologies Europe; Lisso et al., 2011).
Analysis of Root Growth Rate
Seeds of the wild type and raptor1b were surface sterilized before being sown on plates containing 0.5× MS medium and 1% (w/v) agar. The plates were arranged vertically in racks before being transferred to a cold room (4°C) in darkness for 3 d to synchronize germination. After stratification, the plates were transferred to growth chambers with a 16/8-h light/dark cycle (LD). On day 9, seedlings that had developed roots of at least 1 cm length were selected and transferred to a new 120- × 120-mm petri dish filled with solid medium. The selected seedlings were distributed equally on the surface of the medium along a row 3 cm away from the upper edge of the petri dish. After another 2 d in the phytotron, petri dishes were mounted in the PlaRoM platform to analyze root elongation kinetics according to Yazdanbakhsh and Fisahn (2012). Root elongation was measured for six wild-type and six raptor1b plants continuously over a period of 10 d with a sampling rate of 30 min. Root hair numbers were measured from the wild type and raptor1b in 7- and 12-d-old seedlings that were grown on agar plates (0.5× MS medium and 1% [w/v] agar) under LD growth conditions. At least 20 seedlings from each line were used for microscopic analysis, which was performed using the Olympus Epi-Florescence Microscope BX-51, DC View III (5 MP), Soft Imaging System (CellSens).
In Vivo Determination of Photosynthetic Efficiency and Gas-Exchange Measurements
Chlorophyll fluorescence parameters were determined on rosette leaves of wild-type and raptor1b plants at the same developmental stage (10 rosette leaves) with the MAXI-version of the IMAGING-PAM M-series fluorometer (Walz; IMAG K6 unit). Before determination of Fv/Fm, ETRs, qL, and qN, plants were dark adapted for 30 min (Schmitz et al., 2012). Stomatal conductance, assimilation, and photorespiration rates of the wild type and raptor1b were recorded as described previously (Albus et al., 2012). In brief, two portable gas-exchange systems (GFS-3000 with 3010-S standard measuring heads; Walz) served in parallel to quantify photosynthetic parameters under the LD conditions described in the previous section. Two fiber illuminators (FL 400; Walz) provided actinic light at a photon flux density of 150 µmol m‒2 s‒1. A timer (00137251; Hama) adjusted the photoperiod during the gas-exchange measurements to light periods of 16 h (with 8 h of darkness). Because of small leaf areas, the air flow rate within the cuvette was set to only 600 µmol min‒1. Temperature and humidity within the cuvette matched the ambient conditions of 22°C and 60%, respectively, at 350 µL L−1 CO2. Diurnal time series in photosynthetic parameters proceeded with a sampling rate of 10 min. Photorespiration was estimated from the increase in leaf assimilation after flushing the system with nitrogen supplemented with 350 µL L−1 CO2. Gas-exchange measurements were repeated at three different seasons, in May, September, and November 2016, with six to 10 replicates at each time. However, no significant seasonal differences were observed.
Absolute Quantification of ABA from Rosette Leaves
ABA was quantified according to the method described by Pan et al. (2010) with minor modifications. Briefly, frozen ground material (50 mg) from the 10-rosette-leaves developmental stage was homogenized in 0.5 mL of extraction buffer (2-propanol:water:HCl, 2:1:0.002) (v/v/v). Samples were vortexed, incubated for 30 min in a shaker at 4°C, and phase separation was obtained by adding 1 mL of dichloromethane followed by 30 min of incubation in a shaker at 4°C. Samples were centrifuged at 4°C for 5 min at 13,000g. The supernatant from the dichloromethane layer (1 mL) was evaporated, and the residue was redissolved in 0.1 mL of 50% (v/v) methanol/water followed by UPLC-tandem MS analysis using a triple quadrupole/linear ion-trap mass analyzer (Pan et al., 2010). For diurnal measurement of ABA, rosette leaves of the wild type and raptor1b were harvested at the same developmental stage (10 rosette leaves) 1 h before illumination and 1, 2, 4, 8, and 12 h after illumination.
Extraction and Analysis of Metabolites, Proteins, and Other Macromolecules
We used our recently published protocol to extract lipids, primary and secondary metabolites, as well as chlorophyll, protein, and starch contents from one sample (Salem et al., 2016). Frozen homogenized rosette leaves from the 10-rosette-leaves developmental stage (25 mg) were resuspended in 1 mL of extraction buffer (methyl tert-butyl ether:methanol, 3:1, v/v), and the samples were incubated on a shaker for 45 min before a 15-min sonication. Phase separation was achieved by adding 0.65 mL of water:methanol (3:1). Lipids and secondary metabolites were analyzed from the organic and polar phases, respectively, by LC-MS (Giavalisco et al., 2011; Hummel et al., 2011). Primary metabolites were analyzed from the polar phase after derivatization and gas chromatography (GC)-MS analysis (Lisec et al., 2006). Protein and starch were determined in the pellet that remained after metabolite extraction (Bradford, 1976; Rappsilber et al., 2003; Smith and Zeeman, 2006). Lignin was quantified using the thioglycolic acid procedure (Bruce and West, 1989). Briefly, thioglycolic acid and 2 m HCl were added to the pellet that remained after protein/starch extraction, and the samples were incubated at 94°C for 4 h before collecting the pellet by centrifugation. The residues were incubated with 0.5 m NaOH for 12 h, and the samples were centrifuged again. Lignothioglycolic acid complexes were precipitated after acidification of the supernatant with 32% (v/v) HCl, and the samples were incubated at 4°C for 4 h before centrifugation. The absorbance of the samples was measured at 280 nm after resuspending the pellets in 0.5 m NaOH. Lignin content was estimated as based on a standard calibration curve of lignin alkali. The monosaccharide composition was estimated from the cell wall pellet remaining after acid hydrolysis of cell wall material with trifluoroacetic acid (Selvendran et al., 1979). The neutral sugar composition was determined in the hydrolysate by derivatization and GC-MS analysis. Cellulose content was measured in the trifluoroacetic acid-insoluble residues (Updegraff, 1969).
Determination of Total Flavonoid and Anthocyanin Contents
The total flavonoid content of the methanolic extract was estimated using the aluminum chloride assay (Pekal and Pyrzynska, 2014). Briefly, 50 µL of 5% (w/v)sodium nitrite solution was added to 500 µL of the methanolic extract of rosette leaves, and the samples were incubated for 6 min before adding 50 µL of 10% (w/v) aluminum chloride solution. The samples were incubated again for 6 min, and finally 400 µL of 4% (w/v) sodium hydroxide solution was added. The absorbance of the samples was measured at 510 nm after a 15-min incubation at room temperature. Rutin (quercetin-3-O-rutinoside) was used as an external standard, and the total flavonoid content was estimated as rutin equivalents based on a standard calibration curve. Relative anthocyanin levels were estimated spectrometrically (Rabino and Mancinelli, 1986; Nakata et al., 2013). Samples were extracted with 45% (v/v) methanol in 5% (v/v) acetic acid solution. The anthocyanin content was estimated from the absorbance at 530 and 657 nm after removing cell debris by centrifugation.
Estimation of Total and Cell Wall-Bound Phenolics
The total soluble phenolic content was determined with both Folin-Ciocalteu reagent (FCR) and Folin-Denis reagent (FDR; Folin and Ciocalteu, 1927; Singleton et al., 1999; Ainsworth and Gillespie, 2007). Briefly, plant material (50 mg) from the 10-rosette-leaves developmental stage was extracted using 1 mL of 80% (v/v) ice-cold aqueous methanol in the dark at room temperature for 48 h. Samples were centrifuged, 100 µL of the supernatant was diluted with 100 µL of distilled water, and 400 µL of FCR or FDR was added to the plant extract. After a 3-min incubation at room temperature, 200 µL of 1 m sodium carbonate was added to the reaction followed by a further incubation for 45 min on a rotary shaker. The absorbance of the samples was measured at 725 nm. Chlorogenic acid was used as an external standard, and the total soluble phenolic content was estimated as chlorogenic acid equivalents based on a standard calibration curve. Cell wall-bound phenolic content was determined through alkaline hydrolysis (Strack et al., 1988; Eynck et al., 2009). Briefly, 1 m NaOH was added to the pellet remaining after the extraction of total soluble phenolics, and the samples were incubated at 80°C for 1 h before a 12-h incubation at room temperature. Cell wall-bound phenolics were extracted with ethyl acetate after acidification of samples with 86% (v/v) phosphoric acid to pH < 4. The process of extraction using ethyl acetate was repeated twice, the combined ethyl acetate solution was evaporated to dryness, and the remaining pellet was redissolved in methanol. The phenolic content of the methanolic extract was estimated with both FCR and FDR (Folin and Ciocalteu, 1927; Singleton et al., 1999; Ainsworth and Gillespie, 2007).
Measurement of Nitrogen Metabolic Enzyme Activities
For the nitrate reductase activity assay, plant material (50 mg) from the 10-rosette-leaves developmental stage was extracted with 1 mL of nitrate reductase extraction buffer consisting of 0.1 m potassium phosphate and 1 mm EDTA, pH 7.5. Due to nitrate reductaseʼs high sensitivity and instability, additives to protect its activity are supplied in the extraction buffer. Polyvinylpolypyrrolidone is added to protect the enzyme from phenolic compounds, while l-Cys protects sulfhydryl groups that are critical for enzyme structure. After centrifugation at 10,000 rpm/10 min/4°C, assay buffer (25 mm KH2PO4, 0.025 mm EDTA, and 10 mm nitrate, pH 7.5) was added to the plant extract, followed by 2 mm NADH at 30°C. The reaction was stopped after 15 min by adding 1 m zinc acetate. After centrifugation at 5,000 rpm for 5 min, color reagent 1 (1% [w/v] sulfanilamide in 3 n HCl) and color reagent 2 (0.02% [w/v] N-naphthylethylenediamine in deionized water) were added to the supernatant. The absorbance was quantified after 10 min, allowing for color to develop fully at 540 nm (Hageman and Reed, 1980). For nitrite reductase activity, plant material (50 mg) from the 10-rosette-leaves developmental stage was extracted with 1 mL of nitrite reductase extraction buffer consisting of 0.05 m potassium phosphate and 1 mm EDTA, pH 7.5, 10 mm 2-mercaptoethanol, 0.1 mm PMSF, and 1.67% (w/v) polyvinylpyrrolidone. After centrifugation, assay solution (50 mm KH2PO4, 1 mm NaNO2, and 1 mm methyl viologen, pH 7.5) was added to the plant extract followed by 57.4 mm Na2S2O4 solution (in 0.29 m NaHCO3) at 30°C. After adding 1% (w/v) sulfanilamide in 3 n HCl and 0.02% (w/v) N-naphthylethylenediamine in deionized water, absorbance was measured at 540 nm (Takahashi et al., 2001). For Gln synthetase activity, plant material (50 mg) from the 10-rosette-leaves developmental stage was extracted with 1 mL of extraction buffer (50 mm Tris-HCl buffer, pH 8, 1 mm EDTA, 5 mm DTT, 10 mm MgSO4, 10 mm 2-mercaptoethanol, 0.6% [w/v] polyvinylpyrrolidone, and 1 mm Cys). Following centrifugation at 15,000 rpm for 20 min at 4°C, assay buffer (83.3 mm NH2OH, 1.67 mm manganese chloride, 10 mm sodium arsenate, 2.5 mm ADP, 108.3 mm Gln, and 100 mm imidazole, pH 7) at 30°C was added. The reaction was stopped after 15 min by adding 0.6 m ferric chloride in 2.5 m HCl solution. Activity was assessed by measuring A546 (Cruz et al., 2006).
Determination of Ion Contents
Ion contents were determined according to Watanabe et al. (2013). Briefly, frozen ground material (50 mg) from the 10-rosette-leaves developmental stage was homogenized in 1 mL of UPLC-grade water. Samples were vortexed and centrifuged at 4°C for 10 min at 14,000 rpm. The supernatant was transferred to an Amicon Ultra-0.5 centrifugal filter unit with Ultracel-3 membrane (Merck Millipore) and centrifuged for 40 min at 10,000 rpm and 4°C. Samples were analyzed with the Dionex ICS-3000 system with a KOH gradient for nitrate as an anion and a methanesulfonic acid gradient for ammonium as a cation (Dionex).
Histochemical Staining of ROS in Rosette Leaves
Levels of O2•− and H2O2 were assessed by NBT and DAB staining, respectively (Jabs et al., 1996; Thordal-Christensen et al., 1997). Rosettes of the wild type and raptor1b from the 10-rosette-leaves developmental stage were excised from plants grown under LD conditions at the end of the light period. The rosettes were incubated overnight in darkness on petri dishes containing 5 mg mL−1 NBT solution or 1 mg mL−1 DAB solution (pH 3.8). The rosettes were incubated with ethanol until the chlorophyll was bleached and then examined with the microscope.
Microscopic Analysis of Autophagy
Autophagy was analyzed by observing MDC-labeled autophagosomes using fluorescence microscopy (Contento et al., 2005). Roots of 10-d-old seedlings were stained by incubating them for 10 min with 50 µm MDC followed by three washes with PBS. Autophagosomes were observed using a 4′,6-diamino-phenylindole-specific filter of the Olympus Epi-Florescence Microscope BX-51, DC View III (5 MP), Soft Imaging System (CellSens). MDC-derived fluorescence of roots was quantified by confocal microscopy. Confocal images were acquired using the Leica TCS SP8 confocal laser scanning microscope.
S6K1 Activity Assay
The S6K activity assay was performed using the protocol provided from the Enzo Life Sciences kit (p70 S6K activity kit, catalog no. ADI-EKS-470; Sun et al., 2016). Briefly, plant material (50 mg) from the 10-rosette-leaves developmental stage was extracted with 0.5 mL of 50 mm HEPES extraction buffer (pH 7.5), and protein extracts (200 µg) were added to a p70 S6K substrate-coated ELISA microtiter plate. The reaction was initiated by adding 10 µL of ATP solution (1 µg µL−1) followed by a 1-h incubation at 30°C. The reaction was stopped by emptying the wells and adding 40 µL of phospho-specific substrate antibody followed by a 1-h incubation at room temperature. The plate was washed before incubating it for 30 min with 40 µL of diluted solution of horseradish peroxidase-conjugated goat anti-rabbit IgG. The wells were washed and incubated again for 30 min with 60 µL of tetramethylbenzidine substrate solution. The reaction was stopped by adding 20 µL of acid stop solution, and absorbance was measured at 450 nm. Relative kinase activity was calculated for each sample after subtracting the absorbance of blank (kinase assay dilution buffer).
Data Analysis
The obtained raw chromatograms from metabolites and lipids were processed for peak annotation using Xcalibur software version 2.2. Peaks were assigned using an internal Arabidopsis database. Metabolomic and lipidomic data were analyzed further for alignment and chemical noise subtraction using Progenesis QI (version 2.3; Nonlinear Dynamics). A final alignment file of all chromatograms was generated, which contained information about m/z ratio, retention times, and abundance for each annotated peak. Positive and negative data were processed separately and combined prior to downstream analysis. The obtained data were normalized by fresh weight, followed by internal standard normalization. Proteomic data were analyzed using Progenesis QI for Proteomics application (version 3; Nonlinear Dynamics). Proteins were identified from Progenesis-generated .mgf files by using Mascot (version 2.5; Matrix Science), trypsin as the digestion enzyme, carbamidomethyl as a fixed modification, and oxidation as a variable modification, allowing for one missed cleavage. Peptide ion mass tolerance was set to 10 µL L−1, and decoy database search was performed to estimate the false discovery rate for peptide identification. Peptide identifications with false discovery rate less than 0.01 were imported to Progenesis QI. Identified proteins were filtered to contain at least two peptides of score at least 20, of which at least one peptide was unique. Protein abundance was calculated from normalized intensities of the three most abundant peptides. MAPMAN (Thimm et al., 2004) and Cystoscope (Shannon et al., 2003) were used for graphic representation of the proteomic changes after log2 transformation of the average of three replicates from raptor1b mutants to the wild type under each of the specified conditions. Statistical differences between the wild type and raptor1b mutants were analyzed by Student’s t test, and the results were determined to be statistically different at P < 0.05. The log2 fold change of each protein or metabolite abundance was used to generate heat maps. Further data analyses, such as multivariate data analysis, were performed in SIMCA software (version 13; MKS Data Analytics Solutions), following log2 transformation and Pareto scaling (the centered mean is divided by the square root of the sd of each variable). Bar graphs were plotted using either Excel or Sigma Plot 12.5 (http://www.sigmaplot.co.uk). Heat-map representation was performed using MultiExperiment Viewer (MeV version 4.9.0; Saeed et al., 2003). In silico analysis of TOR, RAPTOR1A, and RAPTOR1B transcript accumulation was performed along Arabidopsis developmental stages. Analysis was performed using GENEVESTIGATOR (https://genevestigator.com/gv/; Hruz et al., 2008).
Accession Numbers
Accession numbers are as follows: At3g08850 (Raptor1B, Raptor1) and At5g01770 (Raptor1A, Raptor2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Analysis of RAPTOR gene expression and raptor phenotypes.
Supplemental Figure S2. Phenotyping of the wild type and raptor1b.
Supplemental Figure S3. Rosette phenotypes of raptor1b.
Supplemental Figure S4. Detailed cross sections of leaves of the wild type and raptor1b.
Supplemental Figure S5. Analysis of cell size and number of wild-type and raptor1b leaves.
Supplemental Figure S6. Influence of raptor1b mutation on seedling phenotype.
Supplemental Figure S7. Influence of raptor1b mutation on photosynthetic pigments.
Supplemental Figure S8. Influence of raptor1b mutation on the major carbon sources.
Supplemental Figure S9. Regulation of proteins in raptor1b.
Supplemental Figure S10. Influence of raptor1b mutation on primary metabolites and nitrogen assimilation/recycling.
Supplemental Figure S11. Heat map representing changes in lipid levels in raptor1b compared with the wild type.
Supplemental Figure S12. Heat map representing changes in detected secondary metabolites identified by UPLC-MS in raptor1b compared with the wild type.
Supplemental Figure S13. RAPTOR1B repression or disruption induces autophagy similar to TOR repression.
Supplemental Table S1. Different growth stages of the Arabidopsis wild type and raptor1b mutants.
Supplemental Table S2. Leaf size of the wild type and raptor1b mutants measured from 21-d-old rosette leaves.
Supplemental Table S3. Leaf dimensions of the wild type and raptor1b mutants grown under LD conditions (28 DAS).
Supplemental Table S4. Cell measurements of the wild type and raptor1b mutants.
Supplemental Data Set S1. LC-MS data for proteins identified from the wild type and raptor1b mutants.
Supplemental Data Set S2. Metabolite data for the wild type and raptor1b using GC-TOF-MS.
Supplemental Data Set S3. Lipid data for wild-type and raptor1b analysis using UPLC-FT-MS.
Supplemental Data Set S4. Secondary metabolite data for wild-type and raptor1b analysis using UPLC-FT-MS.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Andrea Leisse, Änne Michaelis, and Gudrun Wolter for excellent technical assistance and Dr. Eugenia Maximova for helping with microscopy. We also thank Dr. Andrew Wiszniewski, Dr. Saleh Alseekh, and Dr. Takayuki Tohge for helpful discussions.
Footnotes
M.A.Sa. gratefully acknowledges the financial support through a PhD scholarship from the GERLS-DAAD program.
Articles can be viewed without a subscription.
References
- Ache P, Bauer H, Kollist H, Al-Rasheid KA, Lautner S, Hartung W, Hedrich R (2010) Stomatal action directly feeds back on leaf turgor: new insights into the regulation of the plant water status from non-invasive pressure probe measurements. Plant J 62: 1072–1082 [DOI] [PubMed] [Google Scholar]
- Ahn CS, Han JA, Lee HS, Lee S, Pai HS (2011) The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 23: 185–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth EA, Gillespie KM (2007) Estimation of total phenolic content and other oxidation substrates in plant tissues using Folin-Ciocalteu reagent. Nat Protoc 2: 875–877 [DOI] [PubMed] [Google Scholar]
- Albus CA, Salinas A, Czarnecki O, Kahlau S, Rothbart M, Thiele W, Lein W, Bock R, Grimm B, Schöttler MA (2012) LCAA, a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol 160: 1923–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GH, Veit B, Hanson MR (2005) The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM. (2003) OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci 8: 151–153 [DOI] [PubMed] [Google Scholar]
- Avin-Wittenberg T, Bajdzienko K, Wittenberg G, Alseekh S, Tohge T, Bock R, Giavalisco P, Fernie AR (2015) Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 27: 306–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Hanson J (2017) Shaping plant development through the SnRK1-TOR metabolic regulators. Curr Opin Plant Biol 35: 152–157 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baretić D, Williams RL (2014) The structural basis for mTOR function. Semin Cell Dev Biol 36: 91–101 [DOI] [PubMed] [Google Scholar]
- Barrero JM, Piqueras P, González-Guzmán M, Serrano R, Rodríguez PL, Ponce MR, Micol JL (2005) A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J Exp Bot 56: 2071–2083 [DOI] [PubMed] [Google Scholar]
- Ben-Sahra I, Manning BD (2017) mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol 45: 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Broeckx T, Hulsmans S, Rolland F (2016) The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J Exp Bot 67: 6215–6252 [DOI] [PubMed] [Google Scholar]
- Bruce RJ, West CA (1989) Elicitation of lignin biosynthesis and isoperoxidase activity by pectic fragments in suspension cultures of castor bean. Plant Physiol 91: 889–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P (2013) Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J 73: 897–909 [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Sabatini DM (2015) Nutrient-sensing mechanisms across evolution. Cell 161: 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy MK, Sullivan JA, Theobald JC, Davies WJ, Gray JC (2008) An Arabidopsis mutant able to green after extended dark periods shows decreased transcripts of seed protein genes and altered sensitivity to abscisic acid. J Exp Bot 59: 3869–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Comparot-Moss S, Kötting O, Stettler M, Edner C, Graf A, Weise SE, Streb S, Lue WL, MacLean D, Mahlow S, et al. (2010) A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol 152: 685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL, Xiong Y, Bassham DC (2005) Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Coruzzi GM. (2003) Primary N-assimilation into amino acids in Arabidopsis. The Arabidopsis Book 2: e0010, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz C, Bio AFM, Domínguez-Valdivia MD, Aparicio-Tejo PM, Lamsfus C, Martins-Loução MA (2006) How does glutamine synthetase activity determine plant tolerance to ammonium? Planta 223: 1068–1080 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Das PK, Geul B, Choi SB, Yoo SD, Park YI (2011) Photosynthesis-dependent anthocyanin pigmentation in Arabidopsis. Plant Signal Behav 6: 23–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Truong HN, Robaglia C, Meyer C (2005) An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem Biophys Res Commun 326: 844–850 [DOI] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C (2007) The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL (2008) The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4: 851–865 [DOI] [PubMed] [Google Scholar]
- Dobrenel T, Marchive C, Azzopardi M, Clément G, Moreau M, Sormani R, Robaglia C, Meyer C (2013) Sugar metabolism and the plant target of rapamycin kinase: a sweet operaTOR? Front Plant Sci 4: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Mancera-Martínez E, Forzani C, Azzopardi M, Davanture M, Moreau M, Schepetilnikov M, Chicher J, Langella O, Zivy M, et al. (2016) The Arabidopsis TOR kinase specifically regulates the expression of nuclear genes coding for plastidic ribosomal proteins and the phosphorylation of the cytosolic ribosomal protein S6. Front Plant Sci 7: 1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Xiong F, Que Y, Wang K, Yu L, Li Z, Ren M (2015) Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front Plant Sci 6: 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM (2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop EA, Tee AR (2014) mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Semin Cell Dev Biol 36: 121–129 [DOI] [PubMed] [Google Scholar]
- Eynck C, Koopmann B, Karlovsky P, von Tiedemann A (2009) Internal resistance in winter oilseed rape inhibits systemic spread of the vascular pathogen Verticillium longisporum. Phytopathology 99: 802–811 [DOI] [PubMed] [Google Scholar]
- Fini A, Brunetti C, Di Ferdinando M, Ferrini F, Tattini M (2011) Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal Behav 6: 709–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. (2013) Abscisic acid synthesis and response. The Arabidopsis Book 11: e0166, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folin O, Ciocalteu V (1927) On tyrosine and tryptophane determinations in proteins. J Biol Chem 73: 627–650 [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7: 373–385 [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Li Y, Matthes A, Eckhardt A, Hubberten HM, Hesse H, Segu S, Hummel J, Köhl K, Willmitzer L (2011) Elemental formula annotation of polar and lipophilic metabolites using 13C, 15N and 34S isotope labelling, in combination with high-resolution mass spectrometry. Plant J 68: 364–376 [DOI] [PubMed] [Google Scholar]
- González A, Hall MN (2017) Nutrient sensing and TOR signaling in yeast and mammals. EMBO J 36: 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C, Nielsen E, Ketelaarc T, Schiefelbein J (2014) Root hairs. The Arabidopsis Book 12: e0172, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Xu Y, Shi M, Lai Y, Wu X, Wang H, Zhu Z, Poethig RS, Wu G (2017) Repression of miR156 by miR159 regulates the timing of the juvenile-to-adult transition in Arabidopsis. Plant Cell 29: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y (2009) Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci USA 106: 7648–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman RH, Reed AJ (1980) Nitrate reductase from higher plants. Methods Enzymol 69: 270–280 [Google Scholar]
- Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110: 177–189 [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Hummel J, Segu S, Li Y, Irgang S, Jueppner J, Giavalisco P (2011) Ultra performance liquid chromatography and high resolution mass spectrometry for the analysis of plant lipids. Front Plant Sci 2: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs T, Dietrich RA, Dangl JL (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273: 1853–1856 [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110: 163–175 [DOI] [PubMed] [Google Scholar]
- Kötting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G (2005) Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves: the phosphoglucan, water dikinase. Plant Physiol 137: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Sabatini DM (2013) A central role for mTOR in lipid homeostasis. Cell Metab 18: 465–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeNoble ME, Spollen WG, Sharp RE (2004) Maintenance of shoot growth by endogenous ABA: genetic assessment of the involvement of ethylene suppression. J Exp Bot 55: 237–245 [DOI] [PubMed] [Google Scholar]
- Lepistö A, Rintamäki E (2012) Coordination of plastid and light signaling pathways upon development of Arabidopsis leaves under various photoperiods. Mol Plant 5: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cai W, Liu Y, Li H, Fu L, Liu Z, Xu L, Liu H, Xu T, Xiong Y (2017) Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc Natl Acad Sci USA 114: 2765–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Lisso J, Schröder F, Fisahn J, Müssig C (2011) NFX1-LIKE2 (NFXL2) suppresses abscisic acid accumulation and stomatal closure in Arabidopsis thaliana. PLoS ONE 6: e26982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC (2010) TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J (2009) Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkowski C, Strack D (2010) Sinapate esters in brassicaceous plants: biochemistry, molecular biology, evolution and metabolic engineering. Planta 232: 19–35 [DOI] [PubMed] [Google Scholar]
- Molendijk AJ, Bischoff F, Rajendrakumar CS, Friml J, Braun M, Gilroy S, Palme K (2001) Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J 20: 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané MH, Menand B (2013) ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J Exp Bot 64: 4361–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M, Azzopardi M, Clément G, Dobrenel T, Marchive C, Renne C, Martin-Magniette ML, Taconnat L, Renou JP, Robaglia C, et al. (2012) Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI (2015) Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol 28: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M (2013) A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25: 1641–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP. (2010) TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol 22: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukarinen E, Nägele T, Pedrotti L, Wurzinger B, Mair A, Landgraf R, Börnke F, Hanson J, Teige M, Baena-Gonzalez E, et al. (2016) Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci Rep 6: 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X (2010) Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc 5: 986–992 [DOI] [PubMed] [Google Scholar]
- Pearson RB, Dennis PB, Han JW, Williamson NA, Kozma SC, Wettenhall RE, Thomas G (1995) The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J 14: 5279–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekal A, Pyrzynska K (2014) Evaluation of aluminium complexation reaction for flavonoid content assay. Food Anal Methods 7: 1776–1782 [Google Scholar]
- Pérez-Pérez ME, Crespo JL (2010) Autophagy in the model alga Chlamydomonas reinhardtii. Autophagy 6: 562–563 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Florencio FJ, Crespo JL (2010) Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 152: 1874–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Lemaire SD, Crespo JL (2012) Reactive oxygen species and autophagy in plants and algae. Plant Physiol 160: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Sabatini S (2010) Analysis of root meristem size development. Methods Mol Biol 655: 177–187 [DOI] [PubMed] [Google Scholar]
- Pu Y, Luo X, Bassham DC (2017) TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Front Plant Sci 8: 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabino I, Mancinelli AL (1986) Light, temperature, and anthocyanin production. Plant Physiol 81: 922–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Ren M, Qiu S, Venglat P, Xiang D, Feng L, Selvaraj G, Datla R (2011) Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol 155: 1367–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Venglat P, Qiu S, Feng L, Cao Y, Wang E, Xiang D, Wang J, Alexander D, Chalivendra S, et al. (2012) Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24: 4850–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Salem MA, Jüppner J, Bajdzienko K, Giavalisco P (2016) Protocol: a fast, comprehensive and reproducible one-step extraction method for the rapid preparation of polar and semi-polar metabolites, lipids, proteins, starch and cell wall polymers from a single sample. Plant Methods 12: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem MA, Li Y, Wiszniewski A, Giavalisco P (2017) Regulatory-associated protein of TOR (RAPTOR) alters the hormonal and metabolic composition of Arabidopsis seeds, controlling seed morphology, viability and germination potential. Plant J 92: 525–545 [DOI] [PubMed] [Google Scholar]
- Santelia D, Kötting O, Seung D, Schubert M, Thalmann M, Bischof S, Meekins DA, Lutz A, Patron N, Gentry MS, et al. (2011) The phosphoglucan phosphatase like Sex Four2 dephosphorylates starch at the C3-position in Arabidopsis. Plant Cell 23: 4096–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14: 1296–1302 [DOI] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 168: 960–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Kobayashi K, Geldreich A, Caranta C, Robaglia C, Keller M, Ryabova LA (2011) Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO J 30: 1343–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA (2013) TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J 32: 1087–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Makarian J, Srour O, Geldreich A, Yang Z, Chicher J, Hammann P, Ryabova LA (2017) GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J 36: 886–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Schöttler MA, Krueger S, Geimer S, Schneider A, Kleine T, Leister D, Bell K, Flügge UI, Häusler RE (2012) Defects in leaf carbohydrate metabolism compromise acclimation to high light and lead to a high chlorophyll fluorescence phenotype in Arabidopsis thaliana. BMC Plant Biol 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal SN. (2003) Sirolimus: its discovery, biological properties, and mechanism of action. Transplant Proc (Suppl) 35: 7S–14S [DOI] [PubMed] [Google Scholar]
- Selvendran RR, March JF, Ring SG (1979) Determination of aldoses and uronic acid content of vegetable fiber. Anal Biochem 96: 282–292 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton VL, Orthofer R, Lamuela-Raventos RM (1999) Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin-Ciocalteu reagent. Methods Enzymol 299: 152–178 [Google Scholar]
- Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC (2006) Quantification of starch in plant tissues. Nat Protoc 1: 1342–1345 [DOI] [PubMed] [Google Scholar]
- Sormani R, Yao L, Menand B, Ennar N, Lecampion C, Meyer C, Robaglia C (2007) Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol 7: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155: 349–361 [DOI] [PubMed] [Google Scholar]
- Strack D, Heilemann J, Momken M, Wray V (1988) Cell wall-conjugated phenolics from Coniferae leaves. Phytochemistry 27: 3517–3521 [Google Scholar]
- Streb S, Zeeman SC (2012) Starch metabolism in Arabidopsis. The Arabidopsis Book 10: e0160, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Yu Y, Hu W, Min Q, Kang H, Li Y, Hong Y, Wang X, Hong Y (2016) Ribosomal protein S6 kinase1 coordinates with TOR-Raptor2 to regulate thylakoid membrane biosynthesis in rice. Biochim Biophys Acta 1861: 639–649 [DOI] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Sasaki Y, Ida S, Morikawa H (2001) Nitrite reductase gene enrichment improves assimilation of NO2 in Arabidopsis. Plant Physiol 126: 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang ZG, Wei YD, Collinge DB (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Tsai AY, Gazzarrini S (2012) AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J 69: 809–821 [DOI] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Canales J, Gutiérrez RA (2014) Nitrogen control of developmental phase transitions in Arabidopsis thaliana. J Exp Bot 65: 5611–5618 [DOI] [PubMed] [Google Scholar]
- Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Balazadeh S, Tohge T, Erban A, Giavalisco P, Kopka J, Mueller-Roeber B, Fernie AR, Hoefgen R (2013) Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol 162: 1290–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Sheen J (2012) Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem 287: 2836–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J (2013) Moving beyond translation: glucose-TOR signaling in the transcriptional control of cell cycle. Cell Cycle 12: 1989–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J (2014) The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol 164: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J (2013) Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Liang S, Kudla J, Luan S (1998) Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant J 15: 511–519 [DOI] [PubMed] [Google Scholar]
- Yazdanbakhsh N, Fisahn J (2012) High-throughput phenotyping of root growth dynamics. Methods Mol Biol 918: 21–40 [DOI] [PubMed] [Google Scholar]
- Yazdanbakhsh N, Sulpice R, Graf A, Stitt M, Fisahn J (2011) Circadian control of root elongation and C partitioning in Arabidopsis thaliana. Plant Cell Environ 34: 877–894 [DOI] [PubMed] [Google Scholar]
- Yu S, Cao L, Zhou CM, Zhang TQ, Lian H, Sun Y, Wu J, Huang J, Wang G, Wang JW (2013) Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2: e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Ap Rees T (1999) Changes in carbohydrate metabolism and assimilate export in starch-excess mutants of Arabidopsis. Plant Cell Environ 22: 1445–1453 [Google Scholar]